Abstract
Wastewater surveillance for monitoring severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an important epidemiologic tool for the assessment of population-wide coronavirus disease 2019 (COVID-19). This tool can be successfully implemented only if SARS-CoV-2 RNA in wastewater can be accurately recovered and quantified. The lack of standardized procedure for wastewater virus analysis has resulted in varying SARS-CoV-2 concentrations for the same sample. This study reports the effect of 4 key factors—sample volume, percentage polyethylene glycol (PEG)-NaCl, incubation period, and storage duration at 4°C—on the recovery of spiked noninfectious SARS-CoV-2 RNA in raw sewage and sludge samples. N1 and N2 genes of SARS-CoV-2 were quantified using the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and digital droplet PCR (RT-ddPCR) techniques. Results indicate that 1) for raw sewage, 50-ml sample volume, 30% PEG-NaCl addition, 6-h incubation, and sample analysis within 24 h of collection can result in much better RNA recovery (RT-qPCR: 72% for N1 and 82% for N2; RT-ddPCR: 55% for N1 and 85% for N2) when compared with commonly used PEG-based method; 2) for sludge, the sample analysis using raw sewage protocol and all other variations of each factor mostly resulted in false negatives for both N1 and N2. The absence of N1 and N2 suggests that sludge samples probably need a pretreatment step that releases RNA entrapped in sludge solids back into bulk solution. In conclusion, our modified PEG-based concentration method can cut down the analysis time at least by half, which in turn helps to implement early detection system for SARS-CoV-2 in wastewater.
Keywords: modified PEG-NaCl concentration, RT-qPCR, RT-ddPCR, COVID-19, virus extraction, wastewater-based epidemiology
INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first detected in Wuhan, China, in December 2019. Since then, the virus has quickly spread globally, causing more than 130 million confirmed infections as of April 2021, with more than 2.9 million deaths. 1 SARS-CoV-2 transmits primarily through close contacts and inhalation of contaminated aerosol droplets causing respiratory tract disease called coronavirus disease 2019 (COVID-19).2 The clinical approach to detecting COVID-19 includes testing the samples from the respiratory tract of the suspected patients using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). 3 However, clinical testing is only capable of partially portraying the community health scenario because of factors including but not limited to testing capacity and cost. Furthermore, testing every member within a community using a clinical approach is unrealistic from an epidemiologic standpoint for monitoring public health. 4 To further complicate the matter, patients with COVID-19 can be both symptomatic and asymptomatic, and the ratio between symptomatic and asymptomatic varies greatly. 5,6
SARS-CoV-2 tends to bind angiotensin-converting enzyme 2 in lungs, oral mucosa, and small intestine, 7,8 and almost 10% of the patients with COVID-19 reported having gastrointestinal symptoms. 9 Thus, the probability of shedding SARS-CoV-2 during excretion is very high, and it has been reported 48.8% of the patients have shed SARS-CoV-2 via fecal matter during the infection period.10 These viruses in fecal matter will eventually end up in wastewater at the wastewater treatment facilities (WWTFs). It has been established that infected individual’s feces contain 104–108 SARS-CoV-2 RNA copies per gram, 11 which can be detected in wastewater (influent, effluent, and sludge) up to 106 RNA copies per liter. 12–15 Detection of these RNA copies in wastewater may provide a holistic view of COVID-19 dynamics within a given community. Furthermore, wastewater testing is capable of capturing virus circulation from both symptomatic and asymptomatic patients. Hence, accurate quantitation of SARS-CoV-2 in wastewater will enable implementation of wastewater-based epidemiology tools for population-wide COVID-19 monitoring. 16,17 This can also complement clinical testing and strengthen the public health decision-making process.
To develop an early virus detection system by testing wastewater, it is imperative to have a reliable method for SARS-CoV-2 RNA analysis that addresses issues from sample collection to data reporting. The general workflow for SARS-CoV-2 testing in wastewater involves sample collection, sample concentration, RNA extraction and analysis, and data reporting. This workflow is also important for gaining insights into the fate and transport of SARS-CoV-2 in WWTFs. 18–20 Therefore, both successful implementation of wastewater-based epidemiology and the fate and transport of SARS-CoV-2 depends on at least one limiting step: sample concentration, which results in collection of viral material from wastewater. The success of subsequent extraction and analysis steps depends on the availability of high-quality viral material obtained by sample concentration.
