Abstract
Surveillance screening at scale to identify people infected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) prior to extensive transmission is key to bringing an end to the coronavirus disease 2019 (COVID-19) pandemic, even though vaccinations have already begun. Here we describe Corona Detective, a sensitive and rapid molecular test to detect the virus, based on loop-mediated isothermal amplification, which could be applied anywhere at low cost. Critically, the method uses freeze-dried reagents, readily shipped without cold-chain dependence. The reaction detects the viral nucleocapsid gene through a sequence-specific quenched-fluorescence readout, which avoids false positives and also allows multiplex detection with an internal control cellular RNA. Corona Detective can be used in 8-tube strips to be read with a simple open-design fluorescence detector. Other methods to use and produce Corona Detective locally in a variety of formats are possible and already openly shared. Detection specificity is ensured through inclusion of positive and negative control reactions to run in parallel with the diagnostic reactions. A simple user protocol, including sample preparation, and a bioinformatics pipeline to ensure that viral variants will still be detectable with SARS-CoV-2 primer sets complete the method. Through rapid production and distribution of Corona Detective reactions, quite inexpensive at scale, daily or weekly surveillance testing of large populations, without waiting for symptoms to develop, is anticipated, in combination with vaccination campaigns, to finally control this pandemic.
Keywords: computational biology, COVID-19, fluorescence, molecular diagnostic techniques, open-source
INTRODUCTION
The recent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has revealed fragility in public health preparedness. The pandemic requires a fair, rapid, large-scale deployment of biologic testing beyond what the existing biomedical infrastructure was able to supply early in 2020.1 Despite enormous efforts across public and private sectors, detection of SARS-CoV-2 in people or in the environment remains cumbersome and expensive.2 Currently used techniques of molecular amplification are costly and require complex infrastructure and biosafety procedures, limiting diagnosis of people everywhere.3 Even with vaccines already being administered, testing remains a crucial factor to control the current pandemic,4, 5 with prompt surveillance testing of populations being particularly important, as 40% of potential carriers are asymptomatic6 and as new virus variants evolve (nextstrain.org). Sensitive alternatives to quantitative reverse transcription PCR, the “gold standard” for molecular diagnostics of this RNA virus, with a simpler reaction methodology and reagents that can be readily shipped and stored without cold-chain dependence, are necessary.2
The Corona Detective described herein is based upon reverse transcription loop-mediated isothermal amplification (RT-LAMP). Loop-mediated isothermal amplification (LAMP) rivals PCR for sensitivity, without requiring thermocycling or even a cold chain.7, 8 Multiple groups have recently developed RT-LAMP assays for detection of SARS-CoV-23, 9 (see review in this journal). Because the majority of these reported methods rely on using nonspecific readout methods, such as intercalating dyes or a change of pH, they cannot readily distinguish spurious amplicons that can arise during isothermal amplification,10, 11 increasing the risk and rate of false positives.
This shortcoming calls for the use of sequence-specific detection methods,12 such as the one used in the first RT-LAMP test to receive an Emergency Use Authorization (EUA) from the U.S. Food and Drug Administration13 (iAMP COVID-19 Detection Kit) or quenching of unincorporated amplification signal reporters (QUASR).14, 15 This method employs a fluorescently tagged primer from the main LAMP set [forward inner primer (FIP)/backward inner primer (BIP) or loop forward (LF)/loop backward (LB)] and a short oligonucleotide complementary to the one tagged, but with a quencher that blocks the fluorescence when bound, allowing for fluorescent signals specifically from produced amplicons. This detection strategy improves specificity and allows multiplexing for the detection of more than one target in a single reaction.14, 15
Over recent years, we democratized LAMP-based QUASR detection reactions for use by members of the general public (aged 5–75) to learn about and to detect genetically modified organisms in open science workshops spearheaded in the GMO Detective project.16 To this end, we optimized speed, sensitivity, and robust use (Aidelberg G, in preparation) and also designed an affordable (less than $2) and easy-to-build open-source hardware fluorescence detector.17 This earlier work enabled quick adaptation of the QUASR method for SARS-CoV-2 detection. As an open science collaboration with Just One Giant Lab and its international team of collaborators, including the Center for Research and Interdisciplinarity (Paris, France) and the community lab, Hackuarium (Lausanne, Switzerland), do-it-together participatory research brought together the Corona Detective project.18 One key innovation of the method, use of freeze-dried reaction components, allows Corona Detective to be shipped anywhere readily, with no cold-chain dependence and in a variety of shelf-stable formats. This means scaling up this method may be truly possible. That the method can be readily adapted to various targets, additionally, means that this work may also provide better pandemic preparedness for the future.
