Summary
Axons in the adult mammalian central nervous system fail to regenerate after spinal cord injury. Neurons lose their capacity to regenerate during development, but the intracellular processes underlying this loss are unclear. We found that critical components of the presynaptic active zone prevent axon regeneration in adult mice. Transcriptomic analysis combined with live-cell imaging revealed that adult primary sensory neurons downregulate molecular constituents of the synapse as they acquire the ability to rapidly grow their axons. Pharmacogenetic reduction of neuronal excitability stimulated axon regeneration after adult spinal cord injury. Genetic gain- and loss-of-function experiments uncovered that essential synaptic vesicle priming proteins of the presynaptic active zone, but not clostridial-toxin-sensitive VAMP-family SNARE proteins, inhibit axon regeneration. Systemic administration of Baclofen reduced voltage-dependent Ca2+ influx in primary sensory neurons and promoted their regeneration after spinal cord injury. These findings indicate that functional presynaptic active zones constitute a major barrier to axon regeneration.
Keywords: axon injury, axon regeneration, spinal cord injury, Munc13, RIM1/2, active zone, Baclofen
Graphical abstract
Highlights
-
•
Axon growth-competent adult neurons downregulate critical components of the synapse
-
•
Pharmacogenetic reduction of neuronal excitability stimulates axon regeneration
-
•
The presynaptic active zone proteins Munc13 and RIM suppress axon growth
-
•
Systemic Baclofen treatment promotes regeneration after adult spinal cord injury
Combining genetic, pharmacogenetic, pharmacological, and transcriptomic approaches with live-cell, super-resolution, and 3-dimensional two-photon imaging, Hilton et al. demonstrate that critical components of the presynaptic transmitter release machinery actively inhibit axon growth. Munc13, a presynaptic active zone protein essential for synaptic transmission, suppresses axon regeneration following adult CNS injury.
Introduction
Mammalian central nervous system (CNS) neurons lose the ability to regenerate their axons during embryonic or early postnatal development (Hilton and Bradke, 2017; Li et al., 2020), limiting recovery following adult injury (Ramer et al., 2014). Neuron-extrinsic and intrinsic processes drive the CNS into this developmental decline of regeneration capacity (Bradbury and Burnside, 2019; Cregg et al., 2014; He and Jin, 2016; Li et al., 2020; Schwab and Strittmatter, 2014). While recent studies uncovered cellular and molecular changes in neuron-extrinsic CNS processes that cause regeneration failure (Dorrier et al., 2021; Li et al., 2020; Zhou et al., 2020), cell-intrinsic processes underlying the loss of neuronal growth capacity remain unclear (Fawcett, 2020; O’Shea et al., 2017; Tran et al., 2018).
Unlike most central neurons, dorsal root ganglion (DRG) neurons re-establish growth competence of their central axons in the adult injured spinal cord following a conditioning peripheral nerve lesion (PNL) (Neumann and Woolf, 1999; Richardson and Issa, 1984; Ylera et al., 2009). This process relies on the induction of a regenerative gene-expression program (Mahar and Cavalli, 2018) that includes the upregulation of regeneration-associated genes, such as activating transcription factor 3 (ATF3) (Renthal et al., 2020; Seijffers et al., 2007), Jun proto-oncogene (c-Jun) (Raivich et al., 2004), and signal transducer and activator of transcription 3 (Stat3) (Bareyre et al., 2011), as well as the downregulation of growth-restricting genes, such as Cacna2d2, which encodes the voltage-gated calcium channel (VGCC) auxiliary subunit Alpha2delta2 (Tedeschi et al., 2016). Despite the identification of various genes that regulate axon growth competence, knowledge of the actual molecular processes is still fragmentary (Fawcett and Verhaagen, 2018; Tedeschi et al., 2019).
We hypothesized that the fundamental processes underlying axon growth ability could be uncovered by searching for a common gene expression signature related to growth and regeneration. Consistent with this hypothesis, bioinformatic analyses revealed epigenetic changes (Palmisano et al., 2019) and transcriptional networks (Chandran et al., 2016) that control gene expression changes following PNL. Intriguingly, adult DRG neurons also re-adapt axon growth competence with prolonged cell culture (Smith and Skene, 1997) in a process that depends on de novo gene expression in the culture dish (Smith and Skene, 1997; Tedeschi et al., 2016). The increasing number of bulk and single-cell RNA sequencing (RNA-seq) databases of the DRG (Faure et al., 2020; Renthal et al., 2020; Sharma et al., 2020; Tedeschi et al., 2016; Usoskin et al., 2015) provides the opportunity to search for whole-transcriptome gene expression changes that might be the basis of axon growth ability during embryonic development and in adulthood.
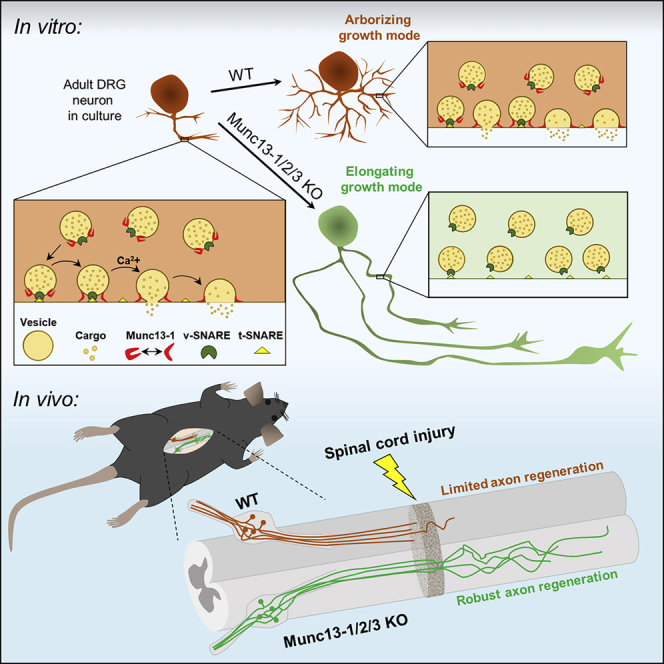
Doing so, we identified a common gene expression signature associated with the axon growth competence of DRG neurons. At its core is the downregulation of presynaptic components that are critical for synaptic vesicle fusion. Pharmacogenetic reduction of neuronal excitability stimulated axon regeneration after spinal cord injury. Genetic gain- and loss-of-function experiments revealed that RIMs and Munc13s, critical synaptic vesicle priming proteins of the presynaptic active zone, suppress axon growth in adult neurons. Our findings reveal that proteins critical for synaptic vesicle priming restrict axon regeneration following spinal cord injury.
Results
Downregulation of genes encoding synaptic proteins in DRG neurons upon acquisition of axon growth competence
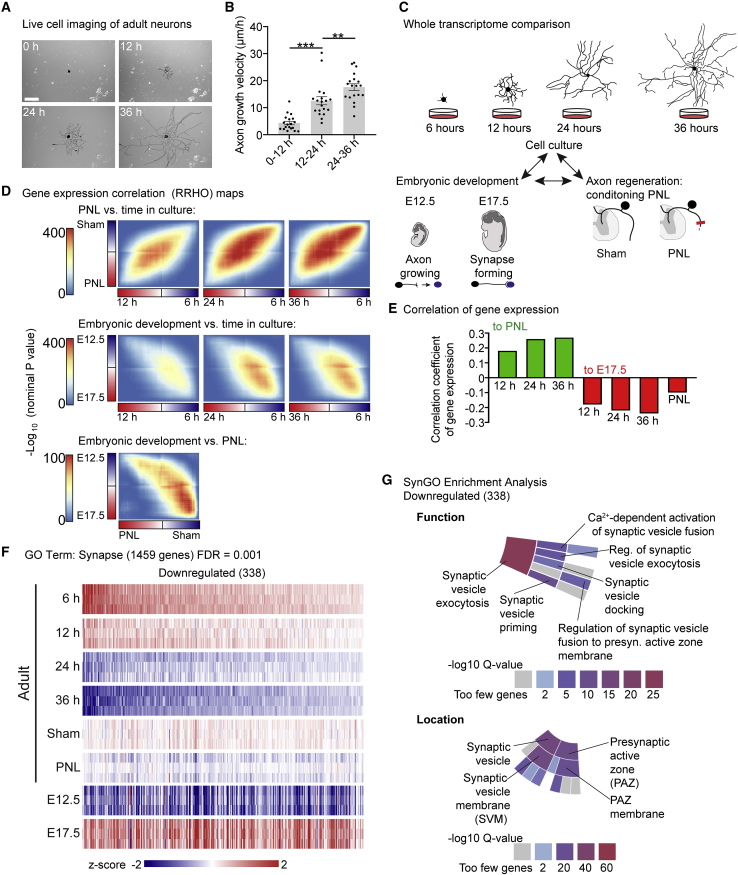
To explore the molecular mechanisms driving axon growth and regeneration, we searched for a common gene expression signature in DRG neurons underlying axon growth competence. DRG neurons subjected to a conditioning PNL prior to culture grow their axons rapidly within the first 12 h of plating (Smith and Skene, 1997), and even adult naive DRG neurons acquire growth competence with prolonged cell culture (Smith and Skene, 1997). Indeed, we found by long-term live imaging that dissociated adult mouse DRG neurons tripled their axon growth rate at 24–36 h versus 6–12 h after plating (Figures 1A and 1B). This raised the possibility that these neurons create an axon-growth-related gene expression signature that shares similarities with the regenerative conditioning-induced gene expression signature.
Figure 1.
DRG neurons downregulate core synaptic transmission genes as they acquire axon growth competence
(A) Live-cell images of a pseudocolored (black) adult DRG neuron 0–36 h after plating. Scale bar, 200 μm.
(B) Axon growth velocity of (A). Values are plotted as mean ± SEM; ∗∗∗p < 0.0001, ∗∗p < 0.01 by one-way ANOVA followed by Tukey’s post hoc test; n = 3 independent experiments with 14, 17, and 19 neurons per experiment.
(C) Scheme of RRHO analysis: comparing whole-transcriptome changes in developing, regenerating, and cultured DRG.
(D) Heatmaps of –log10 p values comparing overlap between whole-genome gene-expression changes in PNL/Sham, E12.5/E17.5, and time in cell culture (6, 12, 24, and 36 h).
(E) Correlation analysis of gene-expression profiles of the 3 paradigms.
(F) Heatmap of differentially expressed genes in DRG at E12.5, E17.5, and adult with PNL, Sham, or after 6–36 h in culture from RNA-seq with GO term “synapse” downregulated from 6 to 36 h (338 genes; FDR = 0.001; FDR-adjusted p < 1 × 10−5).
(G) Sunburst plot insets of SynGO-enrichment analyses showing the locations synaptic vesicle and presynaptic active zone and the function synaptic vesicle exocytosis enriched for in synapse-related genes downregulated in adult DRG neurons after 36 h in cell culture. Color code denotes the –log10 Q-value score.
See also Figures S1–S3 and Tables S1 and S2.
To explore this hypothesis, we performed new analyses on our previously published RNA-seq dataset (Tedeschi et al., 2016). In that study, hierarchical clustering was used to identify the most variable genes across embryonic development and in adult neurons that acquire axon growth competence. However, global trends of transcriptome-level changes were not considered. To compare whole-transcriptome gene expression changes by DRG neurons in cell culture to those that occur following a conditioning PNL (Figure 1C), we used rank-rank hypergeometric overlap (RRHO) analysis, which determines whole-genome correlation between two sets of differentially expressed genes without fixed thresholds (Plaisier et al., 2010). This approach revealed that cultured DRG neurons acquire a gene expression signature that correlates positively with the conditioning-induced gene expression signature. The correlation grew stronger with time in culture, when axons reach their highest growth velocity (c = 0.18, 12 h; c = 0.26, 24 h; c = 0.27 at 36 h; all versus PNL; all p < 1 × 10−149) (Figures 1D and 1E).
We then explored the relationship between these changes in gene expression to those that occur during embryonic development. Using RRHO analysis, we found that with either prolonged cell culture or a conditioning PNL, adult DRG neurons do not globally upregulate genes that are upregulated at embryonic day 12.5 (E12.5), when they are extending axons toward their targets (Kitao et al., 1996; Sharma et al., 2020), relative to E17.5, when their axons have reached targets and formed synapses (Ozaki and Snider, 1997). This may imply that the genes required for developmental axon growth remain upregulated from early development onward and that growth suppression may be mediated by a gene expression program that inhibits axon growth when the axon arrives at its target cells.
From this perspective, cultured adult neurons, as well as conditioned neurons in vivo, would start to regenerate their axons upon downregulation of a growth suppression program. Consistent with this hypothesis, conditioned neurons and adult cultured DRG neurons downregulated genes that were upregulated between E12.5 and E17.5. In cultured neurons, this inverse correlation became progressively stronger with time in culture (c = −0.18, 12 h; c = −0.22, 24 h; c = −0.24, 36 h; c = −0.10, PNL; all versus E17.5; all p < 4.0 × 10−47) (Figures 1D and 1E), as the axons accelerate growth. In total, 1,332 genes were both upregulated at E17.5 relative to E12.5 and downregulated in adult neurons after 36 h in culture relative to 6 h in culture (Figure S1; Table S1). Thus, in adult DRG neurons, the downregulation of genes upregulated between E12.5 and E17.5 represents a possible growth suppressive gene expression signature associated with axon growth competence.
Gene ontology (GO)-enrichment analysis indicated that genes upregulated at E17.5 versus E12.5 and downregulated in growth-competent adult neurons were enriched for functions related to the synapse, including neurotransmitter secretion, synaptic vesicle cycle, and synapse organization (Figure S1). Therefore, we explored how the expression of synapse-related genes changes in cultured neurons. Interestingly, transcriptomic analysis showed that 338 genes with the GO annotation “synapse” were downregulated after 36 h in culture relative to 6 h (FDR threshold = 0.001; FDR-adjusted p < 1 × 10−5) (Figure 1F; Table S1). Consistently, these genes were also globally downregulated after a conditioning lesion relative to sham and were upregulated at E17.5 relative to E12.5. Thus, the global expression of these genes inversely correlates with axon growth capacity in all 3 paradigms (Figure 1F).
Of note, 179 GO-annotated “synapse” genes were upregulated after 36 h in culture relative to 6 h (Table S1). However, these genes showed no discernible pattern when comparing their expression at E12.5 versus E17.5 (Figure S2A). This indicates that their expression is not related to synaptogenesis or the developmental decline in axon regeneration. Indeed, upregulated GO-annotated “synapse” genes are involved in biological processes that are also associated with axon growth, including cell adhesion and trophic factor signaling (Figures S2B–S2E). By contrast, the downregulated genes included key components of the presynaptic release machinery and were enriched for the core synaptic processes of ion gated channel activity and soluble NSF attachment protein receptor (SNARE) binding (Figures S2B–S2E; Table S1).
To specify the functions of these differentially expressed synapse-related genes, we performed GO-enrichment analysis with SynGO, a curated knowledge base of the synapse (Koopmans et al., 2019). GO-annotated “synapse” genes downregulated in cell culture were enriched for proteins localized to the presynaptic active zone, where neurotransmitters are released via exocytosis, and included the core active zone genes RIMS1, RIMS2, and Munc13-1 (Südhof, 2012). Hence, downregulated genes were enriched for the core active zone processes synaptic vesicle docking, priming, and Ca2+-dependent activation of synaptic vesicle fusion (Figures 1G and S3; Table S2). In addition, there was enrichment for genes localized to the synaptic vesicle, including integral components of the synaptic vesicle membrane, such as Vamp1 and Vamp2, as well as extrinsic components such as Syn1, Syn2, and Amph (Südhof, 2012) (Figures 1G and S3). In conclusion, DRG neurons establish a gene expression signature that negatively correlates with axon growth competence. This gene-expression signature indicates that genes encoding proteins critical for synaptic transmission may be involved in suppressing regeneration.
Inhibitory DREADD activation stimulates axon regeneration
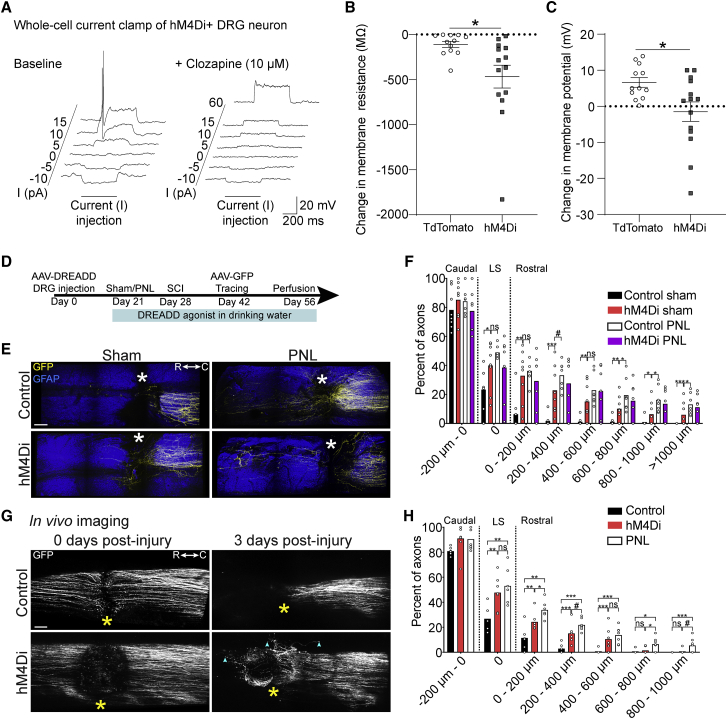
Given that DRG neurons downregulate genes critical for Ca2+-dependent activation of synaptic vesicle fusion as they acquire axon growth competence, we hypothesized that reducing neuronal excitability would promote axon regeneration. To explore this hypothesis, we overexpressed the inhibitory designer receptor exclusively activated by designer drug (DREADD) receptor hM4Di (Roth, 2016) or tdTomato for control in DRG neurons. First, we validated the electrophysiological impact of hM4Di activation in DRG neurons by transfecting these cells with hM4Di-mCherry or tdTomato-expressing plasmids and performing whole-cell patch-clamp recordings. The hM4Di-activating drug clozapine reduced the membrane resistance and membrane potential of hM4Di-mCherry-expressing DRG neurons relative to tdTomato-expressing control neurons (Figures 2A–2C).