Multiple methods, including polyethylene glycol (PEG)-NaCl precipitation, ultrafiltration, AlCl3, flocculation, among others, have been frequently used for virus concentration.3,20 PEG-NaCl precipitation is the most commonly used method because of its higher selectivity and resistance to PCR inhibitors in wastewater. 21 The RNA recovery using the PEG-NaCl method depends on several factors, including sample volume, percentage PEG-NaCl addition, incubation period, and storage duration at 4°C after sample collection. Recent reports indicate that the immediate research focus should be on these factors so that the current PEG-based concentration protocol could be optimized to achieve higher RNA recoveries. 18
The main objective of this study was to evaluate the effect of 4 key factors—sample volume, percentage PEGNaCl, incubation period, and storage duration at 4°C—on recovery of spiked noninfectious SARS-CoV-2 RNA in wastewater samples. For this study, the raw sewage and sludge samples were collected from a local WWTF and used in all experiments. The 4 factors were varied as follows: sample volume (50 ml, 100 ml, 250 ml), PEG-NaCl addition (10%, 20%, and 30% w/v ratio), incubation time at 4°C after 10% (w/v) PEG-NaCl solution addition (6 h, 12 h, and 18 h), and storage duration at 4°C (1 d, 7 d, and 14 d after sampling). The RNA in the samples was quantified using both RT-qPCR and reverse transcription-digital droplet polymerase chain reaction (RT-ddPCR) techniques.
MATERIALS AND METHODS
Reagents
PEG-8000 was purchased from MilliporeSigma ((Missouri, USA). NaCl and RiboLock RNase inhibitors were purchased from Thermo Fisher Scientific (Waltham, MA, USA). PEG-NaCl solution was prepared in Milli-Q ultra-pure water (>18.2 MΩ). NucleoMag DNA/RNA Water kit (744220.1) was purchased from Macherey-Nagel (Düuren, Germany). Luna SARS-CoV-2 RT-qPCR Multiplex Assay Kit was purchased from New England Biolabs (E3006L; Ipswich, MA, USA), COVID-19 primer probes and positive control DNA were purchased from Integrated DNA Technologies (IDT) (2019-nCOV_N_Positive Control, catalog number 10006625) (Coralville, IA, USA), Twist Bioscience (SARS-CoV-2 RNA Control 2, MN908947.3), and One-Step RT-ddPCR Advanced Kit for Probes (186-4021). An enveloped viral control was purchased from ZeptoMetrix [NATSARS(COV2)-ERC#325641, NY, USA].
Sample collection
In total, 2 L of raw sewage and sludge samples was aseptically collected from the Burlington Main WWTP (Burlington, VT, USA) on March 1, 2021. The raw sewage sample was a 24-h flow proportioned composite, and the sludge material was a grab sample collected from the primary sludge tank. After collection, the samples were transported in a cooler box to the laboratory within 30 min, and they were either stored at 4°C or immediately extracted for RNA.
Preparation of stock PEG-NaCl solution
In 100 ml ultrapure water, 8.75 g of NaCl and 50 g PEG-8000 were dissolved. The pH of the solution was adjusted to 7.00 ± 0.02 by adding 0.01 M NaOH and filtered through a 0.2-μm nitrocellulose mixed ester filter (A020A047A, Advantec, Japan) and stored at −20°C until use. 13
SARS-CoV-2 RNA concentration
Based on previously published protocols, 13,15 a modified PEG-NaCl method was developed to concentrate the virus material in the raw sewage and sludge samples. First, a standard protocol for virus concentration was developed, and then the 4 factors in this protocol were varied to study their effect on SARS-CoV-2 RNA recovery in terms of N1 and N2 gene quantities.
