Summary
Centromeric α-satellite repeats represent ∼6% of the human genome, but their length and repetitive nature make sequencing and analysis of those regions challenging. However, centromeres are essential for the stable propagation of chromosomes, so tools are urgently needed to monitor centromere copy number and how it influences chromosome transmission and genome stability. We developed and benchmarked droplet digital PCR (ddPCR) assays that measure copy number for five human centromeric arrays. We applied them to characterize natural variation in centromeric array size, analyzing normal tissue from 37 individuals from China and 39 individuals from the US and UK. Each chromosome-specific array varies in size up to 10-fold across individuals and up to 50-fold across chromosomes, indicating a unique complement of arrays in each individual. We also used the ddPCR assays to analyze centromere copy number in 76 matched tumor-normal samples across four cancer types, representing the most-comprehensive quantitative analysis of centromeric array stability in cancer to date. In contrast to stable transmission in cultured cells, centromeric arrays show gain and loss events in each of the cancer types, suggesting centromeric α-satellite DNA represents a new category of genome instability in cancer. Our methodology for measuring human centromeric-array copy number will advance research on centromeres and genome integrity in normal and disease states.
Keywords: human, centromeres, genome stability, α-satellite, tandem repeats, cancer, α-SIN, copy number
Graphical abstract

Highlights
New digital droplet PCR assays measure copy number of human centromeres
Large variation in copy number and individual-specific fingerprints exist
Centromere array copy number is stable in cultured human cells
Analysis of primary human cancer samples suggest copy number can change in cancer
de Lima et al. develop a PCR method to estimate human centromere size based on a measurement of copy number in centromere arrays. The authors use this method to analyze normal tissue samples across populations, finding that the size of centromeres is highly variable among chromosomes as well as among individuals. They also demonstrate centromeric copy number changes in cancer samples, suggesting a new category of genome instability.
Introduction
Centromeres represent some of the most difficult regions of the human genome to characterize, and until recently, the lack of centromeric sequence in the human reference genome has been a limitation for research on genome integrity and instability. Human centromeres are megabase-sized repetitive regions that are essential for chromosome transmission. To understand genome integrity and pathological states of genome instability, such as cancer, better characterization of human centromere sequence is essential. Centromeres are chromosomal locations in which kinetochore proteins assemble for microtubule attachment and chromosome segregation. Each human centromere is part of a large haplotype,1 which has the potential to bias chromosome-transmission fidelity. Recent advances in long-read-sequencing technology have enabled the assembly of the centromeres of the sex chromosomes and chromosome 8 for a single reference genome.2, 3, 4 However, a single assembly does not begin to capture the diversity in centromere haplotypes and haplotype combinations in the human population.5, 6, 7 Additional technologies to sequence and analyze centromeric DNA are critical for research to characterize how centromeres contribute to genome integrity.
The tandem-repeat structure of centromeric arrays makes it likely to be prone to copy number variation. Increases or decreases in array size may, in turn, affect chromosome transmission. Previous work suggests biases in chromosome transmission could be linked to centromeric DNA. For example, the centromeric histone variant centromere protein A (CENP-A) and kinetochore protein binding can scale with α-satellite content,8 and chromosomes with large kinetochores have an increased surface for potential interaction with microtubules.9 Furthermore, non-random chromosome missegregation has been tracked to centromeres.10,11 The “centromere strength” hypothesis posits that larger centromeric arrays recruit more kinetochore proteins and, therefore, could bias segregation during cell division; size may, therefore, affect chromosome transmission during meiosis and mitosis in mammals.12,13 However, this is a double-edged sword because larger centromeric arrays linked to larger kinetochores have a tendency to establish erroneous merotelic attachments and missegregate during anaphase.9 Centromeric array size may, therefore, affect recruitment of kinetochore proteins or other components of the chromosome segregation machinery, which can lead to a segregation bias. In addition, centromeric array size could affect genome integrity via titration of heterochromatin proteins. Therefore, it is imperative to investigate the stability of centromeric arrays and the mechanisms that prevent instability, given the potential effect on genome integrity.
Cancer is frequently associated with alterations that influence genome integrity. Many types of genome instability events are associated with cancer, from whole-chromosome gain-and-loss events, referred to as chromosomal instability (CIN), to structural variations, such as translocations, gene amplifications, and microsatellite instability (MIN). Copy number variation is rampant in human genomes14 and can occur via chromosomal-repeat expansion and contraction or via extrachromosomal DNA. Copy number changes can significantly alter the phenotype of cells.15 Somatic copy number variation presents an opportunity for selection, dramatically expanding the genetic landscape that can be sampled to achieve a “fit” proliferative cancerous state within a specific environment. The stability of centromeric repeats in cancer is unknown but is an essential question, given the potential effect on genome integrity. Addressing stability is dependent on access to sequences that allow the design of primer pairs that will accurately quantify the copy number of centromeric repeats.
Here, we report the development and application of digital PCR-based assays to measure centromeric-repeat copy number. The development of this method included sequence information and benchmarking against recent linear assemblies of centromeric regions, produced by the T2T (telomere-to-telomere) Consortium. For this study, we developed five centromeric assays and employed additional assays for three tandem gene-repeat arrays and one macrosatellite array on the X chromosome.3 In the future, additional sequence-based experimental tools can be developed based on centromere assemblies. These assays represent a dramatic improvement over existing methods and demonstrate accurate and reproducible measurement of copy number with a requirement of only ∼1 ng of DNA. We applied these droplet digital PCR (ddPCR) assays to assess natural variation in centromeric-array size in the human population, with analyses of normal tissue from 37 individuals from China and 39 individuals from the UK and US. We identify a wide range of copy number variation in individual human centromeric arrays as well as in array combinations. We also applied these assays in four types of cancer, representing the most-comprehensive quantitative analysis of centromeric array stability in cancer to date. We identify centromeric array instability in each of these cancer types, a genome instability event we refer to as α-satellite instability. Future goals include development of tools to detect centromere haplotypes and track haplotype function. To understand the significance of centromeric-array instability in cancer, we must increase the panel of assays and the numbers and types of cancers analyzed to recognize significant cancer-type and centromere-specific signatures and patterns. Moreover, α-satellite instability needs to be placed in the context of broader genome instability signatures. Once these patterns are fully elucidated, researchers can determine their usefulness as biomarkers for predicting therapeutic response. Only once the field has a clear picture of centromeric-array variation, stability, and function can we understand how these important genomic loci act as genetic determinants in human health and disease.
Results
Several methods have been used to analyze human centromere size: (1) sequence assembly, (2) quantitative PCR (qPCR), and (3) pulsed-field gel electrophoresis (PFGE). However, these methods have significant drawbacks. Assembling centromeres from short-read-sequencing data has proven intractable because of the repetitive nature of DNA sequences, making existing cancer-genome data not useful for that purpose. The genome assemblies, to date, which have included assembly of centromic regions, have relied on high-coverage sequencing data using a combination of methods, which is costly, requires intensive computational efforts, and is, therefore, not feasible for a large number of matched tumor-normal samples. qPCR methods rely on standard curves generated from cloned centromere sequences on plasmids; less than 2-fold changes are difficult to detect, normalization to chromosome number has not been performed, and the centromeric-array size estimates from primer pairs have not been benchmarked against a standard genome.16,17 PFGE requires substantial amounts of intact chromosomes and has limited resolution. Our goal was to develop a simple, rapid, accurate method for centromere copy number analysis, which could be easily implemented across laboratories.
