Abstract
Over the course of the coronavirus disease 2019 (COVID-19) pandemic, several severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genetic variants of concern have appeared and spread throughout the world. Detection and identification of these variants are important to understanding and controlling their rapid spread. Current detection methods for a particularly concerning variant, B.1.1.7, require expensive quantitative PCR machines and depend on the absence of a signal rather than a positive indicator of variant presence. Here we report an assay using a pair of molecular beacons combined with reverse transcription loop mediated amplification to allow isothermal amplification from saliva to specifically detect B.1.1.7 and other variants that contain a characteristic deletion in the gene encoding the viral spike protein. This assay is specific and affordable and allows multiplexing with other SARS-CoV-2 loop-mediated amplification primer sets.
Keywords: COVID-19, fluorescence, molecular diagnostic test, alpha variant
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus was first detected in December of 2020 and rapidly spread throughout the world. To date, the virus has infected over 120 million people and caused over 2.5 million deaths. Over the course of the pandemic, several genetic variants of concern have appeared and rapidly increased in frequency. The effects of these mutations remain unclear, but there is concern that these variants may alter virus phenotype, affect detection, and escape immune responses.1–10
Several variants of concern, including the B.1.1.7 variant first identified in the United Kingdom, have a characteristic 6-nucleotide deletion in the spike gene (S1Δ69–70) that appears likely to be an effective target for detection by nucleic-acid testing. Detection of these variants currently requires observation of the dropout of fluorescence from a spike-targeted primer-probe set during reverse transcription (RT)-quantitative PCR (qPCR).1 However, screening using the absence of a signal is prone to falsely label samples near the limit of detection or assay failures as variants, and performing RT-qPCR requires costly qPCR machines. Here, we design a molecular beacon spanning this S1Δ69–70 deletion that allows characterization of virus using simple isothermal RT loop-mediated amplification (LAMP).
MATERIALS AND METHODS
To initiate LAMP reactions, primers are designed such that the ends of the amplified DNA will form 2 dumbbell-like loops of single-stranded DNA. The single-stranded sequence in these loops is dependent on the template DNA and can be completely independent of the primers input to the reaction. Thus this sequence is resistant to creation by artifactual off-target amplification and available for annealing, making it a favorable target for molecular beacons. We used the Primer Explorer software to design a LAMP primer set (S6970) spanning the S1Δ69–70 deletion such that the deletion falls within the backward loop region of the amplification (Table 1) and then designed a molecular beacon (S6970_B117_MB) to both land inside the loop and span the deletion. We also added locked nucleic acids to increase specificity and strengthen hybridization to allow real-time detection at the temperatures used in LAMP. In addition, we designed an additional molecular beacon (S6970_WT_MB) targeting the undeleted S1 69–70 sequence to allow parallel detection of strains containing the pre-existing spike sequence (Fig. 1A).
TABLE 1.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| S6970_F3 | TGTGTTAATCTTACAACCAGAA |
| S6970_B3 | GGAAGCAAAATAAACACCATCA |
| S6970_FIP | AACTGAGGATCTGAAAACTTTGTCA-TCAATTACCCCCTGCATAC |
| S6970_BIP | TTACATTCAACTCAGGACTTGTTCT-TTATCAAACCTCTTAGTACCATTG |
| S6970_LF | GGGTAATAAACACCACGTGTGA |
| S6970_LB | TTTCCAATGTTACTTGGTTCCATGC |
| S6970_B117_MB | Fam-CGGTCCA+T+G+CTA+T+CT+CTGGGACCG-IBFQ |
| S6970_WT_MB | Cy3-GCCCATG+CTATA+CA+T+GT+CT+CTGGGC-IBRQ |
| N2_F3b | ACCAGGAACTAATCAGACAAG |
| N2_B3b | GACTTGATCTTTGAAATTTGGATCT |
| N2_FIPb | TTCCGAAGAACGCTGAAGCGGAACTGATTACAAACATTGGCC |
| N2_BIPb | CGCATTGGCATGGAAGTCACAATTTGATGGCACCTGTGTA |
| N2_LFb | GGGGGCAAATTGTGCAATTTG |
| N2_LB_2b | GGAACGTGGT+T+GACC |
| N2_MBb | Cy5-CCAC+CTTCG+G+GAA+CG+T+GGTGG-IBRQ |
FIGURE 1.
