Abstract
A highly efficient, selective, and sensitive method for analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in raw sewage was developed and tested to illustrate basic characteristics of the procedure. The method uses reverse transcriptase (RT) loop mediated isothermal amplification (LAMP) in a quantitative application, RT qLAMP. The applicability of this procedure to detection of SARS-CoV-2 in clinical samples has been documented in many reports since early 2020. Basic LAMP characteristics depending on the multiple primer design that produce highly selective and sensitive target amplification virtually free of interferences in complex sample media make it ideal for application to target recognition in raw sewage. Three previously described primer sets targeting ORF1a, E- and N-gene regions were selected and tested to define method performance characteristics and performance for SARS-CoV-2 detection in raw sewage samples from a municipal sewage system serving > 600 000, between July and October, 2020. The virus was detected in all samples from each of three independent interceptors near their treatment terminus. Virus quantities varied significantly between samples and between primer targets within samples. Sewage sampling dates corresponded to relatively low COVID-19 incidence rates reported by the local service area health department. The limited number of samples and aggregating downstream sampling locations did not permit resolving concentration differences. The most significant finding was the ability of the RT qLAMP method to detect SARS-CoV-2 in the raw sewage samples directly without preprocessing to isolate or concentrate the virus or to extract and concentrate viral RNA.
INTRODUCTION
The purpose of this report is to describe the ability of loop-mediated isothermal amplification (LAMP), in the form of reverse transcriptase (RT) quantitative (q)LAMP, to detect and quantify severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in raw sewage, directly, that is, without sample processing for virus concentration or RNA extraction. We provide information on the routine application, equipment, and facilities used to illustrate the feasibility of RT qLAMP application for detailed monitoring of SARS-CoV-2 for wastewater-based epidemiology. The most important and novel aspect of this report is demonstration that, even at low reported case rates, for example, 1 to 10/100 000, in a community, the SARS-CoV-2 virus is present in raw sewage at concentrations greater than 1 to 5/μL, sufficient for LAMP-based detection directly, avoiding the quantitative polymerase chain reaction (qPCR) need for cumbersome time-consuming concentration and RNA extraction. Incorporation of this analytic approach will facilitate development of data supporting wastewater-based epidemiology as an important component of policy advice directed to corona virus disease 2019 (COVID-19) control.
The LAMP method is not novel and it is not new. It is a thoroughly demonstrated and well-understood nucleic acid–amplification procedure first described 20 years ago,1 and since developed for largely clinical applications but equally demonstrated for detection of DNA or RNA in a wide variety of viral, microbial, and protozoan pathogens, as well as for identifying gene-specific targets in plants and animals.2,3 The principal characteristics of LAMP include the use of 4 to 6 primers annealing to initially 4, then 6, target sites selected to meet well-established criteria.4 The amplification mechanism is strand extension with a loop formation producing what has been described as a cauliflower-like product, producing a characteristic ladder-band appearance on confirming gels. The multiple primer-target combination gives the process a very high degree of specificity, enabling target detection in crude preparations containing extraneous nucleic acids. The specificity also permits effective multiplex applications. The process uses a polymerase having strand-extension activity, typically Bst, acting at constant temperature in the 60°C to 70°C range. Operation at constant temperature permits amplification with simple means of maintaining constant temperature, such as a water bath, facilitating application in areas of limited laboratory facilities. The process has been found to be insensitive to interferences common to conventional PCR processes applied to analysis of environmental samples. Furthermore, the process is fully as sensitive as conventional PCR and amplification times, eg cycle threshold (Ct), which is typically short, ie, 20 to 40 minutes.3
The relatively slow adoption of the LAMP procedure for environmental monitoring, specifically for water and wastewater, is partly characteristic of processes having few detailed published reports to stimulate the interest of other investigators. Two features of LAMP may be described as disadvantages: (1) the rather intricate process of primer design, testing, and optimization needed to permit routine application; and (2) the very high sensitivity that would permit cross-contamination if not recognized and precluded by proper laboratory procedures and careful techniques.