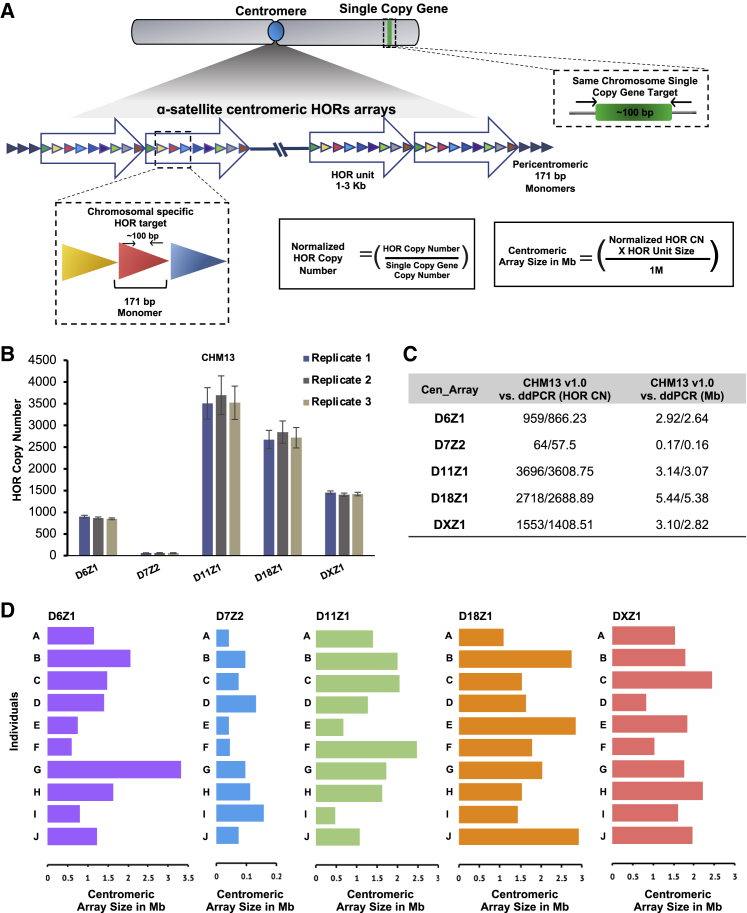
To examine human centromeric repeats, we developed and benchmarked ddPCR-based methods to measure the copy number of five different arrays. Digital PCR works by partitioning the restriction-digested template DNA into thousands of individual parallel PCR reactions, followed by thermocycling to amplify the product, and then dye-based detection of positive and negative droplets. The fraction of positive and negative droplets allows for an absolute count of the number of target molecules in the sample, without the need for standards or endogenous controls. ddPCR is a perfect choice for copy number measurements given its accuracy, reproducibility, the ability to derive an absolute number, scalability, and low template requirements. Centromeric arrays consist of α-satellite DNA, a 171-bp sequence that is only 50%–70% identical between monomers, making it possible to find a unique amplicon within some arrays (Figure 1A). Each array consists of a characteristic number and sequence of monomers that is iterated nearly identically (known as the higher-order repeat [HOR]) to form the array. Multiplying the copy number of a unique amplicon within a single repeat by the size of the HOR in kilobases yields the size of an individual array. Chromosomes may have a single array or multiple arrays, but only one array will be active for chromosome transmission.
Figure 1.
Profiling of centromeric arrays by ddPCR
(A) A schematic overview is presented for the centromeric-array size-measurement workflow by ddPCR. Unique non-overlapping amplicons were identified in GRCh38 for each higher-order repeat (HOR) array analyzed. The array copy number values are normalized by dividing by the copy number of a chromosome-specific single-copy gene. The array size is calculated by multiplying the copy number by the size of a single repeat.
(B) Copy number measurements performed by ddPCR in triplicate for CHM13 for five different centromeric arrays demonstrate high reproducibility. Error bars are calculated by Taylor’s expansion and are based on the standard deviation for each replicate experiment.
(C) The copy number for each HOR and array size in Mb derived from the computational assembly of CHM13 v1.0 (95% amplicon identity by BLAST) are compared with the results generated by ddPCR.
(D) Unique fingerprint profiles for 10 individuals (A–J) are shown. Bars represent the normalized average centromeric array size in Mb for five centromeric arrays in normal breast tissue from 10 individuals.
We designed PCR primers to unique amplicons present in arrays DXZ1, D18Z1, D11Z1, D7Z2, and D6Z1 (Figure 1A; Table S1). The chromosome is specified by the symbol following the “D,” and the array is specified by the number following the “Z.” To design primers, we used the Muscle algorithm18 to run multiple sequence alignments and perform pairwise comparisons for each of the monomers that comprise the HOR, to find unique regions in each HOR, and to identify primers of 17–24 bp that would yield an amplicon of ∼100 bp in the hg38 genome. We next performed in silico PCR using the University of California, Santa Crus (UCSC) browser (https://genome.ucsc.edu/cgi-bin/hgPcr) to confirm that the amplicons all derived from the chromosome with the targeted HOR. The restriction enzymes selected to digest the template surrounding each amplicon are predicted to cut a minimum of twice in each repeat of a given array (Table S1), ensuring that the repeats will be separated for partitioning into droplets. In addition to the amplicons in Figure 1, we include information for the D8Z2 assay previously published (Tables S1 and S2).19 Amplification of a unique single-copy gene on each chromosome enables normalization for chromosome copy number, which is especially important for cancer samples. The error associated with multiple biological-replicate measurements is approximately 10% (Figure 1B). The DNA template for benchmarking was derived from CHM13, the newest and most-complete haploid-reference genome.3,19 The ability to design and validate primers that accurately reflect the size of an array was greatly enhanced by recent centromere assemblies (CHM13 v1.0). However, we cannot report foolproof amplicon design principles at this time, and extensive trial and error and benchmarking will be required to develop additional assays. Furthermore, some chromosomes bear centromeric arrays that are highly similar and are likely indistinguishable by PCR (e.g., 1/5/19, 13/21, 14/22).
To further benchmark the assays, we carried out two types of analysis. First, we compared the size calculated from the copy number to the size estimated by PFGE in four different cell lines (LT690, HAP1, T6012, and CHM13) for DXZ1. In all four cases, the two measurements were concordant.3 The DXZ1 array in LT690 is notable for its small size (∼1,500 kb) relative to the average, which is closer to 3,000 kb, as first reported in 1998 based on PFGE.20 Second, we compared the size calculated from ddPCR to the size estimated for all five computationally assembled arrays (D6Z1, D7Z2, D11Z1, D18Z1, and DXZ1) in the CHM13 genome. CHM13 was derived from a hydatidiform mole—all the chromosomes are paternally derived, so it possesses only a single haplotype of each chromosome, despite being diploid. The Basic Local Alignment Search Tool (BLAST) was used to identify amplicons in the CHM13 genome with 95% identity to calculate the expected copy number in CHM13 and to compare with the copy number obtained experimentally. The estimates by ddPCR are consistent with the number of repeats and the size of the assembled arrays for CHM13 v1.0 (Figure 1C), although the ddPCR method appears to slightly underestimate copy number and array size (see Limitations of the study). However, overall the copy number measurements are accurate and reproducible, only require ∼1 ng of DNA, and can be carried out in a few hours, making ddPCR a dramatic improvement on existing methods to measure centromeric arrays (see Document S1). Efforts are underway to develop and benchmark assays for more centromeric arrays.
Centromeric haplotypes are stably transmitted through the germline at the resolution of PFGE for pedigrees.5,21,22 We used two somatic diploid cell paradigms to analyze the centromeric array copy number before and after differentiation: (1) human foreskin fibroblasts with induced pluripotent stem cells, and (2) human trophoblast stem cells and their corresponding differentiated extravillous trophoblasts (Figure S1). Measurements for the five arrays were similar in the stem cells and the differentiated cells, suggesting that centromeric arrays are stable over differentiation in somatic cells (Table S2). It is important to note that, for diploid cells, measurements represent the average of two haplotypes.
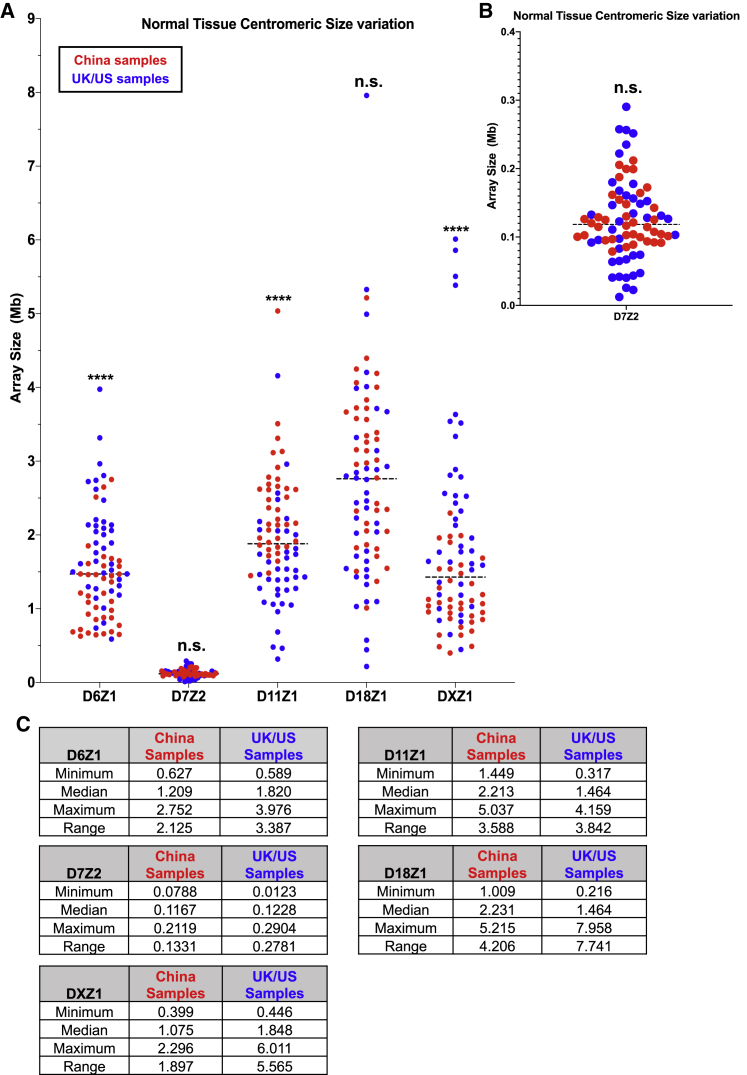
Centromeric array length polymorphisms have been documented in humans.6,7,22 We used ddPCR assays to examine natural variation in centromeric array size in the human population. We analyzed array size for DXZ1, D18Z1, D11Z1, D7Z2, and D6Z1 in normal tissue from 37 individuals from China and from 39 individuals from the UK and the US. Individual arrays show up to 10-fold variation in size and up to 50-fold variation among different arrays (e.g., 0.1–5 Mb), with a statistically significant difference in average array size by country of origin for three of the five arrays, suggesting there may be geographically distinct haplotypes (Figure 2).1,6,23
Figure 2.
