Abstract
Background
A(H1N1)pdm09 influenza viruses replicate efficiently in respiratory epithelia and are transmitted via respiratory droplets and aerosols expelled by infected hosts. The relative onward transmission potential of influenza viruses replicating in the upper and lower respiratory epithelial cells has not been fully defined.
Methods
Wild-type and barcoded A(H1N1)pdm09 viruses that differed by 2 synonymous mutations per gene segment were inoculated into ferrets via intranasal and intratracheal routes. Naive recipients were exposed to the exhaled breath of inoculated donors for 8 hours on day 2 postinoculation. Onward transmission potential of wild-type and barcoded genotypes were monitored by next generation sequencing.
Results
Transmissible airborne particles were respired from the upper but not the lower respiratory epithelial cells of donor ferrets. There was limited mixing of viral populations replicating in the upper and lower respiratory tissues.
Conclusions
The ferret upper respiratory epithelium was mapped as the anatomic site that generated influenza virus-laden particles mediating onward transmission by air. Our results suggest that vaccines and antivirals should aim to reduce viral loads in the upper respiratory tract for prevention of influenza transmission.
Keywords: influenza virus, tropism, transmission, ferrets
This study compared the relative onward transmission potential of A(H1N1)pdm09 influenza viruses replicating in the upper and lower respiratory epithelia. The ferret upper respiratory epithelium was mapped as the anatomic site that generated virus-laden particles mediating onward transmission by air.
Influenza viruses replicate in the airway epithelial cells and are transmitted via respiratory and oral secretions, droplets, and aerosols expelled by infected hosts. Due to differences in particle size, droplets and aerosols demonstrate variable temporal-spatial dispersion potential and may mediate influenza transmission via multiple nonmutually exclusive modes [1, 2]. As influenza virus transmits most efficiently under close contact settings, it has been conventionally perceived that large droplets, which cannot travel a long distance, play an important role in mediating transmission. In reality, a susceptible host in close contact with a contagious host is exposed to a continuum of different-sized respiratory droplets and aerosols, and the significance of short-range airborne transmission via airborne particles may be underestimated [2, 3]. The size of virus-laden airborne particles also determines the initial site of viral infection in the respiratory tract. Through mechanisms including inertial impaction, gravitational sedimentation, and Brownian diffusion, particles at 10–100 µm may deposit at the oropharynx, particles at 5–10 µm may deposit at central airway, and only fine particles < 5 µm may reach small airways and alveoli of the lungs [4–6]. In the human airway, α2,6-linked sialic acids (SA) are abundantly present on the surface of ciliated cells and goblet cells in the upper airway while α2,3-linked SA are abundantly present on the surface of nonciliated bronchoepithelium cells and type II pneumocytes in the lower airway [7–9]. Influenza virus attachment to α2,6-linked or α2,3-linked SA is essential for initiating infection and this has been implicated as a critical viral determinant for transmissibility. Thus, exposure to virus-laden particles of different sizes may determine the anatomic site of infection, whether the virus lands on permissive cells, and the kinetics of viral replication.
For onward transmission, virus-laden particles are formed and released from the infected respiratory epithelial mucosa. In influenza patients, infectious influenza viruses or viral RNA have been detected from cough or exhaled breath aerosols in sizes > 5 μm or ≤ 5 μm [10–13]. With the use of impactors, we have previously evaluated the onward transmission potential of airborne particles by size in the exhaled breath of ferrets infected with A(H1N1)pdm09 or A(H3N2) human seasonal influenza viruses [14]. Airborne particles that mediated influenza transmission between ferrets were those > 1.5 µm in size, which only accounted for less than 25% of all exhaled airborne particles below 20 µm in size. However, there is limited knowledge on the mechanisms driving the formation and release of infectious particles that mediate onward transmission.
Airborne particles in human breath are dominated by those < 1 μm in size, which may be generated via fluid film rupture at the respiratory bronchioles during inhalation [15, 16]. As such, viral replication in lower respiratory tissues, in close proximity to human respiratory bronchioles, may be responsible for generating most of fine particles that confer efficient airborne transmissibility. However, zoonotic avian influenza viruses that preferentially recognize α2,3-linked SA and replicate efficiently in lower respiratory tract do not transmit well among humans. This concept is also in conflict with the experimental approach of yielding airborne-transmissible H5N1, H7N1, or H9N2 variants through sequential viral adaptations in the ferret upper respiratory tract [17–20]. In this study, we aimed to investigate the relative roles of the upper or lower respiratory epithelia in generating the airborne particles that mediate onward transmission in ferrets.
METHODS
Cells
Madin-Darby canine kidney (MDCK) cells and 293T cells were obtained from the American Type Culture Collection and were maintained in minimum essential medium (MEM) and Opti-MEM with 10% and 5% fetal calf serum, respectively.
