Abstract
Conventional reverse transcription quantitative polymerase chain reaction (RT-qPCR) technology has struggled to fulfill the unprecedented need for diagnostic testing created by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. Complexity and cost hinder access to testing, and long turnaround time decreases its utility. To ameliorate these issues, we focus on saliva and introduce several advances to colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) technology; RT-LAMP offers a minimal equipment alternative to RT-qPCR. First, we validated the use of the novel dye LAMPShade Violet (LSV), which improves the visual clarity and contrast of the colorimetric readout. Second, we compared different inactivation conditions on infectivity and RNA yield from saliva. Third, we developed a 10-minute RNA purification protocol from saliva. We call this magnetic bead protocol SalivaBeads. Finally, we developed a magnetic stick, StickLAMP, which provides reliable bead-based RNA purification as well as simple and low-cost access to scalable testing from saliva.
Keywords: COVID-19, SARS-CoV-2, coronavirus, saliva, magnetic beads, loop-mediated isothermal amplification
INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has highlighted many shortcomings in our national response and has driven an enormous increase in our need for in vitro diagnostics. A diagnostic test for SARS-CoV-2 using quantitative real-time polymerase chain reaction (qPCR) technology and nasopharyngeal swabs was developed within weeks of identifying the virus but is suboptimal for serving all diagnostic needs of this global pandemic.
This technology has struggled with turnaround time, supply chain shortages, and cost in the effort to increase the amount of testing worldwide.1 Many entities, commercial and academic alike, have risen to the challenge of adapting and developing molecular technologies to address these shortcomings. Among the myriad efforts that subsequently emerged, reverse transcription loop-mediated isothermal amplification (RT-LAMP) technology may prove to be a viable if not preferable alternative to qPCR technology in addressing at least some of the diagnostic needs of a global pandemic.
LAMP is a nucleic acid detection technology that operates like PCR on the principle of nucleic acid amplification.2 Unlike PCR however, LAMP is an isothermal technology and therefore obviates the need for thermal cyclers that would otherwise gate diagnostics behind equipment that costs several thousand dollars. In brief, LAMP requires at least 4 (and up to 6) different primers: 2 that contain a self-complementary region that generates a perpetually single-stranded loop structure as well as 2 that target the region within the single-stranded loop structure. A strand invasion event with the 2 self-complementing primers initiates production of the initial product containing open loops at either end; exponential amplification can then occur using the loop-targeting primers and a polymerase with strand-displacement ability. In short, LAMP can generate a detectable amount of DNA in an amount of time comparable to that needed for PCR but at a single reaction temperature. The DNA can be detected through several different means, including turbidity or through fluorescence with the inclusion of a fluorescent dye. However, a result interpretable to the naked eye would be optimal for a test to enjoy widespread use.3,4
Earlier efforts have explored the use of colorimetric detection of amplification products using a pH dye, phenol red, or a magnesium indicator, hydroxynaphthol blue.5–9 With phenol red, the color will change from red to yellow as more DNA acidifies the reaction. With hydroxynaphthol blue, the color will change from light blue to dark blue in the presence of magnesium, which is a by-product of amplification. These color changes are adequate but suffer from limited visible contrast and can often exhibit ambiguous color changes. Our work here uses the novel pH dye LAMPShade Violet (LSV) as an alternative to phenol red in RT-LAMP.10 LSV has greater visual contrast between high and low pH and a stronger inflection point, which both eases interpretation and reduces the number of ambiguous events.
Saliva is growing increasingly popular as an alternative respiratory specimen to nasopharyngeal swabs for the detection of SARS-CoV-2. Saliva is much easier to collect, and some report that it is a comparable, even superior, specimen for SARS-CoV-2 diagnostics.11 Previous efforts have identified saliva as compatible with RT-LAMP even in the absence of RNA purification. It was replaced by an inactivation step that normalizes the pH, releases RNA, and inactivates RNases.5 However, saliva is very heterogenous between individuals, which confounds a one-size-fits-all inactivation strategy. We therefore optimized an inactivation protocol with improved success across heterogenous saliva samples when combined with LSV. We suggest that this direct method is suitable for small-scale testing environments where it is easy to resample.
Despite this improvement, many saliva samples were still incompatible with direct input into a colorimetric RT-LAMP reaction. This is prohibitive at scale, at which samples cannot be individually resampled or modified for compatibility. We therefore developed and introduced a magnetic bead-based rapid RNA-purification procedure for saliva, which we term SalivaBeads. It features a saliva-optimized bead binding solution and uses a magnetic stick to isolate RNA-bound magnetic beads, which not only improves processing time and scalability but also resolves issues surrounding saliva compatibility with colorimetric RT-LAMP. The SalivaBeads procedure also takes less than 10 minutes and substantially improves sensitivity over direct input.
This article therefore describes our efforts to develop and optimize this low-cost, scalable, and sensitive saliva RT-LAMP protocol, which also minimizes reliance on specialized equipment. The magnetic stick is an inexpensive and dramatically more scalable alternative for RNA purification from saliva compared with magnetic racks and multichannel pipettes. We also improved several previous efforts with enhanced visual fidelity, sample compatibility, and sensitivity at minimal additional cost. Altogether, our protocol exhibits a limit of detection (LOD) of 3.7 copies/μL in a 200-μL saliva sample, costs less than $5 per sample without pooling or accounting for labor, and takes approximately 1 hour to conduct from beginning to end.
