Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) control in the United States remains hampered, in part, by testing limitations. We evaluated a simple, outdoor, mobile, colorimetric reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay workflow where self-collected saliva is tested for SARS-CoV-2 RNA. From July 16, 2020, to November 19, 2020, surveillance samples (n = 4704) were collected from volunteers and tested for SARS-CoV-2 at 5 sites. Twenty-one samples tested positive for SARS-CoV-2 by RT-LAMP; 12 were confirmed positive by subsequent quantitative reverse-transcription polymerase chain reaction (qRT-PCR) testing, whereas 8 tested negative for SARS-CoV-2 RNA, and 1 could not be confirmed because the donor did not consent to further molecular testing. We estimated the false-negative rate of the RT-LAMP assay only from July 16, 2020, to September 17, 2020 by pooling residual heat-inactivated saliva that was unambiguously negative by RT-LAMP into groups of 6 or fewer and testing for SARS-CoV-2 RNA by qRT-PCR. We observed a 98.8% concordance between the RT-LAMP and qRT-PCR assays, with only 5 of 421 RT-LAMP-negative pools (2493 total samples) testing positive in the more-sensitive qRT-PCR assay. Overall, we demonstrate a rapid testing method that can be implemented outside the traditional laboratory setting by individuals with basic molecular biology skills and that can effectively identify asymptomatic individuals who would not typically meet the criteria for symptom-based testing modalities.
Keywords: point-of-care test, saliva-based, colorimetric LAMP
INTRODUCTION
More than 340 000 000 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) diagnostic tests have been performed in the United States as of February 22, 2021; however, it is estimated that 80% to 95% of infected individuals are not tested.1,2 The availability of diagnostic testing for population surveillance around the United States has been limited because of testing supply shortages and guidelines set by public health officials.3,4 Multiple studies have shown that asymptomatic and presymptomatic individuals infected with SARS-CoV-2 can be as infectious as symptomatic individuals are,5–9 with recent estimates of up to 59% of transmission coming from asymptomatic or presymptomatic individuals.10 Virologic assessments of SARS-CoV-2-positive individuals and patients with coronavirus disease 2019 (COVID-19) further support the reports of asymptomatic transmission, identifying no significant differences in viral loads found in the upper respiratory tracts of asymptomatic and symptomatic individuals.5,7,11–13 Furthermore, Arons et al (2020)5 demonstrated that positive viral cultures can be isolated from presymptomatic patients for up to 6 days before the onset of symptoms.
Delays in reporting test results can prevent timely isolation of infected individuals. Because transmission can occur before symptoms manifest, reporting delays create a major barrier to safely returning to workplaces and schools.14 Therefore, there remains an urgent need for rapid tests that identify presymptomatic and asymptomatic individuals and conserves diagnostic testing reagents. Nondiagnostic point-of-care (POC) testing, used in conjunction with the current clinical diagnostic testing regimen, may improve our ability to identify infectious individuals and limit their exposure to others when they are most contagious and help conserve clinical diagnostic tests for those who require confirmatory testing. Incorporating active surveillance using POC tests as part of mitigation strategies for reopening kindergarten through 12th grade (K-12) schools could have an integral role in reducing SARS-CoV-2 transmission among students, teachers and staff members, families, and the surrounding community.15,16
Loop-mediated isothermal amplification (LAMP) is a low-cost method for rapid, target-specific detection of nucleic acids.17 Moreover, LAMP has long been used as an alternative to the gold standard, quantitative reverse-transcription polymerase chain reaction (qRT-PCR) to surveil populations for a variety of pathogens, especially in resource-limited settings.18–22 Reverse-transcription LAMP (RT-LAMP) assays have recently been developed for rapid SARS-CoV-2 testing.23–29 In addition, RT-LAMP is an appealing candidate for POC SARS-CoV-2 testing because it is inexpensive, circumvents supply shortages by relying on different reagents than current diagnostic tests, requires minimal sample processing, and can be deployed outside of traditional laboratory settings. Recently, a number of studies have shown the correlation between the presence of virus in saliva and nasopharyngeal swabs, demonstrating that saliva specimens are a valid and reliable alternative to nasopharyngeal swab specimens for SARS-CoV-2 testing.30–35 Saliva specimen self-collection is noninvasive, can be done at home, does not require swabs or personal protective equipment, and limits direct contact between test operators and the testing populations. Here, we describe our experience implementing a simple, rapid-turnaround, mobile, non-diagnostic SARS-CoV-2 testing workflow combining self-collected saliva and RT-LAMP in volunteers without symptoms of SARS-CoV-2 infection. Individuals were strongly encouraged to isolate and obtain follow-up diagnostic testing after receiving a positive result by RT-LAMP. This addresses a key knowledge gap of how on-site RT-LAMP testing performs in real-world conditions because virtually all previous studies have only evaluated SARS-CoV-2 RT-LAMP in well-equipped molecular biology laboratories.
MATERIALS AND METHODS
POC Testing Sites
To begin operating voluntary POC testing, we developed a system of color-coded storage bins for equipment and supplies, and we assembled folding tables, chairs, extension cords, and coolers that could be easily decontaminated and packed to fit in a Dodge Caravan (FCA US LLC, Auburn Hills, MI), or another, similarly sized minivan, for transportation between testing sites and our base laboratory facility. On July 16, 2020, we launched our first mobile POC testing sites, which ultimately expanded over 18 weeks to include 2 workplaces, 2 K-12 schools, and an athletics program (Table S1). With the exception of the athletics program, sites were initially outdoors, sometimes under an overhang, but otherwise open to the environment. The athletics site was a climate-controlled, indoor practice field. At all sites, equipment and reagents were transported by minivan, and surfaces were disinfected during assembly, breakdown, and, frequently, throughout testing. Participant consenting and volunteer sample collection were performed on-site but were separated from the sample preparation and assay areas (most commonly on the other side of the building). In an effort to limit contamination, each assay area was set up with 3 separate folding tables: (1) sample heat-inactivation and preparation, (2) preparation of RT-LAMP reagents and assay setup, and (3) RT-LAMP incubation and imaging. Individuals responsible for sample inactivation and for performing assays wore appropriate personal protective equipment, including N95 face masks, face shields, or safety glasses; disposable laboratory coats; and double gloves. In anticipation of wet and cold fall weather, by September 2020, assay workspaces were transitioned to biosafety hoods in a vacant indoor laboratory space for several POC testing locations. In October 2020, we received institutional review board (IRB) approval for obtaining consent for repeat SARS-CoV-2 testing. This allowed us to transition away from consenting participants at each testing time point and, instead, allowed each enrolled participant to consent once, regardless of the number of times they supplied a sample. Following reports that SARS-CoV-2 RNA is stable in saliva at room temperature for prolonged periods,36 we also transitioned away from in-person sample collection at some of the testing sites and, instead, distributed self-collection take-home kits for drop off at designated locations for same-day processing.