Figure 2.
Pharmacogenetic reduction of neuronal excitability stimulates axon regeneration
(A) Representative whole-cell recordings from an hM4Di-mCherry+ adult mouse DRG neuron injected with current before (baseline) and after administration of 10 μM clozapine dihydrochloride.
(B and C) Change in membrane resistance (B) and membrane potential (C) of DRG neurons. Values are plotted as mean ± SEM; ∗p < 0.05 in (B) and (C), tdTomato+ versus hM4Di-mCherry+ by Student’s t test (n = 12 tdTomato+ and 14 hM4Di+ neurons).
(D) Timeline of (E) and (F).
(E) Multiphoton tile scan of GFP+ sensory axons (yellow) and GFAP+ astrocytes (blue) in the unsectioned spinal cord after complete dorsal column SCI in the given conditions. Asterisks indicate lesion centers. Scale bar, 200 μm.
(F) Quantification of (E). Scatterplot with means; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, and #p < 0.1 by permutation test. n = 8, 8, 7, and 7 for control sham, hM4Di sham, control PNL, and hM4Di PNL, respectively.
(G) In vivo tile scan images of GFP+ sensory axons 0 and 3 days after SCI. Asterisks indicate the lesion epicenters; cyan arrowheads point to distal tips of regenerating axons. Scale bar, 100 μm.
(H) Quantification of (G) and comparison with the conditioning paradigm. Scatterplot with means; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, and #p < 0.1 by permutation test. n = 6, 7, and 7 for control, hM4Di, and PNL, respectively.
We then explored the effect of hM4Di activation on the regeneration of adult DRG neurons following spinal cord injury in the naive state and following a conditioning PNL. To this end, we overexpressed hM4Di-mCherry or tdTomato for control in DRG neurons via intraganglionic injection of adeno-associated virus (AAV) into the lumbar 3 (L3), L4, and L5 DRG of adult wild-type mice (Figures 2D and S4). Three weeks later, we performed PNL or sham operations on the mice and immediately thereafter administered clozapine via intraperitoneal injection, with clozapine also administered via drinking water until endpoint. One week later, mice received a thoracic T12 dorsal column spinal cord injury that ablated their dorsal column sensory axons. To trace the axons, we delivered AAV-eGFP into the sciatic nerves of these mice 2 weeks prior to the experiment endpoint. This set of experiments followed by whole-tissue immunostaining and two-photon 3D imaging analysis (Hilton et al., 2019) showed that activation of hM4Di stimulates axon regeneration after spinal cord injury (Figures 2E and 2F; Video S1). Importantly, activation of hM4Di had no additive effect on the central axon regeneration of DRG neurons that had undergone a conditioning PNL (Figures 2E and 2F), indicating that hM4Di activation stimulates the same processes as the conditioning effect.
Movie shows multiphoton tile scan and 3D rendering of GFP+ sensory axons (yellow) and GFAP+ astrocytes (blue) in the injured adult mouse spinal cord in control conditions or with hM4Di activation.
To determine the kinetics of axon growth following hM4Di activation, we performed two-photon in vivo live imaging in a separate cohort of animals. Adult mice received intraganglionic delivery of AAV-eGFP together with AAV-hM4Di-mCherry or AAV-tdTomato for control, with clozapine administered 3 weeks later. We performed live imaging of the axons immediately after injury and 3 days post-injury, with regeneration distances calculated by morphological landmarks such as branched axons caudal to the lesion in each mouse. This in vivo imaging approach of live axons revealed that most regeneration in response to hM4Di activation occurs rapidly, within 3 days after injury (Figures 2G and 2H). Interestingly, we found a similar rapid regenerative response was also induced by a conditioning PNL (Figures 2G and 2H). Together, these data show that reducing the excitability of injured dorsal column axons stimulates their regeneration after spinal cord injury.
Axon growth and regeneration are independent of VAMPs1–3
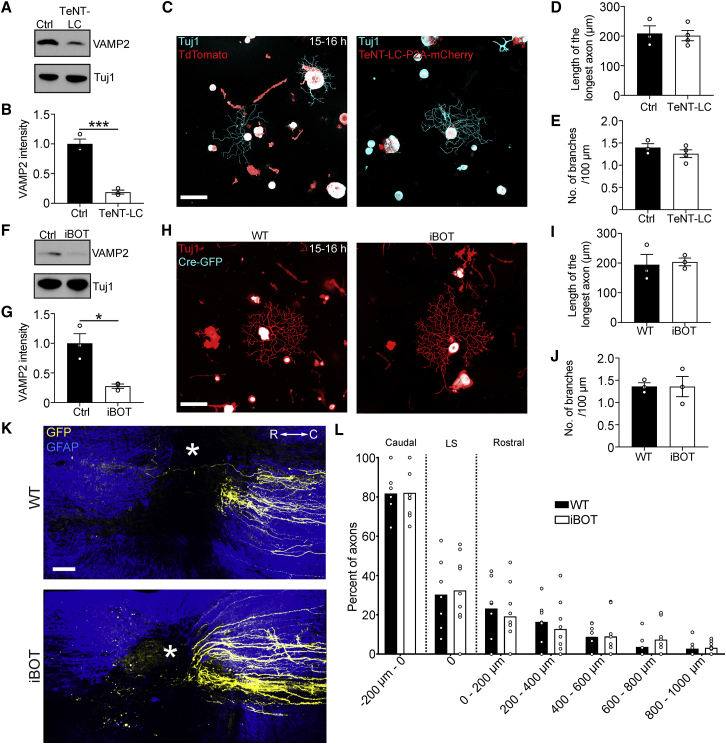
We then genetically manipulated different components of the transmitter release machinery that our transcriptomic analysis identified as being downregulated in axon-growth-competent neurons to explore their effects on axon growth. Among these components were the vesicle-associated membrane protein (VAMP) family members VAMP1 and VAMP2. Therefore, we assessed whether overexpression of tetanus toxin light chain (TeNT-LC), which cleaves VAMP1, -2, and -3 but leaves other VAMP protein family members intact (Hoogstraaten et al., 2020; Humeau et al., 2000; McMahon et al., 1993; Schiavo et al., 1992), would promote axon growth or regeneration. We generated an AAV encoding TeNT-LC separated from mCherry by a P2A peptide, injected it into the sciatic nerves of adult wild-type mice, and plated the neurons 3 weeks later. TeNT-LC reduced VAMP2 in DRG neurons (Figures 3A and 3B) but did not affect axon growth or branching in DRG neurons relative to tdTomato-expressing control neurons (Figures 3C–3E).
Figure 3.
TeNT-LC or BoNT/B overexpression does not promote axon growth in adult DRG neurons
(A) Immunoblot of VAMP2 and Tuj1 in L3–L5 DRG extracts 3 weeks after AAV-TeNT-LC-P2A-mCherry or AAV-tdTomato injection into wild-type mice.
(B) Quantification of (A). Values are plotted as mean ± SEM; ∗∗∗p < 0.001 by Student’s t test; n = 3 independent experiments.
(C) Representative fluorescence images of Tuj1 (cyan) and TeNT-LC-P2A-mCherry/tdTomato (red) immunolabeled DRG neurons 3 weeks after AAV administration and plated for 15–16 h. Scale bar, 100 μm.
(D and E) Length of the longest axon (D) and branching frequency (E) of (C). Values are plotted as mean ± SEM; n = 3–4 independent experiments with 145 tdTomato+ neurons, 133 TeNT-LC-P2A-mCherry+ neurons.
(F) Immunoblot of VAMP2 and Tuj1 in L3–L5 DRG extracts 3 weeks after AAV-Cre-GFP injection into iBOT and wild-type control mice.
(G) Quantification of (F). Values are plotted as mean ± SEM;∗p < 0.05 by Student’s t test. n = 3 independent experiments.
(H) Representative fluorescence images of Tuj1 (red) and Cre-GFP (cyan) immunolabeled DRG neurons 3 weeks after AAV-Cre-GFP administration and plated for 15–16 h. Scale bar, 100 μm.
(I and J) Length of the longest axon (I) and branching frequency (J) of (H). Values are plotted as mean ± SEM; n = 3 independent experiments with 100 Cre-GFP+ wild-type neurons, 164 Cre-GFP+ iBOT neurons.
(K) Multiphoton tile scan of GFP+ sensory axons (yellow) and GFAP+ astrocytes (blue) in the unsectioned spinal cord after complete dorsal column SCI in the given conditions. R, rostral; C, caudal. Asterisks indicate lesion centers. Scale bar, 200 μm.
(L) Quantification of (K). Scatterplot with means. n = 7 WT mice and 9 iBOT mice.
We then explored whether Cre-induced expression of Botulinum neurotoxin serotype B (BoNT/B) in adult DRG neurons of iBOT mice (Slezak et al., 2012) would promote axon growth or regeneration. Similarly to TeNT-LC expression, Cre-induced BoNT/B expression reduced VAMP2 in the DRG (Figures 3F and 3G) but did not affect axon growth or branching in DRG neurons relative to wild-type Cre-expressing mice (Figures 3H–3J). Moreover, BoNT/B overexpression in DRG neurons did not stimulate axon regeneration after spinal cord injury in vivo (Figures 3K–3L). These data show that axon growth and regeneration are independent of VAMPs1–3.
RIM1/2 deletion promotes axon growth
DRG neurons express additional isoforms of VAMP that are insensitive to TeNT or BoNT/B but capable of forming functional SNARE complexes (Hasan et al., 2010) and might compensate for the loss of VAMPs1–3. Moreover, VAMP proteins are not just involved in Ca2+-dependent vesicle fusion but also in other membrane trafficking pathways, including those involved in developmental axon outgrowth (Calakos et al., 1994; Kimura et al., 2003; Osen-Sand et al., 1996). We therefore more specifically targeted key components of the presynaptic active zone (Südhof, 2012).
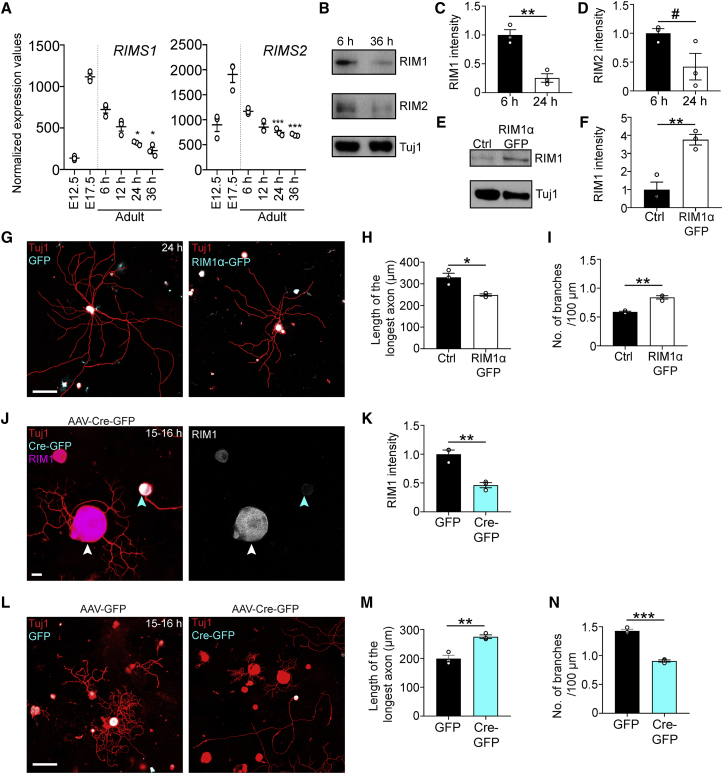
Our transcriptomic analysis identified the genes expressing the active zone proteins Rab3 interacting molecule 1 (RIM1) and RIM2 as being upregulated at E17.5 relative to E12.5 and downregulated in adult DRG with prolonged cell culture (Figure 4A). Immunoblots of cell extracts confirmed that RIM1 protein expression is downregulated (and RIM2 expression trends toward downregulation) in DRG neurons grown for 36 h in cell culture relative to 6 h in culture (Figures 4B–4D). To determine whether RIM suppresses axon growth, we dissociated adult lumbar DRG neurons and electroporated them with either GFP (control) or RIM1α-GFP-expressing plasmids. RIM1α-GFP overexpression inhibited axon growth and enhanced branching in adult DRG neurons after 24 h in culture (Figures 4E–4I). We then determined whether conditional deletion of all RIM1 and RIM2 isoforms would promote axon growth. To this end, we used adult RIM1/2fl/fl mice (Kaeser et al., 2011). Immunocytochemical analysis showed that viral expression of Cre-eGFP in RIM1/2fl/fl mice knocked out RIM1 in DRG neurons (Figures 4J and 4K). RIM1/2-knockout (KO) enhanced axon growth and reduced branching in DRG neurons in vitro (Figures 4L–4N). Thus, RIM1/2 deletion promotes axon growth.
Figure 4.
RIM1/2 deletion promotes axon growth in adult DRG neurons
(A) Normalized expression values of RIMS1 and RIMS2 in DRG at E12.5, E17.5, and adult from 6 to 36 h in culture. ∗∗∗p < 0.001 6 versus 24 or 36 h, ∗p < 0.05 6 versus 24 or 36 h in culture by one-way ANOVA followed by Tukey’s post hoc test; n = 3 technical replicates per condition.
(B) Immunoblot of RIM1, RIM2, and Tuj1 in L3–L5 DRG extracts after 6 or 36 h in culture.
(C and D) Quantification of RIM1 (C) and RIM2 (D) in (B). Values are plotted as mean ± SEM; ∗∗p < 0.01, #p < 0.10 by Student’s t test; n = 3 independent experiments.
(E) Immunoblot of RIM1 and Tuj1 in L3–L5 DRG extracts electroporated with Rim1α-GFP or GFP (control)-expressing plasmids and cultured for 24 h.
(F) Quantification of (E). Values are plotted as mean ± SEM; ∗∗p < 0.01 by Student’s t test. n = 3 independent experiments.
(G) Representative fluorescence images of naive DRG neurons electroporated with RIM1α-GFP or GFP-expressing plasmids and cultured for 24 h. Scale bar, 100 μm.
(H and I) Length of the longest axon (H) and branching frequency (I) of (G). Values are plotted as mean ± SEM; ∗∗p < 0.01, ∗p < 0.05 control GFP versus RIM1α-GFP by Student’s t test; n = 3 independent experiments with 103 control GFP+ neurons, 141 RIM1α-GFP+ neurons.
(J) Representative images of cultured DRG neurons after AAV-Cre-GFP administration into RIM1fl/flRIM2fl/fl mice. Cyan arrowhead points to Cre-GFP+ neuron and white arrowhead points to Cre-GFP– neuron. Scale bar, 15 μm.
(K) Quantification of (J). Values are plotted as mean ± SEM; ∗∗p < 0.01 by Student’s t test; n = 3 independent experiments, 106 GFP+ neurons, and 98 Cre-GFP+ neurons.
(L) Representative fluorescence images of Tuj1 (red) and GFP/Cre-GFP (cyan)-immunolabeled DRG neurons 3 weeks after AAV-GFP or AAV-Cre-GFP administration into RIM1fl/flRIM2fl/fl mice and plated for 15–16 h. The AAV-Cre-GFP image shows the same neurons as in (J). Scale bar, 100 μm.
(M and N) Length of the longest axon (M) and branching frequency (N) of (L). Values are plotted as mean ± SEM; ∗∗∗p < 0.001, ∗∗p < 0.01 by Student’s t test. n = 3 independent experiments, 108 GFP+ neurons, 101 Cre-GFP+ neurons.
