Quantitative bioassay enables the development of next-generation biodegradable polymeric nanoparticles for in vivo mRNA delivery.
Abstract
Nanoparticle-based mRNA therapeutics hold great promise, but cellular internalization and endosomal escape remain key barriers for cytosolic delivery. We developed a dual nanoparticle uptake and endosomal disruption assay using high-throughput and high-content image-based screening. Using a genetically encoded Galectin 8 fluorescent fusion protein sensor, endosomal disruption could be detected via sensor clustering on damaged endosomal membranes. Simultaneously, nucleic acid endocytosis was quantified using fluorescently tagged mRNA. We used an array of biodegradable poly(beta-amino ester)s as well as Lipofectamine and PEI to demonstrate that this assay has higher predictive capacity for mRNA delivery compared to conventional polymer and nanoparticle physiochemical characteristics. Top nanoparticle formulations enabled safe and efficacious mRNA expression in multiple tissues following intravenous injection, demonstrating that the in vitro screening method is also predictive of in vivo performance. Efficacious nonviral systemic delivery of mRNA with biodegradable particles opens up new avenues for genetic medicine and human health.
INTRODUCTION
Recent advances in the synthesis of in vitro transcribed mRNA (1, 2) have spurred a vast amount of research into mRNA-based gene therapies including the development of next-generation vaccines (3). Compared to their plasmid DNA counterparts, mRNA offers safer and more controlled gene expression by virtually eliminating the risk for integration into the host genome (4). mRNA delivery could also lead to more potent expression in cell populations that are largely refractory to DNA transfection, such as T cells, which have been shown to mount immune responses against foreign cytosolic DNA (5, 6). However, because of their size and hydrophilicity, mRNA molecules are membrane-impermeable, making safe and efficient cytosolic mRNA delivery a major obstacle to their clinical utility.
Nonviral nanoparticle (NP) formulations have emerged as promising mRNA delivery vehicles. Many lipid-based (7) and several polymeric (8) mRNA NP systems have recently been reported for protein replacement (9, 10), immune modulation (11, 12), and gene editing applications (13, 14). To fully realize the promise of mRNA therapeutics, NP systems must be engineered to overcome intracellular barriers, such as cellular internalization and escape from endosomal sequestration (15). A study of lipid NPs encapsulating small interfering RNA (siRNA) showed that only an estimated 1 to 2% (16) of internalized siRNA reaches the cytosol, highlighting the need for improved nanomaterials as well as quantitative high-throughput in vitro assays that can measure NP performance at key delivery bottlenecks and improve NP design.
Several image-based methods for quantifying the ability of NPs to overcome endosomal entrapment have been reported. The most common method is assessing the lack of colocalization of fluorescently labeled NPs with the pH-sensitive LysoTracker dye (17, 18), which selectively accumulates in the acidic environment of endosomes. This approach is easy to use and applicable to a wide variety of materials, but only provides an indirect assessment, as it does not indicate effective endosomal escape or disruption. Transmission electron microscopy (TEM) imaging is another widely accepted method for confirming endosomal disruption and escape (16, 19). However, this method is not amenable to high-throughput analysis, cannot be done on living cells, and requires electron-dense labels such as gold NPs, which could alter the properties of the native NP system. More recently, several groups have reported the use of advanced imaging approaches such as high–dynamic range confocal microscopy (20) or superresolution stochastic optical reconstruction microscopy (21), which have yielded important mechanistic data for the intracellular fate of the materials being studied, but lack the high-throughput screening capacity required to evaluate arrays of nanomaterials.
In this study, we used Galectin 8 (Gal8) tracking for high-throughput image-based quantification of endosomal disruption. Gal8 is a β-galactoside carbohydrate-binding protein that selectively binds to glycans found on the inner leaflet of endosomal membranes (22, 23). Using cells genetically engineered to constitutively express a Gal8-mRuby fusion protein, we characterized the endosomal disruption capabilities of nanocarriers by quantifying the fluorescent puncta that formed following Gal8-mRuby clustering on damaged endosomal membranes, building upon the Gal8 recruitment assay using polyethylene glycol -b-[(2-(dimethylamino)ethyl methacrylate)-co-(butyl methacrylate)] [(PEG)]–(DMAEMA-co-BMA)] siRNA NPs by Kilchrist et al. (24). We adapted this approach to a high-throughput, wide-field imaging assay to simultaneously study how cellular internalization and endosomal disruption correlated with nucleic acid delivery efficacy of biodegradable poly(beta-amino ester)s (PBAEs) and other common materials for nucleic acid delivery. For PBAEs specifically, we systematically varied polymer backbone hydrophobicity and polymer end-cap structure to probe structure-function relationships. The predictive capacity of this dual cellular uptake and endosomal disruption assay was compared to that of several polymer and NP physiochemical properties such as polymer nucleic acid binding strength, pH buffering capacity, predicted LogP value, NP hydrodynamic diameter, and zeta potential. The effects of nucleic acid cargo type and cell type for in vitro transfection were investigated. In total, a library of 22 PBAEs with unique chemical structures were screened as well as widely used commercially available transfection reagents such as Lipofectamine 3000, polyethyleneimine (PEI), and poly-l-lysine (PLL). Last, we examined whether our new in vitro screening assays correlated with systemic in vivo delivery efficacy of polymeric NPs encapsulating mRNA upon tail vein injection in mice. The data presented here demonstrate the robustness of this image-based dual NP uptake and endosomal disruption NP screening system across a broad range of materials for mRNA delivery efficacy in vitro and in vivo. Such a quantitative, high-throughput screening platform with high predictive capacity for delivery efficacy has important implications for the standardization of the optimization and testing of novel materials for nonviral gene delivery and genetic medicine.
