Epithelial-mesenchymal plasticity is a prevalent source of intratumoral heterogeneity, driven by diverse molecular programs.
Abstract
Epithelial-mesenchymal plasticity (EMP) contributes to tumor progression, promoting therapy resistance and immune cell evasion. Definitive molecular features of this plasticity have largely remained elusive due to the limited scale of most studies. Leveraging single-cell RNA sequencing data from 266 tumors spanning eight different cancer types, we identify expression patterns associated with intratumoral EMP. Integrative analysis of these programs confirmed a high degree of diversity among tumors. These diverse programs are associated with combinations of various common regulatory mechanisms initiated from cues within the tumor microenvironment. We show that inferring regulatory features can inform effective therapeutics to restrict EMP.
INTRODUCTION
Epithelial-mesenchymal plasticity (EMP) refers to the ability of cells to interconvert between epithelial and mesenchymal phenotypes, dynamically adopting mixed features of these states in response to signals in the cells’ microenvironment (1). Throughout a tissue, cells with phenotypes spanning an epithelial/mesenchymal (E/M) continuum can be observed, emerging in response to specific features of their local environment. At the leading edge of tumors, for example, epithelial architecture becomes progressively disorganized, and the cancer cells express higher levels of mesenchymal-associated genes (2, 3). In cancer, this plasticity has been broadly associated with promoting metastasis, chemoresistance, and immunosuppression (4).
Given its supposed impact on tumor progression and treatment, understanding the molecular mechanisms that drive EMP and developing therapeutic strategies to modulate it have been a priority for years (1, 5–7). Identifying molecular determinants of EMP has largely focused on studying dynamics associated with the epithelial-mesenchymal transition (EMT) induced in experimental settings, through the addition of exogenous cytokines [e.g., transforming growth factor–β (TGF-β1)] or genetic manipulation. Over the past two decades, however, it has become increasingly clear that molecular features of the EMT are highly context specific (1, 8–12). The reliability of even the most canonical EMT genes (e.g., SNAI1, SNAI2, CDH1, CDH2, and VIM) has become unclear, and the reliance on specific genes as molecular markers of EMP has led to controversy (13–17). As a result, recent guidelines from “the EMT International Association” suggest that the primary criteria for defining EMP should focus on changes to cellular properties (e.g., loss of cell-cell junctions and enhanced migratory capacity) (1). Given the increasing number of genes becoming associated with EMP in the literature, this recommendation will be helpful in avoiding erroneous conclusions based on the expression of a small number of genes. In certain settings, however, assessing cellular properties is not particularly reasonable. Many studies depend on retrospective analysis of samples (e.g., data generated from tumor samples), and it may not be feasible to faithfully recapitulate EMP ex vivo. Comprehensive interrogation into the molecular properties of EMP and, importantly, the diversity of features across contexts is critical to enabling reliable interpretation of data from samples that are not amenable to phenotypic assessment. Also, embracing the diversity of EMP gene expression programs will allow for an updated conceptual model that may help explain its involvement in tumor progression and how it may be addressed therapeutically.
The advent of single-cell transcriptomics (scRNA-seq) has enabled the identification of coordinated gene expression patterns within individual cells. Developments to increase the throughput of these assays have allowed for the parallel measurement of gene expression in thousands of single cells, sampling the phenotypic diversity among a population within a tissue (18, 19). Under the assumption that intratumoral EMP is reasonably prevalent, single-cell genomics holds the promise of revealing its intrinsic molecular characteristics. Supporting this assumption, studies applying these methods have independently identified heterogeneous expression of EMT-associated genes in a variety of cancers, including squamous cell carcinoma (SCC) (3, 20, 21), pancreatic ductal adenocarcinoma (PDAC) (22), colorectal (23), and lung (24). Detailed exploration of these programs has been limited, however, and little has been done to compare features of these programs between individual tumors and cancers. Kinker et al. (25) recently assessed sources of heterogeneity in the expression profiles of monolayer-cultured cancer cell lines and identified three recurrent programs consistent with EMP, but the similarity of these programs to those that emerge in solid tumors is unclear.
Here, we leveraged scRNA-seq data from 266 tumor samples spanning eight cancer types to identify coordinated expression programs consistent with intratumoral EMP. While the overall composition of these programs was highly variable, we derived a set of well-conserved genes from these programs that can serve as a general EMP signature. We used this signature to query the pan-cancer data from The Cancer Genome Atlas (TCGA) and found that EMP was associated with reduced progression-free intervals (PFIs) and changes in immune cell proportions within the tumor microenvironment (TME). Inferences of regulatory mechanisms contributing to these EMP programs suggest that the diversity of these programs can arise from common underlying regulatory mechanisms, including ubiquitous activation of TGF-β1/nuclear factor κB (NFκB)/tumor necrosis factor (TNF) signaling but that some programs have notably high mitogen-activated protein kinase (MAPK) or signal transducers and activators of transcription (STAT)/hypoxia signaling activity. We integrated transcriptomic data from kinase inhibitor screens to demonstrate that these common regulatory mechanisms present promising targets for therapeutic restriction of EMP.
RESULTS
A pan-cancer census of EMT-associated gene expression
We first collected droplet-based scRNA-seq data from 17 studies of eight different cancer types, including colorectal (26–28), gastric (29), lung (24, 28, 30–32), nasopharyngeal (33), SCC (20), ovarian (28, 34), PDAC (35), and breast (28, 36, 37). After removing samples with fewer than 100 malignant cells, the data comprise expression profiles of 223,501 cancer cells from 266 tumor samples (Fig. 1A and figs. S1 and S2).
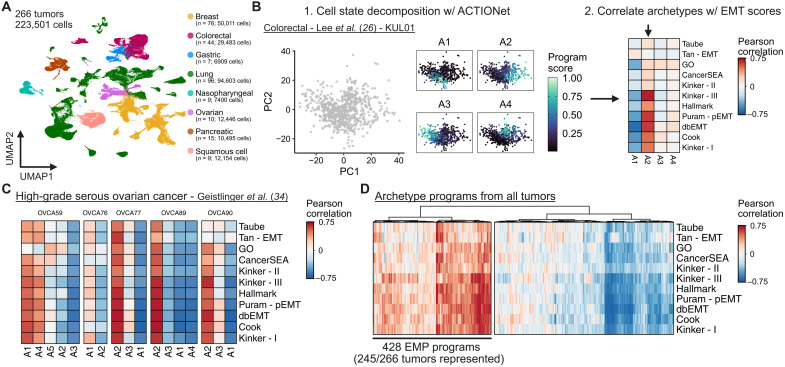
Fig. 1. Using archetypal analysis to learn EMP-associated gene expression programs.
(A) UMAP embedding of malignant cell scRNA-seq data from 266 tumors and 17 different studies. (B) Schematic representation of the analysis strategy to identify sample-specific EMP programs using archetypal analysis and correlating archetype scores with EMT gene set scores. (C) Pearson correlation coefficients of archetype scores with EMT gene set scores of individual cells from five high-grade serous ovarian tumors. (D) Hierarchically clustered heatmap of Pearson correlation coefficients of all archetype scores with EMT gene set scores from the 266 tumors analyzed.
To assess intratumoral heterogeneity of EMT-associated gene expression, we used rank-based scoring of each cell for its relative expression of genes contained in 11 different EMT gene sets, including several curated sets [the “Epithelial-mesenchymal transition” Gene Ontology (GO) Term, MSigDB Hallmark gene set (38), CancerSEA (39), dbEMT1.0 (40), and Tan cancer-specific EMT gene set (41)] and others derived from individual studies [Taube (9), Puram (3), Cook (8), and Kinker I, II, III (25)]. The composition of these gene sets is highly variable, with more than two-thirds of the genes being present in only a single gene set (fig. S3A). While it is possible that these diverse gene sets could represent different subsets of a common underlying biological process, we correlated the gene set scores across all cells and found that this is not uniformly true. Several gene sets were poorly correlated with the others, suggesting that they may only reflect EMT programs under specific contexts. Other gene sets were well correlated despite differences in their composition, suggesting that they likely reflect a common expression program (fig. S3B).
Gene set scores can provide biological insight into gene expression patterns, but they can be influenced by uninteresting features of the data. Specifically, they can be inflated by high expression of a small proportion of the set’s genes that are not necessarily determinants of the queried biological process. Variation in scores across a population of cells may also reflect random fluctuations of the set’s genes and not necessarily a coordinated activation of the process. Matrix factorization approaches have been applied to scRNA-seq data to learn coordinated expression programs heterogeneously expressed across a population. By learning these programs from the data itself, reliance on previously defined gene sets is restricted to only the interpretation of the programs.
