An artificial protein cage can be programmed to disassemble on demand.
Abstract
Engineered protein cages are promising tools that can be customized for applications in medicine and nanotechnology. A major challenge is developing a straightforward strategy for endowing cages with bespoke, inducible disassembly. Such cages would allow release of encapsulated cargoes at desired timing and location. Here, we achieve such programmable disassembly using protein cages, in which the subunits are held together by different molecular cross-linkers. This modular system enables cage disassembly to be controlled in a condition-dependent manner. Structural details of the resulting cages were determined using cryo–electron microscopy, which allowed observation of bridging cross-linkers at intended positions. Triggered disassembly was demonstrated by high-speed atomic force microscopy and subsequent cargo release using an encapsulated Förster resonance energy transfer pair whose signal depends on the quaternary structure of the cage.
INTRODUCTION
Protein cages are well-defined, hollow nanostructures self-assembled from multiple protein subunits. They are widespread in nature and display diverse morphologies and functions, with viral capsids and ferritin being among the most well-known examples (1). Natural protein cages have been functionalized for potential uses across biotechnology and medicine including as nanoreactors (2–4), building blocks to construct nanomaterials (5, 6), and display/delivery vehicles (7–9). Their broad prospects have inspired construction of synthetic equivalents using engineered protein building blocks that do not assemble into cage-like structures in the naturally occurring state (10). Artificial cages have the added advantage that aspects of their structure and functional characteristics can be controlled in a tailored manner (11–13).
A major focus of current artificial protein cage design is to control assembly-disassembly processes such that they can be triggered by stimuli exogenous to the cage and its milieu such as light or addition of chemical agents. Protein cages using native (14) or designed, native-like (15, 16) protein-protein interfaces to drive self-assembly into defined quaternary structures have been produced and, where cages comprised two or more different protein subunits, their formation can be triggered upon mixing (17, 18). In addition, triggered disassembly by exogenous stimuli allows release of encapsulated cargo molecules. We define encapsulation as the placement of a cargo molecule or nanoparticle fully within the protein cage lumen, achieved via one or more types of interaction between the cargo and cage including electrostatic and covalent bonds (e.g., genetic fusion). Metal coordination has been used as part of (19, 20) or the sole (21) bonding chemistry to connect the building blocks of protein cages in a reversible fashion. In the latter case, engineered cytochrome cb562 variants were shown to assemble into dodecameric and hexameric cages with Fe(III) and Zn(II) and disassemble upon addition of the chelating agent EDTA (21).
Tryptophan RNA-binding attenuation protein (TRAP) is a bacterial, ring-shaped homo 11-mer (22). We previously described a modified TRAP [TRAP (K35C/R64S)] that can assemble with Au(I) into a hollow cage-like structure, 22 nm in diameter and consisting of 24 rings, referred to as TRAP-cageAu(I), in which the role of Au(I) is to form linear coordination bonds bridging cysteines of neighboring rings (23–25). An additional modification at position 64 (R64S) was added to reduce positive charge around the central hole of the TRAP ring (23).
In this work, we aimed to produce a protein cage whose disassembly is programmable. In a programmable rather than simple triggerable system, the same cages should be readily customized to respond to different stimuli as desired. To achieve this goal, we imagined a protein cage in which building blocks are held together by cross-linkers that could be easily interchanged. We envisaged that these cross-linkers would be situated in place of Au(I) but carrying out a similar function, i.e., connecting the thiols of opposing cysteines from different TRAP rings (Fig. 1A). Disassembly of such cages would depend on the cleavage characteristics of the cross-linkers used. To test this possibility, we used TRAP (K35C/R64S) as a building block.
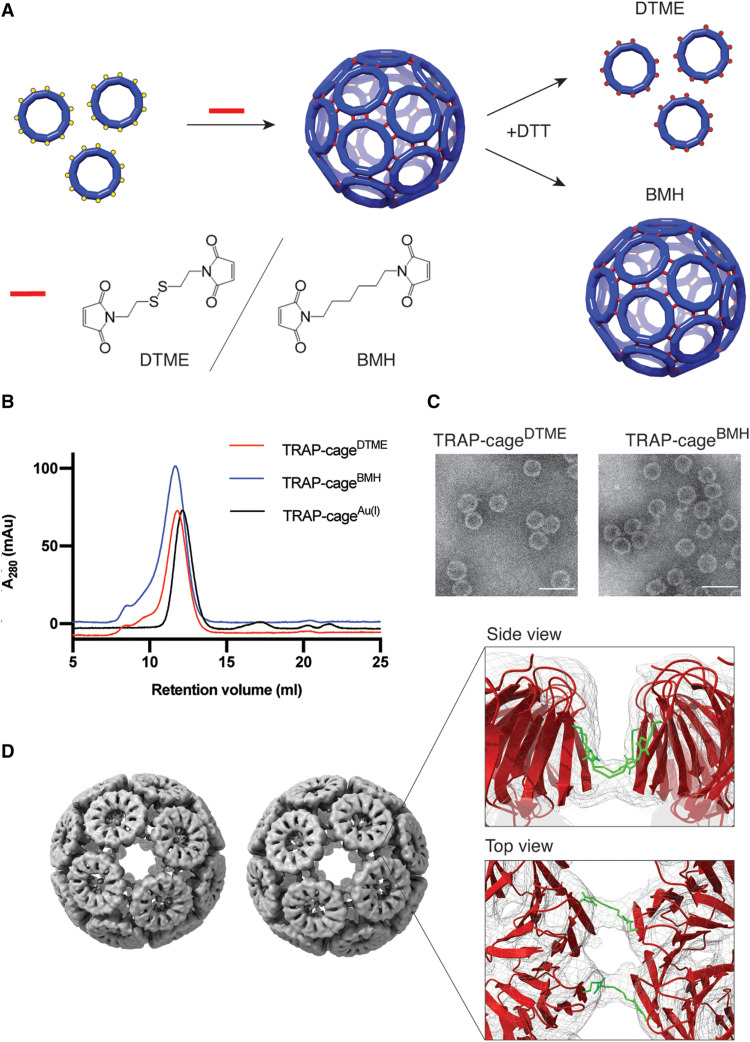
Fig. 1. Molecular cross-link–mediated TRAP-cage formation.
