Abstract
Pseudomonas aeruginosa is an important opportunistic pathogen capable of causing variety of infections in humans. The type III secretion system (T3SS) is a critical virulence determinant of P. aeruginosa in the host infections. Expression of the T3SS is regulated by ExsA, a master regulator that activates the expression of all known T3SS genes. Expression of the exsA gene is controlled at both transcriptional and posttranscriptional levels. Here, we screened a P. aeruginosa transposon (Tn5) insertional mutant library and found rplI, a gene coding for the ribosomal large subunit protein L9, to be a repressor for the T3SS gene expression. Combining real-time quantitative PCR (qPCR), western blotting and lacZ fusion assays, we show that RplI controls the expression of exsA at the posttranscriptional level. Further genetic experiments demonstrated that RplI mediated control of the exsA translation involves 5’ untranslated region (5’ UTR). A ribosome immunoprecipitation assay and qPCR revealed higher amounts of a 24 nt fragment from exsA mRNA being associated with ribosomes in the ΔrplI mutant. An interaction between RplI and exsA mRNA harboring its 24 nt, but not 12 nt, 5’ UTR was confirmed by RNA Gel Mobility Shift and Microscale Thermophoresis assays. Overall, this study identifies the ribosomal large subunit protein L9 as a novel T3SS repressor that inhibits ExsA translation in P. aeruginosa.
Author summary
Ribosomes provide all living organisms the capacity to synthesize proteins. The production of many ribosomal proteins is often controlled by an autoregulatory feedback mechanism. P. aeruginosa is an opportunistic human pathogen and its type III secretion system (T3SS) is a critical virulence determinant in host infections. In this study, by screening a Tn5 mutant library, we identified rplI, encoding ribosomal large subunit protein L9, as a novel repressor for the T3SS. Further exploring the regulatory mechanism, we found that the RplI protein interacts with the 5’ UTR (5’ untranslated region) of exsA, a gene coding for transcriptional activator of the T3SS. Such an interaction likely blocks ribosome loading on the exsA 5’ UTR, inhibiting the initiation of exsA translation. The significance of this work is in the identification of a novel repressor for the T3SS and elucidation of its molecular mechanism. Furthermore, this work provides evidence for individual ribosomal protein regulating mRNA translation beyond its autogenous feedback control.
Introduction
P. aeruginosa is an important opportunistic pathogen capable of causing a variety of infections in humans, particularly in patients with burn wounds, immunodeficiency and cystic fibrosis [1]. P. aeruginosa utilizes multifactorial virulence properties to establish acute and chronic infections. The type III secretion system (T3SS) is a critical virulence determinant of P. aeruginosa in acute infections [2]. Deficiency in T3SS gene expression/function severely attenuates P. aeruginosa virulence in mouse infection models of acute pneumonia, burn wounds, corneas, and neutropenia [3–6].
In P. aeruginosa, the T3SS can be induced by growth in a low Ca2+ environment or direct contact with host cells [3]. ExsA, an AraC-type DNA binding protein, is the master regulator of T3SS that can activate all of the known T3SS genes, including its own operon through the PexsC promoter [3]. Many regulatory factors have been determined to control the expression of T3SS through direct or indirect regulation of the exsA transcription and/or translation [3]. In addition to the exsC promoter (PexsC), transcription of the exsA is also driven by its own promoter PexsA, whose transcriptional activity is much lower than that of the PexsC [7]. Unlike PexsC which is directly bound and activated by ExsA, the PexsA promoter is directly bound and stimulated by the cAMP-Vfr signaling system [7]. Once transcribed, the exsA mRNA is subject to regulatory control at the posttranscriptional level. Translation of the exsA is positively controlled by the RNA helicase DeaD and the RNA-binding protein RsmA, but negatively regulated by a small noncoding RNA, sRNA 0161 [8–11].
In the present report, we identified rplI as a repressor for expression of the T3SS in P. aeruginosa through a genetic screen for isolates with altered T3SS expression in the mPAO1 strain. We show that mutants lacking rplI have increased expression of the T3SS and cytotoxicity toward cultured mammalian cells. We demonstrate that RplI controls expression of the T3SS through repressing the ExsA translation. Further studies revealed that RplI mediated translational repression requires a 24-nucleotide fragment residing upstream of the exsA coding region. EMSA and MST assays demonstrate that RplI can bind to the 24 nt upstream of the exsA start codon. In combination, these data identified a novel posttranscriptional regulatory mechanism that negatively controls the P. aeruginosa T3SS gene expression, elucidating a novel function of the L9 ribosomal protein in gene repression other than feedback control.
Results
Isolation of mutants with elevated PexsC expression
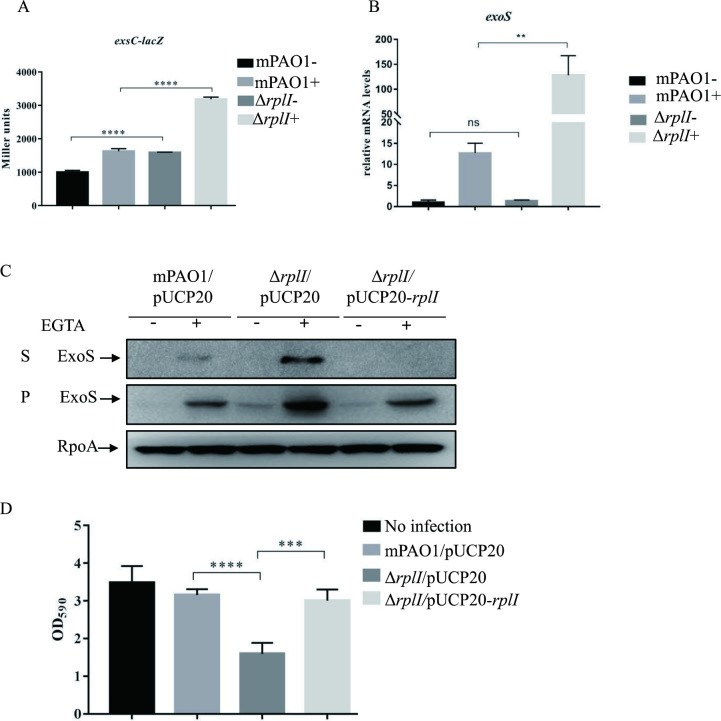
To identify novel repressors of the T3SS in P. aeruginosa, a Tn5 transposon insertion library was generated in mPAO1 strain containing a PexsC-lacZ reporter plasmid. The mPAO1 displays a lower level expression of T3SS compared to PAK strain due to mexT gene mutation and was referred as PAO1 in our previous study [12]. Approximately 10, 000 mutants were screened for dark blue colonies on X-gal plates under Ca2+ depletion conditions. A total of 34 transposon mutants displayed darker blue color compared to the parental mPAO1/PexsC-lacZ strain. These mutants were then subjected to sequence analysis to locate the Tn5 insertion sites. Total 8 different genetic loci on the chromosome were found to have the Tn5 insertions (S1 Table). Of them, 2 mutants had Tn5 insertions at two sites (372 bp/447 bp) on a rplI gene, which encodes 50S ribosomal protein L9. A rplI gene deletion mutant was further generated in the background of mPAO1 and introduced with the PexsC-lacZ reporter plasmid. Liquid β-galactosidase assay results confirmed that the rplI mutant indeed displays an elevated β-galactosidase activity (Fig 1A).
Fig 1. RplI represses T3SS expression and cytotoxicity.
(A) mPAO1 and ΔrplI containing the PexsC-lacZ transcriptional fusion were grown to an OD600 of 1.0 in LB with (+) or without (-) 5 mM EGTA and subjected to β-galactosidase assays. Each assay was performed in triplicate, and the error bars indicate standard deviations. ****P < 0.0001 compared with wild-type mPAO1 by Student’s t test. (B) Relative exoS mRNA levels in the mPAO1 and ΔrplI strains. Total RNA was isolated under T3SS inducing (+) and non-inducing (-) conditions with 5 mM EGTA, and the exoS mRNA levels were determined by real-time qPCR using rpsL as the internal control. ns, not significant, **P < 0.01, by Student’s t test. (C) Expression and secretion of ExoS in mPAO1/pUCP20, ΔrplI/pUCP20 and ΔrplI/pUCP20-rplI. Bacteria were cultured to an OD600 of 1.0 in LB medium with or without 5 mM EGTA. Proteins in supernatants (S) and pellets (P) from equivalent bacterial cells were loaded onto 12% SDS-PAGE gels and probed with an antibody against ExoS or an anti-RpoA antibody. The data shown represent the results from three independent experiments. (D) Cytotoxicity of mPAO1/pUCP20, ΔrplI/pUCP20 and ΔrplI/pUCP20-rplI. HeLa cells were infected with the indicated strains at an MOI of 50. Three hours post infection, cells attached to the 24-well plate were washed with PBS and stained with crystal violet. The cell-associated crystal violet was dissolved in ethanol and quantified by measuring the OD590. HeLa cells with no bacterial infection served as a control. ***P< 0.001, ****P < 0.0001 by Student’s t test.