We next sought to explore heterogeneously expressed programs from malignant populations from each of the 266 tumors independently. To identify these gene expression programs contributing to intratumoral heterogeneity, we performed multiresolution archetypal analysis on each tumor sample using the ACTIONet algorithm (42). This method identifies a set of extremal phenotypes (archetypes) from the distribution of gene expression profiles and represents each cell as a mixture of these archetype programs (Fig. 1B). This revealed multiple programs in each sample, including expected sources of variation, such as cell cycle activity. To identify those associated with EMP, we correlated the cellular activity of each archetype program learned by ACTIONet with the previous EMT gene set scores. We defined EMP programs by hierarchically clustering the archetype programs to identify those well correlated with EMT gene set scores (Fig. 1, C and D, and fig. S4A). Of the 266 tumors, 245 had identifiable EMP programs, suggesting that intratumoral EMP is a ubiquitous feature of solid tumors and readily captured in scRNA-seq experiments.
Defining a conserved signature associated with EMP
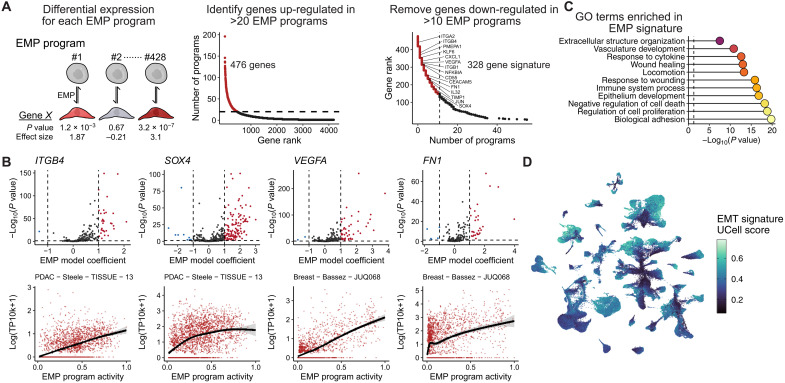
To define the specific genes contributing to EMP in each tumor, we identified genes whose expression is associated with each archetype program learned from ACTIONet. We have previously shown that transcriptional responses of experimentally induced EMTs are highly context specific, but it was unclear whether the same diversity existed in vivo (8). Of the 4187 genes up-regulated in at least one EMP program, the vast majority were associated with a small number of samples. Assessing the frequency at which each gene was differentially expressed, we assembled a general EMP signature from genes differentially expressed in 20 or more EMP programs (Fig. 2A). We further refined this signature by removing genes that were also down-regulated in more than 10 EMP programs, resulting in an EMP signature of 328 genes. We assessed these genes to determine whether the association of their expression with EMP was cancer type specific and found that the majority (251 of 329) showed no specific bias (analysis of variance, P > 0.05). Many of the 78 with cancer-specific patterns differed only in the magnitude of their association (fig. S4B).
Fig. 2. Defining a conserved EMP gene signature.
(A) Schematic showing the strategy for defining a signature of genes most frequently associated with the 428 identified EMP signatures. (B) EMP-associated GO terms significantly enriched in the 289 conserved EMP genes. P values were calculated using Fisher’s exact tests and adjusted using the Benjamini-Hochberg method. (C) Examples of EMP associated genes. Top plots show the distribution of effect sizes (model coefficients) for each gene with all EMP programs. Dashed line represents the mean value. The horizontal dashed line represents a Benjamini-Hochberg adjusted P value of 0.05. (D) UMAP embedding of malignant cells from the 266 tumors analyzed, colored by a gene set score for the signature of 289 EMP-associated genes.
Genes down-regulated upon the activation of EMP programs were even less consistent, with many down-regulated in only a small number of programs (fig. S4C). There was no overwhelming evidence of a suppressed epithelial phenotype, which may further support the growing evidence of hybrid E/M phenotypes being highly prevalent in cancer (3, 43–45). Several repressed genes were consistent, however, with activation of a mesenchymal phenotype, including suppression of STMN1 (46) and PGC (47), along with reduced expression of the proliferation markers TOP2A and MKI67 (48).
Of the 328 genes positively associated with EMP, no individual gene was a perfect indicator of its activity, and in many EMP programs, they show little-to-no activity (Fig. 2B). As a collective, however, the signature represents a set of the most consistent markers of intratumoral EMP. They also enrich for GO terms consistent with a mesenchymal phenotype, including cell motility, regulation of cell adhesion, and response to wounding (Fig. 2C). Many of the canonical EMT genes are not included in this signature, including CDH2, VIM, SNAI1, SNAI2, and ZEB1, although these genes did have variable associations with EMP programs (fig. S5). The signature did, however, include 38 of the 200 EMT Hallmark genes, and many that have previously been implicated in the EMT, including various transcription factors (SOX4, KLF4/5/6/10, and JUND), integrins (ITGB1/4 and ITGA2), secreted factors (VEGFA, IL32, CXCL1, and CXCL8), ECM components (FN1 and COL17A1), and membrane proteins (CD55 and CD59), and more (Fig. 2B and table S1).
The stability and distribution of phenotypes along an EM continuum has gained recent attention, with the relevance of hybrid phenotypes being contrasted to fully epithelial or mesenchymal cells (49). Using gene set scores of the conserved signature as a relative measure of the cells’ EMP, we found that average scores were heterogeneous between samples (Fig. 2D and fig. S6). Intratumoral heterogeneity of scores was higher than expected by chance in all samples (P < 0.01, permutation test with random gene sets) (fig. S6). We also note that most tumors do not have clear multimodal distributions that would be consistent with a model where various states along the phenotypic continuum have elevated stability. Rather, cells span the continuum, forming a distribution with most cells occupying intermediate states and tails spreading to more extreme phenotypes.
A refined, malignant cell–specific EMP signature is associated with poor patient prognosis
Because of their inherent similarities, the ability to distinguish fibroblast and EMP-specific expression patterns has been a challenge. Many EMT gene sets contain genes highly expressed in fibroblast populations, and as a result, “mesenchymal” features of tumors defined from bulk RNA-seq data have been found to often be associated with fibroblast content of the tumor rather than cancer cell plasticity (3, 50, 51). The choice of specific markers used to assess EMP in studies has also led to controversy (1). This confusion has made it challenging to draw conclusions about the involvement of EMP in tumor progression and clinical outcomes. Recently, Tyler and Tirosh (52) identified malignant cell–specific EMP signatures by using scRNA-seq data to assess the specificity of EMT gene sets and genes that correlate with canonical EMT markers. As we have established a conserved EMP signature, we similarly used expression profiles from nonmalignant cells to refine a cancer cell–specific signature.
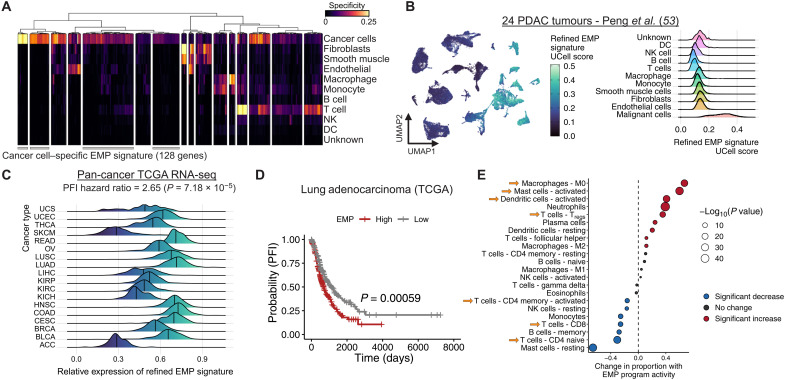
For each tumor sample, we calculated a cell type specificity score for each of the 328 conserved EMP genes and averaged these scores across tumors to get an overview of how specific the markers were to cancer cells (Fig. 3A). Of the 328 genes, 128 were highly specific to cancer cells, whereas the remaining 200 were also expressed in fibroblasts, macrophages, and/or T cells. A signature of cancer cell–specific EMP genes could be valuable for generating EMP activity scores in scRNA-seq data, so we established a refined signature comprising the 128 genes highly specific to cancer cells (table S2). To confirm the specificity of this signature in data that were not used to generate the signature, we scored cells from 24 pancreatic tumors (53) and found heterogeneous activity among cancer cell populations, with minimal activity in nonmalignant cell types (Fig. 3B).
Fig. 3. EMP is associated with worse progression-free survival and an immunosuppressive TME.