RESULTS
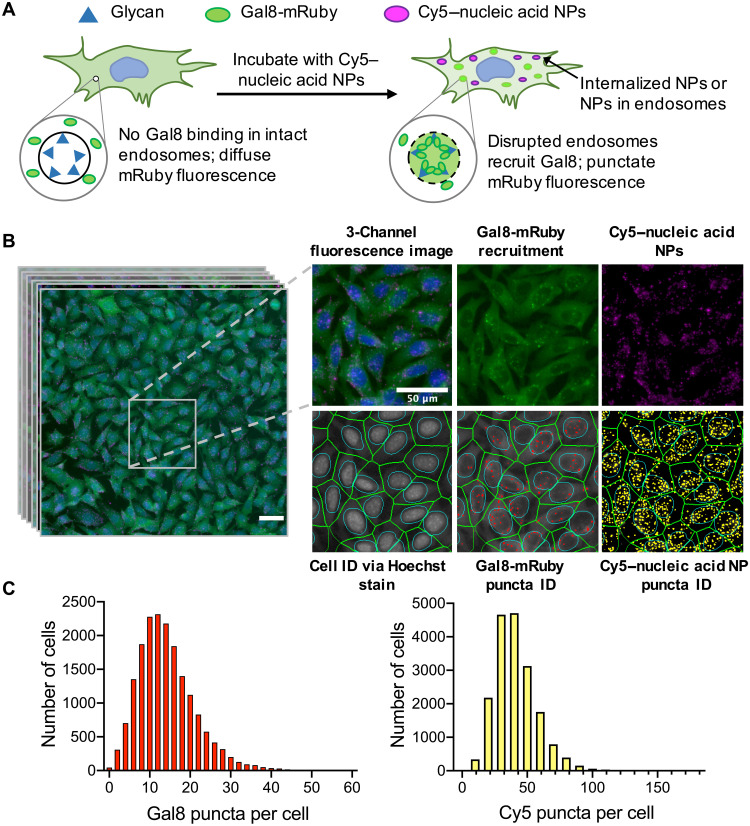
High-content imaging of NP uptake and endosomal disruption
We engineered B16-F10 murine melanoma cells to genetically encode a Gal8-mRuby endosomal disruption sensor to facilitate simultaneous characterization of NP uptake and endosomal disruption. NP uptake was measured by quantifying Cy5 puncta resulting from intracellular delivery of NPs carrying Cy5-labeled nucleic acids; endosomal disruption was measured by quantifying mRuby puncta resulting from Gal8-mRuby clustering at damaged endosomal membranes (Fig. 1A). This dual NP uptake and endosomal disruption assay was performed in a high-throughput manner using a CellInsight CX7 LZR high-content imager capturing 20 fields of view per well of a 96-well plate at 20× magnification. An image analysis algorithm was then optimized and used to identify cells by extrapolating the cell body surrounding Hoechst 33342–stained cell nuclei and provide puncta counts per cell (Fig. 1B). On average, intracellular puncta count was collected for over 15,000 cells per NP formulation.
Fig. 1. Image-based analysis of NP uptake and Gal8 endosomal disruption assay.
(A) Assay overview: Cells genetically encoding a Gal8-mRuby fusion fluorescence protein exhibited diffuse cytosolic mRuby signal in the absence of endosomal disruption. Endosomal disruption caused by NPs carrying Cy5-labeled nucleic acid NPs allows Gal8-mRuby to bind to intra-endosomal glycans, resulting in punctate fluorescent spots. (B) Typical field of view (taken from 80 per NP formulation) imaged by high-throughput fluorescence microscopy of B16-F10 murine melanoma cells after 6-hour exposure to PBAE NPs carrying Cy5-mRNA. Cell identification was done using Hoechst 33342 staining of cell nuclei. Identification of Gal8-mRuby puncta and Cy5 puncta was used to quantify endosomal disruption and NP uptake, respectively. Scale bars, 50 μm. (C) Representative distributions of the Gal8 puncta or Cy5 puncta count per cell obtained from image analysis data.
