Abstract
The outbreak of the COVID-19 pandemic has led to millions of fatalities worldwide. For preventing epidemic transmission, rapid and accurate virus detection methods to early identify infected people are urgently needed in the current situation. Therefore, an electrochemical biosensor based on the trans-cleavage activity of CRISPR/Cas13a was developed in this study for rapid, sensitive, and nucleic-acid-amplification-free detection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Herein, a redox probe conjugated with ssRNA is immobilized on the electrode surface modified with a nanocomposite (NC) and gold nanoflower (AuNF) for enhancing the sensing performance. The SARS-CoV-2 RNA is captured by the Cas13a–crRNA complex, which triggers the RNase function of Cas13a. The enzymatically activated Cas13a–crRNA complex is subsequently introduced to the reRNA-conjugated electrochemical sensor, and consequently cleaves the reRNA. A change in current occurs due to the release of the redox molecule labeled on the reRNA, which is trans-cleaved from the Cas13a–crRNA complex. The biosensor can detect as low as 4.4 × 10−2 fg/mL and 8.1 × 10−2 fg/mL of ORF and S genes, respectively, over a wide dynamic range (1.0 × 10−1 to 1.0 × 105 fg/mL). Moreover, the biosensor was evaluated by measuring SARS-CoV-2 RNA spiked in artificial saliva. The recovery of the developed sensor was found to be in an agreeable range of 96.54–101.21%. The designed biosensor lays the groundwork for pre-amplification-free detection of ultra-low concentrations of SARS-CoV-2 RNA and on-site and rapid diagnostic testing for COVID-19.
Keywords: SARS-CoV-2, CRISPR/Cas13a, Trans-cleavage activity, Electrochemical biosensor
1. Introduction
The COVID-19 pandemic, driven by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has drastically affected people and the global economy (Karlinsky et al., 2021; Antiochia, 2020). According to the World Health Organization (WHO), 205 million people have been infected, and 4 million people have died because of COVID-19 until August 2, 2021 (World Health Organization, 2021). The virus is transmitted from person to person via diverse routes, such as direct contact, air, fomites, and droplets, which enable the rapid spread of this virus (Kutter et al., 2021; Li et al., 2020). In addition, SARS-CoV-2 has high transmissibility (reproduction number, R0 = 3.1) compared with other respiratory viruses, such as the Middle East respiratory syndrome coronavirus (MERS-CoV) (R0 = 0.6), severe acute respiratory syndrome coronavirus (SARS-CoV) (R0 = 0.7), and influenza viruses (R0 = 1.3) (Abdelrahman, 2020). This pandemic situation can be managed by screening suspected COVID-19 cases through rapid diagnosis of the viral infection and isolating infected patients to curb further transmission (Xu et al., 2020;Taleghani et al., 2020).
The real-time reverse transcription-polymerase chain reaction (RT-PCR) technique is currently the gold standard for detecting SARS-CoV-2 RNA with high sensitivity and specificity (Huang et al., 2020; Lee et al., 2021). However, the need for trained personnel and the requirement of a lengthy testing period (3–4 h), which includes sample preparation and gene amplification, limit the applicability of RT-PCR for point-of-care testing (POCT). Therefore, the testing duration for rapid on-site detection must be minimized. Immunodetection methods have emerged as an alternative to SARS-CoV-2 monitoring owing to their potential for achieving rapid testing results; however, false negatives due to the poor accuracy and low sensitivity of these testing methods can exacerbate the spread of SARS-CoV-2 (Cui and Zhou, 2020, Huang et al., 2021)
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) protein systems based on targeting specific nucleotide sequences have been spotlighted as highly effective detection and rapid monitoring strategies (Nouri et al., 2021; Wang et al., 2020). Among the CRISPR systems applied in biosensors, the trans-cleavage activities of Cas13a and Cas12a have been exploited for unspecific single-stranded RNAs and DNAs functionalized with fluorescence dye and quencher-tagged reporter molecules; e.g., Gootenberg, et al. integrated CRISPR with recombinase polymerase amplification (RPA), a nucleic-acid-amplification technique, to detect Zika viral RNA by analyzing fluorescence signals down to 1.0 × 10−1 fg/mL (Gootenberg et al., 2017; Broughton et al., 2020; Wang et al., 2020). Despite the excellent detection limits and specificity provided by this CRISPR-based optical detection strategy, the bulky and expensive optical device can restrict their ability to utilize in POCT-related applications. Meanwhile, electrochemical measurement techniques can be used as alternative methods for quantifying low amounts of a target gene because of their high sensitivity, specificity, simplicity of miniaturization, portability, and cost-effectiveness. These advantages of electrochemical detection have been harnessed to develop a nucleic-acid-amplification-free electrochemical biosensor combined with a CRISPR/Cas12a system to detect human papillomavirus (HPV) and parvovirus at levels as low as 2.8 × 106 fg/mL and 6.0 × 102 fg/mL, respectively (Zhang et al., 2020; Dai et al., 2019). The detection procedure is more simplified thanks to the omission of the pre-amplification step. However, this biosensor exhibits relatively poor performance for monitoring low concentrations of viral RNA, which is essential for early detection of viral infections, compared with those of conventional CRISPR/Cas-based sensing methods, such as specific high sensitivity enzymatic reporter unlocking (SHERLOCK) and DNA endonuclease targeted CRISPR trans reporter (DETECTR) (Kellner et al., 2019; Chen et al., 2018).
