A cell-autonomous 12-hour clock coordinates nuclear speckle LLPS with proteostasis control.
Abstract
Phase separation and biorhythms control biological processes in the spatial and temporal dimensions, respectively, but mechanisms of four-dimensional integration remain elusive. Here, we identified an evolutionarily conserved XBP1s-SON axis that establishes a cell-autonomous mammalian 12-hour ultradian rhythm of nuclear speckle liquid-liquid phase separation (LLPS) dynamics, separate from both the 24-hour circadian clock and the cell cycle. Higher expression of nuclear speckle scaffolding protein SON, observed at early morning/early afternoon, generates diffuse and fluid nuclear speckles, increases their interactions with chromatin proactively, transcriptionally amplifies the unfolded protein response, and protects against proteome stress, whereas the opposites are observed following reduced SON level at early evening/late morning. Correlative Son and proteostasis gene expression dynamics are further observed across the entire mouse life span. Our results suggest that by modulating the temporal dynamics of proteostasis, the nuclear speckle LLPS may represent a previously unidentified (chrono)-therapeutic target for pathologies associated with dysregulated proteostasis.
INTRODUCTION
Most life on earth is governed by biological rhythms that are defined as self-sustained oscillations cycling with a fixed period. Biological clocks enable organisms to keep track of the time of day and to adjust their physiology to recurring daily changes in the external environment, including nutrient and microenvironment status. Our understandings of biological rhythms in mammals have expanded beyond the well-characterized circadian rhythms (~24-hour oscillation) in recent years through the discovery of 12-hour ultradian rhythms in mammals (1, 2). In contrast to earlier hypothesis that these 12-hour rhythms are under the combined regulation of the 24-hour circadian clock and feeding/fasting cues (2, 3), our group identified a cell-autonomous mammalian 12-hour ultradian oscillator that regulates 12-hour rhythms of systemic gene expression and metabolism (4). The 12-hour oscillator is independent from the 24-hour circadian clock but instead is regulated by the unfolded protein response (UPR) transcription factor spliced form of X-Box Binding Protein 1 (XBP1s) (4–7). In mouse, the liver-specific deletion of XBP1s impaired more than 80% of 12-hour transcriptome, while leaving the majority of circadian transcriptome intact (including all known core circadian clock genes) (5, 6). As a result of the 12-hour clock ablation, XBP1s liver-specific knockout (XBP1LKO) mice exhibited markedly accelerated liver aging and fatty liver diseases (5).
Subsequent Gene Ontology (GO) analysis of XBP1s-dependent mouse hepatic 12-hour transcriptome revealed top-enriched genes involved in the entire central dogma information flow (CEDIF) process, ranging from transcription initiation, mRNA processing and export, ribosome biogenesis, translation initiation to protein folding, processing and sorting in the endoplasmic reticulum (ER) and Golgi, as well as various metabolic pathways including lipid and nucleotide metabolism (6). While the regulation of protein and lipid homeostasis by XBP1s is well established (8), the control of mRNA metabolism by XBP1s and the mechanistic link between mRNA and protein homeostasis remain poorly characterized. Therefore, in this study, we aim to uncover the underlying mechanisms of coordinated mRNA and protein metabolism by investigating the 12-hour oscillator. By combining single-cell time-lapse microscopy, cistrome profiling, and mathematical modeling, we unexpectedly identified an XBP1s-SON axis that dictates a cell-autonomous mammalian 12-hour ultradian rhythm of nuclear speckle liquid-liquid phase separation (LLPS) dynamics, which drives rhythmic global 12-hour nuclear speckle–chromatin interactions, uncoupled from the transcriptional state of individual genes. We found that the expression of genes involved in proteostasis, including Xbp1 itself, is hypersensitive to nuclear speckle LLPS dynamics change. We further observed correlative Son and proteostasis gene expression dynamics during the transient response to ER stress and across the entire mouse life span. Functionally, the XBP1s-SON axis can protect cells from proteome stress via transcriptionally amplifying the UPR. Our results thereby uncovered an intrinsic feedforward loop connecting nuclear speckle LLPS and proteostasis control that likely ensures a highly efficient genetic information transfer functioning at multiple temporal scales.
RESULTS
XBP1s regulates a cell-autonomous 12-hour rhythm of nuclear speckle morphology change
We first confirmed that the 12-hour rhythms of mRNA metabolic genes are also observed at the protein level by performing a post hoc analysis of two published hepatic nuclear protein mass spectrometry datasets (fig. S1, A and B, and table S1) (9, 10). GO analysis of 12-hour nuclear proteins in both datasets revealed top enriched GO term “spliceosome” (fig. S1C). Spliceosome is predominantly found in nuclear speckles, which are membraneless organelles enriched in pre-mRNA processing factors, as well as various other proteins and noncoding RNAs (ncRNAs) involved in RNA export, transcription regulation, pre-mRNA cleavage and polyadenylation, and RNA degradation (11, 12). We further observed a robust 12-hour expression of nuclear speckle ncRNA Malat1, which is also a direct transcriptional target gene of XBP1s and exhibits slightly dampened rhythm in XBP1 liver-specific knockout (XBP1LKO) mice from our previously published RNA sequencing (RNA-seq) dataset (fig. S1, D and E) (6).
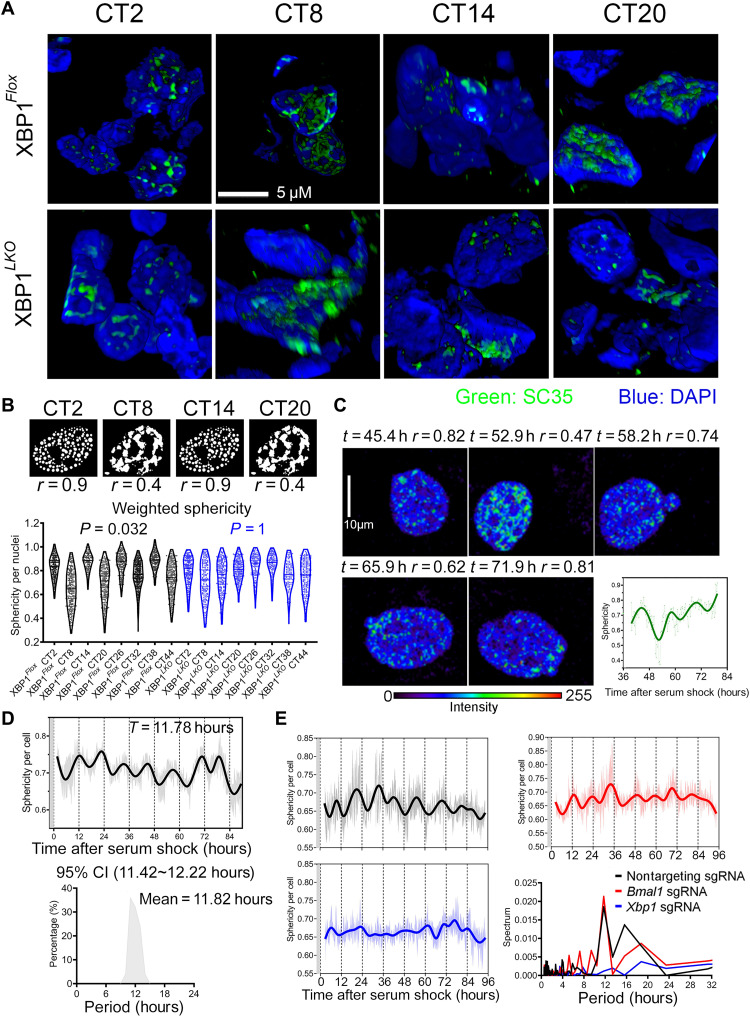
To look for additional evidence of 12-hour mRNA metabolism besides gene expression, we initially asked whether the morphologies of nuclear speckle may exhibit time-of-the-day variation. Nuclear speckles normally are microscopically presented as “punctate” nuclear localization pattern, formed through LLPS because of the prevalence of low complexity domains found in splicing factors (11–13). We performed immunofluorescence against SC35 (SRSF2), one well-established marker of nuclear speckles (12, 13), in the liver section of XBP1Flox and XBP1LKO mice at different circadian time (CTs). Consistent with previous observations (12, 14), SC35-positive loci are associated with particularly low chromatin density, but in close proximity to chromatin, at all times in both XBP1Flox and XBP1LKO mice (fig. S2A). Nonetheless, we observed markedly distinct staining patterns of SC35 in XBP1Flox mouse liver at different times of day. At CT2, CT14, CT26, and CT38, spherical punctate patterns of SC35 found in the majority of nuclei are suggestive of LLPS formed via a process termed “nucleation” (15). By contrast, at CT8, CT20, CT32, and CT44, we observed a much more diffuse and network-like spatial distribution of SC35 staining, reminiscent of LLPS formed via spinodal decomposition (Fig. 1A and fig. S2A) (15). Quantifying these two distinct forms of LLPS by calculating the roundness of nuclear speckle staining as previously described (16) evinced an apparent 12-hour rhythm of nuclear speckle LLPS dynamics in XBP1Flox [P value of 0.032 by Rhythmicity Analysis Incorporating Nonparametric (RAIN) analysis (17)], but not in XBP1LKO mice (P value of 1 by RAIN analysis) (Fig. 1B), which is further confirmed by staining against a different nuclear speckle marker SON (P value of 0.031 and 0.54 in XBP1Flox and XBP1LKO mice by RAIN analysis, respectively) (fig. S2B).
Fig. 1. XBP1s regulates a cell-autonomous 12-hour rhythm of nuclear speckle morphology change.
(A) Three-dimensional reconstruction of immunofluorescence of SC35 costained with 4′,6-diamidino-2-phenylindole (DAPI) in the liver of XBP1Flox and XBP1LKO mice at different CTs. (B) Cartoon showing the different nuclear speckle LLPS patterns at different CTs in XBP1Flox mice (top) and violin plot quantification of weighted sphericity of nuclear speckles in XBP1Flox and XBP1LKO mice liver at different CTs. n = 150 to 400 nuclei from three mice per CT. P values exhibiting statistically significant 12-hour rhythms by RAIN analysis in XBP1Flox and XBP1LKO mice are also shown. (C to E) GFP::SC35 mouse embryonic fibroblasts (MEFs) were serum synchronized and subject to time-lapse imaging. Selective images and quantification of temporal sphericity from one single GFP::SC35 MEF. Narrow line, raw data; thick line, spline fit (C). Temporal sphericity (6 to 10 cells quantified at any given time; gray area, means ± SEM; solid line, spline fit) and period distribution of dominant sphericity rhythms in single cells quantified by the eigenvalue/pencil method (n = 25) (D). Temporal sphericity of GFP::SC35 MEFs expressing control, Bmal1, or Xbp1 single-guide RNA (sgRNA) (6 to 10 cells quantified at any given time; light area, means ± SEM; solid line, spline fit), and periodogram analysis of average sphericity rhythms in the three groups (E). Gray areas in (D) and (E) indicate 2 hours of serum shock.
To determine whether the 12-hour rhythm of nuclear speckle morphology change is cell autonomous, we used CRISPR-CAS9 system to knock in green fluorescent protein (GFP) to the N-terminal region of endogenous SC35 locus in immortalized mouse embryonic fibroblasts (MEFs) and performed time-lapse imaging to track nuclear speckle morphology changes in single cells over time. This system allows us to probe nuclear speckle phase separation dynamics under physiological condition without the overexpression of speckle proteins. Western blot and immunofluorescence confirmed the successful generation of MEFs expressing GFP::SC35 fusion protein (fig. S2, C and D). In MEF synchronized by serum shock, we observed robust in-phase ~12-hour rhythms of nuclear speckle morphology changes alternating between a high roundness punctate state and a low roundness diffuse state in single cells (Fig. 1, C and D, fig. S2E, and movie S1). However, consistent with our previous study (4), this 12-hour rhythm of nuclear speckle morphology changes is independent from the cell cycle (fig. S2, F and G). In unsynchronized MEFs (no prior serum shock), we observed a weak local synchronization for cells within 140 μm of each other, in terms of their nuclear speckle morphology oscillation (fig. S3, A to H), implying the very likely existence of paracrine factors for the local synchronization of 12-hour oscillator in adjacent cells in vitro.