Standard protocol for virus concentration
Homogeneous samples of raw sewage and sludge samples were prepared on an orbital shaker shaking at 150 rpm for 30 min at 4°C. In total, 100 ml of each sample was spiked with 100 μl (concentration: 4:75×107 gene copies per liter) of noninfectious SARS-CoV-2 RNA (ZeptoMetrix NATrol) and mixed thoroughly on the shaker for 30 min. The mixture was then centrifuged (Eppendorf 5804R centrifuge, USA) at 4500 g for 30 min at 4°C to remove the suspended solids from the mixture. After centrifugation, the supernatant was carefully collected and filtered using a 2-μm membrane to remove any remaining suspended solids. Then, 10% (w/v) stock PEG-NaCl solution was added to the filtered sample, mixed thoroughly for 2 min, and incubated for 12 h at 4°C. After incubation, the sample was centrifuged at 8000 g, and supernatant was carefully discarded without disturbing the pellet. The resulting pellet was resuspended in 1 ml of 10% (w/v) PEG-NaCl solution at 4°C and centrifuged (Eppendorf 5415C centrifuge, USA) at 12,000 g for 20 min. After discarding the supernatant, 2.5 μl RNase inhibitor (EO0382; Thermo Fisher Scientific) was added to each pellet and stored at −80°C until further analysis. This sample concentration process is referred to as the standard protocol. Table 1 shows both the factors that were varied and those that were kept constant during each test. The effect of storage duration at 4°C on RNA recovery was evaluated using the above standard protocol.
TABLE 1.
Factors that affect the recovery of SARS-CoV-2 RNA in wastewater
| Sample volume (ml) | PEG-NaCl % (w/v) | Incubation time (h) | Storage duration at 4°C (day) | |
|---|---|---|---|---|
| Sample volume (ml) | 50, 100, and 250a | 10 | 12 | 1 |
| PEG-NaCl (%) | 100 | 10, 20, and 30a | 12 | 2 |
| Incubation time (h) | 100 | 10 | 6, 12, and 18a | 3 |
| Storage duration at 4°C (day) | 100 | 10 | 12 | 1, 7, and 14a |
The test values of each factor compared with the standard protocol.
RNA extraction and analysis
Preserved sample pellets were resuspended in 50–200 μl of RNA-free water, and the RNA was extracted using NucleoMag DNA/RNA kit according to the manufacturer’s instruction (744220.1; Macherey-Nagel). Purified RNA was eluted in 40 μl of RNase-free water. RT-qPCR (CDC-006-00005) and RT-ddPCR methods were used to determine SARS-CoV-2 RNA copy number in wastewater samples. Zeptometrix NATrol SARS-CoV-2 external run control was used as spike positive control. No Template Control was used as a negative control for each run.
RT-qPCR
One-step RT-qPCR was used for the primary determination of SARS-CoV-2 RNA copy number from 5 ml of purified RNA for each sample. Master mix reactions containing primer and probe sets for both the N1 and N2 regions were prepared as outlined by the standard Centers for Disease Control and Prevention (CDC) document (2019-nCov CDC EUA Kit, 10006770; IDT). Standard curves were prepared from 2 positive controls including the Twist Bioscience and the IDT using dilutions of 104–101, 50, and 25 copies. Luna SARS-CoV-2 RT-qPCR Multiplex assay kit was used as the qPCR master mix on the Quant-Studio 6 Flex instrument (Thermo Fisher Scientific) with the protocol described by the CDC. The detection limit of the RT-qPCR is 5 copies per microliter.
RT-ddPCR
Because digital droplet PCR has the benefit of generating discrete molecular enumeration without the use of a standard curve, it was used to determine copy number of SARS-CoV-2 RNA using the same primer probes for N1 and N2 as RT-qPCR. In total, 5 μl of RNA was subjected to the ddPCR protocol with the One-Step RT-ddPCR Advanced Kit for Probes. Although the same IDT primer probes were used for the ddPCR master mix, the N1 and N2 primer probes were modified to include Iowa Black-Zeta as the dye chemistry reporter instead of the standard Black Hole Quencher. Thermocycling protocol was 50°C for 60 min, 95°C for 10 min and 40 cycles of 94°C for 30 s, 55°C for 60 s, and enzyme deactivation at 98°C for 10 min. The detection limit of the RT-ddPCR is 0.4 gene copies per microliter.