To identify the optimal time point to conduct the assay, we performed a time course experiment in which B16-mRuby-Gal8 cells were incubated with PBAE NPs for up to 30 hours and imaged at select time points. We found that the Cy5 and Gal8 puncta counts both peaked at 6 hours after transfection for most nucleic acid cargo types and generally decreased thereafter (fig. S1), guiding us to perform this assay at 6 hours for all remaining experiments. The decreases in Gal8 and Cy5 puncta over time are consistent with expected autophagy timelines for damaged endocytic vesicles (20). We further quantified NP uptake using Cy5 puncta count as well as the overall Cy5 fluorescence in the cell and found that these two metrics correlated positively with each other (fig. S1C; Spearman’s coefficient = 0.98). Although overall Cy5 fluorescence may take into account diffuse fluorescent signal from NPs that have escaped into the endosome, we elected to use Cy5 puncta count to quantify NP uptake to enable direct comparisons between NP uptake and endosomal disruption.
Effects of PBAE backbone hydrophobicity
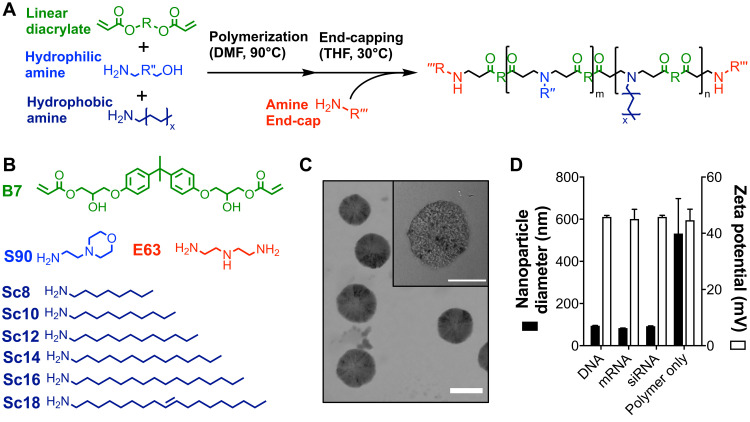
We synthesized two series of PBAE polymers with varying hydrophobic monomer content to investigate the effects of polymer backbone hydrophobicity on NP uptake, endosomal disruption, and transfection capabilities. These were first synthesized as lipophilic PBAE terpolymers consisting of a linear diacrylate (B7) copolymerized with a hydrophilic amine (S90) and a hydrophobic amine (ScX) synthesized via Michael addition reactions (Fig. 2). Polymer hydrophobicity was varied in one series by incorporating hydrophobic amines of varying lipid tail length at 30 mole percent and in a second series by varying the molar content of the Sc12 monomer. Polymers in both series were then end-capped with monomer E63 to create PBAE quadpolymers, and molecular weight was found to be in the range of 4 to 10 kDa. All polymers were found to rapidly self-assemble into NPs with plasmid DNA, mRNA, and siRNA after simple pipette mixing in aqueous buffer. NPs encapsulating nucleic acid cargo were 100 to 400 nm in diameter, with positive zeta potential in the range of 30 to 60 mV (fig. S2).
Fig. 2. Chemical structure and characterization of PBAE NPs.
(A) PBAE synthesis via two-step Michael addition reactions for linear end-capped polymers. (B) Structures of diacrylate (B), hydrophilic side-chain (S), hydrophobic side-chain (Sc), and end-cap (E) monomers used in the synthesis of backbone hydrophobicity variation polymer series. (C) Representative TEM image of 7-90,c12-63, 50%-Sc12, mRNA NPs formulated at 60 (w/w) with 10% DMG-PEG2k and dialyzed into PBS. Scale bar, 100 nm. (D) Dynamic light scattering measurements of z-average NP hydrodynamic diameter and zeta potential of 7-90,c12-63, 50%-Sc12 NPs formed at 60 (w/w) and diluted into PBS. Data are means + SD; n = 3.