Sample Collection and Preparation
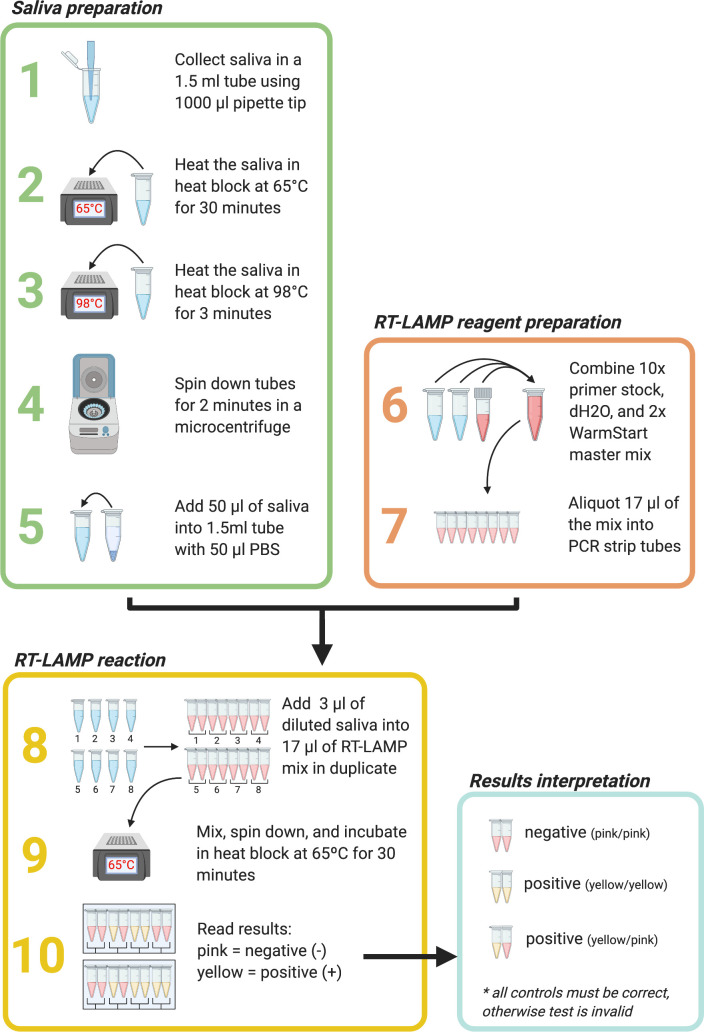
We obtained approval from the University of Wisconsin-Madison IRB (2020-0855 and 2020-1142). Participants were advised to avoid eating or drinking anything, except for water, for 30 minutes before providing a sample. After providing informed consent, volunteers self-collected at least 50 μL of saliva in a 1.5-mL “safe-lock” microcentrifuge tube that used a 1000-μL unfiltered pipette tip to funnel the specimen into the tube. Each volunteer disinfected the outside of the tube with a pre-moistened disinfectant wipe. Samples collected in person were typically processed within 3 hours of collection through our RT-LAMP mobile-testing workflow, whereas samples collected using take-home kits were typically processed within 30 hours (Fig. 1). Samples were first incubated in a preset heat block at 65°C for 30 minutes to inactivate SARS-CoV-2 37 and were then incubated in another preset heat block at 98°C for 3 minutes to improve nucleic acid detection and inactivate salivary enzymes.38 The inactivated saliva was then centrifuged for 2 minutes in a benchtop microcentrifuge. Fifty microliters of the saliva supernatant was then added to 50 μL of 1× phosphate-buffered saline at pH 7.4.
FIGURE 1.
Point-of-care reverse-transcription loop-mediated isothermal amplification (RT-LAMP) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) testing workflow. Steps 1–5: saliva sample preparation. Steps 6–7: RT-LAMP reagent preparation. Steps 8–10: RT-LAMP reactions and results interpretation. A reaction color change from pink/orange to yellow after 30 minutes in at least 1 of 2 sample replicates was scored as a positive result. Figure was created using BioRender.com (Toronto, ON, Canada).
RT-LAMP Reactions
Three microliters of the saliva/phosphate-buffered saline mixture for each sample was added in duplicate to 17 μL of a colorimetric RT-LAMP reaction mix containing WarmStart colorimetric LAMP mastermix (catalogue no. M1800, New England Biolabs, Ipswich, MA), water, and a set of 6 SARS-CoV-2-specific RT-LAMP primers designed against the N gene.38 The SARS-CoV-2 RT-LAMP primer set was previously designed by Broughton et al39 and is currently used in an US Food and Drug Administration (FDA) emergency-use-authorized COVID-19 test by Color Genomics (Table 1).40 Reactions were incubated for 30 minutes at 65°C. A smartphone or tablet was used to record images of each reaction before (time 0) and after (time 30) the incubation period. A color change from pink/orange to yellow in at least 1 of 2 replicates was scored relative to γ-irradiated SARS-CoV-2 specimen (irSARS-CoV-2; BEI Resources, Manassas, VA), which was directly added to the RT-LAMP reactions as a positive control in each batch of reactions, at concentrations ranging from 220 to 3333 copies/μL (2.2 × 105 to 3.33 × 106 copies/mL). The irSARS-CoV-2 specimen was diluted and aliquoted as ready-to-run, positive control standards and was stored at −80°C when not in use. On the day of testing, the positive controls were removed from the freezer and stored on ice at the POC sites. Individuals whose samples were recorded as potentially positive for SARS-CoV-2 by RT-LAMP were contacted by an infectious disease clinician in accordance with the IRB protocol and were urged to obtain a clinical diagnostic test to confirm findings and to self-isolate in accordance with public health recommendations.
TABLE 1.
Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) N-gene primers
|
Primer |
Sequence 5′→3′ |
Concentration, μM |
| Outer forward primer (F3) | AACACAAGCTTTCGGCAG | 0.2 |
| Outer backward primer (B3) | GAAATTTGGATCTTTGTCATCC | 0.2 |
| Forward inner primer (FIP) | TGCGGCCAATGTTTGTAATCAGCCAAGGAAATTTTGGGGAC | 1.6 |
| Backward inner primer (BIP) | CGCATTGGCATGGAAGTCACTTTGATGGCACCTGTGTAG | 1.6 |
| Loop forward primer (LF) | TTCCTTGTCTGATTAGTTC | 0.8 |
| Loop backward primer (LB) | ACCTTCGGGAACGTGGTT | 0.8 |
Limit-of-Detection Estimation Using Contrived Saliva Samples
To estimate the limit of detection (LOD) of the RT-LAMP assay, contrived positive saliva samples were prepared by adding irSARS-CoV-2 initially diluted in nuclease-free water directly into unaltered saliva collected from a total of 20 SARS-CoV-2-negative individuals with the final dilutions ranging from 1 × 104 to 10 copies/μL (1 × 107 to 1 × 104copies/mL). Dilutions were based on independent, in-house qRT-PCR experiments showing that the irSARS-CoV-2 stock concentrations had 8.79 × 106 copies/μL (8.79 × 109 copies/mL). Seven dilutions of irSARS-CoV-2 were prepared for each saliva sample in duplicate. The RT-LAMP reactions were set up as described previously. Negative controls consisting of saliva from each of the donors without addition of irSARS-CoV-2 were also prepared in duplicate. Reactions were called positive if a color change from preamplification to postamplification occurred in at least 1 of 2 replicates that was consistent with that of the positive controls (a clean yellow color).