RESULTS
The effect of 4 key factors—sample volume, percentage PEG-NaCl, incubation period, and storage duration at 4°C—on N1 and N2 gene recoveries and concentration is described below. Overall, RT-qPCR analysis resulted in a mean recovery efficiency of 72% for N1 and 82% for N2. RT-ddPCR resulted in a mean recovery efficiency of 55% for N1 and 85% for N2.
Effect of raw sewage sample volume
Appropriate volume is an indispensable component of the SARS-CoV-2 virus concentration. A literature review suggests that different sample volumes ranging from 30 ml to 250 ml 12,18 have been used for virus concentration. However, a recommendation for optimal sample volume that results in the best SARS-CoV-2 RNA recovery has not been established yet. In this study, the effect of sample volume on recovery of virus RNA was studied using 50 ml, 100 ml, and 250 ml. Although both 50-ml and 100-ml samples produced similar N1 gene copies per liter, the concentration of N1 in 250 ml had decreased by 37% (Fig. 1). Although N2 values were consistently lower than N1, the N2 trend was similar to that of N1. RT-ddPCR analysis consistently produced an order of magnitude lower number of gene copies per liter (Fig. 1B) when compared with the RT-qPCR results (Fig. 1A). This is probably due to the use of the IDT control instead of an RNA control, such as the Twist or ZeptoMetrix. The results from this study are also consistent with the City of Burlington SARS-CoV-2 RNA monitoring data. The city also carried out an independent SARS-CoV-2 wastewater surveillance project through an independent contractor that found the RNA target at appropriately 3.5×105 gene copies per liter in their influent raw sewage sample. 22
FIGURE 1.
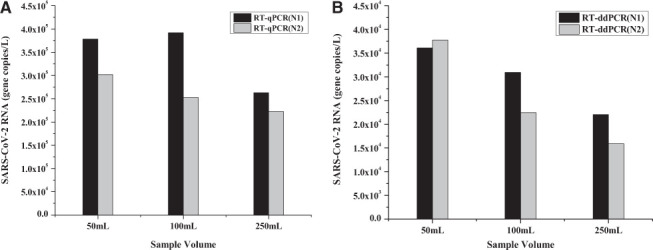
SARS-CoV-2 RNA concentrations (N1 and N2 genes) in different raw sewage sample volumes. These samples were treated with 10% (w/v) PEG-NaCl, incubated for 12 h, and analyzed using both RT-qPCR (A) and RT-ddPCR (B).
Effect of percentage PEG-NaCl solution
The addition of PEG-NaCl solution is central to the viral material precipitation, and in a previous work, 13 10% (w/v) PEG-NaCl was used for virus enrichment. To study the effect of PEG addition on RNA recovery, 10%, 20%, and 30% (w/v) PEG-NaCl solutions were added to the filtered raw sewage sample, and the RNA was quantified using RT-qPCR and RT-ddPCR. Fig. 2 indicates that the overall RNA yield is constant in the presence of 10% and 20% PEG-NaCl. In the presence of 30% PEGNaCl, both N1 and N2 were higher (Fig. 2A). On the contrary, RT-ddPCR analysis revealed an opposite trend for N1 and N2, i.e., N1 increased and N2 decreased, in the presence of 30% PEG-NaCl. A possible reason might be that PCR inhibitors in the sample might have interfered with RNA amplification. 23 Nonetheless, the addition of 30% PEG-NaCl nearly doubled the N1 yield from 3.92×105 gene copies per liter to 6.42×105 gene copies per liter (Fig. 2A). Similarly, the N1 yield also increased from 3.09×104 gene copies per liter to 4.85×104 gene copies per liter (Fig. 2B).
FIGURE 2.
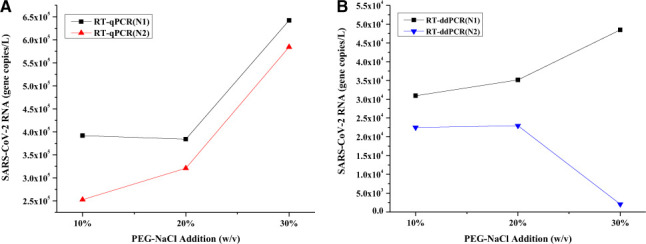
SARS-CoV-2 RNA concentrations (N1 and N2 genes) in 100-ml raw sewage. These samples were treated with different percentages PEG-NaCl, incubated for 12 h, and analyzed using both RT-qPCR (A) and RT-ddPCR (B).