METHODS
RT-LAMP
The polymerases Bst LF and Bst 2.0 and the RTx reverse transcriptase (the latter two enzymes “WarmStart” versions) were obtained from New England Biolabs (Ipswich, MA, USA) and used with provided buffers as per manufacturer instructions, titrating magnesium and adjusting primer ratios to optimize reactions based upon real-time amplification, using SYBR-Safe (Thermo Fisher Scientific, Waltham, MA, USA) intercalation for detection, by diluting the stock solution (10,000×) 1/250 and using 1.6 μl per real-time reaction, as described by Carter et al.8 A plasmid encoding the nucleocapsid (N) gene (10006625; Integrated DNA Technologies, Coralville, IA, USA) and a synthetic RNA control for SARS-CoV-2 (NR-52358; BEI Resources, Manassas, VA, USA) were used as positive controls. Reaction volumes were 20 μl total with 4 μl sample input. Total cell RNA was extracted from inner cheek cells with either the Monarch Kit (New England Biolabs) or RNeasy (Qiagen, Germantown, MD, USA) per manufacturer instructions. Environmental samples (of doorknobs and surfaces) were obtained by wiping with cellulose tissue, and ∼10 mm squares were cut and heated to 95°C for 5 min in 100 μl RNase-free water before use in reactions.
QUASR detection
N-target primers19, 20 were chosen for further use in QUASR detection. The FIP primer was tagged at its 5′ end with fluorescein (FAM), and a short sequence complementary to it, with a quencher (either Black Hole Quencher or Iowa Black FQ Eurofins, Germany and Integrated DNA Technologies, Belgium, respectively) at its 3′ end, was also synthesized (sequence 5′CATTGGC CGCA-Q) for QUASR endpoint detection of the amplification product. RNaseP detection was previously described by Curtis et al.21 and used a HEX fluorescence marker on the LF primer with a complementary 5′CAGCCAT CCACAT-Q quencher. Quencher oligonucleotides were added at 1.5-fold the concentration of their complementary tagged primer. Reactions were cooled to ambient temperature for hybridization (and thus quenching) of unincorporated signal before imaging with blue excitation and an amber filter [with the detector from the GMO Detective17 or with a trans-illuminator or plate reader (fluorimeter) for 96-well plates].
Sample treatments
Validation tests of the QUASR detection reactions were done on inactivated SARS-CoV-2 viral particles (NR-52286; BEI Resources) and also on viral surrogates (Zepto-Metrix, Buffalo, NY, USA), in particular from a “blind” set of samples to test (from the XPRIZE semifinals), with all unknown samples already inactivated.22 The inactivated viral particles were spiked into various matrices (water, PBS, saliva) before subsequent treatments (below). For tests with these samples, the “direct” sample treatment method used 100× TCEP/EDTA solution (0.25 M TCEP, 0.1 M EDTA pH 8), diluted to 1× in the sample for a final concentration of 2.5 mM TCEP/1 mM EDTA, and heated 5 min at 95°C.23 Four microliters of this solution was added to rehydrated Corona Detective reactions and incubated at 55°C for 10 min of reverse transcription, then 64°C for 45 min of isothermal amplification. Reactions were allowed to cool to ambient temperatures before viewing results. More details can be found in the user protocol24 for Corona Detective.
Bioinformatics workflow
To assess sequence evolution as the pandemic continues, particularly in viral genomic regions covered by Corona Detective reaction primer targets, the bioinformatics work-flow is as follows:
Complete and high coverage genomes are collected from the Global Initiative on Sharing All Influenza Data (GISAID) EpiCoV Database (https://www.gisaid.org/).25 Only genomic sequences with lengths of 5000 to 32,000 nucleotides were analyzed. The results reported below are from the most recent 2000 submitted sequences for each geographical region (before April 15, 2021).
Regional multifasta files are aligned using the MAFFT online service.26, 27
The resulting alignment files in CLUSTAL format are analyzed via Python functions. (See the GitHub repository of the project: https://github.com/CoronaDetective/Mutation_Analysis_v1.1.)
RESULTS
Reaction design and optimization
Research to develop Corona Detective was openly documented throughout18 as part of the Just One Giant Lab OpenCovid19 Initiative. Detailed and annotated protocols for making19 and using24 the Corona Detective are available. In brief, RT-LAMP primer sets targeting the N gene, the most abundant viral RNA in infected cells,28 and also primers targeting cellular human mRNAs for use as internal controls were initially compared. The N gene plasmid and a synthetic RNA control for SARS-CoV-2 were used to optimize reaction conditions, in particular relative primer ratios and magnesium concentrations, while comparing reactions with one of two polymerases, Bst LF or Bst 2.0.
Two primer sets were chosen for reaction optimization from these early empirical tests: one termed NM, for the viral N gene from the Mammoth protocol,20 and the other, from the RNaseP POP7 gene, termed RP. Examples of real-time data are presented (Fig. 1) with amplification of the control plasmid N gene (Fig. 1A) and reverse transcription and amplification of the control synthetic RNA (Fig. 1B). The Bst 2.0 enzyme is more rapid than the original Bst LF polymerase, as expected, with faster time to detection threshold (16 vs. 22 min). It also tolerates decreases in magnesium down to 5 mM, unlike Bst LF, which would not amplify target under those conditions. Late nontemplate amplification by Bst 2.0 was evident, however, although this was only rarely observed with Bst LF. Amplification of synthetic target RNA was readily observed, with an apparent limit of detection of about 1 copy per microliter of the Corona Detective reaction or 20 copies (Fig. 1B).