(A) Schematic representation of the cross-linking reaction with dithiobismaleimidoethane (DTME) or bismaleimidohexane (BMH). TRAP(K35C/R64S) rings are covalently connected to each other via reaction between the cysteines (represented by yellow spheres) and the bismaleimide compounds (represented by red lines) to form a cage structure. In the formed cages, 1 of every 11 cysteines is not cross-linked and remains unrepresented in the cage cartoon. Addition of dithiothreitol (DTT) results in disassembly of the DTME-mediated cage (TRAP-cageDTME, top, red dots represent cleaved cross-linkers) but has no effect on cages assembled with BMH (TRAP-cagesBMH, bottom). (B) Size-exclusion chromatography (SEC) profiles of the purified TRAP-cageDTME (red line) and TRAP-cageBMH (blue line). The profile of TRAP-cageAu(I) (black line) is provided as a control. (C) Transmission electron microscopy (TEM) images of TRAP-cageDTME (right) and TRAP-cageBMH (left). Scale bars, 50 nm. (D) Cryo–electron microscopy (cryo-EM) density maps of the left-handed (left) and right-handed (right) forms of TRAP-cageDTME, refined to 4.7 and 4.9 Å resolution, respectively (EMD-11415 and EMD-11414, respectively). Inset: A magnified image of the ring-ring interface with the density map in gray mesh, fitted protein model as a red cartoon, and fitted cross-linker models highlighted as green sticks, side view (top) and top view (bottom).
RESULTS
Cage formation via molecular cross-linkers
We attempted to link the TRAP rings with bifunctional molecular cross-linkers, either dithiobismaleimidoethane (DTME) or bismaleimidohexane (BMH). These were expected to serve as connectors analogous to Au(I) in TRAP-cageAu(I), owing to the thiol-specific reaction at neutral pH. As DTME contains a cleavable disulfide bond but BMH does not, the resulting two cages (TRAP-cageDTME and TRAP-cageBMH) should have contrasting disassembly characteristics when exposed to reducing agents (Fig. 1A).
To obtain the covalently cross-linked cages, TRAP (K35C/R64S) was mixed with either DTME or BMH in an aqueous buffer. Size-exclusion chromatography (SEC) of the resulting reaction mixtures showed a peak at an elution volume similar to that of TRAP-cageAu(I) for both cross-linkers, suggesting successful cage formation (fig. S1). The isolated fractions were further analyzed by SEC (Fig. 1B), dynamic light scattering (DLS) (table S3), and negative-stain transmission electron microscopy (TEM) (Fig. 1C and fig. S2). These results were consistent with the successful formation of monodisperse spherical cage structures ~25 nm in diameter. Assembly appeared to be complete within 60 min (fig. S3). No free cysteines were detected after reaction, while denaturation of the cages resulted in the presence of otherwise undetectable TRAP dimers, in line with the expected thiol-mediated covalent bond formation where TRAP monomers of one ring are linked to monomers of a neighboring ring resulting in covalent dimer formation (fig. S4). Further analysis of the obtained TRAP-cages using SEC coupled with right-angle and low-angle light scattering (SEC-RALS/LALS) indicated the apparent average molecular masses of both particles as ~2.2 MDa, suggesting a 24-ring arrangement (table S4).