Generation of a Barcoded Recombinant A/California/04/2009 (H1N1) Virus That Differed From the Wild-Type Virus by Synonymous Mutations
The recombinant wild-type (WT) A/California/04/2009 virus was generated as described previously [21, 22] using viral RNA of A/California/04/2009 virus passaged 5 times in MDCK cells. To construct the barcoded (BC) recombinant A/California/04/2009 virus, available pandemic H1N1 sequences from the Influenza Virus Resource was accessed in 2015 to identify common synonymous mutations with minimal frequency at 2% (PB2, n = 5114; PB1, n = 5125; PA, n = 5358; HA, n = 11321; NP, n = 5457; NA, n = 8990; M1, n = 7608; M2, n = 7051; NS1, n = 5907; NEP, n = 5537). In each gene segment, 2 nearby common synonymous mutations, with a maximal distance of 33 nucleotides (Table 1), were selected as the barcode sequences that will not alter known splice sites, open reading frame, or packaging signals. Spacing the dual barcodes 33 nucleotides apart has facilitated Illumina short-read sequencing. The computer code used to choose these synonymous mutations is available at https://github.com/jbloomlab/InfluenzaTransmissionHuiLingCollaboration. The synonymous mutations were introduced by site-directed mutagenesis into the pHW2000 plasmids encoding gene segments of WT A/California/04/2009 virus. Recombinant WT and BC viruses were rescued by transfecting 293T cells (TransIT-LT1; Mirus) as previously described [22]. The stock viruses were passaged twice in MDCK cells and stored at −80°C in aliquots. The full genomes of recombinant WT (GISIAD accession number EPI_ISL_2105719) and BC (EPI_ISL_2105720) viruses were verified.
Table 1.
Nucleotide Differences Between Recombinant Wild-Type and Barcode Viruses
| Gene Segment | Site | Wild-Type Sequence | Barcode Sequence | Effect in Known ORF |
|---|---|---|---|---|
| PB2 | 2190 | G | A | PB2 residue 721 (synonymous) |
| 2211 | A | G | PB2 residue 728 (synonymous) | |
| PB1 | 2214 | C | T | PB1 residue 730 (synonymous), PB1-N40 (synonymous), PB1-F2 (out of ORF) |
| 2235 | A | G | PB1 residue 737 (synonymous), PB1-N40 (synonymous), PB1-F2 (out of ORF) | |
| PA | 1815 | G | A | PA residue 597 (synonymous), PA-155 (synonymous), PA-182 (synonymous), PA-x (out of ORF) |
| 1848 | C | T | PA residue 608 (synonymous), PA-155 (synonymous), PA-182 (synonymous), PA-x (out of ORF) | |
| HA | 1440 | C | T | HA residue 470a (synonymous) |
| 1469 | C | T | HA residue 479a (synonymous) | |
| NP | 1407 | A | G | NP residue 454 (synonymous) |
| 1419 | C | T | NP residue 458 (synonymous) | |
| NA | 1241 | T | C | NA residue 407 (synonymous) |
| 1268 | T | C | NA residue 416 (synonymous) | |
| M | 917 | G | A | M1 (out of ORF), M2 residue 68 (synonymous) |
| 950 | A | G | M1 (out of ORF), M2 residue 79 (synonymous) | |
| NS | 729 | G | A | NS1 (out of ORF), NS2 residue 77 (synonymous) |
| 750 | G | A | NS1 (out of ORF), NS2 residue 84 (synonymous) |
Abbreviation: ORF, open reading frame.
aHA residues are numbered sequentially from the N-terminal methionine rather than via the H1 or H3 ectodomain numbering schemes.
Competitive Replication Fitness of Recombinant WT and BC Viruses
MDCK cells were infected at the multiplicity of infection (MOI) of 0.005 plaque forming units (PFU)/cell with a mixture of recombinant WT and BC viruses at 100:0, 80:20, 50:50, 20:80, or 0:100, respectively. Viral RNA was extracted from culture supernatants at 0 hour (inoculum), 24 hours, and 48 hours postinfection and amplified by real-time quantitative polymerase chain reaction (RT-PCR). The proportion of WT and BC sequences were determined using next-generation sequencing (NGS).
Infectivity In Vitro
The 50% tissue culture infectious dose (TCID50) was determined in confluent MDCK cells in 96-well plates. Prior to infection, cells were overlayed with infection media (MEM supplemented with 0.3% bovine serum albumin and 1 μg/mL N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)-trypsin). Cells were incubated with serial half-log diluted ferret nasal wash samples or supernatants from ferret respiratory tissue homogenate (replicates of 4 for each dilution) at 37°C for 72 hours. Hemagglutination assay using 0.5% turkey erythrocytes was performed to determine the end point of infection and virus titers (log10 TCID50/mL) were calculated by the Reed-Muench method [23].
