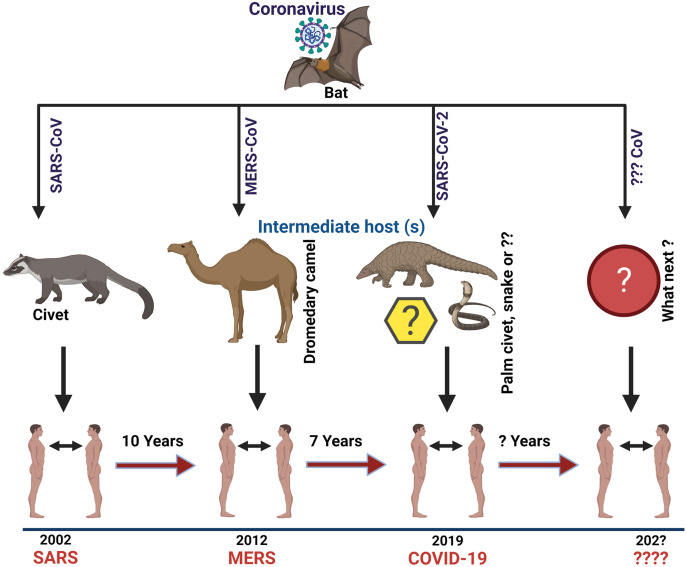
Most of the disease outbreaks witnessed in the past two decades have been attributed to the zoonotic spillover of coronaviruses. In 2003, a unique virus of the Coronaviridae family called SARS-CoV infected over 8,000 people in 29 countries, killing 774 individuals. The outbreak was discovered in November 2002 in the Foshan, Guangdong in China. An outbreak of the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 followed the SARS outbreak. It started in 2012 in Saudi Arabia and has spread to various other countries since then, including the US. Most MERS-CoV patients had severe respiratory symptoms like fever, cough, and breathlessness and recorded a mortality rate of around 35%.
It was not the end of the pandemic era as in late 2019, a new pneumonia i.e. coronavirus disease 2019 (COVID-19), was discovered in Wuhan, Hubei Region, China. There has been substantial debate about the source of the causal virus, SARS-CoV-2, which belongs to the sub-genus Sarbecovirus within the genus Betacoronavirus, and has appeared as the source of a worldwide pandemic. SARS-CoV-2 infections are currently widespread, with more than 0.27 billion persons affected worldwide and 5.3 million deaths as of December 16, 2021 (Worldometer, https://www.worldometers.info/coronavirus/). It is genetically related to SARS-CoV (79% identity) and the MERS-CoV (50% identity).
SARS-CoV-2 is the seventh CoV that has been known to infect human beings as well as animals. SARS-CoV, MERS-CoV, and SARS-CoV-2 can cause severe sickness, whilst the HKU1, NL63, OC43, and 229E produce mild symptoms similar to a common cold. In this letter, we aim to provide a viewpoint on the prominent characteristics of the SARS-CoV-2 genomes and discuss the situations that might have resulted in their evolution so that we can be better prepared to understand, predict and possibly avert such future happenings. Many scientists have observed all notable SARS-CoV-2 features, including the optimized receptor-binding domain (RBD) and polybasic cleavage site, in related coronaviruses in nature which clearly show that the SARS-CoV-2 may not be a laboratory construct or a purposefully manipulated virus [1,2]. These viruses are believed to have evolved in bats and then spread to certain other animals, including man. Molecular evolutionary investigations of the SARS-CoV-2 genomic sequences indicate that perhaps the virus originated in non-human animals, including bats [3]. However, the exact spillover event and emergence process of SARS-CoV-2 is still unclear, and more information from the earliest stage of the epidemic is essential to understand how SARS-CoV-2 came into contact with people [4]. (Fig. 1 ).
Fig. 1.
Origin of SARS, MERS and COVID-19 with their intermediate hosts.