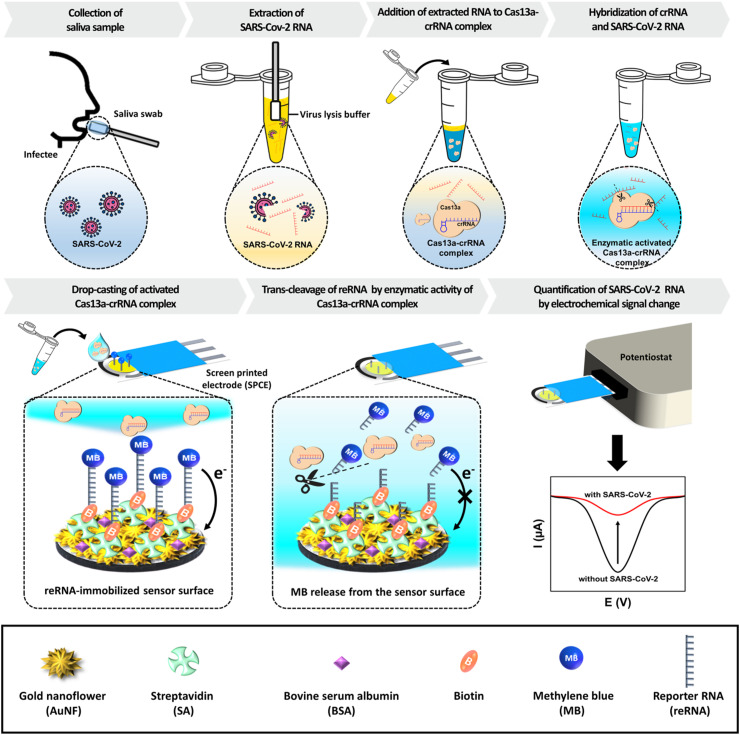
Therefore, an electrochemical biosensor that combines a high-conductivity nanostructured electrode with the trans-cleavage activity of the CRISPR/Cas13a system is developed in this study to overcome the aforementioned limitations of conventional SARS-CoV-2 detection methods and CRISPR/Cas-assisted biosensors; the proposed biosensor enables quantification of extremely low concentrations of SARS-CoV-2 RNA without the nucleic acid amplification step. The electrode is modified with a nanocomposite (NC) and flower-shaped gold nanostructure (AuNF) to increase the conductivity and surface-to-volume ratio of the working electrode (Kashefi-Kheyrabadi et al., 2020; Wang et al., 2021). Subsequently, reporter RNA (reRNA) molecules tagged with methylene blue (MB) and biotin at each terminal end are immobilized on streptavidin (SA)-coated electrode. By utilizing this modified electrode, the sensing strategy for SARS-CoV-2 detection based on Cas13a-mediated trans-cleavage of reRNA is illustrated in Fig. 1 . First, saliva is collected by a saliva swab method, and the viral RNA is extracted by the lysis buffer. The viral RNA is subsequently loaded into a solution of the Cas13a–crRNA complex, which can recognize a specific sequence of SARS-CoV-2 RNA based on the crRNA sequence that complementarily binds to the target region via the proto spacer-flanking site (PFS) sequence (Bruch et al., 2021). Upon the activation of the Cas13a–crRNA complex through the binding to a specific region of the SARS-CoV-2 RNA, the non-specific cleavage of collateral single-stranded RNA (ssRNA) is triggered (van Dongen et al., 2020; Zuo et al., 2017). The Cas13a–crRNA–SARS-CoV-2 RNA ternary complex is subsequently introduced to the sensor surface, resulting in collateral cleavage of the reRNA immobilized on the sensor into short fragments and the release of MB from the biosensing surface. This consequently leads to the elimination of electron transfer from the redox probe to the electrode surface, resulting in a decrease in the peak current. Finally, the SARS-CoV-2 RNA is quantified by converting the decreased peak current. The developed biosensor could detect ORF and S genes of SARS-CoV-2 at levels as low as 4.4 × 10−2 fg/mL and 8.1 × 10−2 fg/mL, respectively. The sensor performance, tracing a low amount of SARS-CoV-2 RNA, shows tremendous promise as a monitoring platform for diagnosing COVID-19 with a highly sensitive analytical performance despite the omission of the pre-amplification step.
Fig. 1.
Schematic illustration of the proposed electrochemical biosensing strategy utilized with the CRISPR/Cas13a for SARS-CoV-2 detection. Viral RNA is extracted from saliva collected from infected patients using the lysis buffer and mixed with a Cas13a–crRNA complex containing solution. This complex binds with the SARS-CoV-2 RNA, resulting in enzymatic activity. The activated Cas13a–crRNA complex is subsequently loaded onto the sensor surface for cleaving the reRNA immobilized on the electrode. The presence of SARS-CoV-2 can be quantified via analysis of the current change.
2. Material and methods
2.1. Reagents and materials
Graphene nanoplatelets (GNPs), chitosan (CHT), gold(III) chloride trihydrate, sodium chloride, 3-mercaptopropionic acid (MPA), 1-ethyl-3-(3-(dimethylamino) propyl) carbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), ethyl alcohol, and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (USA). Tris-Borate-EDTA (TBE) buffer (10 × ), agarose, and UltraPure™ 1 M Tris-HCI Buffer were purchased from ThermoFisher (USA). Molybdenum disulfide nanosheets (MoS2NSs) were provided by Graphene Supermarket (USA). DNA ladder solutions (25/100 base, 25/100 bp, and 1 kb) and LoadingSTAR reagent were acquired from DyneBio (South Korea). CRISPR/Cas13a protein was purchased from MCLAB (USA). RNA Cleanup Kit and HiScribe™ T7 High Yield RNA Synthesis Kit were purchased from Monarch® (USA) and New England BioLabs (England), respectively. The DNA and RNA oligonucleotides used in this study (Table S1) were synthesized and purified by BIONEER (South Korea).
2.2. Apparatus
Disposable screen-printed carbon electrodes (SPCE; C110) were purchased from Dropsens Inc., Spain. Electrochemical experiments were performed using a CHI-650E electrochemical analyzer (CH Instruments, USA). Gel electrophoresis was conducted using a Mupid-exU system (Takara, Japan) and subsequent analysis was performed using a MiniBIS UV-transilluminator (DNR Bio-Imaging Sytems, Israel). Moreover, total RNA purity and concentration were determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). Atomic force microscopy (AFM) imaging and roughness analysis were performed using an NX-10 apparatus (Park Systems, South Korea). Confocal microscopy image analysis was conducted using an LSM 700 (Carl Zeiss, Germany). Fluorescence intensity was measured using a Nanodrop 3300 fluorospectrometer (Thermo Fisher Scientific, USA).