Transmission Experiments in Ferrets
Transmissibility was tested in 4- to 8-month old desexed male ferrets obtained from Sangosho (Wuxi, China). The ferrets used in the experiments were seronegative for influenza A virus NP protein using ID Screen Influenza A Antibody Competition enzyme-linked immunosorbent assay (ELISA) kit (ID.vet) and with hemagglutination inhibition titer ≤ 1:20 against selected seasonal influenza A viruses (A/HK/4801/2014 (H3N2) and A/Michigan/45/2015 A(H1N1)pdm09). All studies were conducted at the BSL2 animal facility at the Centre for Comparative Medicine Research, The University of Hong Kong. The study protocol was reviewed and approved by the Committee on the Use of Live Animals in Teaching and Research, The University of Hong Kong (CULATR No. 3905–16).
To compare the effect of viral replication at the upper and lower respiratory tract on onward transmission potential, donor ferrets were anesthetized with intramuscular injection of ketamine (25 mg/kg) and xylazine (2 mg/kg) and inoculated with WT recombinant A/CA/04/09 virus intranasally (1000 PFU diluted in 50 µL of infection media; 25 µL inoculum per nostril) or intratracheally (1000 PFU diluted in 500 µL of infection media). We used 500 µL for intratracheal inoculation as we aimed to deliver the virus at the bronchus region and to prevent liquid backflow to the upper trachea region. For intratracheal inoculation, lidocaine (Xylocaine® Spray, AstraZeneca) was additionally applied to the vocal cords of anesthetized ferrets to prevent laryngospasm prior to intubation using a 2.5-mm endotracheal tube guided by a laryngoscope. Virus was delivered to the bronchus region using a sterile polyethylene tubing placed inside the endotracheal tube. Each donor was single-housed inside individually ventilated ferret cages (Allentown) after inoculation. Onward transmission potential from 1 inoculated donor ferret to 2 naive ferrets via exhaled/expelled aerosols of various sizes was evaluated at 2 days postinoculation (dpi), using the transmission chambers described previously [14], without the use of impactors. In brief, the transmission chambers maintained a unidirectional airflow at 30 L/min and allowed passing of exhaled breath aerosols from the donor chamber to the recipient chamber. Two naive recipient ferrets were exposed to the exhaled breath of 1 donor ferret for 8 hours, and subsequently all animals were single-housed in individually-ventilated cages. The experiments were independently repeated 2–3 times.
To allow competition between viruses replicated at upper and lower respiratory tract prior to onward transmission, donor ferrets were coinoculated with (1) WT virus intranasally (1000 PFU diluted in 50 µL of infection media) and BC virus intratracheally (1000 PFU diluted in 500 µL of infection media), or (2) BC virus intranasally and WT virus intratracheally. Intratracheal inoculation was performed first and the animals were allowed to rest for 1 hour; afterwards, the animals were anesthetized again and were inoculated intranasally. At 2 dpi, 2 naive recipient ferrets were exposed to the exhaled breath of 1 donor ferret for 8 hours using the transmission chamber described above. All animals were single-housed in individually ventilated cages after exposure.
To monitor viral shedding, nasal washes were collected from donor and recipient ferrets and titrated in MDCK cells. The proportion of WT and BC sequences were determined using NGS. Ferret weight, temperature, and clinical signs were monitored daily. Postexposure sera from exposed naive recipient ferrets were collected at 14 days postexposure (dpe) to monitor seroconversion.
Viral Replication Kinetics in Ferrets
To monitor virus replication in ferrets coinoculated with WT and BC virus, tissues from the respiratory tract (nasal turbinate, soft palate, trachea, bronchus, right upper lung lobe, right lower lung lobe, left upper lung lobe, and left lower lung lobe) were collected at 2 dpi. Influenza M gene segment copy number was determined using real-time quantitative RT-PCR to approximate viral load in tissue samples (forward primer: 5′-GACCRATCCTGTCACCTC TGAC-3′, reverse primer: 5′-AGGGCATTYTGGACAAAKCGTCTA-3′, probe: 5′-FAM-TGC AGTCCTCGCTCACTGGGCACG-BHQ1-3′) following the conditions described [24]. The proportion of WT and BC sequences were determined from the upper respiratory tissue (nasal turbinate) and lower respiratory tissues (bronchus and left lower lung lobe) by NGS.
Next-Generation Sequencing
Cell culture supernatant, nasal washes, and tissue homogenates collected at the indicated time points were analyzed by NGS. Briefly, viral RNA was extracted from 140 μL of samples using QIAamp viral RNA mini kit (Qiagen) and eluted with 60 μL buffer AVE (RNase-free water with 0.04% Sodium azide). Eight μL RNA was transcribed into cDNA with SuperScript III reverse transcriptase (Invitrogen) in a 20-μL reaction with primer 454-Tag1-U12 (5′-GCCGGAGCTCTGCAGATATCAGCRAAAGCAGG-3′). Eight gene segments were amplified by Q5 High Fidelity DNA polymerase (NEB) using a pair of primers (454-Tag1-U12: 5′-GCCGGAGCTCTGCAGATATCAGCRAAAGCAGG-3′; 454-Tag1-U13: 5′-GCCGGAGCTCTGCAGATATCAGTAGAAACAAGG-3′) from 10 μL cDNA template in a 100-μL reaction. PCR products were purified using MinElute Reaction Cleanup Kit (Qiagen). Nextera XT DNA Sample Prep Kit (Illumina) was used for library preparation and Illumina MiSeq or NovaSeq 6000 was used for pair-end sequencing at the Centre for PanorOmic Sciences, The University of Hong Kong. All data were analyzed on a CLC Genomic workbench version 20 (Qiagen) and the minimal variant frequency was set at 1%.