The evolution of SARS-CoV-2 via the accumulation of mutations is slower than that of certain other RNA viruses such as influenza. Nonetheless, the emergence of novel variants of SARS-CoV-2 possess a serious threat against the antiviral therapeutic regimens, and some of these strains are thought to be more infectious than the wild type of virus [5]. RNA viruses have been commonly identified as the most common class of pathogens responsible for new human illnesses, with a rate of 2–3 new viruses unearthed each year. They have a higher probability of infecting new host species due to their incredibly short generation periods and rapid evolutionary levels. They are distinguished by an extraordinary ability to adapt to new habitats, initial selection pressures, and various hosts when the chance arises [6]. Coronaviruses (CoVs) are more stable and have fewer alterations than other RNA viruses because they encode an enzyme that rectifies some of the replication faults [7].
For the prediction of the epidemic trend and infection control, it is crucial to understand if natural selection is effectively influencing the adaptive evolution of SARS-CoV-2 transmissibility and severity during the pandemic. When it is under selection, more investigation is necessary to discover the functional mutations that are related to the developing epidemiological and pathogenic traits [8]. According to recent computational model research, SARS-CoV-2 may further evolve in its human host by a combination of missense, deletion, insertion, and other mutations. For example, various alterations in the Spike (S) protein, which interacts with the ACE2 receptors on human cells to enable viral entry, have resulted in the dramatic variations and strengthening of the existing spike-ACE2 binding affinity [9].
Almost every country on the planet has been affected by cases and mortality caused by the COVID-19, breaking the backbone of the global healthcare system. The emergence of several variants of concern (VOC), including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), along with other variants of interest (VOI) such as Epsilon (B.1.427/B.1.1429), Zeta (P.2), Eta (B.1.525), Theta (P3), Iota (B.1.526), and Kappa (B.1.617.1), have contributed to the global resurgence of the consequent waves of the pandemic [10]. Aside from the control approaches that rely on the use of face masks, social distancing, hand washing, and environmental sterilization [11], the worldwide vaccination effort is in full gear, with the overall aim of mitigating the succeeding waves of COVID-19. However, this is not the end, as we must be prepared for the next plausible pandemic, which would be as unexpected as the previous COVID-19. The evolution of various SARS-CoV-2 variants have also put the vaccine's effectiveness into contention. The major issue remains whether we are prepared for what comes next? Vaccine enhancers have already been deployed for the COVID-19 to restrict the several variants of SARS-CoV-2 [12]. Since we all know, the Delta variant was extremely dangerous and was also responsible for a significant fatality rate in different countries, and now a novel SARS-CoV-2 VOC named Omicron (previously named B.1.1.529), that has been stipulated to be more transmissible than the Delta variant, has been appearing in many regions within the South Africa, especially Gauteng. The fast spread, particularly in the younger age group, in Gauteng, South Africa, has drawn the attention of the World Health Organization (WHO) and worldwide health care systems. The major concerns regarding the omicron variant are whether it is highly infectious or virulent as compared to the other variants and whether it may evade the protection accorded by the COVID-19 vaccines. Whereas the immunological and clinical findings are insufficient to give conclusive proof yet, we may generalize according to what we understand about the Omicron mutations to offer early evidence of disease transmission, lethality, as well as immunological evasion.
Omicron has multiple deletions and over 30 mutations in the spike protein, including some (for example, 69–70del, T95I, G142D/143–145del, K417 N, T478K, N501Y, N655Y, N679K, and P681H) that coincide with those in the alpha, beta, gamma, or delta versions. Such deletions and alterations have been associated with enhanced disease transmission, stronger binding ability of virus, and escape from the antibodies. Other mutations in the Omicron variant with documented effects include those that improve the transmissibility and change the binding affinity [13]. Also, several changes in the receptor-binding domain (RBD) and N-terminal domain (NTD) are also of concern since they have been linked to the resistance to neutralizing antibodies and enhanced transmissibility. As a result, various mitigation methods, including wearing a mask, adequate sanitization, and maintaining social distance, have already been adopted to mitigate the upcoming waves of the COVID-19. The purpose of this letter article is to emphasize the severity and mortality of the next probable pandemic following the Spanish flu, SARS, MERS and the most recent COVID-19.