Centromeric array size variation between two geographical locations
(A) Comparison of centromeric array size in Mb for D6Z1, D7Z2, D11Z1, D18Z1, and DXZ1 in normal tissue from samples divided by geographical origin for 37 individuals from China (red) and 39 individuals from the UK and the US (blue). The distributions in the two groups differed significantly (p < 0.001) in D6Z1, D11Z1, and DXZ1 centromeric arrays using the Wilcoxon-Mann-Whitney two-sample rank-sum test. D18Z1 and D7Z2 centromeric arrays did not show a significant difference.
(B) The y axis for D7Z2 is expanded, relative to the presentation in (A), so that the data can be better visualized, because D7Z2 is much smaller than the other arrays.
(C) The minimum, median, maximum, and range of array sizes are shown in Mb for each array, divided by geographical region of origin.
Each individual has a unique complement of centromeric arrays, or fingerprint, as characterized by measurements of multiple arrays in multiple individuals (Figure 1D; Table S3), suggesting the potential for functional differences, an important topic for future research. We did not detect any correlation among the sizes of the five different centromeric arrays analyzed within a single genome. Given the unique individual signatures, analysis of centromere stability in disease states is entirely reliant on having a matched normal DNA sample, an essential reference that has not been consistently used in previous investigations.17 We obtained matched DNA samples for four different cancer types: head and neck, breast, medulloblastoma, and acute lymphoblastic leukemia (ALL) and compared the copy number for DXZ1, D18Z1, D11Z1, D7Z2, and D6Z1 (Table S3). Medulloblastoma and ALL are predominantly childhood cancers, and the samples included molecular subgroupings based on cytogenetic and molecular characterization (Table S4).
In addition to centromeric arrays, we examined the stability of tandem repeats more broadly. Extensive genomic profiling of ploidy in the medulloblastoma and ALL samples (Table S4), including cytogenetic analysis such as G-banding and fluorescence in situ hybridization (FISH; ALL), and methylation arrays (medulloblastoma)24 enabled the assessment and interpretation of copy number changes of additional tandem repeats in each pediatric cancer sample relative to normal. The X chromosome encodes about 50% of all the cancer-testis (CT) antigen-encoding genes in the human genome,25 so named because they are expressed in testis and cancer. These genes are often encoded in tandem repeats (e.g., CT45, CT47, and GAGE). Low levels of meiotic rearrangements in pedigrees and mitotic rearrangements in solid tumors have been reported.26 DXZ4 is a macrosatellite X-linked tandem array with meiotic instability.27 ddPCR assays for all four arrays were benchmarked against the computational assembly of the X chromosome for CHM13, and results were highly congruent3 (Table S2).
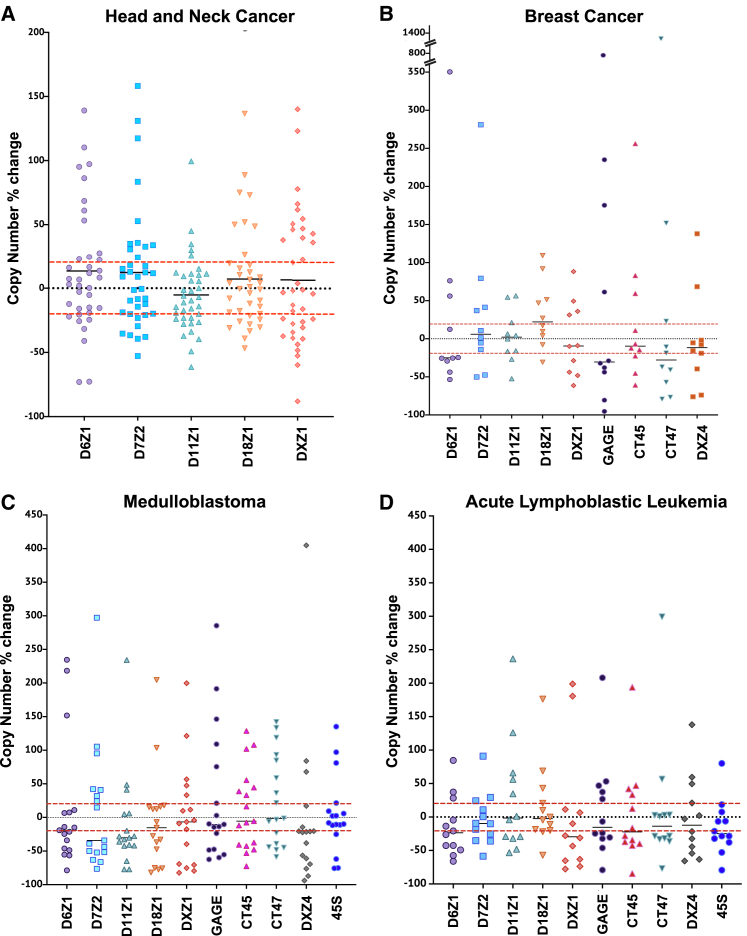
In contrast to the stability observed in cultured cells, we observed dramatic changes in array copy number in all four groups of cancer samples relative to matched normal tissue. We measured the copy number in each matched tumor and normal sample, subtracted the copy number of the tumor from the normal sample, and plotted it as percentage of change in array size for each array measured (Figure 3). All nine arrays display expansions and contractions in all four cancer types, and the variation is comparable once gain-and-loss events are scaled to the size of the starting array (Figure 3). Based on the 10% error of the method, the number of measurements outside a 20%-error window for 380 centromeric arrays and 185 tandemly repeated sequences was evaluated in 76 matched samples. More than 58% of the arrays measured (328/565) fell outside the error window in the cancer sample, indicating widespread tandem-repeat instability.
Figure 3.
Copy number changes in centromeric and tandemly repeated arrays in matched samples
Scatterplots contain points indicating the percentage of change from cancer, relative to normal, for each pair of samples. The absolute copy number difference was scaled to the size of the starting array of the normal tissue.
(A) The scatterplot depicts the percentage of change for the five indicated centromeric arrays in 35 head and neck cancer samples.
(B) The scatterplot depicts the percent change for five indicated centromeric and four tandemly repeated gene arrays (DXZ4, GAGE, CT45, and CT47) for 10 breast cancer samples.
(C) The scatterplot depicts the percentage of change for five indicated centromeric arrays and five tandemly repeated gene families (DXZ4, GAGE, CT45, CT47, and 45S rDNA) for 17 medulloblastoma samples.
(D) The scatterplot depicts the percentage of change for five indicated centromeric arrays in 12 acute lymphoblastic leukemia samples. The red dotted lines indicate the 20% window of error for the method; measurements within that range are considered “not significant.”