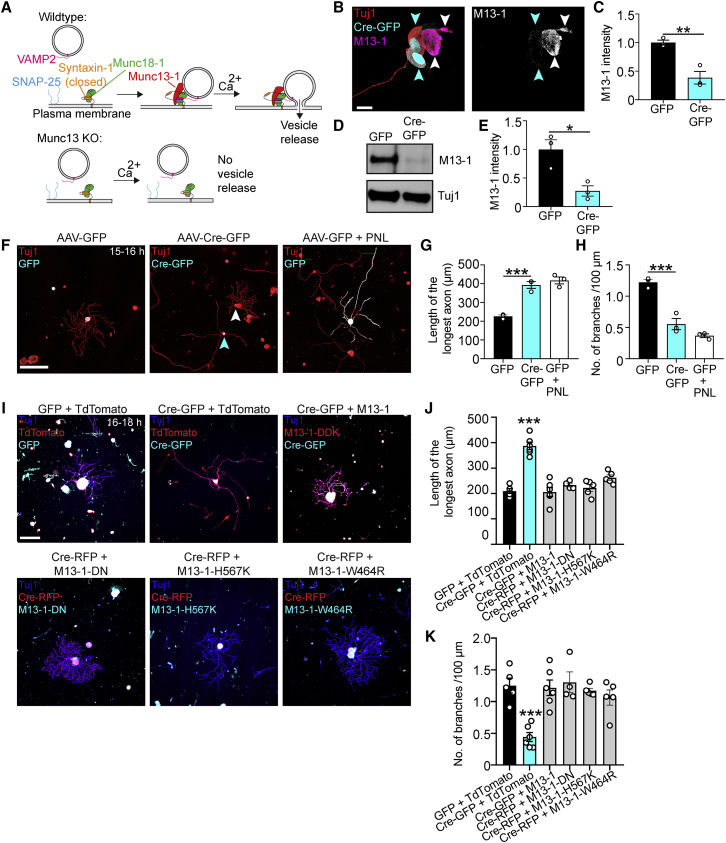
Munc13 suppresses axon growth
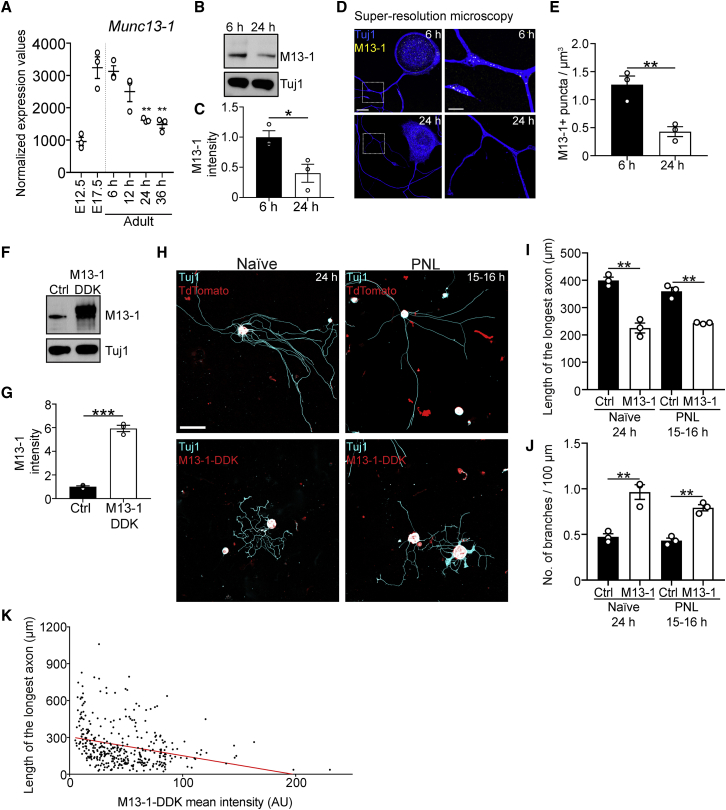
RIM proteins are multi-functional, with roles in Ca2+ channel clustering (Kaeser et al., 2011; Kiyonaka et al., 2007), coupling of vesicles with Ca2+ channels (Han et al., 2011), and vesicle docking/priming (Betz et al., 2001; Han et al., 2011). Given that Alpha2delta2 suppresses axon growth by enhancing Ca2+ influx (Tedeschi et al., 2016), we tested whether synaptic vesicle docking/priming more specifically suppressed axon growth. To this end, we explored the role of the core active zone protein Munc13, the effector through which RIM regulates vesicle docking/priming (Betz et al., 2001) and a protein essential for synaptic vesicle docking, priming, and fusion (Augustin et al., 1999; Imig et al., 2014; Siksou et al., 2009; Varoqueaux et al., 2002). As for RIMS1 and RIMS2, our transcriptomic analysis identified Munc13-1 as one of the genes upregulated at E17.5 relative to E12.5 and downregulated in adult DRG with prolonged cell culture (Figure 5A). Consistently, immunoblots of cell extracts and super-resolution microscopy showed that Munc13-1 protein expression is downregulated in DRG neurons grown for 24 h in cell culture relative to 6 h (Figures 5B–5E). Specifically, super-resolution microscopy revealed the expression of Munc13-positive puncta not only in the axons but also in the cell bodies of DRG neurons (Figure 5D), consistent with the ability of these neurons to undergo somatic Ca2+-dependent exocytosis (Huang and Neher, 1996); both decreased 24 h after plating.
Figure 5.
Munc13-1 suppresses axon growth in adult DRG neurons
(A) Normalized expression values of Munc13-1 in DRG at E12.5, E17.5, and adult from 6 to 36 h in culture. ∗∗p < 0.01 6 versus 24 h, ∗∗p < 0.01 6 versus 36 h in culture by one-way ANOVA followed by Tukey’s post hoc test; n = 3 technical replicates per condition.
(B) Immunoblot of Munc13-1 and Tuj1 in L3-L5 DRG extracts after 6 or 24 h in culture.
(C) Quantification of (B). Values are plotted as mean ± SEM; ∗p < 0.05 by Student’s t test; n = 3 independent experiments.
(D) Representative fluorescence images of adult DRG neurons cultured for 6 or 24 h and stained with Munc13-1 and Tuj1 antibodies. Scale bars, 10 μm in outset and 3 μm in inset.
(E) Quantification of (D). Values are plotted as mean ± SEM; ∗∗p < 0.01 by Student’s t test; n = 21 neurons at 6 h, 18 neurons at 24 h from 3 independent experiments.
(F) Immunoblot of Munc13-1 and Tuj1 in L3-L5 DRG extracts electroporated with Munc13-1 or tdTomato-expressing plasmids and cultured for 24 h.
(G) Quantification of (F). Values are plotted as mean ± SEM; ∗∗∗p < 0.001 by Student’s t test. n = 3 independent experiments.
(H) Representative fluorescence images of uninjured/naive or conditioned/PNL DRG neurons electroporated with Munc13-1-DDK or tdTomato-expressing plasmids. Scale bar, 100 μm.
(I and J) Length of the longest axon (I) and branching frequency (J) of (H). Values are plotted as mean ± SEM; ∗∗p < 0.01 naive tdTomato versus naive Munc13-1-DDK and PNL tdTomato versus PNL Munc13-1-DDK by Student’s t test in (I) and ∗∗p < 0.01 naive tdTomato versus naive Munc13-1-DDK and PNL tdTomato versus PNL Munc13-1-DDK by Student’s t test in (J); n = 3 independent experiments with 94 naive tdTomato+ neurons, 224 naive Munc13-1-DDK+ neurons, 125 PNL tdTomato+ neurons, 145 PNL Munc13-1-DDK+ neurons.
(K) XY plot of Munc13-1-DDK mean intensity (arbitrary units) and length of the longest axon for all Munc13-1-DDK+ neurons analyzed in (H)–(J). Red line shows linear regression (p < 0.0001).
To explore whether Munc13-1 suppresses axon growth, we dissociated adult lumbar DRG neurons and electroporated them with either tdTomato (control) or Munc13-1-expressing plasmids (Figures 5F and 5G). Munc13-1 overexpression prevented adult DRG neurons from transitioning to an elongating growth phenotype after prolonged culture, restricting axon growth and enhancing branching after 24 h (Figures 5H–5J). We then tested whether Munc13-1 overexpression could restrict the growth of conditioned neurons. To this end, we performed conditioning PNLs in adult mice and 3 days later, dissociated L3–L5 DRG, transfected them with tdTomato (control) or Munc13-1-expressing plasmids, and plated the cells for 15–16 h. Even in the context of a conditioning PNL, Munc13-1 overexpression restricted the outgrowth and promoted branching of DRG axons (Figures 5H–5J) dose dependently (Figure 5K). Thus, an increase of Munc13 protein levels can transform growth-competent neurons to become growth incompetent.
Munc13 deletion promotes axon regeneration
We then determined whether loss of Munc13s would enhance axon growth and regeneration. To this end, we used adult mice with a floxed Munc13-1 gene (Banerjee et al., 2020) and homozygous deletions of the Munc13-2 and Munc13-3 genes, which allowed deletion of all relevant Munc13 variants (Augustin et al., 1999, 2001; Varoqueaux et al., 2002) (Figure 6A). Immunoblot and immunocytochemical analyses showed that viral expression of Cre-eGFP in Munc13-1fl/flMunc13-2KO/KOMunc13-3KO/KO mice effectively knocked out Munc13-1 in DRG neurons (Figures 6B–6E). To explore whether Munc13 triple-KO would enhance axon growth, we injected AAV-Cre-eGFP or AAV-eGFP for control into the sciatic nerves of these mice and plated the neurons 3 weeks later. Munc13 triple-KO enhanced axon elongation and reduced branching in vitro, phenocopying the effect size of a conditioning PNL (Figures 6F–6H). This effect depended on loss of Munc13 as re-expression of Munc13-1 in Munc13 triple-KO neurons restored the limited growth and increased branching found in control cells (Figures 6I–6K).
Figure 6.
Munc13 deletion promotes axon growth in adult DRG neurons
(A) Scheme of Munc13’s function.
(B) Representative images of cultured DRG neurons after AAV-Cre-GFP administration into Munc13-1fl/flMunc13-2KO/KOMunc13-3KO/KO mice. Cyan arrowheads point to Cre-GFP+ neurons, and the white arrowhead points to Cre-GFP– neuron. Scale bar, 15 μm.
(C) Quantification of (B). Values are plotted as mean ± SEM; ∗∗p < 0.01 by Student’s t test; n = 3 independent experiments, 18 GFP+ neurons, 22 Cre-GFP+ neurons.
(D) Immunoblot of Munc13-1 and Tuj1 in L3-L5 DRG extracts 3 weeks after AAV-Cre-GFP injection into Munc13-1fl/flMunc13-2KO/KOMunc13-3KO/KO mice.
(E) Quantification of (D). Values are plotted as mean ± SEM; ∗p = 0.0192 by Student’s t test; n = 3 independent experiments.
(F) Representative fluorescence images of Tuj1 (red) and GFP/Cre-GFP (cyan)-immunolabeled DRG neurons 3 weeks after AAV-GFP or AAV-Cre-GFP administration and plated for 15–16 h. Cyan arrowhead points to Cre-GFP+ neuron and white arrowhead points to Cre-GFP– neuron. Scale bar, 200 μm.
(G and H) Length of the longest axon (G) and branching frequency (H) of (F). Values are plotted as mean ± SEM; ∗∗∗p < 0.001 by Student’s t test. n = 3 independent experiments, 102 GFP neurons, 125 Cre-GFP neurons, 116 GFP + PNL neurons.
(I) Representative fluorescence images of Tuj1, GFP/Cre-GFP/Cre-RFP, and tdTomato/Munc13-1-DDK/Munc13-1-DN/Munc13-1-H567K/Munc13-1-W464R immunolabeled DRG neurons from Munc13-1fl/flMunc13-2KO/KOMunc13-3KO/KO mice administered AAV-GFP, AAV-Cre-GFP or AAV-Cre-RFP, transfected with plasmids and cultured for 16–18 h. Scale bar, 200 μm.
(J and K) Length of the longest axon (J) and branching frequency (K) of (I). Values are plotted as mean ± SEM; ∗∗∗p < 0.0001 by one-way ANOVA followed by Tukey’s post hoc test; n = 80 GFP + tdTomato neurons from 5 independent experiments, 64 Cre-GFP + tdTomato neurons from 6 independent experiments, 78 Cre-GFP + Munc13-1 (DDK) neurons from 6 independent experiments, 71 Cre-RFP + Munc13-1-DN neurons from 4 independent experiments, 80 Cre-RFP + Munc13-1-H567K neurons from 5 independent experiments, 81 Cre-RFP + Munc13-1-W464R neurons from 5 independent experiments.
Munc13 proteins contain multiple domains. In addition to the Munc13 homology domain that is sufficient for the basal priming function (Basu et al., 2005), the protein contains a C2B domain that acts as a Ca2+ and PIP2-dependent modulator of short-term plasticity (Shin et al., 2010), a C1 domain that mediates diacylglycerol and phorbol ester binding (Betz et al., 1998; Michelassi et al., 2017), and a Ca2+-Calmodulin-binding region (Lipstein et al., 2013). We therefore explored whether expression of mutant constructs with an inactivated C2B domain (Munc13-1-DN) (Shin et al., 2010), disrupted C1 domain (Munc13-1-H567K) (Betz et al., 1998), or an inactivated Ca2+-Calmodulin-binding region (Munc13-1-W464R) (Junge et al., 2004) would restore the limited axon growth phenotype in Munc13 triple-KO neurons. Expression of each of these mutant constructs restored the arborizing growth phenotype found in control neurons (Figures 6I–6K). Thus, the C2B, C1, and Ca2+-Calmodulin-binding regions of Munc13 and consequently regulation of Munc13 function by second messengers are not necessary for Munc13's role in limiting axon growth. Instead, it is rather the basal priming function of Munc13 or another role as a structural protein important for presynaptic release site formation that mediates the protein’s growth suppressive role.
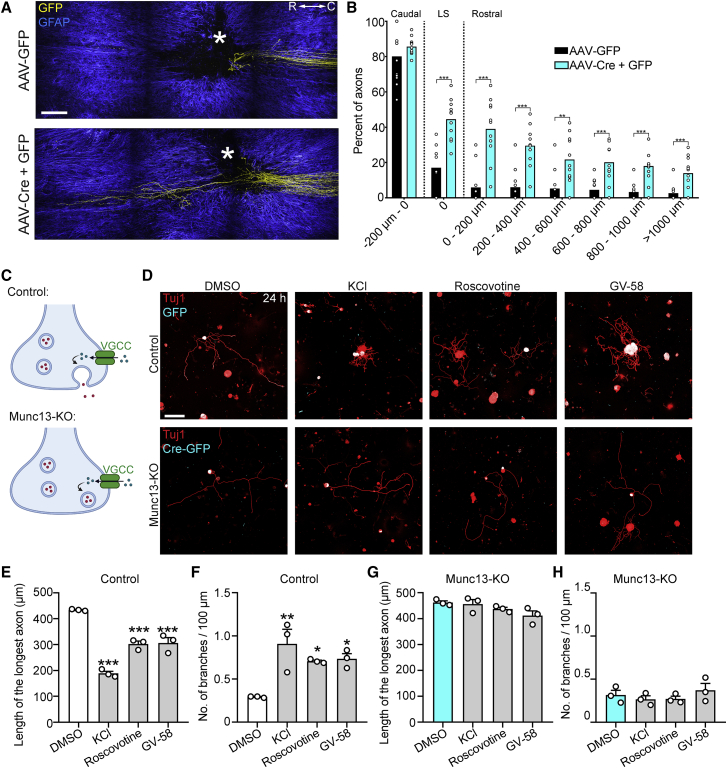
We therefore tested whether loss of Munc13s could promote axon regeneration following spinal cord injury in vivo. To this end, we injected AAV-Cre-eGFP and AAV-eGFP or AAV-eGFP alone into the sciatic nerves of Munc13-1fl/flMunc13-2KO/KOMunc13-3KO/KO mice and performed a T12 dorsal column spinal cord injury 3 weeks later. Whole-mount immunostaining and two-photon imaging revealed that Munc13 triple-KO enhanced regeneration more than 1 mm rostral to the lesion site (Figures 7A and 7B). Thus, ablating Munc13 activity stimulates axon regeneration after adult mouse spinal cord injury. Overall, our data indicate that Munc13 expression acts as a developmental switch that inhibits axon growth and regeneration.
Figure 7.
Munc13 deletion promotes axon regeneration following adult CNS injury
(A) Multiphoton tile scan of GFP+ sensory axons (yellow) and GFAP+ astrocytes (blue) in the unsectioned spinal cord after complete dorsal column SCI in the given conditions. R, rostral; C, caudal. Asterisks indicate lesion centers. Scale bar, 200 μm.
(B) Quantification of (A). Scatterplot with means; ∗∗∗p < 0.001, ∗∗p < 0.01 by permutation test. n = 11 animals per group.
(C) Scheme of VGCC activation, presynaptic Ca2+ influx, and vesicle release.
(D) Representative fluorescence images of Tuj1 (red) and GFP/Cre-GFP (cyan) immunolabeled DRG neurons from Munc13-1fl/flMunc13-2KO/KOMunc13-3KO/KO mice administered AAV-GFP or AAV-Cre-GFP and cultured for 24 h in the presence of DMSO, KCl (40 mM), Roscovotine (20 μM), or GV-58 (20 μM). Scale bar, 200 μm.
(E and F) Length of the longest axon (E) and branching frequency (F) of AAV-GFP+ neurons in (D). Values are plotted as mean ± SEM; ∗∗∗p < 0.001 GFP DMSO versus GFP KCl, GFP Roscovotine, GFP GV-58 in (E) and ∗∗p < 0.01 GFP DMSO versus GFP KCl, ∗p < 0.05 GFP DMSO versus GFP Roscovotine, GFP DMSO versus GFP GV-58 in (F) by one-way ANOVA followed by Tukey’s post hoc test; n = 72 GFP DMSO, 86 GFP GV-58, 37 GFP KCl, 53 GFP Roscovotine-treated neurons from 3 independent experiments.
(G and H) Length of the longest axon (G) and branching frequency (H) of AAV-Cre-GFP+ neurons in (D). Values are plotted as mean ± SEM n = 82 Cre-GFP DMSO, 92 Cre-GFP GV-58, 48 Cre-GFP KCl, 23 Cre-GFP Roscovotine-treated neurons from 3 independent experiments.