The detailed structures of both TRAP-cageDTME and TRAP-cageBMH were determined using cryo–electron microscopy (cryo-EM) single-particle reconstruction. We obtained electron density maps at 4.7 and 4.9 Å resolution for the two types of cages, respectively. These structures revealed that each cage was composed of 24 TRAP rings arranged into two chiral forms, similar to that seen for TRAP-cageAu(I) (25) (Fig. 1D, figs. S5 to S8, and tables S5 and S6). TRAP ring models were fit into the cryo-EM maps, and closer examination of the ring-ring interface revealed two electron densities bridging two opposing subunits, which likely correspond to the bismaleimide cross-linkers (Fig. 1D). The cross-linkers appear to be bent in a horseshoe shape between the opposing cysteine residues.
Stability of the cross-linked TRAP-cages
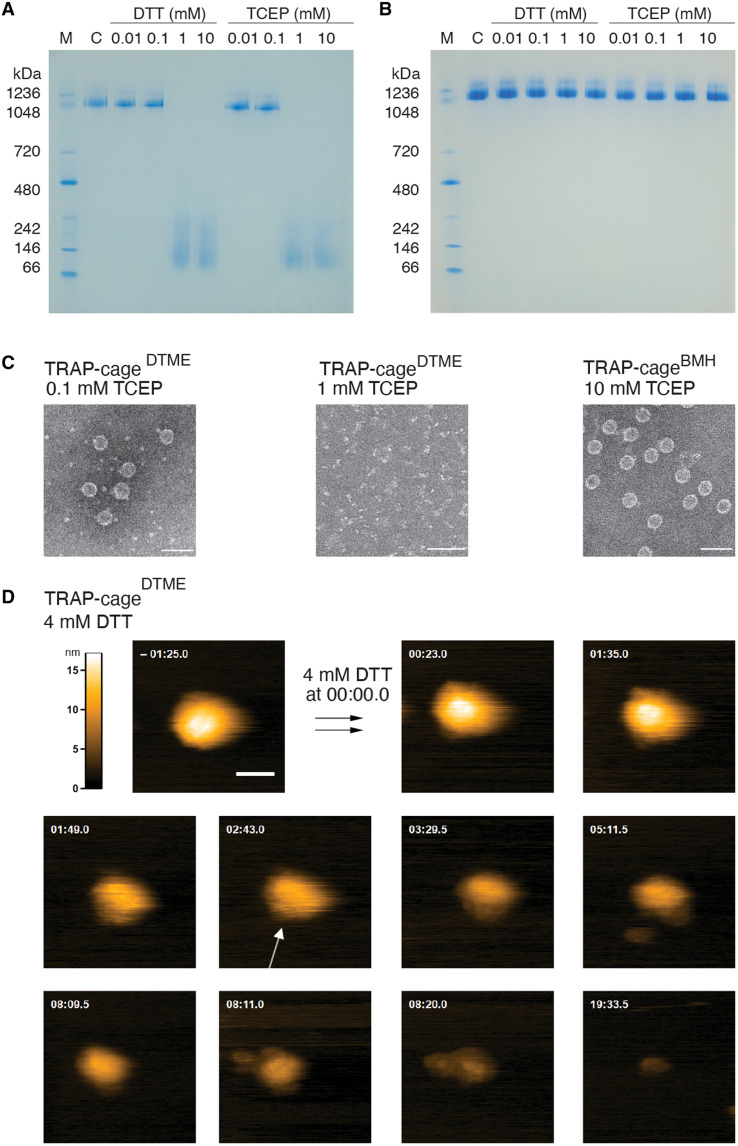
Both TRAP-cageDTME and TRAP-cageBMH showed similarly high stability in response to elevated temperatures, chaotropic agents, and surfactants. Specifically, they displayed no notable morphology change after 10-min incubation at 75°C, pHs in the range of 3 to 11, up to 4 M GdnHCl, up to at least 7 M urea, 7% of SDS, and 10% serum (figs. S9 and S10). However, TRAP-cageDTME readily disassembles, as expected, upon addition of reducing agents tris(2-carboxyethyl) phosphine (TCEP) or dithiothreitol (DTT) (Fig. 2, A and C, and fig. S11). In contrast, TRAP-cageBMH was unaffected by TCEP or DTT (Fig. 2, B and C, and fig. S11). In comparison, TRAP-cageAu(I) was previously shown to have similar but slightly higher levels of stability, being stable for at least 180 min at 95°C, at pHs 2 to 13, >7 M urea, and > 5% SDS. Lower heat stability is also implied by comparison of TEM images of TRAP-cageDTME and TRAP-cageBMH before heating (fig. S2) and after 10-min incubation at 95°C (figs. S9A, bottom, and S10A, bottom, respectively). Postheating shows some deformation and aggregation of cage structures, suggesting that the cages are undergoing denaturation/aggregation processes.
Fig. 2. Redox responsiveness of TRAP-cages.
(A and B) Native polyacrylamide gel electrophoresis (PAGE) analysis of TRAP-cageDTME (A) and TRAP-cageBMH (B) in the presence of DTT and tris(2-carboxyethyl) phosphine (TCEP). “C” in (A) and (B) denote TRAP-cageDTME and TRAP-cageBMH, respectively. M, molecular weight marker. (C) TEM images showing effects of 0.1 mM (left) and 1 mM (middle) TCEP on TRAP-cageDTME and the effect of 10 mM TCEP addition on TRAP-cageBMH (right). Scale bars, 50 nm. (D) Selected frames from a high-speed atomic force microscopy (HS-AFM) movie (movie S1) of TRAP-cageDTME, taken at two frames/s, 200 nm by 200 nm, 200 × 200 pixel, showing the effect of 4 mM DTT addition to TRAP-cageDTME. Time after addition of DTT indicated. A TRAP ring subunit within the TRAP-cage is indicated with a white arrow. Scale bar, 50 nm, in frame t = −01:25.0. Z color scale is set to −1 to 16 nm (lookup table, top left).
To understand more details of the mechanism of DTT-dependent disassembly in TRAP-cageDTME, we further investigated at the single-cage level in real time using high-speed atomic force microscopy (HS-AFM). This confirmed that, in contrast to TRAP-cageBMH, TRAP-cageDTME readily disassembles into discrete patches with TRAP subunits appearing to “peel off” from the cage surface, eventually leading to the opening of the whole structure approximately 2 min after the first ring detachment (Fig. 2D, fig. S12, and movies S1 and S2). Cage disassembly observed in AFM upon reducing agent addition was confirmed to be due to the action of the reducing agent rather than the AFM tip (fig. S13 and movie S3). Such a stepwise disassembly process of the TRAP-cageDTME is markedly different from TRAP-cageAu(I), which showed an almost concerted breaking mechanism on a much shorter time scale (23).