FIGURE 1.
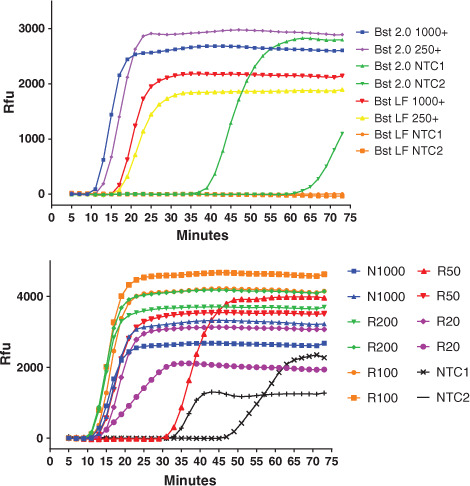
Real-time amplification data demonstrate rapid and sensitive detection. A) Bst 2.0 is more rapid than Bst LF but also more prone to non-template amplification. N250/N1000 reactions have 250/1000 copies of the N plasmid control, respectively. Incubation temperature was at 64°C, and reaction volumes were 20 μl. B) RNA controls are reverse transcribed and amplified for detection down to a single copy per microliter of the reaction. Synthetic RNA (BEI Resources) dilutions were spiked into duplicate reactions at 20, 50, 100, and 200 (R20, R50, R100, and R200) copies, in addition to nontemplate control (NTC) and DNA controls. These reactions used Bst 2.0 and RTx; incubation included an initial 10 min at 55°C for reverse transcription before shifting up to 64°C for the real-time acquisition.RFU - relative fluorescence units.
Optimized reaction conditions from these experiments used primer concentrations of 1.6, 0.8, and 0.2 μM, respectively, for FIP/BIP, LF/LB, and F3/B3, and a magnesium concentration of 7 mM, while aiming to avoid nontemplate control amplification (Table 1).
TABLE 1.
Magnesium optimization of Corona Detective. Concentrations of MgSO4 were varied in reactions with each of the two polymerases (Bst LF or Bst 2.0). The fastest time to threshold in minutes for 1000 copies of the N plasmid (N1000) in a 20 μl reaction is reported.
| MgSO4 (mM) | Bst LF (min) | Bst 2.0 (min) | Notes |
|---|---|---|---|
| 5 | na | 27 | |
| 6 | 35 | 21 | |
| 7 | 31 | 19 | Both NTCs up for Bst 2.0 but not Bst LF |
| 8 | 21 | 16 | Both NTCs up for Bst LF and Bst 2.0 |
NA, no amplification; NTC, nontemplate control.
Sequence-specific fluorescence detection
We designed QUASR primers for the NM primer set (NM*FAM), adding the FAM fluorescent tag to the 5′ end of the FIP primer. A short complementary quencher primer, of 11 bases, with the quencher at its 3′ end was also synthesized. When annealed to the tagged FIP primer, this quencher prevents fluorescence, but when the FIP is incorporated in the amplicon, this tag fluoresces freely, providing the basis for strong QUASR detection. The internal human control, RP, was detected via a QUASR design used by others already,21 with a 13mer attached to a quencher, complementary to the 5′ end of the RP LF primer, tagged with HEX, for the RP primer set RP*HEX, used in the multiplex reaction with NM*FAM.
The QUASR fluorescence cannot be read at amplification temperatures but requires lower temperatures (less ∼35°C) to allow the quencher to anneal to unincorporated tagged primers and prevent fluorescence. It is thus used for endpoint detection. With the simple fluorescence detector from the GMO Detective project,16, 17 illuminated by blue LEDs and with amber filters in front of the tubes, controls were readily detected after 10 min incubation at 55°C and 30–90 min at 64°C (Fig. 2). Furthermore, detection of both targets in the reactions was sensitive and reliable, with a limit of detection of 20 RNA copies per reaction for the reaction with NM*FAM (Fig. 2A). In this detector, the HEX signal is somewhat more orange than the green FAM, particularly by eye, and the multiplex reactions, including the NM*FAM and RP*HEX primer sets, are indeed visible in the simple detector (Fig. 2 B, D). A standard phone camera is sufficient to capture images, another advantage of the QUASR method (Fig. 2).
FIGURE 2.