Next, we used CRISPR-CAS9 to knock out the circadian clock master regulator BMAL1, or XBP1 in GFP::SC35 MEFs (fig. S4, A to C), and found that XBP1, but not BMAL1, is required for the establishment of the 12-hour rhythm of nuclear speckle morphology change (Fig. 1E). Because the ER-localized endoribonuclease IRE1α can act both upstream (via alternatively splicing Xbp1 mRNA to generate Xbp1s) and downstream (Ire1α mRNA exhibits XBP1s-dependent 12-hour rhythm) of XBP1s and was previously proposed to be an integral component of 12-hour oscillator regulatory network (fig. S5, A to C) (7), we further investigated whether IRE1α inhibition can also impair the 12-hour rhythm of nuclear speckle morphology change. Treating GFP::SC35 MEFs with the selective IRE1α inhibitor 4μ8c resulted in a low-amplitude ~8-hour oscillation instead (fig. S5D). In summary, we therein demonstrated an IRE1α/XBP1s-dependent cell-autonomous 12-hour ultradian rhythm of nuclear speckle morphology that alternates between a more punctate and a more diffuse state. This 12-hour ultradian rhythm is further uncoupled from both the cell cycle and the 24-hour circadian clock and exhibits local coupling in otherwise globally unsynchronized cells.
An evolutionarily conserved XBP1s-SON axis controls 12-hour rhythm of cell-autonomous nuclear speckle LLPS dynamics
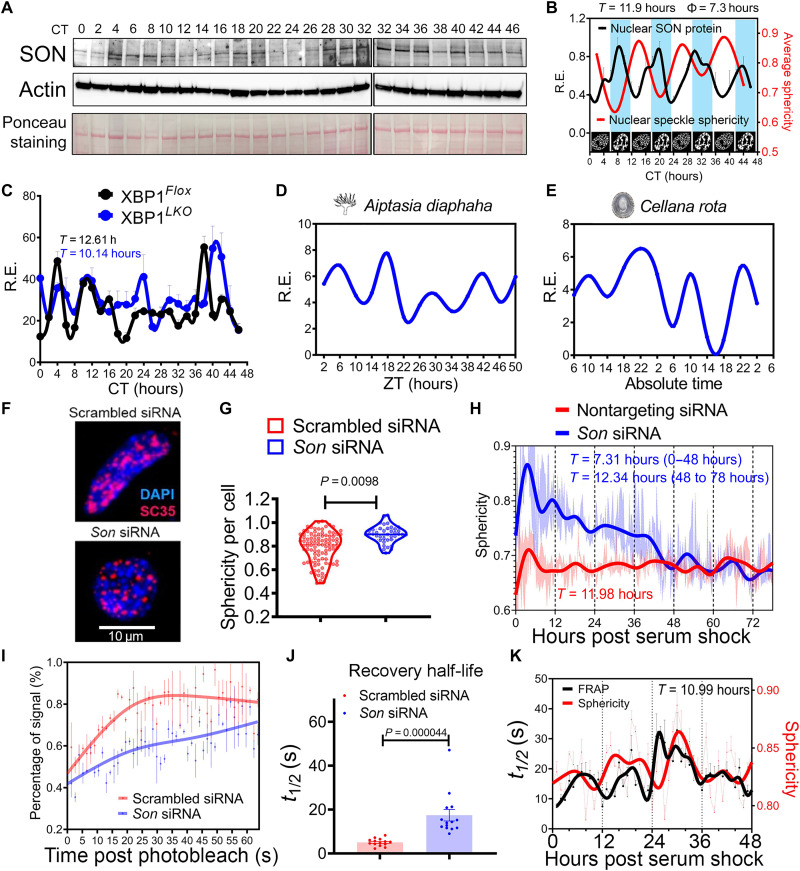
Thus, what may be the mechanism(s) underlying the observed 12-hour rhythm of nuclear speckle morphology change? A recent study on LLPS demonstrated that the concentration of scaffolding protein in the condensates can dictate the way by which LLPS occurs (15, 18). Simply put, under constant valency condition, a higher concentration of scaffolding protein (within the blue region of the phase diagram) will induce LLPS via spinodal decomposition (thus more diffuse), while a lower concentration (within the red region of the phase diagram) will lead to nucleation (thus rounder) (fig. S6, A and B). In nuclear speckles, the protein SON was previously hypothesized to act as a scaffolding protein upon which other RNA processing factors and ncRNA assemble (19). To test the idea that SON level may dictate nuclear speckle morphology, we first performed Western blot analysis to examine the nuclear level of SON in the liver of wild-type mice at different times of day and observed a robust 12-hour oscillation of SON nuclear expression peaking at ~CT8, CT20, CT32, and CT44 (Fig. 2, A and B, and fig. S6C). The oscillation of SON is antiphase with that of nuclear speckle sphericity, and the amplitude of the physiological level of SON oscillation is two- to threefold change (Fig. 2, A and B). Both the phase and amplitude of 12-hour SON oscillation are consistent with the theoretical prediction by the LLPS phase diagram (Fig. 2, A and B, and fig. S6, A to C). The 12-hour rhythm of hepatic SON expression is further validated by the hepatic mass spectrometry dataset (fig. S6, D and E) (9). We next performed quantitative polymerase chain reaction (qPCR) and identified a 12-hour rhythm of Son expression at the mRNA level in XBP1Flox mice (Fig. 2C and fig. S6F). By contrast, the period of Son mRNA oscillation was shortened to ~10-hour in XBP1LKO mice (Fig. 2C and fig. S6F). Cell-autonomous 12-hour Son mRNA expression is further identified in serum synchronized MMH-D3 hepatocytes in vitro (20), maintaining a similar relative phase to that of Bmal1 as in vivo (fig. S6, G and H).
Fig. 2. An evolutionarily conserved XBP1s-SON axis controls 12-hour rhythm of cell-autonomous nuclear speckle LLPS dynamics.
(A and B) Representative Western blot (A) and quantification (B) (n = 2 to 4) of hepatic nuclear SON protein (normalized to the Ponceau S staining intensity), superimposed with the average nuclear speckle sphericity and morphology cartoons at different CTs. The same CT32 sample was run twice to enable comparison across different gels. (C) qPCR analysis of hepatic Son in in the liver of XBP1Flox and XBP1LKO mice at different CTs. Periods calculated by the eigenvalue/pencil method are also shown. (D and E) RNA-seq data of Son ortholog expression in A. diaphaha (D) and C. rota (E). (F and G) Immunofluorescence of SC35 costained with DAPI in scrambled or Son siRNA MEFs (F) and quantification of sphericity (n = 40 to 100) (G). (H to J) GFP::SC35 MEFs were transfected with nontargeting scrambled or Son siRNA. Temporal sphericity (5 to 10 cells quantified at any given time; light area, means ± SEM; solid line, spline fit) and calculated periods by the eigenvalue/pencil method after serum synchronization (H). FRAP analysis with representative recovery curve (I) (data showing quantification from three speckles per cell; means ± SEM for each point; solid line, spline fit) and quantified recovery half-life (J). (K) Calculated temporal sphericity and recovery half-life of nuclear speckles from serum synchronized GFP::SC35 MEFs. n = 7 to 10 cells for FRAP analysis; sphericity was calculated from the average sphericity of each image (n = 6) with ~20 nuclear speckles on each image that are captured before photobleaching. Dash line, raw data; solid line, spline fit.
To determine whether Son is a direct transcriptional target gene of XBP1s, we first examined our previously published hepatic XBP1s chromatin Immunoprecipitation sequencing (ChIP-seq) dataset (6) and found a 12-hour XBP1s chromatin recruitment to the Son gene promoter region (fig. S6I). Consistent with the ChIP-seq result, motif analysis scanning a 1-kb region of the Son gene promoter further identified the XBP1s consensus binding sequence ACGTCA (fig. S6J). In addition, transient overexpression of XBP1s increased the expression of Son mRNA, together with canonical UPR genes Manf and Hyou1 (fig. S6K). While these results strongly indicate that Son is under the direct transcriptional control of XBP1s, it is highly likely that additional transcriptional factors also regulate Son gene expression. Our motif analysis also uncovered putative binding sites for GABPA and NFYA, two additional transcriptional factors involved in the 12-hour oscillator control (fig. S6J) (Gabpa and Nyfa themselves are also direct XBP1s transcriptional target genes) (7). The potential involvement of multiple interlocked transcriptional loops in the regulation of Son gene expression is also likely responsible for its shortened ~10-hour period observed in XBP1LKO mice, as previously seen in circadian period control (21).
We previously proposed that the mammalian 12-hour clock likely evolved from the ancient circatidal clock of marine animals, who adapt their behaviors to the ~12-hour ebb and flow of the tides resulting from the gravitational pull of the moon (1, 22). Evidence supporting this evolutionary origin includes conserved 12-hour rhythms of gene expression between mice and two marine species: the sea anemone Aiptasia diaphana and the limpet Cellana rota (6). In line with this evolutionary conservation, we observed robust 12-hour rhythms of mRNA expression of Son orthologs in A. diaphana (23) and C. rota (Fig. 2, D and E) (24). mRNA processing is the most enriched biological pathway associated with circatidal genes in both marine animals (6). Because the 12-hour rhythm of Xbp1 expression was previously found to be also conserved in marine species (1), these results indicate that the 12-hour rhythm of the XBP1s-SON axis is evolutionarily conserved.
To establish the causality between SON expression and nuclear speckle morphology, we knocked down or overexpressed SON by small interfering RNA (siRNA) or the CRISPR-dCAS9-VPR (VP64-p65-Rta) (25) system in GFP::SC35 MEFs, respectively (fig. S7, A to F). As expected, both manipulations impaired the 12-hour rhythm of cell-autonomous nuclear speckle morphology change, with the former increasing (Fig. 2, F to H, and fig. S7C) and the latter decreasing (fig. S7G) the average roundness of nuclear speckles, again in line with the theoretical prediction by the LLPS phase diagram. This morphology change is ostensibly reversible, as in the later days of imaging, the 12-hour rhythm of nuclear speckle morphology change is restored, likely due to the gradual loss/dilution of Son siRNA (Fig. 2H). To determine whether the morphology changes of nuclear speckle are associated with fluctuations in its dynamics, we performed fluorescence recovery after photobleaching (FRAP) in GFP::SC35 MEFs and found that SON positively regulates the fluidity of nuclear speckles (Fig. 2, I and J; fig. S7, H and I; and movies S2 and S3). We further observed a very robust 12-hour rhythm (with a range of ~8 to ~32 s of recovery half-life) of nuclear speckle fluidity in serum shock–synchronized GFP::SC35 MEFs, well in line with the 12-hour rhythm of nuclear speckle sphericity, with decreased fluidity (longer recovery half-life) coinciding with rounder nuclear speckles (sphericity approaching 1) (Fig. 2K). Thus far, we have demonstrated an evolutionarily conserved XBP1s-SON axis that controls a 12-hour rhythm of cell-autonomous nuclear speckle LLPS dynamics: A higher level of SON expression, observed at early afternoon (CT8) and early morning (CT20), leads to a more diffuse and fluid nuclear speckle morphology, while a lower SON expression, seen at early evening (CT14) and late morning (CT2), renders nuclear speckles to a more spherical and more stagnant state.