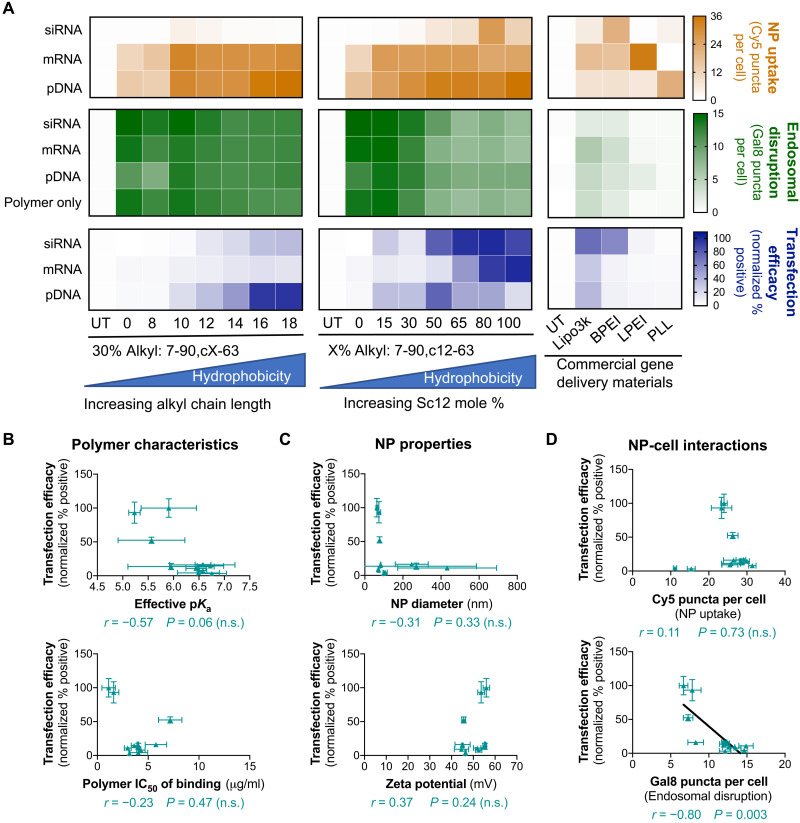
We next assessed NP uptake, endosomal disruption, and gene delivery efficacy. Transfection by siRNA NPs was assessed by siRNA-mediated green fluorescent protein (GFP) knockdown in cells engineered to be GFP+, while transfection by DNA and mRNA NPs was assessed by GFP expression resulting from functional delivery of DNA or mRNA encoding the GFP gene in non-GFP+ cells. All PBAE NP formulations maintained >90% cell viability (fig. S3). In both PBAE polymer series, increasing polymer backbone hydrophobicity generally increased nucleic acid uptake and transfection in all three nucleic acid modalities (Fig. 3A). The opposite was true for Gal8 endosomal disruption, where the polymer containing 100% Sc12 (most hydrophobic) resulted in half of the Gal8-mRuby puncta count compared to the polymer containing 0% Sc12 (least hydrophobic). Commercially available gene delivery materials were used to provide a benchmark for the bioassays. Of the five commercially available materials tested, Lipofectamine 3000 enabled the highest transfection across all nucleic acid types, followed by 25-kDa branched PEI. Transfection with these commercially available materials correlated positively with endosomal disruption (Spearman’s coefficient of 0.68), and no significant correlation with NP uptake was observed. The Gal8 puncta counts for these materials were much lower than those achieved by PBAE NPs even when transfection efficacy was similar, suggesting that the two classes of materials use different mechanisms to enable endosomal disruption.
Fig. 3. Validation of dual NP uptake/Gal8 endosomal disruption assay in PBAE NPs and commercial reagents delivering different nucleic acid cargos to B16-F10 cells.
(A) Heatmaps summarizing NP uptake, Gal8 endosomal disruption, and transfection efficacy data. Uptake and Gal8 data were obtained from high-throughput imaging analysis. Transfection efficacy was assessed by flow cytometry. For DNA and mRNA delivery, GFP fluorescence intensity for each formulation was normalized to the max fluorescence intensity across all treatment conditions. siRNA-mediated GFP knockdown was quantified by normalizing the percent GFP+ cells for siGFP-treated wells to the corresponding formulation delivering the scRNA (scrambled control siRNA) control. Data are presented as the mean of four replicate wells. Transfection efficacy of NPs formed with PBAE polymers encapsulating mRNA was plotted against common predictor readouts such as (B) various polymer characteristics, (C) NP properties, and (D) NP-cell interactions. Correlation significance was assessed for PBAE NPs using Spearman’s method, and datasets with statistically significant correlations were indicated with fitted lines. Data are means ± SD; n = 4. n.s., not significant.