2.3. Fabrication of AuNF/NC/SPCE
The SPCE was cleaned by cyclic voltammetry (CV) technique over the range of 0.1–0.7 V in 0.5 M H2SO4 solution up to uniform voltammograms were acquired. The NC was subsequently prepared by mixing 1 μg/mL of MoS2NSs, 1 mg/mL of GNPs, and 20 μg/mL of CHT in a ratio of 10:5:1, respectively, according to our previously reported protocol (Kashefi-Kheyrabadi et al., 2020). Then, 20 μL of the NC solution was drop casted onto the surface of the working electrode. After the NC deposition, AuNF were formed in 10 mM HAuCl4 solution using a previously reported method involving an amperometric technique conducted at 0.2 V for 600 s.
2.4. Biosensing surface modification
The sensing surface was prepared as follows. First, AuNF/NC/SPCE was treated with 0.1 M MPA for 30 min to enable a layer formation of carboxyl groups on the electrode surface. In order to induce an amide bond between amine groups of SA and carboxyl groups on the electrode surface, 7 μL of mixture solution consist of 0.5 mg/mL SA and EDC/NHS (0.2 M and 0.05 M, respectively) in 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) (pH 4.7) was incubated in the dark for 2 h at 23 °C. For passivation of the remaining active sites against non-specific adsorption, 7 μL of 0.01% BSA solution was placed on the electrode surface for 10 min at 23 °C. Finally, a reRNA mixture containing reRNA (40 μg/mL), RNase inhibitor (4 U/μL), and TRIS-HCl buffer (40 mM) was added to the electrode to immobilize reRNA through SA–biotin binding. At each modification step, the unbound materials were removed by rinsing with deionized water (DI water) and 0.1 M phosphate buffered saline (PBS) (pH 7.4).
2.5. Gel electrophoresis
The Cas13a:crRNA ratio suitable for the formation of the Cas13a–crRNA complex was investigated by 2% agarose electrophoresis. Prior to the addition of the Cas13a–crRNA mixture to the agarose gel, the crRNA was stained with the LoadingSTAR and incubated with Cas13a for 30 min at 37 °C. The electrophoresis was performed in 1 × TBE buffer (pH 8.1) at 45 V for 20 min. The intensity of the line was investigated using the ImageJ software, and the capture efficiency of crRNA by Cas13a was calculated as follows:
The trans-cleavage activity was explored by analyzing the gel results of SARS-CoV-2 RNA by the Cas13a–crRNA complex. The SARS-CoV-2 RNA and crRNA were stained and subsequently mixed with Cas13a. After 30 min of incubation at 37 °C, the mixture was placed on the gel and investigated in 1 × TBE buffer (pH 8.1) at 45 V for 20 min. The images of both electrophoretic results were visualized by MiniBIS UV-transilluminator.
2.6. Cas13a–crRNA-based fluorescent assays
The measurements of fluorescence intensity were performed after incubation of a mixture containing 1 ng/mL of SARS-CoV-2 RNA, 10 μg/mL of ssDNA labeled with a fluorophore (6-carboxyfluorescein; FAM) and a quencher (Iowa Black) at the end of each terminal, and different concentrations of Cas13a and crRNA for 30 min at 37 °C. The fluorescence signal was measured by using NanoDrop 3300 fluorospectrometer.
2.7. Electrochemical measurements
The biosensing surface was characterized by CV at a scan rate of 0.1 V s−1and electrochemical impedance spectroscopy (EIS) in K3[Fe(CN)6] (5.0 × 10−3 M) containing 0.1 M KCl. Nyquist plots were recorded at open-circuit potential and an AC potential of 0.005 V over a frequency range varied between 10 kHz and 0.1 Hz. DPV was employed to quantify SARS-CoV-2 RNA over a range of −0.5 to −0.1 V, with the voltammograms being measured at a pulse amplitude of 0.025 V and a scan rate of 0.05 V s−1.
2.8. Detection of the SARS-CoV-2 RNA
The Cas13a and crRNA were stored at 80 °C until used. Then, just before detecting the SARS-CoV-2 RNA, 0.5 mg/mL of Cas13a and 10 μg/mL of crRNA were thawed at 4 °C and mixed with the SARS-CoV-2 RNA and 4 U/μL of RNase inhibitor in Tris-HCl buffer (40 mM). Subsequently, the 7 μL of the mixture was directly drop-casted on the electrode in an RNase-free environment and the temperature was maintained at 37 °C to trigger of enzymatic activity of the Cas13a–crRNA complex. DPV was conducted at the end of the reaction and the current change (ΔI %) was calculated as follows,
(Background signal: the current after reRNA immobilization, Target signal: the current after treatment of activated Cas13a–crRNA complex). At last, the amounts of SARS-CoV-2 RNA were quantified by analyzing ΔI %.
3. Results and discussion
3.1. Characterization of the biosensing surface
The fabrication steps of the biosensor are illustrated in Fig. 2 a. At the beginning of electrode modification, the NC consisted of MoS2NSs, GNPs, and CHT was applied on the bare electrode to enhance the physicochemical properties of the sensor. To briefly explain the role of each component in NC, MoS2NSs provide an excellent substrate for the well-ordered electro-deposition of metal nanostructures. GNPs offer a high electrical conductivity and the extra-large surface area that increase the immobilization efficiency of the reRNA and subsequently lead to the efficient activity of trans-cleavage. AuNF was subsequently electro-deposited to increase the sensing region and electrode conductivity (Wang et al., 2011). Next, SA and BSA were sequentially coated for immobilizing reRNA and blocking the activated sensor surface, respectively. The reRNA, labeled with biotin and MB playing a role as a redox probe, was immobilized on the electrode by SA-biotin binding. Finally, after capturing the SARS-CoV-2 RNA, the activated Cas13a–crRNA complex was added to the sensor to induce the discharge of the redox molecule (MB). As a result, the current signal decreased.
Fig. 2.