XBP1s regulates 12-hour rhythmic nuclear speckle–chromatin interactions
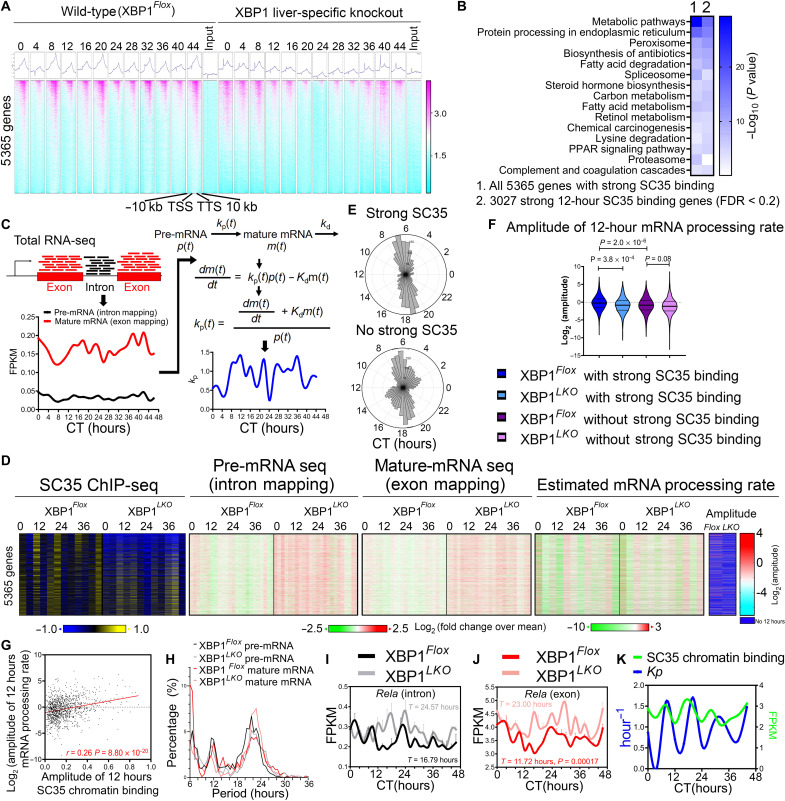
Next, we wondered whether these observed nuclear speckle condensates dynamics lead to a change in their spatial distribution, more specifically, their propensity to associate with chromatin. To detect temporal nuclear speckle–chromatin interactions, we performed ChIP-seq in the liver of XBP1Flox and XBP1LKO mice at 4-hour interval for 2 days at constant darkness, using a monoclonal antibody against mouse SC35 (SRSF2), which has recently been successfully used to characterize splicing condensate–chromatin interactions (26). Consistent with previous SC35 ChIP-seq and Malat1 Capture hybridization analysis of RNA targets (CHART)–seq results (26, 27), nuclear speckle–chromatin interactions are enriched in gene bodies, with a gradual increase in binding intensity toward the 3′ transcription termination site (TTS), as exemplified by very strong binding observed at Neat1 and Malat1 themselves as expected (Fig. 3A and fig. S8A). We observed a dominant global 12-hour rhythm of nuclear speckle–chromatin interactions in XBP1Flox mice cresting at CT8, CT20, CT32, and CT44 (Fig. 3A and fig. S8, A to F), which corresponds to peaking SON expression and nuclear speckle fluidity and diffuseness at the same time (Figs. 1, A and B, and 2, A and B). We identified a total of 5365 genes in wild-type mice that have high-confidence nuclear speckle–chromatin interactions within gene bodies, and 3027 of them have robust ~12-hour rhythmic nuclear speckle–chromatin interaction [with a false discovery rate (FDR) cutoff of 0.2 by RAIN analysis] (Fig. 3A and tables S2 and S3). These genes are very strongly enriched in metabolic and protein homeostasis/ER stress pathways (Fig. 3B). Although below detection threshold for peak calling algorithms, weaker 12-hour rhythmic nuclear speckle–chromatin interactions were also observed in an additional 4130 genes by the eigenvalue/pencil method (table S4 and fig. S8, B, D, and F) (4, 28), which together with the 3027 genes account for more than half of all expressed genes in the wild-type mouse liver. In contrast, ~12-hour rhythms of global nuclear speckle–chromatin interactions were substantially impaired in XBP1LKO mice. Instead, a low-amplitude ~10-hour global binding rhythm was observed after polynomial detrend (Fig. 3A; fig. S8, A to G; and table S4). Notably, this rhythmicity is in line with the ~10-hour Son mRNA oscillation observed in XBP1LKO mice liver (Fig. 2C and fig. S6F).
Fig. 3. XBP1s regulates 12-hour rhythmic nuclear speckle–chromatin interactions.
(A) Heatmap of temporal SC35 ChIP-seq signal as well as input signal for 5365 genes in XBP1Flox and XBP1LKO mice from 10 kb upstream of transcription start site (TSS) to 10 kb downstream of transcription termination site (TTS) for each gene. (B) GO analysis of all 5365 genes or 3207 genes with strong SC35 ChIP signal. PPAR, peroxisome proliferator–activated receptor. (C) Illustration on the workflow to estimate the pre-mRNA and mature-mRNA level, as well the as the mRNA processing rate from temporal RNA-Seq data. (D) Heatmaps of relative integrated SC35 binding signal over gene bodies, pre-mRNA and mature-mRNA expression, and estimated mRNA processing rate (with amplitude for 12-hour rhythm) at different CTs in XBP1Flox and XBP1LKO mice. (E) Polar histogram demonstrating the phase distributions of 12-hour rhythmic mRNA processing rates for genes with (top) or without (bottom) strong SC35 signal in XBP1Flox mice. (F) Log2-transformed amplitude of 12-hour mRNA processing rates for genes with or without strong SC35 binding in XBP1Flox and XBP1LKO mice. (G) Scatter plot of the amplitude of log2-transformed 12-hour mRNA processing rates versus integrated 12-hour SC35 binding signal over gene bodies for 1160 genes having both in XBP1Flox mice. (H) Period distribution of pre-mRNA and mature-mRNA oscillations for 1160 genes in (G). (I to K) Temporal expression of Rela at the pre-mRNA (I) and mature-mRNA (J) level in XBP1Flox and XBP1LKO mice, and integrated SC35 gene body signal and mRNA processing rate in XBP1Flox mice (K). Data are means ± SEM in (I) and (J).
To determine whether the rhythmic nuclear speckle–chromatin interactions are correlated with transcriptional state fluctuations in mouse liver in vivo, we estimated the temporal pre-mRNA and mature-mRNA level of each hepatic gene by, respectively, quantifying the reads mapped to intron and exon regions, using our previously published RNA-seq dataset (6) (Fig. 3C and tables S5 and 6). Consistent with past studies (12, 13, 29), on a global scale, higher daily-average nuclear speckle–chromatin recruitment is strongly associated with higher daily-average gene expression in both XBP1Flox and XBP1LKO mice (fig. S9A). However, on an individual gene level, temporal rhythmic nuclear speckle–chromatin interaction is largely decoupled from temporal gene expression, as not all nuclear speckle–associated genes exhibit 12-hour rhythms of expression (Fig. 3D). We further estimated the mRNA processing rate of each gene via a simple first-order kinetic model of transcription regulation (30) (assuming the mRNA degradation rate remains constant during a day) and found a strong positive correlation between 12-hour rhythmic nuclear speckle–chromatin association and 12-hour rhythmic mRNA processing rate in XBP1Flox mice that both peak around CT8/CT20 (Fig. 3, C to G, and fig. S9B). Compared to XBP1Flox mice, the daily-average mRNA processing rate in XBP1LKO mice liver was slightly reduced (fig. S9C) and exhibited a dominant population of shortened ~10-hour oscillations (Fig. 3D and fig. S9D), again in line with observed ~10-hour rhythm of Son expression (Fig. 2C and fig. S6F) and ~10-hour oscillation of nuclear speckle–chromatin interactions (fig. S8, F and G). For those genes that do maintain ~12-hour mRNA processing rate in XBP1LKO mice, they have lower amplitude compared with their wild-type counterparts (Fig. 3F) and a more diffuse phase distribution (fig. S9E). Twelve-hour rhythmic nuclear speckle–chromatin interaction and subsequent 12-hour mRNA processing rates greatly contribute to the establishment of 12-hour rhythm, but not 24-hour circadian rhythm of gene expression posttranscriptionally in XBP1Flox mice (Fig. 3H), with examples of Rela and Id1 genes only exhibiting 12-hour rhythms at the mature-mRNA level in XBP1Flox mice (Fig. 3, I to K, and fig. S9, F to H). Dominant 24-hour circadian rhythms of nuclear speckle–chromatin interactions and mRNA processing rates were observed on all core circadian clock genes with similar amplitudes in both XBP1Flox and XBP1LKO mice, in phase to their respective temporal gene expression profile (fig. S10, A to D), although some genes (such as Per1 and Nfil3) have weaker superimposed 12-hour rhythmic nuclear speckle–chromatin interactions (also peaking at CT8/CT20), which are lost in XBP1LKO mice (fig. S10C). Because the core circadian gene expressions are not altered in XBP1LKO mice (6), these results suggest that the SON-mediated 12-hour rhythmic nuclear speckle–chromatin interactions are largely dispensable for the establishment of 24-hour core circadian clock gene expression in mice.
Proteostasis gene expressions are hypersensitive to nuclear speckle LLPS dynamic change
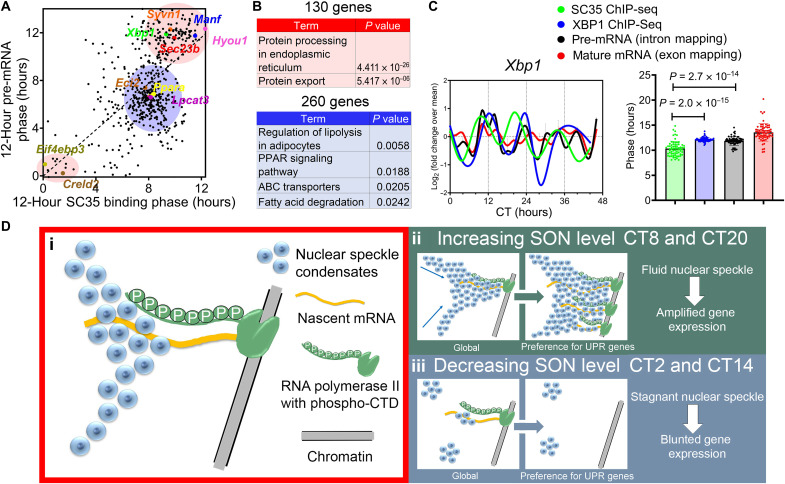
Focusing on 528 genes that exhibit very robust 12-hour rhythms of nuclear speckle–chromatin interactions (FDR = 0.2 among the 5365 genes with strong SC35 binding peaks) and XBP1s-dependent 12-hour rhythms of gene expression (FDR = 0.2 at both the pre-mRNA and mature-mRNA level) (fig. S11, A to D), we identified two major groups of genes with different phase relationship between the two. For 260 genes enriched in lipid metabolism and peroxisome proliferator–activated receptor (PPAR) signaling (blue area in Fig. 4, A and B), nuclear speckle–chromatin interaction peaks at CT8, closely following the peak of nascent mRNA expression at CT6 and matches the peak of mature-mRNA expression (fig. S11, E and F). For the majority of 130 genes enriched in ER stress and protein sorting pathways (red area in Fig. 4, A and B), the peaking times of the nuclear speckle–chromatin interactions precede those of nascent mRNA expression (above the diagonal line in Fig. 4A), which include genes like Manf, Hyou1, and Sec23b (Fig. 4A). For 69 genes with additional 12-hour XBP1s chromatin recruitment (6), the acrophase of nuclear speckle–chromatin interaction can even precede that of XBP1s chromatin binding, with Xbp1 itself as a good example (Fig. 4C and fig. S11, G and H).
Fig. 4. Proteostasis gene expressions are hypersensitive to nuclear speckle LLPS dynamic change.
(A) Scatter plot demonstrating the phases of 12-hour rhythm of nuclear speckle–chromatin interactions (x axis) versus those of 12-hour rhythm of pre-mRNA expression (y axis) for 528 genes. Two major clusters of genes are highlighted in red and blue, respectively. (B) GO analysis of 130 genes highlighted in red and 260 genes highlighted in blue. (C) Quantification of the phases of 12-hour rhythms of nuclear speckle–chromatin interaction, XBP1s chromatin binding, and pre-MRNA and mature-mRNA expression of 69 genes that exhibit all four, with temporal profile for Xbp1 gene also shown. Data are means ± SEM. (D) A simplified model demonstrating that SON can positively regulate nuclear speckle fluidity and their interactions with chromatin, and subsequent gene expression involved in proteostasis and UPR.
These results imply the existence of multitiered mechanisms for regulating nuclear speckle–chromatin interactions. As recently demonstrated, nuclear speckle condensates are thought to be “passively” recruited to chromatin following nascent mRNA transcription, mediated by RNA polymerase II hyperphosphorylation during transcriptional elongation (Fig. 4Di) (26). This mechanism is likely responsible for maintaining the core circadian clock and lipid metabolism gene expression, as the dynamics of their nuclear speckle–chromatin interactions closely follow their temporal gene expression change. The emerging data further suggest an additional layer of nuclear speckle–chromatin interaction control. Rather than responding to the transcriptional state of individual genes, nuclear speckle–chromatin interactions can also be modulated by the SON-mediated nuclear speckle LLPS dynamics on a global scale (Fig. 4D, ii and iii). In this case, the dynamics of nuclear speckle–chromatin interactions can even precede the subsequent gene expression change. These data further suggest that this second mode of “proactive” nuclear speckle–chromatin interaction is strongly implicated in the transcription regulation of proteostasis and UPR genes.