Effect of incubation time
The raw sewage sample incubation period after the addition of 10% (w/v) PEG-NaCl solution is a critical step. The incubation time should be long enough to capture a significant portion of the viral material yet short enough to allow the processing of sufficient sample volume within a reasonable amount of time (e.g., a few hours). The standard incubation time reported in the literature 17,24,25 varied between 12 and 18 h. However, if the incubation time can be shortened without compromising the RNA quality and concentration, more samples can be processed at a reduced time. Fig. 3 shows the effect of incubation time period, including 6 h, 12 h, and 18 h, on RNA recovery. Fig. 3A shows that with RT-qPCR, N1 and N2 levels were similar at all incubation times, except for N2, which appeared to have decreased at the end of 18 h. Fig. 3B shows that with RT-ddPCR, the N1 and N2 were detected at similar levels at the end of 6 h, and thereafter, N1 had slightly decreased but remained at a similar level in 12-h and 18-h samples. On the contrary, N2 exhibited an order of magnitude decrease at the end of 12 h and then increased slightly at the end of 18 h. These RT-ddPCR results necessitate further investigation into the effect of incubation time on the stability of N2 region on the SARS-CoV-2 RNA.
FIGURE 3.
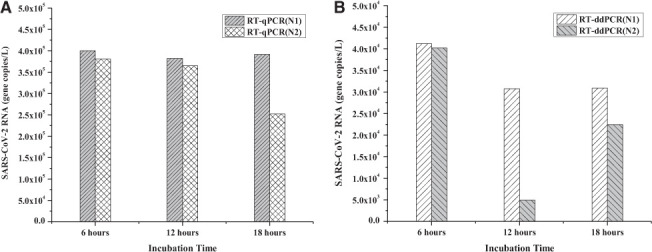
SARS-CoV-2 RNA concentrations (N1 and N2 genes) in 100-ml raw sewage. These samples were treated with 10% (w/v) PEG-NaCl; incubated for 6, 12, and 18 h; and analyzed using both RT-qPCR (A) and RT-ddPCR (B).
Effect of storage duration on RNA at 4°C
The study of SARS-CoV-2 RNA degradation during storage at 4°C is important because multiple factors could affect RNA quality in raw sewage sample. For instance, the distance between sampling location (WWTF) and sample processing laboratory, transportation time, storage temperature, and the time lag between sample collection and processing. For this study, samples were stored at 4°C and processed after 1 d, 7 d, and 14 d to examine the effect of storage duration on RNA quality and recovery. Fig. 4 indicates that the highest gene copy number was observed in the samples that were stored for a day after collection. After 1 wk of storage, RT-qPCR revealed that the concentration of N1 and N2 genes was decreased by 37% and 50%, respectively. When compared with the 1-wk samples, the N1 and N2 concentrations in the 2-wk samples apparently increased by 10% and 80%, respectively. This type of increase is only possible if some of the previously intact SARS-CoV-2 were lysed during storage, releasing their RNA into the bulk solution. The N1 and N2 trends obtained by RT-ddPCR were similar to that of RT-qPCR, and these results are also consistent with the previous reports. 17,24,25
FIGURE 4.
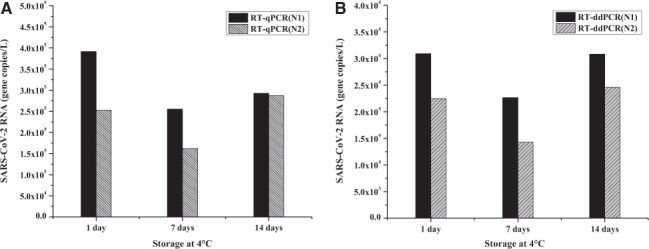
SARS-CoV-2 RNA concentrations (N1 and N2 genes) in 100-ml raw sewage that were stored for 1 d, 7 d, and 14 d at 4°C. The samples were treated with 10% (w/v) PEG-NaCl, incubated for 12 h, and analyzed using both RT-qPCR (A) and RT-ddPCR (B).