LOD Estimation Using Clinical Samples
Deidentified, discarded saliva samples from 38 SARS-CoV-2-positive patients were provided by the University of Wisconsin Hospitals and Clinics (UWHC) for evaluation of the RT-LAMP performance with known-positive saliva samples. Clinical saliva samples were originally collected and stored at 4°C for up to 4 weeks before assessment by RT-LAMP. Additional 10-fold and 100-fold dilutions were prepared for 13 of the positive clinical saliva samples, in additional saliva collected from a negative volunteer. Clinical samples and dilutions of 13 of those samples were prepared as described previously, except that 20 to 50 μL of heat-inactivated sample, dependent on total sample volume, was added to an equal volume of 1× phosphate-buffered saline in a clean 1.5-mL screw-top tube and pipetted gently to mix. For each sample, 3 μL was then added to duplicate colorimetric RT-LAMP reactions. Negative and positive control reactions (described previously) were also prepared in duplicate, except that saliva collected from a negative volunteer was used as the negative control for these reactions. The RT-LAMP reactions were prepared, and images collected as described previously.
Quantitative RT-PCR
POC samples
We measured the viral RNA (vRNA) concentration using a sensitive qRT-PCR in a subset of the inactivated saliva samples described above after initial evaluation using RT-LAMP. From July 16 until September 17, saliva samples that were negative for SARS-CoV-2 by RT-LAMP were pooled into groups of 6 or fewer for qRT-PCR to balance cost effectiveness with reasonable, estimated detection sensitivity. Ten additional, individual RT-LAMP-negative samples were submitted as negative controls alongside samples identified as positive by RT-LAMP. Saliva samples that were identified as positive for SARS-CoV-2 by RT-LAMP were tested by qRT-PCR individually to estimate our POC LOD. RNA was isolated from up to 150 μL saliva and combined with an equivalent volume of nuclease-free water using the Viral Total Nucleic Acid kit for the Maxwell RSC instrument (Promega, Madison, WI), following the manufacturer's instructions. Viral load quantification was performed using a sensitive qRT-PCR assay developed by the Centers for Disease Control and Prevention to detect SARS-CoV-2 (specifically the N1 assay) and commercially available from IDT (Coralville, IA). The assay was run on a LightCycler 96 or LC480 instrument (Roche, Indianapolis, IN) using the Taqman Fast Virus 1-Step Master Mix enzyme (Thermo Fisher Scientific, Waltham, MA). The LOD of this assay is estimated to be 0.2 genome equivalents/μL (200 genome equivalents/mL) of saliva. To determine the vRNA load, samples were interpolated onto a standard curve consisting of serial 10-fold dilutions of the in vitro–transcribed SARS-CoV-2 N gene RNA kindly provided by Nathan Grubaugh (Yale University) and described by Dudley et al.35
Clinical Samples
The qRT-PCR was performed using the conditions described above for each of the 38 SARS-CoV-2–positive saliva samples individually; however, sample volume limitations required that for some samples, only 100 μL saliva was combined with 100 μL of nuclease-free water before RNA isolation. In addition, sample UWHC3 contained a lower volume than the remaining 37 samples, so 50 μL saliva was combined with 50 μL nuclease-free water and used for RNA isolation as described previously. Viral loads in copies per microliter and corresponding cycle threshold (Ct) numbers are reported in Table 2.
TABLE 2.
Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) evaluation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–positive clinical saliva samples from University of Wisconsin Hospitals and Clinics (UWHC)
|
Sample |
Ct (N1 assay) |
Positive by RT-LAMP |
vRNA load (copies/μL) |
Sample |
Ct (N1 assay) |
Positive by RT-LAMP |
vRNA load (copies/μL) |
| UWHC1 | 27.65 | 0/2 | 3.25 × 102 | UWHC20 | 25.80 | 2/2 | 9.48 × 102 |
| UWHC2 | 32.7 | 0/2 | 10.9 | UWHC21 | 20.18 | 2/2 | 4.40 × 104 |
| UWHC3 | 20.98 | 2/2 | 5.17 × 104 | UWHC22 | 28.92 | 0/2 | 1.13 × 102 |
| UWHC4 | 24.07 | 2/2 | 3.57 × 103 | UWHC23 | 21.26 | 2/2 | 2.10 × 104 |
| UWHC5 | 26.53 | 2/2 | 6.81 × 102 | UWHC24 | 29.92 | 0/2 | 57.2 |
| UWHC6 | 30.85 | 1/2 | 37.4 | UWHC25 | 36.71 | 0/2 | 0.796a |
| UWHC7 | 36.96 | 0/2 | 0.701 | UWHC26 | 25.96 | 2/2 | 1.31 × 102 |
| UWHC8 | 26.28 | 1/2 | 8.10 × 102 | UWHC27 | 29.99 | 0/2 | 54.1 |
| UWHC9 | 37.59 | 0/2 | 0.402 | UWHC28 | 24.34 | 2/2 | 2.58 × 103 |
| UWHC10 | 24.01 | 2/2 | 3.72 × 103 | UWHC29 | 20.55 | 2/2 | 4.72 × 104 |
| UWHC11 | 22.39 | 2/2 | 1.10 × 104 | UWHC30 | 33.18 | 0/2 | 7.89 |
| UWHC12 | 35.46 | 0/2 | 1.75 | UWHC31 | 22.87 | 2/2 | 9.57 × 103 |
| UWHC13 | 36.09 | 0/2 | 1.14 | UWHC32 | 23.07 | 2/2 | 8.33 × 103 |
| UWHC14 | 23.11 | 2/2 | 5.96 × 103 | UWHC33 | 26.85 | 2/2 | 6.20 × 102 |
| UWHC15 | 23.38 | 2/2 | 4.95 × 103 | UWHC34 | 20.33 | 0/2 | 5.49 × 104 |
| UWHC16 | 33.86 | 0/2 | 3.99 | UWHC35 | 23 | 2/2 | 8.88 × 103 |
| UWHC17 | n/a | 0/2 | 0 | UWHC36 | 32.26 | 0/2 | 14.9a |
| UWHC18 | 23.02 | 2/2 | 6.34 × 103 | UWHC37 | 33.94 | 0/2 | 4.33 |
| UWHC19 | 37.31 | 0/2 | 0.612 | UWHC38 | 25.96 | 2/2 | 1.74 × 103 |
Abbreviations: Ct, cycle threshold; n/a, sample not available; vRNA, viral RNA.
Sample only positive in 1 qRT-PCR replicate.