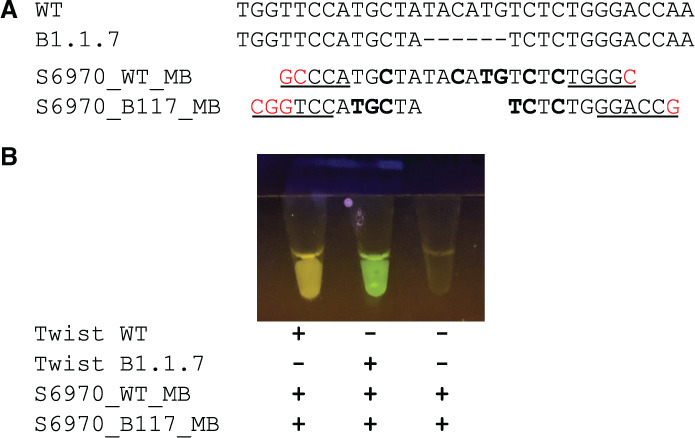
Development of molecular beacons to discriminate SARS-CoV-2 variants with a S1Δ69–70 mutation. A) A comparison of preexisting genomic S1 sequence (WT) and variant B.1.1.7 genomic sequence (B.1.1.7) and molecular beacons targeting these sequences (S6970_WT_MB and S6970_B117_MB, respectively). Dashes indicatetheS1Δ69–70 deletion, bold letters indicate locked nucleic acids, underlined letters indicate bases paired in the stem of the beacon, and red letters indicate extra nucleotides added to form the stem. B) Demonstration of visual detection using the molecular beacons. RT-LAMP reactions with molecular beacons at a final concentration of 0.25 μM were incubated using a heat block, and the end-point fluorescence was visualized using a low-cost viewer (miniPCR p51) and cell-phone camera (Google Pixel 2 “Night Sight” mode).
We then tested the S6970 primers and beacons on synthetic RNA from wild-type (WT; Twist Australia/VIC01/2020) and B.1.1.7 (Twist England/205041766/2020) SARS-CoV-2 variants. For comparison, we also multiplexed S6970 with the N2 primer set11 and molecular beacon.12 For these experiments, S6970_B117_MB was synthesized with a fluorescein (Fam) label, S6970_WT_MB was synthesized with a cyanine (Cy) 3 label, and N2_MB was synthesized with a Cy5 label. All oligonucleotides were obtained from Integrated DNA Technologies.
Reactions were performed using locally produced enzyme mix equivalent to commercial RT-LAMP mix.12 Reactions consisted of 10 μl of 2× RT-LAMP enzyme mix, 2 μl of 10× primer-beacon mix, and 8 μl of water doped with varying amounts of synthetic RNA. The 10× primer-beacon mix consisted of 16 μM forward inner primer (FIP), 16 μM backward inner primer f (BIP), 4 μM loop forward (LF), 4 μM loop backward (LB), 2 μM backward outer primer (B3), 2 μM forward outer primer (F3), 2.5 μM S6970_WT_MB beacon, and 2.5 μM S6970_B117_MB beacon. For multiplexed S6970 and N2 primer-beacon mix, the primer concentrations were halved, and beacon N2_MB was added while maintaining all beacon concentrations at 2.5 μM.
Reactions were incubated for 60 min at 63°C using a QuantStudio 5 qPCR machine for real-time quantification or a simple heat block. Note that the long incubation time was used to verify that no false positives were observed, and most reactions completed within 30 min. We also used a simple and affordable (∼$25) illuminator (miniPCR p51) using blue light–emitting diodes and an orange filter to observe the end-point fluorescence of these assays.
RESULTS
Our goal was to design 2 molecular beacons that could be combined in a single reaction to distinguish SARS-CoV-2 variants containing the S1Δ69–70 deletion. To test our design, we used synthetic SARS-CoV-2 RNA containing either a prototypical genome sequence from early in the pandemic or a genome containing the S1Δ69–70 deletion. We saw that fluorescence of the 2 molecular beacons, S6970_WT_MB and S6970_B117_MB, was easily visible by eye or cell-phone camera after isothermal RT-LAMP and that the distinction between yellow Cy3 and green Fam allowed visual characterization of SARS-CoV-2 variants (Fig. 1B).