RESULTS
Onward Transmission Potential of A(H1N1)pdm09 Virus From Ferrets Inoculated via Intranasal or Intratracheal Routes
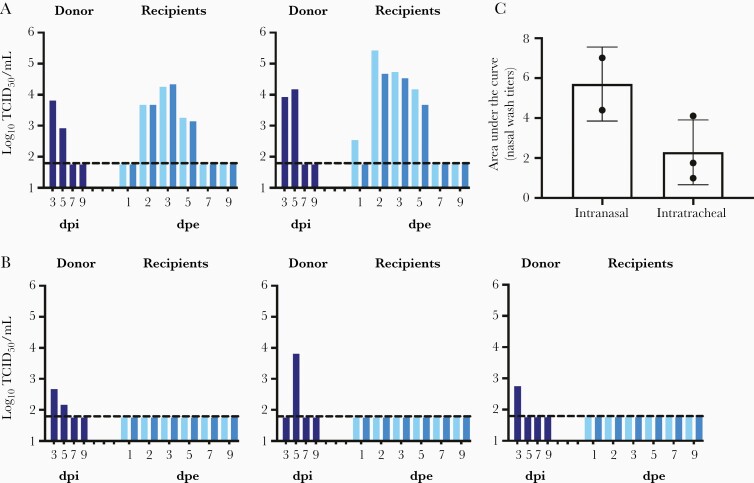
A(H1N1)pdm09 viruses transmit efficiently among ferrets via respiratory droplets [25–27]. We previously reported that A(H1N1)pdm09 viruses replicate efficiently in ferret upper and lower respiratory tissues, with mean viral titers detected at 7.0–7.2 log10 TCID50/mL and 4.6–6.4 log10 TCID50/mL in nasal turbinate and lungs at 3 dpi, respectively [28]. However, there has been limited studies that investigated the onward transmission potential of viruses that replicate at upper and lower respiratory tissues [29, 30]. We compared onward transmission potential of donor ferrets inoculated with 1000 PFU of recombinant A/California/04/2009 virus intranasally or intratracheally (Figure 1). At 2 dpi, 2 naive recipient ferrets were exposed to the exhaled breath of 1 donor ferret for 8 hours and all animals were single-housed in individually ventilated cages subsequently. Donor ferrets inoculated intranasally efficiently transmit the virus to naive recipient ferrets via breath aerosols, with viral shedding detected from the nasal wash of 1/4 recipient at 1 dpe and from 4/4 recipients at 2 dpe. (Figure 1A). In contrast, none of the donor ferrets inoculated intratracheally were able to transmit the virus to the naive recipient ferrets, as no viral shedding or seroconversion were detected from the recipients (Figure 1B). The total amounts of virus shed in donors’ nasal washes were approximated by calculating the area under the curve; donors inoculated intranasally shed higher amounts of virus in the nasal washes than those inoculated intratracheally (Mann-Whitney test, P = .200; Figure 1C). These results highlight the importance of ferret upper respiratory tissues as the initial anatomic site of viral replication for onward aerosol transmission.
Figure 1.
Onward transmission potential of donor ferrets inoculated with A(H1N1)pdm09 intranasally or intratracheally. A, Donor ferrets were inoculated with 1000 PFU in 50 µL of infection media. B, Donor ferrets were inoculated intratracheally with 1000 PFU in 500 µL of infection media. On day 2 postinoculation, 2 naive recipient ferrets were exposed to the exhaled breath of 1 inoculated donor ferret for 8 hours. All animals were single-housed in individually ventilated cages after exposure. Viral loads in the nasal washes (log10TCID50/mL) of the donor (dark blue) and 2 recipients (2 shades of lighter blue) were determined at various time points to monitor transmission and viral shedding. C, Area under the curve to approximate viral loads in the nasal washes of donor ferrets inoculated via intranasal or intratracheal route. Dotted lines indicate the limit of detection at 1.789 log10TCID50/mL. Abbreviations: dpe, days postexposure; dpi, days postinoculation; PFU, plaque forming unit; TCID50, 50% tissue culture infectious dose.