(A) Clustered heatmap of cell type specificity scores for each of the 328 EMP-associated genes. Clusters of genes with high cancer cell specificity were defined as a cancer cell–specific EMP signature. (B) UMAP embedding (left) and density plot (right) showing the distribution of gene set UCell scores for the cancer cell–specific EMP signature in all cell types from 24 pancreatic tumors. (C) PFI hazard ratio and relative expression (average Z score) of the cancer-specific EMP signature in the TCGA’s pan-cancer bulk RNA-seq cohort. The hazard ratio and P value are derived from a Cox proportional hazards model, including variables for the refined EMP signature’s expression, cancer type, stage, age, and tumor purity. (D) Changes in immune cell proportion estimates as a function of EMP signature scores from the TCGA’s pan-cancer cohort. Cell type proportions were modeled using a linear model including the same variables as in (C). The change in proportion reflects the model coefficient for the EMP expression variable. DC, dendritic cell; Tregs, regulatory T cells.
Given this relatively high specificity, the signature could be used as a measure of mesenchymal properties in bulk RNA-seq data without being confounded by expression from stromal cell types. We used the pan-cancer RNA-seq data from TCGA (54) to calculate a relative EMP score for all tumors. Modeling patients’ PFI as a function of this signature activity, tumor type, stage, age at diagnosis, and tumor purity, we found that EMP activity was associated with a reduced PFI (Cox hazard ratio: 2.65; P = 7.18 × 10−5) (Fig. 3, C and D). We then modeled estimates of immune cell proportions for all TCGA samples (55) as a function of this EMP score while controlling for the same covariates as above and found an association with not only higher proportions of inflammatory cell types (activated mast and dendritic cells) but also more immunosuppressive regulatory T cells and fewer effector cell types, including CD8 T cells and naive CD4 T cells (Fig. 3E).
Reconstructing EMP regulatory networks
While unifying molecular signatures are appealing, appreciating the diversity of EMP programs is critical as it likely contributes to functional nuances of the phenotype. These programs may also have varying regulatory dependencies that would warrant different therapeutic approaches. Variation in cell state emerges from complex differences in the cells’ microenvironment that ultimately converge on signal transduction pathways and transcription factor networks. We next sought to use computational approaches to infer how regulation at these levels contribute to the patient-specific EMP programs we learned from scRNA-seq data.
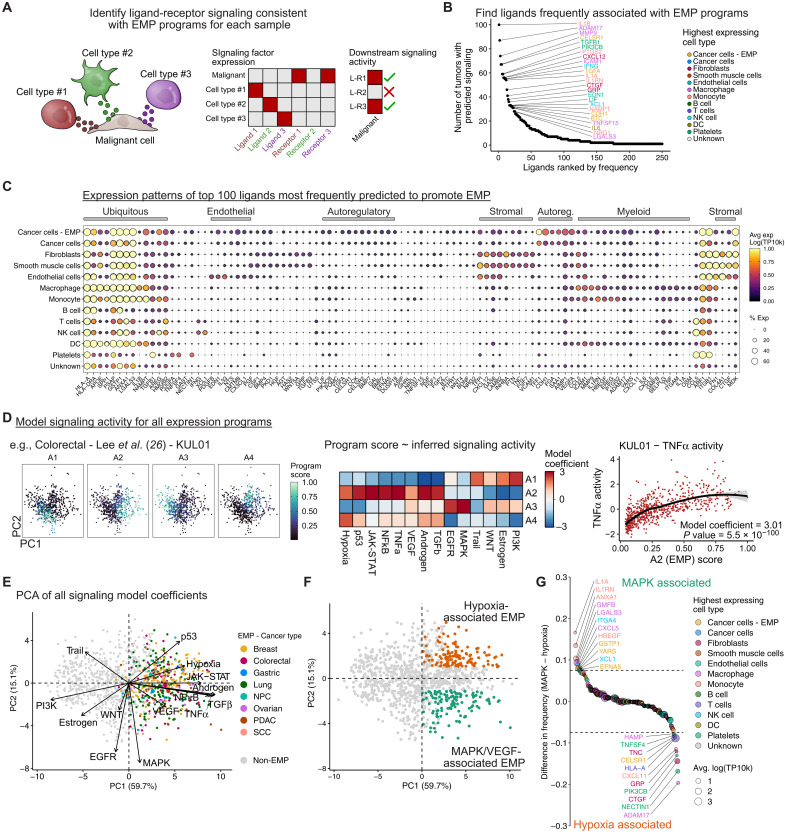
Since the datasets we have used included matched gene expression profiles of stromal cells for all tumors, we used NicheNet (56) to infer cell-cell communication to identify factors from the TME that could contribute to each tumor’s specific EMP program (Fig. 4A). Several cytokines were associated with EMP in many tumors, including TGF-β1, interleukin-6, posttranscriptional gene silencing 2 (PTGS2), interferon-γ (IFN-γ), and others (Fig. 4B). Consistent with previous literature, macrophages and fibroblasts frequently expressed many factors contributing to EMP (Fig. 4C) (3, 4). The majority of stromal cell types contribute implicated factors, such as PDGFB (Platelet-derived growth factor) primarily from endothelial cells; INHBA, FN1, and FGF7 from fibroblasts; IFN-γ and XCL1 from natural killer (NK) cells; and TNF, matrix metallopeptidase 9, and PTGS2 from myeloid cells. Malignant cells also express various ligands predicted to promote EMP, such as VEGFA and bone morphogenetic protein 7, suggesting that they may establish self-regulatory signaling within the TME (Fig. 4C).
Fig. 4. Signaling networks regulating EMP in cancer.
(A) Schematic showing the identification of ligands that may contribute to EMP. Expression patterns of ligands and their cognate receptors are assessed across all cell types. Putative regulatory ligands are identified if malignant cells express a matching receptor and if the sample’s EMP program is consistent with downstream signaling related to that ligand. (B) Potential ligands from the TME regulating EMP. Ligands were ranked by the number of tumors in which they were predicted to promote EMP programs. (C) Expression of the top 100 ligands in cell types of the TME. The gene order is based on hierarchical clustering and manually annotated. Expression values (log-transformed and scaled UMI counts) were averaged across all 266 tumors. (D) Schematic showing the inference of signaling pathway activity associated with EMP programs. Signaling activity scores are inferred for each individual cell using PROGENy, and each archetype score for a given tumor is then modeled as a function of signaling activity using a linear model. (E) Principal components analysis (PCA) of the signaling model coefficients for all archetype programs. Each point represents an archetype program with EMP programs colored by the cancer type they are associated with. Loading vectors for each of the 14 signaling pathways are shown. (F) Sample as (E) but colored to define hypoxia/p53 (orange) and MAPK/VEGF-associated (green) EMP programs. These were defined as programs having PC1 > 0 and either PC2 > 1 (hypoxia) or PC2 ≤ 1 (MAPK/VEGF). (G) Ligands with preferential associations with MAPK/VEGF- or hypoxia/p53-associated EMP. Values represent the difference in the proportion of programs in which each ligand was implicated.
To assess intracellular signaling activity associated with EMP, we used PROGENy (57)—a model of consensus signatures of signaling pathways—to calculate a relative activity score for 14 different signaling pathways in each cell. We then modeled EMP program activity as a function of these signaling activity scores (Fig. 4D). EMP programs were consistently associated with elevated TGF-β, NFκB, and TNFα signaling, but some had distinctly high levels of either MAPK and VEGF signaling or hypoxia and p53 signaling (Fig. 4, E and F). This suggests that hypoxia-associated EMP may have distinct features from plasticity coordinated by MAPK/extracellular signal–regulated kinase (ERK) signaling. This is consistent with findings that treatment of breast cancer cells with epidermal growth factor (EGF) induces an EMT with distinct characteristics from those following a hypoxia-induced EMT (58). These programs likely emerge in response to different interactions with the TME, as we found several ligands preferentially associated with each, such the ERK-activating ITGA4 and HBEGF (Herapin-binding EGF-like growth factor) with MAPK/VEGF programs (Fig. 4G).