RESULTS
LOD Estimation Using Contrived Saliva Samples
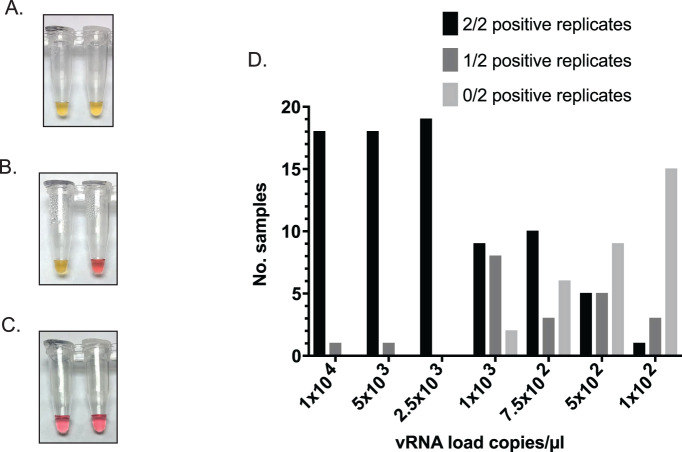
We assessed the LOD for minimally processed saliva samples collected from 20 volunteers by RT-LAMP using irSARS-CoV-2 spiked into negative saliva samples. We detected irSARS-CoV-2 by RT-LAMP in 2 of 2 replicates (Fig. 2A) at 2.5 × 103 copies/μL (2.5 × 106 copies/mL) for 100% of samples, at 1 × 103 copies/μL (1 × 106 copies/mL) for 47.4% of samples and at 500 copies/μL (5 × 105 copies/mL) for 26% of samples (Fig. 2D). When we included samples called positive in at least 1 of 2 replicates (see “Methods” and Fig. 2B), the percentage of contrived samples positive by RT-LAMP at each of the aforementioned dilutions was 100%, 89.5%, and 53%, respectively (Fig. 2D). One sample was omitted from the analysis because it turned yellow-orange at all dilutions before the RT-LAMP reaction incubation began and was, therefore, uninterpretable. Because, in POC testing, we defined a positive RT-LAMP result as an observed postincubation color change to yellow in at least 1 replicate, these results suggested that our 90% LOD is approximately 1 × 103 copies/μL (1 × 106 copies/mL).
FIGURE 2.
Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in contrived saliva samples by direct reverse-transcription loop-mediated isothermal amplification (RT-LAMP). A) Representative example of a positive sample in 2 of 2 replicates. Sample used negative saliva spiked with irSARS-CoV-2. B) Representative example of a sample positive in 1 of 2 replicates C) Representative negative sample showing no colorimetric change in either replicate. D) Bar graph of results of limit of detection (LOD) assessment with contrived saliva samples from 19 volunteers. γ-Irradiated SARS-CoV-2 (irSARS-CoV-2) vRNA load is shown as copies/μL on the x-axis; the number (No.) of samples positive in 2 (black), 1 (dark gray), or zero (light gray) replicates is shown on the y-axis.
LOD Estimation Using Clinical Samples
To assess the performance of SARS-CoV-2 RT-LAMP in known SARS-CoV-2 positive saliva samples as opposed to contrived positive samples, we acquired deidentified, discarded saliva samples that were collected from 38 patients with laboratory-confirmed SARS-CoV-2 from UWHC. Nineteen of 38 undiluted qRT-PCR–confirmed-positive saliva samples were also positive for SARS-CoV-2 in 2 of 2 replicates by RT-LAMP (Fig. 3; Table 2). Two additional samples were positive in 1 of 2 replicates. Quantitative RT-PCR data showed that the viral RNA (vRNA) loads of the positive samples ranged from 131 copies/μL to 5.7 × 104 copies/μL (1.31 × 105 to 5.71 × 107 copies/mL), which was consistent with our LOD for contrived samples (Table 3). Positive clinical saliva samples that were negative by RT-LAMP had estimated vRNA loads ranging from 0.402 × 104 to 5.49 × 104 copies/μL. All of the samples that were negative by RT-LAMP, with the exception of UWHC34 (5.49 × 104 copies/μL), had vRNA loads below our estimated reliable LOD. Furthermore, for the 13 positive clinical saliva samples that were diluted 10-fold and 100-fold in additional saliva collected from a negative volunteer, detection decreased, as expected, with increasing dilution factor (Table 4).
FIGURE 3.
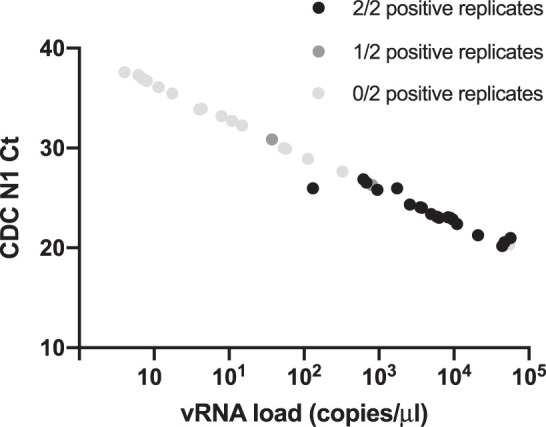
Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 38 clinical saliva specimens by direct reverse-transcription loop-mediated isothermal amplification (RT-LAMP). The viral RNA (vRNA) load of each clinical sample is plotted on the x-axis relative to the in-house Centers for Disease Control and Prevention–based N1 quantitative reverse-transcription polymerase chain reaction (qRT-PCR) assay cycle threshold (Ct) on the y-axis. Black, dark gray, and light gray indicate 2, 1, and zero positive replicates, respectively.
TABLE 3.
Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) results for 10-fold and 100-fold dilutions of 13 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–positive samples from University of Wisconsin Hospitals and Clinics (UWHC)
|
Sample |
1:10 dilution result |
1:100 dilution result |
Undiluted vRNA load (copies/μL) |
| UWHC1 | 1/2 | 0/2 | 3.25 × 102 |
| UWHC2 | 0/2 | 0/2 | 10.9 |
| UWHC3 | 2/2 | 2/2 | 5.17 × 104 |
| UWHC4 | 2/2 | 2/2 | 3.57 × 103 |
| UWHC5 | 1/2 | 0/2 | 6.81 × 102 |
| UWHC6 | 0/2 | 0/2 | 37.4 |
| UWHC7 | 0/2 | 0/2 | 0.701 |
| UWHC8 | 1/2 | 0/2 | 8.10 × 102 |
| UWHC9 | 0/2 | 0/2 | 0.402 |
| UWHC10 | 2/2 | 0/2 | 3.72 × 103 |
| UWHC11 | 2/2 | 1/2 | 1.10 × 104 |
| UWHC12 | 0/2 | 0/2 | 1.75 |
| UWHC13 | 0/2 | 0/2 | 1.14 |
Abbreviation: vRNA, viral RNA.
TABLE 4.
Samples identified as potentially positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse-transcription loop-mediated isothermal amplification (RT-LAMP during point-of-care (POC) testing
|
RT-LAMP-positive sample |
qRT-PCR viral load, copies/μL |
| POC1 | 8.53 |
| POC2 | 2.15 × 104 |
| POC3 | Negative |
| POC4 | Negative |
| POC5 | Negative |
| POC6 | Negative |
| POC7 | 3.62 × 105 |
| POC8 | Negative |
| POC9 | n/aa |
| POC10 | 2.12 × 103 |
| POC11 | Negative |
| POC12 | 1.04 × 103 |
| POC13 | 2.06 × 102 |
| POC14 | Negative |
| POC15 | 52.8 |
| POC16 | 6.02 × 102 |
| POC17 | 87.3 |
| POC18 | 1.17 × 103 |
| POC19 | Negative |
| POC20 | 1.38 × 102 |
| POC21 | 4.07 × 102 |
Abbreviations: n/a, sample not available; qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
Volunteer did not consent to follow-up testing.