RplI represses ExoS expression and reduces bacterial cytotoxicity
We further pursued the role of RplI in the regulation of the T3SS. RplI encodes an L9 protein of the ribosomal 50S large subunit and the rplI deletion mutant displayed no discernible growth rate change when cultured in LB medium (S1A Fig), which suggested that L9 is not essential in P. aeruginosa, similar to that in Salmonella enterica [13]. Consistent with the phenotypes of the original transposon insertion mutant, the mPAO1ΔrplI mutant showed higher mRNA levels of exoS, pcrV and pscF (Figs 1B and S1B), as well as increased expression and secretion of ExoS (Fig 1C) compared to those in the wild type mPAO1. Complementation with a rplI-expressing plasmid decreased the expression and secretion of ExoS (Fig 1C). To further confirm the relationship between rplI and T3SS, the rplI gene was deleted in another PAO1 laboratory strain which has a proficient T3SS. Similar to ΔrplI in mPAO1, this mutant also displayed increased expression and secretion of ExoS compared to the parental wild type PAO1, which could be complemented by rplI expression in trans (S1C Fig). T3SS-mediated cytotoxicity was further measured by crystal violet staining assay. When HeLa cells were infected with wild-type mPAO1/pUCP20 at an MOI (multiplicity of infection) of 50, majority of the cells looked fine and remained attached after 3 hours of infection. Loss of the rplI, however, resulted in a strain that was much more cytotoxic, and complementation with a rplI gene decreased the cytotoxicity (Fig 1D). These results suggested that RplI is a repressor of the T3SS in P. aeruginosa.
Role of RplI in the expression of exsA gene
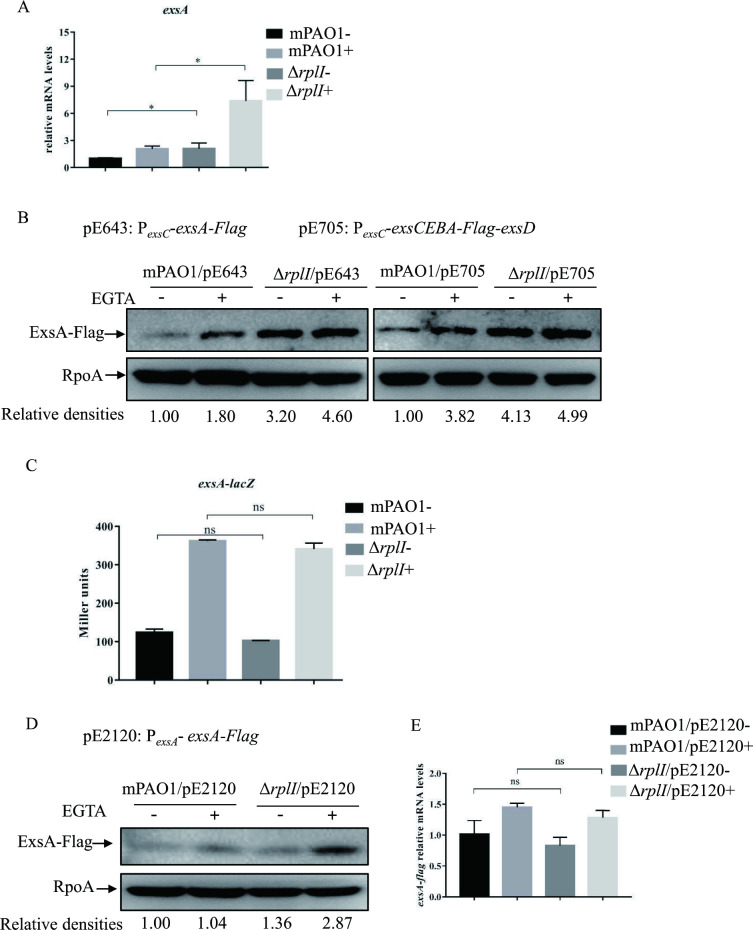
ExsA is the central activator of the T3SS located in the exsCEBA operon [3]. We wanted to know whether the significant increase in ExoS expression in the ΔrplI mutant was due to the upregulation of exsA expression. Real-time qPCR was performed to examine the exsA mRNA level in mPAO1 and its ΔrplI mutant. Consistent with the increased exsC promoter activity (Fig 1A), the mRNA level of exsA in the ΔrplI mutant was significantly higher than that in wild type mPAO1 (Fig 2A). To further confirm the increased expression of ExsA protein, a C-terminal Flag-tagged ExsA (alone/pE643 or in the exsCEBA operon/pE705) [14] driven by the exsC promoter was integrated into the chromosome of P. aeruginosa. Consistent with the elevated exsA transcription, the ΔrplI mutant displayed an increased ExsA-Flag protein level under both T3SS inducing and non-inducing conditions (Figs 2B, S2A, and S2B). These results indicated that RplI directly or indirectly represses the expression of the exsCEBA operon.
Fig 2. Role of RplI in the expression of ExsA.
(A and E) The relative exsA (A) or exsA-flag (E) mRNA levels in mPAO1 and ΔrplI containing pE2120 (E) or not (A). Total RNA was isolated under T3SS inducing (+) and non-inducing (-) conditions with 5 mM EGTA and the exsA or exsA-flag mRNA levels were determined by real-time qPCR using rpsL as the internal control. ns, not significant, *P < 0.05, by Student’s t test. (B and D) Amounts of ExsA-Flag. mPAO1 and ΔrplI containing pE643 or pE705 integrated into the chromosome or pE2120 were grown to an OD600 of 1.0 in LB with (+) or without (-) 5 mM EGTA and collected by centrifugation. Samples from equivalent numbers of bacterial cells were separated by SDS-PAGE and probed with an anti-Flag or an anti-RpoA antibody. The density of each band was determined with Image J. Relative densities represent the density of ExsA-Flag/density of RpoA with the respective first lane as 1. The data shown represent the results from three independent experiments. (C) mPAO1 and ΔrplI containing the PexsA-lacZ transcriptional fusion were grown to an OD600 of 1.0 in LB with (+) or without (-) 5 mM EGTA and subjected to β-galactosidase assays. Each assay was performed in triplicate, and the error bars indicate standard deviations. ns, not significant compared with wild-type mPAO1 by Student’s t test.
ExsA is known to activate its own expression through the promoter of exsC operon [15]; therefore, the upregulation of exsA expression in the ΔrplI mutant might occur at either transcriptional or posttranscriptional level. To address this, a PexsA-lacZ transcriptional fusion reporter plasmid [16] was introduced into the wild type mPAO1 and its ΔrplI mutant. In both strains, the β-galactosidase activities were significantly higher under T3SS inducing condition than that under non-inducing condition. However, mRNA levels displayed no difference between wild type mPAO1 and the ΔrplI mutant under either T3SS inducing or non-inducing condition (Fig 2C). To visualize the ExsA protein expression, pE2120 encoding a C-terminal Flag-Tagged ExsA driven by its own promoter (the promoter of exsA) [16], was introduced into the two bacterial strains. Consistent with the results of PexsA-lacZ transcriptional fusion, the relative mRNA levels of exsA-flag (primers shown in S3 Table) showed no difference in the ΔrplI mutant compared to that in wild type mPAO1 strain. However, an increased ExsA-Flag protein level was observed in the ΔrplI mutant compared to that in the wild type mPAO1 strain (Fig 2D and 2E). These data suggested that RplI likely affects the ExsA expression at the posttranscriptional level.
RplI controls the expression of ExsA at the posttranscriptional level
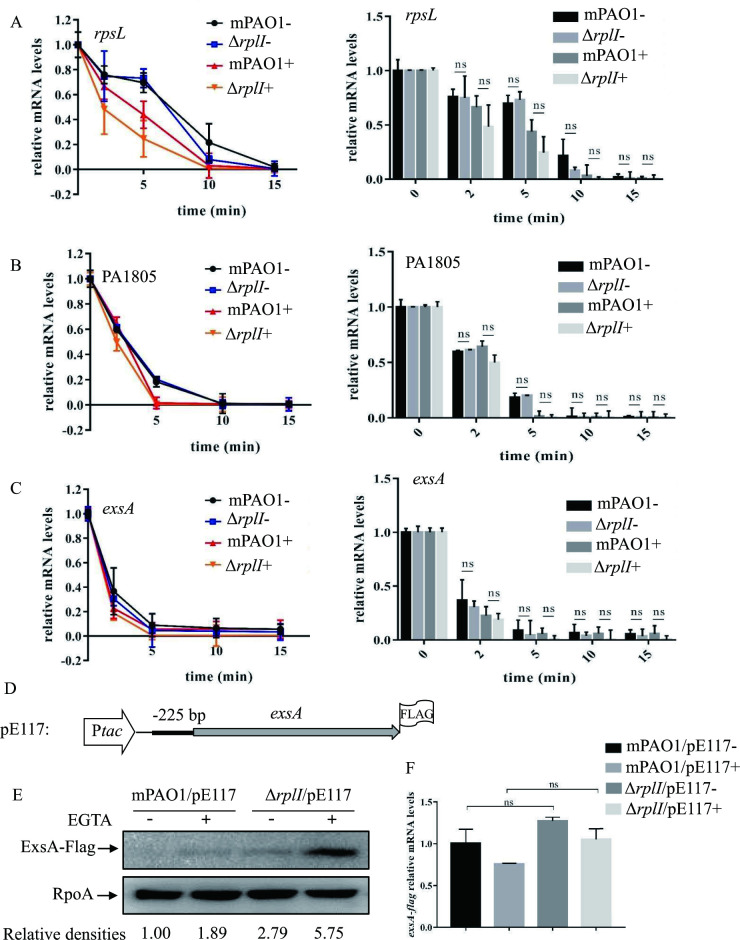
The higher protein level of ExsA in the ΔrplI mutant might be due to an increased exsA mRNA stability or enhanced exsA translation. To address these possibilities, we initially compared the stability of exsA mRNA in wild type mPAO1 and the ΔrplI mutant. Bacteria were cultured to an OD600 of 1.0 under either T3SS inducing or non-inducing condition, and 200 μg/mL rifampicin was added to inhibit further transcription. After 2, 5, 10 or 15 min, total RNAs were collected and examined with real-time qPCR (primers in S3 Table). As shown in Fig 3, the exsA mRNA decay rates displayed no significant difference between mPAO1 and its ΔrplI mutant derivative under either T3SS-inducing or non-inducing condition. These data indicated that the increased ExsA protein level was not due to changes in the exsA mRNA stability.