We further assessed the predictive capacity of various polymer and NP properties on transfection efficacy. The polymer median inhibitory concentration (IC50) of nucleic acid binding, with larger values indicating weaker nucleic acid binding affinity, correlated negatively with DNA transfection but positively with siRNA knockdown (fig. S4A). This may be due to the different intracellular sites of action for each nucleic acid. Plasmid DNA needs to reach the nucleus, and strong initial binding could facilitate nuclear trafficking and maximize likelihood of transfection in each cell. On the other hand, siRNA needs to be released to the cytosol to be active, and thus, weaker polymer–nucleic acid binding could enable quicker and more effective cargo release and activity. mRNA transfection was not observed to correlate significantly with nucleic acid binding affinity in these experiments (Fig. 3B and fig. S6). Standard biophysical characterization measurements of NP size and zeta potential showed no significant correlations with transfection efficiency (Fig. 3C and fig. S4B). The NP-cell interactions quantified by our new high-throughput and high-content imaging-based assay showed that PBAE transfection generally correlated positively with NP uptake and negatively with Gal8 endosomal disruption (Fig. 3D and fig. S4C). The negative correlation between transfection and endosomal disruption levels in this series of PBAE NPs was unexpected, although, even at their lowest, the endosomal disruption levels achieved with the PBAE NPs were significantly higher than those induced by the commercial gene delivery materials. Thus, all PBAE NPs evaluated may be above a critical threshold of endosomal disruption capacity necessary to enable functional nucleic acid delivery that is at least equal to or greater than the endosomal disruption capacity achieved by commercial gene delivery materials. The data indicated that in these experiments, endosomal disruption was not a major transfection bottleneck for PBAEs. Empty PBAE polymeric NPs in the absence of nucleic acids resulted in equivalent levels of endosomal disruption as NPs loaded with nucleic acids (Fig. 3A). This may explain why certain PBAE NP formulations less effective at transfection nonetheless exhibited high levels of endosomal disruption, as these polymers may have formed a larger fraction of empty NPs. These empty PBAE NPs could lead to a high Gal8 puncta count, indicating endosomal disruption, but would do so in a nonproductive manner as no nucleic acids would be delivered to the cytosol. PBAE transfection with this series of polymers also did not correlate significantly with the polymers’ effective pKa (where Ka is the acid dissociation constant), as quantified in the physiologically relevant pH range (Fig. 3B and figs. S4A and S5), which also reinforces that endosomal disruption does not depend significantly on polymer pH buffering capacity. Other polymer properties such as the predicted LogP value, which is a measure of polymer hydrophobicity, showed strong positive correlations with transfection for all three cargo types (fig. S7B), further confirming our hypothesis that increased backbone hydrophobicity improves polymeric gene delivery efficacy.
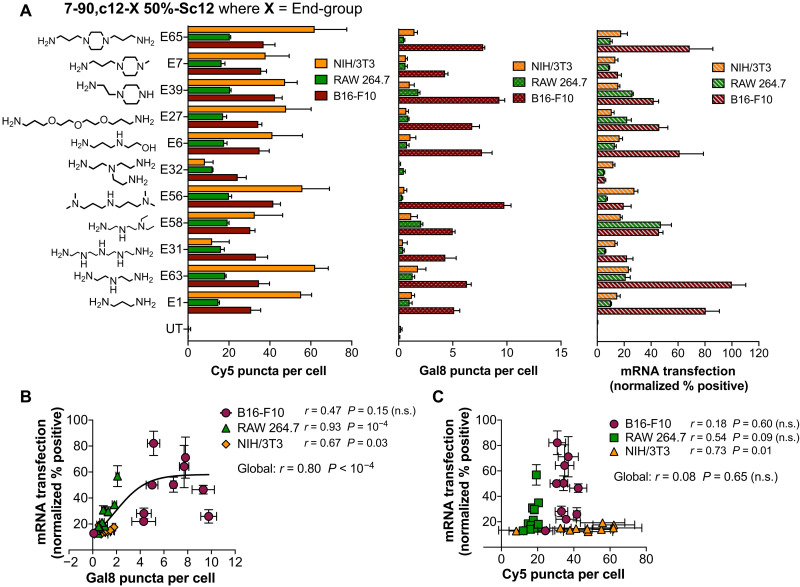
Effects of polymer end groups
We next investigated the effects of polymer end-group structure on NP uptake and endosomal disruption by synthesizing an end-group variation polymer series. This was done by using a moderately hydrophobic PBAE terpolymer backbone (7-90,c12-X, 50%-Sc12) and then independently conjugating 11 different end-group monomers to it (Fig. 4A). Previous work by our laboratory has shown that polymer end-group structure can play an important role in imparting biomaterial-mediated, selective transfection in certain cell types over others (25, 26) and that these effects may be due to changes in NP uptake pathways (27). We hypothesized that our dual NP uptake/endosomal escape assay could be useful in further ascertaining how polymer end-group structure affects NP function in different cell types. To test this