METHODS
Oligonucleotides RT-LAMP reactions
Phenol red experiments were conducted using WarmStart Colorimetric LAMP 2X Master Mix (New England Biolabs, Ipswich, MA; M1800L) in 25-μL reactions with 5 μL of inactivated saliva. LSV experiments were conducted using a buffer consisting of 5-mM Tris pH 8.5, 8 mM magnesium sulfate (MgSO4; MilliporeSigma, St. Louis, MO; 10708976001), 30-mM potassium chloride (KCl; MilliporeSigma, St. Louis, MO; P9541), 0.1% Tween-20, 10-mM deoxynucleoside triphosphates (dNTPs; Thermo Fisher Scientific, Waltham, MA; R1121), 12.5-mM potassium hydroxide (KOH; MilliporeSigma, St. Louis, MO; 221473), 10-mM LSV10 (Luke Lavis, Janelia Research Labs), 0.5-μL WS RTx (New England Biolabs, Ipswich, MA; M0380L), WS BST 2.0 (NEB M0538L; New England Biolabs, Ipswich, MA; M0538L) in 25-μL reactions with 5 μL of inactivated saliva. For magnetic stick experiments, SARS-CoV-2 reactions were conducted in 25-μL reactions, whereas actin reactions were conducted in 20-μL. All reactions were heated to 65°C for 45 minutes and cooled to at least room temperature prior to examination.
Reactions were imaged on an Epson V850 Scanner (Epson, Suwa, Japan).
Primers
Table 1 presents primer sequences and concentrations used to produce a 5× primer solution. The primer sequences are derived from previous work.4,5
TABLE 1.
Primer sequences and concentrations used to produce a 5× primer solution
|
Concentration
|
Component
|
Sequence
|
| 1 μM | E1-F3 | TGAGTACGAACTTATGTACTCAT |
| 1 μM | E1-B3 | TTCAGATTTTTAACACGAGAGT |
| 8 μM | E1-FIP | ACCACGAAAGCAAGAAAAAGAAGTTCGTTTCGGAAGAGACAG |
| 8 μM | E1-BIP | TTGCTAGTTACACTAGCCATCCTTAGGTTTTACAAGACTCACGT |
| 2 μM | E1-LB | GCGCTTCGATTGTGTGCGT |
| 2 μM | E1-LF | CGCTATTAACTATTAACG |
| 1 μM | Orf1a-HMS-F3 | CGGTGGACAAATTGTCAC |
| 1 μM | Orf1a-HMS-B3 | CTTCTCTGGATTTAACACACTT |
| 8 μM | Orf1a-HMS-LF | TTACAAGCTTAAAGAATGTCTGAACACT |
| 8 μM | Orf1a-HMS-LB | TTGAATTTAGGTGAAACATTTGTCACG |
| 2 μM | Orf1a-HMSe-FIP | TCAGCACACAAAGCCAAAAATTTATTTTTCTGTGCAAAGGAAATTAAGGAG |
| 2 μM | Orf1a-HMSe-BIP | TATTGGTGGAGCTAAACTTAAAGCCTTTTCTGTACAATCCCTTTGAGTG |
Control RNA
Contrived samples were created using either heat-inactivated SARS-CoV-2 RNA from BEI (Manassas, VA; NR-52286), or inactivated SARS-CoV-2 viral particles from Zeptometrix (Franklin, MA; NATSARS[COV2]-ST). In order to optimize and evaluate magnetic-bead purification procedures, RNA from BEI was diluted in 1 ng/μL Drosophila RNA to 1000 copies/μL and spiked into inactivated saliva at the stated concentration. To evaluate the full-process performance of the protocol, LOD experiments and experiments intended to evaluate RNA release from viral particles were conducted using Zeptometrix SARS-CoV-2 particles added into raw saliva at the stated concentrations. In the proceeding methods, the control used will be specified.
Saliva Inactivation
A 10× inactivation was prepared with 62.5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP; Goldbio, St. Louis, MO; TCEP1), 10 mM EDTA (Thermo Fisher Scientific, Waltham, MA; 10977015), and 130 mM sodium hydroxide (NaOH; MilliporeSigma, St. Louis, MO; S5881). A 10× inactivation reagent was added to a final concentration of 1× to raw saliva and inverted 10 times, or vortexed for 5 seconds at maximum speed. Saliva samples were then heated to 95°C for 5 minutes and allowed to cool at room temperature for 3 minutes prior to downstream processing. When testing 65°C inactivation, we inactivated for 15 minutes at 65°C and let samples cool to room temperature for 3 minutes prior to downstream processing. During optimization experiments, TCEP concentrations used are as described in Fig. 3 and the “Results” section. During proteinase K testing, the indicated amount of proteinase K (New England Biolabs, Ipswich, MA; P8107S) was added to saliva and heated at 65°C for 5 minutes and inactivated at 95°C for 10 minutes. Virus titers in untreated or treated samples were then determined using Vero E6 cells (grown in 10% fetal bovine serum–Dulbecco's Modified Eagle Medium; Thermo Fisher Scientific, Waltham, MA; 12430054). For plaque assays, cells were fixed with 10% formaldehyde 3 days after infection and stained with crystal violet.
FIGURE 3.