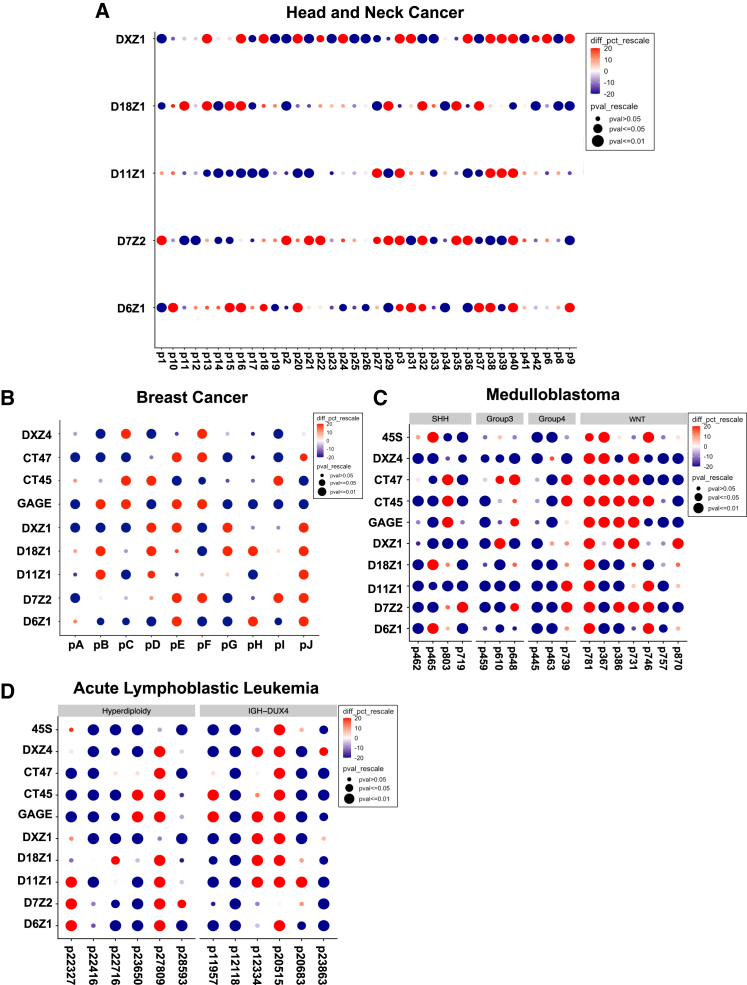
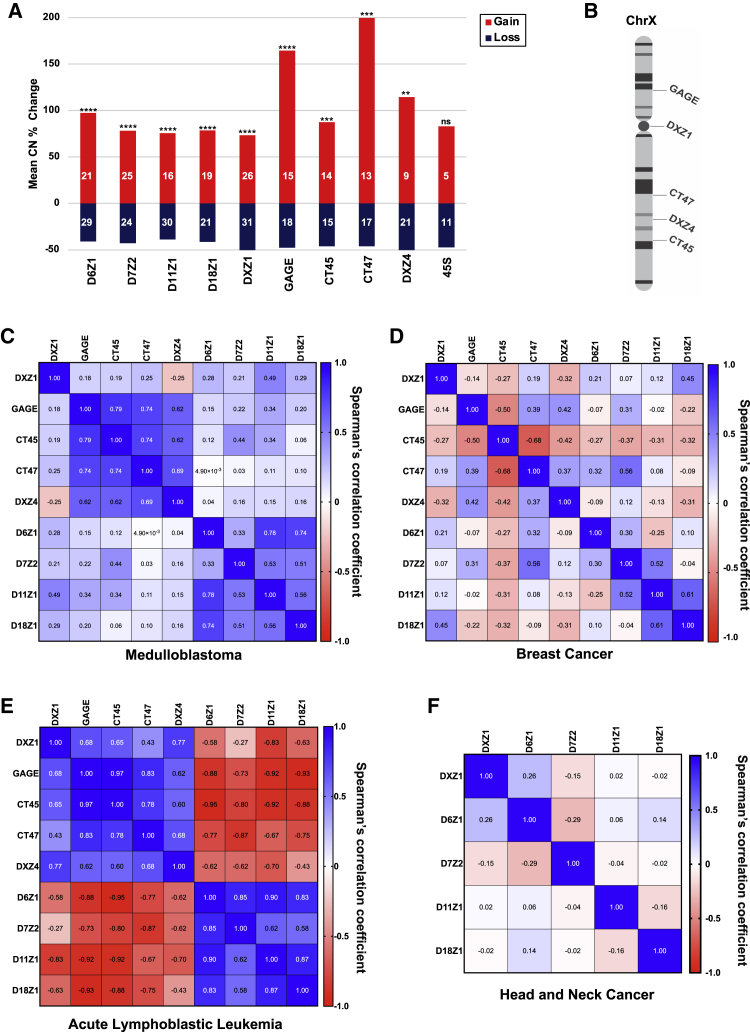
Every individual cancer sample presented one or more significant events, suggesting changes in centromeric α-satellite DNA copy number are a frequent occurrence in cancer (Figure 4). Interestingly, although 1 in 10 (10%) matched breast samples had all gains or all losses in all repeats monitored, ∼40% of the pediatric samples (5/12 in ALL, 7/17 in medulloblastoma) have all gains or all losses, suggesting a more-coordinated pattern. Overall, gain-and-loss events were observed in about equal frequency at all arrays, suggesting both can be tolerated. However, some gains were very large in size (e.g., 1 Mb), and gain magnitude was significantly larger than loss (Figure 5A). We speculate that equally large loss events at centromeres would be incompatible with chromosome transmission and would be lost. Array instability in the adult cancers seemed to occur independently for each array based on poor pairwise correlations between array changes (Figures 5D and 5F).
Figure 4.
Significant changes in arrays in each individual
Gridplots indicate the percentage and significance of changes per matched sample. Relative changes are scaled by the percentage of change (red is gain, and blue is loss) and p value (dot size).
(A) A gridplot is shown for the five indicated centromeric in 35 head and neck cancer samples.
(B) A gridplot is shown for five centromeric and four tandemly repeated gene arrays (DXZ4, GAGE, CT45, and CT47) for 10 breast cancer samples.
(C and D) A gridplot is shown for five indicated centromeric arrays and five tandemly repeated gene arrays (DXZ4, GAGE, CT45, CT47, and 45S rDNA) for 17 medulloblastoma samples (C) and 12 acute lymphoblastic leukemia samples (D), respectively. Medulloblastoma samples are categorized into four subgroups: WNT (n = 7), SHH (n = 4), group 3 (n = 3), and group 4 (n = 3), as previously described.28 85.7% (6/7) WNT samples have chromosome 6 loss. ALL samples are defined by hyperdiploidy (n = 6) or IGH-DUX4 (n = 6). High-hyperdiploid ALL samples exhibit non-random gain of chromosomes, which includes chromosomes 6, 10, 14, 21, and X. Gain of chromosomes 6, 18, and X was frequent in this study (see Table S4). Importantly, the methodology normalizes for chromosome copy number.
Figure 5.
Correlations between changes in copy number of tandemly repeated arrays from cancer genomes
(A) Average size of the significant copy number changes for each array by percentage is depicted and summed over all cancer types. The number in each category is shown on the bar, and whether the bias in gain size is significant is indicated by asterisks based on the Wilcox rank test (∗∗p < 0.05, ∗∗∗p = 0.01, ∗∗∗∗p < 0.001).
(B) Schematic representation of the chromosomal localization of the tandemly repeated arrays on the X chromosome.
(C–F) Heatmap plots representing Spearman’s correlation coefficient for the copy number changes of tandemly repeated arrays by cancer type: (C) breast cancer, (D) medulloblastoma, (E) acute lymphoblastic leukemia, and (F) head and neck cancer.
Male samples contain a single X chromosome and, therefore, a single array haplotype, whereas females have two X chromosomes, and measurements will represent an average of the two array haplotypes. If averaging two haplotypes obscures significant copy number differences, we would expect to find significant differences when stratifying the copy number measurements on the X chromosome (DXZ1, CT45, CT47, GAGE, and DXZ4) by sex. However, when stratified by sex, we identified no statistically significant differences between the copy number on the X chromosome in normal tissue for males and females (Figure S2A). When the percentage of change in copy number between tumor and normal tissue was compared for males and females, there were no significant differences between the sexes (Figure S2B). Based on this analysis, albeit with limited samples, we do not find evidence that haplotype averaging is masking the detection of copy number changes. Ultimately, the field will need tools that can distinguish haplotypes.
Although adult cancers can be related to lifestyle and a have high mutational burden, childhood cancers are fundamentally diseases of dysregulated development and are frequently associated with epigenetic dysregulation in stem or progenitor cells in growing tissues.29 In contrast to the adult samples, pediatric cancers displayed signatures of coordinated instability. Pairwise correlations between array changes identified changes in X-linked tandem repeats correlated with each other in medulloblastoma and ALL but showed low correlation with DXZ1 in medulloblastoma (Figures 5C and 5E). Furthermore, the changes in the X-linked arrays were anti-correlated with changes in centromeric arrays on other chromosomes in ALL. The WNT subgroup of medulloblastoma showed a trend toward gains, although the number of samples is small (Figure 4C). When the ALL samples were further subdivided into males and females, we observed that the gene repeats on the X chromosome were not as correlated with the centromeric DXZ1 array in males as it was in females (Figure S3). Because X-linked arrays in males are present in a single haplotype and the IGH-DUX4 male cancer samples were determined to have normal ploidy, the copy number changes of arrays on the X chromosome in these samples provide the best evidence for the expansion and contraction of individual arrays in cancer genomes (Figure S3C). These fascinating trends of coordinated instability will require additional investigation but suggest selection may operate on these tandem arrays in pediatric cancer. Together these data suggest that tandem repeats generally, and centromeric arrays specifically, represent unstable regions in cancer genomes.