Fig 3. RplI controls the expression of ExsA at the posttranscriptional level by repressing ExsA translation.
(A-C) Degradation of rpsL (A), PA1805 (B), and exsA (C) mRNA in wild type mPAO1 and its ΔrplI mutant under T3SS inducing (+) and non-inducing (-) conditions with 5 mM EGTA. Bacterial cells were treated with rifampicin, collected at 2, 5, 10, and 15 min and mixed with equal numbers of gfp-expressing E. coli cells. Total RNA was purified, and the relative mRNA levels were determined by real-time qPCR. The gfp mRNA level in each sample was used as the internal control for normalization. The right panel is the bar diagram of the respective left panel. ns, not significant by Student’s t test. (D) Construct of exsA-Flag in pE117. (E) Amounts of ExsA-Flag. mPAO1 and ΔrplI containing pE117 were grown to an OD600 of 1.0 in LB with (+) or without (-) 5 mM EGTA and collected by centrifugation. Samples from equivalent numbers of bacterial cells were separated by SDS-PAGE and probed with an anti-Flag or an anti-RpoA antibody. The density of each band was determined with Image J. Relative densities represent the density of ExsA-Flag/density of RpoA with the first lane as 1. The data shown represent the results from three independent experiments. (F) Relative exsA-flag mRNA levels in mPAO1 and ΔrplI containing pE117. Total RNA was isolated under T3SS inducing (+) and non-inducing (-) conditions with 5 mM EGTA, and the exsA-flag mRNA levels were determined by real-time qPCR using rpsL as the internal control. ns, not significant, by Student’s t test.
Next, we examined whether translation of the ExsA was enhanced in the ΔrplI mutant using a previously constructed exsA-Flag fusion plasmid (exsA-Flag-S/pE117) [17]. In this plasmid, the exsA coding sequence with its 225-bp upstream region was driven by a tac promoter, and a Flag-tag was fused at the C-terminus of the ExsA (Fig 3D). Western blot assay revealed an increased ExsA-Flag level in the ΔrplI mutant compared to that in wild type mPAO1 containing the pE117 (Figs 3E and S2C), while the mRNA levels of exsA-flag displayed no difference between the two strains (Fig 3F). These results suggested that RplI represses ExsA translation.
RplI-dependent repression of the ExsA translation requires a -24 bp region of the exsA 5’ UTR
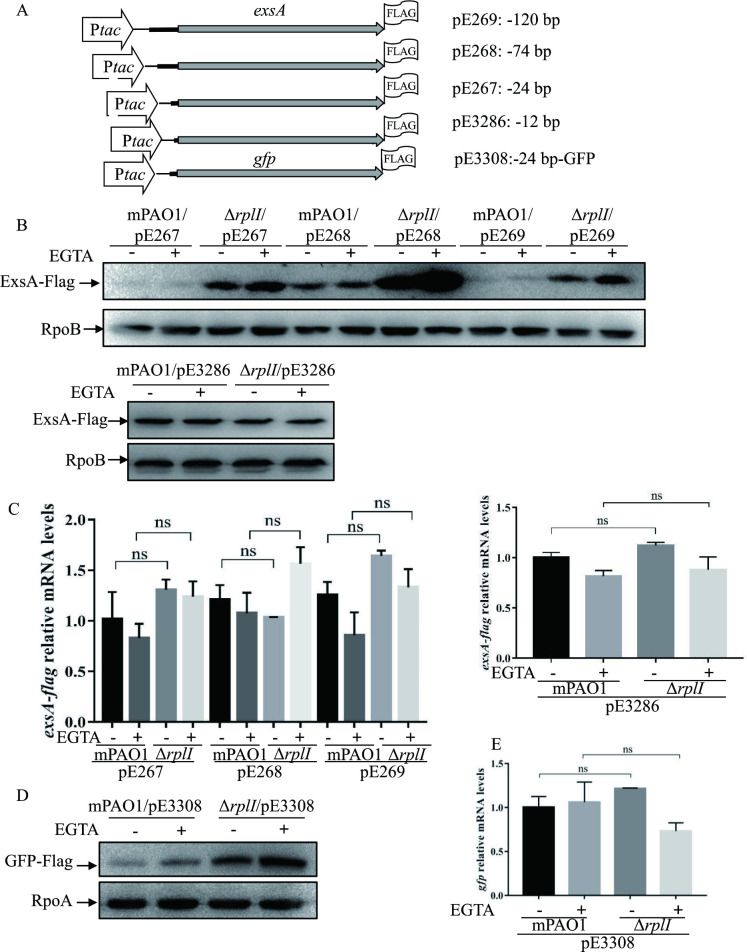
Based on our finding that RplI attenuates ExsA translation, we wanted to know the sequence requirements for the RplI-mediated control. Plasmid constructs, carrying the tac promoter driven exsA-flag fusion gene with varying lengths of the 5’ untranslated region (5’ UTR) of exsA, were introduced into the mPAO1 and ΔrplI strains (Fig 4A). The mRNA levels and protein amounts of ExsA-Flag were examined with real-time qPCR and Western blot analysis, respectively. As shown in Fig 4B and 4C, in the presence of -120, -74, and -24 bp upstream regions, despite of comparative mRNA levels, ExsA-Flag amounts were much higher in the ΔrplI mutant than those in the wild type mPAO1. In contrast, pE3286, which harbors the exsA-Flag and its -12 bp upstream region, rendered mPAO1 and ΔrplI similar amounts of ExsA-Flag. These data suggested that while most of the sequence upstream of the exsA ORF is dispensable for the RplI-mediated regulation, at least a 24-nt untranslated upstream sequence is required.
Fig 4. RplI-dependent repression of ExsA translation requires a 24 bp 5’ untranslated region of exsA.
(A) Constructs of exsA-Flag and gfp-Flag. (B and D) Amounts of ExsA-Flag or GFP-Flag in the indicated strains. Overnight-cultured mPAO1 and ΔrplI containing the indicated constructs were diluted 1:50 into LB medium, grown to an OD600 of 1.0 with (+) or without (-) 5 mM EGTA and collected by centrifugation. Samples from equivalent numbers of bacterial cells were loaded onto an SDS-PAGE gel and probed with an anti-Flag, an anti-RpoB antibody or an anti-RpoA antibody. The data shown represent the results from three independent experiments. (C and E) exsA-flag (C) or gfp (E) mRNA levels in mPAO1 and ΔrplI containing pE267, pE268, pE269 or pE3286 (C) or pE3308 (E). Total RNA was isolated under T3SS inducing (+) and non-inducing (-) conditions with 5 mM EGTA, and the exsA-flag or gfp mRNA levels were determined by real-time qPCR using rpsL as the internal control. ns, not significant, by Student’s t test.
Next, we determined whether the exsA coding sequence was required. The exsA coding sequence in pE267 was replaced with a gfp gene, resulting in pE3308, in which the gfp coding region was fused with the 24-bp 5’ UTR sequence of exsA (Fig 4A). Similar to the pE267 (Ptac-24-exsA-Flag), gfp-flag fusion with the 24 bp 5’ UTR of exsA resulted in a higher GFP-Flag protein level in the ΔrplI mutant than that in wild type mPAO1, although their mRNA levels for gfp were similar (Fig 4D and 4E). This finding suggested that the 24 bp upstream region of the exsA coding sequence plays a critical role in the RplI-mediated translational repression of ExsA.
Enhanced ribosome binding to the exsA mRNA in the ΔrplI mutant
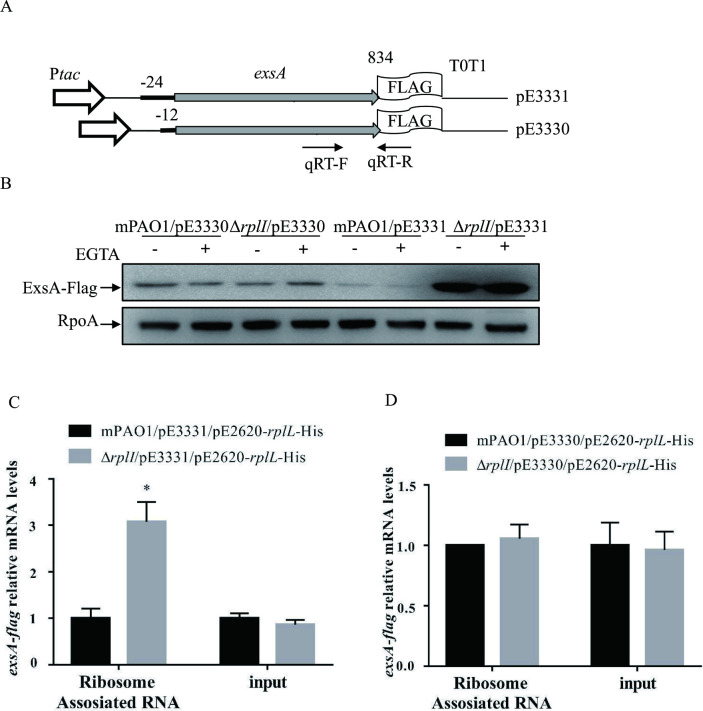
Based on our finding that ExsA translation is increased in a ΔrplI mutant and the fact that rplI encodes the L9 protein of the 50S subunit of ribosomes, we hypothesized that more ribosomes might associate with the 24 nt 5’ UTR of exsA in the absence of RplI. To test this possibility, a modified RNA-binding protein immunoprecipitation was coupled with real-time qPCR to directly determine the amount of ribosome-associated exsA mRNA as previously described [18]. First, using pE267 plasmid as a template, we cloned the exsA-flag with its 24 bp 5’ UTR together with the tac promoter and two consecutive transcription terminators (tac promoter and T0T1 from pFlag-CTC, Sigma) into pE1553 [16], resulting in pE3331 (Fig 5A). The RNA transcribed from this construct harbors a single ribosome binding site for exsA; thus, ribosomes can only bind to the exsA mRNA. The C-terminal Flag ensures that exsA mRNA transcribed from the genomic locus will not be detected by real-time qPCR (primer set shown in Fig 5A). Indeed, no amplicon could be observed when the detection primer set (S3 Table) was used with mPAO1 genomic DNA as template in the PCR. A control plasmid pE3330, in which the 24-bp 5’ UTR was replaced by a 12-bp 5’ UTR, was also constructed (Fig 5A). The ExsA-Flag amounts were then examined in mPAO1 and the ΔrplI mutant containing pE3331 or pE3330. As the results shown in Fig 5B, similar to pE267 and pE3286, the 24 nt 5’ UTR of exsA rendered an increased amount of ExsA-Flag protein in ΔrplI, while the 12 nt 5’ UTR of exsA displayed similar amounts of ExsA-Flag between wild type mPAO1 and the ΔrplI strain.