SON transcriptionally amplifies the UPR and protects against proteome stress
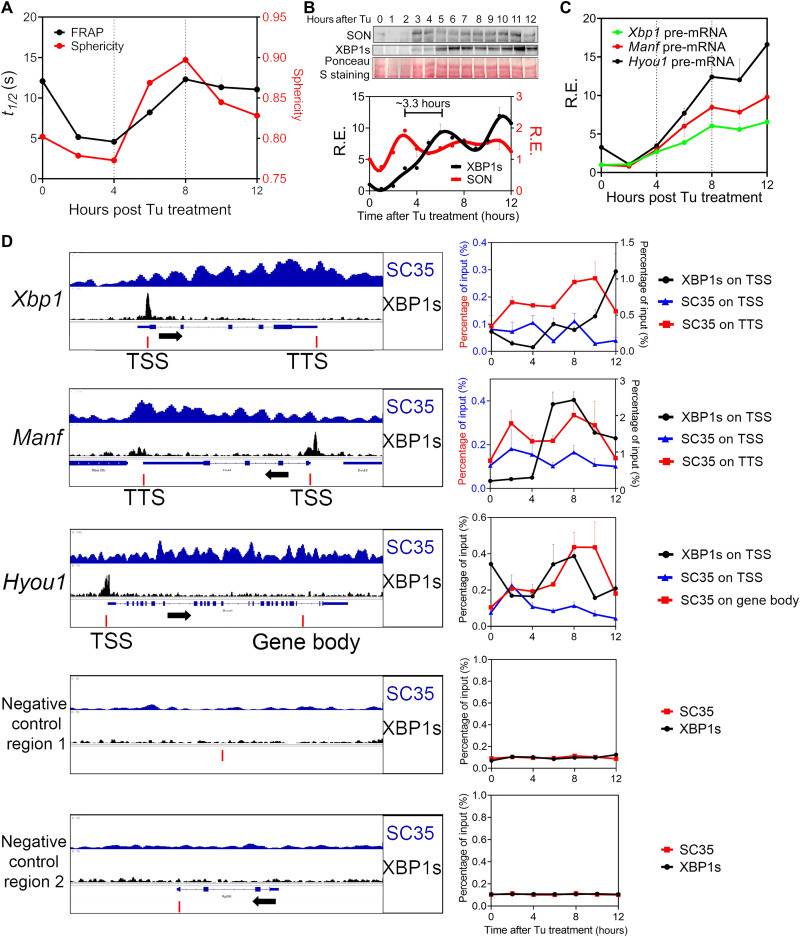
Before the discovery of the mammalian 12-hour oscillator, UPR was classically studied as a transient response to an insult to the ER (also known as ER stress). Transmitting a cascade of signals from ER to the nucleus, UPR ultimately activates three main transcription factors: XBP1s, activating transcription factor 4 (ATF4), and ATF6 (31). To seek more support for the causal role of SON and nuclear speckle LLPS on UPR gene regulation, we further examined the temporal kinetics of canonical UPR in response to a very low dose of ER stress inducer tunicamycin (Tu) (100 ng/ml) in MEFs. We noted that starting from 2 hours after Tu treatment, an immediate early increase of SON expression was concomitant with increasing nuclear speckle fluidity and diffuseness and increasing nuclear speckle–chromatin interactions in the gene bodies and/or TTSs of Xbp1, Manf, and Hyou1 genes (Fig. 5, A to D). This immediate early SON-mediated nuclear speckle LLPS dynamics change precedes the increase in XBP1s promoter recruitment as well as Xbp1, Manf, and Hyou1 gene expression with a phase advance of ~3.3 hours (Fig. 5, B to D), similar to what was observed in mouse liver in vivo (fig. S6E). We further observed a second wave of nuclear speckle–chromatin interactions peaking at 8 hours after Tu treatment that follows XBP1s promoter recruitment (Fig. 5D). This second wave is not associated with increased nuclear speckle fluidity and thus reflects “passive” chromatin recruitment of nuclear speckles during transcription elongation (Fig. 5A). This reduced nuclear speckle fluidity during the second wave is likely a result of a more stable interaction between nuclear speckle and chromatin during mRNA processing.
Fig. 5. Temporal kinetics of UPR in response to low level of ER stress recapitulates endogenous 12-hour oscillator.
MEFs were treated with Tu (100 ng/ml) for different hours. (A) FRAP assay and quantification of nuclear speckle sphericity. (B) Western blot and quantification (normalized to total Ponceau S staining intensity) of SON and XBP1s expression. (C) qPCR analysis of pre-mRNA level of different UPR genes. (D) Selected genes aligned for SC35 and XBP1s ChIP-seq signal from CT12 in XBP1Flox mice (left) and ChIP-qPCR of XBP1s and SC35 on selected regions (indicated by red bars) (right). Data are means ± SEM.
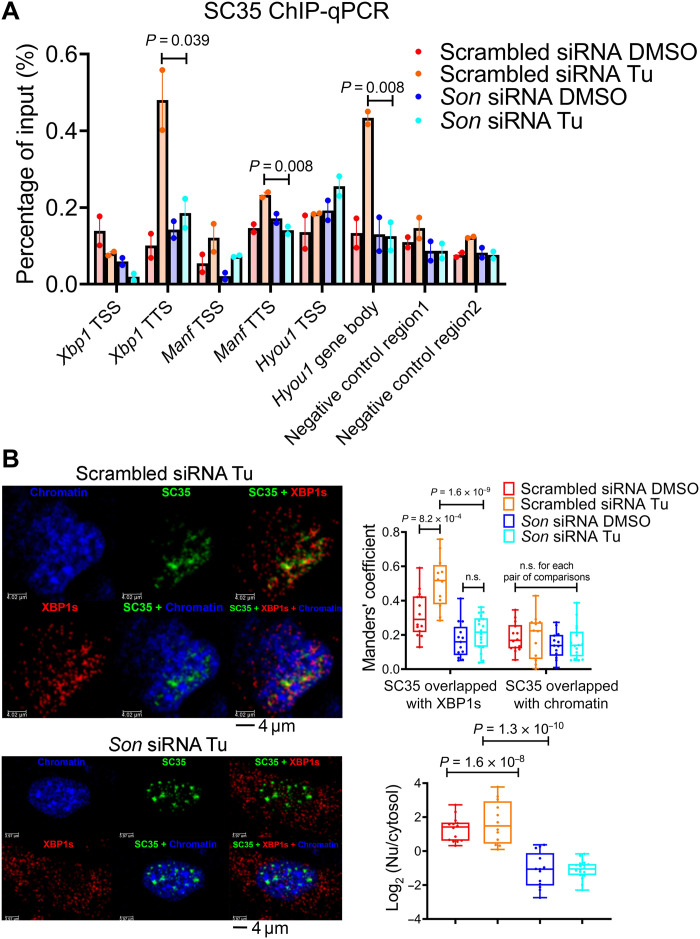
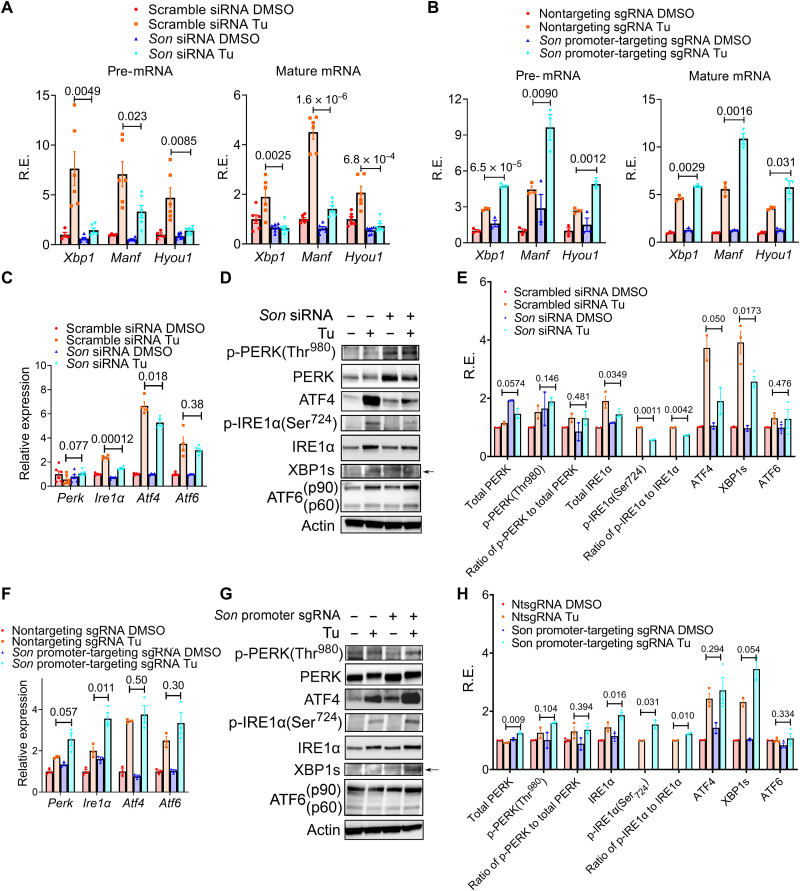
To confirm the ChIP-qPCR results, we performed immunofluorescence and observed a dose-dependent increase in colocalization of XBP1s with nuclear speckles in response to ER stress in MEFs (fig. S12, A to D). siRNA-mediated knocking down of SON greatly reduces ER stress–induced nuclear speckle–chromatin interactions (Fig. 6A), disrupts XBP1s and nuclear speckle colocalization (Fig. 6B), and subsequently greatly blunts Tu-induced UPR at both the pre-mRNA and mature-mRNA level, while not affecting core circadian clock genes expression (Fig. 7A and fig. S13, A and B). By contrast, both stable and transient dCAS9-VPR–mediated overexpression of endogenous SON substantially amplifies UPR at the transcriptional level (Fig. 7B and fig. S13, C to F). UPR genes under SON control include not only ER stress–responsive output genes such as Manf and Hyou1 but also core regulatory genes in the XBP1 and ATF4 branches of the UPR: Xbp1 itself, Ire1α, and Atf4 (Fig. 7, C to H), the latter of which appears to respond to a narrower range of SON level, as overexpression of SON has little effect on Atf4 gene expression (Fig. 7F). By contrast, SON does not significantly affect Atf6 expression (Fig. 7, C and F). The dichotomy of the effects of SON on Atf4 and Atf6 gene expression during transient ER stress in MEFs is consistent with their differential temporal gene expression profiles in mouse liver in vivo: While both Atf4 and Atf6 pre-mRNA exhibit 12-hour rhythms in wild-type mouse liver, only the former exhibits a robust 12-hour rhythm of XBP1s-dependent nuclear speckle–chromatin interaction and dampened 12-hour rhythm of expression in XBP1 LKO mice (fig. S13, G to J). These UPR gene expression changes in response to SON manipulation are similarly conserved at the protein level (Fig. 7, D, E, G, and H) and largely recapitulated with a different ER stress inducer, dithiothreitol (DTT) (fig. S14, A and B). Last, we ruled out the possibility that protein synthesis is involved in the transcriptional regulation of the early stage of UPR by SON, as neither changes of protein synthesis rate (fig. S15, A and B) nor the relative amount of phospho-PERK (protein kinase R-like endoplasmic reticulum kinase) (Thr980) levels (Fig. 7, D, E, G, and H) was observed under SON knocking down or overexpression conditions. Together, our data strongly indicate that SON can rapidly amplify XBP1s and ATF4 (although to a lesser extent)–mediated UPR, while having very modest effects on the ATF6 branch at the transcriptional level.
Fig. 6. SON is required for colocalization of nuclear speckles with XBP1s during ER stress.
MEFs were transfected with control or Son siRNAs and treated with Tu (100 ng/ml) for 6 hours. (A) ChIP-qPCR of SC35 on TSS and TTS of gene bodies of selective UPR genes. Data are means ± SEM. (B) Immunofluorescence of anti-XBP1s (red), GFP signal (green) from GFP::SC35 fusion protein and DAPI nuclei staining (blue), as well as merged images of either two or all three channels. Representative images (left), Manders’ coefficient quantification of colocalization of SC35/XBP1s and SC35/chromatin signals (top right), and quantification of log2-transformed ratio of nuclear to cytosol level of XBP1s (bottom right). Box and whiskers plot showing minimum to maximum values. n.s., not significant.
Fig. 7. SON amplifies the UPR transcriptionally.
(A) MEFs were transiently transfected with scramble control or Son siRNA and treated with Tu (100 ng/ml) for 6 hours, and qPCR analysis was performed on selective genes. (B) Control MEFs or MEFs with dCAS9-VPR–mediated stable overexpression of Son were treated with Tu (100 ng/ml) for 6 hours, and qPCR analysis was performed on selective genes. (C to E) MEFs were transiently transfected with scramble control or Son siRNA and treated with Tu (100 ng/ml) for 6 hours and qPCR analysis (C) and representative Western blot images (D) and quantification (E). (F to H) Control MEFs or MEFs with dCAS9-VPR–mediated stable overexpression of Son were treated with Tu (100 ng/ml) for 6 hours and qPCR analysis (F) and representative Western blot images (G) and quantification (H). For p-IRE1α, DMSO condition expression is too low to be accurately quantified. Data are means ± SEM.