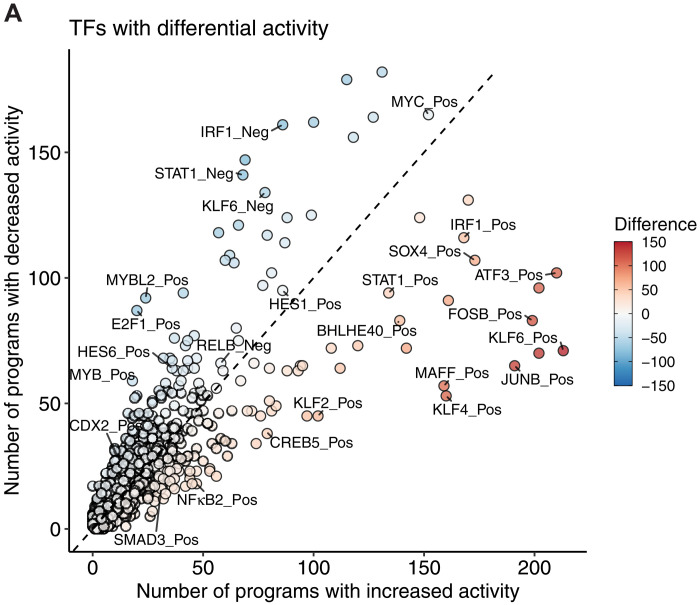
Last, as this regulatory signaling converges on transcription factor networks to modulate gene expression, we used SCENIC (59) to construct gene regulatory networks for each tumor sample and then assessed how transcription factor activity changes in each EMP program. The most frequently associated transcription factors include many noncanonical factors, such as AP-1 (JUNB and FOSB), SOX4, KLF2/4/6, and STAT1 (Fig. 5A). Fewer transcription factors had consistent deactivation upon activation of EMP programs; however, several factors were repressed fairly commonly, including MYBL2, E2F1, and HES6 (Fig. 5A).
Fig. 5. Transcription factor regulons associated with EMP.
(A) Transcription factors predicted to have differential activity in EMP programs. Each point represents a group of coexpressed genes coregulated by a given transcription factor (a regulon).
Pharmacologic restriction of EMP
EMP has been implicated in both chemoresistance and immune cell evasion (4). Given this, we predict that nonlethal restriction of EMP could be a promising therapeutic approach to sensitize tumors to orthogonal treatments and elicit immune cell killing. Diversity of EMP programs could introduce challenges for effectively preventing these cell dynamics, but the dependence of EMP on factors from the cells’ microenvironment suggests that the diversity likely arises from combinatorial effects from the relatively limited number of signal transduction pathways. Therefore, we suspect that diverse EMP programs may be susceptible to common pathway perturbations, and rational treatments could be devised by inferring signaling activity associated with EMP in a given tumor.
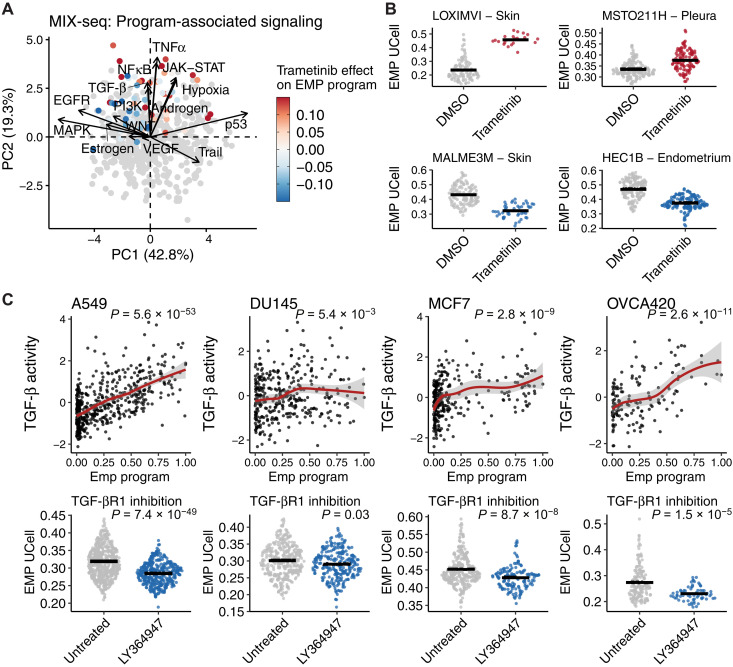
To begin to test this prediction, we first explored the MIX-Seq dataset comprising scRNA-seq profiles of more than 100 cancer cell lines treated with various drugs, including the MAPK kinase (MEK) inhibitor trametinib (60). We used ACTIONet to define cell line–specific EMP programs from untreated expression profiles, identifying high-confidence EMP programs in 46 of the 99 lines we assessed (lines with >100 cells; fig. S7). Many others had programs that correlated well with individual EMT gene sets and may represent EMP, but out of caution, we did not annotate them as such. We then inferred changes in signaling pathway activity associated with all archetype programs and found that, like in the tumor samples, TGF-β/NFκB/TNFα activity was consistently higher in EMP programs, but programs could be distinguished by high MAPK or hypoxia/p53 signaling (Fig. 6A).
Fig. 6. Regulatory predictions can infer strategies to therapeutically restrict EMP.
(A) PCA of signaling model coefficients for all archetype programs of the 99 cell lines in the MIX-seq dataset. Gray dots represent programs not associated with EMP. EMP programs are colored by the effect trametinib has on its activity. (B) Examples of cell lines whose specific EMP program is enhanced (top) or limited (bottom) by trametinib. DMSO, dimethyl sulfoxide. (C) Effect of LY364947-mediated TGF-βR1 inhibition on sample-specific EMP programs in A549, DU145, MCF7, and OVCA420 cell lines. P values were calculated from linear models for each condition and were all corrected with the Benjamini-Hochberg method.
To determine whether MEK inhibition preferentially limits MAPK-associated EMP, we used the MIX-Seq dataset to assess the effects of trametinib on all EMP programs. As expected, trametinib impaired EMP in the programs inferred to be associated with high MAPK signaling (Fig. 6, A and B). Kinase inhibition can promote a mesenchymal phenotype in breast cancer cells under hypoxic conditions (58), and we observe that this is generalizable across the diverse cancer lines included in the MIX-seq dataset, with trametinib enhancing EMP activity in programs associated with hypoxia/p53 signaling (Fig. 6, A and B). Given the consistent association of EMP with TGF-β1 and NFκB signaling, we would also predict that inhibition of these pathways could restrict EMP. While TGF-β1 and NFκB inhibitors were not included in the MIX-Seq dataset, we have previously published scRNA-seq data of four cancer cell lines (A549, DU145, MCF7, and OVCA420) treated with the TGF-βR1 inhibitor LY364947. Performing the same analysis on these data, we found that inhibition of TGF-βR1 in cells cultured in control conditions led to repression of most EMP programs, proportional to the inferred level of TGF-β1 activity associated with each line’s intrinsic EMP (Fig. 6C and fig. S7B). Together, this supports that EMP programs with highly diverse molecular features may still have common dependencies, and effective strategies for restricting EMP can be found by inferring these regulatory mechanisms.
DISCUSSION
EMP has long been appreciated as a prominent source of intratumoral heterogeneity that ultimately promotes tumor progression and hinders effective treatment. While select molecular patterns associated with EMP have been known for decades, variability in their involvement across contexts has raised confusion, and as a result, high-level conceptual models to understand the molecular basis of this plasticity have been lacking. Here, we used scRNA-seq data of 266 tumor samples to identify gene expression programs consistent with EMP, compared features of these programs between tumor samples, inferred regulatory networks driving them, and explored therapeutic options to limit their activity. We have found that, consistent with experimental studies, intratumoral EMP–associated expression is highly variable. Unique genetic aberrations of each tumor likely contribute to this diversity but are challenging to assess. Experimental strategies to promote an EMT in both normal and cancer cell lines with distinct inducers can elicit unique EMT responses, suggesting that the diversity cannot solely be due to genetic variation (8, 61). It is likely that this diversity is the product of multiple regulatory variables, including genetic variation, the epigenetic profile moulded by the cell’s developmental history, spatial proximity to stromal cell types, and physical properties of the TME.
EMP programs identified in each sample were consistently associated with higher expression of genes associated with mesenchymal traits (e.g., motility and cellular rearrangement), yet there was no consistent reduction of epithelial genes upon activation of these programs. This supports studies demonstrating the presence of hybrid phenotypes in cancer with distinct functional traits (43–45). It is possible that reduction of epithelial traits is achieved through internalization of epithelial membrane proteins rather than transcription repression, as has been seen with hybrid E/M phenotypes in pancreatic cancer (49). However, the generalizability of posttranslational regulation to the broader scope of phenotypically relevant epithelial proteins is still unclear. This could, however, reconcile the presence of various epithelial membrane proteins in EMP programs, perhaps acting as a feedback mechanism to counter this regulation.