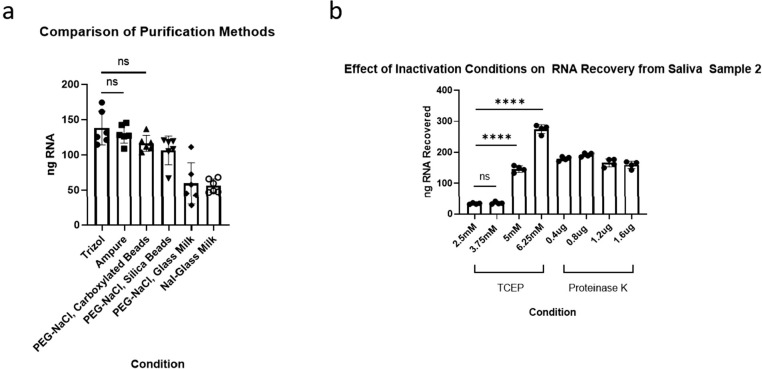
Investigating and optimizing saliva for purification. (a) A comparison of select purification methods on total RNA recovered from saliva measured by Qubit fluorimetry. (b) A comparison of RNA recovered from polyethylene glycol–sodium chloride (PEG-NaCl), carboxylated beads following inactivation methods using different concentrations of Tris(2-chloroethyl) phosphate (TCEP) or proteinase K.
Purification
Commercial Ampure XP beads (Beckman-Coulter Life Sciences, Indianapolis, IN; A63881) were used according to protocol at 2× volumes and eluted in 30 μL Milli-Q water (H2O).
Unmodified homemade magnetic bead buffer was prepared using 100 mM sodium chloride (NaCl; MilliporeSigma, St. Louis, MO; S9888), 20% PEG-8000 wt/vol (MilliporeSigma, St. Louis, MO; 1546605), 10 mM Tris-HCl, pH 8, 1-mM EDTA (Thermo Fisher Scientific, Waltham, MA; 10977015), and Milli-Q H2O to 50 mL. 1000 μL Sera-mag Speedbeads (Thermo Fisher Scientific, Waltham, MA; 09-981-123) were washed twice in 1 mL of 10-mM Tris pH 8, 1-mM EDTA, and added to the bead buffer.
The final SalivaBeads recipe uses the same materials as listed herein, but with 700 mM NaCl, 14% PEG-8000 wt/vol, 10 mM Tris-HCl, pH 8, 1 mM EDTA, 200 μL Sera-mag Speedbeads, and Milli-Q H2O to 50 mL.
Commercial Ampure XP beads and unmodified homemade magnetic bead buffer was used according to commercial Ampure XP protocols.
For SalivaBeads, 2 volumes of SalivaBeads were added to inactivated saliva samples and inverted 5 times. The bead+saliva mix was incubated at room temperature for 3 minutes. A magnetic stick with tip was added to the bead+saliva mix for 2 minutes, with agitation at 1 minute and before removing. Magnetic stick with bound beads were dipped in 100 μL Milli-Q H2O prepared in a PCR strip tube up and down 5 times, then left to incubate for 20–25 seconds. The magnetic stick was then removed from the water and placed in a SARS-CoV-2 reaction mix for 60 seconds. After 60 seconds, the magnetic stick was removed from the SARS-CoV-2 reaction mix and placed in an actin reaction mix for 30 seconds, then removed and the tip discarded.
For purifying RNA from wash steps, ethanol was added to each sample to a final concentration of 75% and a final volume of 200 μL. Sodium acetate (Invitrogen, Waltham, MA; AM9740) was added to a final concentration of 0.3 M. Samples were left to precipitate overnight at −20°C. The next day, samples were spun down for 30 minutes at 18 000 relative centrifugal forces. Samples were washed once in 80% ethanol and eluted into 20 μL of Milli-Q H2O, 5 μL of which was subsequently used in qPCR reactions.
Quantitative Polymerase Chain Reaction
The qPCR was done using the Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs, Ipswich, MA; E3006L) and the Centers for Disease Control and Prevention N1 primer (Integrated DNA Technologies, Coralville, IA; 10006713). Reactions and cycling conditions were prepared according to manufacturer's protocol.
3D Printing
Magnetic sticks and tips were printed using Siraya Tech Blu (Siraya Tech, San Gabriel, CA) on an Epax X10 UV LCD 3D Printer with an 8.9-inch 4K mono LCD (EPAX, Morrisville, NC) using the following settings: 0.05-mm layer height, 8 bottom layers, 3.2-second exposure time, 12.4-second bottom exposure time, 7-mm lift, 35 mm/min lift speed, 125 mm/min retract speed. The magnets used were 2.54-mm diameter, 0.600-in long N50 magnets (SuperMagnetMan, Pelham, AL; Cyl0072-20).
RESULTS
We present 2 different SARS-CoV-2 protocols. The first, the direct assay, is conducted on inactivated, unpurified saliva; it is suitable for low-throughput testing in low-resource environments. However, high-frequency or large population testing is likely to prove problematic. This is in part due to the pH variation exhibited by different sources of saliva. To address this issue, we developed a second version, the purified assay, which adds a novel and rapid purification step. It normalizes saliva pH from different sources while improving sensitivity.
Both protocols begin identically: The crude saliva samples are inactivated by addition of a TCEP, EDTA, and NaOH solution. The samples are then heated to 95°C for 5 minutes and allowed to cool at room temperature for at least 3 minutes. In the direct assay, 5 μL of inactivated saliva is then added to 2 RT-LAMP reactions, 1 targeting SARS-CoV-2 and the other actin. In the purified assay, 2 volumes of SalivaBeads—described below—are added for 5 minutes before being removed by a magnetic stick. The stick-bound beads are washed in water for 30 seconds and then eluted twice sequentially, first directly into a SARS-CoV-2 RT-LAMP reaction and then into an actin RT-LAMP reaction. In both assays, RT-LAMP reactions are incubated at 65°C for 45 minutes.