The worldwide spread of the SARS-CoV-2 infection and COVID-19 disease throughout the human population has stimulated massive effort to develop and improve the ability to detect and to monitor the virus.5 The standard method being applied at the beginning of the first quarter of 2021 to both clinical detection and environmental monitoring of sewage is PCR, typically qRT-PCR.6,7 However, building on previous clinical applications, many variations of LAMP-based procedure have been reported.3 Because it does not require a thermocycler, the LAMP process lends itself to both scale-up and miniaturization and can be combined with increasingly sophisticated technology and downstream refinements, including sequencing. Numerous reports of LAMP-based assays for SARS-CoV-2 detection in clinical samples are available,8–10 providing information and encouragement for the development of LAMP-based assays for SARS-CoV-2 in raw sewage.
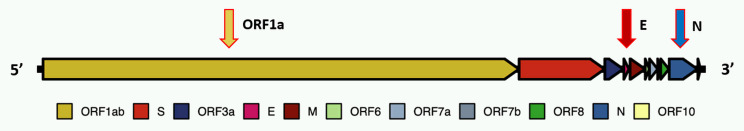
The potential for successful application of LAMP to SARS-CoV-2 monitoring in raw sewage is illustrated by previous experiences in applying a multiplex LAMP to the detection of Cryptosporidium spp. and Giardia spp. in surface water samples.11,12 That work showed that (1) target organisms were detectable in the complex untreated water and wastewater matrix; (2) LAMP could be multiplexed for detection of both simultaneously; (3) organisms are detectable at low concentration of approximately 1 to 5/10 L; (4) detection was not affected by extraneous components in a complex sample concentrate; and (5) quantification using a qPCR instrument (Roche Light Cycler 480) was possible. From early reports on monitoring SARS-CoV-2 in raw sewage,13–15 calculation of likely virus concentrations at a sewage treatment plant serving a population having a COVID-19 daily reported cases in the range of 5 to 10/100 000 of population, suggested that the virus would be detectable without concentration and that LAMP would not be affected by extraneous sewage components. To test that potential, taking advantage of many well-described LAMP primer sets reported for clinical application since February 2020, we selected primers for 3 potential targets (Fig. 1)—ORF-1a,16 E-gene, and N-gene17,18 (Table 1), assembled essential materials, and arranged to obtain raw sewage samples with the local wastewater agency, East Bay Municipal Utility District Special District 1 (EBMUD SD1).
FIGURE 1.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome and subunit arrangement with primer locations.
TABLE 1.
Primer sequences used for direct raw sewage reverse-transcriptase quantitative loop-mediated isothermal amplification (RT qLAMP)
|
Gene |
Primer |
Sequence 5′→3′ |
No. of bases |
| Lamb et al,16 2020, 63°C; Color Genomics,5 2020, 65°C | |||
| ORF 1a | F3 | TCCAGATGAGGATGAAGAAGA | 21 |
| ORF 1a | B3 | AGTCTGAACAACTGGTGTAAG | 21 |
| ORF 1a | FIP | AGAGCAGCAGAAGTGGCACAGGTGATTGTGAAGAAGAAGAG | 41 |
| ORF 1a | BIP | TCAACCTGAAGAAGAGCAAGAACTGATTGTCCTCACTGCC | 40 |
| ORF 1a | LF | CTCATATTGAGTTGATGGCTCA | 22 |
| ORF 1a | LB | ACAAACTGTTGGTCAACAAGAC | 22 |
| Broughton et al,17,18 2020, 62°C; Color Genomics,5 2020, 65°C | |||
| N | F3 | AACACAAGCTTTCGGCAG | 18 |
| N | B3 | GAAATTTGGATCTTTGTCATCC | 22 |
| N | FIP | TGCGGCCAATGTTTGTAATCAGCCAAGGAAATTTTGGGGAC | 41 |
| N | BIP | CGCATTGGCATGGAAGTCACTTTGATGGCACCTGTGTAG | 39 |
| N | LF | TTCCTTGTCTGATTAGTTC | 19 |
| N | LB | ACCTTCGGGAACGTGGTT | 18 |
| Broughton et al,17,18 2020, 62°C; Color Genomics,5 2020, 65°C | |||
| E | F3 | CCGACGACGACTACTAGC | 18 |
| E | B3 | AGAGTAAACGTAAAAAGAAGGTT | 23 |
| E | FIP | ACCTGTCTCTTCCGAAACGAATTTGTAAGCACAAGCTGATG | 41 |
| E | BIP | CTAGCCATCCTTACTGCGCTACTCACGTTAACAATATTGCA | 41 |
| E | LF | TCGATTGTGTGCGTACTGC | 19 |
| E | LB | TGAGTACATAAGTTCGTAC | 19 |
Abbreviations: B3, backward outer; F3, forward outer; BIP, backward inner primer; FIP, forward inner primer; LB, loop backward; LF, loop forward.