Packaging of protein cargoes in the cross-linked TRAP-cages
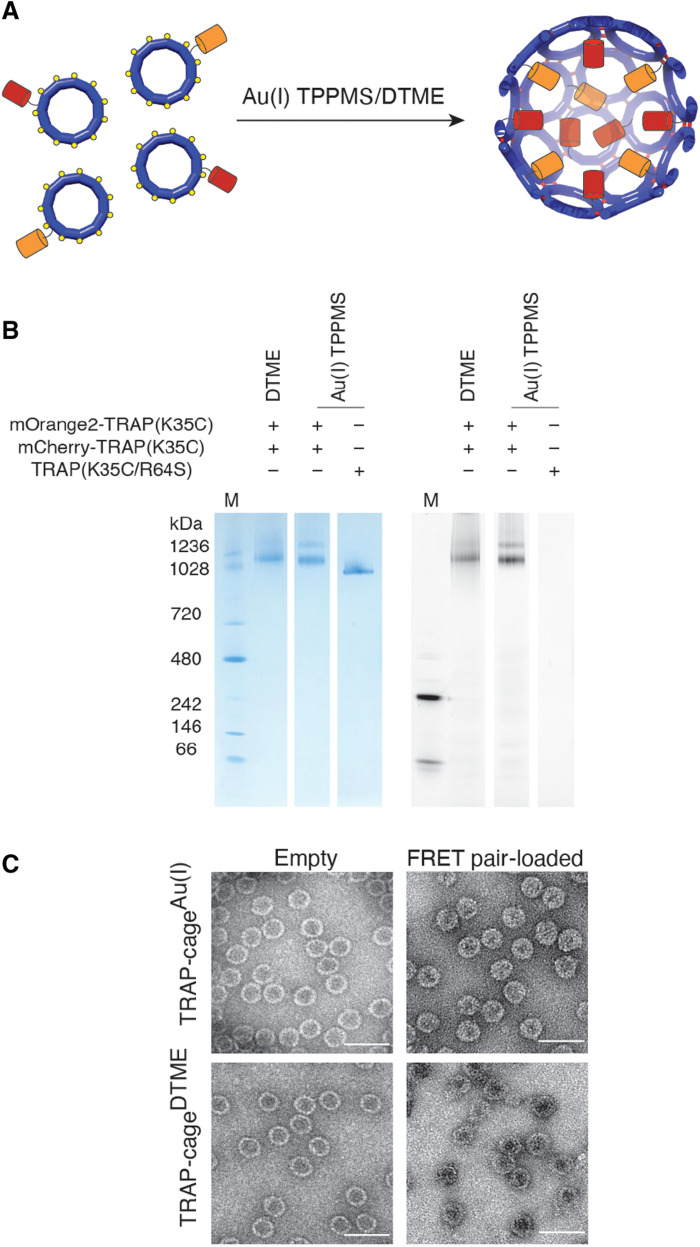
To facilitate cage stability tests against various thiol-containing reagents, we used a spectroscopic method (26) for real-time monitoring of bulk cage disassembly in solution. In this system, we encapsulated two fluorescent proteins, mOrange2 and mCherry, serving as a Förster resonance energy transfer (FRET) donor and acceptor, respectively. These fluorescent proteins are genetically fused to the TRAP N terminus, which faces the cage interior in the assemblies and were coproduced with unmodified TRAP to form “patchwork” rings, thus avoiding steric hindrance during cage formation (27). Such covalent fusion to a shell-forming protein is a widely used strategy for encapsulation of desired guests in protein cages (1, 27, 28). The resulting TRAP ring-fluorescent protein fusions were then assembled into cages using either Au(I) or DTME (Fig. 3A). After purification using SEC, isolated particles were analyzed by native polyacrylamide gel electrophoresis (PAGE) combined with fluorescence imaging (Fig. 3B and fig. S14) and TEM imaging (Fig. 3C), which confirmed encapsulation of the FRET labels in the lumen of monodisperse spherical cages. Guest protein stoichiometry was determined using absorbance ratio at 280/548 nm or 280/587 nm, showing that approximately 12 ± 3 mOrange2 and 8 ± 1 mCherry proteins were encapsulated per cage (see the details in Materials and Methods) (29).
Fig. 3. Packaging protein cargoes in TRAP-cages.
(A) Schematic representation of TRAP-cage loading with fluorescent proteins. Patchworked TRAP rings fused with either mCherry (red cylinders) or mOrange2 (orange cylinders) at the N terminus were mixed together with either DTME or triphenylphosphine monosulfate (TPPMS)–Au(I)-Cl. (B) Native PAGE showing the fluorescent properties of purified TRAP-cages associated with the fluorescent cargoes. The gel was visualized using InstantBlue protein staining (left) and fluorescence using excitation at 532 nm and emission at 610 nm (right). (C) TEM images of empty (left) TRAP-cages and those filled with fluorescent proteins (right), assembled using either Au(I) (top) or DTME (bottom). Scale bars, 50 nm.
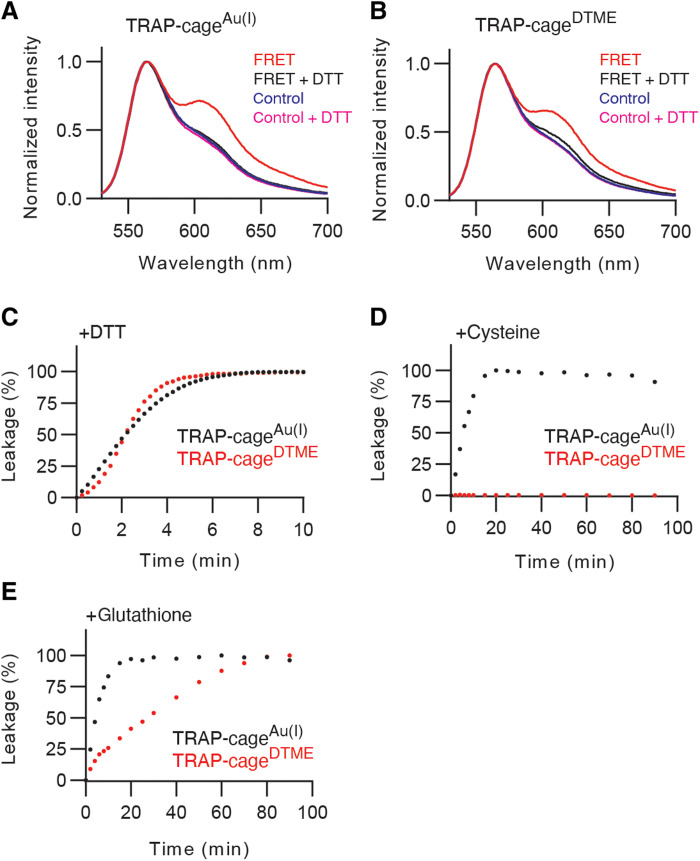
The presence of both proteins in the constrained volume of the TRAP-cage lumen should enable efficient FRET (28, 30). Such intracage energy transfer should result, upon mOrange2 excitation, in decrease in the donor fluorescence and increase in the acceptor mCherry signal (compared to when free in solution). Cage disassembly by stimulus, however, should lead to release of these guests from the spatial confinement to the surrounding environment, cancelling the FRET effect (26). All the spectra in this study are normalized at mOrange2 fluorescence peak to judge their colocalization in TRAP-cages by the relative values of fluorescence intensity ratios (28). The processed fluorescence spectra of TRAP-cageAu(I) and TRAP-cageDTME copackaging the FRET pair showed an approximately 1.5-fold higher signal in mCherry emission at 610 nm, compared to the corresponding control samples containing cages encapsulating only mCherry or mOrange2 mixed in solution (Fig. 4, A and B). However, the addition of DTT, which induces disassembly for both types of cages, led to FRET cancellation as observed in the resulting spectra where fluorescence levels were reduced to nearly the same as that of the corresponding control samples (Fig. 4, A and B). These results indicated efficient energy transfer between the FRET labels by encapsulation and its cancellation by TRAP dissociation, indicating that the assay can be used to monitor the TRAP-cage disassembly process.