Both assays rely on the color difference caused by successful DNA amplification. A positive test is indicated by both reactions turning clear; a negative test is indicated by the actin reaction turning clear and the SARS-CoV-2 reaction remaining purple; and an inconclusive or unsuccessful test is indicated by the actin reaction remaining purple. Comparing the color changes in the 2 reactions is critical in the direct assay, wherein baseline color may be affected by the pH of the input saliva sample. However, a careful comparison is less important in the purified assay, wherein all samples exhibit comparable baseline pH values following purification.
What follows describes our efforts in developing and optimizing the parameters of our protocol.
LSV and Modifications to Avoid Heterogeneity to Avoid Saliva pH Variation
In our initial tests, we used a previously described 100× inactivation reagent consisting of 2.5-M TCEP, 100-mM EDTA, and 1.2-M NaOH.5 The NaOH concentration is critical, because the colorimetric readout varies with pH. As in this publication,5 we initially used the colorimetric RT-LAMP reagent provided by NEB, which uses phenol red as a pH sensor. Phenol red changes from red to yellow upon acidification by successful amplification.
Although 1.2 M NaOH was sufficient in most cases, many samples were still too acidic and caused the reaction to turn prematurely positive, meaning even prior to incubation. To address this problem, we took 2 approaches, which were evaluated on 4 different samples from 4 individuals (Fig. 1A). First, we increased the NaOH concentration to 1.4 M and 1.6 M and observed the color changes pre- and postincubation. Second, we used a different pH-sensitive dye—LSV.10 It changes from purple to clear upon successful amplification. Because LSV has sharper contrast and fewer intermediate color changes than phenol red, we hypothesized that LSV may help with the interpretation of samples from saliva with outlier pH values.
FIGURE 1.
Two comparisons of phenol red against LAMPShade Violet for detection of amplification products in reverse transcription loop-mediated isothermal amplification (RT-LAMP). (a) A comparison of color change fidelity and consistency before and after incubation in 4 varied saliva samples as a function of sodium hydroxide (NaOH). (b) A comparison of the sensitivity of the commercial phenol red RT-LAMP mix and our in-house reaction mix with LAMPShade Violet judged by their ability to detect 20 total copies of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA.
We found that 1.4-M NaOH was the best concentration to accommodate samples across different pH values. Although a color difference was observed between positive and negative samples, more alkaline samples were still ambiguous with phenol red. LSV in contrast was superior in distinguishing positive and negative samples.
To compare our in-house RT-LAMP reaction with NEB-supplied reagents, we used them both to detect 20 copies of SARS-CoV-2 RNA, which is near the threshold for success (Fig. 1B). They were qualitatively similar: Our in-house version was positive 5 of 8 times, whereas the commercial was positive 4 of 8 times. Yet we prefer LSV due to its superior contrast and performance with samples of heterogenous pH values.
Viral Inactivation at 65°C Is Effective
The current protocol involves virus inactivation at 95°C followed by a 65°C RT-LAMP incubation. To simplify the protocol and further reduce equipment demands, we assayed virus inactivation at 65°C or even at room temperature with or without inactivation reagent.
Whereas 5 minutes at room temperature was insufficient to eliminate biological activity even with the addition of inactivation reagent, 5 minutes at 65°C with or without reagent addition reduced activity to undetectable (i.e., by at least 5–6 log units; Fig. 2A).
FIGURE 2.
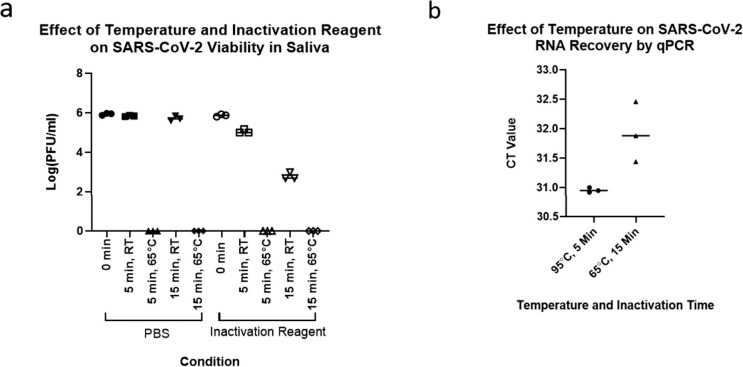
Evaluation of inactivation of saliva samples at 65°C. (a) A comparison of room temperature and 65°C incubations on the viability of (severe acute respiratory syndrome coronavirus 2) SARS-CoV-2 infected Vero E-6 cells with or without inactivation reagent. (b) A comparison of RNA recovery from encapsulated inactivated SARS-CoV-2 virions in saliva at 95°C for 5 minutes and 65°C for 10 minutes.
We also compared SARS-CoV-2 RNA yield from a 65°C vs. a 95°C incubation. To create specimens that most closely resembled clinical samples, we contrived saliva samples using inactivated but intact SARS-CoV-2 virions from Zeptometrix (Fig. 2B). A 65°C inactivation for 15 minutes led to a significant 1-cycle, approximately 2-fold loss in RNA yield compared with a 95°C inactivation for 5 minutes (t4 = 3.274, P = .0307). This modest decrease suggests that 65°C inactivation is an acceptable alternative in environments that can only afford minimal equipment or are otherwise averse to near-boiling temperatures. However, the improvement in yield at 95°C suggests that it is preferred in environments that can accommodate this temperature. Further efforts described in the next section use this inactivation temperature.