MATERIALS AND METHODS
Development, selection, and optimization of amplification conditions for each set of primers, ORF-1a, E-gene, and N-gene, was described in the original references.16,17 Primers were applied here using all of the concentration and amplification conditions established in the original references (ORF1a,16 E-gene and N-gene17). To facilitate testing the approach we used off-the-shelf materials, where possible. Materials used included:
Primers prepared with standard desalting (Integrated DNA Technologies, Coralville, IA)
Master mix: WarmStart LAMP Kit E1700, E2019 (New England Biolabs [NEB], Ipswich, MA)
Control: Synthetic SARS-CoV-2 RNA, control 6 (MT118835) (Twist Bioscience, South San Francisco, CA)
Raw sewage: EBMUD (Oakland, CA)
Reactions for RT-qLAMP were 25 μL total volume, according to proportions listed in Table 2.
TABLE 2.
Direct raw sewage reverse-transcriptase quantitative loop-mediated isothermal amplification (RT qLAMP) reaction mix components
|
Reaction mix component |
Volume |
| WarmStart master mix (1700 or 2019) | 12.5 μL, includes dNTPs at 1.4 mM; RTx, 8 mM MgSO4 |
| 10× primers | 2.5 μL (1.6 μM FIP/BIP, 0.2 μM F3/B3, 0.4 μM loop F/B) |
| NEB green fluorescent dye with E1700 kit | 0.5 μL (@0.5 μM) |
| Nuclease-free H2O | 4.5–0 μL (adjust w/ template to 25 μL total rxn volume) |
| Template: control or raw sewage | 5–9.5 μL |
| Total reaction mix | 25 μL |
Abbreviations: RTx, reverse transcriptase; BIP, backward inner primer; dNTP, deoxynucleoside triphosphate; F/B, forward and backward loop; FIP, forward inner primer; rxn, reaction.
Upon receiving fresh sewage samples, representative aliquots were distributed, along with standards and no-template controls as appropriate, with reaction components, into 0.2-mL PCR reaction tubes on ice during preparation. All reactions were conducted in triplicate. Reactions were prepared in a PCR hood using routine laboratory techniques designed to preclude potential cross-contamination. Amplifications were conducted using a Rotor-Gene Q (QIAGEN, Hilden, Germany) programmed according to the manufacturer's protocol for constant temperature (65°C; 63°C after initial runs), for 30 to 40 minutes, followed by high-resolution melting. Reaction tubes were not opened after run completion and were frozen to permit future analysis.
Raw sewage was obtained from the EBMUD SD1 serving a population of approximately 650 000 largely in Alameda County, CA. Separate regions of the service area contribute flows to the 3 interceptor sewers: North (N), Adeline (ADA), and South (S), which terminate at the single treatment plant (Fig. 2a). Samples of 1 L in total volume were collected from each interceptor individually at a point just before the treatment plant. Samples were 24-hour composites representative of the period 9:00 am to 9:00 am (Fig. 2c). Samples were transported on ice to the laboratory for processing and analysis and kept refrigerated without preservatives until analysis. Sampling dates were July 29, 2020; September 9, 2020; September 22, 2020; and October 1, 2020.
FIGURE 2.
Left, East Bay Municipal Utility District Special District 1 (EBMUD SD1) service area (mauve) with north (blue), Adaline (red), and south (green) interceptors. Center, Alameda County Health District COVID-19 monitoring by zip code. Right, EBMUD SD1 staff retrieving 24-hour samples for compositing.