Fig 5. Expression of ExsA-Flag and quantification of ribosome-associated exsA mRNAs.
(A) Constructs of Ptac-24 bp-exsA-Flag-T0T1 (pE3331) and Ptac-12 bp-exsA-Flag-T0T1 (pE3330). The positions of qPCR primers are represented by arrows. (B) The protein levels of ExsA-Flag in the indicated strains. mPAO1 and ΔrplI containing pE3330 or pE3331 were grown to an OD600 of 1.0 in LB with (+) or without (-) 5 mM EGTA and collected by centrifugation. Samples from equivalent numbers of bacterial cells were loaded onto an SDS-PAGE gel and probed with an anti-Flag or an anti-RpoA antibody. The data shown represent the results from three independent experiments. (C and D) mPAO1 and ΔrplI containing pE2620-rplL-His and pE3331 (C) or pE3330 (D) were lysed with sonication and subjected to Ni-NTA chromatography or not (input), followed by RNA purification. The relative mRNA levels of exsA-Flag were determined by real-time qPCR with 16S ribosome RNA (PA0668.1) as the internal control. *, P < 0.05 by Student’s t test.
A plasmid construct with another 50S ribosomal protein L7/L12 coding gene rplL fused with his-tag (pE2620-rplL-His) was further introduced into the wild type mPAO1 and the ΔrplI mutant containing either pE3330 or pE3331. Ribosomes were then purified from the cell extracts of these strains by Ni-NTA chromatography, and the associated RNA was isolated and subjected to real-time qPCR assay. The mRNA levels of exsA transcribed from both plasmids were similar between mPAO1 and the ΔrplI mutant (Fig 5C and 5D). However, many more exsA mRNAs with 24 nt 5’ UTR, but not with 12 nt 5’ UTR, were associated with the ribosome isolated from the ΔrplI mutant (Fig 5C and 5D). In combination, these results suggested that increased amounts of ribosomes were associated with the 24-nt 5’ UTR of exsA in the ΔrplI mutant.
Sr0161 is not involved in the RplI mediated control of ExsA translation
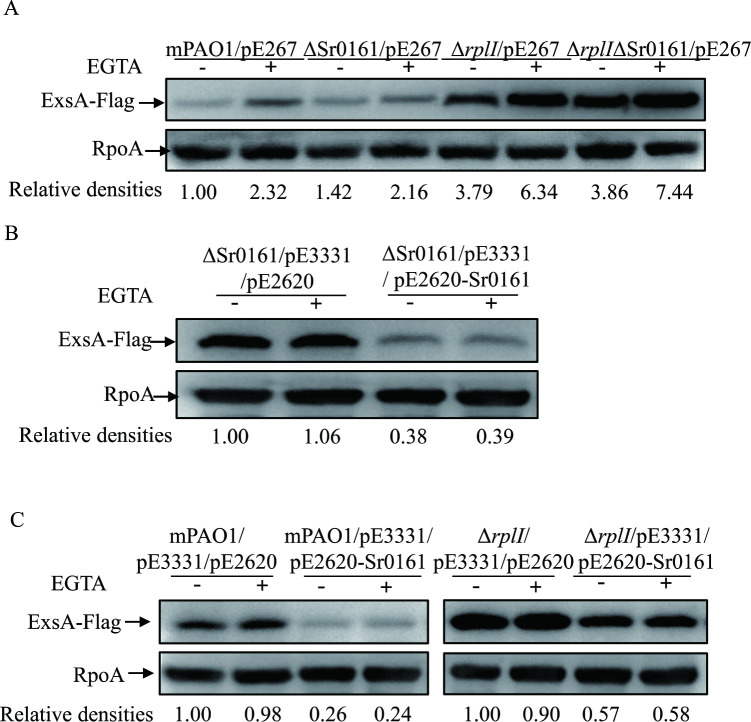
A recent study showed that a small regulatory RNA Sr0161 interacts with the 5’ UTR of exsA mRNA to repress ExsA synthesis at the posttranscriptional level, and overexpression of Sr0161 decreased the expression of the T3SS in P. aeruginosa [9,10]. The 5’ UTR of exsA, between -24 nt and -14 nt, was predicted to base pair with Sr0161 [9,10]. Because 24 bp, but not 12 bp, 5’ UTR of exsA was needed for the RplI-mediated translational repression of exsA, we tested whether Sr0161 was also involved in the RplI mediated control of ExsA translation. The Sr0161 gene was deleted in mPAO1 and the ΔrplI mutant backgrounds, and then pE267 (-24 bp-exsA-Flag construct) was introduced into the resulting mutant strains. Consistent with a previous report [9], no increase in ExsA-Flag protein level was observed in the ΔSr0161 compared to that of mPAO1 (Fig 6A). Similarly, the amount of ExsA-Flag in ΔrplI was the same as that in ΔrplIΔSr0161.
Fig 6. Sr0161 is not involved in the RplI mediated control of ExsA translation.
The protein levels of ExsA-Flag in mPAO1, ΔSr0161, ΔrplI and ΔrplIΔSr0161 containing pE267 (A); ΔSr0161 containing pE3331 and empty vector (pE2620) or pE2620-Sr0161 (B); mPAO1 and ΔrplI containing pE3331 and empty vector (pE2620) or pE2620-Sr0161 (C). Bacteria cultured overnight were diluted 1:50 into LB medium with 1 mM IPTG (B and C) or without IPTG (A), grown to an OD600 of 1.0 with (+) or without (-) 5 mM EGTA, and collected by centrifugation. Samples from equivalent numbers of bacterial cells were separated by SDS-PAGE and probed with an anti-Flag or an anti-RpoA antibody. The density of each band was determined with Image J. Relative densities represent the density of ExsA-Flag/density of RpoA with the respective first lane as 1. The data shown represent the results from three independent experiments.
To further test the hypothesis, we examined the influence of Sr0161 overexpression on the expression of ExsA. We cloned Sr0161 into pE2620 (pMMB67EH, Gm) and introduced it into ΔSr0161 harboring pE3331 (-24 bp-exsA-Flag). Consistent with the previous study [9], overexpression of Sr0161 reduced the amounts of ExsA-Flag in ΔSr0161 (Fig 6B). Next, the pE2620-Sr0161 and -24 bp-exsA-Flag (pE3331) were co-introduced into mPAO1 and ΔrplI. Similar to that in ΔSr0161, overexpression of the Sr0161 decreased the amount of ExsA-Flag in the wild type mPAO1 strain (Fig 6C). Again, the reduced amount of ExsA-Flag was observed in ΔrplI with overexpression of the Sr0161 (Fig 6C). Together, these data indicated that Sr0161 was not involved in the RplI mediated repression of the ExsA.
RplI binds to the 24 nt 5’ UTR of the exsA mRNA
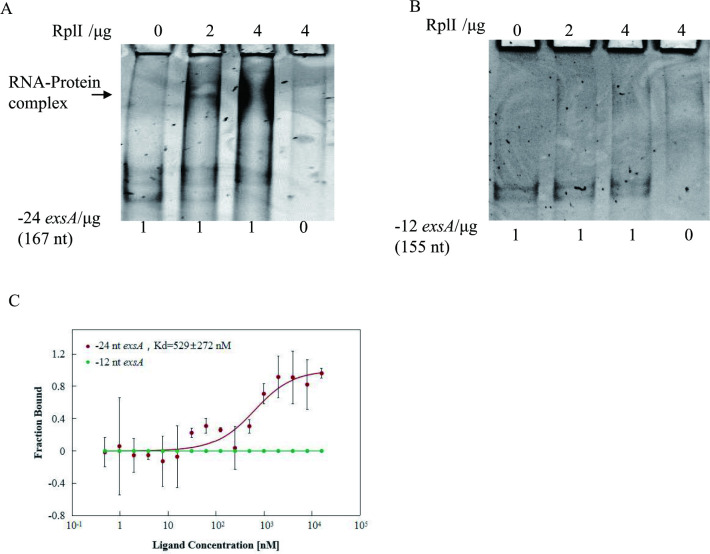
It has been reported that some ribosomal proteins, such as primary rRNA-binding proteins S1, S7, S8 and L1, interact with mRNAs of their own or others in the operons to inhibit the translation of target genes in E. coli [19–22]. Since L9 is the primary 23S rRNA-binding protein in the 50S ribosome, we asked if L9 repressed the translation of exsA through direct binding to the 24-nt 5’ UTR of exsA. To address this, we examined the RplI interaction with the 5’ UTR of exsA by EMSA. The RNA of -24 nt-exsA or -12 nt-exsA of P. aeruginosa was transcribed with specific primers (S3 Table) from corresponding PCR products in vitro and incubated with the indicated amounts of purified C-terminal 6×His tagged RplI of P. aeruginosa. The -24 nt/12 nt -exsA probe is comprised of the native 24-nt/12-nt 5’ UTR and 143 nt downstream of the start codon for exsA. As shown in Fig 7A and 7B, RplI of P. aeruginosa retarded the migration of RNA containing partial exsA with its 24 nt 5’ UTR but not that with the 12 nt 5’ UTR, indicating a direct interaction with the 24 nt 5’ UTR of exsA.