Voltage-gated Ca2+ channel activation suppresses axon growth through Munc13s
Previous work showed that Ca2+ signaling inhibits axon growth and regeneration of DRG neurons (Enes et al., 2010; Tedeschi et al., 2016). This, together with our present data, raised the possibility that Ca2+ signaling suppresses axon growth by triggering vesicle fusion activity (Figure 7C). Alternatively, Munc13s can also affect Ca2+ signaling by regulating VGCCs (Calloway et al., 2015). To explore the relationship between VGCC activation and Munc13 in suppressing axon growth, we depolarized neurons with KCl or treated them with GV-58, a P/Q-type VGCC agonist (Liang et al., 2012), or Roscovotine, a cyclin-dependent kinase 5 (cdk5) inhibitor that is also an agonist of Cav2 VGCCs (Yan et al., 2002), and cultured them for 24 h. Consistent with previous results (Enes et al., 2010; Tedeschi et al., 2016), these treatments suppressed axon growth and increased branching in control neurons (Figures 7D–7F). By contrast, Munc13 triple-KO neurons were not affected by these treatments and showed persistently increased growth and decreased branching (Figures 7D, 7G, and 7H). Thus, suppression of axon growth by VGCC activation or depolarization requires Munc13.
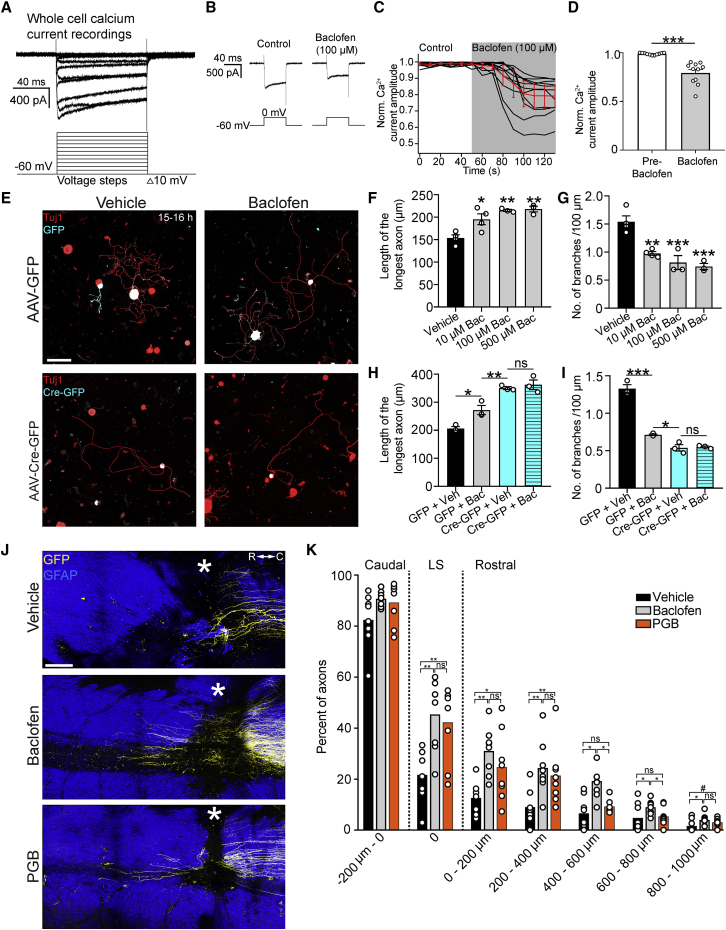
Baclofen promotes axon regeneration
If VGCC-activation suppresses axon growth in a Munc13-dependent manner, for instance, by promoting Ca2+-triggered exocytosis at active zones, we reasoned that reducing presynaptic Ca2+ influx could promote regeneration by attenuating Munc13-mediated inhibition of axon growth. To test this in a clinically relevant manner, we explored the regenerative efficacy of the GABAB receptor agonist Baclofen, an anti-spasticity medication that depresses synaptic transmission (Fox et al., 1978). First, we determined whether Baclofen administration could indeed reduce voltage-gated presynaptic Ca2+ influx in adult DRG neurons by performing whole-cell voltage-clamp recordings of dissociated DRG neurons and isolating their Ca2+ currents using a combination of pharmacological blockers and ion substitution (Dolphin and Scott, 1986; Husch et al., 2008). In adult DRG neurons, Baclofen treatment reduced the amplitude of voltage-gated Ca2+ currents to 79.3% ± 11.3% of baseline values (Figures 8A–8D).
Figure 8.
Baclofen promotes axon regeneration following spinal cord injury
(A) Whole-cell recording of voltage-activated calcium currents in a cultured DRG neuron one day in vitro. The holding potential was −60 mV, and the calcium current was evoked by 100 ms voltage steps from −60 to 50 mV in 10 mV increments.
(B) Raw traces of calcium currents in cultured DRG neurons before and after bath application of Baclofen (100 μM).
(C) Plot of all DRG neurons indicating the reduction of the voltage-activated calcium current in response to Baclofen administration.
(D) Quantification of (C). Values are plotted as mean ± SEM; ∗∗∗p = 0.001 by Wilcoxon matched-pairs signed-rank test; n = 11 neurons.
(E) Representative fluorescence images of Tuj1 (red) and GFP/Cre-GFP (cyan)-immunolabeled DRG neurons from Munc13-1fl/flMunc13-2KO/KOMunc13-3KO/KO mice administered AAV-GFP or AAV-Cre-GFP and cultured for 15–16 h in the presence of vehicle or Baclofen (100 μM). Scale bar, 100 μm.
(F and G) Length of the longest axon (F) and branching frequency (G) of DRG neurons cultured for 15–16 h in the presence of vehicle or Baclofen (10 μM, 100 μM, or 500 μM). Values are plotted as mean ± SEM; ∗p < 0.05 vehicle versus 10 μM, ∗∗p < 0.01 vehicle versus 100 μM, vehicle versus 500 μM by one-way ANOVA followed by Tukey’s post hoc test in (F); ∗∗p < 0.01 vehicle versus 10 μM, ∗∗∗p < 0.001 vehicle versus 100 μM, vehicle versus 500 μM by one-way ANOVA followed by Tukey’s post hoc test in (G); n = 235 vehicle, 136 10 μM, 122 100 μM, 156 500 μM Baclofen-treated neurons from 3–4 independent experiments.
(H and I) Length of the longest axon (H) and branching frequency (I) of (E). Values are plotted as mean ± SEM; ∗p < 0.05 GFP + vehicle versus GFP + Baclofen, ∗∗p < 0.01 GFP + Baclofen versus Cre-GFP + vehicle, p = 0.9024 Cre-GFP + vehicle versus Cre-GFP + Baclofen in (H), ∗∗∗p < 0.0001 GFP + vehicle versus GFP + Baclofen, ∗p < 0.05 GFP + Baclofen versus Cre-GFP + vehicle in (I) by one-way ANOVA followed by Tukey’s post hoc test; n = 68 GFP + vehicle, 73 GFP + Baclofen, 62 Cre-GFP + vehicle, 58 Cre-GFP + Baclofen neurons from 3 independent experiments.
(J) Multiphoton tile scan of GFP+ sensory axons (yellow) and GFAP+ astrocytes (blue) in the unsectioned spinal cord after complete dorsal column SCI in the given conditions. R, rostral; C, caudal. Asterisks indicate lesion centers. Scale bar, 200 μm.
(K) Quantification of (J). Scatterplot with means; ∗∗p < 0.01, ∗p < 0.05, #p < 0.1 by permutation test. n = 8 animals per group.
See also Figure S5.
We next tested whether Baclofen could promote axon growth by culturing adult DRG neurons and exposing them to 10–500 μM of the drug starting 1 h after plating. At the concentrations tested, Baclofen enhanced DRG axon outgrowth and reduced branching relative to vehicle-control-treated neurons (Figures 8E–8G). To determine whether Baclofen promotes axon growth by attenuating Munc13 inhibition of axon growth, we virally delivered eGFP or Cre-eGFP to Munc13-1fl/flMunc13-2KO/KOMunc13-3KO/KO transgenic mice and plated their lumbar DRG neurons in the presence of Baclofen or vehicle-control 3 weeks later. While Baclofen stimulated axon growth in GFP+ control neurons, it had no additive effect on the outgrowth or branching of Munc13 triple-KO neurons (Figures 8E–8I). Thus, promotion of axon growth by Baclofen requires the presence of Munc13 protein.
To determine whether systemic Baclofen administration could promote axon regeneration after spinal cord injury, we lesioned the dorsal column sensory axons at T12 in adult mice. We began Baclofen administration 1 h after injury by intraperitoneally injecting the mice with 10 mg/kg of the drug or vehicle for control and administered Baclofen or vehicle via drinking water to the mice for 4 weeks total, with AAV-eGFP injected into the sciatic nerves of these mice starting 2 weeks after injury to trace their sensory axons. Consistent with previous results (Paredes and Agmo, 1989), we observed decreased mobility in mice treated with Baclofen. Whole-mount immunostaining and two-photon imaging revealed that systemic delivery of Baclofen-stimulated sensory axons to regenerate after spinal cord injury (Figure S5).
Previous work from our lab demonstrated that the blockade of Alpha2delta2 via administration of Pregabalin (PGB) could promote axon regeneration after spinal cord injury (Tedeschi et al., 2016). Therefore, we tested how the effect of PGB compared to that of Baclofen in promoting regeneration following injury. To this end, we performed concurrent dorsal column spinal cord injury lesions and sciatic nerve AAV-eGFP injections in adult mice and treated them twice daily with intraperitoneal injections of 46 mg/kg PGB, 5 mg/kg Baclofen, or vehicle control commencing 1 h after spinal cord injury. The dose of Baclofen used is within the human equivalent dose range used clinically (Cragg et al., 2019; Heetla et al., 2016; Nair and Jacob, 2016). Two weeks after spinal cord injury, both PGB and Baclofen stimulated sensory axon regeneration relative to vehicle-injected control mice (Figures 8J and 8K). At the lesion site and at short distances rostrally, the extent of axon regeneration induced by PGB and Baclofen was not different. However, at 400–600 and 600–800 μm rostral to injury, Baclofen administration stimulated more axon regeneration than PGB administration. Together, these data show that systemic administration of Baclofen stimulates sensory axon regeneration after adult spinal cord injury.
Discussion
We discovered a critical role for core components of the presynaptic active zone in suppressing the regenerative capacity of neurons in adulthood. As Munc13 proteins are essential for neurotransmitter release (Augustin et al., 1999; Varoqueaux et al., 2002) but dispensable for neurite growth and synaptogenesis in developing hippocampal neurons (Broeke et al., 2010; Sigler et al., 2017; Varoqueaux et al., 2002), our data indicate that axon regeneration and the maturation of functional synaptic circuitry rely on antagonistic molecular programs.
Identification of a gene expression signature that inversely correlates with axon growth and regeneration
Our findings indicate that the ability of neurons to effectively grow axons requires a process of “de-maturation” and loss of synaptic function. Immature or newly born neurons have an immense capacity to grow axons (Flynn et al., 2012; Goldberg et al., 2002), even in the injured adult central nervous system (Lu et al., 2012; Wictorin et al., 1990), but adult neurons do not. It was thought that mature neurons need to reactivate their growth program by re-expressing the genes that drive axon growth during development (Harel and Strittmatter, 2006; Hilton and Bradke, 2017). Interestingly, however, the acquisition of growth competence in adult DRG neurons is not strictly associated with the upregulation of genes that are also upregulated during developmental axon growth. Thus, growth-competent adult neurons might already contain the gene set necessary to drive axon growth and regeneration, at least on the effector level.
This could imply either that axon growth in developing neurons and in mature neurons operates through very different effector mechanisms or that a maturation program is initiated in adult neurons that inhibits further growth once synaptic connections are established. Consistent with the latter view, our analyses revealed that a common gene expression signature associated with the reacquisition of axon growth competence in adulthood is the downregulation of genes that are typically upregulated during the maturation of synaptic circuitry. In the context of both prolonged cell culture as well as following peripheral nerve injury, DRG neurons downregulate genes that are upregulated when axons reach their peripheral and central target fields (Hasegawa et al., 2007; Kitao et al., 1996; Ozaki and Snider, 1997; Sharma et al., 2020). This view is supported by the finding that adult DRG neurons, which stop growing their axons upon KCl-induced depolarization, initiate growth upon parallel inhibition of transcription (Enes et al., 2010), which would reduce the expression of growth inhibitory genes. In culture, the switch to axon growth competence is reflected by a transition from arborizing to elongating growth, where branch sites likely reflect the formation of presynaptic sites (Hoersting and Schmucker, 2021). Indeed, in vivo imaging of developing retinal ganglion cells showed that axon branch formation and synaptogenesis occur simultaneously during development, with new axon branches initiating from synapses (Meyer and Smith, 2006).
Consistent with this view, target interaction of developing axons leads to elongation arrest and pre-synapse assembly (Sanes and Lichtman, 1999; Sanes and Zipursky, 2020; Südhof, 2018). Interestingly, recent work in the developing mouse showed that target innervation is likely critical for DRG transcriptional maturation (Sharma et al., 2020). Specifically, disruption of target-derived nerve growth factor (NGF) prevents subtype-specific gene expression underlying the maturation of developing DRG neurons (Sharma et al., 2020). It is even possible that the loss of target innervation following peripheral injury leads to the loss of neuronal identity, which is then only re-specified following target reinnervation (Renthal et al., 2020). Together, our work supports a model where neuronal maturation broadly, and target innervation more specifically, restricts axon growth by expressing a set of genes that blocks further axon growth and, thereby, also regeneration. This gene expression signature involves, in fact, key components of the presynaptic active zone.
The relationship between axon growth and the active zone proteins RIM and Munc13
Our results indicate that RIM and Munc13 proteins inhibit axon regeneration. Previous work from our lab demonstrated that the VGCC auxiliary subunit Alpha2delta2 inhibits axon regeneration by enhancing Ca2+ influx (Tedeschi et al., 2016). RIM proteins cluster VGCCs (Kaeser et al., 2011), and there is evidence that Munc13 can modestly enhance Ca2+ influx (Calloway et al., 2015). Thus, we cannot unequivocally rule out the possibility that Munc13 and RIM proteins suppress axon growth via processes that converge on Ca2+ signaling. However, overexpression of Munc13-1 had a stronger effect in suppressing axon growth and enhancing branching than did overexpression of RIM1α, and Munc13 triple-KO had a stronger effect in promoting axon growth and reducing branching than did RIM1/2 ablation. In view of these data, and given the principal involvement of RIM proteins in tethering VGCCs (Kaeser et al., 2011) as compared to a rather minor role of Munc13s in Ca2+ signaling (Calloway et al., 2015), it suggests that RIM and Munc13 restrict axon growth via their functions in vesicle priming, docking, and fusion and not by affecting Ca2+ signaling.
Because axon growth and regeneration were not affected by the loss of VAMPs1–3, we postulate that such RIM- and Munc13-dependent and axon-growth-related secretory processes either do not involve VAMPs1–3 or can use other v-SNAREs instead. For example, Munc13 and RIM proteins are required for dense core vesicle fusion (Persoon et al., 2019; van de Bospoort et al., 2012), whose blockade might reduce autocrine signaling by trophic factors that affect axon growth. Furthermore, Munc13-family proteins are known to control lysosomal fusion processes in immune cells (Feldmann et al., 2003; Neeft et al., 2005), and it is possible that Munc13s1–3 have related functions in DRG neurons that affect axon-growth-related processes. Moreover, Munc13 and RIM might suppress axon growth by shifting membrane trafficking from constitutive to regulated exocytosis (Bloom and Morgan, 2011). Given that re-expression of Munc13-1 in Munc13-KO neurons restored the limited growth phenotype found in control neurons, it is likely that Munc13 suppresses regeneration cell-autonomously instead of relying on target-derived factors. This view is supported by our finding that Baclofen stimulates growth in cultured DRG neurons, given that monocultures of adult mouse DRG neurons do not form synapses (Malin et al., 2007). In this context, it is worth mentioning that the inhibitory effect is either independent of the released factor or occurs in an autocrine fashion, as growth effects are only observed in Munc13-KO neurons and not in adjacent Munc13-expressing neurons in the same culture dishes.
Axon regeneration and neuronal activity
The role of neuronal activity in axon growth following injury is complex. For example, sensory axons can be immobilized by synapse-like contacts on non-neuronal cells after injury (Di Maio et al., 2011; Filous et al., 2014). Moreover, an intact axonal process can suppress the regeneration of an injured axonal process (Lorenzana et al., 2015). In addition to Alpha2delta2, the L-type voltage-gated Ca2+ channel Cav1.2 also suppresses axon regeneration (Enes et al., 2010). Interestingly, while neurotransmitters and electrical activity elevate intracellular Ca2+ and suppress neurite outgrowth in molluscan (Helisoma trivolvis) neurons (Cohan and Kater, 1986; Haydon et al., 1984), high concentrations of Ca2+ channel blockers also suppress axon elongating and growth cone motility (Mattson and Kater, 1987), indicating that there is a “set point” for Ca2+ levels in the neuron that are optimal for axon growth (Kater et al., 1988). Additionally, pharmacogenetic or electrical excitation of DRG neurons can also promote axon regeneration (Gordon et al., 2009; Wu et al., 2020), as can housing mice in an enriched environment (Hutson et al., 2019), highlighting a growth promoting role of enhancing neuronal activity. One possibility underlying the differences between these studies may relate to the complex role of Ca2+ signaling. Specifically, global changes in Ca2+ signaling are likely to have different signaling effects in the neuron compared to changes in Ca2+ influx at the presynaptic active zone (West et al., 2001). Given that DRG neurons downregulate active zone components as they acquire growth competence, future studies will investigate how other regeneration-promoting strategies alter the function of the presynaptic active zone. We anticipate that the disruption of the presynaptic active zone may be a common feature underlying the promotion of axon regeneration in other paradigms (Blackmore et al., 2012; Liu et al., 2010).