(a) Schematic representation for fabrication of the electrochemical biosensor for SARS-CoV-2 RNA detection. Characterization of various modification steps of the biosensing surface by (b) CV and (c) EIS (insert shows Randles equivalent circuit) for (ⅰ) AuNF/NC/SPCE, (ⅱ) SA/AuNF/NC/SPCE, (ⅲ) BSA/SA/AuNF/NC/SPCE, (ⅳ) reRNA/BSA/SA/AuNF/NC/SPCE, and (ⅴ) cleaved reRNA/BSA/SA/AuNF/NC/SPCE. (d) Representation of the change in current obtained by DPV for steps (ⅳ) and (ⅴ).
Various modification steps for the sensor surface were characterized by CV and EIS (Fig. 2b and c). The AuNF/NC/SPCE exhibited a couple of redox peaks along with a high background current, revealing a high conductivity of the improved sensor surface (Fig. 2b, curve (ⅰ)). The observation of a lower faradaic peak current after SA immobilization on the surface indicated that SA layer hinders the electron transfer between the [Fe(CN)6]3− redox probe and the sensing surface (Fig. 2b, curve (ⅱ)). Subsequent blocking of the surface with BSA for the prevention of unspecific adsorption further alleviated the peak current (Fig. 2b, curve (ⅲ)). A similar trend in decreasing current was identified upon reRNA immobilization through the SA-biotin binding on the sensing surface (Fig. 2b, curve (ⅳ)). However, the loading of the Cas13a–crRNA–target RNA on the surface and the subsequent cleavage of reRNA led to a slight increase in the current (Fig. 2b, curve (ⅴ)) because of disintegration of the materials that hinders the electron transfer to the electrode. The characterization of the biosensing surface was validated by EIS as well (Fig. 2c). The EIS was measured between 100 kHz and 0.1 Hz. The values of Rs, constant phase elements (C1 and C2), and charge transfer resistance (Rct) were determined by fitting the experimental data to the Randles equivalent circuit as shown in the inset of Fig. 2c. At high frequencies, no semicircle was observed in the AuNF/NC/SPCE, presenting negligible resistance of electron-transfer at the electrode (Fig. 2c, curve (ⅰ)). As a consequence of the sequential immobilization of SA and BSA on the electrode surface, a layer formed on the surface repulsed the [Fe(CN)6]3- redox molecule and it resulted in an escalate of the electron-transfer resistance (Fig. 3 c, curves (ⅱ) and (ⅲ)). By immobilizing reRNA on the electrode surface led to the distribution of a stack for negative charges onto the surface. The increase in electron-transfer resistance originated from the significant repelling of the [Fe(CN)6]3− redox molecule against the negatively charged surface (Fig. 2c, curve (ⅳ)). Finally, the treatment of the sensing surface with the activated Cas13a–crRNA complex decreased the interfacial electron-transfer resistance by alleviating the hindrance related to the access of the [Fe (CN)6]3− redox molecule to the biosensing surface (Fig. 2c, curve (ⅴ)). The results of CV and EIS were in compliance with each other, showing that the surface fabrication for biosensing was satisfactorily conducted. Additionally, the change in current by the trans-cleavage effect on the reRNA was also characterized through DPV signals (Fig. 2d). The cleavage of the reRNA induced the escape of MB molecules from the electrode surface and consequently decreased the current.
Fig. 3.
(a) Atomic force microscopy (AFM) micrographs and (b) cross-section profiles of AuNF/NC/SPCE, SA/AuNF/NC/SPCE, and BSA/SA/AuNF/NC/SPCE. (c) Confocal microscopic images of BSA/SA/AuNF/NC/SPCE, reRNA/BSA/SA/AuNF/NC/SPCE, and cleaved reRNA/BSA/SA/AuNF/NC/SPCE.
The modification states of the electrode surface from bare SPCE to the AuNF/NC/SPCE were identified by the scanning electron microscope (SEM) analysis (Fig. S1). The surface image of the bare SPCE showed a typical feature of the carbon-ink-printed with a rough and uneven layer (Fig. S1a). After the NC was treated on bare SPCE, a flaky layer due to the presence of the GNPs and MoS2NSs was identified (Fig. S1b). Moreover, the uniformly formed flower-shaped morphology was shown on the surface after the electro-deposition of HAuCl4 onto NC/SPCE electrode (Fig. S1c). Next, the roughness changes owing to the different modification steps were also defined as well through the AFM measurement. The AFM results were compared with the electrochemical results at each modification step. Fig. 3a (a) and (b) display the measurement result of the AFM micrographs acquired from each modification step up to BSA blocking and their accordant cross-sectional description. The rough topography was examined for the AuNF/NC/SPCE with an average roughness (Rq) of 128.3 ± 12.5 nm. The immobilization of SA on the AuNF/NC/SPCE increased the Rq to 370.5 ± 17.4 nm, illustrating the successful formation of the SA layer on the evenly formed AuNF structure. However, after the surface blocking, the Rq value of the BSA/SA/AuNF/NC/SPCE was 206.1 ± 13.2 nm lower than that of the SA/AuNF/NC/SPCE, possibly owing to the filling of gaps between the immobilized SA molecules by BSA. The AFM measurement was also conducted on the reRNA/BSA/SA/AuNF/NC/SPCE and cleaved reRNA/BSA/SA/AuNF/NC/SPCE; however, no significant changes in the roughness were observed (data not shown), due to the small size of reRNA (10 nt) and the resolution in the non-contact mode of the AFM measurement (Marrese et al., 2017). Therefore, the validation of reRNA immobilization and the trans-cleavage activity were investigated by image analysis using cyanine (Cy5) (fluorescent dye) tagged reRNA. The fluorescence signal was derived on the sensor surface by the immobilization of the reRNA on the BSA/SA/AuNF/NC/SPCE (Fig. 3c, left and middle). By contrast, the fluorescence signal decreased after the treatment of the activated Cas13a–crRNA complex binding with the target RNA because of the trans-cleavage effect of Cas13a (Fig. 3c, right). The observed results confirm the genuine immobilization of reRNA and the cleavage of reRNA by Cas13a. Furthermore, the AFM and fluorescence results were consistent with the electrochemical results of each modification step of the electrode surface.