Fig 7. RplI interacts with exsA mRNA containing a 24 nt 5’ UTR but not a 12 nt 5’ UTR.
(A and B) Binding of RplI to the indicated RNAs was examined by EMSA. Increasing amounts of purified P. aeruginosa RplI-His were incubated with 167 nt/155 nt (A/B) exsA mRNA respective corresponding to the 24 nt/12 nt 5’ UTR and its first 143 nt mRNA on ice for 30 min. The mixtures were electrophoresed on an 8% native polyacrylamide gel, and the bands were visualized by staining with Gel-red for 10 min. (C) MST assay to test the binding capability of purified P. aeruginosa RplI-His to the indicated RNA (same as A, B) transcribed in vitro. The error bar represents the standard deviation from triplicate assays for each sample.
To further confirm the direct interaction, a microscale thermophoresis assay (MST) was carried out using NT-647-NHS labeled RplI as an aptamer probe. As the results shown in Fig 7C, binding between RplI and the -24 nt-exsA (same RNA probe as in EMSA) was detected with a Kd of 529 ± 272 nM, while interaction between the -12 nt-exsA (same RNA probe as in EMSA) and RplI was undetectable. These results further confirmed that RplI interacts with the 24 nt 5’ UTR of exsA directly.
RplI inhibits exsA translation via its 24 nt 5’ UTR
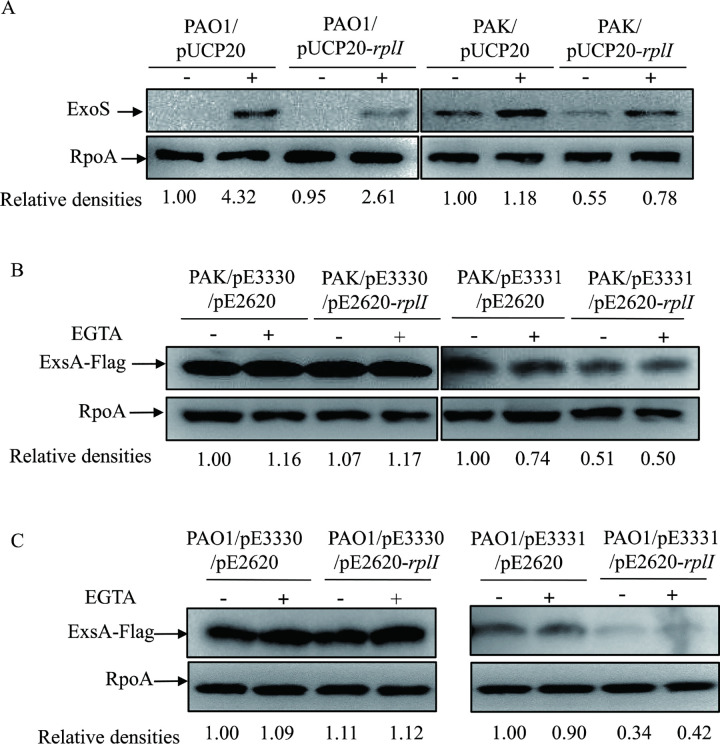
If RplI represses exsA translation by binding to its 24 nt 5’ UTR, overexpression of rplI in wild type P. aeruginosa strains should decrease the expression of ExoS, as well as the translation of exsA with its 24 nt 5’ UTR, but not with its 12 nt 5’ UTR. To test these, RplI was overexpressed in the T3SS-proficient P. aeruginosa laboratory strains PAK and another PAO1 strain. Overexpression of rplI indeed decreased the expression of ExoS in both PAK and PAO1 (Figs 8A and S3A). Furthermore, pE2620-rplI was introduced into the PAO1 or PAK strain containing pE3331 (24 nt 5’ UTR-exsA) or pE3330 (12 nt 5’ UTR-exsA). Consistent with the expression of ExoS, overexpression of rplI decreased the ExsA-Flag amounts in the presence of its 24 nt 5’ UTR in both PAK and PAO1 strains. However, overexpression of rplI in PAK or PAO1 had no effect on the expression of ExsA-Flag in the presence of its 12 nt 5’ UTR (Figs 8B, 8C, S3B and S3C). These results further confirmed that RplI represses the expression of T3SS in P. aeruginosa.
Fig 8. RplI represses the expression of ExsA and ExoS in PAK and PAO1 strains.
(A) Expression of ExoS in another PAO1 laboratory strain and PAK containing empty vector pUCP20 or pUCP20-rplI. (B and C) Expression of ExsA-Flag in PAK (B) and PAO1 (C) containing pE3330 or pE3331 with empty vector pE2620 or pE2620-rplI. Bacteria cultured overnight were diluted 1:50 into fresh LB medium, grown to an OD600 of 1.0 with (+) or without (-) 5 mM EGTA and collected by centrifugation. Proteins from equivalent bacterial cells were separated by SDS-PAGE gels and probed with an antibody against ExoS/Flag or an anti-RpoA antibody. The density of each band was determined with Image J. Relative densities represent the density of ExoS or ExsA-Flag/density of RpoA with the respective first lane as 1. The data shown represent the results from three independent experiments.
Discussion
The bacterial rplI gene encodes ribosomal large subunit (50S) protein L9 which is a primary 23S rRNA binding protein with N- and C-terminal globular domains, both containing a predicted RNA binding site connected by an exposed α-helix [23]. It was speculated that L9 functions to serve as a molecular strut, likely playing an architectural role in ribosome assembly and/or maintaining the catalytically active conformation of ribosomal RNA in the large ribosomal subunit by binding and positioning two regions of the 23S rRNA [24,25]. In addition to its role as a ribosomal 23S rRNA-binding protein, L9 has been reported to play an important role in reading frame maintenance in Salmonella enterica serovar Typhimurium [13], ribosomal “hopping” over a 50 nucleotide region within the mRNA of bacteriophage T4 gene 60 in E. coli, and response to starvation stress in E. coli [26]. Although bacterial L9 possesses a highly conserved secondary and tertiary structure and contains several invariant amino acids, L9 is not essential, and rplI deletion strains seem to grow fine under normal conditions in S. enterica [13].
In this study, we identified that L9 interacts with the 5’ UTR of exsA and serves as a translational repressor, suppressing the T3SS in P. aeruginosa. Small RNA regulators (sRNAs) often sequester the ribosome binding site (RBS) of target genes via intramolecular base-pairing with the 5’ UTR of target mRNAs to downregulate translation initiation. Sr0161 was recently reported to directly target the exsA leader region through imperfect base pairing to repress the T3SS [9,10]. It has also been demonstrated that Sr0161 inhibits ExsA synthesis at the posttranscriptional level [10]. As the RplI mediated repression of ExsA translation also involves the exsA 5’ untranslated region, a possible model to account for the inhibitory activity of RplI is that imperfect base pairing of Sr0161with the exsA leader region prevents the loading of wild-type ribosomes, but not RplI-deficient ribosomes. However, our finding that overexpression or deletion of Sr0161 has the same influence on the expression of ExsA in both mPAO1 and the ΔrplI mutant (Fig 6) suggests independence of RplI from Sr0161-mediated regulation of the T3SS in P. aeruginosa. While both RplI and Sr0161 control ExsA translation via the 5’ UTR of exsA, RplI works independent of Sr0161, indicating the possibility that they may function as T3SS repressors but respond to different environmental cues.
There are several regulators that have been shown to regulate the expression of T3SS genes at the posttranscriptional level. Intile. et. al. reported that the RNA helicase DeaD directly stimulates exsA translation to promote the expression of T3SS in P. aeruginosa [8]. DeaD likely functions by relaxing an inhibitory mRNA secondary structure in the exsA 5’ untranslated region to enhance ribosomal recruitment [8]. Our previous study showed that the suhB mutation is linked to a defective translation of the ExsA [17]. RsmA, a CsrA family RNA binding protein, exerts a positive effect on the synthesis of ExsA through an unknown mechanism [11]. In addition, Sr0161 directly targets the leader region through base pairing and inhibits ExsA synthesis at the posttranscriptional level [9,10]. Recently, a CspA family protein (CspC) was demonstrated to control the translation of exsA through its 74 nt 5′ UTR [27]. Here, we further identified the ribosomal large subunit protein RplI to inhibit ExsA synthesis at the translational level.
Genes coding for ribosomal proteins usually form operons in bacteria. It has been demonstrated that numerous ribosomal proteins in E. coli autogenously repress the translation of their own mRNAs or others in the operons by direct binding to their respective mRNAs [28]. These proteins include both small subunit ribosomal proteins, such as S4 and S7 [29], and those of large subunit ribosomal proteins, such as L1, L10 and L7/L12 [30,31]. Such a feedback regulation is derived from the capacities of these ribosomal proteins to function not only as primary rRNA binding components but also as highly specific repressors for their own mRNAs when they are produced in excess over the rRNA available for the ribosome assembly [28–32]. In E. coli, the translational repression by ribosomal proteins may occur either through competing with and hampering ribosome binding to mRNA or by entrapment of the ribosome in its nonproductive complex [28]. Since a much higher amount of ribosome association with the exsA mRNA was detected in the ΔrplI mutant, it is possible that RplI interacts with the 5’ UTR and prevents ribosome binding, thus inhibiting the initiation of exsA translation. To the best of our knowledge, this is the first report on ribosomal protein inhibiting mRNA translation beyond its own mRNA via direct binding.