Axon regeneration: Successive versus parallel combinations
Axon regeneration is only one critical step for functional recovery following spinal cord injury (Ramer et al., 2014). Indeed, the subsequent consolidation and maturation of new synaptic circuits are equally important, and efforts to stimulate regeneration in humans with spinal cord injury will need to occur in the context of rehabilitation therapy (Courtine and Sofroniew, 2019; Fouad and Tetzlaff, 2012). Given the importance of Munc13 for synaptic vesicle release itself, we postulate that regenerating lost circuits following adult injury will require a process consisting of axon regenerating treatments prior to activity-based therapies to consolidate newly grown axons into functional circuits. This idea is well supported by the demonstration that asynchronous therapy with a growth-promoting therapy being instigated prior to and separate from rehabilitation therapy is maximally efficacious at promoting functional recovery following stroke (Wahl et al., 2014). In this regard, Baclofen may have therapeutic potential as a growth-promoting therapy. Baclofen is a GABAB receptor agonist used primarily to treat spasticity following spinal cord injury, cerebral palsy, and other neurological disorders (Albright et al., 1991; Loubser et al., 1991). A recent retrospective analysis found that even after adjusting for injury severity, Baclofen use was associated with higher rates of marked neurologic recovery in individuals with spinal cord injury (Cragg et al., 2019). Although such recovery of motor function might relate to the effect of Baclofen on muscle spasms, its association with enhanced function in pinprick and light touch scores indicates that it might enhance the functionality of injured DRG neurons.
STAR★Methods
Key resources table
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Tubulin β 3 (TUBB3) | Sigma-Aldrich | T2200; RRID: AB_262133 |
| Chicken anti-GFP | Abcam | ab13970; RRID: AB_300798 |
| Mouse anti-tubulin β 3 (Tuj-1) | Biolegend | 801201; RRID: AB_2313773 |
| Mouse anti-VAMP2 | Synaptic Systems | 104211; RRID: AB_887811 |
| Rabbit anti-Munc13-1 | Synaptic Systems | 126103; RRID: AB_887733 |
| Rabbit anti-RIM1 | Synaptic Systems | 140103; RRID: AB_887774 |
| Rabbit anti-RIM2 | Synaptic Systems | 140 103; RRID: AB_88776 |
| Rabbit anti-Glial Fibrillary Acidic Protein (GFAP) | DAKO | Z0334; RRID: AB_10013382 |
| Rabbit anti-RFP | Rockland | 600-401-379; RRID: AB_2209751 |
| Mouse anti-DDK (FLAG) | Origene | TA50011-100; RRID: AB_2622345 |
| Guinea Pig anti-NeuN | Merck Millipore | ABN90P; RRID: AB_2341095 |
| Donkey anti-chicken IgY IgG H+L Alexa Fluor 488 | Jackson Immunoresearch | 703-545-155; RRID: AB_2340375 |
| Donkey anti-rabbit IgG H+L Alexa Fluor 594 | Jackson Immunoresearch | 711-586-152; RRID: AB_2340622 |
| Donkey anti-guinea pig IgG H+L Alexa Fluor 647 | Jackson Immunoresearch | 706-605-148; RRID: AB_2340476 |
| Goat anti-rabbit IgG H+L Alexa Fluor 568 | Thermo Fisher | A-11036; RRID: AB_10563566 |
| Goat Anti-chicken IgY H&L Alexa Fluor 488 | Abcam | ab150169; RRID: AB_2636803 |
| Goat anti-rabbit IgG H&L Alexa Fluor 647 | Abcam | ab150079; RRID: AB_2722623 |
| Goat anti-Mouse IgG H+L Alexa Fluor 647 | Thermo Fisher | A-21235; RRID: AB_2535804350 |
| Goat anti-Mouse IgG H+ Alexa Fluor 594 | Thermo Fisher | A-11032; RRID: AB_2534091 |
| Bacterial and virus strains | ||
| AAV5.CAG-tdTomato (codon diversified) | A gift from Edward Boyden | Addgene #59462-AAV5 |
| AAV1.CMV.PI.EGFP.WPRE.bGH | A gift from James M. Wilson | Addgene #105530-AAV1 |
| AAV1.CMV.HI.eGFP-Cre.WPRE.SV40 | A gift from James M. Wilson | Addgene #105545-AAV1 |
| AAV1.CAG.Cre.mCherry | SignaGen | SL101117 |
| AAV5.hSyn.hM4D(Gi)-mCherry | A gift from Bryan Roth | Addgene #50475-AAV5 |
| AAV1.CMV-TeNT-LC-P2A-mCherry | Sirion Biotech | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| HBSS | GIBCO | 14025-053 |
| HEPES | GIBCO | 15630-56 |
| DMEM/F-12, HEPES | Life Technologies | 11330032 |
| Fetal bovine serum | Thermo Scientific | Cat#10500064 |
| Penicillin/Steptomycin (PenStrep) antibiotics | GIBCO | 15140122 |
| Collagenase IV | Sigma | C1889 |
| Trypsin | Worthington | LS003703 |
| DNase | Worthington | LS002007 |
| Neurobasal medium | GIBCO | 12349-015 |
| B-27 supplement | GIBCO | 17504-044 |
| L-glutamine | GIBCO | 25030-024 |
| Bovine serum albumin | Sigma | A3294 |
| Horse serum | Pan Biotech | P30-0712 |
| Poly-L-lysine | Sigma | P2636 |
| Laminin | Roche | 11243217001 |
| Phosphate buffer solution 0.1 M | Sigma | P5244 |
| Phosphate buffered saline (PBS) | AppliChem | A0965,9050 |
| Sucrose | Fluka | 84100 |
| Paraformaldehyde | Merck Millipore | 104005 |
| Dimethyl sulfoxide (DMSO) | Sigma | D5879 |
| Fluoromount | Sigma | F4680-25 |
| Phosphatase inhibitor | Roche | 04906837001 |
| Protease inhibitor | Roche | 11836170001 |
| Bradford reagent | AppliChem | A6932 |
| Ponceau S | AppliChem | A2935 |
| FastGreen | Sigma | 68724 |
| Goat serum | Invitrogen | 16210064 |
| Donkey serum | Jackson ImmunoReasearch | JIM-017-000-121 |
| Triton X-100 | Sigma | X100 |
| Baclofen | Sigma | B5399 |
| Pregabalin | Tocris | 3775 |
| Critical commercial assays | ||
| P3 Primary Cell 4D X Kit S (32 RCT) | Lonza | V4XP-3032 |
| ECL SuperSignal Dura West | Thermo Fisher Scientific | 10220294 |
| ECL SuperSignal Pico West | Thermo Fisher Scientific | 15513766 |
| Experimental models: Organisms/strains | ||
| Mouse: Munc13-1fl/fl | Banerjee et al., 2020 | N/A |
| Mouse: Munc13-2−/− (unc13btm1Rmnd) | Varoqueaux et al., 2002 | RRID: IMSR_EM:11739 |
| Mouse: Munc13-3−/− (unc13ctm1Bros) | Augustin et al., 2001 | RRID: IMSR_EM:11617 |
| Mouse: RIM1fl/fl | Kaeser et al., 2008 | RRID: JAX:015832 |
| Mouse: RIM2fl/fl | Kaeser et al., 2011 | RRID: IMSR_JAX:01 5833 |
| Mouse: iBOT | Slezak et al., 2012 | RRID: IMSR_JAX:018056 |
| Recombinant DNA | ||
| ef1a-GFP-RIM1α | Müller et al., 2020 | N/A |
| pmaxGFP | Lonza | Cat#V4XP-3024 |
| Unc13a (Myc-DDK-tagged) | Origene | MR225156 |
| Munc13-1-H567K | Betz et al., 1998 | N/A |
| Munc13-1-DN | Shin et al., 2010 | N/A |
| Munc13-1-W464R | Lipstein et al., 2013 | N/A |
| pCSCMV:tdTomato | Waldner et al., 2009 | Addgene # 30530 |
| Software and algorithms | ||
| ImageJ | NIH, USA | N/A |
| AxioVision | Zeiss | N/A |
| ZEN | Zeiss | N/A |
| SoftWoRx 3.5.0 | Applied Precision | N/A |
| MetaMorph Microscopy Automation and Image Analysis Software | Molecular Devices | N/A |
| Imaris 9.1 | Bitplane | N/A |
| Other | ||
| 35 mm glass bottom dish, No. 1.5 Coverslip, 20 mm, Uncoated | MatTek | Cat#P35G-1.5-20-C |
| 6-well plates | Thermo Scientific | Cat#140675 |
| 4-well plates | Thermo Scientific | Cat#179820 |
| 13 mm coverslip | Marienfeld | Cat#01-11530 |
| Immobilon-PSQ PVDF Membrane | Millipore | Cat#ISEQ00010 |
| X-ray film | Thermo Scientific | Cat#34076 |
| ImmEdge pen | Vector laboratories | Cat#H-4000 |
| Fluorescence microscope | Zeiss | Axio Observer D1 |
| Live cell microscope | Applied Precision | DeltaVision RT |
| Confocal microscope | Zeiss | LSM700 |
| 2-photon microscope | Zeiss | LSM 7MP |
| Super-Resolution confocal microscope | Zeiss | LSM980 |
| Photometrics CoolSnap HQ camera | Roper Scientific | N/A |
| CCD camera | Zeiss | N/A |
| Heating System | Ibidi | Cat#10918 |
| Heating Insert μ-Dish 35 mm high for ibidi Heating System | Ibidi | Cat#10934 |
| Gas Incubation System for CO2 | Life Imaging Services | N/A |
| Cryostat | Leica | CM3050S |
| CaWomat | Cawo | 2000IR |
| Pneumatic picopump (picospritzer) | WPI | Cat#SYS-PV820 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Frank Bradke (frank.bradke@dzne.de).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact without restriction.
Experimental model and subject details
Animals
Experiments were conducted in accordance with the guidelines of the Animal Welfare act and the Landesamt für Natur, Umwelt und Verbraucherschutz (LANUV). Experiments were also performed in accordance with the guidelines of animal research: reporting in vivo experiments (ARRIVE) (Kilkenny et al., 2010). Adult female wild-type (WT, C57BL/6J, Charles River) and transgenic mice of both sexes (8-12 weeks old) were used. Conditional Munc13-1 KO mice were generated by introducing loxP sites upstream and downstream of exon 21 of the Munc13-1 gene in embryonic stem cells by homologous recombination and Munc13-1fl/fl mice were crossed onto a homozygous Munc13-2 KO (Varoqueaux et al., 2002) and Munc13-3 KO (Augustin et al., 2001) background as previously described in detail (Banerjee et al., 2020). iBOT B6.FVB-Tg(CAG-boNT/B,-EGFP)U75-56Fwp/J mice were purchased from the Jackson Laboratories (#018056). RIM1fl/flRIM2fl/fl mice were provided by the coauthors Prof. Dr. Susanne Schoch-McGovern and Dr. Johannes Alexander Müller. Animals were randomized into treatment and control groups and analyses were conducted blinded to treatment group.
Method details
Peripheral nerve lesion (PNL)
Mice were anesthetized with isoflurane (5% for induction and 1.5% for maintenance). The left sciatic nerve was exposed at the mid-thigh level, ligated with Ethilon 5-0 suture and cut with Iris scissors distal to the ligation site. The nerve was then placed back in the leg and the skin closed with Reflex 7-mm skin-closing clips. In sham-operated control mice, the left nerve was exposed and then placed back in the leg.
AAV transduction
Mice were anesthetized with a mixture of ketamine and xylazine (100 mg/kg and 10 mg/kg respectively). The mid and lower back of the mouse was shaved and disinfected with a combination of 70% ethanol and betadine. L3, L4, and/or L5 DRG were exposed and injected with 0.5-1 μl of AAV combined 10:1 with Fast Green FCF using a Picospritzer equipped with a pulled glass micropipette (pressure: 30 psi, pulse duration: 8 ms). Alternatively, mice were anesthetized with isoflurane (5% for induction and 1.5% for maintenance). The left leg was shaved and disinfected as above, the sciatic nerve was exposed at the mid-thigh level and a pulled glass micropipette was placed into the sciatic nerve along its longitudinal axis. 1-2 μl of AAV mixed 10:1 with Fast Green FCF was injected into the sciatic nerve. AAV particles were injected at a titer of 0.4-1∗1013 genome copies (GC)/ml and titer matched in individual experiments. AAV5-hSyn-hM4D(Gi)-mCherry was a gift from Bryan Roth (Addgene plasmid # 50475-AAV5 ; http://addgene.org/50475; RRID:Addgene_50475). AAV5-CAG-tdTomato (codon diversified) was a gift from Edward Boyden (Addgene plasmid # 59462-AAV5; http://addgene.org/59462; RRID:Addgene_59462). AAV1.CMV.PI.EGFP.WPRE.bGH was a gift from James M. Wilson (Addgene plasmid #105530-AAV1 ; http://addgene.org/105545; RRID:Addgene_105530). AAV1.CMV.HI.eGFP-Cre.WPRE.SV40 was a gift from James M. Wilson (Addgene plasmid #105545-AAV1; http://addgene:105545; RRID:Addgene_105545). AAV1.CAG.Cre.mCherry was purchased from SignaGen (SL101117). Immediately after injection, the skin was closed with 7 mm skin clips.
For in vitro studies of the effect of Munc13-KO on axon outgrowth, the left and right sciatic nerves were injected three weeks prior to culture. For the in vivo DREADD experiment, AAV-hM4Di or tdTomato was injected into the left lumbar L3, L4, and L5 DRG of adult WT mice three weeks prior to PNL and four weeks prior to SCI. Two weeks after SCI, AAV-GFP was injected into the left sciatic nerve to trace dorsal column sensory axons. For in vivo imaging, AAV-hM4Di or tdTomato was co-injected with AAV-GFP (1:1 mixture) into the L4 DRG of adult WT mice. For the Munc13-KO in vivo experiment, AAV-Cre-GFP was co-injected with AAV-GFP (1:1 mixture) or AAV-GFP alone was injected into the left sciatic nerves of Munc13-1fl/fl Munc13-2KO/KO Munc13-3KO/KO mice.
TeNT-P2A-mCherry construct
pAAV.CMV.PI.TeTxLC.P2A.mCherry.WPRE.bGH was cloned using pAAV.CMV.PI.EGFP.WPRE.bGH plasmid (gift from James M. Wilson ; Addgene plasmid #105530 ; http://addgene:105530 ; RRID:Addgene_105530) with GFP being replaced by PCR products of TeTxLC-P2A and mCherry. The backbone (digested with NotI and HindIII to remove GFP) was assembled with the PCR products using the NEBuilder HiFi DNA Assembly Master Mix (NEB, #E2621L). TeTxLC-P2A was amplified with upstream overhanging ends from the backbone in the forward primer (CCTTTCTCTCCACAGGTGTCCAGGC) and downstream overhanging ends for mCherry in the reverse primer (GTTATCCTCCTCGCCCTTGCTCACCAT) from pAAV.hSyn.FLEX.TeLC.P2A.dTomato (gift from Sandeep Datta ; Addgene plasmid #159102; http://addgene.org/159102; RRID:Addgene_159102; Pashkovski et al., 2020; forward primer: CCTTTCTCTCCACAGGTGTCCAGGCATGCCGATCACCATCAACAACTTCC ; reverse primer: GTTATCCTCCTCGCCCTTGCTCACCATAGGACCGGGGTTTTC). MCherry was amplified with upstream overhanging ends for P2A in the forward primer (GAGACGTGGAAGAAAACCCCGGTCCT) and downstream overhanging ends from the backbone in the reverse primer (GTAATCCAGAGGTTGATTGGATCCA; forward primer: GAGACGTGGAAGAAAACCCCGGTCCTATGGTGAGCAAGGGCGAGG ; reverse primer: GTAATCCAGAGGTTGATTGGATCCATTACTTGTACAGCTCGTCCATGC).
Spinal cord injury (SCI)
Mice were anesthetized via intraperitoneal injection of ketamine (100 mg/kg body weight) and xylazine (10 mg/kg body weight). SCI was performed similarly to as previously described (15). Briefly, a T11-T12 laminectomy was performed and the spinal cord was crushed with modified #5 forceps (Dumont, FST) to sever dorsal column axons completely. Mice were transcardially perfused with 20 mL of PBS containing 10 U/ml Heparin followed by 40 mL of 4% paraformaldehyde one to four weeks following injury. Tissue was then post-fixed in 4% paraformaldehyde overnight and washed 5 times in PBS.