Fig. 4. FRET label release upon TRAP-cages disassembly.
(A and B) Normalized emission spectra of TRAP-cagesAu(I) (A) and TRAP-cagesDTME (B) loaded with both mOrange2 and mCherry upon excitation at 510 nm before (red lines) and after addition of 10 mM DTT (black lines). mOrange2 emission peak is visible at 568 nm, mCherry emission peak at 610 nm. The blue and magenta lines indicate spectra of cages loaded only with mOrange2 or mCherry proteins mixed together immediately before measurement in the absence or presence of DTT, respectively. (C to E) Time-dependent disassembly of TRAP-cagesAu(I) (black circles) and TRAP-cagesDTME (red circles) after addition of 10 mM DTT (C), 2.5 mM cysteine (Cys) (D), or 50 mM glutathione (GSH) (E). A total of 100% leakage denotes the highest donor intensity after 10 min of 10 mM DTT treatment.
Kinetics of cross-linked TRAP-cage disassembly
We next measured the disassembly kinetics of both TRAP-cageAu(I) and TRAP-cageDTME upon DTT addition. The change in the fluorescence intensity ratio at 568/610 nm was exploited to track the time-dependent disassembly process (Fig. 4C). For both cages, the increase in the fluorescence ratio reached a plateau approximately 5 min after DTT addition, indicating complete cargo liberation. However, the mechanism of this process appeared to be different as indicated by the curve shapes. The pseudo-exponential curve for TRAP-cageAu(I) suggests that there is a notable rate acceleration over the bond breaking reactions at the single ring and cage levels. In contrast, the sigmoidal curve obtained for TRAP-cageDTME with DTT indicates that individual bond cleavage events are less cooperative compared to the TRAP-cageAu(I) case (see the detailed discussion in fig. S15). The hypothesis of two distinct cage-breaking mechanisms is greatly supported by the HS-AFM results, showing a concerted or a stepwise disassembly of TRAP-cageAu(I) and TRAP-cageDTME, respectively (movie S1).
Protein cages able to carry cargo and disassemble in the presence of reducing agents have potential applications in intracellular delivery. Ideally, such nanovehicles should remain intact under extracellular conditions but disassemble upon exposure to intracellular environments, liberating their cargoes. We assessed this possibility by monitoring cage disassembly kinetics in the presence of cysteine and glutathione (GSH) used as model thiol-containing compounds.
A time-dependent increase in FRET cancellation was observed for TRAP-cageAu(I) upon cysteine addition, plateauing (indicating complete cargo release) after approximately 15 min when 2.5 mM cysteine was used and much more quickly at higher cysteine concentrations (Fig. 4D and fig. S16A). In contrast, TRAP-cageDTME containing the same FRET pair showed essentially no change in fluorescence, suggesting that the cage did not disassemble under these conditions (Fig. 4D and fig. S16A). The observed stability toward cysteine is likely due to the slow kinetics of the covalent bond cleavage reaction catalyzed by free thiol, in contrast to likely a much faster ligand exchange for TRAP-cageAu(I). We found that disassembly of TRAP-cageDTME can be induced by increasing the concentration and redox potential of free thiol-containing compound: The reduced form of GSH could open TRAP-cageDTME despite the disassembly rate being notably slower than for TRAP-cageAu(I) (Fig. 4E and fig. S16B).
DISCUSSION
With naturally occurring protein cages serving as an inspiration, artificial cages are being developed with customized capabilities (10, 11). This study demonstrates that connecting protein building blocks with molecular cross-linkers provides a straightforward mechanism to produce artificial protein cages with programmable disassembly properties and that are free of both protein-protein interactions and metals as a means of connecting the building blocks. The increased tolerance of TRAP-cageDTME to cysteine highlights its potential utility as a therapeutic delivery vehicle designed to survive in extracellular environments. Incorporation of a FRET pair cargo exemplifies the potential of TRAP-cages to act as efficient, generalized containers for protein guests, while it also presents itself as a rigorous and potentially universal method for monitoring cage disassembly. We expect that the modular design strategy presented here will lay the groundwork for further development of protein nanocontainers having specific cargo packaging and release properties.
MATERIALS AND METHODS
Plasmids
pET21b_TRAP-K35C R64S (25) was used for expression of the TRAP(K35C/R64S) variants. The tetracycline-inducible protein expression vectors, pACTet_H-mOrange-TRAP-K35C or pACTet_H-mCherry-TRAP-K35C, were derived from pACTet_H-mOrange or pACTet_H-mCherry, respectively. These parent plasmids were gifts from D. Hilvert (ETH Zürich). pET21b_TRAP-K35C was synthesized by BioCat GmbH. All the molecular cloning details, as well as the primer (table S1), plasmid, and protein sequences (table S2), used in this study are described in the Supplementary Materials.
Protein expression and purification
TRAP(K35C/R64S) was produced using Escherichia coli strain BL21(DE3) cells that were transformed with pET21b_TRAP-K35C R64S as described previously (25). The protein was purified by ion-exchange chromatography using HiTrap Q FF column, followed by SEC on a HiLoad 26/600 Superdex 200 pg column. To produce patchwork TRAP rings, E. coli strain BL21(DE3) cells were cotransformed with either pACTet_H-mOrange-TRAP-K35C or pACTet_H-mCherry-TRAP-K35C and pET21_TRAP-K35C (see table S2). Protein expression was induced by addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside and tetracycline (10 ng/ml) in the case of pACTet_H-mCherry-TRAP-K35C or tetracycline (30 ng/ml) in the case of pACTet_H-mOrange-TRAP-K35C. Patchwork TRAP rings were then isolated using Ni–nitrilotriacetic acid (NTA) affinity chromatography, followed by SEC using a Superdex 200 Increase 10/300 GL column (GE Healthcare). Protein purity was checked by SDS-PAGE, and protein concentration was determined by absorbance measured using extinction coefficients: ε mCherry 587 = 72,000 M−1 cm−1, ε mOrange 548 = 58,000 M−1 cm−1 (31), and ε TRAP 280 = 8250 M−1 cm−1 (http://expasy.org/tools/protparam.html). The detailed procedure of protein production is described in the Supplementary Materials.