We analyzed how array instability correlated with chromosomal instability for the pediatric cancers because we had ploidy information. The ALL samples fall into two subtypes: hyperdiploid ALL characterized by the non-random gain of chromosomes, including 6, 10, 14, 21, and X,30 and IGH-DUX4 ALL, which typically has a euploid genome with low incidence of chromosomal aneuploidy, chromothripsis, or large chromosomal abnormalities.31 Importantly, for our analysis, chromosomal abnormalities were absent in all IGH-DUX4 cases in this study, and hyperdiploid cases had recurrent gain of chromosomes 6, 18, and X. The medulloblastoma samples included four distinct genetic subgroups (WNT, n = 7; SHH, n = 4; group 3, n = 3; and group 4, n = 3), with 6/7 of the WNT samples showing loss of chromosome 6. Combining all the pediatric samples together, we found no correlation between aneuploidy events and repeat instability on the corresponding chromosome (Figure S4), suggesting the copy number changes detected are structural variations, distinct from numerical chromosome gain or loss. Nonetheless, we speculate that a major loss event in an active centromeric array could result in the loss of the corresponding chromosome, an event we would not detect because the chromosome would be lost from the population. When we grouped centromeric array changes in the two distinct subtypes of ALL (Figure S5A), there were no statistically significant differences, again consistent with copy number changes occurring independently from numerical chromosome gain/loss. Furthermore, we compared D6Z1 array changes between WNT medulloblastoma samples which have lost chromosome 6 (6/7), to the rest of the medulloblastoma samples, and found no statistical difference. Our observations support the idea that centromeric array size changes are independent from changes in the ploidy of the corresponding chromosome.
We further analyzed how array instability correlated with cancer type or stage, acknowledging that the sample size for several of the comparisons is small. We combined all the centromeric array changes for each medulloblastoma subgroup and compared the subgroups. We find that the WNT subgroup has less overall array loss than the other three subgroups (Figure S5B), but more samples are needed to explore this further. The head and neck cancer samples were categorized into four stages, from early (stage I) to late (stage IV) based on previous analysis.32 When we grouped centromeric array changes by stage and compared them, we found no significant differences among stages (Figure S5C). The limited sample size of the current study does not provide sufficient power to identify significant associations with stages. Further studies with large sample sizes of individuals across cancer stages and types are needed to identify these associations and to inform how centromeric array changes may be used prognostically.
In contrast to the 171-bp poorly transcribed centromeric repeats, ribosomal DNA encodes the most highly transcribed genes in the genome in a very large 45-kb repeat. Ribosomal DNA tandem repeats encoding the 45S gene are located on the short arm of the five acrocentric chromosomes (Chr13, 14, 15, 21, and 22) and display both meiotic and mitotic instability.33 Furthermore, 45S gene repeats are recombinational hotspots in cancer.34 Several groups reported that 45S repeats are lost in many types of cancer.32,35, 36, 37, 38 We measured 45S copy number in medulloblastoma (Figure 3C) and ALL (Figure 3D) samples because information regarding ploidy allowed us to normalize appropriately, even though the 45S repeats are spread across five chromosomes. We observed loss in 11/29 or 38% of cases of ALL and medulloblastoma (Figures 4C, 4D, and 5A). However, five cases (17%) had significantly increased copies. Together these data confirm the observation that 45S gene repeat copy number is plastic in cancer genomes39 and extend it to pediatric cancers.
Discussion
In this study, we develop simple quantitative ddPCR assays for measuring copy number of five centromeric arrays, three tandem gene repeat arrays, and one macrosatellite array on the X chromosome. We applied these assays to population samples, demonstrating the wide range of copy number variation in individual human centromeric arrays and the enormity of array combinations present in the human population. The stability of centromeric arrays observed in normal tissue cell culture conditions contrasts starkly with the instability in primary human cancer samples. This work also represents the most-comprehensive quantitative analysis of centromeric array stability in cancer to date and demonstrates centromeric array instability in cancer.
The repeats in this study are distinct in size from microsatellite repeats (2–5 bp) and fall into broad categories of gene repeats (e.g., 45S, CT45, CT47, and GAGE), macrosatellite repeats (e.g., DXZ4), and α-satellite centromeric repeats. We suggest terms analogous to MIN (microsatellite instability) to refer to the instability of these sequence categories, for example “GIN” for “gene repeat instability” and “α-SIN” for “α-satellite instability.” We observed a seemingly random pattern of instability in the adult cancer samples but coordinated copy number variation for α-satellite DNA (DXZ1), gene repeats, and microsatellite repeats on the X chromosome in pediatric cancers, signatures that warrant further investigation. Our understanding of these patterns is in its infancy because these sequences are only now accessible to copy number evaluation but represent an exciting frontier for discovery of genomic instability events that have gone undetected, and potential biomarkers that have been unexplored. Some types of instability events are driven by loss of function, in particular, molecular-maintenance processes under pathological conditions. For example, the MIN signature in cancer is often driven by loss of mismatch repair. Future efforts to understand what drives copy number variation in different categories of repeats will be important to understand the mechanisms that maintain the integrity of these chromosomal regions.
Human centromeric sequence has been difficult to characterize, and there remains much to learn. Excitingly, these new ddPCR assays will allow researchers to analyze centromeric copy number in many more disease states and samples with a high degree of accuracy and speed. With measuring stick in hand, we can begin to contemplate the functional relevance of changes in centromeric arrays. For example, experiments in hybrid mice have demonstrated that centromere size can act as a meiotic driver.13,40 Some centromeric array epialleles may act more efficiently than others for chromosome transmission.8 Centromere-localized proteins may normally protect centromeric arrays from undergoing recombination events,41 but many of these proteins become misregulated in cancer,42 suggesting recombination events could be elevated. Future efforts to examine centromeric array stability and heterogeneity will be greatly aided by the straightforward, accurate, and rapid assays described herein.
Limitations of the study
There are technical limitations and knowledge gaps related to this study. The design of primers to detect a representative amplicon to measure copy number is based on an assumption that the repeats that comprise an array share high sequence identity among individuals. This is a reasonable assumption because satellite DNA sequences, in general, arise by concerted evolution. This evolutionary pattern results in the homogenization of repeats within a genome and fixation in members of reproductive populations.43 Human centromeric arrays can be defined by chromosome-specific homogeneous arrays of higher-order repeats that are largely invariant among individuals.44 Random mutation and transposable element insertions occur, but the major differences among individuals are expected to be expansions and contractions, with a nucleotide spectrum that is reasonably stable.
Another type of sequence variation that may occur with greater likelihood than single-nucleotide variation is higher-order repeat variants. Canonical repeats constitute more than 92% of the DXZ1 array in CHM13,3 making DXZ1 relatively straightforward to measure with ddPCR. However, repeat variants can occur in centromeric arrays.4,7 The frequency of higher-order repeat variants in centromeric arrays in the human population is just beginning to emerge.7 Variant repeats may complicate calculations of array size in Mb from ddPCR data. Although variant repeats are only a small fraction of the overall array for the arrays analyzed in the present study, based on the CHM13 v1.0 release, amplicon copy number may not correlate directly to the total array size in Mb if variant repeats are different in size from the canonical higher-order repeat. For example, one higher-order repeat variant in DXZ1 is 2,000 bp whereas the canonical repeat is 2,200 bp. Importantly, the copy number of each amplicon can still be accurately measured and compared for matched tumor and normal samples for each individual. Given these limitations, future efforts to design amplicons to measure the copy number and size of additional arrays should take into account the evidence for variants and will continue to require rigorous benchmarking.