POC SARS-CoV-2 RT-LAMP Testing
From July 16, 2020, to November 19, 2020, SARS-CoV-2 RT-LAMP was used to test a total of 4704 samples collected from 5 locations. Participants were enrolled into the study regardless of their SARS-CoV-2 symptom status on the day of testing. Seventy-one percent of the samples were obtained from individuals at 2 research facilities, 11% from 2 K-12 schools, and 18% from an athletics program (Table S1). A total of 21 samples were identified as positive for SARS-CoV-2 by RT-LAMP based on a colorimetric change from pink/orange to yellow in at least 1 of 2 sample replicates (see Fig. 2B for example). Similar to our experience with our contrived LOD samples, about 0.40% (19/4704) of samples collected during POC testing exhibited a color change to yellow before RT-LAMP assay amplification and were, therefore, uninterpretable. Follow-up qRT-PCR testing was conducted on each sample that appeared positive after the 30-minute amplification reaction throughout the study to determine vRNA load. Twelve of the 21 samples called positive in RT-LAMP had detectable SARS-CoV-2 RNA by qRT-PCR. Viral RNA loads of these samples ranged from 8.58 copies/μL to 3.62 × 105 copies/μL (8.58 × 103 copies/mL to 3.62 × 108 copies/mL) with a median of 504.5 copies/μL (5.04 × 105 copies/mL) (Table 4). Eight of the saliva samples identified as positive by RT-LAMP were negative by qRT-PCR, suggesting that they were false-positive RT-LAMP results (approximately 40% of samples called positive by RT-LAMP; 0.17% of total samples tested). One RT-LAMP-positive sample was not tested by qRT-PCR because the participant did not consent to additional molecular testing. For volunteers who consented to additional research testing from July 16 to September 17, qRT-PCR testing was conducted for pools of 6 or fewer for all residual, heat-inactivated samples that appeared unambiguously negative by RT-LAMP. A total of 421 RT-LAMP-negative pools (2493 samples) were tested to estimate the number of SARS-CoV-2–positive samples missed by RT-LAMP. The qRT-PCR tests detected SARS-CoV-2 nucleic acids in 5 pools of RT-LAMP-negative samples. Four of 5 of the positive pools contained levels of SARS-CoV-2 that were below the estimated LOD range for RT-LAMP using crude samples with vRNA load estimates of 0.236, 0.444, 0.460, 37.5, and 142 copies/μL (236 × 104, 444 × 104, 460 × 104, 3.75 × 104, and 1.42 × 105 copies/mL). Taken together, the low prevalence of SARS-CoV-2 in our volunteer testing population (0.36%, including RT-LAMP-negative, qRT-PCR-positive pools) and the low vRNA load of pools positive by follow-up qRT-PCR, suggest that these 5 pools likely contained only a single positive sample each and suggests a false-negative rate of 0.02% (5/2493 pools) (Table 4).
DISCUSSION
Strategic surveillance testing of asymptomatic individuals has been suggested as an important mitigation strategy for places at high risk for close-contact, indoor SARS-CoV-2 transmission: schools, workplaces, places of worship, and prisons, among others. Decentralized, mobile RT-LAMP-based POC testing workflows can provide same-day results, which can enable people with potential SARS-CoV-2 infections to quickly self-isolate and then obtain confirmatory diagnostic testing. The low per-test cost (approximately $7 per sample tested in duplicate) allows for repeated testing to identify incident infections and reduce the duration of a potentially infected individual's exposure to others. Although RT-LAMP is not as sensitive as diagnostic qRT-PCR tests in laboratory testing, qRT-PCR tests require centralized laboratories, which, in turn, lead to lengthy turnaround times. During a period of 18 weeks, we performed 4704 SARS-CoV-2 tests across 5 sites using a simple, saliva-based, direct RT-LAMP assay. This work demonstrates the scalability of decentralized, mobile RT-LAMP–based testing and addresses a key knowledge gap of how POC RT-LAMP testing performs outside of well-equipped molecular biology laboratories.
Our experiment using direct RT-LAMP with contrived saliva samples from a total of 20 donors demonstrated an approximate LOD of 1 × 103 copies/μL (89.5% in at least 1 replicate). Overall, our data suggest that the actual LOD for RT-LAMP without RNA isolation may be dependent on the individual sample because of heterogeneity in saliva pH and composition.41–43 The RT-LAMP results for 38 clinical saliva samples obtained from SARS-CoV-2–positive individuals at the UWHC were consistent with those for the contrived samples. We recognize that more clinical samples are required for a comprehensive clinical validation, but the LOD observed in clinical samples is further supported by the low vRNA loads obtained from qRT-PCR–confirmed SARS-CoV-2–positive samples identified in our volunteer population (Table 4). The performance of our RT-LAMP POC testing workflow demonstrates that inexpensive, mobile testing can be successfully performed outdoors or in other nontraditional laboratory settings to identify SARS-CoV-2–positive individuals regardless of whether or not symptoms are present. Our observed SARS-CoV-2 RT-LAMP positivity rate was 0.25% (12/4704) for samples confirmed by follow-up qRT-PCR. Interestingly, the positivity rate of 0.25% in our volunteer population was lower than expected, given that the disease activity in our region during this period was listed as critically high, particularly between September 1, 2020, and November 19, 2020, when the county had a 5.42% positivity rate (19 031 positive tests of 350 722).44,45 The low positivity rate in our volunteer population may be partly explained by the fact that 71% of tested saliva specimens came from 2 research facilities in which mask wearing and physical distancing guidelines were implemented early in the pandemic and followed relatively stringently (Supplemental Table S1). Volunteers for nonsymptomatic research testing might also have a different risk profile from that of the overall population.