We acknowledge that our ability to define EMP programs may be limited by the sparsity and sensitivity constraints of scRNA-seq assays. It is very possible that some phenotypically relevant expression patterns were not reliably detected in our analysis for technical reasons. For example, we found that some of the canonical EMT transcription factors (e.g., SNAI1, SNAI2, and ZEB1) were poorly detected, and it is unclear whether this is a feature of these samples or a technical limitation. However, other approaches have supported that the involvement of canonical transcription factors is inconsistent (11), and in some settings, their expression could be attributed to the presence of cancer-associated fibroblasts rather than EMP (52). Moreover, given the relevance of posttranscriptional regulation of EMP (49), it is feasible that some of the most conserved features of EMP reside beyond cells’ expression profiles. Advances in morphological and protein-based assays (e.g., high-content imaging, imaging mass cytometry, etc.) will allow for the discovery of additional features of EMP.
A general observation worth noting is that the E/M phenotypes observed within a single sample seem to span a continuous, often monomodal distribution. There has been much speculation about the stability of phenotypes along an E/M continuum and the presence of discrete stable states (62). Modeling frameworks suggest their existence (63, 64), but it is not clear that they are compatible with the distributions of phenotypes observed in these samples: The dominant E/M state between tumors is variable, the extent of phenotypic variation within individual samples is highly variable, and very few samples show the multimodality that should coincide with varying stability along a phenotypic continuum. Although, considering these findings, it is intriguing to question whether it is more relevant to a tumor’s biology that its malignant cells are more mesenchymal on average or whether they span a wider range of E/M phenotypes.
We identified a signature of 328 genes that were commonly associated with EMP programs and a refined signature of 128 genes with high specificity to malignant cells, which could be useful for quantifying EMP in gene expression data, as we did here with TCGA data. In no way do we argue that this signature represents the most biologically relevant components of EMP gene expression programs. Enrichment of GO terms related to mesenchymal functions suggests that they likely contribute to the phenotype, but we found a total of 4187 genes associated with EMP across the various samples. Even under highly controlled experimental conditions, EMT responses are vastly context specific (8, 10, 61). The phenotypic changes associated with EMP are likely an emergent property of these many changes, and variation in expression programs may provide phenotypic nuances that we are unaware of. Hence, rather than try to reduce EMP to some consistent phenomenon, we think it is important to recognize context specificity and begin building conceptual models and experimental designs to understand its importance.
The ability to therapeutically restrict plasticity in tumors could greatly improve the efficacy of existing treatment options. One could imagine several strategies for accomplishing this: interfering with the cues that initiate plasticity, therapeutically impairing transcriptional regulatory mechanisms (e.g., inhibiting histone modifying proteins), or targeting effector proteins associated with a given cell state (e.g., neutralizing immunosuppressive cytokines released from mesenchymal cells). The latter may be challenging due to the diversity of EMP phenotypes but is perhaps the most direct strategy for preventing undesirable features of mesenchymal cells. In this study, we have begun to explore strategies for blocking stimulatory pathways, restricting diverse programs with common signaling cues. We computationally inferred that EMP programs were consistently associated with TGF-β and NFκB signaling, whereas some had notably high levels of either MAPK or hypoxia/p53 signaling. Leveraging transcriptomic data from various drug screens, we confirmed that targeting these active pathways led to reduced activity of sample-specific EMP programs despite the diversity of the programs themselves. While this diversity introduces challenges for effective therapeutics, this observation suggests that these diverse programs may have common dependencies that can be inferred from their molecular features and exploited therapeutically.
In summary, we have used scRNA-seq data from 266 tumors spanning eight cancer types to identify molecular features associated with EMP. We find that EMP is a ubiquitous source of intratumoral heterogeneity but—consistent with previous findings—is highly context specific. We identify a cancer cell–specific signature of the most common genes positively associated with EMP and demonstrate its utility as a general-use gene set, using the TCGA pan-cancer RNA-seq data to associate EMP with worse PFIs and a more immunosuppressive TME. We use computational approaches to infer regulatory features of EMP across hundreds of samples and highlight that diversity may emerge from common regulatory mechanisms that can be inferred and used to rationalize therapeutic strategies.
METHODS
Processing and annotating scRNA-seq data
A summary of scRNA-seq datasets used in this study are described in table S3. To avoid comparing data collected from vastly different technologies, only droplet-based scRNA-seq data were used in the analysis. Raw UMI (Unique Molecular Identifier) count matrices and cell metadata were collected from the various sources (table S3). Several datasets included matched normal tissue samples. In these cases, we removed the normal samples and only proceeded with the tumor samples.
Initial quality control was performed independently for each dataset using the R package Seurat v4.0 (65). Cells with fewer than 200 detected genes were removed, and only genes detected in more than three cells were included in the analysis. Data points representing potential cell doublets within individual samples were removed using scDblFinder v1.6.0 using default parameters (https://github.com/plger/scDblFinder). Cells with a high percentage (>20%) of mitochondrial transcripts were also removed. However, for several datasets, the original authors prefiltered the data at a lower threshold, and for the breast cancer samples from Wu et al. (36), the original authors had filtered cells with >30% due to the higher average values, but data quality was not apparently affected. In these cases, the original filtering thresholds were used. Table S1 summarizes these thresholds.
UMI counts were normalized with standard library size scaling and log transformation. The data were then processed with principal components analysis (PCA), a nearest neighbor graph was generated on the first 30 PCs, and the data were clustered at a fairly low resolution (FindClusters, resolution = 0.2) using the Louvain algorithm. For visualizations presented in the figures, UMAP (Uniform manifold approximation and projection) embeddings were generated from the first 30 PCs.
The identity of Individual cells for each sample was annotated using SingleR v1.6.1 (66) and the Human Primary Cell Atlas from the R package celldex v1.2.0. Only expected cell types from the TME were included as references in the annotation. This includes fibroblasts, monocytes, endothelial cells, T cells, NK cells, B cells, macrophages, smooth muscle cells, dendritic cells, platelets, and epithelial cells. Following automated annotation, malignant epithelial cells were identified by inferring copy number variations across the annotated cell types with inferCNV. Epithelial populations with copy number aberrations were considered malignant cells in downstream analysis. While mesenchymal markers may be expressed in fibroblasts, we found that global gene expression profiles were sufficiently different such that cells annotated as fibroblasts formed distinct clusters, well separated from malignant cells.
To determine the quality of the annotation, we assessed the purity of annotations within clusters of cells with similar gene expression profiles. We found strong concordance between clustering patterns and annotations (fig. S1). Similarly, we find that annotations match expression patterns of canonical cell type markers. In some cases, clusters were annotated as two closely related cell types (e.g., fibroblasts and smooth muscle cells). We found that these mixed annotations could be fairly uniformly distributed within a cluster and were not distributed among groups of cells that could be separated by higher-resolution clustering. Although we suspect that these cells are truly a single cell type/state, we used the automated annotations for downstream analysis to ensure consistency. This issue did not occur for epithelial populations, so it does not affect the majority of the analysis.
Identifying latent EMP expression programs with archetypal analysis
To ensure that we had a sufficient number of cells to identify heterogeneous expression patterns within samples, we removed samples with <100 annotated malignant cells from the analysis. Multiresolution archetypal analysis (42) was performed independently of the cancer cells from all 266 tumors using ACTIONet v2.1.7 (42) to decompose cells’ gene expression profiles into a small set of latent expression programs that are heterogeneously expressed throughout the population. Reduced kernel matrices were first computed with the reduce.ace() function implemented in the R package ACTIONet with the parameter reduced_dim=20. Given that each population represented a single cell type, ACTIONet was then run with the k_max=10 option to reduce the maximum depth of decompositions and with min_cells_per_arch=5 to prevent archetypes driven by a small number of cells.
Resulting archetype footprints (program activities) were correlated with gene set scores for 11 EMT gene sets. Clustering of the Pearson correlation coefficients allowed us to define EMP-associated programs as clusters with high correlation values. We then used linear models to identify genes whose expression is associated with all archetype programs. To only recover reliable changes, differential expression was limited to the top 2000 variable genes with a minimum detection frequency of 5% in each sample’s malignant cell population. Normalized (scaled and log-transformed) counts were used for differential expression, modeling each gene’s expression as a function of program activity, and program-associated genes were defined as those with a Benjamini-Hochberg–corrected P value of <0.05 and a model coefficient (effect size) of >1.
Gene set scoring
Gene set scoring was performed using the R package UCell v1.0.0 (67). UCell scores are based on the Mann-Whitney U statistic, which evaluates the rank of each query genes’ expression level in individual cells. Because it is rank-based, the scores are independent of the cellular composition of the dataset and are interpretable as the relative ranking of the gene set within the cell’s transcriptome. For gene set scoring of the EMP signature, we also performed 100 permutations of random gene sets of an equivalent size to define the distribution of scores expected chance.