Generation of Recombinant WT and BC A/California/04/2009 Viruses
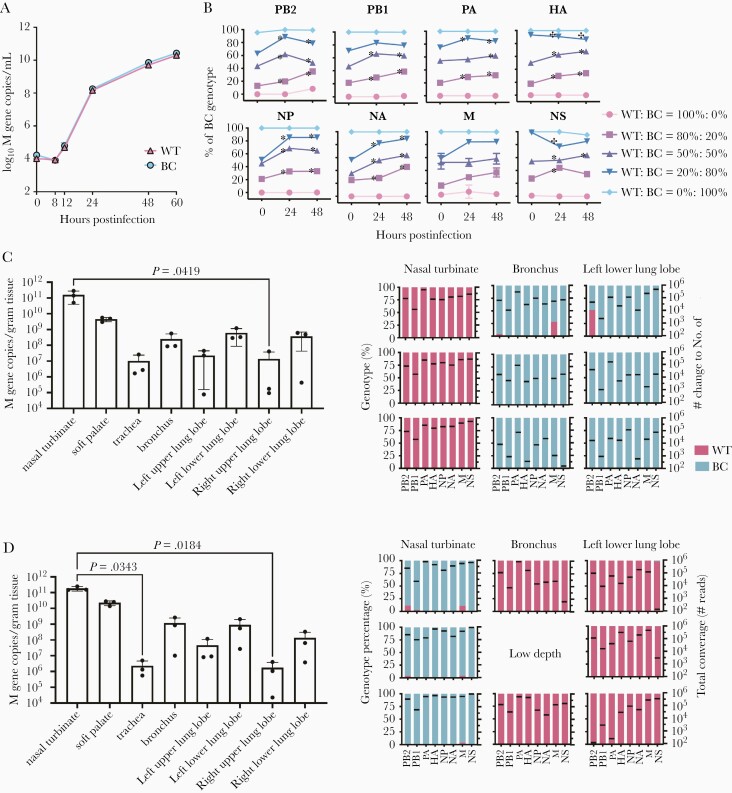
Influenza virus may spread from the initial anatomic site of replication to other regions of the respiratory tract over the course of infection. To confirm the importance of upper respiratory tropism for onward aerosol transmissibility, we generated recombinant WT and BC A/California/04/2009 viruses that differed by 2 synonymous mutations at each gene segment, with a maximal distance of 33 nucleotides between the 2 mutations (Table 1). The synonymous mutations were chosen so that they introduced nucleotides present in other circulating A(H1N1)pdm viruses, under the assumption that such mutations are likely to be neutral with respect to fitness. Comparable viral loads of WT and BC viruses were detected in MDCK cells infected with MOI of 0.001 (Figure 2A), suggesting that there was minimal effect of the synonymous mutations on viral growth in vitro. To further compared the competitive fitness of WT and BC viruses, MDCK cells were infected with mixed WT and BC viruses at different ratios and the percentages of WT and BC genotypes in culture supernatants were monitored using NGS. Compared to the inoculum at 0 hour, increased proportions of PB2, NP, and NA gene segments of the BC genotype were consistently noted at 24 and 48 hours postinfection using mixed inoculum of WT and BC viruses at 80%:20%, 50%:50%, and 20%:80% (Figure 2B). On the other hand, increased proportion of WT genotype was only transiently observed for the HA and NS gene segments. The results suggest that the PB2, NP, and NA gene segments of BC virus showed minor growth advantages over those of the WT virus in MDCK cells.
Figure 2.
Competitive replication kinetics of WT and BC viruses in vitro and in ferret respiratory tissues. A, MDCK cells were infected with WT and BC viruses at MOI = 0.001, culture supernatants were collected at indicated time points, and influenza virus M gene copy numbers were determined (log10 M gene copies/mL). B, MDCK cells were infected with WT and BC viruses mixed at different ratios (at MOI = 0.005), and the genotype percentages of WT and BC viruses were determined at 0 hour (inoculum), 24, and 48 hours postinfection using NGS. Compared to the inoculum at 0 hour, there was a significantly increased percentage of BC genotype at 24 or 48 hours (indicated by asterisks) and a significantly increased percentage of WT genotype (indicated by crosses; P < .05, 2-way ANOVA followed by Tukey multiple comparison test). C, Respiratory tissues were collected at 2 dpi from ferrets coinoculated with WT (intranasally) and BC (intratracheally) viruses (n = 3). D, Respiratory tissues were collected at 2 dpi from ferrets coinoculated with BC (intranasally) and WT (intratracheally) viruses (n = 3). Viral loads were determined by RT-PCR (M gene copies/g tissue, mean ± SD). Kruskal-Wallis test and Dunn multiple comparison test were used to compare viral loads detected in different tissues; P values <.05 are shown. Genotype percentage of WT and BC viruses in respiratory tissues were determined by NGS. The mean sequence reads of each gene segment are indicated by black lines. Abbreviations: BC, barcoded; MDCK, Madin-Darby canine kidney cells; MOI, multiplicity of infection; NGS, next-generation sequencing; RT-PCR, real-time quantitative polymerase chain reaction; WT, wild type.