Testing Means of Purifying RNA From Saliva
To further improve sensitivity and sample compatibility, we sought to develop a rapid RNA purification protocol optimized for saliva. We first measured RNA recovery using fluorimetry with a Qubit device and compared various bead-based methods with Trizol purification (Fig. 3A). They included 2 volumes of Ampure Beads XP, 2 volumes of magnetic silica beads in an NaCl/PEG-8000 solution, 2 volumes of magnetic carboxylated beads in an NaCl/PEG-8000 solution, glass milk in a NaCl/PEG-8000 solution as well as glass milk in a sodium iodide (NaI) solution as previously described.5,12 Ampure XP beads as well as the NaCl/PEG-8000–based bead mixes were comparable to Trizol (Fig. 3A). Due to lower cost and the advantage of being suitable for scale and automation, we decided to use the NaCl/PEG-8000–based bead mix with carboxylated beads for further optimization of RNA purification from saliva.
Because current inactivation procedures were developed for direct input of saliva into an RT-LAMP reaction, we further optimized inactivation for input into bead-based RNA purification. Different concentrations of TCEP as well as proteinase K were assayed, the latter with a 5 minute 65°C incubation followed by a 5-minute 95°C incubation (Fig. 3C).
Although proteinase K was effective in increasing RNA yield compared with the initial condition of 2.5-mM TCEP, 6.25-mM TCEP was even better and used below.
Optimizing Salt and Salt Concentration
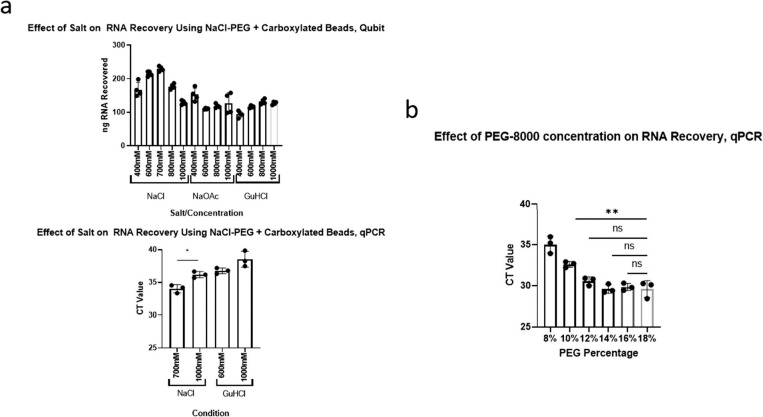
To optimize the NaCl/Peg-8000 bead mix, we examined the effects of salt type, salt concentration, and PEG-8000 concentration on RNA yield from saliva. For reference, the original purification recipe calls for 1-M NaCl, and 18% PEG-8000.12 Different concentrations of NaCl and different concentrations of sodium acetate (NaOAc) were assayed; the latter is also commonly used in nucleic acid precipitation. Different concentrations of guanidine hydrochloride (GuHCl)—another reagent commonly used in nucleic acid purification—were also assessed (Fig. 4A). Although all salts at all concentrations tested could purify RNA, NaCl at a reduced concentration of 0.7 M produced a significant higher yield compared with the original recipe at 1 M (P < 1 × 10−4, 1-way analysis of variance [ANOVA], Tukey honestly significant difference [HSD]) and compared with the other salts.
FIGURE 4.
Optimizing the concentrations of salt and polyethylene glycol (PEG) for RNA purification from saliva. (a) A comparison of multiple salts at multiple concentrations on RNA recovery from saliva with 18% PEG. Top: Measurement of recovery by Qubit fluorimetry. Bottom: Measurement of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA recovery by N1 quantitative polymerase chain reaction (qCPR) from contrived saliva samples. (b) A comparison of the effect of multiple PEG concentrations with 700-mM sodium chloride (NaCl) on RNA recovery, measured by SARS-CoV-2 RNA recovery by N1 qPCR from contrived saliva samples.
In other tests, the addition of PEG-8000 to RT-LAMP increased the false-positive rate (data not shown). Although we did not see a similar change with the purified assay, it is possible that a modestly increased rate becomes relevant with large-scale testing. We therefore decided to reduce the concentration of PEG-8000 until it affected yield. We found no significant difference between 18% PEG-8000 and 12% PEG-8000 (Fig. 4B). Because visual examination of the data suggested a slight loss of sensitivity between 14% and 12% (data not shown), we adjusted the buffer to 14% PEG-8000. Our final adjusted recipe therefore uses 0.7-M NaCl and 14% PEG- 8000.
Optimizing Elution and Binding Times
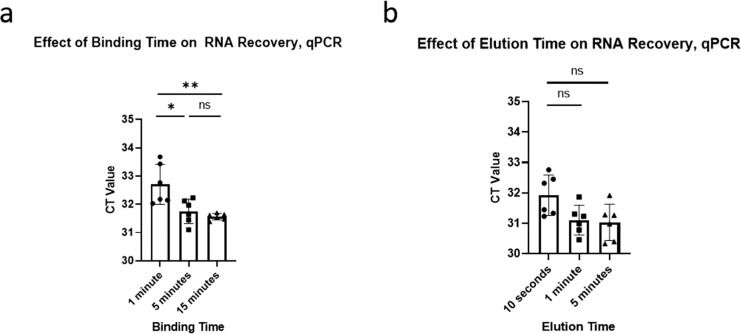
In our purification protocol, the bead mix is left to bind RNA for a certain amount of time before being removed by a magnet, henceforth referred to as binding time. At the end of the protocol, the beads are left to release RNA in the reaction mix for a certain amount of time, henceforth referred to as elution time. Our earlier development procedures used a 5-minute binding time and a 1-minute elution time. To minimize the test processing time, we determined the minimal binding and elution times without sacrificing sensitivity.