DREADD experiments
Three weeks after DRG injections, mice were anesthetized with isoflurane and a left PNL or sham operation performed as described above. Immediately following PNL or sham operation, the mouse was intraperitoneally injected with 0.1 mg/kg clozapine dihydrochloride (HB6129; HelloBio) and returned to their homecage where clozapine (1 mg / 200 ml) was delivered via drinking water. The drinking water containing clozapine was replenished every 3-4 days until endpoint.
Baclofen SCI experiments
Two different SCI experiments were conducted to assess the regenerative efficacy of Baclofen. In both experiments, adult mice were randomized into treatment groups and T12 dorsal column lesions were conducted as above. In the first experiment, one hour after injury, mice were injected intraperitoneally with 10 mg/kg Baclofen (B5399, Sigma) or saline control. In Baclofen injected mice, Baclofen was then administered via drinking water at a dosage of 1 mg/ml for four weeks until endpoint with water refreshed every 3-4 days. Sciatic nerve tracing with AAV-eGFP was conducted as above 2 weeks after injury and 2 weeks prior to endpoint. In the second experiment, sciatic nerve tracing with AAV-eGFP was conducted as above immediately after T12 dorsal column lesion. One hour after injury, mice were injected intraperitoneally with 5 mg/kg Baclofen (B5399, Sigma), 46 mg/kg Pregabalin (PGB; 3775, Tocris), or saline control. Mice were then injected with 5 mg/kg Baclofen, 46 mg/kg PGB, or saline every 10-14 h morning and evening for 2 weeks post-injury and then sacrificed.
Whole-mount immunostaining
The dura of isolated adult mouse spinal cords was dissected off and the cords cut 5 mm rostral and 5 mm caudal to the lesion site. Samples were permeabilized in PBS containing 0.2% Triton X-100, 2.3% Glycine, and 20% DMSO overnight at 37°C with 100 rpm shaking, and then blocked in PBS containing 6% Normal Donkey Serum, 10% DMSO and 0.2% Triton X-100 overnight at 37°C with 100 rpm shaking. Samples were then incubated in primary antibody solution: 1x PBS containing 0.2% Tween-20, 0.01 mg/ml Heparin, 5% DMSO, 3% Donkey serum, 1:500 Chicken anti-GFP (Abcam #ab13970) and 1:500 Rabbit anti-GFAP (DAKO #Z0334) at 37°C with 100 rpm shaking for 3 days. Samples were washed 5x overnight in PBS containing 0.2% Tween-20, 0.01 mg/ml Heparin and incubated in secondary antibody solution containing 1:300 Donkey anti-Chicken AlexaFluor 488 and 1:300 Donkey anti-Rabbit Alexafluor 594 at 37°C with 100 rpm shaking for 2 days. Samples were then washed 5x overnight in 1x PBS containing 0.2% Tween-20, 0.01 mg/ml Heparin and imaged in washing media.
Neuronal culture
L3-L5 DRG were dissected, collected and rinsed in ice-cold Hank’s balanced salt solution (HBSS, GIBCO) supplemented with 7 mM HEPES. Ganglia were incubated in collagenase type IV (C1889, 1.25 mg/ml, Sigma) at 36.5°C for 90 min, washed three times in DMEM/F12, and incubated in Trypsin (0.05%, GIBCO) at 36.5°C for 10 min. Ganglia were then washed three times in DMEM/F12 containing 5% fetal calf serum and 1% Penicillin/Streptavidin, then dissociated by repeated pipetting. For live-cell imaging, cells were transferred to a 15 mL tube containing 2 mL of 15% BSA in DMEM/F12 and centrifuged at 900 rpm for 10 min. For all other cell culture experiments, immediately after dissociation the cell suspension was filtered through a nylon cell strainer (70 μm, BD Falcon) and centrifuged at 900 rpm for 5 minutes. For electroporation experiments, after centrifugation, the media was replaced with 20 μl of P3 primary cell nucleofector solution containing plasmid DNA and electroporated with the 4D Amaxa Nuclefector system (program DC-100). Dissociated DRG neurons were electroporated with sequence verified CMV-tdTomato (4 μg; Addgene plasmid # 30530; http://addgene.org/30530; RRID:Addgene_30530), pmaxGFP (4 μG, Lonza # V4XP-3024), CMV-Munc13-1-DDK (4 μg; Origene plasmid #MR225156), CMV-Munc13-1-DN-GFP (4 μg), CMV-Munc13-1-H567K-GFP (4 μg), or CMV-Munc13-1-W464R-GFP (4 μg) expressing plasmid DNA. Cells were then resuspended in DMEM/F12 supplemented with B-27, Glutamine, and Pen/Strep and plated at low density on poly-L-lysine (1 mg/ml in borate buffer, Sigma) and laminin (5 μg/ml, Roche) coated dishes or coverslips and incubated at 36.5°C in a humidified chamber containing 5% CO2. For live-cell imaging, cells were plated on 35 mm glass bottom dishes (MatTek). For immunocytochemistry, cells were plated on 13 mm glass coverslips (Marienfeld). Cell media was refreshed 1-2 h after plating. In a subset of experiments, Baclofen (10-500 μM, Sigma, B5399), KCl (40 mM, Mettler Toledo, 51350072), Roscovitine (20 μM, Sigma, R7772), GV-58 (20 μM, Sigma, SML1551), or vehicle (DMSO, Carl Roth, 4720.4) were added during cell media replenishment 1 h after plating.
Immunocytochemistry
For morphometric analysis of DRG axon outgrowth and branching, cultures were fixed with 4% paraformaldehyde, 4% sucrose for 15 min, washed 3x with PBS, quenched for 10 min with 50 mM NH4Cl, washed 3 times with PBS, and extracted with 0.1% Triton X-100 in PBS for 3 min. After being washed 3 times with PBS, the cells were blocked with 2% BSA, 2% FCS, and 0.2% fish gelatin in PBS at room temperature for 1 h. After blocking, cells were incubated in the following antibodies depending on the experiment: Rabbit anti-βIII tubulin (1:500, Tuj1, MMS-435P, Covance), Mouse anti- βIII tubulin (1:500, TUBB3, 801201, BioLegend), chicken anti-GFP (1:500, ab13970, Abcam); Rabbit anti-Red Fluorescent Protein (RFP, 1:1000, 600-401-379, Rockland), Rabbit anti-RIM1 (1:200, 140 003, Synaptic Systems), Rabbit anti-RIM2 (1:200; 140 103, Synaptic Systems) and/or mouse anti-DDK (FLAG) (1:500, TA50011-100, OriGene) in PBS containing 10% blocking solution at room temperature for 1 h. Cells were then washed 3x with PBS and incubated in secondary antibodies (1:500) at room temperature for 30 min. Cells were then washed 3x with PBS, washed once with dH2O, dried, and mounted in Fluouromount (Sigma) on microscope slides.
For super-resolution microscopy, cultures were washed 3x with PBS and fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (PB) at room temperature for 30 min. Coverslips were washed 3x with 0.1 M PB and incubated in blocking buffer (0.1 M PB, 10% donkey serum, 0.3% Triton X-100, 0.1% fish gelatin) for 45 min. Coverslips were then incubated in 0.1 M PB, 5% donkey serum, 0.1% Triton X-100 containing primary antibodies for 1 h, washed 3x with 0.1 M PB, and incubated in 0.1 M PB, 5% donkey serum, 0.1% Triton X-100 containing secondary antibodies for 30 min. Coverslips were then washed 3x with 0.1 M PB and mounted in Fluouromount onto slides. The following primary antibodies were used: rabbit anti-Munc13-1 (1:1000, Synaptic Systems #126 103), mouse anti-βIII tubulin (1:500, Tuj1, MMS-435P, Covance).
Immunohistochemistry on tissue sections
Fixed tissues were incubated in 30% sucrose at 4°C for 3 days. Tissues were embedded in optimum cutting temperature (OCT) compound, frozen, cut into 20 μm sections (CM3050S, Leica) onto SuperFrost glass slides, and stored at −80°C. Frozen slides were thawed at room temperature (RT) for one h. Sections were washed in PBS for 5 min, and then blocked at RT with 10% normal donkey serum (NDS) in PBS containing 0.1% Triton X-100 (PBST) for 1 h. Sections were incubated in PBST containing primary antibodies at room temperature overnight. After washing the samples 3 × 5 min with PBS, sections were incubated with AlexaFluor conjugated secondary antibodies (1:200, Invitrogen) for 2 h and then washed 3 × 5 min with PBS. Samples were then mounted in Fluromount and stored at 4°C until imaging. Images were taken using an LSM700 confocal microscope (Zeiss). The following primary antibodies were used: Rabbit anti-RFP (1:1000, 600-401-379, Rockland), chicken anti-GFP (1:500, ab13970, Abcam), Guinea pig anti-NeuN (1:200, ABN90P, Merck Millipore).
Microscopy
For in vivo imaging, mice were anesthetized via intraperitoneal injection of ketamine (100 mg/kg body weight) and xylazine (10 mg/kg body weight). Vertebral level T12 was identified as the apex of the vertebral curvature, and small incisions were made parallel to the spinal cord on both sides of the dorsal processes. A laminectomy was performed to expose the spinal cord under the T12 vertebrae and the spinal cord was stabilized with spinal cord holders (Narishige) fixed to a custom-made 3D printed insert and clamped at the vertebrae rostral and caudal to the laminectomy. A small incision of the spinal cord to a depth of 0.3 mm was made with 2 mm Cutting Edge Vannas Spring Scissors (Fine Science Tools; #15000-03). Immediately thereafter (15-60 min after injury), intravital imaging was performed with a multiphoton microscope (LSM 7MP, Zeiss) using a 0.80 NA 16x objective to verify that axons were lesioned. At 3 days post-injury, the lesion site was re-exposed and intravital imaging repeated.
For live-cell imaging, cells were mounted on an Apotome.2 (Zeiss) microscope equipped with an Ibidi heating system to maintain the cells at 37°C and 5% CO2 incubation. Images were taken with a 10x objective lens using the microscope’s differential interference contrast (DIC) function every 10 min for 36 h. For morphological analysis of DRG axon growth and branching, images were taken using an LSM 700 confocal microscope (Zeiss) with a 10x objective lens. For whole mount tissue imaging analysis, spinal cords were imaged with an LSM 7MP (Zeiss) mounted with a 16x objective lens as previously described (Hilton et al., 2019). For super-resolution microscopy, images were taken with an LSM980 system (Zeiss) equipped with a 40x objective lens using the system’s Airyscan 2 function.
Electrophysiology
Whole cell patch-clamp recordings were used to characterize the impact of clozapine dihydrochloride on hM4Di-mCherry+ or tdTomato+ dissociated DRG neurons. Cells were electroporated as above with 4 μg of hSyn-hM4Di-mCherry or CMV-tdTomato expressing constructs and plated on PLL and laminin-coated coverslips in DMEM/F12 for 2-7 days, with media refreshed every 2 days until endpoint. Whole-cell current-clamp recordings were performed with a Multi-Clamp 700B patch-clamp amplifier (Axon Instruments) and an Axon Digidata 1550B (Axon Instruments) digitizer controlled by Multiclamp and Clampex (MolecularDevices) running under Windows. The electrophysiological data were sampled at intervals of 100 μs (10 kHz). The recordings were low pass filtered at 2 kHz with a 4-pole Bessel-Filter. The cells were transferred from the culture medium to a Ringer solution containing (mM) 145 NaCl, 3 KCl, 1 MgCl, 1.5 CaCl2, 10 HEPES, 10 D-glucose (adjusted to pH 7.3 with NaOH). Freshly pulled borosilicate glass pipettes (3-6 MΩ) were filled with an intracellular solution containing (mM) 135 K-gluconate, 10 KCl, 10 HEPES, 0.1 EGTA, 2 MgCl2, 3 K-ATP, 0.3 Na-GTP (adjusted to pH 7.2 with KOH). Recordings were carried out at 31-32°C controlled by Temperature Controller VII (Luigs-Nemann) in flowing Ringer solution. Resting membrane potential was measured in current clamp mode. Membrane input resistance was calculated based on voltage deflection in response to negative current injections.
Whole-cell voltage-clamp recordings were used to characterize the impact of Baclofen on voltage-activated calcium currents of dissociated adult mouse DRG neurons. Cells were plated on PLL and laminin-coated coverslips in DMEM/F12 as above. To isolate the Ca2+ currents, a combination of pharmacological blockers and ion substitution was used. Transient voltage-gated Na+ currents were blocked by tetrodotoxin (TTX) (10−7–10−4 M; T-550; Alomone). 4-Aminopyridine (4-AP) (4 × 10−3 M; A78403; Aldrich) was used to block transient K+ currents (IA) and tetraethylammonium (TEA) (2 × 10−2 M; T2265; Sigma-Aldrich) was used to block sustained K+ currents (IK(V)) as well as Ca2+-activated outward currents (IO(Ca)). A test pulse to 0 mV every 10 s was executed to visualize the amplitude over time, Baclofen (100 μM, Sigma) was applied, and test pulses were applied to assess Ca2+ current amplitude. Calcium current amplitudes were normalized to the average amplitude during the 10 s prior to Baclofen administration. The linear time dependent rundown of the calcium current amplitudes was compensated by subtracting a linear fit from the amplitudes over time.
Protein extraction and immunoblotting
DRG were dissected, rinsed in ice-cold HBSS supplemented with 7 mM HEPES and snap frozen in liquid nitrogen. The samples were ground with a micropistille in liquid nitrogen and lysed on ice in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.25% sodium deoxycholate, 0.1% sodium dodecyle sulfate) containing phosphatase (phosSTOP, Roche) and protease inhibitors (Complete, Roche), then centrifuged. 5 μg of protein lysates were fractionated by 10%–12% SDS-polyacrylamide gel electrophoresis (PAGE). The samples were then transferred to a polyvinylidene difluoride (PVDF, Millipore) membrane and stained with Ponceau S (Sigma) to confirm loading mass consistency between samples. The samples were then blocked with 5% non-fat milk and 5% normal goat serum in TBS-T or 3% BSA in TBS-T at RT for 40 min, probed with Rabbit anti-Munc13-1, RIM1, RIM2, or Mouse anti-VAMP2 antibody as well as Rabbit anti-TUJ1 antibody at RT for 3 h, washed 3x in TBS-T and incubated with horseradish peroxidase conjugated secondary antibodies (1:20000, GE Healthcare) for 40 min. For protein detection, the membrane was incubated with SuperSignal West Pico PLUS Chemiluminescent Substrate or SuperSignal West Dura Extended Duration Substrate solution. The following primary antibodies were used: anti-VAMP2 (1:1000, Synaptic Systems #104 211, mouse monoclonal), anti-Munc13-1 (1:2000, Synaptic Systems #126 103, rabbit polyclonal), anti-RIM1 (1:500; Synaptic Systems #140 003, rabbit polyclonal), anti-RIM2 (1:500; Synaptic Systems #140 103, rabbit polyclonal), anti-tuj1 (1:10000, Sigma #T2200, rabbit polyclonal).
Quantification and statistical analysis
Transcriptomic analysis
Transcriptomic analysis was performed on bulk RNA-Seq of the DRG using a previously published dataset (GEO: GSE66128) (Tedeschi et al., 2016). Rank-rank hypergeometric overlap analysis was conducted with RRHO2 (Cahill et al., 2018) on the datasets to compare gene expression patterns with conditioning (PNL versus Sham), embryonic development (E12.5 versus E17.5), and time in culture (6 h versus 12 h, 24 h, 36 h). Genes encompassed in the Gene Ontology (GO):0045202 term “synapse” (n = 1459) were quantified and triplicate experiments were statistically compared at 06h versus 36 h in culture. The normalization of original expression matrix and differential gene expression analysis were conducted using DESeq2 (Love et al., 2014). Differentially expressed genes (n = 517) were screened with a cut-off of FDR-adjusted p < 1∗10−5 and a threshold of FDR = 0.001. To visualize expression changes in each of the three paradigms: 6 h, 12 h, 24 h, and 36 h in culture; PNL versus sham; and E12.5 versus E17.5, the normalized expression matrix was processed with a regularized-logarithm transformation to stabilize variance across the mean and the mean was subtracted from the transformed expression value. The associated heatmaps were generated with pheatmap (Kolde, 2015). Gene ontology enrichment analysis was performed on the genes found to be upregulated at E17.5 versus E12.5 and downregulated at 36 h in culture versus 6 h in culture using clusterProfiler (Yu et al., 2012). p value cutoff was 0.01 and q-value cutoff was 0.05. To reduce the redundancy of enriched GO terms, clusterProfiler::simplify was used with cutoff = 0.6. The above analysis and data visualization were conducted in R version 4.0.0 (https://www.r-project.org/). Gene ontology enrichment analysis was also performed on synaptic genes upregulated and downregulated at 6 h versus 36 h in culture using SynGO (https://syngoportal.org/) using brain expressed genes as background (Koopmans et al., 2019).