In conclusion, the COVID-19 pandemic has not been over yet, and there is an urgent need to ramp up the vaccination in humans as well as animals, as these are also susceptible in a considerable proportion. Besides investing in the global efforts aimed at mitigation, we must gain a valuable lesson from the continuing COVID-19 pandemic and prepare for any future occurrence of novel pandemic.
Ethical approval
This article does not require any human/animal subjects to acquire such approval.
Sources of funding
This study received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author contribution
Om Prakash Choudhary: Conceptualization, Data Curation, Visualization, Writing - Original Draft, Writing - review & editing. Priyanka: Conceptualization, Writing - Original Draft, Writing - review & editing. Rezhna Kheder Ali: Writing - Original Draft, Writing - review & editing. Sazan Qadir Maulud: Writing - Original Draft, Writing - review & editing. Manish Dhawan: Writing - Original Draft, Writing - review & editing. Teroj A. Mohammed: Writing - Original Draft, Writing - review & editing. All authors critically reviewed and approved the final version of the manuscript.
Research registration Unique Identifying number (UIN)
Name of the registry: Not applicable.
Unique Identifying number or registration ID: Not applicable.
Hyperlink to your specific registration (must be publicly accessible and will be checked): Not applicable.
Guarantor
Om Prakash Choudhary, Assistant Professor, Department of Veterinary Anatomy and Histology, College of Veterinary Sciences and Animal Husbandry, Central Agricultural University (I), Selesih, Aizawl-796015, Mizoram, India. Tel: +91–9928099090; Email: dr.om.choudhary@gmail.com.
Provenance and peer review
Not commissioned, internally peer-reviewed.
Declaration of competing interest
All authors report no conflicts of interest relevant to this article.
Acknowledgements
All the authors acknowledge and thank their respective Universities and Institutes. The figure has been created with BioRender (https://biorender.com/).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijsu.2021.106208.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes E.C., Goldstein S.A., Rasmussen A.L., Robertson D.L., Crits-Christoph A., Wertheim J.O., Anthony S.J., Barclay W.S., Boni M.F., Doherty P.C., Farrar J., Geoghegan J.L., Jiang X., Leibowitz J.L., Neil S., Skern T., Weiss S.R., Worobey M., Andersen K.G., Garry R.F., Rambaut A. The origins of SARS-CoV-2: a critical review. Cell. 2021;184(19):4848–4856. doi: 10.1016/j.cell.2021.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Upadhyay V., Lucas A., Panja S., Miyauchi R., Mallela K.M. Receptor binding, immune escape, and protein stability direct the natural selection of SARS-CoV-2 variants. J. Biol. Chem. 2021;297(4):10120. doi: 10.1016/j.jbc.2021.101208. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J., Lai S., Gao G.F., Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- 5.Thoradeniya T., Jayasinghe S. COVID-19 and future pandemics: a global systems approach and relevance to SDGs. Glob. Health. 2021;17:59. doi: 10.1186/s12992-021-00711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmonds P. RNA viruses-evolution in action. Microbiol. Today. 2004;31:163–165. [Google Scholar]
- 7.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2-what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 8.Singh D., Soojin V.Y. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021;53:537–547. doi: 10.1038/s12276-021-00604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day T., Gandson S., Lion S., Otto S.P. On the evolutionary epidemiology of SARS-CoV-2. Curr. Biol. 2020;30(15):R849–R857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhary O.P., Dhawan M., Priyanka Omicron variant (B.1.1.529) of SARS-CoV-2: threat assessment and plan of action. Int. J. Surg. 2021;97:106187. doi: 10.1016/j.ijsu.2021.106187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Priyanka, Choudhary O.P., Singh I., Patra G. Aerosol transmission of SARS-CoV-2: the unresolved paradox. Trav. Med. Infect. Dis. 2020;37:101869. doi: 10.1016/j.tmaid.2020.101869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhary O.P., Priyanka, Mohammed T.A., Singh I. Intranasal COVID-19 vaccines: is it a boon or bane? Int. J. Surg. 2021;94:106119. doi: 10.1016/j.ijsu.2021.106119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.