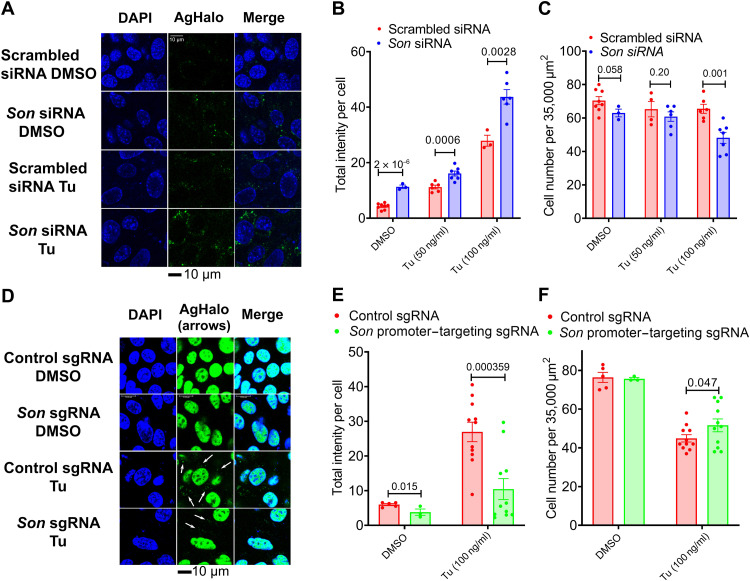
To determine the functional importance of SON in regulating proteostasis in the ER, we went on to detect and quantify ER proteome stress in response to SON manipulation in MEFs. We used a previously published Halo-tag mutant (K73T/L172Q) prone to ER-localized aggregation (AgHaloER) (32). The AgHaloER sensor was labeled with solvatochromic fluorogenic probe (P1), which turns on fluorescence only upon its misfolding and aggregation (33). As expected, live cell imaging of MEFs expressing AgHaloER labeled by P1 probe demonstrated that the AgHaloER was well folded with little green fluorescence signal under basal conditions and formed granular green fluorescent structures after 16-hour Tu treatment (Fig. 8A). Under both basal and Tu conditions, a notable increase in AgHaloER staining intensity was observed in SON-depleted MEFs (Fig. 8, A and B), which is concomitant with reduced cell number in response to a higher concentration of Tu treatment under SON-depleted condition (Fig. 8C). Conversely, dCAS9-VPR–mediated SON overexpression significantly reduced AgHaloER misfolding and protected against ER stress–induced cell death (Fig. 8, D to F). Collectively, these data indicate that by amplifying the UPR, SON can protect cells against proteome stress.
Fig. 8. SON protects cells against proteome stress.
(A to C) MEFs were transiently transfected with AgHaloER plasmid and then further transfected with control or Son siRNAs and treated with different concentrations of Tu for 16 hours before being subjected to confocal live imaging. Representative confocal images of DAPI staining and AgHaloER-probe conjugates in response to Tu (C). Quantification of averaged AgHalo-probe intensity per cell (D) and averaged cell number per area of 35,000 μm2 (E). Each data point in (B) is averaged intensity per cell calculated from each image of 35,000 μm2 area. (D to F) dCAS9-VPR GFP::SC35 MEFs expressing control or Son promoter–targeting sgRNA were transiently transfected with AgHaloER plasmid and then treated with tunicamycin (Tu; 100 ng/ml) for 16 hours before subject to confocal live imaging. Representative confocal images of DAPI staining and ER-targeting AgHaloER-probe conjugates in response to Tu (100 ng/ml) (D). White arrows indicate ER-targeting AgHaloER-probe conjugates. Quantification of averaged AgHalo-probe intensity (nonnuclear portion) per cell (E) and averaged cell number per area of 35,000 μm2 (F). Each data point in (E is averaged intensity per cell calculated from each image of 35,000 μm2 area. Data are means ± SEM.
Consistent with its role in amplifying UPR during transient ER stress, Son knockdown further dampened the 12-hour oscillation of Manf promoter–driven luciferase activity, while having no apparent effects on circadian Bmal1 promoter–driven luciferase rhythm (fig. S16, A to F). Together, these results strongly support a positive causal role of SON and nuclear speckle LLPS on the transcriptional regulation of proteostasis, both for UPR in response to a transient ER stress and the cell-autonomous 12-hour oscillator.
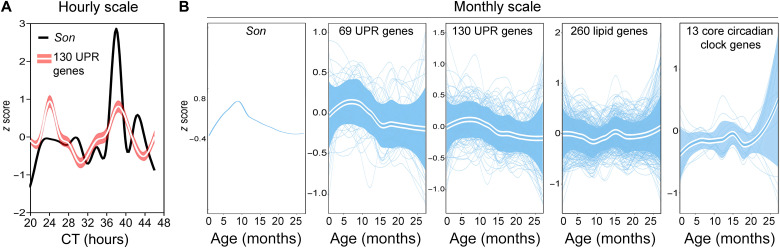
Correlative SON and UPR gene expression dynamics across mouse life span
To determine whether the hourly Son and UPR gene expression dynamics can be extrapolated to a longer temporal scale, we analyzed a recently published RNA-seq data in multiple mouse tissues across a 27-month life span (34). We observed strong correlative Son and UPR mRNA expression dynamics in the liver, skin, heart, pancreas, and bone across the entire mouse life span (Fig. 9, A and B, and fig. S17, A to D). In addition to the liver, 12-hour rhythms of Son and UPR mRNA were also observed in mouse skin (35), heart (36), and pancreas (37) (fig. S17, A to C), with the latter exhibiting a very intriguing fractal feature of antiphase oscillations of Son and UPR gene expression at the periods of 12 hours and ~16 months (fig. S17C). The anticorrelation between Son and UPR mRNA in mouse pancreas could be due to a large discordance between the mRNA and protein level of SON and warrants further investigation. In contrast to UPR genes, no strong correlation was observed between the gene expression dynamics of Son, and lipid and core circadian clock genes in different tissues across the mouse life span (Fig. 9, A and B, and fig. S17, A to D).
Fig. 9. Correlative hepatic Son and UPR gene expression dynamics is observed across the mouse life span.
Expression of Son and UPR genes in a 28-hour time window (A) or across the entire mouse life span (B) in the mouse liver. Solid line, mean; shaded area, 95% confidence interval.
DISCUSSIONS
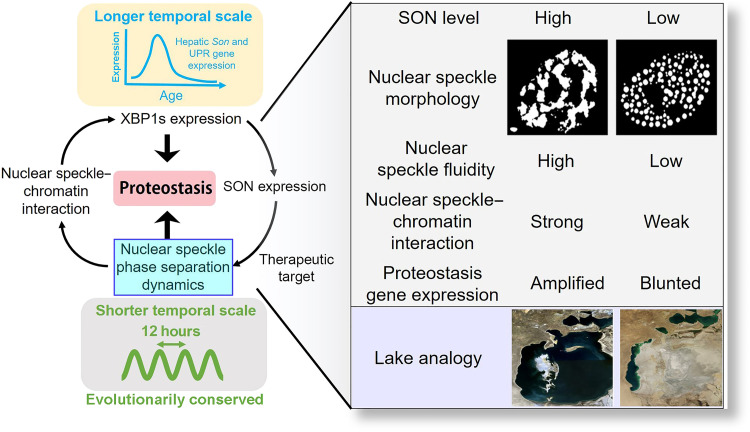
While considerable progress has been made toward understanding the biophysical properties and the biological functions of biomolecular condensates and LLPS (38), it remains elusive whether LLPS dynamics are under the control of “autonomous clocks” (39). Our study hereby demonstrates the existence of an evolutionarily conserved XBP1s-SON axis that integrates the 12-hour oscillator with nuclear speckle LLPS to spatiotemporally program proteostasis (Fig. 10). As the scaffolding protein of nuclear speckle condensates (19), SON level can dictate the mechanisms by which nuclear speckles phase separate. Nuclear speckles with a lower concentration of SON exhibit features reminiscent of punctate nucleation and growth, while those with higher SON expressions exhibit early-stage coarsening morphologies—connected network-like condensates—associated with spinodal decomposition (15). While the coarsening of condensates will eventually result in the coalescence of all droplets into a single large sphere in an ideal situation driven by surface tension, the chromatin likely greatly slows down this process so that an intermediate connected network-like nuclear speckle morphology can occur (40). Both the phase and amplitude of SON oscillation further align with the prediction by the phase separation diagram. More fascinatingly, nuclear speckles with lower SON expression are also more stagnant, whereas much more fluid dynamics are found in nuclear speckles with higher SON expression. Whether the dynamics of nuclear speckle LLPS is intrinsic to the way by which LLPS occurs (nucleation or spinodal decomposition) remains to be determined. One possibility is that nucleation-mediated LLPS will result in many individually isolated condensates, and the barriers of liquid droplets would limit molecular diffusion between dense and dilute phases, while during spinodal decomposition, the connected network-like condensate morphology would greatly favor rapid molecular diffusion within the condensates. The latter would also facilitate the rapid delivery of splicing factors to transcription foci during ER stress to amplify the UPR at the cotranscriptional splicing/transcription elongation stage (18). Conversely, lower SON expression–associated stagnant nuclear speckles are sequestered away from chromatin and thereby would greatly dampen the UPR (Figs. 4D and 10). It is worth noting that besides SON, we are not ruling out the possibility that other nuclear speckle proteins and/or RNAs may also regulate the 12-hour nuclear speckle LLPS dynamics, with the Pickering agent, which has recently been found to be regulating P granules coarsening, being a tantalizing candidate (41). Our results further reconcile the debate on the exact roles of nuclear speckles in gene regulation: whether the nuclear speckles mainly function as the “storage facility” of mRNA processing factors away from chromatin, or they can actively participate in gene regulation process via physical engagement with chromatin (42). We showed here that nuclear speckles can function as both, but these two functions are probably temporally separated because of the oscillation of SON expression and the according changes in their LLPS dynamics and propensity to associate with chromatin.
Fig. 10. XBP1s-SON axis spatiotemporally controls nuclear speckle LLPS to regulate proteostasis.
An analogy would be the water level fluctuations in a lake: Nuclear speckles with higher SON level is like a lake filled with free-flowing water with fast dynamics, while nuclear speckles with reduced SON expression resemble a nearly dried-out lake with a few disconnected pools of stagnant water. Our study further indicates that the nuclear speckle LLPS may be a previously unidentified therapeutic target for pathologies that arise as a result of dysregulated proteostasis. Please refer to Discussions for details.
Thus, what could be the biological advantages of having a 12-hour rhythm of nuclear speckle LLPS dynamics? Because the acrophases (the time period in a cycle during which the cycle peaks) of 12-hour rhythms of gene expression are strongly biased toward dawn and dusk, we previously proposed a vehicle-cargo hypothesis to elucidate the distinct functions of 12-hour versus the circadian rhythms (6). We argued that the 12-hour rhythm accommodates increased demands for gene expression/processing at the two biological “rush hours” (dawn and dusk) by elevating the global traffic capacity of the CEDIF. This connects and tunes rates of mRNA, protein, and lipid metabolism to the 12-hour cycle of metabolic stress (thus acting as the vehicle). The circadian clock, on the other hand, dictates the particular genes/gene products processed at each rush hour (thus acting as the cargo). We posit that having increased nuclear speckle fluidity at early morning/early afternoon enables the in-land animals to anticipate and, subsequently, to rapidly turn on UPR genes to cope with heightened metabolic stress associated with transition periods later at dawn and dusk. This feature is likely co-opted from the circatidal clock of marine animals, who probably used a similar system to adapt to the 12-hour environmental cues resulting from tidal changes. The hypersensitivity of proteostasis gene expression to nuclear speckle LLPS dynamics would ensure tightly coupled mRNA and protein metabolic processes, which, in turn, can entail a highly efficient genetic information transfer across multiple compartments within the cell. The fact that manipulating the LLPS dynamics of nuclear speckle is sufficient to alter the transcriptional output of proteostasis genes further suggests that the nuclear speckle LLPS may represent a previously unidentified (chrono)-therapeutic target for pathologies associated with dysregulated proteostasis (Fig. 10). A recent study found that the nuclear speckle can also amplify p53-mediated gene expression (43). Because p53 is known to be part of the DNA damage response, it is reasonable to conjecture that boosting nuclear speckle function via modulating its LLPS may exert beneficial effects via simultaneously augmenting multiple adaptive stress responses, thereby potentially enhancing the overall antiaging hormesis (44).
On a more philosophical note, our study is a good example of how seemingly unrelated biological processes can be tightly connected through the time dimension, in this case, their 12 hours of rhythmicity. In this study, we showed that one can use frequency spectrum similarity to interrogate genetic interactions within a cell. We hope that our study will make the scientific community think more deeply about the temporal dimension of their biological problems and facilitate the achievement of the ultimate goal of “mapping the four-dimensional atlas” of biological processes.