Imaging analysis
For live-cell imaging, the longest axon of each neuron was identified at t = 36 h and its growth rate analyzed by comparing the length at each time point. Axon growth velocity was binned into 12 h increments for each neuron. For morphological analysis of DRG axon growth and branching, the length of the longest axon and frequency of branches were calculated using ImageJ (NIH) and the Simple Neurite Tracer plugin (Longair et al., 2011). Puncta detection was performed using Imaris software (Bitplane, Zurich, Switzerland), versions 8 or 9 similarly as to previously described in detail (Reitz et al., 2021). Super-resolution image stacks were imported into Imaris and in the fluorescence channel corresponding to Munc13-1 immunolabeling, a spots detection function was applied with an XY diameter of 0.2 μm and background subtraction selected. The volume of the cell was calculated by applying a surface function in the fluorescence channel corresponding to Tuj1 immunolabelling, and the # of puncta / μm3 was calculated by dividing the total number of Munc13+ puncta by the Tuj1+ volume. For whole mount tissue imaging analysis, tiled z stack images were 3D rendered using Imaris 9.1.2 (Bitplane). The injury site was identified as the middle of the GFAP negative area and axons were manually annotated using the Imaris spot function. The number of regenerating axons at different distances from the lesion site was normalized to the number of annotated axons 200 μm – 400 μm caudal to the injury site. Mice with incomplete lesions based on axon morphology and the absence of a GFAP- area were excluded from analysis. For in vivo imaging, axon branchpoints were established as landmarks in the caudal spinal cord and used to verify the lesion epicenter to measure axon regeneration. For super-resolution microscopy, Z stack images were 3D rendered using Imaris 9.1.2 (Bitplane). 3D cell volume was calculated by using Imaris’ Surface function based on Tuj1 immunoreactivity.
Statistical analysis
Except when indicated, statistical tests were performed using GraphPad Prism version 8.0 software. Homogeneity of variance was tested for using Bartlett’s test. Normality was tested for using the Shapiro–Wilk test. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey’s post-test or by using a two-tailed unpaired Student’s t test or a Wilcoxon matched-pairs signed rank test as indicated in the figure legends. For in vivo regeneration experiments, statistical analysis was performed by permutation test using a custom script implemented in Python (2.7.3 version) (https://pypi.org/pypi/permutation_test). Statistical significance was set at p < 0.05.
Acknowledgments
We thank Volker Haucke, Jeffrey Macklis, Thorben Pietralla, Carla Shatz, Gaia Tavosanis, and Wolfram Tetzlaff for critically reading and discussing the manuscript; Jessica Gonyer, Liane Meyn, Anh-Tuan Pham, and Blanca Randel for technical assistance; and the DZNE Animal Research Facility (ARF), the Image and Data Analysis Facility (IDAF), and the Light Microscope Facility (LMF) for their support. We also thank Noa Lipstein (FMP) for providing plasmids. B.J.H. is supported by a Wings for Life (WfL) Aguayo-Tator Mentoring Fellowship and a non-stipendiary European Molecular Biology Organization (EMBO) Long-Term Fellowship (ALTF 28-2017). J.K. was supported by a Harvard College summer grant via the Macklis laboratory at Harvard University. S.S. is supported by grants from the DFG (SCHO 820/4-1, SCHO 820/6-1, SCHO 820/7-1, SCHO 820/5-2, SPP1757, SFB1089) and the BONFOR program of the University of Bonn Medical Center. N.B. is supported by an ERC Advanced Grant (SynPrime). F.B. is supported by the Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), the International Foundation for Research in Paraplegia, WfL, the Deutsche Forschungsgemeinschaft, ERANET AXON REPAIR, and ERANET RATER SCI. F.B. is a member of the excellence cluster ImmunoSensation2 (EXC2151–390873048) and the SFB 1089 and is a recipient of the Roger de Spoelberch Prize.
Author contributions
B.J.H., N.B., and F.B. conceived of the project and designed experiments. B.J.H., A.H., B.S., E.R.B., M.S., S.D., and J.K. performed the experiments. B.J.H., B.S., A.H., T.-C.L., and J.K. analyzed the data. J.A.M., C.I., S.S., and N.B. provided mutant mice and plasmids. B.J.H. and F.B. wrote the paper. All of the authors provided feedback and contributed to editing the manuscript.
Declaration of interests
The authors declare no competing interests. N.B. is a member of the Neuron advisory board.
Inclusion and diversity
One or more of the authors of this paper self-identifies as living with a disability.
Published: October 26, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.neuron.2021.10.007.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code. Permutation tests were performed using a custom script “permutation_test 0.18” implemented in Python (2.7.3 version) including Pandas and NumPy libraries. The script is available for download in the Python Package Index (https://pypi.python.org/pypi/permutation_test). The sunburst plots in Figures 1 and S3 were generated using the SynGO Portal (https://www.syngoportal.org/). Figure 7C was created using BioRender (https://biorender.com).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Albright A.L., Cervi A., Singletary J. Intrathecal baclofen for spasticity in cerebral palsy. JAMA. 1991;265:1418–1422. [PubMed] [Google Scholar]
- Augustin I., Rosenmund C., Südhof T.C., Brose N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 1999;400:457–461. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- Augustin I., Korte S., Rickmann M., Kretzschmar H.A., Südhof T.C., Herms J.W., Brose N. The cerebellum-specific Munc13 isoform Munc13-3 regulates cerebellar synaptic transmission and motor learning in mice. J. Neurosci. 2001;21:10–17. doi: 10.1523/JNEUROSCI.21-01-00010.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Imig C., Balakrishnan K., Kershberg L., Lipstein N., Uronen R.-L., Wang J., Cai X., Benseler F., Rhee J.S. Molecular and functional architecture of striatal dopamine release sites. bioRxiv. 2020 doi: 10.1101/2020.11.25.398255. Published online November 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareyre F.M., Garzorz N., Lang C., Misgeld T., Büning H., Kerschensteiner M. In vivo imaging reveals a phase-specific role of STAT3 during central and peripheral nervous system axon regeneration. Proc. Natl. Acad. Sci. USA. 2011;108:6282–6287. doi: 10.1073/pnas.1015239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J., Shen N., Dulubova I., Lu J., Guan R., Guryev O., Grishin N.V., Rosenmund C., Rizo J. A minimal domain responsible for Munc13 activity. Nat. Struct. Mol. Biol. 2005;12:1017–1018. doi: 10.1038/nsmb1001. [DOI] [PubMed] [Google Scholar]
- Betz A., Ashery U., Rickmann M., Augustin I., Neher E., Südhof T.C., Rettig J., Brose N. Munc13-1 is a presynaptic phorbol ester receptor that enhances neurotransmitter release. Neuron. 1998;21:123–136. doi: 10.1016/s0896-6273(00)80520-6. [DOI] [PubMed] [Google Scholar]
- Betz A., Thakur P., Junge H.J., Ashery U., Rhee J.-S., Scheuss V., Rosenmund C., Rettig J., Brose N. Functional interaction of the active zone proteins Munc13-1 and RIM1 in synaptic vesicle priming. Neuron. 2001;30:183–196. doi: 10.1016/s0896-6273(01)00272-0. [DOI] [PubMed] [Google Scholar]
- Blackmore M.G., Wang Z., Lerch J.K., Motti D., Zhang Y.P., Shields C.B., Lee J.K., Goldberg J.L., Lemmon V.P., Bixby J.L. Krüppel-like Factor 7 engineered for transcriptional activation promotes axon regeneration in the adult corticospinal tract. Proc. Natl. Acad. Sci. USA. 2012;109:7517–7522. doi: 10.1073/pnas.1120684109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom O.E., Morgan J.R. Membrane trafficking events underlying axon repair, growth, and regeneration. Mol. Cell. Neurosci. 2011;48:339–348. doi: 10.1016/j.mcn.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Bradbury E.J., Burnside E.R. Moving beyond the glial scar for spinal cord repair. Nat. Commun. 2019;10:3879. doi: 10.1038/s41467-019-11707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeke J.H., Roelandse M., Luteijn M.J., Boiko T., Matus A., Toonen R.F., Verhage M. Munc18 and Munc13 regulate early neurite outgrowth. Biol. Cell. 2010;102:479–488. doi: 10.1042/BC20100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill K.M., Huo Z., Tseng G.C., Logan R.W., Seney M.L. Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci. Rep. 2018;8:9588. doi: 10.1038/s41598-018-27903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calakos N., Bennett M.K., Peterson K.E., Scheller R.H. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 1994;263:1146–1149. doi: 10.1126/science.8108733. [DOI] [PubMed] [Google Scholar]
- Calloway N., Gouzer G., Xue M., Ryan T.A. The active-zone protein Munc13 controls the use-dependence of presynaptic voltage-gated calcium channels. eLife. 2015;4:e07728. doi: 10.7554/eLife.07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V., Coppola G., Nawabi H., Omura T., Versano R., Huebner E.A., Zhang A., Costigan M., Yekkirala A., Barrett L., et al. A systems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron. 2016;89:956–970. doi: 10.1016/j.neuron.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan C.S., Kater S.B. Suppression of neurite elongation and growth cone motility by electrical activity. Science. 1986;232:1638–1640. doi: 10.1126/science.3715470. [DOI] [PubMed] [Google Scholar]
- Courtine G., Sofroniew M.V. Spinal cord repair: advances in biology and technology. Nat. Med. 2019;25:898–908. doi: 10.1038/s41591-019-0475-6. [DOI] [PubMed] [Google Scholar]
- Cragg J.J., Tong B., Jutzeler C.R., Warner F.M., Cashman N., Geisler F., Kramer J.L.K. A Longitudinal Study of the Neurologic Safety of Acute Baclofen Use After Spinal Cord Injury. Neurotherapeutics. 2019;16:858–867. doi: 10.1007/s13311-019-00713-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J.M., DePaul M.A., Filous A.R., Lang B.T., Tran A., Silver J. Functional regeneration beyond the glial scar. Exp. Neurol. 2014;253:197–207. doi: 10.1016/j.expneurol.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Maio A., Skuba A., Himes B.T., Bhagat S.L., Hyun J.K., Tessler A., Bishop D., Son Y.-J. In vivo imaging of dorsal root regeneration: rapid immobilization and presynaptic differentiation at the CNS/PNS border. J. Neurosci. 2011;31:4569–4582. doi: 10.1523/JNEUROSCI.4638-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolphin A.C., Scott R.H. Inhibition of calcium currents in cultured rat dorsal root ganglion neurones by (-)-baclofen. Br. J. Pharmacol. 1986;88:213–220. doi: 10.1111/j.1476-5381.1986.tb09489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrier C.E., Aran D., Haenelt E.A., Sheehy R.N., Hoi K.K., Pintarić L., Chen Y., Lizama C.O., Cautivo K.M., Weiner G.A., et al. CNS fibroblasts form a fibrotic scar in response to immune cell infiltration. Nat. Neurosci. 2021;24:234–244. doi: 10.1038/s41593-020-00770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enes J., Langwieser N., Ruschel J., Carballosa-Gonzalez M.M., Klug A., Traut M.H., Ylera B., Tahirovic S., Hofmann F., Stein V., et al. Electrical activity suppresses axon growth through Ca(v)1.2 channels in adult primary sensory neurons. Curr. Biol. 2010;20:1154–1164. doi: 10.1016/j.cub.2010.05.055. [DOI] [PubMed] [Google Scholar]
- Faure L., Wang Y., Kastriti M.E., Fontanet P., Cheung K.K.Y., Petitpré C., Wu H., Sun L.L., Runge K., Croci L., et al. Single cell RNA sequencing identifies early diversity of sensory neurons forming via bi-potential intermediates. Nat. Commun. 2020;11:4175. doi: 10.1038/s41467-020-17929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J.W. The struggle to make CNS axons regenerate: why has it been so difficult? Neurochem. Res. 2020;45:144–158. doi: 10.1007/s11064-019-02844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J.W., Verhaagen J. Intrinsic determinants of axon regeneration. Dev. Neurobiol. 2018;78:890–897. doi: 10.1002/dneu.22637. [DOI] [PubMed] [Google Scholar]
- Feldmann J., Callebaut I., Raposo G., Certain S., Bacq D., Dumont C., Lambert N., Ouachée-Chardin M., Chedeville G., Tamary H., et al. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3) Cell. 2003;115:461–473. doi: 10.1016/s0092-8674(03)00855-9. [DOI] [PubMed] [Google Scholar]
- Filous A.R., Tran A., Howell C.J., Busch S.A., Evans T.A., Stallcup W.B., Kang S.H., Bergles D.E., Lee S.I., Levine J.M., Silver J. Entrapment via synaptic-like connections between NG2 proteoglycan+ cells and dystrophic axons in the lesion plays a role in regeneration failure after spinal cord injury. J. Neurosci. 2014;34:16369–16384. doi: 10.1523/JNEUROSCI.1309-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn K.C., Hellal F., Neukirchen D., Jacob S., Tahirovic S., Dupraz S., Stern S., Garvalov B.K., Gurniak C., Shaw A.E., et al. ADF/cofilin-mediated actin retrograde flow directs neurite formation in the developing brain. Neuron. 2012;76:1091–1107. doi: 10.1016/j.neuron.2012.09.038. [DOI] [PubMed] [Google Scholar]
- Fouad K., Tetzlaff W. Rehabilitative training and plasticity following spinal cord injury. Exp. Neurol. 2012;235:91–99. doi: 10.1016/j.expneurol.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Fox S., Krnjević K., Morris M.E., Puil E., Werman R. Action of baclofen on mammalian synaptic transmission. Neuroscience. 1978;3:495–515. doi: 10.1016/0306-4522(78)90016-7. [DOI] [PubMed] [Google Scholar]
- Goldberg J.L., Klassen M.P., Hua Y., Barres B.A. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science. 2002;296:1860–1864. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- Gordon T., Udina E., Verge V.M., de Chaves E.I.P. Brief electrical stimulation accelerates axon regeneration in the peripheral nervous system and promotes sensory axon regeneration in the central nervous system. Mot. Contr. 2009;13:412–441. doi: 10.1123/mcj.13.4.412. [DOI] [PubMed] [Google Scholar]
- Han Y., Kaeser P.S., Südhof T.C., Schneggenburger R. RIM determines Ca2+ channel density and vesicle docking at the presynaptic active zone. Neuron. 2011;69:304–316. doi: 10.1016/j.neuron.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel N.Y., Strittmatter S.M. Can regenerating axons recapitulate developmental guidance during recovery from spinal cord injury? Nat. Rev. Neurosci. 2006;7:603–616. doi: 10.1038/nrn1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan N., Corbin D., Hu C. Fusogenic pairings of vesicle-associated membrane proteins (VAMPs) and plasma membrane t-SNAREs--VAMP5 as the exception. PLoS ONE. 2010;5:e14238. doi: 10.1371/journal.pone.0014238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H., Abbott S., Han B.-X., Qi Y., Wang F. Analyzing somatosensory axon projections with the sensory neuron-specific Advillin gene. J. Neurosci. 2007;27:14404–14414. doi: 10.1523/JNEUROSCI.4908-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon P.G., McCobb D.P., Kater S.B. Serotonin selectively inhibits growth cone motility and synaptogenesis of specific identified neurons. Science. 1984;226:561–564. doi: 10.1126/science.6093252. [DOI] [PubMed] [Google Scholar]
- He Z., Jin Y. Intrinsic Control of Axon Regeneration. Neuron. 2016;90:437–451. doi: 10.1016/j.neuron.2016.04.022. [DOI] [PubMed] [Google Scholar]
- Heetla H., Proost J., Molmans B., Staal M., Van Laar T. Wiley Online Library; 2016. A Pharmacokinetic–Pharmacodynamic Model for Intrathecal Baclofen in Patients with Severe Spasticity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton B.J., Bradke F. Can injured adult CNS axons regenerate by recapitulating development? Development. 2017;144:3417–3429. doi: 10.1242/dev.148312. [DOI] [PubMed] [Google Scholar]
- Hilton B.J., Blanquie O., Tedeschi A., Bradke F. High-resolution 3D imaging and analysis of axon regeneration in unsectioned spinal cord with or without tissue clearing. Nat. Protoc. 2019;14:1235–1260. doi: 10.1038/s41596-019-0140-z. [DOI] [PubMed] [Google Scholar]
- Hoersting A.-K., Schmucker D. Axonal branch patterning and neuronal shape diversity: roles in developmental circuit assembly: Axonal branch patterning and neuronal shape diversity in developmental circuit assembly. Curr. Opin. Neurobiol. 2021;66:158–165. doi: 10.1016/j.conb.2020.10.019. [DOI] [PubMed] [Google Scholar]
- Hoogstraaten R.I., van Keimpema L., Toonen R.F., Verhage M. Tetanus insensitive VAMP2 differentially restores synaptic and dense core vesicle fusion in tetanus neurotoxin treated neurons. Sci. Rep. 2020;10:10913. doi: 10.1038/s41598-020-67988-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.-Y., Neher E. Ca(2+)-dependent exocytosis in the somata of dorsal root ganglion neurons. Neuron. 1996;17:135–145. doi: 10.1016/s0896-6273(00)80287-1. [DOI] [PubMed] [Google Scholar]
- Humeau Y., Doussau F., Grant N.J., Poulain B. How botulinum and tetanus neurotoxins block neurotransmitter release. Biochimie. 2000;82:427–446. doi: 10.1016/s0300-9084(00)00216-9. [DOI] [PubMed] [Google Scholar]
- Husch A., Hess S., Kloppenburg P. Functional parameters of voltage-activated Ca2+ currents from olfactory interneurons in the antennal lobe of Periplaneta americana. J. Neurophysiol. 2008;99:320–332. doi: 10.1152/jn.00719.2007. [DOI] [PubMed] [Google Scholar]