hypothesis, and to further evaluate the robustness of our new high-throughput and high-content bioassay, we evaluated these polymers on three different cell types induced to express the Gal8-mRuby construct: B16-F10 murine melanoma cells, RAW 264.7 murine macrophages, and NIH/3T3 murine fibroblasts. These cell lines were chosen to validate our assay in a diverse range of cell types. In particular, B16-F10 cells were included as a model cancer cell line that is frequently used for mouse tumor studies (28, 29); RAW macrophages were included to investigate gene delivery to immune cells, which many studies have demonstrated pose additional intracellular barriers to transfection (30, 31); and 3T3 fibroblasts were included as a noncancerous, healthy control cell line. Our results showed highest mRNA transfection levels in B16-F10 cells, medium transfection in RAW 264.7 macrophages, and lowest transfection levels in NIH/3T3 fibroblasts (Fig. 4A). Endosomal disruption showed positive correlations with mRNA transfection levels in RAW and 3T3 cells, but not B16 cells, with a significant positive correlation when all three cell lines were evaluated together (Fig. 4B). This effect is particularly notable for more difficult-to-transfect cell types such as RAW 264.7 and NIH/3T3 cells (Spearman’s coefficient of 0.92 and 0.67, respectively), which suggests that mRNA transfection efficacy in difficult-to-transfect cells may largely be attributable to barriers in endosomal escape. The highest NP uptake levels were observed in NIH/3T3 cells, which demonstrated the lowest levels of mRNA transfection, and in general, mRNA transfection did not show significant correlations with PBAE NP uptake among these cell types (Fig. 4C). Collectively, these results suggest that for PBAEs with the same polymer backbone (and similar hydrophobicity), end-group structure plays an important role in endosomal disruption. These results also indicate that for these PBAEs, endosomal disruption, rather than NP uptake, is acting as a greater bottleneck for effective mRNA delivery. Differing levels of resistance to endosomal disruption among different cell types may also at least partially explain the differential transfection levels observed among these cells.
Fig. 4. Effects of polymer end-group structure on mRNA transfection efficacy in multiple cell lines.
(A) NP uptake, Gal8 puncta count, and mRNA delivery efficacy of polymer end-group variation PBAE library on three different cell lines. Transfection efficacy was plotted against (B) Gal8 puncta count indicating endosomal disruption or (C) Cy5 puncta count indicating NP uptake. Data are means ± SD; n = 4. Correlation significance in (B) and (C) was calculated using Spearman’s method; a hyperbolic curve was fitted in (B) to indicate a statistically significant correlation.
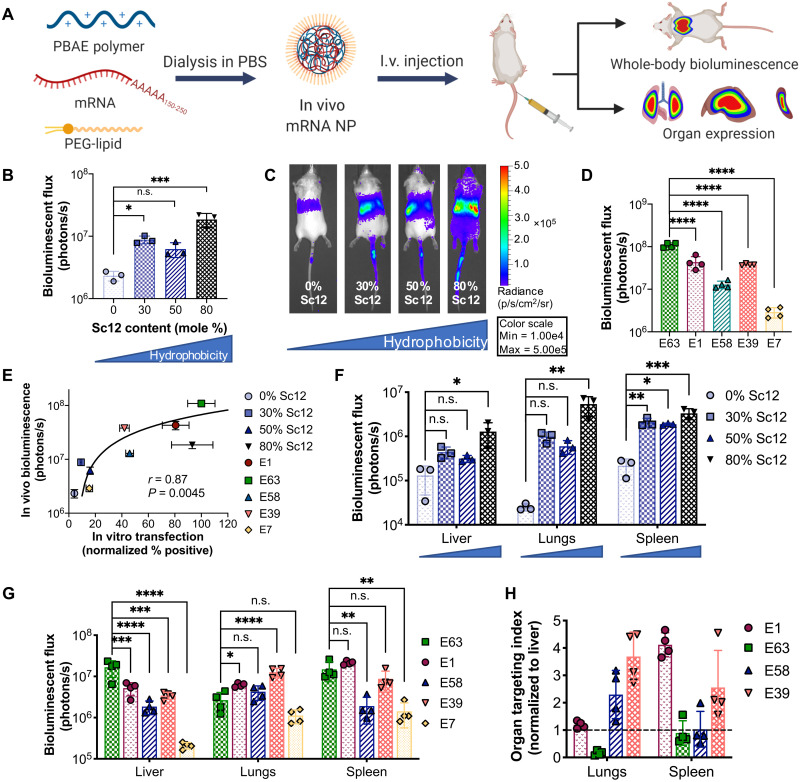
In vivo mRNA delivery: Whole-body and organ-level expression
We next characterized the in vivo mRNA delivery capabilities of NPs after intravenous administration of NPs encapsulating mRNA encoding firefly luciferase (fLuc) to mice. For in vitro experiments, NPs were formulated by admixing PBAE polymer and nucleic acid cargo (termed PBAE NPs) to enable facile self-assembly and rapid screening of large NP libraries. For in vivo experiments, NPs were formulated with the PEG-lipid DMG-PEG2k (1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000) and dialyzed in phosphate-buffered saline (PBS) (termed PBAE + PEG-lipid NPs; Fig. 5A). PEG-lipids were added to PBAE NP formulations to enhance NP serum stability, and previous studies have shown that incorporation of PEG-lipids into related PBAE NPs improved in vivo mRNA expression (32, 33). PEG-lipids are a common component in gene delivery NPs and have most recently been used in coronavirus disease 2019 (COVID-19) mRNA vaccines (34). Previous studies demonstrated that in lipid NPs delivering siRNA, PEG-lipids containing alkyl chains with greater than 14 carbons inhibited hepatic siRNA delivery due to inadequate lipid desorption from NPs (35); thus, DMG-PEG2k (containing 14 carbon alkyl chains) was used in our studies. Incorporation of DMG-PEG2k into the PBAE quadpolymers was observed to decrease NP size and neutralize surface charge. PEG-lipid coating did not significantly change transfection efficacy in vitro, although NP uptake and endosomal disruption were reduced. Upon in vivo administration, PEG-coated and dialyzed NPs enabled significantly higher mRNA expression compared to NPs without PEG coating, and this increased expression was predominately due to increased expression in the liver (fig. S8).