To characterize the sensitivity and replicability of this assay, we attempted to amplify across a range of 10,000 to 39 genome copies per reaction (Fig. 2A). The S6970 beacons were specific to their intended target with only minimal fluorescence detected from S6970_WT_MB when Twist B.1.1.17 was amplified and only minimal fluorescence detected from S6970_B117_MB when wild-type Twist was amplified. In contrast, in the presence of their intended targets, the beacons were strongly fluorescent. For comparison, the S6970 primer set was also multiplexed with the sensitive N2 primer set and molecular beacon. The N2 beacon was highly fluorescent for all positive samples (Fig. 2B). In contrast, the S6970 primer set was less sensitive only detecting 11 of 12 reactions containing 625 genomic copies, 10 of 12 containing 312 copies, 9 of 12 containing 156 copies, 6 of 12 containing 78 copies, and 5 of 12 containing 39 copies. No false positives were observed in any beacon.
FIGURE 2.
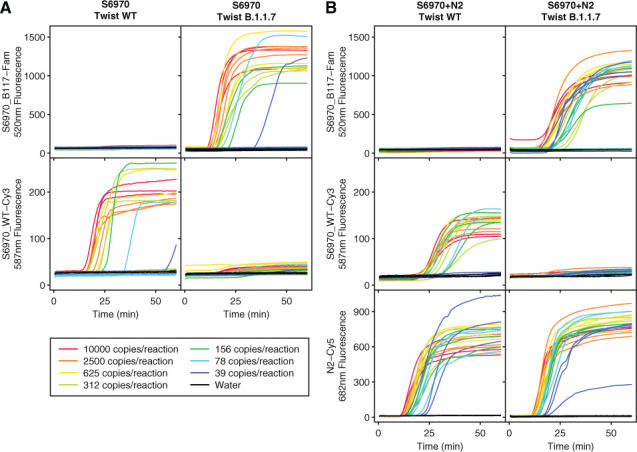
Testing of the S6970 primer set and molecular beacons on synthetic SARS-CoV-2 RNA. Fluorescence was monitored over time for LAMP isothermal amplification using the S6970 primer set alone (A) or S6970 multiplexed with N2 primer (B) for Twist Australia/VIC01/2020 (Twist WT) and Twist England/205041766/2020 (Twist B.1.1.7). The amount of synthetic SARS-CoV-2 RNA in each reaction was varied from 10,000 to 39 copies (indicated by line color) or water control (black). All fluorescence is given as relative fluorescence units (RFUs) divided by 1000.
Estimating the probability of detecting a single copy of target RNA as p(positive|copies,detectionRate) = 1 − (1 − detectionRate)copies, the maximum likelihood estimate for the detection of a single copy for S6970 was 0.7%, and the copy number necessary to give a 95% detection rate was estimated at 430 copies. With 8 μl of saliva input to each reaction, this translates to a detection at 54 copies per μl of saliva. This is less sensitive than some previously reported SARS-CoV-2 primer sets, such as the N2 primer set used here, so it would not be optimal for use as the only primer set in first-line screening. But the primer set would be well suited to a follow-up test of potential positives or multiplexed with more sensitive primer sets (Fig. 2B). Because of the dual-beacon design, the S6970 assay gives a clear readout of either wild-type SARS-CoV-2 detection, S1Δ69–70 detection or failure to detect either. Failures to detect SARS-CoV-2 in samples that are believed to be positive can be followed up with larger or more numerous reactions to give a higher probability of detection.
DISCUSSION
Here we show that molecular beacons and RT-LAMP provide an effective method for detecting SARS-CoV-2 variants containing the S1Δ69–70 deletion. Using simple isothermal amplification of unpurified saliva and end-point fluorescence, the genotype of a viral sample can be determined visually without expensive equipment. With locally purified enzymes,12 the reagent costs can be reduced to pennies per test. Combined with the easily parallelizable nature of isothermal incubation, scaling is effectively limited only by sample collection and processing. Although the large deletion found in this variant was particularly well suited for detection, molecular beacons are often used to detect even single-nucleotide polymorphisms.13 This pilot study showcases the capabilities of RT-LAMP and molecular beacons and promises further improvements in the monitoring and control of variants of concern through the design of additional beacons targeting diagnostic genetic signatures.
ACKNOWLEDGMENTS
This work was funded by the Penn Center for Research on Coronaviruses and Other Emerging Pathogens. We thank Young Hwang, Aoife Roche, and members of the COVID-19 Screening Assessment for Exposure (COVID-SAFE) laboratory for help and suggestions.
Footnotes
This work was funded by the Penn Center for Research on Coronaviruses and Other Emerging Pathogens. No conflicts of interest to report .
No human or animal subjects were involved in this research.