Previous work has shown that a 778 nucleotide RNA fragment ranging from nucleotide 1999 to 2776 of the 23 S rRNA contains two RNA binding sites of ribosomal protein L9 in E. coli [24]. With the toeprinting assay, they also revealed that the N- and C-terminal domains of L9 respectively interact with nucleotides just 5’ to nucleotides 2231 and 2179 of the 23S rRNA [24]. The interaction between RplI and the mRNA fragment of exsA in the presence of its 24-nt 5’ untranslated region was detected, but not in the presence of the 12-nt 5’ UTR, which suggested a specific interaction between the region from the 24 nt to 12 nt 5’ UTR. It is possible that RplI binds to the 24 nt 5’ UTR and affects the binding of ribosomes to exsA mRNA. However, no obvious primary sequence homolog was found at -24 nt-exsA when compared to 23S rRNA. Since the mRNA secondary structure linking sequences upstream and downstream of the RBS of rpsM was important for the S4 mediated translational repression [33,34], it is possible that the stem-loop structure of 24 nt upstream of the coding sequence of exsA (S4 Fig predicted with Mfold) is responsible for its binding and repression by the L9.
Taken together, our study showed that RplI binds to the 5’ UTR of and inhibits exsA translation to repress expression of the T3SS in P. aeruginosa. Further studies are required to understand the global regulatory role of the L9 and their detailed regulatory mechanisms in P. aeruginosa.
Materials and methods
Bacterial strains and plasmids
Bacteria and plasmids used in this study are listed in S2 Table. All strains were cultured in LB medium (10 g/L tryptone, 5 g/L yeast extract and 5 g/L NaCl) or on L-agar plates at 37°C. When needed, antibiotics were used at the following concentrations (μg/mL): for E. coli, ampicillin 100, gentamicine 10, tetracycline 10; for P. aeruginosa, carbenicillin 150, gentamicin 100, tetracycline 50. When needed, IPTG (isopropyl β-D-1-thiogalactopyranoside) at the indicated concentrations was added into the culture medium. Primers used to generate constructs and in real-time qPCR are listed in S3 Table.
For complementation of the rplI gene, the open reading frame of rplI and its putative SD region was PCR amplified with mPAO1 chromosomal DNA as the template. The PCR product was ligated into the BamHI-HindIII sites of pUCP20 [35], resulting in pUCP20-rplI. pE2620-Sr0161 was constructed with similar manipulation. To generate pE2620, a Gmr fragment was amplified from pUC18T-miniTn7T [36] with primers pE2620F and pE2620R and cloned into the PvuI site of pMMB67EH (ATCC). The rplI deletion construct was made by cloning 1034 bp upstream and 1075 bp downstream fragments of the rplI gene into the SacI-HindIII sites of plasmid pEX18Tc [37]. pEX18-Sr0161 were constructed with similar manipulation. To generate the -12 bp-exsA-Flag construct, the ORF of exsA (without a stop codon) and its 12 bp upstream fragment was cloned into the HindIII-KpnI sites of RBS-free pFlag-CTC, in which RBS was removed by PCR amplification with primers pFlag-CTCF and pFalg-CTCR (S3 Table) from pFlag-CTC (Sigma) followed by cloning into its BamHI-HindIII sites. The resulting construct was inserted into the BamHI site of pDN19 [38], generating pE3286. pE269, pE268, pE267 and pE3308 were constructed with similar manipulations. To construct pE3330 and pE3331, -12 bp-exsA-Flag and -24 bp-exsA-flag with the tac promoter of pFlag-CTC were PCR amplified with pE3286 and pE267 as templates using primers PtacF and PterR (S3 Table), and then cloned into the SacI-BamHI sites of pE1553 [16]. pE1553 was used in a previous study [16] and generated by removing the promoter of pUCP20 [35] via point mutation using PCR with primers pE1553F and pE1553R (S3 Table) followed by digestion with EcoRI and self-ligation.
Western blot assay
Bacteria cultured overnight were diluted 1:50 into fresh LB medium and grown to an OD600 of 1.0 with or without 5 mM EGTA. After separation by centrifugation at 13, 000 × g for 2 min, supernatant and/or pellet samples from an equal number of cells were separated on a 12% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE), transferred onto a PVDF (polyvinylidene difluoride) membrane (Millipore), and probed with a rabbit polyclonal antibody against ExoS or mouse monoclonal antibody against RpoA, RpoB (BioLegend) and Flag (Sigma). The RNA polymerase alpha/beta subunit (RpoA/RpoB) was used as a loading control. Signals were detected with the ECL-plus kit (Millipore).
Cytotoxicity assay
Bacterial cytotoxicity was determined by examining the detachment of HeLa cells after P. aeruginosa infection as previously described [14]. 1.4 × 105 HeLa cells were seeded into each well of a 24-well plate and cultured in DMEM (Dulbecco’s modified Eagle’s medium, Corning, USA) containing 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Gibco, USA) at 37°C with 5% CO2 the night before infection. Three hours before infection, cells were washed twice with phosphate-buffered saline (PBS) and further cultured in DMEM with 10% FBS. Before infection, log phase bacteria grown in LB medium were collected, washed twice with PBS, and resuspended in DMEM with 10% FBS. Then HeLa cells were infected with log phase bacteria at a multiplicity of infection (MOI) of 50 for 3 hours. After that, the culture medium was removed, and the remaining attached cells were washed twice with PBS and stained with 0.1% crystal violet for 15 min at 37°C. Then, each well was washed twice with 1 mL distilled water. The stained crystal violet was dissolved in 95% ethanol and measured at a wavelength of 590 nm.
RNA isolation and real-time qPCR
RNA purification and real-time qPCR were carried out following the manufacturer’s instructions with minor modifications as described below. Bacteria cultured overnight were diluted 1:50 into fresh LB medium and grown to an OD600 of 1.0 at 37°C with or without 5 mM EGTA. Total RNA was extracted using an RNAprep Pure Cell/Bacteria Kit (Tiangen Biotech, Beijing, China). cDNA was synthesized using random primers and PrimeScript Reverse Transcriptase (TaKaRa, Dalian, China). Real-time qPCR was performed with the indicated primers (shown in S3 Table) and SYBR Premix ExTaq II (TaKaRa, Dalian, China). rpsL, a 30S ribosomal protein encoding gene, was used as an internal control.
mRNA stability assay
mRNA stability assay was performed as described before with minor modifications [39]. P. aeruginosa strains cultured overnight were diluted 1:50 into fresh LB medium containing 5 mM EGTA or not, grown to an OD600 of 1.0, and treated with 200 μg/mL rifampicin. Equal numbers of bacterial cells were collected at each indicated time point. Then each sample was mixed with an equivalent number of gfp-expressing E. coli cells. Total RNA was isolated, and the mRNA levels of rpsL, PA1805 (the peptidyl-prolyl cis-trans isomerase D encoding gene) and exsA were examined by real-time qPCR with gfp as an internal control.
Detection of exsA mRNA associated with ribosome
The amount of ribosome-associated exsA mRNA was determined by a modified RNA-binding protein immunoprecipitation procedure followed by real-time qPCR [18,40,41]. Flag-tagged exsA coding sequence with its 24 bp upstream region followed by transcription terminators T0 and T1 was PCR amplified from pE267 plasmid and then cloned into pE1553, resulting in construct pE3331. pE3330 containing -12 bp-exsA-Flag-T0T1 was generated with the same procedure. Wild-type mPAO1 and its ΔrplI mutant containing pE2620-rplL-His and pE3331 or pE3330 were grown to an OD600 of 1.0 and collected by centrifugation. Ribosome was isolated by affinity chromatography as described previously with modifications [42]. The collected bacteria were suspended in lysis buffer (150 mM NaCl, 20 mM Tris-HCl, 10 mM imidazole, 3 mM β-mercaptoethanol, 0.5% NP-40, pH 8.0) and lysed by sonication. Cell debris was removed by centrifugation, and the supernatants were incubated with Ni-NTA agarose beads at 4°C for 1 h. After that, the beads were washed five times with lysis buffer, followed by RNA purification with an RNAprep Pure Cell/Bacteria kit (Tiangen Biotech, China). The amount of exsA-Flag mRNA was determined by real-time qPCR with specific primers (qexsAFlagF and qexsAFlagR in S3 Table) and 16S ribosomal RNA (PA0668.1) as the internal control for normalization.
Expression and purification of proteins from E. coli
The rplI coding region was amplified by PCR using mPAO1 chromosome DNA as the template with specific primers (S3 Table). The gene was cloned into the NcoI-XhoI sites of plasmid pET28a, resulting in rplI translational fusion with the His tag at the C-terminus. E. coli strain BL21 (DE3) carrying the plasmid pET28a-rplI was cultured at 37°C to an OD600 of 0.4 to 0.6, and expression of His-tagged RplI was induced by the addition of IPTG at a final concentration of 100 μM for 16~18 h at 16°C. Bacteria from 100 mL culture were collected by centrifugation at 4,000 × g for 20 min, resuspended in 10 mL lysis buffer (46.6 mM Na2HPO4, 3.4 mM NaH2PO4, 0.3 M NaCl, 8 M urea, pH 8.0) and lysed by sonication on ice. The lysate was centrifuged at 12,000 × g for 20 min at room temperature. Then, the supernatant was mixed with Ni-NTA agarose (Qiagen) and incubated at room temperature for 1 h. After that, the lysate-resin mixture was loaded into an empty column and washed twice with lysis buffer containing 50 mM imidazole. The RplI-His protein was finally eluted with l mL lysis buffer containing 300 mM imidazole. To remove urea and imidazole, proteins were extensively dialyzed in PBS with various concentrations of urea (4, 2, and 1 M) and finally in PBS.