Cell type specificity scoring
Specificity scores were calculated for EMP signature genes using the R package genesorteR v0.4.3 (68). Specificity scores represent the exclusivity of a gene to a given cluster and extent to which it is expressed (proportion of cells with detection greater than population median levels). Values range from 0 to 1, with 1 corresponding to genes that are exclusive to a given cluster and detected in all cells.
Pan-cancer TCGA analysis
Bulk RNA-seq profiles and clinical outcomes were accessed from https://gdc.cancer.gov/node/905, tumor purity estimates were acquired from Aran et al. (69), and immune cell proportion estimates were accessed from Thorsson et al. (70).
RNA-seq count data for each sample were normalized to counts per million and log-transformed. EMP signature scores were calculated for each sample by computing the average gene-level Z score of the genes from the 128 cancer cell–specific genes. The association of EMP signature activity with PFI was assessed using a Cox proportional hazards model, including tumor type, purity, stage, and age along with continuous EMP scores as covariates in the model. Changes in immune cell proportions were assessed by independently using linear models to model each immune cell type’s predicted proportion as a function EMP signature score, including tumor type as a covariate to account for differences simply attributable to cancer type. We used the Benjamini-Hochberg method to correct P values to account for multiple comparisons.
Inferring EMP-associated signaling activity
The R package PROGENy v1.11.2 (57, 71, 72) was used to infer the activity of 14 signaling pathways in each individual cell. For each tumor sample, normalized expression values were used to calculate pathway activity with the progeny() function with the top=500 parameter set to use the top 500 genes of each pathway in PROGENy’s model for the activity calculation. We used simple linear models to identify changes in signaling pathway activity with archetype program activity.
Cell communication inference
The R package nichenetr (NicheNet; v1.0.0) (56) was used to infer cell communication within the TME that could contribute to a sample’s specific EMP program. Given the diversity of EMP programs, this was performed on all 266 tumors independently. For each sample, cancer cells annotated by ACTIONet as the EMP-associated archetype were defined as the “receiver” population, and all cell types were considered as potential “sender” cells. The “gene set of interest” was defined as that sample’s specific EMP program, and expressed genes were defined as those with a detection rate of at least 5%. For each sample, we considered top ligands as the top 10 ligands inferred to promote expression of the EMP-associated genes.
Assessing effects of small-molecule inhibitors on EMP
ScRNA-seq data from epithelial cancer cell lines in control and drug-treated culture conditions were acquired from McFarland et al. (60) and Cook and Vanderhyden (8). Initial data processing was performed identically to tumor samples, and cell lines with fewer than 100 measured cells were removed. EMP programs were defined for each cell line using ACTIONet with only data from control cultured cells. Genes associated with each program were defined and used as a sample-specific EMP gene set for scoring both control and drug-treated cells. We then used a linear model to compare the effects of MEK and TGF-βR inhibition on EMP in each line.
Acknowledgments
We extend gratitude to the many developers and maintainers of open-source software that keep this field running. We thank P. Robineau-Charette for helpful discussions and feedback.
Funding: This work was supported by an NSERC Discovery Grant (RGPIN 2018-0653.8). D.P.C. was supported by a CIHR Frederick Banting and Charles Best Canada Graduate Scholarships Doctoral Award.
Author contributions: D.P.C. and B.C.V. conceived the study and wrote the manuscript. D.P.C. performed all analysis.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Sources for all data analyzed are available in table S3. Code required to reproduce all findings is publicly available at https://github.com/dpcook/emp_programs or https://zenodo.org/record/5636711#.YYMwemDMI2w. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Legends for tables S1 and S2
Table S3
Figs. S1 to S7
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.Yang J., Antin P., Berx G., Blanpain C., Brabletz T., Bronner M., Campbell K., Cano A., Casanova J., Christofori G., Dedhar S., Derynck R., Ford H. L., Fuxe J., Garcia de Herreros A., Goodall G. J., Hadjantonakis A.-K., Huang R. J. Y., Kalcheim C., Kalluri R., Kang Y., Khew-Goodall Y., Levine H., Liu J., Longmore G. D., Mani S. A., Massagué J., Mayor R., McClay D., Mostov K. E., Newgreen D. F., Nieto M. A., Puisieux A., Runyan R., Savagner P., Stanger B., Stemmler M. P., Takahashi Y., Takeichi M., Theveneau E., Thiery J. P., Thompson E. W., Weinberg R. A., Williams E. D., Xing J., Zhou B. P., Sheng G.; EMT International Association (TEMTIA) , Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabbert H., Wagner R., Moll R., Gerharz C. D., Tumor dedifferentiation: An important step in tumor invasion. Clin. Exp. Metastasis 3, 257–279 (1985). [DOI] [PubMed] [Google Scholar]
- 3.Puram S. V., Tirosh I., Parikh A. S., Patel A. P., Yizhak K., Gillespie S., Rodman C., Luo C. L., Mroz E. A., Emerick K. S., Deschler D. G., Varvares M. A., Mylvaganam R., Rozenblatt-Rosen O., Rocco J. W., Faquin W. C., Lin D. T., Regev A., Bernstein B. E., Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dongre A., Weinberg R. A., New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Horn L. A., Fousek K., Palena C., Tumor plasticity and resistance to immunotherapy. Trends Cancer Res. 6, 432–441 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramesh V., Brabletz T., Ceppi P., Targeting EMT in cancer with repurposed metabolic inhibitors. Trends Cancer 6, 942–950 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Bhatia S., Wang P., Toh A., Thompson E. W., New insights into the role of phenotypic plasticity and EMT in driving cancer progression. Front. Mol. Biosci. 7, 71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook D. P., Vanderhyden B. C., Context specificity of the EMT transcriptional response. Nat. Commun. 11, 2142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taube J. H., Herschkowitz J. I., Komurov K., Zhou A. Y., Gupta S., Yang J., Hartwell K., Onder T. T., Gupta P. B., Evans K. W., Hollier B. G., Ram P. T., Lander E. S., Rosen J. M., Weinberg R. A., Mani S. A., Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A. 107, 15449–15454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peixoto P., Etcheverry A., Aubry M., Missey A., Lachat C., Perrard J., Hendrick E., Delage-Mourroux R., Mosser J., Borg C., Feugeas J.-P., Herfs M., Boyer-Guittaut M., Hervouet E., EMT is associated with an epigenetic signature of ECM remodeling genes. Cell Death Dis. 10, 205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stemmler M. P., Eccles R. L., Brabletz S., Brabletz T., Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 21, 102–112 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Williams E. D., Gao D., Redfern A., Thompson E. W., Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer 19, 716–732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer K. R., Durrans A., Lee S., Sheng J., Li F., Wong S. T. C., Choi H., El Rayes T., Ryu S., Troeger J., Schwabe R. F., Vahdat L. T., Altorki N. K., Mittal V., Gao D., Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X., Carstens J. L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C.-C., LeBleu V. S., Kalluri R., Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aiello N. M., Brabletz T., Kang Y., Nieto M. A., Weinberg R. A., Stanger B. Z., Upholding a role for EMT in pancreatic cancer metastasis. Nature 547, E7–E8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye X., Brabletz T., Kang Y., Longmore G. D., Nieto M. A., Stanger B. Z., Yang J., Weinberg R. A., Upholding a role for EMT in breast cancer metastasis. Nature 547, E1–E3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer K. R., Altorki N. K., Mittal V., Gao D., Fischer et al. reply. Nature 547, E5–E6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M., Trombetta J. J., Weitz D. A., Sanes J. R., Shalek A. K., Regev A., McCarroll S. A., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein A. M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D. A., Kirschner M. W., Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji A. L., Rubin A. J., Thrane K., Jiang S., Reynolds D. L., Meyers R. M., Guo M. G., George B. M., Mollbrink A., Bergenstråhle J., Larsson L., Bai Y., Zhu B., Bhaduri A., Meyers J. M., Rovira-Clavé X., Hollmig S. T., Aasi S. Z., Nolan G. P., Lundeberg J., Khavari P. A., Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 1661–1662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma A., Cao E. Y., Kumar V., Zhang X., Leong H. S., Wong A. M. L., Ramakrishnan N., Hakimullah M., Teo H. M. V., Chong F. T., Chia S., Thangavelu M. T., Kwang X. L., Gupta R., Clark J. R., Periyasamy G., Gopalakrishna Iyer N., DasGupta R., Longitudinal single-cell RNA sequencing of patient-derived primary cells reveals drug-induced infidelity in stem cell hierarchy. Nat. Commun. 9, 4931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. Raghavan, P. S. Winter, A. W. Navia, H. L. Williams, A. DenAdel, R. L. Kalekar, J. Galvez-Reyes, K. E. Lowder, N. Mulugeta, M. S. Raghavan, A. A. Borah, S. A. Väyrynen, A. D. Costa, R. W. S. Ng, J. Wang, E. Reilly, D. Y. Ragon, L. K. Brais, A. M. Jaeger, L. F. Spurr, Y. Y. Li, A. D. Cherniack, I. Wakiro, A. Rotem, B. E. Johnson, J. M. McFarland, E. T. Sicinska, T. E. Jacks, T. E. Clancy, K. Perez, D. A. Rubinson, K. Ng, J. M. Cleary, L. Crawford, S. R. Manalis, J. A. Nowak, B. M. Wolpin, W. C. Hahn, A. J. Aguirre, A. K. Shalek, Transcriptional subtype-specific microenvironmental crosstalk and tumor cell plasticity in metastatic pancreatic cancer. doi:10.1101/2020.08.25.256214.