Potential drawbacks of colorimetric RT-LAMP–based surveillance for SARS-CoV-2, as described here, include that minimally processed saliva can result in variable-reaction color changes without the presence of the target RNA. However, modifications of RT-LAMP–based SARS-CoV-2 assays to reduce saliva sample variability, improve result ambiguity, and increase throughput have recently been reported elsewhere and may improve the implementation of RT-LAMP–based assays for POC use.46–50 In addition, we relied on a manual RT-LAMP format during POC testing. Reading assays “by eye” inevitably results in a somewhat-subjective determination of positives. Reducing false-positive results in our POC volunteer populations required consistent use of duplicate reactions for each individual, which reduced assay throughput and increased the per-sample cost. Even with our efforts to reduce calling false-positive results in our volunteer populations, we still were unable to confirm approximately 40% of RT-LAMP–positive samples by follow-up qRT-PCR. Whether these false-positives resulted from the individual sample variability across saliva donors or from temporary storage of the samples before follow-up qRT-PCR is unclear but because volunteers with a potential positive finding were strongly encouraged to receive follow-up, confirmatory diagnostic testing, we chose to err on the side of caution when interpreting direct RT-LAMP results. Furthermore, the testing landscape changed dramatically during the months we performed RT-LAMP testing. The first noninstrumented antigen test, the Abbott BinaxNOW COVID-19 Ag CARD, received FDA emergency-use-authorized approval in the United States on August 26, 2020.51 Although the sensitivity of RT-LAMP is broadly comparable to the Abbott BinaxNOW antigen test (reported as 1.6 × 104 to 4.3 × 104 vRNA copies; Ct, 30.3–28.8), because the former is technically straightforward and can be used as a SARS-CoV-2 diagnostic at testing sites operating under a Clinical Laboratory Improvement Amendments waiver, it is likely a better choice for rapid turnaround, on-site testing in most circumstances.52 However, even with the existence of antigen tests, RT-LAMP surveillance programs still have a place as part of a comprehensive SARS-CoV-2 risk-mitigation strategy, especially in areas in which access to antigen tests is limited.
There are advantages to continuing saliva-based RT-LAMP surveillance testing. Importantly, the supply of diagnostic antigen tests remains tightly constrained, and in the United States, these tests are available only through government contracts. Widespread testing of individuals without symptoms with such a scarce resource may not be a wise use of these limited tests. Furthermore, recent studies have shown that antigen-test performance may differ between asymptomatic and symptomatic populations. Compared with qRT-PCR, the sensitivity of FDA-approved antigen tests, BinaxNOW and the Quidel Sofia SARS Antigen Fluorescent Immunoassay, were 35% and 41% in asymptomatic individuals and 64% and 80% in symptomatic individuals, respectively.53,54 BinaxNOW is currently only approved for use in symptomatic individuals, within 7 days of symptom onset, and samples are required to be tested within an hour of collection.55 In contrast, RT-LAMP reagents do not require a government contract and can be acquired readily from commercial and noncommercial sources, and they can also be used more flexibly for surveillance purposes because RT-LAMP is not limited to use in symptomatic individuals.56 Additionally, user acceptance of testing may also favor saliva-based RT-LAMP because it is less invasive than nasal-swab–based tests. Although an individual BinaxNOW test is rapid, performing several tests during a single day could cumulatively take as long as processing a batch of tests by RT-LAMP. For these reasons, RT-LAMP may still be the preferred testing method to incorporate into a local program. In Madison, WI, 2 local schools have implemented RT-LAMP surveillance programs modeled on the program described here. Implementation of each program required approximately 50 hours of hands-on training by our group. School staff members were trained in adherence to regulations pertaining to nondiagnostic testing and to competently perform RT-LAMP assays. Each school also needed time and resources to acquire the modest laboratory infrastructure necessary to perform RT-LAMP. In addition, a larger saliva-based RT-LAMP surveillance program has been successfully implemented in school districts in the greater Chicago suburbs.57,58
A looming question for both RT-LAMP and antigen testing programs is whether the real-world effectiveness of frequently testing individuals without symptoms mirrors the theoretical benefits. Several important considerations that we factored into the design of RT-LAMP testing remain true: nonsymptomatic individuals account for up to 59% of all transmission (24% asymptomatic and 35% presymptomatic); low-sensitivity tests are able to effectively identify those with high levels of virus shedding, and individuals with high viral loads are likely to be responsible for a significant fraction of onward community transmission; and the duration of peak infectiousness is short, so lengthy lags in reporting test results could miss a critical window of high transmissibility.10,59 Conversely, high-quality, exceptionally well-resourced testing programs, such as those at the White House and among intercollegiate athletic programs, have failed to stop outbreaks.60 The latter deserves special note: outbreaks in these programs occurred in spite of 100% adherence to daily testing. Data from daily sampling of individuals with incident SARS-CoV-2 infection suggests that the mean duration of time from infection to peak viral shedding is approximately 3 days, but some individuals potentially reach peak viral shedding in less than 1 day.61 The potential for an extremely rapid increase in viral load, which likely parallels shedding of infectious virus, means that, in some cases, even daily testing might be insufficient to protect a community from someone who is newly infected.
Importantly, the benefit of frequent testing of individuals without symptoms with RT-LAMP or other assays may be substantially undermined by risk disinhibition. When people are tested frequently, they may both underestimate their own risk of becoming infected in the interval between tests and overestimate the possibility that their similarly tested contacts are uninfected; anecdotal evidence of this phenomenon is perhaps most vividly seen in the September 26, 2020, White House Rose Garden reception for Justice Amy Coney Barrett, in which many attendees were photographed not wearing masks and not following guidelines for physical distancing.62 If infections among people without symptoms are rare (approximately 0.4% of tests in this study, when combining RT-LAMP and pooled qRT-PCR positives), but 10% of the tested population views testing as a license for increased risk-tasking, is frequent testing of symptomless people a net positive? Appropriate messaging to the community is essential with any testing program to ensure the population understands the meaning of a test result. Such issues will require an optimization of messaging to mitigate the effect of risk disinhibition to the extent possible.
Ultimately, this study provides POC and guidance for how decentralized rapid testing could be implemented in a mobile-testing scenario, which may be especially useful in resource-limited settings. Despite the caveats presented above, we identified 12 SARS-CoV-2–positive individuals and likely prevented onward transmission from those individuals who, otherwise, would not have known they were positive for the disease. Rapid tests, although less sensitive than qRT-PCR, have shorter turnaround times and could bridge the gap between SARS-CoV-2 surveillance and diagnostic testing. Although POC testing can be effective for identifying asymptomatic individuals, it must be used in conjunction with appropriate messaging and other mitigation strategies to effectively reduce SARS-CoV-2 transmission.
Supplementary Material
Acknowledgments
This work was made possible by financial support through National Institutes of Health Rapid Acceleration of Diagnostics (3 U54 EB027690-02S1). Additional funding was provided, in part, by the office of the director, National Institutes of Health, under award number P51OD011106 to the Wisconsin National Primate Research Center (WNPRC), University of Wisconsin-Madison. NIH RADx (3 U54 EB027690-0S1), NIH P51 (OD011106 to WNPRC), RFIP (RR15459-01 and RR020141-01 to WNPRC), WARF COVID-19 Challenge grant, NIH T32 (A|007414 to A.S.H. and C.M.C.). This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. A.S.H. and C.M.C. have been supported by the National Institutes of Health National Research Service Award T32 AI007414. We are grateful for the assistance of both the University of Wisconsin Institutional Biosafety Committee and the Health Sciences Institutional Review Board at University of Wisconsin-Madison. We also acknowledge Christopher E. Mason, PhD, Cornell-Weill Medical School, and Alison Kriegel, PhD, Medical College of Wisconsin, as well as public employees from Racine, WI, for their partnership to this project. We would like to thank Abigail Johnson, Abby Weaver, Abdel Daoud, Allison Eierman, Ben Boerigter, Clarissa Tjoanda, Julia Pulokas, Julie Chen, Ryan Anderson, WIll Vuyk, Jenny Lee, Max Bobholz, Sam Havlicek, Hunter Ries, Nicole Minerva, Emma Boehm, Elizabeth Brown, and Jayden Lee for their help preparing and organizing supplies for POC testing. David Beebe, Nate Grubaugh, and Kristian Andersen provided useful technical discussions. We thank Eli O'Connor for assistance with POC label generation and template preparation. We are grateful to the study volunteers and organizations that allowed us to evaluate this mobile testing strategy.