We first tested a 1-, 5-, and 15-minute binding time with a 1-minute elution. We observed a significant difference between 1-minute and a 5-minute binding time (P < .01, 1-way ANOVA, Tukey HSD) but no significant difference between 5 and 15 minutes (Fig. 5A). We therefore chose a 5-minute binding time. We then compared 10-second, 1-minute, and 5-minute elution time (Fig. 5B). We found no significant differences and so chose an elution time of 30 seconds for improved operational consistency.
FIGURE 5.
Optimizing the binding and elution times for magnetic bead-based purification using carboxylated beads with 700-mM sodium chloride (NaCl) and 14% polyethylene glycol (PEG), as measured by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA recovery through N1 qPCR from contrived saliva samples. (a) The effect of binding time on RNA recovery, with a 1-minute elution. (b) The effect of elution time on RNA recovery, with a 5-minute binding time.
Optimizing Wash Buffers
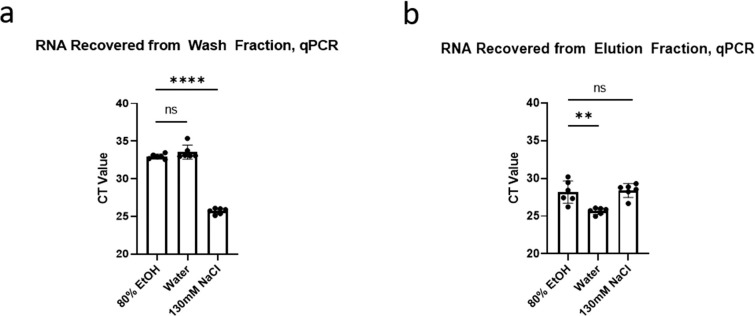
The majority of bead-based RNA purification protocols feature a 70%–80% ethanol wash to remove nonspecifically bound contaminants without removing nucleic acids. Because ethanol is incompatible with many downstream enzymatic reactions, protocols involving an ethanol wash typically require a long drying step to remove all traces of ethanol. To eliminate this drying step, we compared this standard 80% ethanol wash to water and to 130-mM NaCl, which was used in a cellulose dipstick-based purification assay.13 We therefore washed the beads for 30 seconds in 200 μL of the indicated wash buffers and then eluted in 50 μL of deionized H2O for 30 seconds.
We then ethanol-precipitated the RNA under standard conditions (see the “Methods” section) and assayed recovery by qPCR.
There was no significant difference between RNA levels in the 80% ethyl alcohol (EtOH) and in the water wash fractions (Fig. 7A). However, 130-mM NaCl washed off significantly more RNA (P < 1 × 10−4, 1-way ANOVA, Tukey HSD). In the eluate, there was no significant difference between RNA recovered from the 130-mM NaCl-washed beads and the 80% EtoH-washed beads, but the H2O-washed beads exhibited a significant 1-cycle improvement (P < 1 × 10−3, 1-way ANOVA, Tukey HSD; Fig. 7C). We therefore decided that water was the most suitable wash reagent for quick purification, also because water is compatible with the RT-LAMP reaction and requires no drying step following washing.
FIGURE 7.
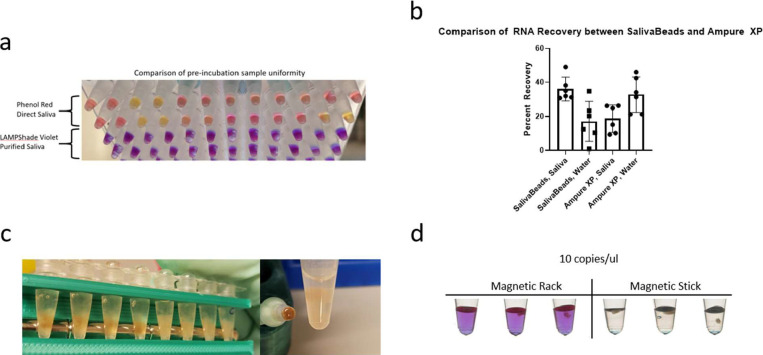
Several features of SalivaBeads purification. (a) A comparison of preincubation sample color uniformity in 12 samples, with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and actin reactions side by side. Direct input of inactivated saliva in a commercial phenol red-based reaction is compared to a SalivaBeads-purified RNA input into our LAMPShade Violet-based reaction. (b) A comparison of SARS-CoV-2 RNA recovered from contrived saliva samples by N1 quantitative polymerase chain reaction (qPCR), measured by number of copies recovered by qPCR divided by number of copies contrived into saliva. (c) A visual comparison of debris bound to magnetic beads when isolating beads by magnetic rack (left) or a magnetic stick (right). (d) A comparison of sensitivity between SalivaBeads purified via magnetic rack (left) and magnetic stick (right), by RT-LAMP with LAMPShade Violet.
FIGURE 6.
A comparison of 3 washing conditions for RNA purification from saliva using carboxylated beads with 700-mM sodium chloride (NaCl) and 14% polyethylene glycol (PEG), as measured by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA recovery by N1 qPCR from contrived saliva samples. (a) A measurement of RNA recovered from a 30-second wash in the indicated wash buffer. (b) A measurement of RNA recovered from a 30-second elution in water from samples washed using the indicated wash buffer.
These changes and optimizations and our bead mix are herein referred to as SalivaBeads.
StickLAMP
Most nucleic acid isolation procedures that use magnetic beads also use an elution step, which releases the purified RNA into an intermediate buffer such as water or Tris prior to a reaction. We sought to simplify this strategy by eluting RNA from SalivaBeads directly into the RT-LAMP mix. However, the sensitivity was poor compared with the direct assay. Both assays were able to routinely detect 100 copies/μL of SARS-CoV-2 RNA, but only the direct assay was able to detect 25 copies/μL. (Fig. 7A).