- Hutson T.H., Kathe C., Palmisano I., Bartholdi K., Hervera A., De Virgiliis F., McLachlan E., Zhou L., Kong G., Barraud Q., et al. Cbp-dependent histone acetylation mediates axon regeneration induced by environmental enrichment in rodent spinal cord injury models. Sci. Transl. Med. 2019;11:eaaw2064. doi: 10.1126/scitranslmed.aaw2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imig C., Min S.-W., Krinner S., Arancillo M., Rosenmund C., Südhof T.C., Rhee J., Brose N., Cooper B.H. The morphological and molecular nature of synaptic vesicle priming at presynaptic active zones. Neuron. 2014;84:416–431. doi: 10.1016/j.neuron.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Junge H.J., Rhee J.-S., Jahn O., Varoqueaux F., Spiess J., Waxham M.N., Rosenmund C., Brose N. Calmodulin and Munc13 form a Ca2+ sensor/effector complex that controls short-term synaptic plasticity. Cell. 2004;118:389–401. doi: 10.1016/j.cell.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Kaeser P.S., Kwon H.-B., Chiu C.Q., Deng L., Castillo P.E., Südhof T.C. RIM1α and RIM1β are synthesized from distinct promoters of the RIM1 gene to mediate differential but overlapping synaptic functions. J. Neurosci. 2008;28:13435–13447. doi: 10.1523/JNEUROSCI.3235-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser P.S., Deng L., Wang Y., Dulubova I., Liu X., Rizo J., Südhof T.C. RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell. 2011;144:282–295. doi: 10.1016/j.cell.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater S.B., Mattson M.P., Cohan C., Connor J. Calcium regulation of the neuronal growth cone. Trends Neurosci. 1988;11:315–321. doi: 10.1016/0166-2236(88)90094-x. [DOI] [PubMed] [Google Scholar]
- Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Mizoguchi A., Ide C. Regulation of growth cone extension by SNARE proteins. J. Histochem. Cytochem. 2003;51:429–433. doi: 10.1177/002215540305100404. [DOI] [PubMed] [Google Scholar]
- Kitao Y., Robertson B., Kudo M., Grant G. Neurogenesis of subpopulations of rat lumbar dorsal root ganglion neurons including neurons projecting to the dorsal column nuclei. J. Comp. Neurol. 1996;371:249–257. doi: 10.1002/(SICI)1096-9861(19960722)371:2<249::AID-CNE5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Kiyonaka S., Wakamori M., Miki T., Uriu Y., Nonaka M., Bito H., Beedle A.M., Mori E., Hara Y., De Waard M., et al. RIM1 confers sustained activity and neurotransmitter vesicle anchoring to presynaptic Ca2+ channels. Nat. Neurosci. 2007;10:691–701. doi: 10.1038/nn1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. 2015. Pheatmap: pretty heatmaps. R Package Version 1.0.10.(CRAN)https://CRAN.R-project.org/package=pheatmap [Google Scholar]
- Koopmans F., van Nierop P., Andres-Alonso M., Byrnes A., Cijsouw T., Coba M.P., Cornelisse L.N., Farrell R.J., Goldschmidt H.L., Howrigan D.P. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron. 2019;103:217–234. doi: 10.1016/j.neuron.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., He X., Kawaguchi R., Zhang Y., Wang Q., Monavarfeshani A., Yang Z., Chen B., Shi Z., Meng H., et al. Microglia-organized scar-free spinal cord repair in neonatal mice. Nature. 2020;587:613–618. doi: 10.1038/s41586-020-2795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Tarr T.B., Bravo-Altamirano K., Valdomir G., Rensch G., Swanson L., DeStefino N.R., Mazzarisi C.M., Olszewski R.A., Wilson G.M., et al. Synthesis and biological evaluation of a selective N- and p/q-type calcium channel agonist. ACS Med. Chem. Lett. 2012;3:985–990. doi: 10.1021/ml3002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipstein N., Sakaba T., Cooper B.H., Lin K.-H., Strenzke N., Ashery U., Rhee J.-S., Taschenberger H., Neher E., Brose N. Dynamic control of synaptic vesicle replenishment and short-term plasticity by Ca(2+)-calmodulin-Munc13-1 signaling. Neuron. 2013;79:82–96. doi: 10.1016/j.neuron.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Liu K., Lu Y., Lee J.K., Samara R., Willenberg R., Sears-Kraxberger I., Tedeschi A., Park K.K., Jin D., Cai B., et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat. Neurosci. 2010;13:1075–1081. doi: 10.1038/nn.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair M.H., Baker D.A., Armstrong J.D. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- Lorenzana A.O., Lee J.K., Mui M., Chang A., Zheng B. A surviving intact branch stabilizes remaining axon architecture after injury as revealed by in vivo imaging in the mouse spinal cord. Neuron. 2015;86:947–954. doi: 10.1016/j.neuron.2015.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubser P.G., Narayan R.K., Sandin K.J., Donovan W.H., Russell K.D. Continuous infusion of intrathecal baclofen: long-term effects on spasticity in spinal cord injury. Paraplegia. 1991;29:48–64. doi: 10.1038/sc.1991.7. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Wang Y., Graham L., McHale K., Gao M., Wu D., Brock J., Blesch A., Rosenzweig E.S., Havton L.A., et al. Long-distance growth and connectivity of neural stem cells after severe spinal cord injury. Cell. 2012;150:1264–1273. doi: 10.1016/j.cell.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahar M., Cavalli V. Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 2018;19:323–337. doi: 10.1038/s41583-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin S.A., Davis B.M., Molliver D.C. Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat. Protoc. 2007;2:152–160. doi: 10.1038/nprot.2006.461. [DOI] [PubMed] [Google Scholar]
- Mattson M.P., Kater S.B. Calcium regulation of neurite elongation and growth cone motility. J. Neurosci. 1987;7:4034–4043. doi: 10.1523/JNEUROSCI.07-12-04034.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., Ushkaryov Y.A., Edelmann L., Link E., Binz T., Niemann H., Jahn R., Südhof T.C. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature. 1993;364:346–349. doi: 10.1038/364346a0. [DOI] [PubMed] [Google Scholar]
- Meyer M.P., Smith S.J. Evidence from in vivo imaging that synaptogenesis guides the growth and branching of axonal arbors by two distinct mechanisms. J. Neurosci. 2006;26:3604–3614. doi: 10.1523/JNEUROSCI.0223-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelassi F., Liu H., Hu Z., Dittman J.S. A C1-C2 module in Munc13 inhibits calcium-dependent neurotransmitter release. Neuron. 2017;95:577–590. doi: 10.1016/j.neuron.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J.A., Betzin J., Oprisoreanu A.-M., Engholm-Keller K., Mayer A., Paulußen I., Schönhense E., Gulakova P., McGovern T.D., Gschossmann L.J., et al. A Presynaptic Phosphosignaling Hub for Lasting Homeostatic Plasticity. SSRN. 2020 doi: 10.2139/ssrn.3728948. [DOI] [PubMed] [Google Scholar]
- Nair A.B., Jacob S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016;7:27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeft M., Wieffer M., de Jong A.S., Negroiu G., Metz C.H., van Loon A., Griffith J., Krijgsveld J., Wulffraat N., Koch H., et al. Munc13-4 is an effector of rab27a and controls secretion of lysosomes in hematopoietic cells. Mol. Biol. Cell. 2005;16:731–741. doi: 10.1091/mbc.E04-10-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S., Woolf C.J. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron. 1999;23:83–91. doi: 10.1016/s0896-6273(00)80755-2. [DOI] [PubMed] [Google Scholar]
- O’Shea T.M., Burda J.E., Sofroniew M.V. Cell biology of spinal cord injury and repair. J. Clin. Invest. 2017;127:3259–3270. doi: 10.1172/JCI90608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osen-Sand A., Staple J.K., Naldi E., Schiavo G., Rossetto O., Petitpierre S., Malgaroli A., Montecucco C., Catsicas S. Common and distinct fusion proteins in axonal growth and transmitter release. J. Comp. Neurol. 1996;367:222–234. doi: 10.1002/(SICI)1096-9861(19960401)367:2<222::AID-CNE5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Ozaki S., Snider W.D. Initial trajectories of sensory axons toward laminar targets in the developing mouse spinal cord. J. Comp. Neurol. 1997;380:215–229. [PubMed] [Google Scholar]
- Palmisano I., Danzi M.C., Hutson T.H., Zhou L., McLachlan E., Serger E., Shkura K., Srivastava P.K., Hervera A., Neill N.O., et al. Epigenomic signatures underpin the axonal regenerative ability of dorsal root ganglia sensory neurons. Nat. Neurosci. 2019;22:1913–1924. doi: 10.1038/s41593-019-0490-4. [DOI] [PubMed] [Google Scholar]
- Paredes R., Agmo A. Stereospecific actions of baclofen on sociosexual behavior, locomotor activity and motor execution. Psychopharmacology (Berl.) 1989;97:358–364. doi: 10.1007/BF00439451. [DOI] [PubMed] [Google Scholar]
- Pashkovski S.L., Iurilli G., Brann D., Chicharro D., Drummey K., Franks K.M., Panzeri S., Datta S.R. Structure and flexibility in cortical representations of odour space. Nature. 2020;583:253–258. doi: 10.1038/s41586-020-2451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persoon C.M., Hoogstraaten R.I., Nassal J.P., van Weering J.R.T., Kaeser P.S., Toonen R.F., Verhage M. The RAB3-RIM pathway is essential for the release of neuromodulators. Neuron. 2019;104:1065–1080. doi: 10.1016/j.neuron.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisier S.B., Taschereau R., Wong J.A., Graeber T.G. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivich G., Bohatschek M., Da Costa C., Iwata O., Galiano M., Hristova M., Nateri A.S., Makwana M., Riera-Sans L., Wolfer D.P., et al. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Ramer L.M., Ramer M.S., Bradbury E.J. Restoring function after spinal cord injury: towards clinical translation of experimental strategies. Lancet Neurol. 2014;13:1241–1256. doi: 10.1016/S1474-4422(14)70144-9. [DOI] [PubMed] [Google Scholar]
- Reitz S.J., Sauerbeck A.D., Kummer T.T. SEQUIN: An imaging and analysis platform for quantification and characterization of synaptic structures in mouse. STAR protocols. 2021;2:100268. doi: 10.1016/j.xpro.2020.100268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W., Tochitsky I., Yang L., Cheng Y.-C., Li E., Kawaguchi R., Geschwind D.H., Woolf C.J. Transcriptional reprogramming of distinct peripheral sensory neuron subtypes after axonal injury. Neuron. 2020;108:128–144. doi: 10.1016/j.neuron.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P., Issa V. Peripheral injury enhances central regeneration of primary sensory neurones. 1984;309:791–793. doi: 10.1038/309791a0. [DOI] [PubMed] [Google Scholar]
- Roth B.L. DREADDs for Neuroscientists. Neuron. 2016;89:683–694. doi: 10.1016/j.neuron.2016.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J.R., Lichtman J.W. Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- Sanes J.R., Zipursky S.L. Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell. 2020;181:536–556. doi: 10.1016/j.cell.2020.04.008. [DOI] [PubMed] [Google Scholar]
- Schiavo G., Benfenati F., Poulain B., Rossetto O., Polverino de Laureto P., DasGupta B.R., Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- Schwab M.E., Strittmatter S.M. Nogo limits neural plasticity and recovery from injury. Curr. Opin. Neurobiol. 2014;27:53–60. doi: 10.1016/j.conb.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seijffers R., Mills C.D., Woolf C.J. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J. Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N., Flaherty K., Lezgiyeva K., Wagner D.E., Klein A.M., Ginty D.D. The emergence of transcriptional identity in somatosensory neurons. Nature. 2020;577:392–398. doi: 10.1038/s41586-019-1900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin O.-H., Lu J., Rhee J.-S., Tomchick D.R., Pang Z.P., Wojcik S.M., Camacho-Perez M., Brose N., Machius M., Rizo J., et al. Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis. Nat. Struct. Mol. Biol. 2010;17:280–288. doi: 10.1038/nsmb.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler A., Oh W.C., Imig C., Altas B., Kawabe H., Cooper B.H., Kwon H.-B., Rhee J.-S., Brose N. Formation and maintenance of functional spines in the absence of presynaptic glutamate release. Neuron. 2017;94:304–311. doi: 10.1016/j.neuron.2017.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siksou L., Varoqueaux F., Pascual O., Triller A., Brose N., Marty S. A common molecular basis for membrane docking and functional priming of synaptic vesicles. Eur. J. Neurosci. 2009;30:49–56. doi: 10.1111/j.1460-9568.2009.06811.x. [DOI] [PubMed] [Google Scholar]
- Slezak M., Grosche A., Niemiec A., Tanimoto N., Pannicke T., Münch T.A., Crocker B., Isope P., Härtig W., Beck S.C., et al. Relevance of exocytotic glutamate release from retinal glia. Neuron. 2012;74:504–516. doi: 10.1016/j.neuron.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Smith D.S., Skene J.H. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J. Neurosci. 1997;17:646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T.C. The presynaptic active zone. Neuron. 2012;75:11–25. doi: 10.1016/j.neuron.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T.C. Towards an understanding of synapse formation. Neuron. 2018;100:276–293. doi: 10.1016/j.neuron.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A., Dupraz S., Laskowski C.J., Xue J., Ulas T., Beyer M., Schultze J.L., Bradke F. The calcium channel subunit Alpha2delta2 suppresses axon regeneration in the adult CNS. Neuron. 2016;92:419–434. doi: 10.1016/j.neuron.2016.09.026. [DOI] [PubMed] [Google Scholar]
- Tedeschi A., Dupraz S., Curcio M., Laskowski C.J., Schaffran B., Flynn K.C., Santos T.E., Stern S., Hilton B.J., Larson M.J.E., et al. ADF/Cofilin-Mediated Actin Turnover Promotes Axon Regeneration in the Adult CNS. Neuron. 2019;103:1073–1085. doi: 10.1016/j.neuron.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran A.P., Warren P.M., Silver J. The biology of regeneration failure and success after spinal cord injury. Physiol. Rev. 2018;98:881–917. doi: 10.1152/physrev.00017.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D., Furlan A., Islam S., Abdo H., Lönnerberg P., Lou D., Hjerling-Leffler J., Haeggström J., Kharchenko O., Kharchenko P.V., et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- van de Bospoort R., Farina M., Schmitz S.K., de Jong A., de Wit H., Verhage M., Toonen R.F. Munc13 controls the location and efficiency of dense-core vesicle release in neurons. J. Cell Biol. 2012;199:883–891. doi: 10.1083/jcb.201208024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F., Sigler A., Rhee J.-S., Brose N., Enk C., Reim K., Rosenmund C. Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc. Natl. Acad. Sci. USA. 2002;99:9037–9042. doi: 10.1073/pnas.122623799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A.S., Omlor W., Rubio J.C., Chen J.L., Zheng H., Schröter A., Gullo M., Weinmann O., Kobayashi K., Helmchen F., et al. Neuronal repair. Asynchronous therapy restores motor control by rewiring of the rat corticospinal tract after stroke. Science. 2014;344:1250–1255. doi: 10.1126/science.1253050. [DOI] [PubMed] [Google Scholar]
- Waldner C., Roose M., Ryffel G.U. Red fluorescent Xenopus laevis: a new tool for grafting analysis. BMC Dev. Biol. 2009;9:37. doi: 10.1186/1471-213X-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.E., Chen W.G., Dalva M.B., Dolmetsch R.E., Kornhauser J.M., Shaywitz A.J., Takasu M.A., Tao X., Greenberg M.E. Calcium regulation of neuronal gene expression. Proc. Natl. Acad. Sci. USA. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wictorin K., Brundin P., Gustavii B., Lindvall O., Björklund A. Reformation of long axon pathways in adult rat central nervous system by human forebrain neuroblasts. 1990;347:556–558. doi: 10.1038/347556a0. [DOI] [PubMed] [Google Scholar]
- Wu D., Jin Y., Shapiro T.M., Hinduja A., Baas P.W., Tom V.J. Chronic neuronal activation increases dynamic microtubules to enhance functional axon regeneration after dorsal root crush injury. Nat. Commun. 2020;11:6131. doi: 10.1038/s41467-020-19914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Chi P., Bibb J.A., Ryan T.A., Greengard P. Roscovitine: a novel regulator of P/Q-type calcium channels and transmitter release in central neurons. J. Physiol. 2002;540:761–770. doi: 10.1113/jphysiol.2001.013376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylera B., Ertürk A., Hellal F., Nadrigny F., Hurtado A., Tahirovic S., Oudega M., Kirchhoff F., Bradke F. Chronically CNS-injured adult sensory neurons gain regenerative competence upon a lesion of their peripheral axon. Curr. Biol. 2009;19:930–936. doi: 10.1016/j.cub.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wahane S., Friedl M.-S., Kluge M., Friedel C.C., Avrampou K., Zachariou V., Guo L., Zhang B., He X., et al. Microglia and macrophages promote corralling, wound compaction and recovery after spinal cord injury via Plexin-B2. Nat. Neurosci. 2020;23:337–350. doi: 10.1038/s41593-020-0597-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie shows multiphoton tile scan and 3D rendering of GFP+ sensory axons (yellow) and GFAP+ astrocytes (blue) in the injured adult mouse spinal cord in control conditions or with hM4Di activation.
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code. Permutation tests were performed using a custom script “permutation_test 0.18” implemented in Python (2.7.3 version) including Pandas and NumPy libraries. The script is available for download in the Python Package Index (https://pypi.python.org/pypi/permutation_test). The sunburst plots in Figures 1 and S3 were generated using the SynGO Portal (https://www.syngoportal.org/). Figure 7C was created using BioRender (https://biorender.com).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.