Footnotes
Author contributions C.M.N., M.D.R., R.W.W., D.M.D., C.G.S., D.H.O., and S.L.O. contributed to assay development and optimization. D.M.D., M.T.M., R.W.W., C.M.N., M.R.S., A.M.W., M.I.B., K.N.F., M.D.R., L.A.H., O.E.H., R.V.M., C.M.C., S.L.O., M.R.R., T.C.F., T.M.P., E.D.S., L.M.S., and E.K.N. contributed to POC testing and PCR confirmation. C.M.N., M.D.R., D.M.D., and D.H.O. contributed to data analysis, interpretation, and writing. J.A.K., D.H.O., S.L.O., H.E.B., T.C.F., M.T.M., A.K.H., L.A.H., C.M.C., K.L.H., C.B.B., and K.N.F. contributed to logistics and organization of POC testing. C.B.B. and K.L.H. contributed to obtaining IRB approval and worked closely with the Institutional Biosafety Committee on other regulatory responsibilities. M.A.A., A.S.H., and W.M.R. contributed to providing residual SARS-CoV-2–positive saliva samples and sample information from the UWHC. All authors contributed to editing the manuscript.
Conflicts of interest C.M.N., D.M.D, and A.M.W provided consulting services to Salus Discovery LLC.
Regulatory oversight This work was performed under approved UW-Madison Health Sciences IRB nos. 2020-0855 and 2020-1142.
REFERENCES
- 1.Silverman JD, Hupert N, Washburne ADUsing influenza surveillance networks to estimate state-specific prevalence of SARS-CoV-2 in the United States. Sci Transl Med. 2020. 12:eabc1126. [DOI] [PMC free article] [PubMed]
- 2. Havers FP, Reed C, Lim T, et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23–May 12, 2020. JAMA Intern Med. 2020;180:1576–1586. doi: 10.1001/jamainternmed.2020.4130. [DOI] [PubMed] [Google Scholar]
- 3. Esbin MN, Whitney ON, Chong S, Maurer A, Darzacq X, Tjian R Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schuetz AN, Hemarajata P, Mehta N, et al. When should asymptomatic persons be tested for COVID-19 [editorial] J Clin Microbiol. 2020;59:e02563–20. doi: 10.1128/JCM.02563-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arons MM, Hatfield KM, Reddy SC, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quicke K, Gallichote E, Sexton N, et al. Longitudinal surveillance for SARS-CoV-2 RNA among asymptomatic staff in five Colorado skilled nursing facilities: epidemiologic, virologic and sequence analysis. MedRxiv. 2020. Preprint. Posted online June 9. [DOI]
- 9. Furukawa NW, Brooks JT, Sobel J Evidence supporting transmission of severe acute respiratory syndrome coronavirus 2 while presymptomatic or asymptomatic. Emerg Infect Dis. 2020;26:e201595. doi: 10.3201/eid2607.201595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johansson MA, Quandelacy TM, Kada S, et al. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw Open. 2021;4:e2035057. doi: 10.1001/jamanetworkopen.2020.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lavezzo E, Franchin E, Ciavarella C, et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vò. Nature. 2020;584:425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PubMed] [Google Scholar]
- 12. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee S, Kim T, Lee E, et al. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern Med. 2020;180:1–6. doi: 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mina MJ, Parker R, Larremore DB Rethinking Covid-19 test sensitivity—a strategy for containment. N Engl J Med. 2020;383:e120. doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 15.Rivers C, Silcox C, Potter C, et al. Risk Assessment and Testing Protocols for Reducing SARS-CoV-2 Transmission in K-12 Schools. Washington, ED: Duke Margolis Center for Health Policy;; 2020. pp. 1–18. [Google Scholar]
- 16. Dibner KA, Schweingruber HA, Christakis DA Reopening K-12 schools during the COVID-19 pandemic: a report from the National Academies of Sciences, Engineering, and Medicine. JAMA. 2020;324:833–834. doi: 10.1001/jama.2020.14745. [DOI] [PubMed] [Google Scholar]
- 17. Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mahony J, Chong S, Bulir D, Ruyter A, Mwawasi K, Waltho D Multiplex loop-mediated isothermal amplification (M-LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen-to-result diagnosis in 40 min with single genome copy sensitivity. J Clin Virol. 2013;58:127–131. doi: 10.1016/j.jcv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 19. McKenna JP, Cox C, Fairley DJ, Burke R, Shields MD, Watt A, Coyle PV Loop-mediated isothermal amplification assay for rapid detection of Streptococcus agalactiae (group B streptococcus) in vaginal swabs—a proof of concept study. J Med Microbiol. 2017;66:294–300. doi: 10.1099/jmm.0.000437. [DOI] [PubMed] [Google Scholar]
- 20. Gonçalves DDS, Hooker DJ, Dong Y, et al. Detecting wMel Wolbachia in field-collected Aedes aegypti mosquitoes using loop-mediated isothermal amplification (LAMP) Parasit Vectors. 2019;12:404. doi: 10.1186/s13071-019-3666-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romero Romero JL, Carver GD, Arce Johnson P, Perry KL, Thompson JR A rapid, sensitive and inexpensive method for detection of grapevine red blotch virus without tissue extraction using loop-mediated isothermal amplification. Arch Virol. 2019;164:1453–1457. doi: 10.1007/s00705-019-04207-y. [DOI] [PubMed] [Google Scholar]
- 22. Calvert AE, Biggerstaff BJ, Tanner NA, Lauterbach M, Lanciotti RS Rapid colorimetric detection of Zika virus from serum and urine specimens by reverse transcription loop-mediated isothermal amplification (RT-LAMP) PLoS One. 2017;12:e0185340. doi: 10.1371/journal.pone.0185340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baek YH, Um J, Antigua KJC, et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg Microbes Infect. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lamb LE, Bartolone SN, Ward E, Chancellor MB Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One. 2020;15:e0234682. doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang WE, Lim B, Hsu CC, et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kitagawa Y, Orihara Y, Kawamura R, et al. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J Clin Virol. 2020;129:104446. doi: 10.1016/j.jcv.2020.104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu R, Wu X, Wan Z, Li Y, Jin X, Zhang C A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int J Mol Sci. 2020;21:2826. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu R, Wu X, Wan Z, et al. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol Sin. 2020;35:344–347. doi: 10.1007/s12250-020-00218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan C, Cui J, Huang L, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagura-Ikeda M, Imai K, Tabata S, et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol. 2020;58:e01438–20. doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wyllie AL, Fournier J, Casanovas-Massana A, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. To KK, Tsang OT, Yip CC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokota I, Shane PY, Okada K, et al. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin Infect Dis. 2020. Accepted manuscript. Published online September 25. [DOI] [PMC free article] [PubMed]
- 35. Dudley DM, Newman CM, Weiler AM, et al. Optimizing direct RT-LAMP to detect transmissible SARS-CoV-2 from primary nasopharyngeal swab samples. PLoS One. 2020;15:e0244882. doi: 10.1371/journal.pone.0244882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott IM, Strine MS, Watkins AE, et al. Simply saliva: stability of SARS-CoV-2 detection negates the need for expensive collection devices. MedRxiv. 2020. Preprint. Posted online August 4. [DOI]
- 37. Kim YI, Casel MAB, Kim SM, et al. Development of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) thermal inactivation method with preservation of diagnostic sensitivity. J Microbiol. 2020;58:886–891. doi: 10.1007/s12275-020-0335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lalli MA, Langmade SJ, Chen X, et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP. MedRxiv. Clin. Chem. 2020;2021;57:415–424. doi: 10.1101/2020.05.07.20093542. Preprint. Posted online August 6. . Now published as Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Color Genomics Inc. SARS-CoV-2 LAMP Diagnostic Assay. 2020. Published May 19. Available at: https://www.color.com/wp-content/uploads/2020/05/LAMP-Diagnostic-Assay.pdf.