MATERIALS AND METHODS
Animals
XBP1Flox mice were as previously described (45). XBP1LKO mice were generated by crossing albumin-CRE mice with XBP1Flox mice. All mice are in C57BL/6 background, male, and between 3 and 4 months of age. Mice were first entrained under 12-hour light/12-hour dark conditions for 2 weeks before they were transferred to constant darkness for 24 hours. Mice were then euthanized at a 2-hour interval for a total of 48 hours. Mice were fed ad libitum during the entire experiment. The animal studies were carried out in accordance with the National Institutes of Health guidelines and were granted formal approval by the University of Pittsburgh’s Institutional Animal Care and Use Committee (approval numbers IS00013119 and IS00013119).
Plasmids
Mouse nontargeting, Bmal1 and Xbp1 single-guide RNAs (sgRNAs) were cloned into the lentiCRISPRv2 plasmid as previously described (46), which is a gift from T. Finkel. The sgRNA sequences (in bold) are as follows: nontargeting control, caccgAAATGTGAGATCAGAGTAAT; Bmal1, caccgCCCACAGTCAGATTGAAAAG; and Xbp1, caccgGGAGCAGCAAGTGGTGGATT. dCAS9-VPR plasmid (47) was a gift from G. Church (Addgene, plasmid no. 63798; http://n2t.net/addgene:63798; RRID:Addgene_63798). Mouse nontargeting, Son promoter–targeting sgRNAs were cloned into the pLenti-SpBsmBI-sgRNA-Hygro plasmid (47), which was a gift from R. Maehr (Addgene, plasmid no. 62205; http://n2t.net/addgene:62205; RRID: Addgene_62205). The sgRNA sequences are as follows: nontargeting control, aaatgtgagatcagagtaat; Son promoter-targeting sgRNA1, atggcggccgagttcgtgcg; and Son promoter–targeting sgRNA2, taggagtccccgcaggctga. XBP1s overexpression plasmid PHAGE-Flag-XBP1s was as previously described (4).
siRNA/sgRNA transient transfections
MEFs were transfected with 10 μM of different siRNAs for 24 to 48 hours with Lipofectamine RNAiMAX reagents (Life Technologies) per the manufacturer’s instructions. Sources of siRNA are as follows: siGENOME Non-Targeting siRNA pool (Dharmacon, D-001206-1305) and siGENOME SMARTpool SON siRNA (Dharmacon, L-059591-01-0005). For transient transfection of Son promoter–targeting sgRNAs, nontargeting sgRNA or Son promoter–targeting sgRNA1 and sgRNA2 were synthesized in vitro by the EnGen sgRNA Synthesis Kit, S. pyogenes per the manufacturer’s protocol. MEFs stably expressing dCAS9-VPR were transfected with 10 μM of different sgRNAs and cotreated with Tu (100 ng/ml) for 6 hours.
Cell culture
MEFs were isolated from male SRC-2fl/fl mice and immortalized by SV40 T antigen as previously described (48). For dexamethasone (Dex) treatment, MEFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (glucose, 4.5 g/liter) supplemented with 10% fetal bovine serum (FBS) and treated with 100 nM Dex for 30 min and then washed with 1× phosphate-buffered saline (PBS) before being cultured in the same medium. For serum shock, MEFs were cultured in DMEM (glucose, 4.5 g/liter) supplemented with 10% FBS and treated with 50% horse serum for 2 hours and then washed with 1× PBS before being cultured in the same medium. For Tu treatment, MEFs were treated with Tu (100 ng/ml) [in dimethyl sulfoxide (DMSO)] for different times, unless it is otherwise indicated. For all cell culture experiments, cells were cultured at 37°C with 5% CO2.
Establishing stable cell line
For sgRNA-mediated Bmal1/XBP1 knockout MEFs, lentiviruses packaged in human embryonic kidney (HEK) 293T cells with cotransfection of lentiCRISPRv2, pMD2.G, and psPAX2 plasmids were used to infect MEFs with a multiplicity of infection of 3. Stable MEFs were selected in the presence of puromycin (4 μg/ml). For SON trans-activation MEFs, MEFs were first transfected with dCAS9-VPR plasmid, and those stably expressing dCAS9-VPR were selected in the presence of G418 (200 μg/ml). dCAS9-VPR stably expressing MEFs were further infected with lentiviruses packaged from HEK293T cells transfected with pLenti-SpBsmBI-sgRNA-Hygro, pMD2.G, and psPAX2 plasmids. Final SON trans-activation MEFs (with Son promoter–targeting sgRNA1) were selected in the presence of G418 (200 μg/ml) and hygromycin (200 μg/ml).
Generation of GFP::SC35 cells
The CRIS-PITCh (v2) system as described in (49) was used to generate an GFP::SC35 knock-in cell line. The CRIS-PITCh (v2) system requires an “all-in-one” expressing CAS9, a CRISPR guide strand targeting a cut site in the desired genomic locus, and a CRISPR guide strand targeting the CRIS-PITCh (v2). The CRIS-PITCh (v2) contains a sequence designed to recombine into the desired locus and insert a sequence [Puro–T2A–enhanced GFP (EGFP)]. All-in-one vector: Oligos (Thermo Fisher Scientific) (forward) 5′-CACCTGTCCGGGGCGTTAGGGTCT-3′ and (reverse) 5′-AAACAGACCCTAACGCCCCGGACA-′3 were annealed using Nuclease Free Duplex Buffer (Integrated DNA Technologies 11-01-03-01). The plasmid pX330_1x2_addgene-58766-sequence-202009 (Addgene, www.addgene.org/58766/) was digested with BbsI-HF [New England Biolabs (NEB), R3539S] (Antarctic Phosphatase M0289S) and gel purified. The resulting fragment was ligated to the annealed oligos using T4 ligase (M0202S). The Pitch guide RNA was cleaved form the Pitch_Cas9_addgene-63670-pX330S-2-Cas9PITCh-106070 (Addgene, www.addgene.org/browse/article/16395/) plasmid using BsaI-HFv2 (NEB, R3733S). This fragment was gel purified and ligated to BsaI BsaI-HFv2 (NEB, R3733S) cleaved and purified SRSF2 guide RNA vector. The resulting vector contained sequences to express the SRSF2 locus-specific guide RNA and the Pitch guide RNA. CRIS-PITCh (v2): The Puro-T2A-EGFP region was amplified from the mp132-psicor-puro-t2a-egfp plasmid [ViraCore at the University of California, San Francisco (UCSF), viracore.ucsf.edu] using the following primers (Thermo Fisher Scientific): Q5 High-Fidelity 2X Master Mix (NEB, M0492S) (forward) 5′-CCGCGTTACATAGCATCGTACGCGTACGTGTTTGGTGTCCGGGGCGTTAGGGTCTATGACCGAGTACAAGCCC-3′ and (reverse) 5′-CAGCATTCTAGAGCATCGTACGCGTACGTGTTTGGGGCGGGCGGCCGTAGCTCATGGATCCGGGCTTGTACAGCTCGTCCATG-3′. The resulting PCR fragment was gel purified and joined to the Mlu I (NEB, R0198S) digested pCRIS-PITChv2-FBL (www.addgene.org/63672/) using Gibson Assembly (NEB, R3539S). The all-in-one vector (1.2 μg) and the CRIS-PITCh (v2) (0.6 μg) were transfected into a 100-mm dish of low-passage number MEFs using Lipofectamine 3000 (Invitrogen, 100022052) and P3000TM Reagent (Invitrogen, 100022058) and Optimem (Gibco, 31985-070). The transfected MEFs were cultured [DMEM (Gibco, 21013-024), 10% FBS (HyClone, SH30910.03), 1% penicillin-streptomycin (Gibco, 15140-122), 1% sodium pyruvate (Gibco, 11360-070)] in increasing concentrations [puromycin, 2 to 5 μg/ml (Gibco, A11138-03)] for selection. Colonies were diluted using 15-cm cell culture plates. Colonies were examined for EGFP fluorescence and were subsequently cloned using cloning circles (Sigma-Aldrich, Z370789) and trypsin EDTA (Gibco, 25200072).
Real-time luminescence assay
Stable Manf-dluc (6) or Bmal1-dluc MEFs (6) were cultured in DMEM (glucose, 4.5 g/liter) supplemented with 10% FBS and treated with 50% horse serum in DMEM for 2 hours or 100 nM for 30 min before being subjected to real-time luminescence assay using a LumiCycle (Actimetrics) as previously described (6). Briefly, after serum shock treatment, MEFs were washed with 1× PBS and cultured with DMEM (glucose, 4.5 g/liter) supplemented with 0.1 mM luciferin and 10 mM Hepes buffer in 35-mm tissue culture dishes in the absence of serum and transferred immediately to LumiCycle for real-time luminescence analysis. Periods of oscillation were identified by embedded Periodogram function. For siRNA-treated MEFs, MEFs were transfected with nontargeting or Son siRNA for 48 hours before being subjected to serum shock and real-time luminescence assay as described above.
Immunofluorescence
Immunofluorescence was performed as previously described (50). Briefly, the liver Optimal cutting temperature (OCT) sections or cells cultured in chamber slide were fixed in cold acetone for 10 min at −20°C. The sections were then air dried, rehydrated with PBS, and permeabilized with PBS + 0.1% Triton X-100. The sections were then blocked with 10% goat serum at room temperature for 1 hour. Primary antibodies against SC35 (Abcam, ab11826), XBP1s (BioLegend, 658802), and SON (Abcam, 121033) were conjugated to Alexa Fluor 488 or Alexa Fluor 555, respectively, per the manufacturer’s protocol and added to the OCT section at 1:1000 dilution overnight at 4°C. Next day, sections were washed five times with PBS and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) before mounting (with ProLong Gold Glass) and imaging using Leica SP8 lightening confocal microscope (Leica Microsystems). Three-dimensional construction from z-stack images was performed using Volume Viewer from ImageJ.
Image analysis
All image analyses were performed in Cell Profiler (version 3.1.5). Quantification of the shape of nuclear speckles was performed with customarily written pipelines in Cell Profiler. For speckle i, the sphericity is defined as in Eq. 1
| (1) |
so that a perfect circle will have a sphericity of 1, and a line will have a sphericity of 0. To calculate the average sphericity of a single cell that have k speckles, we calculated the area-weighted average as described in Eq. 2
| (2) |
Time-lapse microscopy
Time-lapse imaging was performed using SP8 lightening confocal microscope (Leica Microsystems) with Okolab stage top incubator to maintain constant CO2 (5%), temperature (37°C), and humidity (90%). Cells were plated into an eight-well chamber slide in full DMEM, and images were taken every 15 min using autofocus function. For imaging of cells in multiple wells simultaneously, Mark and Find feature was used to ensure accurate capture of the same cells.
Fluorescence recovery after photobleaching
FRAP was performed using a Leica SP8 lightening confocal microscope (Leica Microsystems) with 488-nm laser. Bleaching was performed using 100% laser power with 3.6 ms per pixel dwell time for five cycles, and images were collected every 1.29 s for 50 frames after bleaching. Fluorescence intensity at the bleached spot, a control unbleached spot, and background was measured using the embedded FRAP Profiler. Background intensity was subtracted, and values are reported relative to the unbleached spot to control for photobleaching during image acquisition. The recovery half-life (t1/2) was calculated by the online easyFRAP tool (https://easyfrap.vmnet.upatras.gr/) (51) by fitting to single or double exponential equations with the better fit (larger R-square values). For temporal FRAP analysis, different cells were selected for FRAP at each time point after Tu treatment or serum synchronization to minimize phototoxicity to cells due to repeated photobleaching.
ER proteostasis assay
MEFs were seeded in an eight-well chamber slide and transiently transfected with plasmid expressing cytomegalovirus (CMV) promoter–driven ER-localized aggregation-prone Halo-tag mutant (K73T/L172Q) (AgHaloER) (32). After 24 hours, the same cells were transfected with scrambled or Son siRNA for another 24 hours. Then, cells were treated with DMSO vehicle control or Tu (50 ng/ml or 100 ng/ml) for 16 hours. After that, cells were replaced with fresh DMEM containing 1 μM P1 to label AgHaloER protein for 30 min and then costained with Hoechst 33342/DAPI for nuclei. dCAS9-VPR GFP::SC35 MEFs expressing control or Son promoter–targeting sgRNA were transiently transfected with AgHaloER plasmid and then treated with Tu (100 ng/ml) for 16 hours before being subjected to confocal live imaging. Confocal images were obtained using a Leica SP8 confocal microscope (Leica Microsystems). The P1 signal was visualized with a blue argon laser (488 nm), and the Hoechst/DAPI signal was visualized using an ultraviolet laser (405 nm). Quantification of intensity was performed with Cell Profiler (version 3.15). For GFP::SC35 MEFs, only green signals that do not overlap with Hoechst staining (nucleus) were quantified.