To address whether this was due to a poor RNA yield from SalivaBeads, we compared this yield with that from Ampure XP beads. This was done from saliva as well as from purified RNA in water (Fig. 7B). SalivaBeads had superior yield to Ampure XP beads from saliva but inferior to water. Moreover, the SalivaBeads yield from saliva was comparable with the Ampure XP yield from water. This indicates that the reduced sensitivity is not due to poor yield but rather from some sort of contaminant carryover or other incompatibility.
Indeed, we noticed that considerable debris clung to the SalivaBeads that persisted through the wash step and contaminated the RT-LAMP mix (Fig. 7C, left). We therefore developed a magnetic stick that would remove the beads prior to downstream processing (Fig. 7D). Important, the stick appeared to be selective for removing the beads without most of the saliva debris (Fig. 7C, right). Consistent with this observation, magnetic stick purification could faithfully detect 10 copies/μL, whereas magnetic rack purification could not (Fig. 7E).
Performance Analysis of StickLAMP
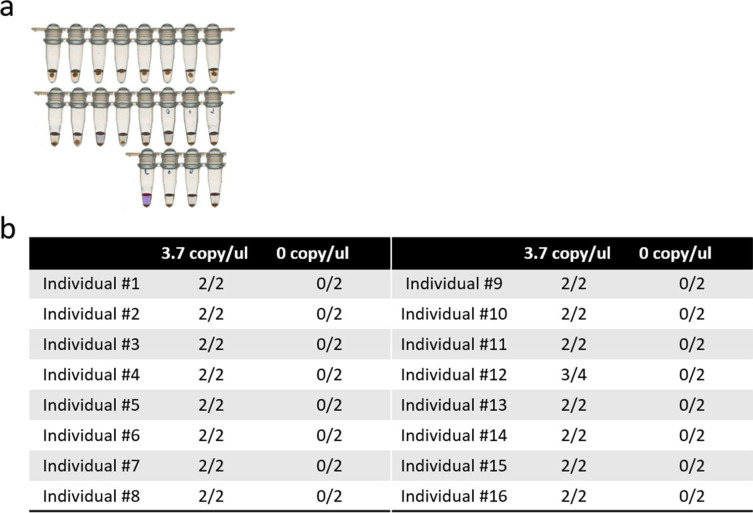
To evaluate the performance of StickLAMP purification, we determined the copy number at which 95% of reactions score positive, henceforth referred to as the LOD. Our LOD was at least 3.7 copies/μL (i.e., 19 of 20 contrived samples with 3.7 copies of SARS-CoV-2/μL in 200-μL saliva scored positive; Fig. 8A).
FIGURE 8.
Performance evaluation of SalivaBeads and StickLAMP. (a) Limit of detection experiment: ability of StickLAMP to detect 3.7 copies/μL of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA from 200 μL of contrived saliva in 20 replicates. (b) Testing the ability of StickLAMP to detect 3.7 copies/μL in 200 μL saliva from 16 different samples.
To ensure that this LOD was similar across a wide variety of saliva types, we collected saliva from 16 different individuals and created 2 contrived samples per individual with 3.7 copies/μL of SARS-CoV-2 (Fig. 8B). For 15 of 16 individuals, both samples were positive. For the 16th individual, only 1 sample was positive, but both samples were positive upon retesting. The 3.7 copies/μL LOD is therefore robust across different individuals.
DISCUSSION
We present 2 protocols for the detection of SARS-CoV-2 RNA from saliva, which feature several innovations. First, LSV is an attractive alternative to phenol red for the colorimetric detection of SARS-CoV-2 RNA in saliva; LSV improves visual fidelity without sacrificing sensitivity. Second, 65°C is completely successful at viral inactivation in saliva. Compared with the more standard 95°C, 65°C enhances safety and reduces testing time and equipment demands with only somewhat reduced detection sensitivity. Third, we present a rapid purification protocol. It adds less than 10 minutes to the processing time, costs less than 20 cents per sample, and is optimized for saliva. The purification improves sensitivity over 10-fold and normalizes sample pH for downstream colorimetric detection.
Purification uses a magnetic stick as an integral tool for rapid nucleic acid purification. Saliva contains substantial and variable levels of debris. It sticks to nucleic acid binding media like beads and inhibits the LAMP colorimetric assay. One previous effort also focused on this assay circumvented the debris issue with centrifugation.5 However, this solution is undesirable for scaled testing because centrifuges are difficult to introduce into an automated workflow. They are also expensive and are contrary to the goal of minimizing equipment requirements. The magnetic stick selectively binds magnetic beads over saliva debris and therefore substitutes for centrifugation. The magnetic stick also has several advantages over the other traditional method of bead separation, magnetic rack purification.