Fig. 5. In vivo validation of PEG-coated PBAE NPs delivering mRNA.
(A) Schematic depicting the experimental workflow. PBAE polymers were dialyzed with fLuc mRNA and the PEG-lipid DMG-PEG2k in PBS to form PEG-coated mRNA NPs, which were administered intravenously (I.v.). fLuc expression was assessed 24 hours after NP injection. (B) Whole-body bioluminescence was assessed for NPs formulated with PBAEs with differential backbone hydrophobicity and (C) representative IVIS images (N = 3). (D) Whole-body bioluminescence quantification for NPs formulated with 50%-Sc12 PBAEs with different end groups (N = 4). (E) In vivo transfection efficacy [from (B) and (D)] was plotted against in vitro transfection in B16 cells. Spearman’s correlation was used to measure the strength of association between the two variables. Organ bioluminescence in the most highly expressing organs when varying (F) polymer backbone hydrophobicity (blue triangles below organ labels indicate increasing backbone hydrophobicity) and (G) polymer end-group structure. Statistical significance was determined using one-way ANOVA with Dunnett’s post hoc analysis comparing against the least hydrophobic polymer (0% Sc12) in (B) and (F) and against end-group E63 in (D) and (G). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (H) The organ targeting index as calculated by normalizing bioluminescent flux in each nonliver organ against that of the liver was calculated for the lungs and spleen in high-expressing polymers of the polymer end-group variation series (E7 was excluded because of minimal expression observed). Dotted black line indicates liver expression level. N = 4. Data are means ± SD in all bar graphs.
Four polymers with 0 to 80% Sc12 content in the polymer backbone and five polymers with different polymer end groups were chosen to assess the effects of polymer backbone and end-group structure on in vivo expression. On the whole-body level, increased backbone hydrophobicity generally resulted in increased mRNA expression (Fig. 5, B and C, and fig. S9), while polymer end-group variation resulted in differential in vivo expression levels (Fig. 5D and fig. S10). Overall in vivo expression correlated positively with in vitro transfection of B16-F10 cells (Fig. 5E) but did not correlate significantly with NP uptake or endosomal disruption as measured by our dual Cy5/Gal8 assay (fig. S11), indicating that in vitro screening had limited predictive capacity for in vivo performance with these nanomaterials. At the level of individual organs, increasing backbone hydrophobicity increased expression in all of the organs evaluated (Fig. 5F), while polymer end group played a major role in targeting NP expression to specific organs (Fig. 5G). When expression in the lungs and spleen was normalized to that in the liver, polymer E1 showed preferential expression in the spleen, polymer E63 showed preferential expression in the liver, polymer E58 showed preferential expression in the lungs, and polymer E39 was almost equally split between the lungs and spleen (Fig. 5H). To assess the in vivo safety profile of mRNA NPs, liver aspartate aminotransferase (AST), and alanine aminotransferase (ALT) levels were measured at 24 hours and 7 days following a single intravenous NP administration, and body mass was monitored for 7 days (fig. S12). PEG-lipid–coated 7-90,c12-63, 80%-Sc12 NPs were used, as this was one of the most effective formulations for in vivo mRNA expression and was used for all subsequent experiments. NP administration did not result in elevated liver enzymes or significantly decreased body mass.