RESULTS
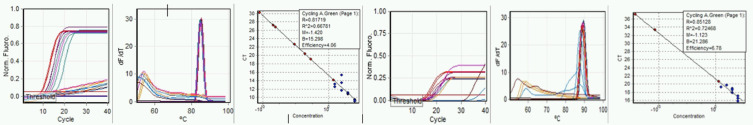
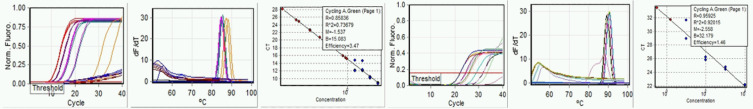
Initial testing of RT LAMP to determine basic performance characteristics was begun using primers to the ORF1a gene. Testing consisted of 5-logs reference standard diluted from 104 to 100, including raw sewage. No acceptable standard curve could be obtained, although raw sewage produced consistent amplification with Ct of about 20 to 35. Performance of the RT qLAMP was then compared for E-gene and N-gene primers (Fig. 3a–f). Standard curve quality was improved but remained low, and significant differences in synthetic control amplification were apparent between E-gene and N-gene amplifications. We continued to include raw sewage with continued apparent amplification. Using melt curves as indicator, raw sewage SARS-CoV-2 RNA amplification appears specific. Not all replicates of either standards or sewage produced product, and generally, standards of less than 102 copies gave inconsistent reproducibility.
FIGURE 3.
Run: September 9, 2020. Left to right, a) E-gene amplification, b) E-gene melt, c) E-gene standard curve, d) N-gene amplification, e) N-gene melt, and f) N-gene standard curve.
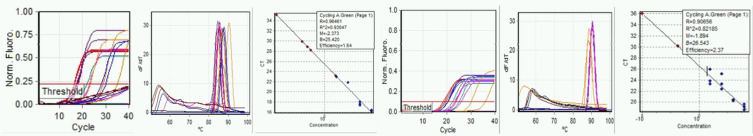
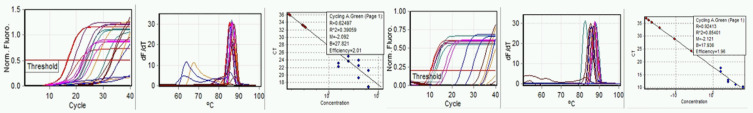
Further comparison of E-gene and N-gene performance was made examining the effect of the reaction mix components, continuing to include raw sewage in amplification runs. Direct comparison between the previous run on September 9, 2020, without alteration of conditions, was made on September 23, 2020. Standard curve quality for both E- and N-gene primers was improved. Amplification of synthetic RNA standards was less consistent, although more consistent amplification from the sewage was observed (Fig. 4a–f).
FIGURE 4.
Run A: September 23, 2020. Left to right, a) E-gene amplification, b) E-gene melt, c) E-gene standard curve, d) N-gene amplification, e) N-gene melt, and f) N-gene standard curve.
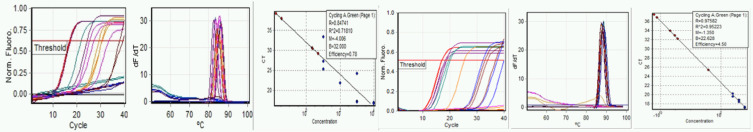
In companion runs on the same day (Fig. 5a–f), different mix components were used; NEB reaction mix 2019 was used instead of NEB reaction mix 1700.
FIGURE 5.
Run B: September 23, 2020. Run using NEB E2019 reaction mix. Left to right, a) E-gene amplification, b) E-gene melt, c) E-gene standard curve, d) N-gene amplification, e) N-gene melt, and f) N-gene standard curve.