We further compared the replication efficiency of WT and BC viruses in ferret respiratory tissues. The first group of ferrets were inoculated intranasally with WT virus and intratracheally with BC virus (Figure 2C). Conversely, the second group of ferrets were inoculated intranasally with BC virus and intratracheally with WT virus (Figure 2D). Comparing group 1 and 2 ferrets, viral loads detected from each respiratory tissue (nasal turbinate, soft palate, trachea, bronchus, and 4 lung lobes) were comparable at 2 dpi, with the highest viral loads detected from nasal turbinate. Importantly, viral loads detected in the lower respiratory tissues (bronchus and lung lobes) were not significantly different from those detected from the upper respiratory tissues (nasal turbinate, soft palate) (Figure 2C and 2D), suggesting that WT and BC viruses may replicate to comparable titers at both upper and lower respiratory tissues. We further analyzed the proportion of WT and BC genotypes replicated at the upper (nasal turbinate) and lower respiratory tissues (bronchus and left lower lung lobes). We noted that the genotype detected from the nasal turbinate matched the virus inoculated intranasally while the genotype detected from the bronchus and left lower lung lobes matched the virus inoculated intratracheally. There was minimal mixing of WT and BC viruses at the upper and lower respiratory tract.
Onward Transmission Potential of Ferrets Coinoculated With WT and BC Virus via Intranasal or Intratracheal Routes
To confirm the importance of upper respiratory tropism for onward aerosol transmissibility, 3 donor ferrets (43C01, 43E01, and 43F01) were coinoculated with WT and BC viruses via intranasal and intratracheal routes, respectively. At 2 dpi, 1 or 2 naive recipients were exposed to the exhaled breath of 1 inoculated donor for 8 hours. Viral shedding were detected from nasal washes of all recipient ferrets postexposure demonstrating efficient transmission (Figure 3). We analyzed the proportion of WT and BC genotypes in donors’ and recipients’ nasal washes at the peak titer days by NGS. WT genotype inoculated intranasally dominated in the nasal wash of donor 43C01 at 5 dpi; WT genotype was also detected from the nasal washes of 2 recipients 43C03 and 43C04 exposed to 43C01 at 5 dpe (Figure 3A). For donor 43E01, WT genotype dominated in the nasal wash at 3 dpi but BC genotype gained dominance over WT at 5 dpi, indicating the spread of BC virus from the lower respiratory tract (ie, the initial site of replication) to upper respiratory tract as infection progressed over time. WT genotype was detected from the nasal washes of the recipient ferrets 43E03 and 43E04 at 2 dpe and 5 dpe (Figure 3B), suggesting that transmission occurred from the upper airways prior to spread of the BC genotype from the lower to upper respiratory tract. For donor 43F01, WT genotype dominated in the nasal washes at 3 dpi and 5 dpi. WT genotype was also detected in the nasal wash of the recipient 43F03 at 5 dpe (Figure 3C).
Figure 3.
Onward transmission potential of donor ferrets coinoculated with WT (intranasally) and BC (intratracheally) A(H1N1)pdm09 viruses, in 3 independently repeated experimental groups (A, B, C). On day 2 postinoculation, 2 naive recipient ferrets were exposed to the exhaled breath of 1 inoculated donor ferret for 8 hours. All animals were single-housed in individually ventilated cages after exposure. Viral loads in the nasal washes (log10TCID50/mL) of donors (dark blue) and recipients (two shades of lighter blue) were determined at various time points to monitor transmission and viral shedding. The dotted line indicates the limit of detection at 1.789 log10TCID50/mL. Genotype percentage of WT (pink) and BC (blue) viruses in nasal washes were determined by NGS on peak titer days. Black lines indicate mean sequence reads of each gene segment. Abbreviations: BC, barcoded; dpe, days postexposure; dpi, days postinoculation; NGS, next-generation sequencing; TCID50, 50% tissue culture infectious dose; WT, wild type.
The experiment was repeated with the virus types and sites reversed, that is by co-inoculating 2 donor ferrets (48A01 and 48B01) with BC and WT viruses via intranasal and intratracheal routes, respectively. Transmission to all recipient ferrets was noted by detecting viral shedding in the nasal washes. The percentages of WT and BC genotypes in donors’ and recipients’ nasal washes at the peak titer days were analyzed by NGS. For both donors 48A01 and 48B01, BC genotype dominated in the nasal washes at 3 dpi and 5 dpi (Figure 4). In the nasal washes of exposed recipients, BC genotype also dominated at 2 dpe, 3 dpe, or 5 dpe. Taken together, these results confirm that onward transmission is mediated by viruses replicated at the upper respiratory epithelial cells.
Figure 4.