- 41. Humphrey SP, Williamson RT A review of saliva: normal composition, flow, and function. J Prosthet Dent. 2001;85:162–169. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- 42. Tanaka M, Masuda M Concentration and individual variation of inorganic ions in unstimulated whole saliva. Kokubyo Gakkai Zasshi. 2000;67:46–51. doi: 10.5357/koubyou.67.46. [DOI] [PubMed] [Google Scholar]
- 43. Varga G Physiology of the salivary glands. Surgery. 2012;30:578–583. [Google Scholar]
- 44.Public Health Madison and Dane County. Dashboard: Weekly COVID-19 Core Data by Date. 2021. Available at: https://www.publichealthmdc.com/coronavirus/forward-dane/data.
- 45.University of Wisconsin-Madison. COVID-19 Response Dashboard. 2020. 2021. Available at: https://covidresponse.wisc.edu/dashboard-2020/
- 46. Howson ELA, Kidd SP, Armson B, et al. Preliminary optimisation of a simplified sample preparation method to permit direct detection of SARS-CoV-2 within saliva samples using reverse-transcription loop-mediated isothermal amplification (RT-LAMP) J Virol Methods. 2021;289:114048. doi: 10.1016/j.jviromet.2020.114048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Janíková M, Hodosy J, Boor P, Klempa B, Celec P Loop-mediated isothermal amplification for the detection of SARS-CoV-2 in saliva. Microb Biotechnol. 2021;14:307–316. doi: 10.1111/1751-7915.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diaz L, Johnson BE, Jenkins DMReal-time optical analysis of a colorimetric LAMP assay for SARS-CoV-2 in saliva with a handheld instrument improves accuracy compared to endpoint assessment. MedRxiv. 2021. Preprint. Posted online January 15. [DOI] [PMC free article] [PubMed]
- 49.Brown TA, Schaefer KS, Tsang A, et al. Direct detection of SARS-CoV-2 RNA using high-contrast pH-sensitive dyes. MedRxiv. 2021. Preprint. Posted online January 11. [DOI] [PMC free article] [PubMed]
- 50. Yang Q, Meyerson NR, Clark SK, et al. Saliva TwoStep for rapid detection of asymptomatic SARS-CoV-2 carriers. MedRxiv. 2021;10:e65113. doi: 10.1101/2020.07.16.20150250. Preprint. Posted online February 16, 2021. . Now published as eLife. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.US Food and Drug Administration. COVID-19 Update: FDA Authorizes First Diagnostic Test Where Results Can Be Read Directly From Testing Card. 2020. Published August 26. Available at: https://www.fda.gov/news-events/press-announcements/covid-19-update-fda-authorizes-first-diagnostic-test-where-results-can-be-read-directly-testing-card.
- 52. Pilarowski G, Lebel P, Sunshine S, et al. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. J Infect Dis. 2021;223:1139–1144. doi: 10.1093/infdis/jiaa802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Prince-Guerra JL, Almendares O, Nolen LD, et al. Evaluation of Abbott BinaxNOW for SARS-CoV-2 Infection at two community-based testing sites—Pima County, Arizona, November 3–17, 2020. MMWR Morb Mortal Wkly Rep. 2021;70:100–105. doi: 10.15585/mmwr.mm7003e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pray IW, Ford L, Cole D, et al. Performance of an antigen-based test for asymptomatic and symptomatic SARS-CoV-2 testing at two university campuses—Wisconsin, September–October 2020. MMWR Morb Mortal Wkly Rep. 2021;69:1642–1647. doi: 10.15585/mmwr.mm695152a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.US Food and Drug Administration. BinaxNOW COVID-19 AgCARD. 2020. Published online December 12. Available at: https://www.fda.gov/media/141570/download.
- 56. Alekseenko A, Barrett D, Pareja-Sanchez Y, et al. Direct detection of SARS-CoV-2 using non-commercial RT-LAMP reagents on heat-inactivated samples. Sci Rep. 2021;11:1820. doi: 10.1038/s41598-020-80352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon S, How AChicago dad developed a coronavirus testing program for his son's school. Transcript. Weekend Edition Saturday. National Public Radio. 2020. September 26. Available at: https://www.npr.org/2020/09/26/917185895/how-a-chicago-dad-developed-a-coronavirus-testing-program-for-his-sons-school.
- 58.Chappell KCoronavirus saliva screening program expands at suburban schools. NBC Chicago. 2020. Published November 18. Available at: https://www.nbcchicago.com/news/local/coronavirus-saliva-screening-program-expands-at-suburban-schools/2374812/
- 59. Lee EC, Wada NI, Grabowski MK, Gurley ES, Lessler J The engines of SARS-CoV-2 spread. Science. 2020;370:406–407. doi: 10.1126/science.abd8755. [DOI] [PubMed] [Google Scholar]
- 60.Ryan S5 Big Ten football takeaways after week 6, including the messy league rules of playing during COVID-19 and who has a shot at the championship. Chicago Tribune. 2020. Published November 30. Available at: https://www.chicagotribune.com/sports/college/ct-cb-big-ten-takeaways-ohio-state-covid-19-20201130-zjxnilkmujhtrop53ytthg7i7i-story.html.
- 61. Kissler SM, Fauver JR, Mack C, et al. Viral dynamics of SARS-CoV-2 infection and the predictive value of repeat testing. PLoS Biol. 2021;19:e3001333. doi: 10.1101/2020.10.21.20217042. Preprint. Published online MedRxiv. . Now published as Viral dynamics of acute SARS-CoV-2 infection and applications to diagnostic and public health strategies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buchanan L, Gamio L, Leatherby L, Stein R, Triebert CInside the White House event now under covid-19 scrutiny. New York Times. 2020. Published September 26. Available at: https://www.nytimes.com/interactive/2020/10/03/us/rose-garden-event-covid.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.