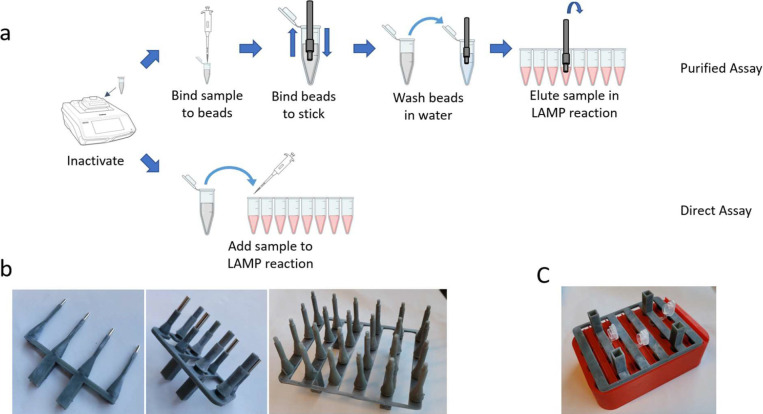
First, the magnetic stick has superior scaling potential. Multichannel pipettes are typically used in magnetic rack purification to improve throughput and sample processing time in both manual and automated workflows. Important to note, however, the number of channels is constrained by mechanical considerations in multichannel pipettes, whereas the magnetic stick simplicity enables the number of simultaneous “channels” to scale well beyond traditional limitations. Indeed, we developed 4-, 8-, and 24-channel versions for the purpose of large-scale multiplex purification reactions, as well as a custom rack designed for a 24-channel magnetic stick to elute directly into a 96- or 384-well plate (Fig. 9B, C).
FIGURE 9.
An overview of StickLAMP and further considerations. (a) A visual overview of the StickLAMP protocol. (b) A 4-channel magnetic stick (left), 8-channel (middle), and 24-channel (right). (c) A demonstration of a 24-channel magnetic stick in 3-dimensional-printed hardware designed to hold 24 Eppendorf tubes, 1.5 mL each.
Second, this superior scaling potential also reduces the additional processing time required by more samples. The addition of every 8 samples increases the required processing time with an 8-channel multipipette, but a 24-channel magnetic stick increases processing time only beyond 24 samples. The processing time is further reduced because 2–3 pipetting steps are replaced with a single dipping step.
Third, the cost of a magnetic stick is dramatically less than a multichannel pipette. A 24-channel magnetic stick costs less than $5 to produce, and each tip is less than 5 cents, about $6 in total. An 8-channel multipipette, in contrast, costs well over a thousand dollars.
Our purification optimization results also suggest several unexplored directions for future saliva-based nucleic acid diagnostics. The optimal NaCl concentrations were well below the theoretical limit required for purifying nucleic acids, suggesting that uncharacterized minerals or salts present in saliva are aiding nucleic acid binding to carboxylated beads.14 In addition, all saliva samples benefitted from additional TCEP, greater than previously published concentrations.5 However, the degree to which they benefit and even the amount of RNA purified from different saliva samples varied. This suggests potential heterogeneity of RNase levels and/or RNA content between samples, which future saliva-based diagnostic test efforts should consider to make further improvements. We are nonetheless confident in our reported LOD, given its consistency across the wide variety of tested samples.
In summary, these new SARS-CoV-2 detection protocols are inexpensive, cost less than $5 per test without considering labor and sample pooling, and offer improved scalability over existing tests without sacrificing sensitivity. They are especially suited for workplaces or schools with modest numbers of employees and students, from single digits to the low thousands. The minimal equipment requirements and low cost also make them well-suited for low resource environments, which still might be able to mount a medium-complexity Clinical Laboratory Improvements Amendments (CLIA) lab. We note in this context that low-resource and underserved environments have been disproportionately vulnerable to the spread of SARS-CoV-2.15
Acknowledgments
This work was supported by the Howard Hughes Medical Institute. Research using human saliva was approved by the institutional review board at Brandeis University. We express our thanks to the gLAMP consortium for their willingness to create an open dialogue and for engaging in helpful discussions and ideas in an exceptionally collaborative space. We would like to thank Brian Rabe and Connie Cepko of Harvard Medical School for sharing the reagents and ideas that initiated this project. We also express enormous gratitude to Tim Brown, Luke Lavis, and Ronald D. Vale for sharing LAMPShade Violet with us, as well helping us through constructive dialogue and general camaraderie. We would like to thank Kate Abruzzi for reading, editing, and generally helpful discussions. Finally, we would like to thank the Brandeis facilities staff for helping facilitate this endeavor.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med . 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res . 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun . 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Ren G, Buss J, et al. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques . 2020;69:178–185. doi: 10.2144/btn-2020-0078. [DOI] [PubMed] [Google Scholar]
- 5.Rabe BA, Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci U S A . 2020;117:24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thi VLD, Herbst K, Boerner K, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020. 12:eabc7075. [DOI] [PMC free article] [PubMed]
- 7.Lalli MA, Langmade SJ, Chen X, et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP. medRxiv . 2020. [DOI] [PMC free article] [PubMed]
- 8.Zhang Y, Odiwuor N, Xiong J, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv . 2020. [DOI]
- 9.Thi VLD, Herbst K, Boerner K, et al. Screening for SARS-CoV-2 infections with colorimetric RT-LAMP and LAMP sequencing. medRxiv . 2020. [DOI]
- 10.Grimm JB, Brown TA, Tkachuk AN, Lavis LD. General synthetic method for si-fluoresceins and si-rhodamines. ACS Cent Sci . 2017;3:975–985. doi: 10.1021/acscentsci.7b00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyllie AL, Fournier J, Casanovas-Massana A, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV- 2. N Engl J Med . 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res . 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellner MJ, Ross JJ, Schnabl J, et al. A rapid, highly sensitive and open-access SARS-CoV-2 detection assay for laboratory and home testing. bioRxiv . 2020. [DOI] [PMC free article] [PubMed]
- 14.Lis JT, Schleif R. Size fractionation of double-stranded DNA by precipitation with polyethylene glycol. Nucleic Acids Res . 1975;2:383–390. doi: 10.1093/nar/2.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ML, Behrman P, Dulin A, et al. Addressing inequities in COVID-19 morbidity and mortality: research and policy recommendations. Transl Behav Med . 2020;10:516–519. doi: 10.1093/tbm/ibaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]