Although ddPCR can measure the average copy number for a given array of repeats, individuals have two centromeric haplotypes for each chromosome. There is evidence for a high degree of haplotypic diversity in centromeric arrays at the populational level.1,6,7 Our method measures average haplotype copy number and cannot distinguish maternal and paternal haplotypes, except for DXZ1 in male samples. In addition, tumor cell populations can be heterogeneous and may contain heterogeneity in repetitive DNA among cells. Given the huge amount of short-read sequence data available for analyses from human genomes and cancer genome projects, it would be beneficial to develop computational algorithms that could mine existing data for array size and stability. Future challenges in the human centromere field are to develop tools to resolve haplotypes and to quantify centromere size in short-read sequencing data and single cells. Moreover, the assays developed and arrays measured in this study need to be augmented with additional assays and data to achieve a genome-wide view of the stability of different categories of tandemly repeated sequences generally and their place in broader mutational signatures in cancer and other human disease contexts.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Matched Tumor/Normal Breast Cancer samples | KU Med Biorepository | N/A |
| Matched Tumor/Normal head and neck samples | Hospital of Stomatology, Sun Yat-sen University | N/A |
| Matched Tumor/Normal ALL samples | Newcastle University | N/A |
| Matched Tumor/Normal medulloblastoma samples | Newcastle University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| AluI | New England Biolabs | R0137 |
| HaeIII | New England Biolabs | R0108 |
| Critical commercial assays | ||
| DNeasy Blood & Tissue Kit | QIAGEN | Cat. no. 69504 |
| QX200 ddPCR EvaGreen Supermix | Biorad | #1864034 |
| QX200 Droplet Generation Oil for EvaGreen | Biorad | #1864006 |
| Qubit dsDNA HS Assay Kit | ThermoFisher | Cat. no. Q32851 |
| Experimental models: Cell lines | ||
| Human:CHM13 | Magee-Womens Hospital (Pittsburgh, PA), ATCC | |
| Human: HFF-1 | ATCC | SCRC-1041 |
| Human: DYS0100 | ATCC | ACS-1019 |
| Human trophoblast stem cells CT29-male | Arima lab | Okae et al.45 |
| Human trophoblast stem cells CT30-female | Arima lab | Okae et al.45 |
| Oligonucleotides | ||
| D6Z1F: 5′ – GCGTTGAACTCACCGTCTT – 3′ | This paper | N/A |
| D6Z1R: 5′ – TCCAAAGAATGCCTCCAAGG – 3′ | This paper | N/A |
| D7Z2F: 5′ – CGACTTTGTGATGTGTGCATTC – 3′ | This paper | N/A |
| D7Z2R: 5′ – CCTTATCCGCAATGGTCCTAAA – 3′ | This paper | N/A |
| D8Z2F: 5′-GACATTTGGAGGGCTTTGTA-3′ | Logsdon et al., 2021 | N/A |
| D8Z2R: 5′-TCAACTAACTGTGCTGAACATTTC-3′ | Logsdon et al., 2021 | N/A |
| D11Z1F: 5′ – CTTCCTTCGAAACGGGTATATCT – 3′ | This paper | N/A |
| D11Z1R: 5′ – GCTCCATCAGCAGGATTGT – 3′ | This paper | N/A |
| D18Z1F: 5′ – TGGGAAACGGGATTGTCTTC – 3′ | This paper | N/A |
| D18Z1R: 5′ – CTGCTCTACCAAAGGGAATGT – 3′ | This paper | N/A |
| DXZ1F: 5′ – TGATAGCGCAGCTTTGACAC – 3′ | Miga et al.3 | N/A |
| DXZ1R: 5′ – TTCCAACACAGTCCTCCA – 3′ | Miga et al.3 | N/A |
| DXZ4F: 5′ - CACTTCTACCACCACGAGTAA - 3′ | Miga et al.3 | N/A |
| DXZ4R: 5′ - GGGATGACATTCAACTGGGA - 3′ | Miga et al.3 | N/A |
| GAGEF: 5′ - GTAACGGAGGTCGTGGATTA - 3′ | Miga et al.3 | N/A |
| GAGER: 5′- CGCACTGAGAATAAGGGAGT - 3′ | Miga et al.3 | N/A |
| CT45F: 5′ - CATCAGCCATGGTGGAGTAT - 3′ | Miga et al.3 | N/A |
| CT45R: 5′ - TGCGGTGTTTCCCTGTT - 3 | Miga et al.3 | N/A |
| CT47F: 5′ - GAGATCGGACCCGATGATTC - 3′ | Miga et al.3 | N/A |
| CT47R: 5′ - CCAGTAAATCTCCCACCCAA - 3′ | Miga et al.3 | N/A |
| 45SF: 5′-AACGTGAGCTGGGTTTAG-3′ | Xu et al.35 | N/A |
| 45SR: 5′-CTCGTACTGAGCAGGATTAC-3′ | Xu et al.35 | N/A |
| TBP1F: 5′ – GATATGAGACTGTGGGTAAGT – 3′ | Xu et al.35 | N/A |
| TBP1R: 5′ – GATCCTTTGAACACCCTAATG – 3′ | Xu et al.35 | N/A |
| TECPR1F: 5′ – GTGCAGTCACCATCATCAAC – 3′ | This paper | N/A |
| TECPR1R: 5′ – CTGCACCCTCCTACAACA – 3′ | This paper | N/A |
| MTUS1F: 5′-TCAGAGGCTGGATAGGTGGT-3′ | Logsdon et al.19 | N/A |
| MTUS1R: 5′-CTCTGAGGTGCTCCCAGTC-3′ | Logsdon et al.19 | N/A |
| Cllorf16F: 5′ – TCCCTGACCATCTGGAAGAA – 3′ | This paper | N/A |
| Cllorf16R: 5′ – TGATTGGCCCTAGCAGAGA – 3′ | This paper | N/A |
| MROF: 5′ – TAGTAGGTAACACCGAGTGC – 3′ | This paper | N/A |
| MROR: 5′ – TCAGGGTTGTCGCAAGTA – 3′ | This paper | N/A |
| HPRT1F: 5′ – AAGGTGCTGGTCTCCTTTAC – 3′ | Miga et al.3 | N/A |
| HPRT1R: 5′ – GCACCAATGATTCTCTCCCT – 3′ | Miga et al.3 | N/A |
| Software and algorithms | ||
| MUSCLE v4 | https://www.drive5.com/muscle/downloads.htm | Edgar18 |
| PRISM9 | https://www.graphpad.com/scientific-software/prism/ | N/A |
| RStudio version 4.1.1 | https://www.rstudio.com/ | RStudio: Integrated Development for R. RStudio, PBC, Boston, MA |
| R-4.1.2 | https://www.R-project.org/ | R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria |
| Bowtie2.4.1 | Langmead and Salzberg, 201246 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Other | ||
| Human reference genome CHM13 v1.0 | https://www.ncbi.nlm.nih.gov/assembly/GCA_009914755.3 | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Jennifer Gerton, jeg@stowers.org.
Materials availability
This study generated sets of primers for amplicons in D6Z1 (F5′-GCGTTGAACTCACCGTCTT, R5′-TCCAAAGAATGCCTCCAAGG), D7Z2 (F5′-CGACTTTGTGATGTGTGCATTC, R5′-CCTTATCCGCAATGGTCCTAAA), D11Z1 (F5′-CTTCCTTCGAAACGGGTATATCT, R5′-GCTCCATCAGCAGGATTGT), D18Z1 (F5′-TGGGAAACGGGATTGTCTTC, R5′-CTGCTCTACCAAAGGGAATGT) and single copy reference genes TECPR1 (F5′-GTGCAGTCACCATCATCAAC, R5′-CTGCACCCTCCTACAACA), Cllorf16 (F5′-TCCCTGACCATCTGGAAGAA, R5′-TGATTGGCCCTAGCAGAGA) and MRO (F5′-TAGTAGGTAACACCGAGTGC, R5′-TCAGGGTTGTCGCAAGTA). Other primer sets have been previously reported3,19,35 and all are listed in the Key resource table.
Experimental model and subject details
Human cell lines
Human cell lines were used in this study. Male human foreskin fibroblasts HFF-1 (ATCC SCRC-1041) were grown in DMEM supplemented with 15% FBS. The corresponding human induced pluripotent stem (iPS) cells DYS0100 (ATCC ACS-1019) were cultured on CellMatrix Gel (ATCC ACS-3035) - coated dishes without a feeder layer in Pluripotent Stem Cell SFM XF/FF medium (ATCC ACS-3002). CHM13 cells (homozygous diploid with two X chromosomes) were originally grown in culture from a hydatidiform mole isolated at Magee-Womens Hospital (Pittsburgh, PA) as part of a research study (IRB MWH-20-054). Cells from this culture were transformed using the human telomerase reverse transcriptase (TERT) gene to develop the CHM13hTERT cell line, which has a stable karyotype based on chromosome spread analysis. CHM13hTERT cells were grown in DMEM-F12 medium supplemented with 10%FBS, 1x Gutamax (ThermoFisher - 35050061), 1xNEAA (ThermoFisher 11140050), 1mM Sodium Pyruvate (ThermoFisher – 11360070), 1x Insulin-Transferrin-Selenium (ThermoFisher - 41400045).
Human trophoblast stem (TS) cell lines CT29-male and CT30-female were obtained from the Arima lab,45 care of Dr. Michael Soares, University of Kansas Medical School. Cells were maintained in TS medium (TS basal medium, VPA and Inhibitor Cocktail). For differentiation to extravillous trophoblasts (EVTs), 100 mm dish was coated with PBS containing 1 μg/ml Col IV for 1.5 h at 37°C. Plates were washed with PBS two times and hTS cells were plated at either 5.0 e5 (Expt1) or 1.5 e5 (Expt2) in 10 mL EVT medium and 240 μL Matrigel was added per dish. After 3 days of differentiation, EVT medium without NRG1 was added and 30 μL of Matrigel was added. After 6 days of differentiation, EVT medium without NRG1 and KSR was added and 30 μL of Matrigel was added. Cells were collected at day 8 for DNA extraction and frozen in −80°C.
DNA from de-identified human patients
DNA derived from human patient samples was used in this study. Information for each patient such as age, gender, source institution, and cancer type are provided in Table S4. All aspects of this work were done in accordance with the SIMR ethical and procedural guidelines. The human primary materials in this study have been de-identified and were provided and collected under protocols approved by the Institutional Review Board or the equivalent body of each organization. Breast cancer samples (n = 10) were collected at the University of Kansas Medical Center, USA, and were obtained from the Biospecimen Repository. Head and neck cancer samples (n = 37) were collected at the Hospital of Stomatology at Sun Yat-sen University, China. Medulloblastoma and ALL samples were collected at the Newcastle University Centre for Cancer, Newcastle upon Tyne, UK (more details below). Matched normal samples were collected from adjacent tissues, in head and neck and breast cancer patients, blood samples in medulloblastoma patients, and bone marrow aspirates for acute lymphoblastic leukemia patients.