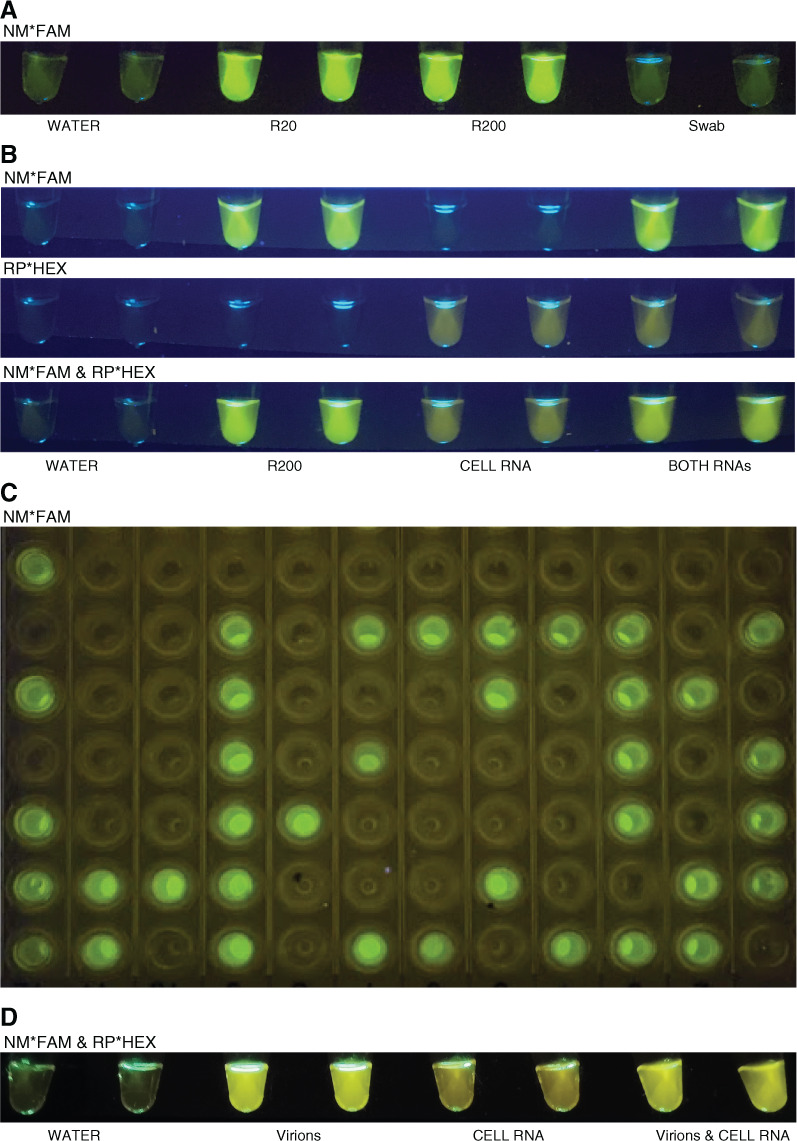
QUASR detection for Corona Detective reactions is affordable and unambiguous. A) Corona Detective gives positive results down to 20copies of the synthetic RNA (R20; BEI Resources) per reaction,but no positive reactions have yet been observed from environmental swabs. NM*FAM (the NM primer set including the FIP primer tagged with FAM and the complementary quencher). B) The multiplex reactions (NM*FAM and RP*HEX, bottom) sum the single reactions, above, and are distinguishable even in this simple ∼$1 do-it-yourself open fluorescence detector. Above-NM primers labeled with FAM; middle-RP primers labeled with HEX; bottom-multiplex reactions withboth primer sets. C) A 96-wellplate imaged onan affordable bluelightgel box transilluminator,1 mo after reactionswere run. D) Freeze-dried reaction tubes for the multiplex run with minimally treated inactivated virus input (NR-52286; BEI Resources), human cell RNA, and both together. Images acquired with standardsmartphones. Reactions runin duplicate with water (nontemplate control), R20 and R200 (20 or 200 copies of synthetic RNA), swab (environmental surface samples), cell RNA (extracted human cheek cell RNA), both RNAs (R200+Cell RNA), and virions (40 copies of heat inactivated virus).
The most encouraging finding from these multiplex QUASR results was that the signals came up well with high sensitivity, but without problems of false positives, even though the Bst 2.0 enzyme was used for these reactions, rather than the slower Bst LF, and even with incubation of up to 2 h. Overall, complete elimination of false positives (to date, in over 200 negative control reactions) provides great confidence in this QUASR detection. Furthermore, results are stable for at least 1 month when stored in a cool dark place (Fig. 2C). An additional strength of this detection method includes its affordability, with cost per reaction of about $1 or even less at scale. As mentioned, this can be done using an open-source29 do-it-yourself fluorescence detector for 8-strip tubes17 (Fig. 2A, B and D), which costs only $1–$2 to make. For whole 96-well plates, off-the-shelf blue light transilluminators affordably and easily detect the fluorescent signal (e.g., https://iorodeo.com/or https://www.minipcr.com/ (Fig. 2C), or if available, a standard gel imaging system or qPCR machines can also be used, all at room temperature.