REFERENCES
- 1.Bal A, Destras G, Gaymard A, Stefic K, Marlet J, Eymieux S, Regue H, Semanas Q, d’Aubarede C, Billaud G, Laurent F, Gonzalez C, Mekki Y, Valette M, Bouscambert M, Gaudy-Graffin C, Lina B, Morfin F, Josset L, COVID-Diagnosis HCL Study Group Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveill . 2021;26:2100008. doi: 10.2807/1560-7917.ES.2021.26.3.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin DP, Weaver S, Tegally H, San EJ, Shank SD, Wilkinson E, Giandhari J, Naidoo S, Pillay Y, Singh L, Lessells RJ, Gupta RK, Wertheim JO, Nekturenko A, Murrell B, Harkins GW, Lemey P, MacLean OA, Robertson DL The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape. 2021. medRxiv. [DOI] [PMC free article] [PubMed]
- 3.Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, Navarro MJ, Bowen JE, Tortorici MA, Walls AC, King NP, Veesler D, Bloom JD. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell . 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kemp SA, Meng B, Ferriera IA, Datir R, Harvey WT, Papa G, Lytras S, Collier DA, Mohamed A, Gallo G, Thakur N, Carabelli AM, Kenyon JC, Lever AM, Marco AD, Saliba C, Culap K, Cameroni E, Piccoli L 2020. Recurrent emergence and transmission of a SARS-CoV-2 spike deletion H69/V70. bioRxiv. [DOI] [Google Scholar]
- 5.Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, Roberts DJ, Chandra A, Temperton N, Sharrocks K, Blane E, Modis Y, Leigh KE, Briggs JAG, van Gils MJ, Smith KGC, Bradley JR, Smith C, Doffinger R, Ceron-Gutierrez L, Barcenas-Morales G, Pollock DD, Goldstein RA, Smielewska A, Skittrall JP, Gouliouris T, Goodfellow IG, Gkrania-Klotsas E, Illingworth CJR, McCoy LE, Gupta RKCITIID-NIHR BioResource COVID-19 CollaborationCOVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 evolution during treatment of chronic infection. Nature . 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collier DA, De Marco A, Ferreira IATM, Meng B, Datir RP, Walls AC, Kemp SA, Bassi J, Pinto D, Silacci-Fregni C, Bianchi S, Tortorici MA, Bowen J, Culap K, Jaconi S, Cameroni E, Snell G, Pizzuto MS, Pellanda AF, Garzoni C, Riva A, Elmer A, Kingston N, Graves B, McCoy LE, Smith KGC, Bradley JR, Temperton N, Ceron-Gutierrez L, Barcenas-Morales G, Harvey W, Virgin HW, Lanzavecchia A, Piccoli L, Doffinger R, Wills M, Veesler D, Corti D, Gupta RKCITIID-NIHR BioResource COVID-19 CollaborationCOVID-19 Genomics UK (COG-UK) Consortium Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature . 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- 7.Muik A, Wallisch AK, Sänger B, Swanson KA, Mühl J, Chen W, Cai H, Maurus D, Sarkar R, Türeci Ö, Dormitzer PR, Şahin U. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GBE, Bennett H, Boyoglu-Barnum S, Shi W, Graham BS, Carfi A, Corbett KS, Seder RA, Edwards DK. 2021. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. [DOI] [Google Scholar]
- 9.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe . 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, van Zandvoort K, Silverman JD, Diaz-Ordaz K, Keogh R, Eggo RM, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. 2020. medRxiv. [DOI] [PMC free article] [PubMed]
- 11.Zhang Y, Ren G, Buss J, Barry AJ, Patton GC, Tanner NA. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques . 2020;69:178–185. doi: 10.2144/btn-2020-0078. [DOI] [PubMed] [Google Scholar]
- 12.Sherrill-Mix S, Hwang Y, Roche AM, Glascock A, Weiss SR, Li Y, Haddad L, Deraska P, Monahan C, Kromer A, Graham-Wooten J, Taylor LJ, Abella BS, Ganguly A, Collman RG, Van Duyne GD, Bushman FD. Detection of SARS-CoV-2 RNA using RT-LAMP and molecular beacons. Genome Biol . 2021;22:169. doi: 10.1186/s13059-021-02387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varona M, Anderson JL. Visual detection of single-nucleotide polymorphisms using molecular beacon loop-mediated isothermal amplification with centrifuge-free DNA extraction. Anal Chem . 2019;91:6991–6995. doi: 10.1021/acs.analchem.9b01762. [DOI] [PubMed] [Google Scholar]