Raw sewage was collected again on October 1, 2020. Samples (1 L each from interceptors N, ADA, and S) were composited from 24-hour discrete samples. The 1-L samples were iced for transport to Cel Analytical (San Francisco, CA) and refrigerated for analysis on October 2, 2020. Analysis consisted of preparing 25-μL reactions in triplicate in 4 separate runs, each run consisting of triplicate standards (Twist, M 118835) at dilutions 104 to 101, plus triplicate 7-μL raw sewage from each of the 3 interceptors. Each of the 4 separate runs was conducted using different primer sets: run 13 (Fig. 6a–c; E-gene primers17), run 14 (Fig. 6d–f; N-gene primers17), run 15 (Fig 7a–c), ORF1a primers16), and run 16 (Fig. 7d–f, NEB E2019 Kit N2-gene + E1-gene19).
FIGURE 6.
Runs 13–14: October 2, 2020. Left to right, run 13: a) E-gene amplification, b) E-gene melt curve, and c) E-gene standard; run 14: d) N-gene amplification, e) N-gene melt curve, and f) N-gene standard curve.
FIGURE 7.
Runs 15–16: Left to right, run 15: a) ORF1a-gene amplification, b) ORF1a-gene melt curve, and c) ORF1a-gene standard curve; run 16: d) ORF1a-gene NEB N1+E1 amplification, e) ORF1a-gene NEB N1+E1 melt curve, and f) ORF1a-gene NEB N1+E1 standard curve.
The amplification efficiency for the Twist standards ranged by more than a factor of 2, with the N-gene16 (Fig. 6d–f) as the least efficient, and the E-gene16 (Fig. 6a–c) as the most efficient. Note that the offset (intercept, B) also ranged by a factor of about 2, with N-gene as highest and the E-gene as the lowest. A significant difference in the time to initiation of amplification was observed with the E-gene amplifying in as little as 10 minutes; the N-gene was slowest, not amplifying until after 20 minutes, with the ORF1a and NEB N2+E1 in between.
Amplifications using the ORF1a primers were least consistent in both amplification and melting temperature, with no acceptable standard curve. Amplification using the NEB combined N1+E1 primers was observed to initiate at times between that of the Broughton E-gene17 and the Broughton N-gene17 primers (Fig. 7d). The minor overlap (slight peak separation) in melt curves of the NEB primers suggested comparable amplification by both N1 and E1 primers (Fig. 7e).
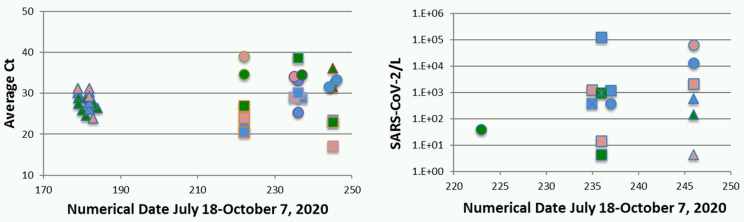
As noted above, detection of SARS-CoV-2 in raw sewage was apparent throughout testing of RT LAMP and RT qLAMP performance. Analysis includes 4 sets of raw sewage samples from each of the 3 EBMUD SD1 interceptor sewers. Samples from July 29, 2020, produced only qualitative results because of an inability to produce a standard curve using the ORF1a primers. Accordingly, all raw sewage amplification results are summarized in terms of the average Ct for each of the 4 sampling dates (July 29, 2020; September 9, 2020; September 22, 2020; and October 1, 2020; Fig. 8a). Based on more-acceptable performance resulting from amplifications summarized above (Figs. 3–7), the quantitative product was determined based on a 7-μL raw sewage component of the 25-μL reaction mix, expressed as SARS-CoV-2/L (Fig. 8b).
FIGURE 8.
a) Raw sewage amplification product, average cycle threshold (Ct), samples from July 29, 2020; September 9, 2020; September 22, 2020; and October 1, 2020. b) Representative calculated concentration, copies/L, samples from September 9, 2020; September 22, 2020; and October 1, 2020. Symbol key: site, N, blue; Adeline, red; S, green; gene: ORF1a, triangle; N, circle; E, square; mix: 1700, filled; 2019, open. Note: some data points are plotted offset from actual dates because of overlap.