Sludge sample analysis
All sludge samples were analyzed using the methods described above. Interestingly, the results from the standard protocol using RT-qPCR showed that the sludge samples contained 2.06 ×105 N1 gene copies per liter but no N2 genes. The RT-ddPCR analysis showed nondetects for both N1 and N2. When various factors were varied, as shown in Table 1, both PCR techniques showed nondetect for N1 and N2. The absence of N2 with RT-qPCR and the absence of both N1 and N2 with RT-ddPCR were interpreted as false negatives.
DISCUSSION
Noninfectious SARS-CoV-2 RNA from ZeptoMetrix was used in this study as a synthetic control for matrix spike experiments to determine RNA recovery. The RT-qPCR results showed N1 and N2 gene copies were consistently an order of magnitude higher than that of RT-ddPCR. In this study, the recovery efficiencies were 5- to 8-fold higher than those reported in other studies. For instance, reported recoveries were in the range of 3%–10% for surrogate viruses, such as porcine coronavirus, murine hepatitis virus, bovine coronavirus, and vesicular stomatitis virus.24–28 In the RT-qPCR analysis, the mean threshold count (Ct) number indicates the required number of cycles for PCR fluorescent signal needed to exceed the background level for a positive detection. 29 In this study, the Ct for N1 and N2 genes were 31.18 ± 0.47 and 32.47 ± 0.46, respectively, and it has been reported that data with a Ct < 40 are acceptable as a positive detection. 12,15,18 However, it is the experience of the authors that any data with a Ct > 37.0 should be carefully considered and scrutinized. It is widely accepted that lower Ct values indicate a higher abundance of RNA as well as possibly better quality of RNA. Therefore, lower Ct values indicate higher abundance of SARS-CoV-2 RNA in the samples, which in turn correlates with the extent of COVID-19 within a community. 21,30
Results showed that using a smaller sample volume, such as 50 ml or 100 ml, for the PEG-NaCl reaction can yield a higher gene copy number per unit volume than 250 ml (Fig. 1). Multiple factors could be responsible for reduced RNA copies in higher sample volumes: for example, an insufficient addition of the PEG-NaCl reagent, inefficient mixing of sample contents, and/or increased activity of RNases unable to be inhibited by the RNase inhibitor added during this study. More research is required to determine the effect of the ratio of RNase inhibitor and raw sewage volume on RNA recovery efficiency.
RNA recovery was also significantly improved when the percentage of PEG-NaCl increased. Addition of 30% PEG-NaCl to the raw sewage increased the RNA yield by 2-fold when compared with the 10% PEG-NaCl (Fig. 2). Because SARS-CoV-2 is a lipid-based enveloped virus and hydrophobic,31 it has higher affinity towards the PEG-NaCl, and thereby gets enriched in the PEG layer.32 Theoretically, addition of a higher percentage of PEG-NaCl solution should yield higher gene copies of the virus per liter.
Results also revealed that the sample incubation period could have a significant effect on N1 and N2 concentrations (Fig. 3). For instance, although the incubation period (6, 12, or 18 h) had negligible effect on the N1 concentration, the N2 concentration appeared to have significantly decreased in the samples stored for 18 h. The apparent decrease of N2 concentration can be attributed to 1) mismatch of melting temperatures between N2 primer and N2 region on the RNA 33,34 and/or 2) deterioration of RNA quality over time wherein the N2 primer is biased to the 3′ end of the genome. 35
Results showed that sample storage duration at 4°C can have a significant effect on RNA quality and recovery (Fig. 4). The highest concentrations of N1 and N2 were observed in samples stored for a day. After 1 wk of storage, the concentrations of N1 and N2 were decreased by 37% and 50%, respectively. This result suggests that N1 is either less susceptible to degradation or has a better primer design than N2. After 2 wk of storage, N1 and N2 have apparently increased by 10% and 80% when compared with the 1-wk samples. These N1 and N2 increases in the samples at the end of 2 wk could be attributed to an additional RNA release due to lysis (via natural degradation) from previously intact viruses. Further work is needed to verify this hypothesis.