- 23.Ganesh K., Basnet H., Kaygusuz Y., Laughney A. M., He L., Sharma R., O’Rourke K. P., Reuter V. P., Huang Y.-H., Turkekul M., Emrah E., Masilionis I., Manova-Todorova K., Weiser M. R., Saltz L. B., Garcia-Aguilar J., Koche R., Lowe S. W., Pe’er D., Shia J., Massagué J., L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat. Cancer. 1, 28–45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laughney A. M., Hu J., Campbell N. R., Bakhoum S. F., Setty M., Lavallée V.-P., Xie Y., Masilionis I., Carr A. J., Kottapalli S., Allaj V., Mattar M., Rekhtman N., Xavier J. B., Mazutis L., Poirier J. T., Rudin C. M., Pe’er D., Massagué J., Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat. Med. 26, 259–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinker G. S., Greenwald A. C., Tal R., Orlova Z., Cuoco M. S., McFarland J. M., Warren A., Rodman C., Roth J. A., Bender S. A., Kumar B., Rocco J. W., Fernandes P. A. C. M., Mader C. C., Keren-Shaul H., Plotnikov A., Barr H., Tsherniak A., Rozenblatt-Rosen O., Krizhanovsky V., Puram S. V., Regev A., Tirosh I., Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 52, 1208–1218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee H.-O., Hong Y., Etlioglu H. E., Cho Y. B., Pomella V., Van den Bosch B., Vanhecke J., Verbandt S., Hong H., Min J.-W., Kim N., Eum H. H., Qian J., Boeckx B., Lambrechts D., Tsantoulis P., De Hertogh G., Chung W., Lee T., An M., Shin H.-T., Joung J.-G., Jung M.-H., Ko G., Wirapati P., Kim S. H., Kim H. C., Yun S. H., Tan I. B. H., Ranjan B., Lee W. Y., Kim T.-Y., Choi J. K., Kim Y.-J., Prabhakar S., Tejpar S., Park W.-Y., Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 52, 594–603 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Uhlitz F., Bischoff P., Peidli S., Sieber A., Obermayer B., Blanc E., Trinks A., Lüthen M., Ruchiy Y., Sell T., Mamlouk S., Arsie R., Wei T.-T., Klotz-Noack K., Schwarz R. F., Sawitzki B., Kamphues C., Beule D., Landthaler M., Sers C., Horst D., Blüthgen N., Morkel M., Mitogen-activated protein kinase activity drives cell trajectories in colorectal cancer. EMBO Mol. Med. 13, e14123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian J., Olbrecht S., Boeckx B., Vos H., Laoui D., Etlioglu E., Wauters E., Pomella V., Verbandt S., Busschaert P., Bassez A., Franken A., Bempt M. V., Xiong J., Weynand B., van Herck Y., Antoranz A., Bosisio F. M., Thienpont B., Floris G., Vergote I., Smeets A., Tejpar S., Lambrechts D., A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 30, 745–762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sathe A., Grimes S. M., Lau B. T., Chen J., Suarez C., Huang R. J., Poultsides G., Ji H. P., Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin. Cancer Res. 26, 2640–2653 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwé H., Pircher A., Van den Eynde K., Weynand B., Verbeken E., De Leyn P., Liston A., Vansteenkiste J., Carmeliet P., Aerts S., Thienpont B., Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Kim N., Kim H. K., Lee K., Hong Y., Cho J. H., Choi J. W., Lee J.-I., Suh Y.-L., Ku B. M., Eum H. H., Choi S., Choi Y.-L., Joung J.-G., Park W.-Y., Jung H. A., Sun J.-M., Lee S.-H., Ahn J. S., Park K., Ahn M.-J., Lee H.-O., Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 11, 2285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu F., Fan J., He Y., Xiong A., Yu J., Li Y., Zhang Y., Zhao W., Zhou F., Li W., Zhang J., Zhang X., Qiao M., Gao G., Chen S., Chen X., Li X., Hou L., Wu C., Su C., Ren S., Odenthal M., Buettner R., Fang N., Zhou C., Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 12, 2540 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y.-P., Yin J.-H., Li W.-F., Li H.-J., Chen D.-P., Zhang C.-J., Lv J.-W., Wang Y.-Q., Li X.-M., Li J.-Y., Zhang P.-P., Li Y.-Q., He Q.-M., Yang X.-J., Lei Y., Tang L.-L., Zhou G.-Q., Mao Y.-P., Wei C., Xiong K.-X., Zhang H.-B., Zhu S.-D., Hou Y., Sun Y., Dean M., Amit I., Wu K., Kuang D.-M., Li G.-B., Liu N., Ma J., Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. 30, 1024–1042 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geistlinger L., Oh S., Ramos M., Schiffer L., LaRue R. S., Henzler C. M., Munro S. A., Daughters C., Nelson A. C., Winterhoff B. J., Chang Z., Talukdar S., Shetty M., Mullany S. A., Morgan M., Parmigiani G., Birrer M. J., Qin L.-X., Riester M., Starr T. K., Waldron L., Multiomic analysis of subtype evolution and heterogeneity in high-grade serous ovarian carcinoma. Cancer Res. 80, 4335–4345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steele N. G., Carpenter E. S., Kemp S. B., Sirihorachai V. R., The S., Delrosario L., Lazarus J., Amir E.-A. D., Gunchick V., Espinoza C., Bell S., Harris L., Lima F., Irizarry-Negron V., Paglia D., Macchia J., Chu A. K. Y., Schofield H., Wamsteker E.-J., Kwon R., Schulman A., Prabhu A., Law R., Sondhi A., Yu J., Patel A., Donahue K., Nathan H., Cho C., Anderson M. A., cV. Sahai, Lyssiotis C. A., Zou W., Allen B. L., Rao A., Crawford H. C., Bednar F., Frankel T. L., di Magliano M. P., Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat. Cancer. 1, 1097–1112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S. Z., Roden D. L., Wang C., Holliday H., Harvey K., Cazet A. S., Murphy K. J., Pereira B., Al-Eryani G., Bartonicek N., Hou R., Torpy J. R., Junankar S., Chan C.-L., Lam C. E., Hui M. N., Gluch L., Beith J., Parker A., Robbins E., Segara D., Mak C., Cooper C., Warrier S., Forrest A., Powell J., O’Toole S., Cox T. R., Timpson P., Lim E., Liu X. S., Swarbrick A., Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J. 39, e104063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bassez A., Vos H., Van Dyck L., Floris G., Arijs I., Desmedt C., Boeckx B., Vanden Bempt M., Nevelsteen I., Lambein K., Punie K., Neven P., Garg A. D., Wildiers H., Qian J., Smeets A., Lambrechts D., A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 27, 820–832 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P., The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan H., Yan M., Zhang G., Liu W., Deng C., Liao G., Xu L., Luo T., Yan H., Long Z., Shi A., Zhao T., Xiao Y., Li X., CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 47, D900–D908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao M., Kong L., Liu Y., Qu H., dbEMT: An epithelial-mesenchymal transition associated gene resource. Sci. Rep. 5, 11459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan T. Z., Miow Q. H., Miki Y., Noda T., Mori S., Huang R. Y.-J., Thiery J. P., Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 6, 1279–1293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohammadi S., Davila-Velderrain J., Kellis M., A multiresolution framework to characterize single-cell state landscapes. Nat. Commun. 11, 5399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., De Clercq S., Minguijón E., Balsat C., Sokolow Y., Dubois C., De Cock F., Scozzaro S., Sopena F., Lanas A., D’Haene N., Salmon I., Marine J.-C., Voet T., Sotiropoulou P. A., Blanpain C., Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Kröger C., Afeyan A., Mraz J., Eaton E. N., Reinhardt F., Khodor Y. L., Thiru P., Bierie B., Ye X., Burge C. B., Weinberg R. A., Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 116, 7353–7362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pastushenko I., Blanpain C., EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29, 212–226 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Williams K., Ghosh R., Giridhar P. V., Gu G., Case T., Belcher S. M., Kasper S., Inhibition of stathmin1 accelerates the metastatic process. Cancer Res. 72, 5407–5417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosales M. A. B., Shu D. Y., Iacovelli J., Saint-Geniez M., Loss of PGC-1α in RPE induces mesenchymal transition and promotes retinal degeneration. Life Sci. Alliance 2, e201800212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai J. H., Yang J., Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 27, 2192–2206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aiello N. M., Maddipati R., Norgard R. J., Balli D., Li J., Yuan S., Yamazoe T., Black T., Sahmoud A., Furth E. E., Bar-Sagi D., Stanger B. Z., EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell. 45, 681–695.