DISCUSSION
Finding that RNA from SARS-CoV-2 in raw sewage can be amplified directly without a concentrating pretreatment using RT LAMP may seem surprising. However, several carefully considered factors support the strength of the findings. First, the RT LAMP process has been widely and successfully applied to detection of SARS-CoV-2 in clinical samples beginning early this year, 2020, in response to COVID-19 monitoring needs.5,9 The specific primers used in the work described here were selected from recent and thorough work to develop effective diagnostic tools. Second, accumulating information from both clinical assays and wastewater monitoring,15 indicate that early COVID-19 infection, likely preceding onset of symptoms, results in fecal shedding bursts, estimated in the 1012/d range. Based on that level, shedding by a single individual in 100 000 would contribute to a SARS-CoV-2 concentration at the sewage treatment plant of about 2.5 × 105/L or about 1.5 in a 5-μL template volume for a 25-μL LAMP reaction. Third, LAMP, as a process, has been widely demonstrated to be sufficiently sensitive to amplify template at this level, that is 1 to 10 target copies/μL. Fourth, because of the multiple primer design and isothermal-action polymerase, the LAMP process has selectivity permitting specific amplification in the presence of extraneous nucleic acids and other components of environmental media—sewage specifically—that interfere with more-common PCR analytic methods.
The work described here should be considered preliminary although it provides clear evidence of SARS-CoV-2 detection. The poor performance of the ORF1a primers limiting quantification may have been partly due to its timing early in the sequence of testing. Although we made no procedural changes in succeeding runs, the quality of the LAMP performance does appear to improve over the course of the 6-week testing period. Continuing work is in progress to retest the ORF1a gene primer performance.
Amplification characteristics observed differed between E-gene and N-gene primers (Figs. 4–6). Initiation of amplification appeared somewhat earlier with E-gene primers and appreciably more product resulted. Variation in melt curve peaks that was observed with E-gene amplifications was less apparent in N-gene results. The peak melting temperature for the N-gene product was 2°C to 3°C higher than for E-gene product.
The reactions in Figure 5 were conducted using the NEB E2019 reaction mix, which differs from the E1700 mix used in the Figure 4 reactions by inclusion of deoxythymidine triphosphate (dTTP), deoxyuridine triphosphate (dUTP), and a thermolabile uracil DNA glycosylase (UDG). With consistent use in a sequence of amplification runs and incorporation of deoxyuridine (dU) into amplification products, the presence of UDG will prevent potential carryover from previous reactions but will not affect amplification of the subsequent run because of complete inactivation at 65°C. Our reactions had no predecessors using the E2019 mix, so that effects observed would have been due to the action of the additional mix components on amplification of both control synthetic RNA and components in the raw sewage. Whatever the mechanism, use of the mix increased product formation from both E- and N-gene primers and appreciably improved the efficiency of the N-gene reactions. It is important to note that although the NEB E2019 kit is supplied with E1-gene and N2-gene primers (both sets differ from the E- and N-gene primers used in our work; Table 1), except for run 16 (October 1, 2020; Fig. 7d-f), we did not use the NEB primers. All other reactions used only the primers listed in Table 1.
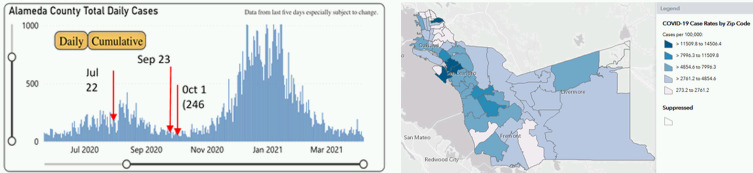
The ability to detect SARS-CoV-2 in sewage is fundamentally dependent on the extent of the COVID-19 infection in the community. A reasonably detailed record of infection history in the EBMUD SD1 service area is maintained by the Alameda County Health Department.20 From the end of July 2020 through September 2020, corresponding to the period of our sewage analysis, the daily reported cases averaged approximately 150/d or, for a population of slightly more than 150,000, about 100/100 000/d (Fig. 9). The data show highly differentiated rates of infection among areas of the county and sewerage service area, cataloged by zip code.
FIGURE 9.