Finally, the results from sludge sample analysis showed the presence of N1 and absence of N2 genes under standard protocol conditions with RT-qPCR analysis. Each factor when varied from the standard protocol resulted in nondetects for both N1 and N2. This apparent absence of N1 and N2 in the samples is probably due to the entrapped RNA in the sludge solids that is not readily available for PCR reaction and detection. A specific protocol to process sludge samples needs to be developed that includes a mechanism to break up and release RNA into bulk solution prior to PEG-based concentration.
Conclusions
Overall, this study demonstrated the effect of 4 key factors—sample volume, percentage PEG, incubation time, and storage duration at 4°C—on RNA enrichment and recovery from raw sewage samples. Better RNA recoveries in terms of N1 and N2 were obtained with the following set of conditions: smaller sample volume (50 to 100 ml), 30% (w/v) PEG-NaCl, shorter incubation time (≤12 h), and ≤24 h of storage duration. RT-qPCR always resulted in RNA concentrations at least one order of magnitude higher than that of RT-ddPCR. On the contrary, both RT-qPCR and RT-ddPCR showed that RNA is mostly absent in the sludge samples under all test conditions, which is a false-negative result. These false negatives indicate that the sludge samples likely need a pretreatment step, for example, sonication to release entrapped RNA into surrounding bulk solution, which can then be enriched and recovered for analysis. Finally, the modified PEG-based concentration method with the set of conditions identified above can reduce the wait time for results by half, which in turn helps to implement early detection system for SARS-CoV-2 in wastewater.
ACKNOWLEDGMENTS
The authors thank the Burlington wastewater facilities manager Mr. Matt Dow for supporting the research by providing samples, Mr. Benjamin Joseph Page for assistance with sample collection, and the Office of the Vice President for Research (OVPR), the University of Vermont, for funding the study.
REFERENCES
- 1.Johns Hopkins University 2021. Available at: https://coronavirus.jhu.edu/map.html Access Date: 04/15/2021.
- 2.CDC 2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/how-covid-spreads.html Access Date: 04/15/2021.
- 3.Dumke R, de la Cruz Barron M, Oertel R, Helm B, Kallies R, Berendonk TU, Dalpke A. Evaluation of two methods to concentrate SARS-CoV-2 from untreated wastewater. Pathogens . 2021;10:195. doi: 10.3390/pathogens10020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cevik M, Bamford CGG, Ho A. COVID-19 pandemic-a focused review for clinicians. Clin Microbiol Infect . 2020;26:842–847. doi: 10.1016/j.cmi.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hata A, Honda R. Potential sensitivity of wastewater monitoring for SARS-CoV-2: comparison with norovirus cases. ACS Publications; Japan: 2020. [DOI] [PubMed] [Google Scholar]
- 6.Murakami M, Hata A, Honda R et al. Letter to the editor: wastewater-based epidemiology can overcome representativeness and stigma issues related to COVID-19. Environ Sci Technol . 2020;54:5311–5311. doi: 10.1021/acs.est.0c02172. [DOI] [PubMed] [Google Scholar]
- 7.Bivins A, North D, Ahmad A. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. ACS Publications Indiana; United States: 2020. [DOI] [PubMed] [Google Scholar]
- 8.Larsen DA, Wigginton KR. Tracking COVID-19 with wastewater. Nat Biotechnol . 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polo D, Quintela-Baluja M, Corbishley A, Jones DL, Singer AC, Graham DW, Romalde JL. Making waves: Wastewater-based epidemiology for COVID-19 - approaches and challenges for surveillance and prediction. Water Res . 2020;186:116404. doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morone G, Palomba A, Iosa M, Caporaso T, De Angelis D, Venturiero V, Savo A, Coiro P, Carbone D, Gimigliano F, Iolascon G, Paolucci S. Incidence and Persistence of Viral Shedding in COVID-19 Post-acute Patients With Negativized Pharyngeal Swab: A Systematic Review. Front Med (Lausanne) . 2020;7:562. doi: 10.3389/fmed.2020.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar-On YM, Flamholz A, Phillips R, Milo R. SARS-CoV-2 (COVID-19) by the numbers. eLife . 2020;9:e57309. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ . 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kocamemi BA, Kurt H, Sait A. SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. 2020 medRxiv. [Google Scholar]