3.2. Effects of the Cas13a:crRNA ratio on the trans-cleavage of SARS-CoV-2 RNA
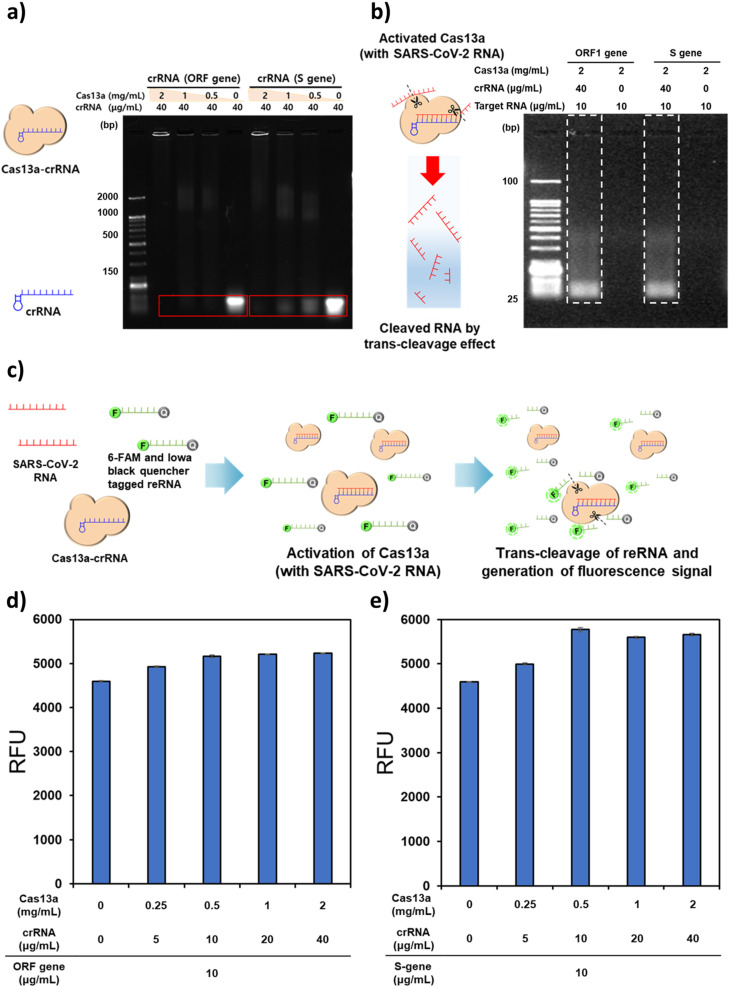
According to the mechanism of the CRISPR-based sensing, the cleaving function toward ssRNA is activated by the combination of the Cas13a–crRNA complex with SARS-CoV-2 RNA (Wang et al., 2020). For applying this CRISPR-based sensing mechanism to the electrochemical measurement, it is essential to determine the ratios between Cas13a and crRNA to form the Cas13a–crRNA complex that can elicit high trans-cleaving activity. In order to explore the formation of the complex, gel electrophoresis was conducted after the incubation of the mixture of Cas13a protein and crRNAs hybridizing with the specific region on the ORF and S sequence of SARS-CoV-2 (Fig. 4 a). A previously reported sequence of SARS-CoV-2 was used as a target sequence in this research (Zhang et al., 2020). Different concentrations of the Cas13a protein (0.5–2.0 mg/mL by two-fold and 0 mg/mL as a control) were mixed with a fixed concentration of crRNA (40 μg/mL), and the mixed solutions were added to the gel. The intensity of the lower band in the electrophoresis result was examined; a weaker signal intensity indicates more effective binding of the crRNA and Cas13a. To quantify the capture efficiency of crRNA from Cas13a, the Image J software analysis was accomplished by subtracting the intensity of unbound Cas13a–crRNA from the initial intensity of crRNA and dividing by the initial intensity of crRNA (Figs. S5a and b). The crRNA targeting the ORF gene (ORF gene_crRNA) was fully captured above the concentration of Cas13a was 0.5 mg/mL. However, with respect to the crRNA targeting the S gene (S gene_crRNA), 100% capture efficiency was achieved only at 2 mg/mL. Therefore, 2 mg/mL of Cas13a per 40 μg/mL of crRNA was selected as the ratio for targeting both these sequences of the SARS-CoV-2 RNA. Afterward, the identification of SARS-CoV-2 RNA cleavage reaction was evaluated using gel electrophoresis (Fig. 4b). Each SARS-CoV-2 gene was mixed with the Cas13a–crRNA complex solution and incubated for 1 h. In the case of the SARS-CoV-2 RNA added to the Cas13–crRNA solution, the gradation observed in the electrophoretic result indicates that the trans-cleavage activity of Cas13a cleaves the remaining fragment of the SARS-CoV-2 RNA after the cis-cleavage of a specific site on the viral RNA. Finally, the concentration of the Cas13a–crRNA complex for maximizing the enzymatic function of Cas13a was investigated using the methods of the SHERLOCK applied with 6-FAM and Iowa black quencher tagged ssRNA (Fig. 4c) (de Puig et al., 2021; Fozouni et al., 2021). Since the RNase activity of Cas13a cleaves the ssRNA, thus increasing the distance between the quencher and the fluorescent dye and then, the fluorescence signal is induced. At fixed amounts of the S gene, ORF gene, and the ssRNA, different concentrations of Cas13a and crRNA were mixed and incubated at 37 °C for 2 h. In both genes, the fluorescence signal gradually increased up to 5 mg/mLCas13a and 10 μg/mL crRNA in both genes and saturated (Fig. 4d, e). Therefore, 0.5 mg/mL and 10 μg/mL were estimated to be the appropriate concentrations of Cas13a and crRNA, respectively. In addition, we investigated the cross-reactivity of the Cas13a–crRNA complexes targeting the ORF and S genes, respectively. The 100 μg/mL of ORF gene was mixed with Cas13a–crRNA targeting S gene and then, this mixture was reacted with fluorophore and quencher tagged ssRNA. Subsequently, the fluorescence signal was analyzed, right after the reaction was completed. The value of fluorescence intensity induced by cross-reaction between the ORF gene and its non-target Cas13a–crRNA complex was 198 RFU (relative fluorescence unit). The S gene was also tested with Cas13a–crRNA targeting the ORF gene by the same method above mentioned. The value of 162 RFU was obtained by cross-reaction between the S gene and its non-target Cas13a–crRNA complex. We compared these values with the fluorescence signals induced from the reactions between the target Cas13-crRNA complexes and the blank samples, which did not contain the target genes. The values of the blank sample were 179 RFU in the Cas13a–crRNA complex targeting the ORF gene and 169 RFU in the Cas13a–crRNA complex targeting the S gene. The results suggest that the ORF and S genes with their corresponding Cas13a–crRNA complexes can derive specific electrochemical responses without obvious cross-reactivity.