COVID-19 cases reported daily, June 2020 through March 2021 with sewage sampling dates, and cumulative cases per 100,000 of population by postal code, Alameda County, CA.
The total cases reported from March 15, 2020, to September 30, 2020 (200 days) was 21 240, an average of 106/d or 70/100 000/d. Cumulative case rates among zip-code areas ranged from about 300/100 000 to nearly 2000/100 000. Understanding approximate incidence rates is important to the utility of monitoring sewage for understanding COVID-19 dynamics.
Considering how monitoring for SARS-CoV-2 in sewage may be of use in the control of transmission, several factors must be taken into account. These considerations have a direct bearing on the LAMP method described here and on the needs for its refinement. A critical factor is the shedding rate in early, perhaps presymptomatic, infections. Calculations described here indicate that detectable virus would be present in sewage for shedding at 1012/d for a single infection/100 000 in population. However, detection at that level is not a challenge to current detection methods. Many reports have shown that ample SARS-CoV-2 can be measured at the sewage treatment plant, indicating only that infection is amply distributed in the community. To be useful for infection control, the need is to be able to identify and, if possible, isolate and trace contacts of the small number of early, high, perhaps superspreading, infections. Accordingly, monitoring focused on population concentrations of 100 to 1000, such as in institutions, hotels, multistory apartment buildings, and industrial sites, will increase the sensitivity to detect by factors of 100 to 1000 in relation to the original assumption of 1 infection/100 000 in population.
Recognizing that the real value of a method is its ability to apply it to focused, upstream sampling serves as a guide to features of the analytic method needing refinement and optimization. The most challenging problem of an RT qLAMP is refinement at a minimal target concentration. Although theoretically capable of amplification from a single copy, that is, a single SARS-CoV-2 in a volume compatible with a 25-μL reaction mix for example, that is, 5 to 10 μL if the concentration in the sewage being sampled is that low. Poisson statistics dictate that a single copy will be present in only a minor proportion of the replicated 5- to 10-μL aliquots analyzed. However, as suggested above, upstream sampling magnifies the concentration from a single shedder in direct proportion to the reduced population in the target sewage source. Thus, refinement of RT qLAMP procedure for lowest limit of detection would not be important. Demonstration of consistently reproducible amplification at moderate concentrations is essential. Such a demonstration should be readily achievable. Additional features require further attention, including treatment of samples to make viral RNA available for amplification. Pretreatment of samples to release and denature viral RNA is important. It is important to optimize the Mg++ concentration as a reaction mix component. The primer-polymerase combination is sensitive to the total Mg++ concentration, optimized for our primers at 8 mM. Raw sewage includes Mg++; in our raw sewage samples, the amount of Mg++ was about 33 mg/L. We did not adjust or reoptimize to take that into account for the work described here. Future work must account for that component, likely to vary widely among communities. Finally, the samples prepared by EBMUD SD1 staff were 24-hour composite samples. Samples collected at upstream sources are more likely to be grab samples and may be subject to variation during the typical 24-hour cycle of human activities that affect sewage composition around the clock.
Features of an RT qLAMP that are attractive for the type of application outlined above are those referenced in virtually all publications describing its advantages: LAMP is faster, cheaper, highly specific, insensitive to interferences, and more flexible than the RT qPCR procedure currently used almost exclusively for sewage monitoring of SARS-CoV-2. Sewage samples collected in the morning and returned to the laboratory can be combined with reaction mix components immediately and amplified, using the same thermocycler (or a simple water bath) programmed for constant temperature, producing interpretable results in less than an hour. No preprocessing for virus concentration, RNA extraction, or interference mitigation is needed, saving time, effort, and materials.
Acknowledgment
Assistance of the EBMUD engineering and sampling staff was essential to providing of sewage samples and understanding collection system details. This work was made possible using the facilities of Cel Analytical Inc (San Francisco CA; ELAP 2647, EPA ID CA 01529) and with the advice and consultation of Dr Yeggie Dearborn, Cel Analytical owner. This project was conducted without external funding, promises, or commitments.