Diagnostic bone marrow samples from 12 childhood (0-18 years old) ALL patients with a) hyperdiploidy (n = 6) or b) IGH-DUX4 fusion (n = 6), were included in this study. Remission samples were used as a matched germline reference in all patients; remission was defined as bone marrow aspirates from time-points at which there was no detectable level (> 0.01%) of minimal residual disease. Chromosomal analysis, fluorescence in situ hybridization (FISH) and genome-wide copy number array analysis was performed on ALL patients (n = 12) at diagnosis, as previously described.47,48 Low hyperdiploidy and high hyperdiploidy were diagnosed in patients with 47-50 and 51-67 chromosomes, respectively, or gain identified by specific FISH probes located to recurrently gained chromosomes, as previously described.49 Karyotyping and genome-wide copy number array analysis of IGH-DUX4 patients identified no large chromosomal abnormalities, consistent with previous studies.31
Tumor material from 17 patients (2-18 years old at diagnosis) with childhood medulloblastoma were included in this study. All tumors assayed had a confirmed histopathological diagnosis of medulloblastoma, with a high tumor cell content. Blood samples were included as a matched germline reference for each patient. DNA methylation array analysis was performed on medulloblastomas sampled at diagnosis, as previously described.28 Methylation-dependent subtyping was used to classify individual patients as WNT, SHH, Group 3 or Group 4. Copy number alterations (whole chromosome/chromosome arm aberrations) were identified as previously described.28
Method details
DNA extraction
Genomic DNA was isolated using DNeasy Blood & Tissue Kit (QIAGEN) in accordance with the manufacturer’s instructions and all DNA samples were quantified using the Qubit Fluorometer with Qubit dsDNA HS Assay (Invitrogen). The Qubit Fluorometer is a DNA quantification device based on the fluorescence intensity of fluorescent dye binding to double-stranded DNA (dsDNA). Qubit is considered useful for checking DNA quality because it measures intact dsDNA.
Centromeric and tandemly repeated array quantification by ddPCR
To perform the quantification to measure the copy number of different centromeric α-satellite DNA repeats we developed a droplet digital PCR based method (see detailed information in Supplemental protocol S1). We used centromeric HORs and tandemly repeated gene families sequences assembled in GRCh38 to design unique non-overlapping primers for amplicons in arrays DXZ1, D18Z1, D11Z1, D7Z2, D6Z1, DXZ4, GAGE, CT45 and 45S rDNA (Table S1). ddPCR reactions were performed using the manufacturer’s protocol (Bio-Rad). Each reaction consists of 10 uL 2x ddPCR QX200 Evagreen Supermix, 0.2 uL of restriction enzyme for fragmentation, 1 uL 10 uM primer mix, 1 uL of 0.1-1 ng DNA template and 7.8 uL with nuclease free water. All DNA templates were digested with either AluI or HaeIII restriction enzymes (New England Biolabs). Both restriction enzymes cleavage sites were located within ∼100 bp flanking the target amplicons. Digestion prevents the amplification of more than one unique target site per HOR fragment. Mastermixes were simultaneously prepared for centromeric and the respective single copy gene, which were then incubated for 15 minutes to allow for restriction digestion. Mastermixes were then emulsified with Evagreen droplet generator oil (Bio-Rad) using a QX200 droplet generator according to the manufacturer’s instructions and transferred to a 96-wells plate, which was then heat-sealed with pierceable sealing foil sheets (Thermo Fisher Scientific). After droplet generation, thermocycling was performed with the following parameters: 10 min at 95°C, 40 cycles consisting of a 30 s denaturation at 94°C and a 60 s extension at 59°C, followed by 10 min at 98°C and a hold at 4°C. Control reactions without DNA were performed to rule out non-specific amplification. Following PCR amplification, the 96-well plate was transferred to a QX200 droplet reader (Bio-Rad). Each well was queried for fluorescence to determine the quantity of positive droplets. Positive droplets were distinguished based on fluorescence amplitude whereas negative droplets (no fluorescence) were compared to the strong fluorescence signal from droplets with amplified target sequences. Positive droplets were automatically determined by the QuantaSoft software. The number of targets per droplet follows a Poisson distribution and the total number of targets in the reaction can be calculated based on the proportion of positive droplets. Concentrations reported were copies/μL of the final ddPCR reaction and were adjusted according to the respective single copy gene. The copy number values for each centromeric/tandemly repeated array was calculated as follows: [(tandemly repeated target copies/μL)/(single copy gene copies/μL)] x 10
Quantification and statistical analysis
The ddPCR values obtained in the study were normalized as previously described and the error was calculated using Taylor’s expression.3 The statistical test performed is indicated in the figure legend. The p value, ddPCR value, and sample number is indicated on the figure and legend each time it was deemed statistically significant (p value less than 0.05). All plots were generated using the ggplot2 packages within the RStudio integrated development environment for the R statistical programming language and Prism8 software.
To compare the number of higher order repeated predicted by computational assembly for CHM13 to the ddPCR results, for each array analyzed, we mapped the predicted amplicons described in Table S1 by BLASTn in the T2T CHM13 v1.0 assembly files for each respective chromosome (https://github.com/nanopore-wgs-consortium/chm13). Only sequences with 95%–100% identity were considered hits.
Acknowledgments
Funding for this work was provided by a pilot grant from University of Kansas Cancer Center and Stowers Institute for Medical Research (J.L.G.), Cancer Research UK (J.C., S.C., S.L.R., S.C.C., and C.J.H.),Blood Cancer UK (C.J.H.), NHGRI (R01HG011274, K.H.M.), the National Natural Science Foundation of China (81771056 and 81972533), and Guangdong Financial Fund for High-Caliber Hospital Construction (174-2018-XMZC-0001-03-0125/D-13), and the Fundamental Research Funds for the Central Universities, Sun Yat-sen University (20ykzd09) (B.X.).
Author contributions
Conceptualization, L.G.L., E.H., and S.L.R.; data curation, L.G.L., S.L.R., J.C., S.C., and S.C.C.; formal analysis, L.G.L., H.L., S.L.R., and S.C.; funding acquisition, J.L.G., S.L.R., C.J.H., and S.C.C.; investigation, L.G.L., E.H., and S.L.R.; methodology, L.G.L., E.H., S.L.R., J.C., and S.C.; project administration, S.L.R.; resources: V.P.S., T.P., B.X., K.H.M., S.L.R., S.C.C., and C.J.H.; supervision, K.H.M.; visualization, L.G.L.; writing – original draft, L.G.L., S.L.R., and J.L.G.; writing – review & editing, L.G.L., S.L.R., J.L.G., S.C.C., and C.J.H.
Declaration of interests
The authors declare no competing interests.
Published: December 8, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2021.100064.
Supplemental information
Data and code availability
All ddPCR data derived from de-identified human patient DNA samples is included in the supplemental tables. Original raw data files for ddPCR are publicly available and can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1657
This paper does not report original code.