Fig. 4.
Gel electrophoresis conducted to determine (a) the ratio of crRNA at different concentrations of Cas13a in the formation of the Cas13a–crRNA complex and (b) the performance of trans-cleavage of SARS-CoV-2 RNA by the activated Cas13a–crRNA complex. (c) Schematic drawing of the Cas13a–crRNA based fluorescent assays. The fluorescence intensities were examined with different concentrations of the Cas13a–crRNA complex at fixed concentrations of the (d) S and (e) ORF genes.
3.3. Optimization of detection conditions
The current signals measured through CV in the [Fe(CN)6]3− redox solution were analyzed to set up the incubation time of 0.1 M MPA for maximizing the coating efficiency of the SA and the concentration of SA for enhancing the immobilization of reRNA. The current signals were sequentially diminished by treatment of the MPA followed by the SA since the electron transport was disrupted step by step due to the formation of a self-assembled monolayer (SAM) and the electrode coating of SA (Figs. S2a and S3a). As the incubation time of the MPA on the sensor surface increased, the anodic peak current (IPa) gradually decreased, finally reaching a stable state at 30 min. So, we determined 30 min as the optimal incubation time for the formation of Au-MPA SAM (Fig. S2b). Fig. S3b shows that IPa values decreased as the concentration of SA increased and settled at 0.5 mg/mL, suggesting the complete reaction of peptide bonds between MPA and SA. Thus, the SA concentration of 0.5 mg/mL was used to follow the next steps for the sensor fabrication.
In order to determine the optimal incubation time and concentration of reRNA immobilized on the sensor surface, DPV was employed to measure the reduction signal of the MB. First, the peak currents were explored after treatment with various concentrations of reRNA on the surface of the BSA/SA/AuNF/NC/SPCE for 2 h. Fig. S4a shows that the DPV signal was drastically elevated with the increase in the concentrations of reRNA and stabilized at 100 μg/mL, reflecting the complete immobilization of the reRNA on the BSA/SA/AuNF/NC/SPCE. Thus, the reRNA concentration of 100 μg/mL was determined for the sensor fabrication. In addition, the sequential time variations for reRNA immobilization from 0.25 to 8 h were analyzed (Fig. S4b). The reduction current increased as the immobilization period increased and reached a plateau after 4 h, indicating that 4 h-incubation can be an appropriate time for reRNA anchoring on the electrode surface.
In our system, the detection of SARS-CoV-2 RNA was conducted by drop-casting the mixture of each material (Cas13a, crRNA, and SARS-CoV-2 RNA) onto the reRNA immobilized sensor surface. Since the enzymatic activity of the complex of Cas13a–crRNA is almost negligible at room temperature (Nalefski et al., 2021), the mixture does not react with reRNA before the sensor is subject to the incubation at 37 °C. Once the sensor was placed into the incubator (37 °C), the trans-cleavage reaction was expected to be activated. The incubation time sustaining trans-cleavage activity of Cas13a–crRNA complex on the electrode surface was investigated and presented in Fig. S6. The reRNA/BSA/SA/AuNF/NC/SPCE was immersed in the activated Cas13a–crRNA complex solution with 1.0 × 106 fg/mL of S gene for various periods of incubation (0.25–4 h). The ΔI rosed with the increase in the reaction time and saturated at 1.5 h, suggesting that it can be the reaction time optimized for the detection of SARS-CoV-2.
3.4. Analytical performance of the electrochemical sensor
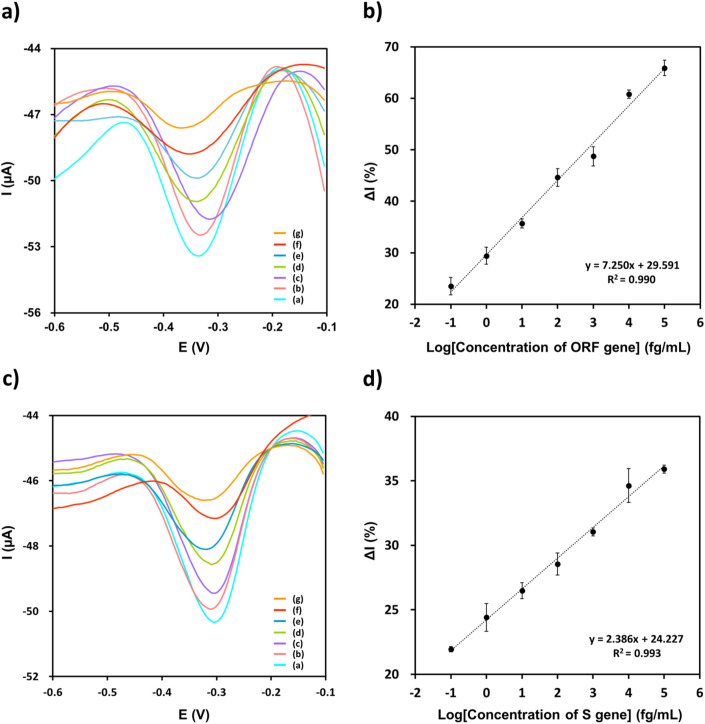
Under the optimized experimental conditions, the analytical performance of the designed sensor was evaluated by conducting DPV experiments on serially diluted ORF and S genes of SARS-CoV-2 in 0.1 M PBS containing 0.1 M KCl. As hown in Fig. 5 a, c, the faradaic peak currents obtained by DPV gradually decreased with declining concentrations of the ORF and S genes. Owing to the cut-off on MB labeled reRNA by trans-cleavage activity from Cas13a–crRNA complex, results of the reduction current were diminished. Calibration plots of the ORF and S genes were obtained in a range of 1.0 × 10−1 to 1.0 × 105 fg/mL (Fig. 5b, d). The current change was linearly related to the logarithms of each gene concentration and mapped with the correlation: and (. The limits of detection (LODs) of the constructed sensor were estimated to be 4.4 × 10−2 fg/mL of ORF and 8.1 × 10−2 fg/mL of S genes with respect to the sum of the change in the current of the blank sample and three standard deviations. All the experiments at different concentrations were performed in triplicate. Considering that the LOD value of the sensor derived from the optimal condition was lower than the reported SARS-CoV-2 RNA concentration in saliva (from 1.0 × 103 fg/mL to 1.0 × 107 fg/mL), an additional experiment was performed using the blank value and the minimum RNA concentration in saliva, 1.0 × 103 fg/mL (Bar-On et al., 2020; Zhu et al., 2020) by reducing the duration of trans-cleavage to 30 min for rapid detection of the viral RNA. The blank values of the ΔI % of S gene and ORF gene were 17.06% and 19.22%, respectively, and 67.91% and 32.51% were obtained at the concentration of 1.0 × 106 fg/mL (data not shown) indicating