Cage assembly and characterization
DTME and BMH were purchased from Thermo Fisher Scientific. The molecular cross-linkers were reconstituted in dimethyl sulfoxide (DMSO) to 20 mM stock concentration before use. TRAP(K35C/R64S) (100 to 500 μM) in 2× phosphate-buffered saline (PBS) containing 2 mM EDTA, referred to as 2× PBS-E was mixed with fivefold molar excess of either DTME or BMH and stirred at room temperature for 1 hour. Final DMSO concentration in solution was kept at no greater than 12.5%. After the reaction, the insoluble fraction, likely due to low solubility of cross-linkers in aqueous solution, was removed by centrifugation for 5 min at 12,000g. Supernatants were then purified by SEC using a Superose 6 Increase 10/300 GL column (GE Healthcare) at a flow rate of 0.5 ml/min on an ÄKTA purifier fast protein liquid chromatography (GE Healthcare). Fractions containing cross-linked TRAP-cages were pooled and concentrated using Amicon Ultra-4 (100 kDa molecular mass cut-off) centrifugal filter units. Approximately 4 mg of TRAP(K35C/R64S) was used for cage formation. After purification and concentration, typically, 1 mg of cross-linker induced cages was obtained, resulting in a yield of approximately 25%. Formation and purification of Au(I)-induced TRAP-cages were performed as previously described (25). Cage formation with fusion proteins was performed using the same protocols as described for both cross-linked and Au(I)-induced cages with an additional Ni-NTA purification step before SEC to purify the sample away from partially assembled cages (His-tagged mCherry and mOrange2 that are not fully protected inside the cages bind to Ni-NTA column). The protein concentration and ratio of encapsulated guests were estimated using the absorbance ratio at 280/548 nm or 280/587 nm using an analogous method to the one previously reported (29, 32). Extinction coefficients used for calculations were ε mCherry 587 = 72,000 M−1 cm−1, ε mOrange2 548 = 58,000 M−1 cm−1, and ε TRAP 280 = 8250 M−1 cm−1. Because of spectral overlap between mCherry and mOrange2, to properly calculate the concentrations of both encapsulated guests, mCherry extinction coefficients was also estimated at 548 nm (ε mCherry 548 = 42,500 M−1 cm−1) using the absorbance ratio at 548/587 nm of mCherry without fusion to TRAP. Likewise, the extinction coefficients of mCherry and mOrange2 at 280 nm were experimentally determined as ε mCherry 280 = 56,700 M−1 cm−1 and ε mOrange2 280 = 52,200 M−1 cm−1, respectively. Chemical cross-linking was assessed by tricine–SDS-PAGE, and the morphological fidelity of assembled cages was confirmed by DLS, RALS/LALS, negative stain TEM, and native PAGE analysis. The detailed procedure of the individual method is provided in the Supplementary Materials.
High-speed atomic force microscopy
Before the HS-AFM experiment, TRAP-cageDTME and TRAP-cageBMH were diluted to 10 to 30 μg/ml with a 10 mM phosphate buffer (pH 7.4) containing 400 mM NaCl and 3 mM (for TRAP-cageDTME) or 10 mM (for TRAP-cageBMH) KCl. Two to three microliters of the sample was applied to the freshly cleaved mica. HS-AFM (SS-NEX, Research Institute of Biomolecule Metrology, Japan) (33, 34) with a laboratory-built phase-shift amplitude detector (35) and an automated force controller (36) was used. Ultra-Short Cantilevers (USC-F1.2-k0.15, NanoWorld, Switzerland), with nominal spring constant of 0.15 N/m, were excited at 550 to 650 kHz with a free amplitude of 2 to 3 nm (peak to peak). Images were taken with 150 × 150 pixel at 300 nm by 300 nm at two frames/s for TRAP-cageBMH and 200 × 200 pixel at 200 nm by 200 nm at two frames/s or 200 × 200 pixel at 600 nm by 600 nm at one frame/s for TRAP-cageDTME. DTT (4 or 10 mM as the final concentration) was added to the observation buffer by pipetting during HS-AFM observation. For TRAP-cageDTME, single cages were monitored before and after DTT addition. For TRAP-cageBMH, the scan area was changed several times after DTT addition to distinguish between cage damage induced by HS-AFM applied tapping forces or DTT addition. The obtained sequential HS-AFM images were analyzed and processed using laboratory-made Igor Pro (WaveMetrics, USA)–based functions. First, the frames were contrast adjusted, and then the minor lateral drift was corrected using an iterative subpixel registration algorithm (37).
Cage stability
Stability of TRAP-cages against chemicals and heat was tested using a similar method to that described previously (25). All agents used for the assays (DTT, TCEP, SDS, Gdn-HCl, urea, and rat serum) were reconstituted or diluted in PBS, mixed with TRAP-cage samples, and incubated at room temperature or 37°C (for rat serum only) overnight. Thermal stability assessment was performed by heating samples at different temperatures for 10 min. The samples were then subjected to native PAGE. These experiments were repeated twice, each giving uniform results.