Onward transmission potential of donor ferrets coinoculated with BC (intranasally) and WT (intratracheally) A(H1N1)pdm09 viruses, in 2 independently repeated experimental groups (A, B). On day 2 postinoculation, 2 naive recipient ferrets were exposed to the exhaled breath of 1 inoculated donor ferret for 8 hours. All animals were single-housed in individually ventilated cages after exposure. Viral loads in the nasal washes (log10TCID50/mL) of donors (dark blue) and recipients (two shades of lighter blue) were determined at various time points to monitor transmission and viral shedding. The dotted line indicates the limit of detection at 1.789 log10TCID50/mL. Genotype percentage of WT (pink) and BC (blue) viruses in nasal washes were determined by NGS on peak titer days. Black lines indicate the mean sequence reads of each gene segment. Abbreviations: BC, barcoded; dpe, days postexposure; dpi, days postinoculation; NGS, next-generation sequencing; TCID50, 50% tissue culture infectious dose; WT, wild type.
Discussion
In the present study, we evaluated the onward transmission potential of A(H1N1)pdm09 influenza virus replicating at ferret upper or lower respiratory epithelial cells. Transmissible airborne particles were respired from the upper respiratory epithelial cells of donor ferrets that were either infected at the upper respiratory tract alone (inoculated intranasally) or were infected at both the upper and lower respiratory tract (inoculated intranasally and intratracheally). The results have implications in the use of antivirals or vaccines to reduce viral loads in the upper respiratory tissues to prevent influenza transmission [31, 32].
We observed limited mixing of viral populations replicating in the upper and lower respiratory tissues, at least at early days postinoculation. Our results are consistent with the findings of a previous study of similar design [33]. Ferret respiratory tissues collected at 2 dpi from ferrets coinoculated with WT and BC viruses showed limited spread of the inoculated genotypes from the respective respiratory tissues. Among 5 donors coinoculated with WT and BC viruses, the genotype inoculated intranasally was detected in donors’ nasal washes and was transmitted to recipient ferrets exposed to the donors at 2 dpi (Figure 3 and Figure 4). Interestingly, 1 donor (43E01) initially shed the genotype inoculated intranasally at 3 dpi and was found to shed the genotype inoculated intratracheally at 5 dpi (Figure 3B), demonstrating the upward spread potential of viruses replicating at the lower respiratory tract as infections progress over time. However, we and others have demonstrated that influenza onward transmissibility decreased over time; by 5 dpi, there was limited onward transmission by air or by direct contact [14, 34]. These results further highlight the importance of the upper respiratory tissues as the initial anatomic site of infection. Although large droplets are perceived to be deposited in the upper respiratory tract due to inertia, fine aerosols are also capable of infecting upper respiratory epithelial cells, as high viral load can be detected in the nasal washes of ferrets exposed to fine aerosols < 1.5 µm in size generated using a Collison nebulizer [14].
Our study is limited by the time points that the recipient ferrets were exposed to the inoculated donors. Recipient ferrets were only exposed to the donors for 8 hours at 2 dpi. While our work was being performed, Richard et al performed a similar study permitting continuous exposure of the recipient ferrets to the donors starting from 4 hours postinoculation until 9 dpi. Their study similarly demonstrated the significance of viral infection in the nasal epithelial cells for onward transmissibility [30]. Our results are also in line with a previous study that identified ferret soft palate as an important anatomic site of adaptation for transmissible influenza viruses [29]. Although it is challenging to collect samples from human upper and lower respiratory tissues, parallel studies in humans, if performed, will further advance our knowledge in this area. Previous studies on influenza transmission bottleneck in humans used nasal and throat swabs samples to monitor viral transmission within households [35–37].
Taken together, the ferret upper respiratory tissue is mapped as the anatomic site generating virus-laden particles that mediate influenza onward transmission. The mechanisms limiting transmission of aerosols generated from the lower respiratory tract warrant further studies, including the size [38], quantity, and infectivity of expelled aerosols, as well as other potential host and environmental factors.
Notes
Acknowledgments. We thank colleagues at the Centre for Comparative Medicine Research, The University of Hong Kong for the excellent animal husbandry support; and Mr Nick Chi-Ho Lin at Centre for PanorOmic Sciences, The University of Hong Kong for assistance in next-generation sequencing data analyses.
Financial support . This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant number HHSN272201400006C); and the Research Grants Council, Hong Kong SAR, China General Research Fund (grant number 17148416).
Potential conflicts of interest. All authors: No reported potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Options X for the Control of Influenza, Singapore, 28 August to 1 September 2019.