In vitro transcription and RNA gel mobility shift assay
The -24 nt-exsA/ -12 nt-exsA RNA was synthesized using the Riboprobe System-T7 (Promega) from PCR product amplified from mPAO1 chromosomal DNA with the specific primers (listed in S3 Table) according to the manufacturer’s instructions. The RNA was purified by isopropanol precipitation, heated at 90°C for 10 min and then refolded by cooling naturally at room temperature for 30 min. One microgram of the purified RNA was mixed with the indicated amounts of purified RplI-His in binding buffer (10 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 50 mM KCl, 10% glycerol, 1 U recombinant RNase inhibitor [TaKaRa]) and incubated for 30 min on ice. Fifteen microliters of each sample was loaded onto a nondenaturing 8% polyacrylamide gel. Electrophoresis was performed at 100 V for 145 min in 1 × TBE buffer (Tris-borate-EDTA: 89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.3) on ice. After that, the gel was stained with Gel-red (Biotium) in 1 × TBE for 10 min and the RNA bands were visualized in a ChemiDoc XRS+ molecular imager (Biorad, CA, USA). To avoid RNase contamination, all buffers were prepared with DEPC-treated water.
Microscale thermophoresis measurements
The purified RplI-His was labeled with the RED fluorescent dye NT-647-NHS (NanoTemper Technologies, Munich, Germany) following the manufacturer’s instructions. 200 nM labeled RplI-His was incubated with a 2-fold dilution series of unlabeled indicated RNA (with final concentrations ranging from 15.9 μM to 0.485 nM) in a twenty microliter system on ice for at least 30 min [43]. Following incubation, the samples were loaded into standard treated silicon capillaries (Monolith NT.115 series capillaries; catalog no. MO-K025). The measurements were carried out using a Monolith NT.115 instrument (NanoTemper Technologies GmbH) at room temperature using a 20% light-emitting diode (LED) and 60% MST power. The dissociation constants (Kd) were calculated as described previously [44]. Data analyses were performed using MO affinity analysis software (NanoTemper Technologies). The whole procedure was carried out in triplicate for each sample.
Other methods
Tn5 mutagenesis was conducted as previously described [45,46]. Chromosomal gene deletion was carried out by homologous recombination as previously described [37]. β-galactosidase activity was measured to determine the indicated promoter transcriptional activity as described before [47].
Supporting information
(DOCX)
(DOCX)
(DOC)
Growth curve of mPAO1 and ΔrplI in LB medium (A), relative mRNA levels of pcrV and pscF in mPAO1 and ΔrplI strains (B), secretion and expression of ExoS in PAO1/pUCP20, PAO1ΔrplI/pUCP20 and PAO1ΔrplI/pUCP20-rplI (C). (B) Total RNA was isolated under T3SS inducing (+) and non-inducing (-) conditions with 5 mM EGTA, and the relative mRNA levels of pcrV and pscF were determined by real-time qPCR using rpsL as the internal control. ns, not significant, *P < 0.05, ***P < 0.001 by Student’s t test. (C) Bacteria were cultured to an OD600 of 1.0 in LB with (+) or without (-) 5 mM EGTA. Proteins in supernatants (S) or pellets (P) from equivalent bacterial cells were separated by 12% SDS-PAGE gels and probed with an antibody against ExoS or RpoA.
(TIF)
The relative grayscale represents the density of the sample/density of the loading control with the respective first lane as 1. ** P < 0.01, ***P< 0.001, ****P < 0.0001, by Student’s t test. The data shown represent the results from three independent experiments.
(TIF)
The relative grayscale represents the density of the sample/density of the loading control with the respective first lane as 1. ns, not significant, ***P< 0.001, ****P < 0.0001, by Student’s t test. The data shown represent the results from three independent experiments.
(TIF)
The region from -24 nt to -12 nt was boxed.
(TIF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was supported by the National Science Foundation of China (31970680, 31870130 and 82061148018 to FB, 32170177 to WW, 31970179 and 32170199 to ZC), National Key Research and Development Project of China (2017YFE0125600, 2021YFE0201300 to FB, 2021YFE0101700 to WW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67(3):351–68. Epub 2007/03/06. doi: 10.2165/00003495-200767030-00003 . [DOI] [PubMed] [Google Scholar]
- 2.Hauser AR. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol. 2009;7(9):654–65. Epub 2009/08/15. doi: 10.1038/nrmicro2199 ; PubMed Central PMCID: PMC2766515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams McMackin EA, Djapgne L, Corley JM, Yahr TL. Fitting Pieces into the Puzzle of Pseudomonas aeruginosa Type III Secretion System Gene Expression. Journal of bacteriology. 2019;201(13). Epub 2019/04/24. doi: 10.1128/JB.00209-19 ; PubMed Central PMCID: PMC6560140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee EJ, Cowell BA, Evans DJ, Fleiszig SM. Contribution of ExsA-regulated factors to corneal infection by cytotoxic and invasive Pseudomonas aeruginosa in a murine scarification model. Invest Ophthalmol Vis Sci. 2003;44(9):3892–8. Epub 2003/08/27. doi: 10.1167/iovs.02-1302 . [DOI] [PubMed] [Google Scholar]
- 5.Hauser AR, Kang PJ, Engel JN. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Molecular microbiology. 1998;27(4):807–18. Epub 1998/03/27. doi: 10.1046/j.1365-2958.1998.00727.x . [DOI] [PubMed] [Google Scholar]
- 6.Holder IA, Neely AN, Frank DW. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns. 2001;27(2):129–30. Epub 2001/02/28. doi: 10.1016/s0305-4179(00)00142-x . [DOI] [PubMed] [Google Scholar]
- 7.Marsden AE, Intile PJ, Schulmeyer KH, Simmons-Patterson ER, Urbanowski ML, Wolfgang MC, et al. Vfr Directly Activates exsA Transcription To Regulate Expression of the Pseudomonas aeruginosa Type III Secretion System. Journal of bacteriology. 2016;198(9):1442–50. Epub 2016/03/02. doi: 10.1128/JB.00049-16 ; PubMed Central PMCID: PMC4836234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Intile PJ, Balzer GJ, Wolfgang MC, Yahr TL. The RNA Helicase DeaD Stimulates ExsA Translation To Promote Expression of the Pseudomonas aeruginosa Type III Secretion System. Journal of bacteriology. 2015;197(16):2664–74. Epub 2015/06/10. doi: 10.1128/JB.00231-15 ; PubMed Central PMCID: PMC4507347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YF, Han K, Chandler CE, Tjaden B, Ernst RK, Lory S. Probing the sRNA regulatory landscape of P. aeruginosa: post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Molecular microbiology. 2017;106(6):919–37. Epub 2017/10/05. doi: 10.1111/mmi.13857 ; PubMed Central PMCID: PMC5738928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssen KH, Corley JM, Djapgne L, Cribbs JT, Voelker D, Slusher Z, et al. Hfq and sRNA 179 Inhibit Expression of the Pseudomonas aeruginosa cAMP-Vfr and Type III Secretion Regulons. mBio. 2020;11(3). Epub 2020/06/18. doi: 10.1128/mBio.00363-20 ; PubMed Central PMCID: PMC7298702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Intile PJ, Diaz MR, Urbanowski ML, Wolfgang MC, Yahr TL. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. Journal of bacteriology. 2014;196(2):357–66. Epub 2013/11/05. doi: 10.1128/JB.01199-13 ; PubMed Central PMCID: PMC3911257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Y, Yang H, Qiao M, Jin S. MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa. Journal of bacteriology. 2011;193(2):399–410. Epub 2010/11/16. doi: 10.1128/JB.01079-10 ; PubMed Central PMCID: PMC3019812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leipuviene R, Björk GR. Alterations in the two globular domains or in the connecting alpha-helix of bacterial ribosomal protein L9 induces +1 frameshifts. Journal of bacteriology. 2007;189(19):7024–31. Epub 2007/07/31. doi: 10.1128/JB.00710-07 ; PubMed Central PMCID: PMC2045208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Y, Zhang M, Zhu F, Peng Q, Weng Y, Zhao Q, et al. NrtR Regulates the Type III Secretion System Through cAMP/Vfr Pathway in Pseudomonas aeruginosa. Front Microbiol. 2019;10:85. Epub 2019/02/15. doi: 10.3389/fmicb.2019.00085 ; PubMed Central PMCID: PMC6363681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yahr TL, Frank DW. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. Journal of bacteriology. 1994;176(13):3832–38. Epub 1994/07/01. doi: 10.1128/jb.176.13.3832-3838.1994 ; PubMed Central PMCID: PMC205579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X, Li M, Pan X, Zheng R, Liu C, Chen F, et al. Fis Regulates Type III Secretion System by Influencing the Transcription of exsA in Pseudomonas aeruginosa Strain PA14. Front Microbiol. 2017;8:669. Epub 2017/05/05. doi: 10.3389/fmicb.2017.00669 ; PubMed Central PMCID: PMC5395579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li K, Xu C, Jin Y, Sun Z, Liu C, Shi J, et al. SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. mBio. 2013;4(6):e00419–13. Epub 2013/10/31. doi: 10.1128/mBio.00419-13 ; PubMed Central PMCID: PMC3809559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Jin Y, Bian T, Li K, Sun Z, Cheng Z, et al. SuhB is a novel ribosome associated protein that regulates expression of MexXY by modulating ribosome stalling in Pseudomonas aeruginosa. Molecular microbiology. 2015;98(2):370–83. Epub 2015/07/17. doi: 10.1111/mmi.13126 . [DOI] [PubMed] [Google Scholar]