Sample preparation and reaction specificity and sensitivity
For rapid, affordable, and simple screening especially in lower resource settings, we decided to focus on so-called “direct” saliva sample processing methods.5 The TCEP method23 for rapidly inactivating and opening virions and reducing viscosity of saliva samples worked best. Benefits of the TCEP strategy include the low volume needed, which prevents sample dilution (1:100), use of only one additional temperature (5 min at 95°C), and the buffer’s room temperature stability. A single copy of the synthetic RNA (BEI Resources) per microliter of reaction was readily detected for these minimally processed samples in Corona Detective assays. The limit of detection for this assay was also determined with viral surrogate particles (ZeptoMetrix) in various matrices (saliva, PBS) and with actual inactivated viral particles (BEI Resources) added prior to TCEP treatment to either PBS or saliva. Limits of detection of 2 copies of SARS-CoV-2 viral particles per microliter of reaction were consistently observed, i.e., 10 copies/μl in the original sample (Fig 2D). For a panel of 30 other viral RNA controls [Twist synthetic RNAs (Twist, South San Francisco, CA, USA), e.g., measles, mumps, influenza, and other coronaviruses], no cross-reactivity was observed, in concordance with the EUA reports30, 31 for the NM primer set.
Improved sensitivity may be achieved by using larger input volumes of treated sample per reaction or preconcentrated viral RNA from samples, using commercial kits32 or magnetic beads.33, 34 An affordable glass milk method23 did not scale well to the 96-well microplate format in our hands. To note: sample collection has its own set of issues, and additional biosafety concerns exist until the samples are at least inactivated.
Freeze drying and manufacture
To reduce costs associated with transport, storage, and manipulations and to allow use even where no cold chain is available, reaction components were lyophilized. Briefly, glycerol-free enzymes, along with dNTPs, primer mixes, and trehalose as protein stabilizer,35 were freeze-dried.8, 19 The rehydration buffer containing isothermal amplification buffer, MgSO4, and water is also stable at room temperature (for more detail see Aidelberg and Aronoff19), and provided together with the reaction tubes. These reaction tubes were found to retain sensitivity and specificity for at least 2 months when sealed with a desiccant and stored in a cool dark location (Fig 2D). Freeze-dried batches have already been utilized after shipping to Sri Lanka at ambient temperatures, with control and contrived sample reactions giving expected results, even after over a month on the shelf.
Bioinformatics
In order to make sure prevalent strains of SARS-CoV-2 will still be detectable by the NM primer set, a bioinformatics pipeline has been developed by the team. A 3-step protocol checks for mutations that might affect recognition by diagnostic primer sets. Results of these analyses, comparing geographical regions, to determine whether any prevalent viral variant may present problems for Corona Detective reactions are shown in Fig. 3.
FIGURE 3.
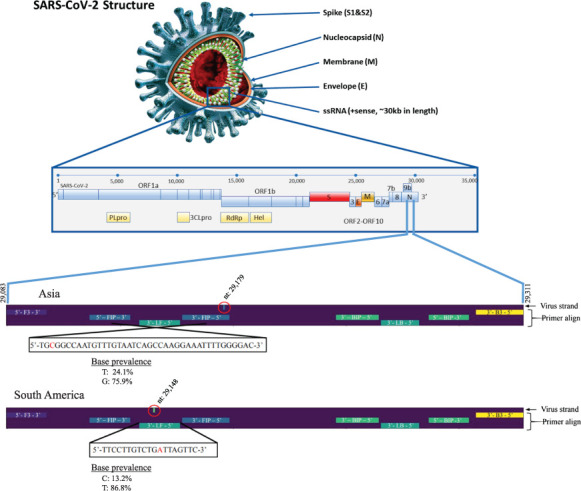
Geographic mutational analysis of sequence variation reveals rare changes in two NM primer targets. The SARS-CoV-2 virion structure and genomic organization is shown above (adapted from Kubina and Dziedzic44 under Creative Commons Attribution 4.0 International). Bioinformatic analyses from the collected GISAID sequences identified mutations in the N gene of Asian and South American sequenced variants, which are included in two different NM primers, FIP and LF. Site numbering and genome structure use Wuhan-Hu-1/2019 (GenBank: MN908947.3) as the reference sequence. Red circles highlight the position of the nucleotide changes in the viral strand. Base prevalence of mutant and wild-type viral sequences are reported as percentages of the number of sequences analyzed (1908 for Asia, 1930 for South America).
The NM primer set target sequences are conserved in over 90% of sequenced isolates from GISAID,25 with the exceptions of the Asian and South American SARS-CoV-2 subsets, where a single nucleotide was mutated in two different NM primer recognition sites. For the most recent analyses (database from April 15, 2021), in the Asian sequences a G is mutated to T in the FIP primer target in 24% of sequenced genomes, whereas for the South American sequences a T is mutated to C in the LF primer target in 13% of sequenced genomes (Fig. 3). The rest of the geographic regions analyzed did not present any relevant mutations (over 10% prevalence) for the Corona Detective primers in local variant strains.