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Izar B., Tirosh I., Stover E. H., Wakiro I., Cuoco M. S., Alter I., Rodman C., Leeson R., Su M.-J., Shah P., Iwanicki M., Walker S. R., Kanodia A., Melms J. C., Mei S., Lin J.-R., Porter C. B. M., Slyper M., Waldman J., Jerby-Arnon L., Ashenberg O., Brinker T. J., Mills C., Rogava M., Vigneau S., Sorger P. K., Garraway L. A., Konstantinopoulos P. A., Liu J. F., Matulonis U., Johnson B. E., Rozenblatt-Rosen O., Rotem A., Regev A., A single-cell landscape of high-grade serous ovarian cancer. Nat. Med. 26, 1271–1279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isella C., Terrasi A., Bellomo S. E., Petti C., Galatola G., Muratore A., Mellano A., Senetta R., Cassenti A., Sonetto C., Inghirami G., Trusolino L., Fekete Z., De Ridder M., Cassoni P., Storme G., Bertotti A., Medico E., Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 47, 312–319 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Tyler M., Tirosh I., Decoupling epithelial-mesenchymal transitions from stromal profiles by integrative expression analysis. Nat. Commun. 12, 2592 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng J., Sun B.-F., Chen C.-Y., Zhou J.-Y., Chen Y.-S., Chen H., Liu L., Huang D., Jiang J., Cui G.-S., Yang Y., Wang W., Guo D., Dai M., Guo J., Zhang T., Liao Q., Liu Y., Zhao Y.-L., Han D.-L., Zhao Y., Yang Y.-G., Wu W., Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 29, 725–738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoadley K. A., Yau C., Hinoue T., Wolf D. M., Lazar A. J., Drill E., Shen R., Taylor A. M., Cherniack A. D., Thorsson V., Akbani R., Bowlby R., Wong C. K., Wiznerowicz M., Sanchez-Vega F., Robertson A. G., Schneider B. G., Lawrence M. S., Noushmehr H., Malta T. M.; Cancer Genome Atlas Network, Stuart J. M., Benz C. C., Laird P. W., Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thorsson V., Gibbs D. L., Brown S. D., Wolf D., Bortone D. S., Yang T.-H. O., Porta-Pardo E., Gao G. F., Plaisier C. L., Eddy J. A., Ziv E., Culhane A. C., Paull E. O., Sivakumar I. K. A., Gentles A. J., Malhotra R., Farshidfar F., Colaprico A., Parker J. S., Mose L. E., Vo N. S., Liu J., Liu Y., Rader J., Dhankani V., Reynolds S. M., Bowlby R., Califano A., Cherniack A. D., Anastassiou D., Bedognetti D., Mokrab Y., Newman A. M., Rao A., Chen K., Krasnitz A., Hu H., Malta T. M., Noushmehr H., Pedamallu C. S., Bullman S., Ojesina A. I., Lamb A., Zhou W., Shen H., Choueiri T. K., Weinstein J. N., Guinney J., Saltz J., Holt R. A., Rabkin C. S.; Cancer Genome Atlas Research Network, Lazar A. J., Serody J. S., Demicco E. G., Disis M. L., Vincent B. G., Shmulevich I., The immune landscape of cancer. Immunity 48, 812–830.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Browaeys R., Saelens W., Saeys Y., NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Schubert M., Klinger B., Klünemann M., Sieber A., Uhlitz F., Sauer S., Garnett M. J., Blüthgen N., Saez-Rodriguez J., Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cursons J., Leuchowius K.-J., Waltham M., Tomaskovic-Crook E., Foroutan M., Bracken C. P., Redfern A., Crampin E. J., Street I., Davis M. J., Thompson E. W., Stimulus-dependent differences in signalling regulate epithelial-mesenchymal plasticity and change the effects of drugs in breast cancer cell lines. Cell Commun. Signal. 13, 26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aibar S., González-Blas C. B., Moerman T., Huynh-Thu V. A., Imrichova H., Hulselmans G., Rambow F., Marine J.-C., Geurts P., Aerts J., van den Oord J., Atak Z. K., Wouters J., Aerts S., SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McFarland J. M., Paolella B. R., Warren A., Geiger-Schuller K., Shibue T., Rothberg M., Kuksenko O., Colgan W. N., Jones A., Chambers E., Dionne D., Bender S., Wolpin B. M., Ghandi M., Tirosh I., Rozenblatt-Rosen O., Roth J. A., Golub T. R., Regev A., Aguirre A. J., Vazquez F., Tsherniak A., Multiplexed single-cell transcriptional response profiling to define cancer vulnerabilities and therapeutic mechanism of action. Nat. Commun. 11, 4296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McFaline-Figueroa J. L., Hill A. J., Qiu X., Jackson D., Shendure J., Trapnell C., A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition. Nat. Genet. 51, 1389–1398 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nieto M. A., Huang R. Y.-J., Jackson R. A., Thiery J. P., EMT: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Zadran S., Arumugam R., Herschman H., Phelps M. E., Levine R. D., Surprisal analysis characterizes the free energy time course of cancer cells undergoing epithelial-to-mesenchymal transition. Proc. Natl. Acad. Sci. U.S.A. 111, 13235–13240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jia D., Jolly M. K., Tripathi S. C., Den Hollander P., Huang B., Lu M., Celiktas M., Ramirez-Peña E., Ben-Jacob E., Onuchic J. N., Hanash S. M., Mani S. A., Levine H., Distinguishing mechanisms underlying EMT tristability. Cancer Converg. 1, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M., Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zagar M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. B., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data.bioRxiv, 2020.10.12.335331, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aran D., Looney A. P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R. P., Wolters P. J., Abate A. R., Butte A. J., Bhattacharya M., Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andreatta M., Carmona S. J., UCell: Robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19, 3796–3798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ibrahim M. M., Kramann R., genesorteR: Feature ranking in clustered single cell data. bioRxiv , 676379 (2019). [Google Scholar]
- 69.Aran D., Sirota M., Butte A. J., Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thorsson V., Gibbs D. L., Brown S. D., Wolf D., Bortone D. S., Yang T.-H. O., Porta-Pardo E., Gao G. F., Plaisier C. L., Eddy J. A., Ziv E., Culhane A. C., Paull E. O., Sivakumar I. K. A., Gentles A. J., Malhotra R., Farshidfar F., Colaprico A., Parker J. S., Mose L. E., Vo N. S., Liu J., Liu Y., Rader J., Dhankani V., Reynolds S. M., Bowlby R., Califano A., Cherniack A. D., Anastassiou D., Bedognetti D., Mokrab Y., Newman A. M., Rao A., Chen K., Krasnitz A., Hu H., Malta T. M., Noushmehr H., Pedamallu C. S., Bullman S., Ojesina A. I., Lamb A., Zhou W., Shen H., Choueiri T. K., Weinstein J. N., Guinney J., Saltz J., Holt R. A., Rabkin C. S.; Cancer Genome Atlas Research Network, Lazar A. J., Serody J. S., Demicco E. G., Disis M. L., Vincent B. G., Shmulevich I., The immune landscape of cancer. Immunity 51, 411–412 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Holland C. H., Szalai B., Saez-Rodriguez J., Transfer of regulatory knowledge from human to mouse for functional genomics analysis. Biochim. Biophys. Acta Gene Regul. Mech. 1863, 194431 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Holland C. H., Tanevski J., Perales-Patón J., Gleixner J., Kumar M. P., Mereu E., Joughin B. A., Stegle O., Lauffenburger D. A., Heyn H., Szalai B., Saez-Rodriguez J., Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 21, 36 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends for tables S1 and S2
Table S3
Figs. S1 to S7
References
Tables S1 and S2