- 19.Mattheakis L, Vu L, Sor F, Nomura M. Retroregulation of the synthesis of ribosomal proteins L14 and L24 by feedback repressor S8 in Escherichia coli. Proc Natl Acad Sci U S A. 1989;86(2):448–52. Epub 1989/01/01. doi: 10.1073/pnas.86.2.448 ; PubMed Central PMCID: PMC286487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skouv J, Schnier J, Rasmussen MD, Subramanian AR, Pedersen S. Ribosomal protein S1 of Escherichia coli is the effector for the regulation of its own synthesis. J Biol Chem. 1990;265(28):17044–9. Epub 1990/10/05. . [PubMed] [Google Scholar]
- 21.Thomas MS, Nomura M. Translational regulation of the L11 ribosomal protein operon of Escherichia coli: mutations that define the target site for repression by L1. Nucleic Acids Res. 1987;15(7):3085–96. Epub 1987/04/10. doi: 10.1093/nar/15.7.3085 ; PubMed Central PMCID: PMC340717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito K, Mattheakis LC, Nomura M. Post-transcriptional regulation of the str operon in Escherichia coli. Ribosomal protein S7 inhibits coupled translation of S7 but not its independent translation. J Mol Biol. 1994;235(1):111–24. Epub 1994/01/07. doi: 10.1016/s0022-2836(05)80020-8 . [DOI] [PubMed] [Google Scholar]
- 23.Hoffman DW, Davies C, Gerchman SE, Kycia JH, Porter SJ, White SW, et al. Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein. EMBO J. 1994;13(1):205–12. Epub 1994/01/01. ; PubMed Central PMCID: PMC394794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adamski FM, Atkins JF, Gesteland RF. Ribosomal protein L9 interactions with 23 S rRNA: the use of a translational bypass assay to study the effect of amino acid substitutions. J Mol Biol. 1996;261(3):357–71. Epub 1996/08/23. doi: 10.1006/jmbi.1996.0469 . [DOI] [PubMed] [Google Scholar]
- 25.Lillemoen J, Cameron CS, Hoffman DW. The stability and dynamics of ribosomal protein L9: investigations of a molecular strut by amide proton exchange and circular dichroism. J Mol Biol. 1997;268(2):482–93. Epub 1997/05/02. doi: 10.1006/jmbi.1997.0982 . [DOI] [PubMed] [Google Scholar]
- 26.Pei H, Han S, Yang S, Lei Z, Zheng J, Jia Z. Phosphorylation of bacterial L9 and its functional implication in response to starvation stress. FEBS Lett. 2017;591(20):3421–30. Epub 2017/09/13. doi: 10.1002/1873-3468.12840 . [DOI] [PubMed] [Google Scholar]
- 27.Li S, Weng Y, Li X, Yue Z, Chai Z, Zhang X, et al. Acetylation of the CspA family protein CspC controls the type III secretion system through translational regulation of exsA in Pseudomonas aeruginosa. Nucleic acids research. 2021;49(12):6756–70. Epub 2021/06/18. doi: 10.1093/nar/gkab506 ; PubMed Central PMCID: PMC8266623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zengel J, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Progress in nucleic acid research and molecular biology. 1994;47:331–70. doi: 10.1016/s0079-6603(08)60256-1 . [DOI] [PubMed] [Google Scholar]
- 29.Nomura M, Yates JL, Dean D, Post LE. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein MRNA. Proc Natl Acad Sci U S A. 1980;77(12):7084–8. Epub 1980/12/01. doi: 10.1073/pnas.77.12.7084 ; PubMed Central PMCID: PMC350445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baughman G, Nomura M. Translational regulation of the L11 ribosomal protein operon of Escherichia coli: analysis of the mRNA target site using oligonucleotide-directed mutagenesis. Proc Natl Acad Sci U S A. 1984;81(17):5389–93. Epub 1984/09/01. doi: 10.1073/pnas.81.17.5389 ; PubMed Central PMCID: PMC391709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukuda R. Autogenous regulation of the synthesis of ribosomal proteins, L10 and L7/12, in Escherichia coli. Mol Gen Genet. 1980;178(2):483–6. Epub 1980/01/01. doi: 10.1007/BF00270505 . [DOI] [PubMed] [Google Scholar]
- 32.Aseev LV, Koledinskaya LS, Boni IV. Regulation of Ribosomal Protein Operons rplM-rpsI, rpmB-rpmG, and rplU-rpmA at the Transcriptional and Translational Levels. Journal of bacteriology. 2016;198(18):2494–502. Epub 2016/07/07. doi: 10.1128/JB.00187-16 ; PubMed Central PMCID: PMC4999927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang CK, Draper DE. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989;57(4):531–6. Epub 1989/05/19. doi: 10.1016/0092-8674(89)90123-2 . [DOI] [PubMed] [Google Scholar]
- 34.Tang CK, Draper DE. Evidence for allosteric coupling between the ribosome and repressor binding sites of a translationally regulated mRNA. Biochemistry. 1990;29(18):4434–9. Epub 1990/05/08. doi: 10.1021/bi00470a025 . [DOI] [PubMed] [Google Scholar]
- 35.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148(1):81–6. Epub 1994/10/11. doi: 10.1016/0378-1119(94)90237-2 . [DOI] [PubMed] [Google Scholar]
- 36.Choi KH, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nature protocols. 2006;1(1):153–61. Epub 2007/04/05. doi: 10.1038/nprot.2006.24 . [DOI] [PubMed] [Google Scholar]
- 37.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212(1):77–86. Epub 1998/07/14. doi: 10.1016/s0378-1119(98)00130-9 . [DOI] [PubMed] [Google Scholar]
- 38.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. Journal of bacteriology. 1990;172(6):2911–9. Epub 1990/06/01. doi: 10.1128/jb.172.6.2911-2919.1990 ; PubMed Central PMCID: PMC209088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen R, Weng Y, Zhu F, Jin Y, Liu C, Pan X, et al. Polynucleotide Phosphorylase Regulates Multiple Virulence Factors and the Stabilities of Small RNAs RsmY/Z in Pseudomonas aeruginosa. Front Microbiol. 2016;7:247. Epub 2016/03/15. doi: 10.3389/fmicb.2016.00247 ; PubMed Central PMCID: PMC4773659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nature protocols. 2006;1(1):302–7. Epub 2007/04/05. doi: 10.1038/nprot.2006.47 . [DOI] [PubMed] [Google Scholar]
- 41.Köster T, Staiger D. RNA-binding protein immunoprecipitation from whole-cell extracts. Methods Mol Biol. 2014;1062:679–95. Epub 2013/09/24. doi: 10.1007/978-1-62703-580-4_35 . [DOI] [PubMed] [Google Scholar]
- 42.Ederth J, Mandava CS, Dasgupta S, Sanyal S. A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli. Nucleic Acids Res. 2009;37(2):e15. Epub 2008/12/17. doi: 10.1093/nar/gkn992 ; PubMed Central PMCID: PMC2632923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moon MH, Hilimire TA, Sanders AM, Schneekloth JS Jr. Measuring RNA-Ligand Interactions with Microscale Thermophoresis. Biochemistry. 2018;57(31):4638–43. Epub 2018/01/13. doi: 10.1021/acs.biochem.7b01141 ; PubMed Central PMCID: PMC6341465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lippok S, Seidel SA, Duhr S, Uhland K, Holthoff HP, Jenne D, et al. Direct detection of antibody concentration and affinity in human serum using microscale thermophoresis. Anal Chem. 2012;84(8):3523–30. Epub 2012/03/09. doi: 10.1021/ac202923j . [DOI] [PubMed] [Google Scholar]
- 45.Larsen RA, Wilson MM, Guss AM, Metcalf WW. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Archives of microbiology. 2002;178(3):193–201. Epub 2002/08/22. doi: 10.1007/s00203-002-0442-2 . [DOI] [PubMed] [Google Scholar]
- 46.Wu W, Badrane H, Arora S, Baker HV, Jin S. MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. Journal of bacteriology. 2004;186(22):7575–85. Epub 2004/11/02. doi: 10.1128/JB.186.22.7575-7585.2004 ; PubMed Central PMCID: PMC524895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller J. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, New York, NY. 1972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOC)
Growth curve of mPAO1 and ΔrplI in LB medium (A), relative mRNA levels of pcrV and pscF in mPAO1 and ΔrplI strains (B), secretion and expression of ExoS in PAO1/pUCP20, PAO1ΔrplI/pUCP20 and PAO1ΔrplI/pUCP20-rplI (C). (B) Total RNA was isolated under T3SS inducing (+) and non-inducing (-) conditions with 5 mM EGTA, and the relative mRNA levels of pcrV and pscF were determined by real-time qPCR using rpsL as the internal control. ns, not significant, *P < 0.05, ***P < 0.001 by Student’s t test. (C) Bacteria were cultured to an OD600 of 1.0 in LB with (+) or without (-) 5 mM EGTA. Proteins in supernatants (S) or pellets (P) from equivalent bacterial cells were separated by 12% SDS-PAGE gels and probed with an antibody against ExoS or RpoA.
(TIF)
The relative grayscale represents the density of the sample/density of the loading control with the respective first lane as 1. ** P < 0.01, ***P< 0.001, ****P < 0.0001, by Student’s t test. The data shown represent the results from three independent experiments.
(TIF)
The relative grayscale represents the density of the sample/density of the loading control with the respective first lane as 1. ns, not significant, ***P< 0.001, ****P < 0.0001, by Student’s t test. The data shown represent the results from three independent experiments.
(TIF)
The region from -24 nt to -12 nt was boxed.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.