To note: potential bias in sequencing clusters, globally, means these analyses are certainly not a definitive measure of all the virus evolving currently.
Since May 12, the last run in GitHub, many more variants are detected, additionally, and NEB has a new on-line tool (https://primer-monitor.neb.com/) for similar analyses, in which the NM primer set is called ‘Broughton N’ with BIP/FIP swapped (as in the reference).
DISCUSSION
Done without complex equipment and with a simple fluorescence readout, Corona Detective is an easy-to-use, sensitive, robust, and highly specific SARS-CoV-2 detection method. The RT-LAMP primers chosen for the multiplex detection (including the cellular mRNA target as an extraction control) have already been approved for use in two EUAs from the U.S. Food and Drug Administration30, 31 based on RT-LAMP, but with different detection strategies (CRISPR–based and pH-colorimetric, respectively). Drawbacks of these methods, for instance, risks of cross-contamination and sensitivity to buffer effects, are avoided with the sequence-specific quenched-fluorescence–based detection of the QUASR method. Even if the reaction is left to incubate longer than 45 min, false positives in non-template controls are avoided. This is crucial for testing in resource-strapped or point-of-care settings.
Controlling for primer reliability through the described pipeline is so far only done in silico but should help ensure robust detection worldwide. Potential problems in amplification for new variants means that vigilance to follow their evolution with bioinformatic algorithms, as we describe here, is highly recommended. Primer sets may require redesign, eventually, to ones that target more preserved genomic regions. Alternatively, two methods can be considered: 1) degenerate or “wobble base” nucleotides, if substituted into problematic positions of useful primer sequences, could allow them to still interact with the target; 2) the addition of a small amount of a high-fidelity polymerase may allow the Corona Detective to be less sensitive to mismatches, as has been described.36 Additional methods could be used, if, on the contrary, being able to distinguish different strains is desirable.37, 38
Use of the freeze-dried reactions after shipment to Sri Lanka under ambient conditions was successful, and further tests for reaction reliability are in progress. More batches of tubes and 96-well plates will be sent soon to Switzerland, Cameroon, Ghana, Chile, Sri Lanka, and potentially more collaborators, made at the Center for Research and Interdisciplinarity in Paris for clinical validation in a variety of contexts. Possibilities for production at scale are already being explored, also in the United States. To further improve Corona Detective, work has begun that aims to utilize in-house–produced, enhanced,39 and/or off patent enzymes (both polymerases and reverse transcriptases34,40, 41). Additionally, in silico optimization studies are in progress by our open science team to increase heat stability and salt tolerance of the polymerase. A point mutation in the tight-binding pocket, anticipated to be more tolerant to modified nucleotides and particularly important for cross-contamination prevention through dUTP incorporation and UTP-glycosidase treatment, will also be examined in vitro.
Bottlenecks to rapid scale-up of testing include administrative delays relating to intellectual property and the conservative production infrastructure designed to efficiently supply market needs with limited capacity. These limitations could in principle be overcome through a testing supply chain based upon open-source principles in a distributed, decentralized, and local way.42 The tools for high-volume Corona Detective production19 cost less than an average quantitative PCR machine and could even incorporate locally produced enzymes34, 40, 41 or nucleotides.43 Adopting the open Corona Detective in its various formats (8-well, 96-well, 384-well) could help scale up SARS-CoV-2 surveillance, particularly in low-resource settings, sooner rather than later.
ACKNOWLEDGMENTS
We sincerely thank all participants of the Just One Giant Lab (JOGL) nucleic acid amplification group and in particular our colleagues at the Sri Lanka Institute of Nanotechnology (SLINTEC), who enabled the post-shipment testing of Corona Detective. We are also grateful to the other JOGL community members, especially Thomas Landrain, the founder, for his active support, and to members of the open science hardware (GOSH), Open Bioeconomy, and Global Community Biosummit networks for inspiration and counsel and to those who participated in the weekly Global LAMP calls, hosted by Chris Mason at Weill Cornell, for valuable insight and feedback. This work was supported by microgrants from the JOGL Community and Hackuarium. G.A. and A.B.L. also thank the Fondation Bettencourt Schueller and Paris City Hall for their support to the Open Nucleic Acid Detection project. K.H. thanks TU Darmstadt for lab, computing grant, and IANUS Peacelab minigrant. The following reagents were obtained from BEI Resources and National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH): Quantitative Synthetic RNA from SARS-Related Coronavirus 2, NR-52358; and Heat inactivated, Novel Coronavirus, 2019-nCoV/USA-WA1/2020, NR-52286.