Cryo–electron microscopy
Preparation of samples in vitreous ice was carried out using 4 μl of protein samples at ~1 mg/ml in PBS. After applying the samples on EM grids (Quantifoil 1.2/1.3, Cu, 300 mesh), they were plunge-frozen in liquid ethane using a FEI Vitrobot with the following parameters: blot force, 0; blot time, 4 s; wait time, 0 s; and drain time, 0 s. Micrographs were collected using a FEI TitanKrios cryo-microscope with 300-kV operation and a Falcon III camera at 75k magnification. A total of 4942 and 10,169 micrographs were collected for TRAP-cageDTME and TRAP-cageBMH, respectively. All micrographs were motion corrected using MotionCorr2 (38), and contrast transfer function estimation was performed using CTFFIND4 (39). Particles were picked and extracted using cryoSPARC v2.12.4 (40) first in manual mode (about 4000 particles), followed by automated mode, where initial two-dimensional (2D) classes served as a template. Extracted particles were 2D classified again to select best particles for subsequent reconstruction steps. 3D reconstruction was performed by the heterogeneous refinement protocol using EMD-4443 and EMD-4444 [TRAP-cageAu(I)] as searching models (25).
Fluorescence measurements
Fluorescent spectra were acquired at room temperature using 70 nM mOrange2 in 2× PBS-E in a 1-cm light-pass-length polystyrene cuvette on an RF-6000 Fluorescence Spectrofluorometer (Shimadzu). The proteins were excited at 510 nm, and emissions were scanned over a wavelength range from 530 to 700 nm. DTT was subsequently added to the same samples to a final concentration of 10 mM. Samples were then incubated for 10 min at room temperature, and spectra were remeasured. As previously discussed (28), the molecular environment of guest proteins can be altered by external conditions, which may lead to subtle changes in fluorophore properties even if they retain correct folding and packaging. Therefore, all the spectra in this study were normalized relative to the mOrange2 fluorescence peak to judge their colocalization in TRAP-cages by the relative values of fluorescence intensity ratios. For kinetic measurements, Cys (final, 2.5, 10, and 25 mM) or GSH (final, 10, 25, and 50 mM) was added to samples which were excited at 510 nm, and emission at 568 and 610 nm was measured at various time points over 20 to 90 min. After each measurement, 10 mM DTT was added to the samples to trigger complete cages disassembly. Time-dependent disassembly was observed as a change in the fluorescence intensity ratio at 568/610 nm and as percentage of the leakage seen for completely disassembled cages treated with DTT. Kinetics measurements were repeated twice.
Acknowledgments
Funding: This research was supported in part by PL-Grid Infrastructure. We acknowledge the MCB Structural Biology Core Facility (supported by the TEAM TECH CORE FACILITY/2017-4/6 grant from the Foundation for Polish Science) for valuable support. We also acknowledge Polish National Science Centre (Symfonia grant no. 2016/20/W/NZ1/00095) (I.S., J.G.H., and A.P.B.) and Polish National Science Centre (Maestro grant no. 2019/34/A/NZ1/00196) (J.G.H. and A.P.B). Work in the Scheuring laboratory is funded by National Institutes of Health grants NIH NCCIH DP1AT010874 and NIH NINDS – RO1NS110790. The open-access publication of this article was funded by the BioS Priority Research Area under the program “Excellence Initiative – Research University” at the Jagiellonian University in Krakow.
Author contributions: Conceptualization: J.G.H. and A.P.B. Experimental design: I.S., Y.A., A.P.B., and M.I. Performing experiments: I.S., Y.A., A.P.B., M.I., and O.W. Methodology: I.S., Y.A., and A.P.B. Funding acquisition: J.G.H. and S.S. Supervision: J.G.H., S.S., and E.P. Figure preparation: I.S. and A.P.B. Writing—original draft: I.S., Y.A., A.P.B., M.I., S.S., and J.G.H. Carried out experiments: D.P.M.
Competing interests: The authors declare the following competing interests: The authors are named on the following patents that are pending: P391 (J.G.H.), P504 (J.G.H., A.P.B., Y.A., and I.S.), P505 (J.G.H. and Y.A.), P506 (J.G.H., A.P.B., Y.A., and I. S.). J.G.H. is also the founder of and holds equity in nCage Therapeutics LLC, which aims to commercialise protein cages for therapeutic applications. Y.A. is also a member of the scientific advisory board of nCage Therapeutics. I.S. is an employee of nCage Therapeutics. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Materials and Methods
Figs. S1 to S16
Tables S1 to S6
Legends for movies S1 to S3
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
REFERENCES AND NOTES
- 1.Aumiller W. M., Uchida M., Douglas T., Protein cage assembly across multiple length scales. Chem. Soc. Rev. 47, 3433–3469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu A., Traulsen C. H. H., Cornelissen J. J. L. M., Nitroarene reduction by a virus protein cage based nanoreactor. ACS Catal. 6, 3084–3091 (2016). [Google Scholar]
- 3.Jordan P. C., Patterson D. P., Saboda K. N., Edwards E. J., Miettinen H. M., Basu G., Thielges M. C., Douglas T., Self-assembling biomolecular catalysts for hydrogen production. Nat. Chem. 8, 179–185 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Maity B., Fujita K., Ueno T., Use of the confined spaces of apo-ferritin and virus capsids as nanoreactors for catalytic reactions. Curr. Opin. Chem. Biol. 25, 88–97 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Lach M., Künzle M., Beck T., Free-standing metal oxide nanoparticle superlattices constructed with engineered protein containers show in crystallo catalytic activity. Chem. A Eur. J. 23, 17482–17486 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Chakraborti S., Korpi A., Kumar M., Stȩpień P., Kostiainen M. A., Heddle J. G., Three-dimensional protein cage array capable of active enzyme capture and artificial chaperone activity. Nano Lett. 19, 3918–3924 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Douglas T., Protein nanocage architectures for the delivery of therapeutic proteins. Curr. Opin. Colloid Interface Sci. 51, 101395 (2021). [Google Scholar]