Quantification of protein synthesis rate
The Click-iT HPG Alexa Fluor 594 protein synthesis HCS kit (Thermo Fisher Scientific) was used to measure protein synthesis in vitro. Briefly, MEFs cultured in chamber slides were treated with DMSO or Tu (100 ng/ml) for 5.5 hours before being pulsed with 50 μM methionine analog l-homopropargylglycine in methionine-free medium for 0.5 hours. Cells were fixed with 2% formaldehyde and permeabilized by 0.5% Triton X-100 and then underwent a ligation reaction for 30 min in the dark. Nuclei were further counterstained with Hoechst. Representative photomicrographs were obtained with a Leica SP8 confocal microscope (Leica Microsystems), and cell average intensity of Alexa Fluor 594 signal was measured using Cell Profiler software (version 3.15). Only cytosolic Alexa Fluor 594 signals (those not overlapped with nuclear Hoechst staining) were measured.
Immunoblot
Nuclear extracts were made from liver according to previously published protocol (52). Protein concentrations were determined by Bradford assays (Bio-Rad), and aliquots were snap frozen in liquid nitrogen and stored at −80°C until usage. Immunoblot analyses were performed as described previously (53). Briefly, 25 μg of proteins separated by 4 to ~20% gradient SDS–polyacrylamide gel electrophoresis gels (Bio-Rad) were transferred to nitrocellulose membranes, blocked in TBST buffer supplemented with 5% bovine serum albumin or 5% fat-free milk, and incubated overnight with primary anti-SON antibody (Abcam, 121033), anti-BMAL1 antibody (Abcam, 3350), anti-PERK (Cell Signaling Technology, no. 3192), anti–phospho-PERK (Thr908) (Thermo Fisher Scientific, MA5-15033), anti-ATF4 (Cell Signaling Technology, no. 11815), anti-IRE1α (Cell Signaling Technology, no. 3294), anti–phospho-IRE1α (Ser724) (ABclonal, AP0878), anti-XBP1s (BioLegend, 658802), anti-ATF6 (Santa Cruz Biotechnology, sc-166659), and β-actin (Cell Signaling Technology, no. 12620) at 4°C. Blots were incubated with an appropriate secondary antibody coupled to horseradish peroxidase at room temperature for 1 hour and reacted with Enhanced chemiluminescence (ECL) reagents per the manufacturer’s (Thermo Fisher Scientific) suggestion and detected by the Bio-Rad ChemiDoc MP Imaging System.
Quantitative reverse transcription polymerase chain reaction
Total mRNA was isolated from MEFs or mouse liver with a PureLink RNA mini kit (Life Technologies) with additional on-column deoxyribonuclease digestion step to remove genomic DNA per the manufacturer’s instructions. Reverse transcription was carried out using 5 μg of RNA using SuperScript III (Life Technologies) per the manufacturer’s instructions. For gene expression analyses, cDNA samples were diluted 1/30-fold (for all other genes except for 18S RNA) and 1/900-fold (for 18S RNA). qPCR was performed using the SYBR Green system with sequence-specific primers. All data were analyzed with 18S or β-actin as the endogenous control. qPCR primer sequences are as follows, and all primers span introns, except for primers for quantifying pre-mRNAs: mouse total Xbp1, (forward) gggtctgctgagtcc and (reverse) cagactcagaatctgaagagg; mouse total Xbp1 pre-mRNA, (forward) GTTAAGAACACGCTTGGGAATG and (reverse) TGGAGGTCCAGAACACAAAC; mouse Arntl, (forward) gccccaccgacctactct and (reverse) tgtctgtgtccatactttcttgg; mouse Per1, (forward) tcctcctcctacactgcctct and (reverse) ttgctgacgacggatcttt; mouse Per2, (forward) caacacagacgacagcatca and (reverse) tcctggtcctccttcaacac; mouse Cry2, (forward) gcagagcctggttcaagc and (reverse) gccactggatagtgctctgg; mouse Sec23b, (forward) tgaccaaactggacttctgga and (reverse) aaagaatctcccatcaccatgt; mouse Son, (forward) ttccgggaaatacaacagga and (reverse) gggtggatttgtttcaccat; mouse Manf, (forward) gacagccagatctgtgaactaaaa and (reverse) tttcacccggagcttcttc; mouse Manf pre-mRNA, (forward) AGGGTATGCAGAGATGGTAGA and (reverse) GATCTGTGAGAAGCTGAAGAAGA; mouse Hyou1, (forward) GAGGCGAAACCCATTTTAGA and (reverse) GCTCTTCCTGTTCAGGTCCA; mouse Hyou1 pre-mRNA, (forward) ACCGCTACAGCCATGATTT and (reverse) ATCATCTGGCAGGCACAC; mouse Atf4, (forward) CCACTCCAGAGCATTCCTTTAG and (reverse) CTCCTTTACACATGGAGGGATTAG; mouse Atf6, (forward) CATGAAGTGGAAAGGACCAAATC and (reverse) CAGCCATCAGCTGAGAATTAGA; mouse Ire1α, (forward) TCCTAACAACCTGCCCAAAC and (reverse) TCTCCTCCACATCCTGAGATAC; mouse 18S RNA, (forward) ctcaacacgggaaacctcac and (reverse) cgctccaccaactaagaacg; and mouse β-actin, (forward) aaggccaaccgtgaaaagat and (reverse) gtggtacgaccagaggcatac.
ChIP and ChIP-seq
ChIP for SC35 was performed using anti-SC35 antibody (Abcam, ab11826) as previously described (53). Briefly, mouse liver samples were submerged in PBS + 1% formaldehyde, cut into small (~1 mm3) pieces with a razor blade, and incubated at room temperature for 15 min. Fixation was stopped by the addition of 0.125 M glycine (final concentration). The tissue pieces were then treated with a TissueTearer and, lastly, spun down and washed twice in PBS. Chromatin was isolated by the addition of lysis buffer, followed by disruption with a Dounce homogenizer. The chromatin was enzymatically digested with MNase. Genomic DNA (input) was prepared by treating aliquots of chromatin with ribonuclease (RNase) and proteinase K and heated for reverse cross-linking, followed by ethanol precipitation. Pellets were resuspended, and the resulting DNA was quantified on a NanoDrop spectrophotometer. An aliquot of chromatin (10 μg) was precleared with protein A agarose beads (Invitrogen). Genomic DNA regions of interest were isolated using 4 μg of antibody. Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Cross-linking was reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation. The DNA libraries were prepared at the University of Pittsburgh and sequenced at Gene by Gene Ltd. per the standard protocols. DNA libraries were prepared with an Ovation Ultralow V2 DNA-Seq library preparation kit (NuGen) using 1 ng of input DNA. The size selection for libraries was performed using SPRIselect beads (Beckman Coulter), and purity of the libraries was analyzed using the High Sensitivity DNA chip on Bioanalyzer 2100 (Agilent). The prepared libraries were pooled and sequenced using NovaSeq 6000 (Illumina), generating ~40 million 101–base pair single-end reads per samples. ChIP-qPCR for MEFs were essentially performed the same way as previously described with anti-SC35 (Abcam, ab11826) and anti-XBP1s antibody (BioLegend, 658802), except that the MEFs were directly fixed with 1% formaldehyde before subject to nuclei isolation and chromatin immunoprecipitation. The primers used for ChIP-qPCR are as follows: negative control region 1, (forward) GCAACAACAACAGCAACAATAAC and (reverse) CATGGCACCTAGAGTTGGATAA; negative control region 2, (forward) GCAGTATAACTTCTCACCCAAGT and (reverse) AACATGGTGTCTGTTTGCTTTC; Xbp1 TSS region, (forward) GGCCACGACCCTAGAAAG and (reverse) GGCTGGCCAGATAAGAGTAG; Xbp1 TTS region, (forward) CTTTCTCCACTCTCTGCTTCC and (reverse) ACACTAGCAAGAAGATCCATCAA; Manf TSS region, (forward) ACAGCAGCAGCCAATGA and(reverse) CAGAAACCTGAGCTTCCCAT; Manf TTS region, (forward) CAACCTGCCACTAGATTGAAGA and (reverse) AGGCATCCTTGTGTGTCTATTT; Hyou1 TSS region, (forward) GACTTCGCAATCCACGAGAG and (reverse) GACTTCTGCCAGCATCGG; and Hyou1 gene body region, (forward) TGGAAGAGAAAGGTGGCTAAAG and (reverse) TCCCAAGTGCTGGGATTAAAG.
ChIP-seq analysis
Replicates were pooled at each time for subsequent ChIP-seq analysis. The sequences identified were mapped to the mouse genome (UCSC mm10) using BOWTIE function in Galaxy. Only the sequences uniquely mapped with no more than two mismatches were kept and used as valid reads. PCR duplicates were also removed. Peak calling was carried out by MACS2 (version 2.1.1.20160309) in Galaxy (options --mfold 5, 50 --pvalue 1e-4), on each ChIP-seq file against the input in XBP1Flox or XBP1LKO mice using the broad region function. To account for the different sequencing depths between samples, the signal files generated from MACS2 were reads per kilobase of transcript per million reads mapped normalized to sequencing depth. A total of 5365 genes with at least one peak identified in the gene body region (from TSS to TTS) in at least one CT in XBP1Flox mice were identified.
RNA-seq quantification
RNA-seq data were as previously reported (6). Raw RNA-seq FASTQ files were analyzed by FastQC for quality control. Adaptors and low-quality reads were filtered by Trimmomatic (54). Then, the processed reads were aligned by HISAT2 (55) against mouse reference mm10. For gene-level intron/exon quantification, bedtools software (56) was used to collect and count reads that aligned to any intron/exon of the given gene. If one read spans across multiple exons of the same gene, then it will only be counted once. If one read spans intron/exon junction, then it will only be counted as intron. The intron/exon count was normalized by gene length and total reads for Fragments Per Kilobase of transcript per Million mapped reads normalization.
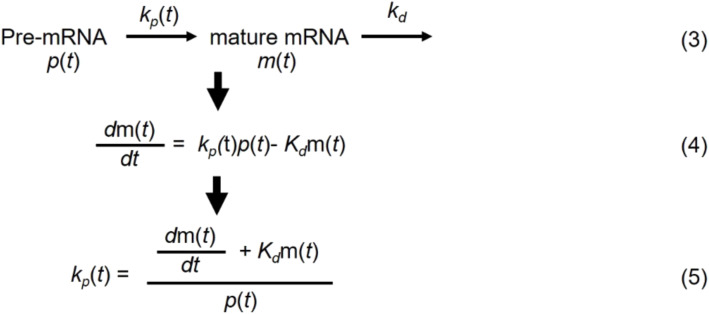
Estimation of mRNA processing rate
The mRNA processing rate was estimated by the simple kinetic model (Eq. 3), where pre-mRNA [p(t)] was converted to mature mRNA [m(t)] with the mRNA processing rate kp, which is also a function of time. We assume that the mature mRNA is subject to decay with a constant decay rate kd. Therefore, the mRNA processing rate kp(t) can be derived as Eq. 5. We used mRNA degradation rate for mouse genes reported in (57), and for gene without reported mRNA degradation rate, we used the mean value of 0.1 as a rough estimate. Because the original temporal gene expression data are fairly sparse (at 2-hour interval), to more accurately estimate the first derivative of mature mRNA [m(t)/dt], we performed a spline regression to obtain a more dense temporal dataset at 0.25-hour interval, and the first derivative at given time t is calculated as [m(t + 0.25) − m(t)]/0.25. Data analysis was performed in MATLAB and Excel.
Identification of oscillations from temporal dataset
Three orthogonal methods were used to identify oscillations from temporal dataset. Periodogram power spectral density was generated using MATLAB with the pxx = periodogram (x) function. Eigenvalue/pencil analysis was performed in MATLAB with customarily written code as previously described (4, 28). Criteria for circadian genes are period between 21 and 25 hours, and decay rate between 0.8 and 1.2; criteria for ~12-hour genes are period between 10.5 and 13.5 hours, and decay rate between 0.8 and 1.2. The FDR rate was calculated with a permutation-based method as previously described (6). RAIN analysis was performed as previously described in Bioconductor (3.4) (www.bioconductor.org/packages/release/bioc/html/rain.html) (17). For temporal SC35 ChIP-seq data in XBP1LKO mice, a polynomial detrend (n = 2) was first applied before being subjected to oscillation-identification algorithms. For all time-lapse microscopy data, the raw data, rather than the spline regression fit, were used to identify oscillations.
Acknowledgments
We want to thank T. Finkel for sharing the lentiCRISPRv2 plasmid, as well as valuable input and discussion during the manuscript preparation. We further want to thank L. Abdurehman and H. Li for the technical support on immunofluorescence and qPCR.
Funding: This study was supported by the National Institute of Health grant DP2GM140924 (B.Z.), National Institute of Health grant T32AG021885 (H.B.), American Diabetes Association grant 1-18-JDF-025 (B.Z.), National Institute of Health grant P30DK120531 (B.Z. and S.L.), EW Biomedical Scholars Program 00033066 (X.Z.), and National Science Foundation grant CHE-1944973 (X.Z.).