REFERENCES
- 1.MacKay MJ, Hooker AC, Afshinnekoo E, Salit M, Kelly J, Feldstein JV, Haft N, Schenkel D, Nambi S, Cai Y, Zhang F, Church G, Dai J, Wang CL, Levy S, Huber J, Ji HP, Kriegel A, Wyllie AL, Mason CE. The COVID-19 XPRIZE and the need for scalable, fast, and widespread testing. Nat Biotechnol . 2020;38:1021–1024. doi: 10.1038/s41587-020-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vandenberg O, Martiny D, Rochas O, van Belkum A, Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat Rev Microbiol . 2020;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Augustine R, Hasan A, Das S, Ahmed R, Mori Y, Notomi T, Kevadiya BD, Thakor AS. Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology (Basel) . 2020;9:182. doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colomer MÀ, Margalida A, Alòs F, Oliva-Vidal P, Vilella A, Fraile L. Modeling of vaccination and contact tracing as tools to control the COVID-19 outbreak in Spain. Vaccines (Basel) . 2021;9:386. doi: 10.3390/vaccines9040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastos ML, Perlman-Arrow S, Menzies D, Campbell JR. The sensitivity and costs of testing for SARS-CoV-2 infection with saliva versus nasopharyngeal swabs: a systematic review and meta-analysis. Ann Intern Med . 2021;174:501–510. doi: 10.7326/M20-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med . 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thai HTC, Mai QL, Cuong DV, Parida M, Minekawa H, Notomi T, Hasebe F, Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol . 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter C, Akrami K, Hall D, Smith D, Aronoff-Spencer E. Lyophilized visually readable loop-mediated isothermal reverse transcriptase nucleic acid amplification test for detection Ebola Zaire RNA. J Virol Methods . 2017;244:32–38. doi: 10.1016/j.jviromet.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moehling TJ, Choi G, Dugan LC, Salit M, Meagher RJ. LAMP diagnostics at the point-of-care: emerging trends and perspectives for the developer community. Expert Rev Mol Diagn . 2021;21:43–61. doi: 10.1080/14737159.2021.1873769. [DOI] [PubMed] [Google Scholar]
- 10.Gonçalves DDS, Hooker DJ, Dong Y, Baran N, Kyrylos P, Iturbe-Ormaetxe I, Simmons CP, O’Neill SL. Detecting wMel Wolbachia in field-collected Aedes aegypti mosquitoes using loop-mediated isothermal amplification (LAMP) Parasit Vectors . 2019;12:404. doi: 10.1186/s13071-019-3666-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang YS, Bhadra S, Li B, Wu YR, Milligan JN, Ellington AD. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal Chem . 2015;87:3314–3320. doi: 10.1021/ac504387c. [DOI] [PubMed] [Google Scholar]
- 12.Becherer L, Borst N, Bakheit M, Frischmann S, Zengerle R, von Stetten F. Loop-mediated isothermal amplification (LAMP) – review and classification of methods for sequence-specific detection. Anal Methods . 2020;12:717–746. [Google Scholar]
- 13.Atila BioSystems, Inc iAMP-COVID19-100: Instructions for Use. Version 2.1. December 2020. Available at: https://www.fda.gov/media/136870/download Accessed May 1, 2021.
- 14.Ball CS, Light YK, Koh C-Y, Wheeler SS, Coffey LL, Meagher RJ. Quenching of unincorporated amplification signal reporters in reverse-transcription loop-mediated isothermal amplification enabling bright, single-step, closed-tube, and multiplexed detection of RNA viruses. Anal Chem . 2016;88:3562–3568. doi: 10.1021/acs.analchem.5b04054. [DOI] [PubMed] [Google Scholar]
- 15.Priye A, Bird SW, Light YK, Ball CS, Negrete OA, Meagher RJ. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci Rep . 2017;7:44778. doi: 10.1038/srep44778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GMO Detective Available at: https://gmodetective.com/ Accessed January 12, 2021.
- 17. GMODetective-Detector. Available at: https://github.com/MakerLabCRI/GMODetective-Detector Accessed January 15, 2021.
- 18.Lombadero FJQ, Aidelberg G, Aronoff R. Do-It-Together SARS CoV-2 Detective. https://app.jogl.io/project/181 Accessed January 12, 2021.
- 19.Aidelberg G, Aronoff R. Freeze-drying (Lyophilization) and manufacturing of Corona Detective assay v.2. November 19, 2020. Available at https://www.protocols.io/view/freeze-drying-lyophilization-and-manufacturing-of-bpv4mn8w Accessed May 1, 2021.
- 20.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan CY, Guevara H, Wadford DA, Chen JS, Chiu CY. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol . 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtis KA, Morrison D, Rudolph DL, Shankar A, Bloomfield LSP, Switzer WM, Owen SM. A multiplexed RT-LAMP assay for detection of group M HIV-1 in plasma or whole blood. J Virol Methods . 2018;255:91–97. doi: 10.1016/j.jviromet.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Bektaş A, Covington MF, Aidelberg G, Arce A, Matute T, Núñez I, Walsh J, Boutboul D, Delaugerre C, Lindner AB, Federici F, Jayaprakash AD. Accessible LAMP-enabled rapid test (ALERT) for detecting SARS-CoV-2. Viruses . 2021;13:742. doi: 10.3390/v13050742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabe BA, Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci USA . 2020;117:24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aronoff R, Aidelberg G. Corona Detective user protocol V2.0. November 20, 2020. Available at https://www.protocols.io/view/corona-detective-user-protocol-v2-0-bpwzmpf6 Accessed July 15, 2021.