- 8.Vujadinovic M., Vellinga J., Progress in adenoviral capsid-display vaccines. Biomedicine 6, 81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das S., Zhao L., Crooke S. N., Tran L., Bhattacharya S., Gaucher E. A., Finn M. G., Stabilization of near-infrared fluorescent proteins by packaging in virus-like particles. Biomacromolecules 21, 2432–2439 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Majsterkiewicz K., Azuma Y., Heddle J. G., Connectability of protein cages. Nanoscale Adv. 2, 2255–2264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stupka I., Heddle J. G., Artificial protein cages – inspiration, construction, and observation. Curr. Opin. Struct. Biol. 64, 66–73 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Edwardson T. G. W., Mori T., Hilvert D., Rational engineering of a designed protein cage for siRNA delivery. J. Am. Chem. Soc. 140, 10439–10442 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Marcandalli J., Fiala B., Ols S., Perotti M., de van der Schueren W., Snijder J., Hodge E., Benhaim M., Ravichandran R., Carter L., Sheffler W., Brunner L., Lawrenz M., Dubois P., Lanzavecchia A., Sallusto F., Lee K. K., Veesler D., Correnti C. E., Stewart L. J., Baker D., Loré K., Perez L., King N. P., Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell 176, 1420–1431.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padilla J. E., Colovos C., Yeates T. O., Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl. Acad. Sci. U.S.A. 98, 2217–2221 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai Y.-T., Cascio D., Yeates T. O., Structure of a 16-nm cage designed by using protein oligomers. Science 336, 1129–1129 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Hsia Y., Bale J. B., Gonen S., Shi D., Sheffler W., Fong K. K., Nattermann U., Xu C., Huang P.-S., Ravichandran R., Yi S., Davis T. N., Gonen T., King N. P., Baker D., Design of a hyperstable 60-subunit protein icosahedron. Nature 535, 136–139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King N. P., Bale J. B., Sheffler W., McNamara D. E., Gonen S., Gonen T., Yeates T. O., Baker D., Accurate design of co-assembling multi-component protein nanomaterials. Nature 510, 103–108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bale J. B., Gonen S., Liu Y., Sheffler W., Ellis D., Thomas C., Cascio D., Yeates T. O., Gonen T., King N. P., Baker D., Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huard D. J. E., Kane K. M., Tezcan F. A., Re-engineering protein interfaces yields copper-inducible ferritin cage assembly. Nat. Chem. Biol. 9, 169–176 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Cristie-David A. S., Marsh E. N. G., Metal-dependent assembly of a protein nano-cage. Protein Sci. 28, 1620–1629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golub E., Subramanian R. H., Esselborn J., Alberstein R. G., Bailey J. B., Chiong J. A., Yan X., Booth T., Baker T. S., Tezcan F. A., Constructing protein polyhedra via orthogonal chemical interactions. Nature 578, 172–176 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antson A. A., Otridge J., Brzozowski A. M., Dodson E. J., Dodson G. G., Wilson K. S., Smith T., Yang M., Kurecki T., Gollnick P., The structure of trp RNA-binding attenuation protein. Nature 374, 693–700 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Imamura M., Uchihashi T., Ando T., Leifert A., Simon U., Malay A. D., Heddle J. G., Probing structural dynamics of an artificial protein cage using high-speed atomic force microscopy. Nano Lett. 15, 1331–1335 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Malay A. D., Heddle J. G., Tomita S., Iwasaki K., Miyazaki N., Sumitomo K., Yanagi H., Yamashita I., Uraoka Y., Gold nanoparticle-induced formation of artificial protein capsids. Nano Lett. 12, 2056–2059 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Malay A. D., Miyazaki N., Biela A., Chakraborti S., Majsterkiewicz K., Stupka I., Kaplan C. S., Kowalczyk A., Piette B. M. A. G., Hochberg G. K. A., Wu D., Wrobel T. P., Fineberg A., Kushwah M. S., Kelemen M., Vavpetič P., Pelicon P., Kukura P., Benesch J. L. P., Iwasaki K., Heddle J. G., An ultra-stable gold-coordinated protein cage displaying reversible assembly. Nature 569, 438–442 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Zschoche R., Hilvert D., Diffusion-limited cargo loading of an engineered protein container. J. Am. Chem. Soc. 137, 16121–16132 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Azuma Y., Herger M., Hilvert D., Diversification of protein cage structure using circularly permuted subunits. J. Am. Chem. Soc. 140, 558–561 (2018). [DOI] [PubMed] [Google Scholar]
- 28.O’Neil A., Prevelige P. E., Basu G., Douglas T., Coconfinement of fluorescent proteins: Spatially enforced communication of GFP and mCherry encapsulated within the P22 capsid. Biomacromolecules 13, 3902–3907 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Azuma Y., Zschoche R., Tinzl M., Hilvert D., Quantitative packaging of active enzymes into a protein cage. Angew. Chem. Int. Ed. 55, 1531–1534 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Dashti N. H., Abidin R. S., Sainsbury F., Programmable in vitro coencapsidation of guest proteins for intracellular delivery by virus-like particles. ACS Nano 12, 4615–4623 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Shaner N. C., Lin M. Z., McKeown M. R., Steinbach P. A., Hazelwood K. L., Davidson M. W., Tsien R. Y., Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frey R., Mantri S., Rocca M., Hilvert D., Bottom-up construction of a primordial carboxysome mimic. J. Am. Chem. Soc. 138, 10072–10075 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Ando T., Uchihashi T., Fukuma T., High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Prog. Surf. Sci. 83, 337–437 (2008). [Google Scholar]
- 34.Ando T., Uchihashi T., Scheuring S., Filming biomolecular processes by high-speed atomic force microscopy. Chem. Rev. 114, 3120–3188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyagi A., Scheuring S., A novel phase-shift-based amplitude detector for a high-speed atomic force microscope. Rev. Sci. Instrum. 89, 083704 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Miyagi A., Scheuring S., Automated force controller for amplitude modulation atomic force microscopy. Rev. Sci. Instrum. 87, 053705 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Thevenaz P., Ruttimann U. E., Unser M., A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Schägger H., Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22 (2006). [DOI] [PubMed] [Google Scholar]
- 42.C. Vallance, “Introduction to Complex Reactions” in An Introduction to Chemical Kinetics (IOP Publishing, 2017); 10.1088/978-1-6817-4664-7, chap. 5. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Figs. S1 to S16
Tables S1 to S6
Legends for movies S1 to S3
References
Movies S1 to S3