that the biosensor can be utilized for SARS-CoV-2 screening in a short time. In comparison with the other nucleic acid amplification-based detection methods, the established sensor exhibits a remarkable capability for detecting low amounts of SARS-CoV-2 genes and a broad linear range without the gene amplification technique. Moreover, the intra- and inter-assay variations were assessed to examine the reproducibility of the biosensor. Under the optimal conditions, the relative standard deviation (RSD) was evaluated four times using 1.0 × 103 fg/mL of ORF and S genes. The RSDs of the intra- and inter-assay were estimated to be 3.14% (n = 4) and 2.52% (n = 4) for the ORF gene and 2.47% (n = 4) and 1.74% (n = 4) for the S gene, respectively. Hence, the obtained results confirm the reliable reproducibility of the proposed strategy.
Fig. 5.
(a) DPV responses of the reRNA/BSA/SA/AuNF/NC/SPCE in the presence of different concentrations of ORF gene [a: 1.0 × 10−1, b: 1.0 × 100, c: 1.0 × 101, d: 1.0 × 102, e: 1.0 × 103, f: 1.0 × 104, and g: 1.0 × 105 fg/mL] in 0.1 M PBS at pH 7.4 containing 0.1 M KCl solution, and (b) the corresponding calibration curve of the sensor. (c) DPV responses of the reRNA/BSA/SA/AuNF/NC/SPCE in the presence of different concentrations of S gene [a: 1.0 × 10−1, b: 1.0 × 100, c: 1.0 × 101, d: 1.0 × 102, e: 1.0 × 103, f: 1.0 × 104, and g: 1.0 × 105 fg/mL] in 0.1 M PBS at pH 7.4 containing 0.1 M KCl solution, and (d) the corresponding calibration curve of the sensor.
3.5. Applicability test using spiked SARS-CoV-2 RNA in artificial saliva for on-site detection
The sensing accuracy was validated to enable the application of the biosensor for SARS-CoV-2 RNA detection in saliva samples. The amounts of SARS-CoV-2 RNA in patient saliva have been reported as 1.0 × 104 to 1.0 × 108 copies/mL; this range of copy number could be converted into 1.0 × 103 to 1.0 × 107 fg/mL. Regarding this concentration range of SARS-CoV-2 RNA, the SARS-CoV-2 RNA was serially diluted to minimum and medium levels (1.0 × 103 fg/mL and 1.0 × 105 fg/mL) using artificial saliva and quantified by the developed biosensor under optimal conditions. As shown in Table 1 , the recovery for the different ORF and S gene concentrations varied in the 109.42–111.33% and 96.54–101.21% ranges, respectively, which reveal within the acceptable range. The recovery of the spiked samples was calculated by dividing the amount of SARS-CoV-2 RNA detected using the biosensor by the amount added to the artificial saliva samples. The results demonstrate that the constructed biosensing system for the detection of SARS-CoV-2 RNA is feasible for the application in a saliva sample matrix with high accuracy.
Table 1.
Quantitative analysis of spiked SARS-CoV-2 RNA levels in artificial saliva.
| SARS-CoV-2 gene | Spiked concentration (fg/mL) | Detected concentration (fg/mL) | Recovery (%) |
|---|---|---|---|
| ORF gene | 1.00 × 103 | 1.09 ± 0.30 × 103 | 109.42 |
| 1.00 × 105 | 1.11 ± 0.13 × 105 | 111.33 | |
| S gene | 1.00 × 103 | 0.97 ± 0.12 × 103 | 96.54 |
| 1.00 × 105 | 1.01 ± 0.17 × 105 | 101.21 |
4. Conclusions
The clinical unmet need in the COVID-19 pandemic era is to provide the rapid and precise diagnostic tools to general public sectors as well as hospitals. However, we are still facing difficulties in the development of a novel biosensor to identify the virus in real time. Herein, the novel electrochemical biosensor utilizing the trans-cleavage activity of Cas13a was presented for the sensitive quantification of SARS-CoV-2 RNA without a nucleic acid amplification step. A nanocomposite and AuNF were sequentially deposited on the electrode surface to enhance the sensing performance by improving conductivity and enlarging the sensing surface area. Optimal combinations of Cas13a–crRNA in the complex formation were determined to maximize the signal enhancement of the sensor by adjusting the proper ratio of Cas13a and crRNA in the complex and measuring the trans-cleavage activity after the accommodation of the SARS-CoV-2 RNA. Our CRISPR/Cas13a-assisted electrochemical sensor could detect the ORF and S genes in a wide linear dynamic range from 1.0 × 10−1 to 1.0 × 105 fg/mL with LOD of 4.4 × 10−2 fg/mL for ORF gene and 8.1 × 10−2 fg/mL for S gene. To the best of our knowledge, the quantification under fg/mL of the SARS-CoV-2 RNA has been for the first time achieved with our proposed sensing platform. Furthermore, the observed recovery (96.5–101.21%) indicates the excellent applicability of our electrochemical sensor in a saliva matrix and offers the feasibility for the direct use of saliva samples without the viral RNA purification. In the future, our sensing platform will be integrated into an all-in-one type cartridge including sequential procedures from the saliva collection and pre-mix for activation of Cas13a to electrochemical detection for viral RNA quantification, allowing the realization of point-of-care-testing (POCT) by the direct usage of the salivary sample. Additionally, its utility will be able to expand onto the various applications of on-site detection of infectious diseases, conferring an invaluable promise as a monitoring platform with a highly sensitive and analytical performance despite excluding the pre-amplification of nucleic acids.