- 25.Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall . 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. MAFFT version 7: multiple alignment program for amino acid or nucleotide sequences. Available at: https://mafft.cbrc.jp/alignment/software/closelyrelatedviralgenomes.html Accessed January 13, 2021.
- 27.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform . 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, Tamir H, Achdout H, Stein D, Israeli O, Beth-Din A, Melamed S, Weiss S, Israely T, Paran N, Schwartz M, Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature . 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 29.Maia Chagas A, Molloy JC, Prieto-Godino LL, Baden T. Leveraging open hardware to alleviate the burden of COVID-19 on global health systems. PLoS Biol . 2020;18:e3000730. doi: 10.1371/journal.pbio.3000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.U.S. Food and Drug Administration Emergency Use Authorization (EUA) Summary for the Color SARS-CoV-2 RT-LAMP Diagnostic Assay. Updated April 14, 2021. Available at: https://www.fda.gov/media/138249/download Accessed May 1, 2021.
- 31.U.S. Food and Drug Administration Accelerated Emergency Use Authorization (EUA) Summary: SARS-CoV-2 RNA Detectr Assay. Available at: https://www.fda.gov/media/139937/download Accessed May 1, 2021.
- 32.Centers for Disease Control and Prevention CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel: Instructions for Use. Available at: https://www.fda.gov/media/134922/download Accessed May 1, 2021.
- 33.Oberacker P, Stepper P, Bond DM, Hüohn S, Focken J, Meyer V, Schelle L, Sugrue VJ, Jeunen GJ, Moser T, Hore SR, von Meyenn F, Hipp K, Hore TA, Jurkowski TP. Bio-On-Magnetic-Beads (BOMB): open platform for high-throughput nucleic acid extraction and manipulation. PLoS Biol . 2019;17:e3000107. doi: 10.1371/journal.pbio.3000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kellner MJ, Ross JJ, Schnabl J Scalable, rapid and highly sensitive isothermal detection of SARS-CoV-2 for laboratory and home testing. bioRxiv. Published online June 23, 2020:2020.06.23.166397. [DOI]
- 35.Colaço C, Sen S, Thangavelu M, Pinder S, Roser B. Extraordinary stability of enzymes dried in trehalose: simplified molecular biology. Biotechnology (N Y) . 1992;10:1007–1011. doi: 10.1038/nbt0992-1007. [DOI] [PubMed] [Google Scholar]
- 36.Lu R, Wu X, Wan Z, Li Y, Jin X, Zhang C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int J Mol Sci . 2020;21:E2826. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gill P, Hadian Amree A. AS-LAMP: a new and alternative method for genotyping. Avicenna J Med Biotechnol . 2020;12:2–8. [PMC free article] [PubMed] [Google Scholar]
- 38.Varona M, Anderson JL. Advances in mutation detection using loop-mediated isothermal amplification. ACS Omega . 2021;6:3463–3469. doi: 10.1021/acsomega.0c06093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maranhao A, Bhadra S, Paik I, Walker D, Ellington AD. An improved and readily available version of Bst DNA polymerase for LAMP, and applications to COVID-19 diagnostics. Published online October 5, 2020. [DOI] [Google Scholar]
- 40.Open Bioeconomy Lab. Open Enzyme Collection. Available at: https://openbioeconomy.org/projects/open-enzyme-collections/ Accessed April 21, 2020.
- 41.Alekseenko A, Barrett D, Pareja-Sanchez Y, Howard RJ, Strandback E, Ampah-Korsah H, Rovšnik U, Zuniga-Veliz S, Klenov A, Malloo J, Ye S, Liu X, Reinius B, Elsässer SJ, Nyman T, Sandh G, Yin X, Pelechano V. Direct detection of SARS-CoV-2 using non-commercial RT-LAMP reagents on heat-inactivated samples. Sci Rep . 2021;11:1820. doi: 10.1038/s41598-020-80352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomez-Marquez J, Hamad-Schifferli K. Distributed biological foundries for global health. Adv Healthc Mater . 2019;8:e1900184. doi: 10.1002/adhm.201900184. [DOI] [PubMed] [Google Scholar]
- 43.Loan TD, Easton CJ, Alissandratos A. DNA amplification with in situ nucleoside to dNTP synthesis, using a single recombinant cell lysate of E. coli. Sci Rep . 2019;9:15621. doi: 10.1038/s41598-019-51917-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubina R, Dziedzic A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics (Basel) . 2020;10:E434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]