REFERENCES
- 1. Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saharan P, Khatri P, Dingolia S, Duhan JS, Gahlawat SK Rapid detection of viruses using LAMP: a review In Salar R, Gahlawat S, Siwach P, et al. eds Biotechnology: Prospects and Applications New Delhi, India: Springer; 2013. 287 306 [Google Scholar]
- 3. Becherer L, Borst N, Bakheit M, Frischmann S, Zengerle R, von Stetten F Loop-mediated isothermal amplification (LAMP)—review and classification of methods for sequence specific detection. Anal Methods. 2020;12:717–746. [Google Scholar]
- 4.Primer Design. Tokyo, Japan: Eiken Chemical; 2020. Eiken Chemical. Available at: http://loopamp.eiken.co.jp/e/lamp/primer.html. Accessed July 15, 2020. [Google Scholar]
- 5.Color Genomics. SARS-CoV-2 LAMP Diagnostic Assay. Version 1.0 5.19.20. San Francisco, CA: Color Genomics. 2020. Available at: https://www.color.com/wp-content/uploads/2020/05/LAMP-Diagnostic-Assay.pdf. Accessed July 15, 2020.
- 6.[CDC] Centers for Disease Control and Prevention. Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes. 2020. Updated June 6. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. Accessed July 12, 2020.
- 7.[WHO] World Health Organization. Protocol: Real-time RT-PCR Assays for the Detection of SARS-CoV-2. Available at: https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2. Accessed July 20, 2020.
- 8. Dong Y, Wu X, Li S, et al. Comparative evaluation of 19 reverse transcriptase loop-mediated isothermal amplification assays for detection of SARS-CoV-2. MedRxiv. 2021;11:2936. doi: 10.1101/2020.07.22.20159525. Preprint. Posted online July 24, 2020. . Now published as Sci Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thompson D, Lei Y Mini review: recent progress in RT-LAMP enabled COVID-19 detection. Sens Actuators Rep. 2020;2:100017. doi: 10.1016/j.snr.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaouch MLoop-mediated isothermal amplification (LAMP): an effective molecular point-of-care technique for the rapid diagnosis of coronavirus SARS-CoV-2. [published online ahead of print, January 21, 2021] Rev Med Virol. [DOI] [PMC free article] [PubMed]
- 11. Gallas-Lindemann C, Sotiriadou I, Plutzer J, Karanis P Prevalence and distribution of Cryptosporidium and Giardia in wastewater and the surface, drinking and ground waters in the Lower Rhine, Germany. Epidemiol Infect. 2013;141:9–21. doi: 10.1017/S0950268812002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ongerth JE, Saaed FMAMultiplex loop-mediated isothermal amplification (LAMP) for simplified monitoring of Cryptosporidium and Giardia in surface water. bioRxiv. 2020. Preprint. Posted online August 20. 256826. [DOI]
- 13. Ahmed W, Bertsch PM, Bivins A, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wurtzer S, Marechal V, Mouchel JM, et al. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. Preprint. Posted online May 6, 2020. MedRxiv. 2000776. 10.1101/2020.04.12.20062679. Now published as Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill. 2020;25:2000776. doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu F, Xiao A, Zhang J, et al. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of COVID-19 cases. Preprint. Published online on July 6, 2020. MedRxiv. 150121. 10.1101/2020.06.15.20117747. Now published as SARS-CoV-2 RNA concentration in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci Total Environ. 2020;805:150121. doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamb LE, Bartlone SN, Ward E, Chancellor MB Rapid detection of novel coronavirus (COVID-19) by reverse transcription loop-mediated isothermal amplification. Preprint. Published online February 24, 2020. MedRxiv. 10.1101/2020.02.19.20025155. Now published as Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One. 2020;15:e0234682. doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broughton JP, Deng X, Yu G, et al. Rapid detection of the 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. 2020. Preprint. Published online March 27. [DOI]
- 18. Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:860–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Odiwuor N, Xiong J, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. MedRxiv. 2020. Preprint. Published online February 29. 20028373. [DOI]
- 20.[ACHD] Alameda County Health Department. COVID-19 Data: Real-Time Data of the Impact of COVID-19. Available at: https://covid-19.acgov.org/data.page. Accessed August 24, 2020.