- 14.La Rosa G, Iaconelli M, Mancini P, Bonanno Ferraro G, Veneri C, Bonadonna L, Lucentini L, Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ . 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medema G, Heijnen L, Elsinga G. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett . 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 16.Daughton CG. Wastewater surveillance for population-wide Covid-19: The present and future. Sci Total Environ . 2020;736:139631. doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez R, Curtis K, Bivins A, Bibby K, Weir MH, Yetka K, Thompson H, Keeling D, Mitchell J, Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res . 2020;186:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitajima M, Ahmed W, Bibby K, Carducci A, Gerba CP, Hamilton KA, Haramoto E, Rose JB. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci Total Environ . 2020;739:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel M, Chaubey AK, Pittman CU, Jr, Mlsna T, Mohan D. Coronavirus (SARS-CoV-2) in the environment: Occurrence, persistence, analysis in aquatic systems and possible management. Sci Total Environ . 2021;765:142698. doi: 10.1016/j.scitotenv.2020.142698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pecson BM, Darby E, Haas CN, Amha YM, Bartolo M, Danielson R, Dearborn Y, Di Giovanni G, Ferguson C, Fevig S, Gaddis E, Gray D, Lukasik G, Mull B, Olivas L, Olivieri A, Qu Y, SARS-CoV-2 Interlaboratory Consortium Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ Sci (Camb) . 2021;7:504–520. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar M, Kuroda K, Patel AK, Patel N, Bhattacharya P, Joshi M, Joshi CG. Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with Upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques. Sci Total Environ . 2021;754:142329. doi: 10.1016/j.scitotenv.2020.142329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burlington Co 2021. Available at: https://coronavirus-response-burlingtonvt.hub.arcgis.com/pages/wastewater-monitoring Access Date: 05/24/2021. [Google Scholar]
- 23.Rački N, Dreo T, Gutierrez-Aguirre I, Blejec A, Ravnikar M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods . 2014;10:42. doi: 10.1186/s13007-014-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Aoust PM, Mercier E, Montpetit D, Jia JJ, Alexandrov I, Neault N, Baig AT, Mayne J, Zhang X, Alain T, Langlois MA, Servos MR, MacKenzie M, Figeys D, MacKenzie AE, Graber TE, Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res . 2021;188:116560. doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res . 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuevas-Ferrando E, Pérez-Cataluña A, Allende A, Guix S, Randazzo W, Sánchez G. Recovering coronavirus from large volumes of water. Sci Total Environ . 2021;762:143101. doi: 10.1016/j.scitotenv.2020.143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miura Y, Hoshino Y, Seto H. Glycopolymer nanobiotechnology. Chem Rev. 2016;116:1673–1692. doi: 10.1021/acs.chemrev.5b00247. [DOI] [PubMed] [Google Scholar]
- 28.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Zhou X, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med . 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wisconsin-Madison Uo Available at: https://www.wvdl.wisc.edu/wp-content/uploads/2013/01/WVDL.Info_.PCR_Ct_Values1.pdf Access Date: 05/24/2021.
- 30.Chang MC, Hur J, Park D. Interpreting the COVID-19 test results: a guide for physiatrists. Am J Phys Med Rehabil . 2020;99:583–585. doi: 10.1097/PHM.0000000000001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Liu M, Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc Natl Acad Sci USA . 2020;117:13967–13974. doi: 10.1073/pnas.2008209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aydin H, Al-Khooly D, Lee JE. Influence of hydrophobic and electrostatic residues on SARS-coronavirus S2 protein stability: insights into mechanisms of general viral fusion and inhibitor design. Protein Sci . 2014;23:603–617. doi: 10.1002/pro.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bustin S, Huggett J. qPCR primer design revisited. Biomol Detect Quantif . 2017;14:19–28. doi: 10.1016/j.bdq.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogels CBF, Brito AF, Wyllie AL et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol . 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brisco MJ, Morley AA. Quantification of RNA integrity and its use for measurement of transcript number. Nucleic Acids Res . 2012;40:e144–e144. doi: 10.1093/nar/gks588. [DOI] [PMC free article] [PubMed] [Google Scholar]