CRediT authorship contribution statement
Woong Heo: Conceptualization, Methodology, Validation, Visualization, Formal analysis, Writing – original draft. Kyungyeon Lee: Conceptualization, Methodology, Validation, Visualization, Formal analysis, Writing – original draft. Sunyoung Park: Methodology, Validation, Visualization, Formal analysis, Funding acquisition, Writing – review & editing. Kyung-A Hyun: Conceptualization, Methodology, Validation, Writing – review & editing. Hyo-Il Jung: Supervision, Project administration, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No.2020R1A5A1018052, No. 2021R1A2C301125411), Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, the Ministry of Food and Drug Safety) (Project Number: 1711139115, KMDF_PR_20210527_0008), Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET), funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (320035031HD030).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113960.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abdelrahman Z., Li M., Wang X. Front. Immunol. 2020;11:552909. doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antiochia R. Biosens. Bioelectron. 2020;173:112777. doi: 10.1016/j.bios.2020.112777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y.M., Flamholz A., Phillips R., Milo R. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruch R., Johnston M., Kling A., Mattmüller T., Baaske J., Partel S., Madlener S., Weber W., Urban G.A., Dincer C. Biosens. Bioelectron. 2021;177:112887. doi: 10.1016/j.bios.2020.112887. [DOI] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Zhou H.S. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Somoza R.A., Wang L., Welter J.F., Li Y., Caplan A.I., Liu C.C. Angew. Chem. 2019;131(48):17560–17566. doi: 10.1002/anie.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Puig H., Lee R.A., Najjar D., Tan X., Soenksen L.R., Angenent-Mari N.M., Donghia N.M., Weckman N.E., Ory A., Ng C.F. Sci. Adv. 2021;7(32) doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozouni P., Son S., de León Derby M.D., Knott G.J., Gray C.N., D'Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I. Cell. 2021;184(2):323–333. doi: 10.1016/j.cell.2020.12.001. e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Ding L., Zhou J., Chen S., Chen F., Zhao C., Xu J., Hu W., Ji J., Xu H. Biosens. Bioelectron. 2021;171:112685. doi: 10.1016/j.bios.2020.112685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsky A., Kobak D. 2021. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashefi-Kheyrabadi L., Kim J., Chakravarty S., Park S., Gwak H., Kim S.-I., Mohammadniaei M., Lee M.-H., Hyun K.-A., Jung H.-I. Biosens. Bioelectron. 2020;169:112622. doi: 10.1016/j.bios.2020.112622. [DOI] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. Nat. Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter J.S., de Meulder D., Bestebroer T.M., Lexmond P., Mulders A., Richard M., Fouchier R.A., Herfst S. Nat. Commun. 2021;12(1):1–8. doi: 10.1038/s41467-021-21918-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., Kim J., Kim N.H., Kim B.-T., Kim H.G. Biosens. Bioelectron. 2021;171:112715. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang Y., Ji M., Pei F., Zhao Q., Zhou Y., Hong Y., Han S., Wang J., Wang Q. Front. Cell Dev. Biol. 2020;8:618. doi: 10.3389/fcell.2020.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrese M., Guarino V., Ambrosio L. J. Funct. Biomater. 2017;8(1):7. doi: 10.3390/jfb8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski E.A., Patel N., Leung P.J.Y., Islam Z., Kooistra R.M., Parikh I., Marion E., Knott G.J., Doudna J.A., Ny A.M.L. Madan. D. 2021;24(9):102996. doi: 10.1016/j.isci.2021.102996. iScience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri R., Tang Z., Dong M., Liu T., Kshirsagar A., Guan W. 2021. Biosensors and Bioelectronics 113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleghani N., Taghipour F. 2020. Biosensors and Bioelectronics 112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J.E., Berendsen J.T., Steenbergen R.D., Wolthuis R.M., Eijkel J.C., Segerink L.I. Biosens. Bioelectron. 2020;166:112445. doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chen X., Wang X., Han X., Liu S., Zhao C. Biosens. Bioelectron. 2011;30(1):151–157. doi: 10.1016/j.bios.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang R., Li J. Biosens. Bioelectron. 2020:112430. doi: 10.1016/j.bios.2020.112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2021. WHO Coronavirus (COVID-19) Dashboard.https://covid19.who.int/ URL. [Google Scholar]
- Xu L., Li D., Ramadan S., Li Y., Klein N. Biosens. Bioelectron. 2020:112673. doi: 10.1016/j.bios.2020.112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Yan Y., Que H., Yang T., Cheng X., Ding S., Zhang X., Cheng W. ACS Sens. 2020;5(2):557–562. doi: 10.1021/acssensors.9b02461. [DOI] [PubMed] [Google Scholar]
- Zhang F., Abudayyeh O.O., Gootenberg J.S. 2020. A Protocol for Detection of COVID-19 Using CRISPR Diagnostics 8. [Google Scholar]
- Zhu J., Guo J., Xu Y., Chen X. J. Infect. 2020;81(3):e48–e50. doi: 10.1016/j.jinf.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X., Fan C., Chen H.-Y. Nature Biomedical Engineering. 2017;1(6):1–2. doi: 10.1038/s41551-017-0128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.