Author contributions: Conceptualization: B.Z. Methodology: B.Z., H.B., and X.Z. Investigation: B.Z., H.B., W.D., S.I., C.E., Y.P., J.Z., M.S., and J.L. Visualization: B.Z. and WD. Formal analysis: B.Z., W.D., J.L., and S.L. Funding acquisition: B.Z. and X.Z. Resources: B.Z. Software: B.Z. Validation: B.Z., W.D., and H.B. Supervision: B.Z. Writing, original draft: B.Z. Writing, review and editing: all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) under accession number GSE174170. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S17
Legends for tables S1 to S6
Legends for movies S1 to S3
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
Movies S1 to S3
REFERENCES AND NOTES
- 1.Zhu B., Dacso C. C., O'Malley B. W., Unveiling "Musica Universalis" of the cell: A brief history of biological 12-hour rhythms. J. Endocr. Soc. 2, 727–752 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes M. E., DiTacchio L., Hayes K. R., Vollmers C., Pulivarthy S., Baggs J. E., Panda S., Hogenesch J. B., Harmonics of circadian gene transcription in mammals. PLOS Genet. 5, e1000442 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cretenet G., Le Clech M., Gachon F., Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 11, 47–57 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Zhu B., Zhang Q., Pan Y., Mace E. M., York B., Antoulas A. C., Dacso C. C., O'Malley B. W., A cell-autonomous mammalian 12 hr clock coordinates metabolic and stress rhythms. Cell Metab. 25, 1305–1319.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng H., Gonzales N. M., Lonard D. M., Putluri N., Zhu B., Dacso C. C., York B., O’Malley B. W., XBP1 links the 12-hour clock to NAFLD and regulation of membrane fluidity and lipid homeostasis. Nat. Commun. 11, 6215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Y., Ballance H., Meng H., Gonzalez N., Kim S.-M., Abdurehman L., York B., Chen X., Schnytzer Y., Levy O., Dacso C. C., McClung C. A., O’Malley B. W., Liu S., Zhu B., 12-h clock regulation of genetic information flow by XBP1s. PLOS Biol. 18, e3000580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu B., Decoding the function and regulation of the mammalian 12h-clock. J. Mol. Cell Biol. 12, 752–758 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sriburi R., Jackowski S., Mori K., Brewer J. W., XBP1: A link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167, 35–41 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Song L., Liu M., Ge R., Zhou Q., Liu W., Li R., Qie J., Zhen B., Wang Y., He F., Qin J., Ding C., A proteomics landscape of circadian clock in mouse liver. Nat. Commun. 9, 1553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Mauvoisin D., Martin E., Atger F., Galindo A. N., Dayon L., Sizzano F., Palini A., Kussmann M., Waridel P., Quadroni M., Dulic V., Naef F., Gachon F., Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 25, 102–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saitoh N., Spahr C. S., Patterson S. D., Bubulya P., Neuwald A. F., Spector D. L., Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15, 3876–3890 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spector D. L., Lamond A. I., Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3, a000646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fei J., Jadaliha M., Harmon T. S., Li I. T. S., Hua B., Hao Q., Holehouse A. S., Reyer M., Sun Q., Freier S. M., Pappu R. V., Prasanth K. V., Ha T., Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell Sci. 130, 4180–4192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin Y., Chang Y. C., Lee D. S. W., Berry J., Sanders D. W., Ronceray P., Wingreen N. S., Haataja M., Brangwynne C. P., Liquid nuclear condensates mechanically sense and restructure the genome. Cell 176, 1518 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Bracha D., Walls M. T., Wei M. T., Zhu L., Kurian M., Avalos J. L., Toettcher J. E., Brangwynne C. P., Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J., Han K. Y., Khanna N., Ha T., Belmont A. S., Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci. 132, jcs226563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thaben P. F., Westermark P. O., Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhat P., Honson D., Guttman M., Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Rev. Mol. Cell Biol. 22, 653–670 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Sharma A., Takata H., Shibahara K., Bubulya A., Bubulya P. A., Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell 21, 650–663 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atwood A., DeConde R., Wang S. S., Mockler T. C., Sabir J. S., Ideker T., Kay S. A., Cell-autonomous circadian clock of hepatocytes drives rhythms in transcription and polyamine synthesis. Proc. Natl. Acad. Sci. U.S.A. 108, 18560–18565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rijo-Ferreira F., Takahashi J. S., Genomics of circadian rhythms in health and disease. Genome Med. 11, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballance H., Zhu B., Revealing the hidden reality of the mammalian 12-h ultradian rhythms. Cell. Mol. Life Sci. 78, 3127–3140 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorek M., Schnytzer Y., Ben-Asher H. W., Caspi V. C., Chen C. S., Miller D. J., Levy O., Setting the pace: Host rhythmic behaviour and gene expression patterns in the facultatively symbiotic cnidarian Aiptasia are determined largely by Symbiodinium. Microbiome 6, 83 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnytzer Y., Simon-Blecher N., Li J., Ben-Asher H. W., Salmon-Divon M., Achituv Y., Hughes M. E., Levy O., Tidal and diel orchestration of behaviour and gene expression in an intertidal mollusc. Sci. Rep. 8, 4917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chavez A., Tuttle M., Pruitt B. W., Ewen-Campen B., Chari R., Ter-Ovanesyan D., Haque S. J., Cecchi R. J., Kowal E. J. K., Buchthal J., Housden B. E., Perrimon N., Collins J. J., Church G., Comparison of Cas9 activators in multiple species. Nat. Methods 13, 563–567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y. E., Manteiga J. C., Henninger J. E., Sabari B. R., Dall'Agnese A., Hannett N. M., Spille J. H., Afeyan L. K., Zamudio A. V., Shrinivas K., Abraham B. J., Boija A., Decker T. M., Rimel J. K., Fant C. B., Lee T. I., Cisse I. I., Sharp P. A., Taatjes D. J., Young R. A., Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West J. A., Davis C. P., Sunwoo H., Simon M. D., Sadreyev R. I., Wang P. I., Tolstorukov M. Y., Kingston R. E., The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 55, 791–802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoulas A. C., Zhu B., Zhang Q., York B., O'Malley B. W., Dacso C. C., A novel mathematical method for disclosing oscillations in gene transcription: A comparative study. PLOS ONE 13, e0198503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J., Khanna N., Belmont A. S., Transcription amplification by nuclear speckle association. bioRxiv 2019, 604298 (2019). [Google Scholar]
- 30.Wang J., Symul L., Yeung J., Gobet C., Sobel J., Luck S., Westermark P. O., Molina N., Naef F., Circadian clock-dependent and -independent posttranscriptional regulation underlies temporal mRNA accumulation in mouse liver. Proc. Natl. Acad. Sci. U.S.A. 115, E1916–E1925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Yu J., Li T., Liu Y., Wang X., Zhang J., Wang X., Shi G., Lou J., Wang L., Wang C. C., Wang L., Phosphorylation switches protein disulfide isomerase activity to maintain proteostasis and attenuate ER stress. EMBO J. 39, e103841 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Fares M., Dunham N. P., Gao Z., Miao K., Jiang X., Bollinger S. S., Boal A. K., Zhang X., AgHalo: A facile fluorogenic sensor to detect drug-induced proteome stress. Angew. Chem. Int. Ed. Engl. 56, 8672–8676 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Schaum N., Lehallier B., Hahn O., Hosseinzadeh S., Lee S. E., Sit R., Lee D. P., Losada P. M., Zardeneta M. E., Pálovics R., Fehlmann T., Webber J., McGeever A., Zhang H., Berdnik D., Tan W., Zee A., Tan M., Pisco A., Karkanias J., Neff N. F., Keller A., Darmanis S., Quake S. R., Wyss-Coray T., The murine transcriptome reveals global aging nodes with organ-specific phase and amplitude. bioRxiv 662254 (2019). [Google Scholar]
- 35.Geyfman M., Kumar V., Liu Q., Ruiz R., Gordon W., Espitia F., Cam E., Millar S. E., Smyth P., Ihler A., Takahashi J. S., Andersen B., Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc. Natl. Acad. Sci. U.S.A. 109, 11758–11763 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., Hogenesch J. B., A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 111, 16219–16224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perelis M., Marcheva B., Moynihan Ramsey K., Schipma M. J., Hutchison A. L., Taguchi A., Peek C. B., Hong H., Huang W., Omura C., Allred A. L., Bradfield C. A., Dinner A. R., Barish G. D., Bass J., Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350, aac4250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyon A. S., Peeples W. B., Rosen M. K., A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mofatteh M., Echegaray-Iturra F., Alamban A., Dalla Ricca F., Bakshi A., Aydogan M. G., Autonomous clocks that regulate organelle biogenesis, cytoskeletal organization, and intracellular dynamics. eLife 10, e72104 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee D. S. W., Wingreen N. S., Brangwynne C. P., Chromatin mechanics dictates subdiffusion and coarsening dynamics of embedded condensates. Nat. Phys. 17, 531–538 (2021). [Google Scholar]
- 41.Folkmann A. W., Putnam A., Lee C. F., Seydoux G., Regulation of biomolecular condensates by interfacial protein clusters. Science 373, 1218–1224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasenson S. E., Shav-Tal Y., Speculating on the roles of nuclear speckles: How RNA-protein nuclear assemblies affect gene expression. Bioessays 42, e2000104 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Alexander K. A., Coté A., Nguyen S. C., Zhang L., Gholamalamdari O., Agudelo-Garcia P., Lin-Shiao E., Tanim K. M. A., Lim J., Biddle N., Dunagin M. C., Good C. R., Mendoza M. R., Little S. C., Belmont A., Joyce E. F., Raj A., Berger S. L., p53 mediates target gene association with nuclear speckles for amplified RNA expression. Mol. Cell 81, 1666–1681.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gems D., Partridge L., Stress-response hormesis and aging: "That which does not kill us makes us stronger". Cell Metab. 7, 200–203 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H., Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492–1496 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelson T., Heckl D., Ebert B. L., Root D. E., Doench J. G., Zhang F., Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chavez A., Scheiman J., Vora S., Pruitt B. W., Tuttle M., Iyer E. P. R., Lin S., Kiani S., Guzman C. D., Wiegand D. J., Ter-Ovanesyan D., Braff J. L., Davidsohn N., Housden B. E., Perrimon N., Weiss R., Aach J., Collins J. J., Church G. M., Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stashi E., Lanz R. B., Mao J., Michailidis G., Zhu B., Kettner N. M., Putluri N., Reineke E. L., Reineke L. C., Dasgupta S., Dean A., Stevenson C. R., Sivasubramanian N., Sreekumar A., Demayo F., York B., Fu L., O'Malley B. W., SRC-2 is an essential coactivator for orchestrating metabolism and circadian rhythm. Cell Rep. 6, 633–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakuma T., Nakade S., Sakane Y., Suzuki K.-I. T., Yamamoto T., MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 11, 118–133 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Zhu B., Khozoie C., Bility M. T., Ferry C. H., Blazanin N., Glick A. B., Gonzalez F. J., Peters J. M., Peroxisome proliferator-activated receptor β/δ cross talks with E2F and attenuates mitosis in HRAS-expressing cells. Mol. Cell. Biol. 32, 2065–2082 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koulouras G., Panagopoulos A., Rapsomaniki M. A., Giakoumakis N. N., Taraviras S., Lygerou Z., EasyFRAP-web: A web-based tool for the analysis of fluorescence recovery after photobleaching data. Nucleic Acids Res. 46, W467–W472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malovannaya A., Lanz R. B., Jung S. Y., Bulynko Y., Le N. T., Chan D. W., Ding C., Shi Y., Yucer N., Krenciute G., Kim B. J., Li C., Chen R., Li W., Wang Y., O'Malley B. W., Qin J., Analysis of the human endogenous coregulator complexome. Cell 145, 787–799 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu B., Gates L. A., Stashi E., Dasgupta S., Gonzales N., Dean A., Dacso C. C., York B., O'Malley B. W., Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol. Cell 60, 769–783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharova L. V., Sharov A. A., Nedorezov T., Piao Y., Shaik N., Ko M. S., Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 16, 45–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshida H., Oku M., Suzuki M., Mori K., pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172, 565–575 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H., Korennykh A. V., Behrman S. L., Walter P., Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc. Natl. Acad. Sci. U.S.A. 107, 16113–16118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DuRose J. B., Tam A. B., Niwa M., Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol. Biol. Cell 17, 3095–3107 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S17
Legends for tables S1 to S6
Legends for movies S1 to S3
References
Tables S1 to S6
Movies S1 to S3