References
- 1.Langley S.A., Miga K.H., Karpen G.H., Langley C.H. Haplotypes spanning centromeric regions reveal persistence of large blocks of archaic DNA. eLife. 2019;8:e42989. doi: 10.7554/eLife.42989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain M., Olsen H.E., Turner D.J., Stoddart D., Bulazel K.V., Paten B., Haussler D., Willard H.F., Akeson M., Miga K.H. Linear assembly of a human centromere on the Y chromosome. Nat. Biotechnol. 2018;36:321–323. doi: 10.1038/nbt.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A., et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020;585:79–84. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logsdon G.A., Vollger M.R., Hsieh P., Mao Y., Liskovykh M.A., Koren S., Nurk S., Mercuri L., Dishuck P.C., Rhie A., et al. The structure, function, and evolution of a complete human chromosome 8. bioRxiv. 2020 doi: 10.1101/2020.09.08.285395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahtani M.M., Willard H.F. Pulsed-field gel analysis of alpha-satellite DNA at the human X chromosome centromere: high-frequency polymorphisms and array size estimate. Genomics. 1990;7:607–613. doi: 10.1016/0888-7543(90)90206-a. [DOI] [PubMed] [Google Scholar]
- 6.Miga K.H., Newton Y., Jain M., Altemose N., Willard H.F., Kent W.J. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 2014;24:697–707. doi: 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki Y., Myers E.W., Morishita S. Rapid and ongoing evolution of repetitive sequence structures in human centromeres. Sci. Adv. 2020;6:eabd9230. doi: 10.1126/sciadv.abd9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldrup-MacDonald M.E., Kuo M.E., Sullivan L.L., Chew K., Sullivan B.A. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 2016;26:1301–1311. doi: 10.1101/gr.206706.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drpic D., Almeida A.C., Aguiar P., Renda F., Damas J., Lewin H.A., Larkin D.M., Khodjakov A., Maiato H. Chromosome segregation is biased by kinetochore size. Curr. Biol. 2018;28:1344–1356.e5. doi: 10.1016/j.cub.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumont M., Gamba R., Gestraud P., Klaasen S., Worrall J.T., De Vries S.G., Boudreau V., Salinas-Luypaert C., Maddox P.S., Lens S.M., et al. Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features. EMBO J. 2020;39:e102924. doi: 10.15252/embj.2019102924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Worrall J.T., Tamura N., Mazzagatti A., Shaikh N., van Lingen T., Bakker B., Spierings D.C.J., Vladimirou E., Foijer F., McClelland S.E. Non-random mis-segregation of human chromosomes. Cell Rep. 2018;23:3366–3380. doi: 10.1016/j.celrep.2018.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwata-Otsubo A., Dawicki-McKenna J.M., Akera T., Falk S.J., Chmátal L., Yang K., Sullivan B.A., Schultz R.M., Lampson M.A., Black B.E. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr. Biol. 2017;27:2365–2373.e8. doi: 10.1016/j.cub.2017.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chmátal L., Gabriel S.I., Mitsainas G.P., Martínez-Vargas J., Ventura J., Searle J.B., Schultz R.M., Lampson M.A. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarrei M., MacDonald J.R., Merico D., Scherer S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 15.Peter J., De Chiara M., Friedrich A., Yue J.X., Pflieger D., Bergström A., Sigwalt A., Barre B., Freel K., Llored A., et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 2018;556:339–344. doi: 10.1038/s41586-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contreras-Galindo R., Fischer S., Saha A.K., Lundy J.D., Cervantes P.W., Mourad M., Wang C., Qian B., Dai M., Meng F., et al. Rapid molecular assays to study human centromere genomics. Genome Res. 2017;27:2040–2049. doi: 10.1101/gr.219709.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha A.K., Mourad M., Kaplan M.H., Chefetz I., Malek S.N., Buckanovich R., Markovitz D.M., Contreras-Galindo R. The genomic landscape of centromeres in cancers. Sci. Rep. 2019;9:11259. doi: 10.1038/s41598-019-47757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logsdon G.A., Vollger M.R., Hsieh P., Mao Y., Liskovykh M.A., Koren S., Nurk S., Mercuri L., Dishuck P.C., Rhie A., et al. The structure, function and evolution of a complete human chromosome 8. Nature. 2021;593:101–107. doi: 10.1038/s41586-021-03420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahtani M.M., Willard H.F. Physical and genetic mapping of the human X chromosome centromere: Repression of recombination. Genome Res. 1998;8:100–110. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- 21.McNulty S.M., Sullivan B.A. Alpha satellite DNA biology: finding function in the recesses of the genome. Chromosome Res. 2018;26:115–138. doi: 10.1007/s10577-018-9582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wevrick R., Willard H.F. Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proc. Natl. Acad. Sci. USA. 1989;86:9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oakey R., Tyler-Smith C. Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics. 1990;7:325–330. doi: 10.1016/0888-7543(90)90165-q. [DOI] [PubMed] [Google Scholar]
- 24.Schwalbe E.C., Hicks D., Rafiee G., Bashton M., Gohlke H., Enshaei A., Potluri S., Matthiesen J., Mather M., Taleongpong P., et al. Minimal methylation classifier (MIMIC): A novel method for derivation and rapid diagnostic detection of disease-associated DNA methylation signatures. Sci. Rep. 2017;7:13421. doi: 10.1038/s41598-017-13644-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross M.T., Grafham D.V., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Killen M.W., Taylor T.L., Stults D.M., Jin W., Wang L.L., Moscow J.A., Pierce A.J. Configuration and rearrangement of the human GAGE gene clusters. Am. J. Transl. Res. 2011;3:234–242. [PMC free article] [PubMed] [Google Scholar]
- 27.Tremblay D.C., Moseley S., Chadwick B.P. Variation in array size, monomer composition and expression of the macrosatellite DXZ4. PLoS ONE. 2011;6:e18969. doi: 10.1371/journal.pone.0018969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwalbe E.C., Lindsey J.C., Nakjang S., Crosier S., Smith A.J., Hicks D., Rafiee G., Hill R.M., Iliasova A., Stone T., et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: A cohort study. Lancet Oncol. 2017;18:958–971. doi: 10.1016/S1470-2045(17)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filbin M., Monje M. Developmental origins and emerging therapeutic opportunities for childhood cancer. Nat. Med. 2019;25:367–376. doi: 10.1038/s41591-019-0383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsson K., Johansson B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009;48:637–660. doi: 10.1002/gcc.20671. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., McCastlain K., Yoshihara H., Xu B., Chang Y., Churchman M.L., Wu G., Li Y., Wei L., Iacobucci I., et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat. Genet. 2016;48:1481–1489. doi: 10.1038/ng.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi S., Luo H., Wang L., Li H., Liang Y., Xia J., Wang Z., Cheng B., Huang L., Liao G., et al. Combined inhibition of RNA polymerase I and mTORC1/2 synergize to combat oral squamous cell carcinoma. Biomed. Pharmacother. 2021;133:110906. doi: 10.1016/j.biopha.2020.110906. [DOI] [PubMed] [Google Scholar]
- 33.Stults D.M., Killen M.W., Pierce H.H., Pierce A.J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008;18:13–18. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stults D.M., Killen M.W., Williamson E.P., Hourigan J.S., Vargas H.D., Arnold S.M., Moscow J.A., Pierce A.J. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 2009;69:9096–9104. doi: 10.1158/0008-5472.CAN-09-2680. [DOI] [PubMed] [Google Scholar]
- 35.Xu B., Li H., Perry J.M., Singh V.P., Unruh J., Yu Z., Zakari M., McDowell W., Li L., Gerton J.L. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017;13:e1006771. doi: 10.1371/journal.pgen.1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Udugama M., Sanij E., Voon H.P.J., Son J., Hii L., Henson J.D., Chan F.L., Chang F.T.M., Liu Y., Pearson R.B., et al. Ribosomal DNA copy loss and repeat instability in ATRX-mutated cancers. Proc. Natl. Acad. Sci. USA. 2018;115:4737–4742. doi: 10.1073/pnas.1720391115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M., Lemos B. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 2017;13:e1006994. doi: 10.1371/journal.pgen.1006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salim D., Bradford W.D., Freeland A., Cady G., Wang J., Pruitt S.C., Gerton J.L. DNA replication stress restricts ribosomal DNA copy number. PLoS Genet. 2017;13:e1007006. doi: 10.1371/journal.pgen.1007006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valori V., Tus K., Laukaitis C., Harris D.T., LeBeau L., Maggert K.A. Human rDNA copy number is unstable in metastatic breast cancers. Epigenetics. 2020;15:85–106. doi: 10.1080/15592294.2019.1649930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akera T., Chmátal L., Trimm E., Yang K., Aonbangkhen C., Chenoweth D.M., Janke C., Schultz R.M., Lampson M.A. Spindle asymmetry drives non-Mendelian chromosome segregation. Science. 2017;358:668–672. doi: 10.1126/science.aan0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giunta S., Funabiki H. Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc. Natl. Acad. Sci. USA. 2017;114:1928–1933. doi: 10.1073/pnas.1615133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W., Mao J.H., Zhu W., Jain A.K., Liu K., Brown J.B., Karpen G.H. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun. 2016;7:12619. doi: 10.1038/ncomms12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dover G.A. Molecular drive in multigene families: How biological novelties arise, spread and are assimilated. Trends Genet. 1986;2:159–165. [Google Scholar]
- 44.Miga K.H. Centromeric satellite DNAs: Hidden sequence variation in the human population. Genes (Basel) 2019;10:E352. doi: 10.3390/genes10050352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okae H., Toh H., Sato T., Hiura H., Takahashi S., Shirane K., Kabayama Y., Suyama M., Sasaki H., Arima T. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22:50–63.e6. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrison C.J., Moorman A.V., Barber K.E., Broadfield Z.J., Cheung K.L., Harris R.L., Jalali G.R., Robinson H.M., Strefford J.C., Stewart A., et al. Interphase molecular cytogenetic screening for chromosomal abnormalities of prognostic significance in childhood acute lymphoblastic leukaemia: A UK Cancer Cytogenetics Group Study. Br. J. Haematol. 2005;129:520–530. doi: 10.1111/j.1365-2141.2005.05497.x. [DOI] [PubMed] [Google Scholar]
- 48.Bashton M., Hollis R., Ryan S., Schwab C.J., Moppett J., Harrison C.J., Moorman A.V., Enshaei A. Concordance of copy number abnormality detection using SNP arrays and multiplex ligation-dependent probe amplification (MLPA) in acute lymphoblastic leukaemia. Sci. Rep. 2020;10:45. doi: 10.1038/s41598-019-56972-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison C.J., Haas O., Harbott J., Biondi A., Stanulla M., Trka J., Izraeli S., Biology and Diagnosis Committee of International Berlin-Frankfürt-Münster study group Detection of prognostically relevant genetic abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: recommendations from the Biology and Diagnosis Committee of the International Berlin-Frankfürt-Münster study group. Br. J. Haematol. 2010;151:132–142. doi: 10.1111/j.1365-2141.2010.08314.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ddPCR data derived from de-identified human patient DNA samples is included in the supplemental tables. Original raw data files for ddPCR are publicly available and can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1657
This paper does not report original code.