References
- 1. Tellier R, Li Y, Cowling BJ, Tang JW. Recognition of aerosol transmission of infectious agents: a commentary. BMC Infect Dis 2019; 19:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen WZ, Zhang N, Wei JJ, Yen HL, Li YG. Short-range airborne route dominates exposure of respiratory infection during close contact. Build Environ 2020; 176:106859. [Google Scholar]
- 3. Tang JW, Bahnfleth WP, Bluyssen PM, et al. Dismantling myths on the airborne transmission of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). J Hosp Infect 2021; 110:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fernández Tena A, Casan Clarà P. Deposition of inhaled particles in the lungs. Arch Bronconeumol 2012; 48:240–6. [DOI] [PubMed] [Google Scholar]
- 5. Thomas RJ. Particle size and pathogenicity in the respiratory tract. Virulence 2013; 4:847–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roy CJ, Milton DK. Airborne transmission of communicable infection–the elusive pathway. N Engl J Med 2004; 350:1710–2. [DOI] [PubMed] [Google Scholar]
- 7. Ibricevic A, Pekosz A, Walter MJ, et al. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol 2006; 80:7469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nicholls JM, Bourne AJ, Chen H, Guan Y, Peiris JS. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res 2007; 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature 2006; 440:435–6. [DOI] [PubMed] [Google Scholar]
- 10. Gralton J, Tovey ER, McLaws ML, Rawlinson WD. Respiratory virus RNA is detectable in airborne and droplet particles. J Med Virol 2013; 85:2151–9. [DOI] [PubMed] [Google Scholar]
- 11. Lindsley WG, Noti JD, Blachere FM, et al. Viable influenza A virus in airborne particles from human coughs. J Occup Environ Hyg 2015; 12:107–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Milton DK, Fabian MP, Cowling BJ, Grantham ML, McDevitt JJ. Influenza virus aerosols in human exhaled breath: particle size, culturability, and effect of surgical masks. PLoS Pathog 2013; 9:e1003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan J, Grantham M, Pantelic J, et al. ; EMIT Consortium . Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc Natl Acad Sci U S A 2018; 115:1081–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou J, Wei J, Choy KT, et al. Defining the sizes of airborne particles that mediate influenza transmission in ferrets. Proc Natl Acad Sci U S A 2018; 115:E2386–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson GR, Morawska L. The mechanism of breath aerosol formation. J Aerosol Med Pulm Drug Deliv 2009; 22:229–37. [DOI] [PubMed] [Google Scholar]
- 16. Johnson GR, Morawska L, Ristovski ZD, et al. Modality of human expired aerosol size distributions. J Aerosol Sci 2011; 42:839–51. [Google Scholar]
- 17. Herfst S, Schrauwen EJ, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 2012; 336:1534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012; 486:420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sorrell EM, Wan H, Araya Y, Song H, Perez DR. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci U S A 2009; 106:7565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sutton TC, Finch C, Shao H, et al. Airborne transmission of highly pathogenic H7N1 influenza virus in ferrets. J Virol 2014; 88:6623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong DD, Choy KT, Chan RW, et al. Comparable fitness and transmissibility between oseltamivir-resistant pandemic 2009 and seasonal H1N1 influenza viruses with the H275Y neuraminidase mutation. J Virol 2012; 86:10558–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 2000; 97:6108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg 1938; 27:493–7. [Google Scholar]
- 24. World Health Organization. WHO information for the molecular detection of influenza viruses.https://www.who.int/influenza/gisrs_laboratory/WHO_information_for_the_molecular_detection_of_influenza_viruses_20171023_Final.pdf. Accessed 12 May 2021.
- 25. Itoh Y, Shinya K, Kiso M, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 2009; 460:1021–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maines TR, Jayaraman A, Belser JA, et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 2009; 325:484–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Munster VJ, de Wit E, van den Brand JM, et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 2009; 325:481–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yen HL, Liang CH, Wu CY, et al. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A 2011; 108:14264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lakdawala SS, Jayaraman A, Halpin RA, et al. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 2015; 526:122–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richard M, van den Brand JMA, Bestebroer TM, et al. Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat Commun 2020; 11:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee LYY, Zhou J, Frise R, et al. Baloxavir treatment of ferrets infected with influenza A(H1N1)pdm09 virus reduces onward transmission. PLoS Pathog 2020; 16:e1008395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lowen AC, Steel J, Mubareka S, Carnero E, García-Sastre A, Palese P. Blocking interhost transmission of influenza virus by vaccination in the guinea pig model. J Virol 2009; 83:2803–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richard M, Herfst S, Tao H, Jacobs NT, Lowen AC. Influenza A virus reassortment is limited by anatomical compartmentalization following coinfection via distinct routes. J Virol 2018; 92:e02063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Inagaki K, Song MS, Crumpton JC, et al. Correlation between the interval of influenza virus infectivity and results of diagnostic assays in a ferret model. J Infect Dis 2016; 213:407–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCrone JT, Woods RJ, Martin ET, Malosh RE, Monto AS, Lauring AS. Stochastic processes constrain the within and between host evolution of influenza virus. Elife 2018; 7:e35962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Poon LL, Song T, Rosenfeld R, et al. Quantifying influenza virus diversity and transmission in humans. Nat Genet 2016; 48:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xue KS, Bloom JD. Reconciling disparate estimates of viral genetic diversity during human influenza infections. Nat Genet 2019; 51:1298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu H, Yen HL, Li YG. Deposition of bronchiole-originated droplets in the lower airways during exhalation. J Aerosol Sci 2020; 142:105524. [Google Scholar]