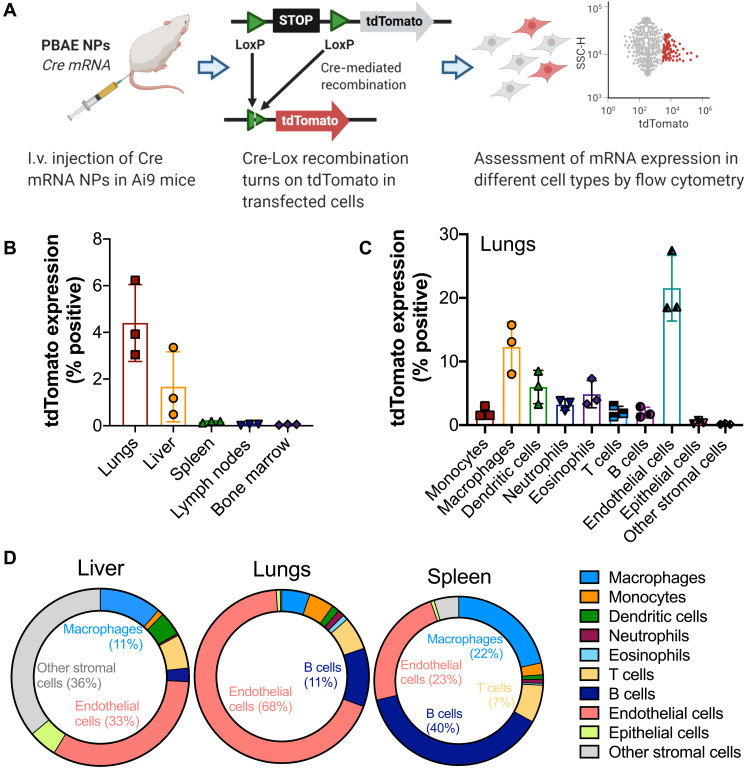
In vivo mRNA delivery: Expression in different cell types
We further probed the cell populations that were transfected in each organ using the Ai9 mouse model, which contains a floxed expression stop cassette upstream of a tdTomato reporter gene. NPs encapsulating Cre mRNA were administered via tail vein injection into Ai9 mice, and transfected cells underwent Cre-Lox recombination, resulting in tdTomato expression that was measured by flow cytometry 3 days after injection (Fig. 6A). For this study, we used 7-90,c12-63, 80%-Sc12 NPs as they were found to enable high in vivo mRNA expression levels from fLuc mRNA experiments. We found that 7-90,c12-63, 80%-Sc12 NPs systemically administered transfected nearly 0.2% of the cells in the spleen, 2% of the cells in the liver, and 4% of the cells in the lungs, with minimal transfection levels seen in any of the other organs evaluated (Fig. 6B). Over 20% of endothelial cells in the lungs were transfected following systemic injection, which is consistent with previous reports for related PBAE structures (32), in addition to significant populations of macrophages and dendritic cells in the lungs (Fig. 6C). Endothelial cells also made up a large fraction of the transfected cells in the liver (33%) and spleen (23%) (Fig. 6D).
Fig. 6. Assessment of in vivo mRNA transfection in different cell types.
(A) Experimental workflow: Ai9 mice were injected with PEG-coated 7-90,c12-63, 80%-Sc12 NPs encapsulating Cre mRNA, and single cell–level transfection could be detected by tdTomato expression, which was quantified 3 days after injection using flow cytometry. (B) tdTomato+ cells as a percentage of the total cell population in each of several major organs. (C) tdTomato expression in the lungs in different cell types (tdTomato+ cells as a percentage of the overall population of each cell type). (D) Distribution of tdTomato+ cells across different cell types in liver, lungs, and spleen. N = 3. Data presented as means ± SD in bar graphs.
DISCUSSION
To realize the full therapeutic potential of mRNA therapeutics, a high-throughput, standardized NP screening platform capable of quantitatively evaluating intracellular delivery steps with great predictive capacity for transfection efficacy is needed. In this study, we developed a high-throughput, high-content, imaging-based screening platform designed to simultaneously assess the cellular internalization and endosomal disruption capabilities of nucleic acid delivery NPs, requiring only wide-field, epifluorescence microscopy to enable full assessment of the cytosolic compartment. This bioassay was developed to be implemented in multiwell plates, enabling the evaluation of many intracellular events per cell, in thousands of replicate cells per condition, with up to 96 conditions per plate. Endosomal sequestration has long been identified as a major bottleneck to functional RNA delivery in multiple NP systems (36, 37), but quantitative evaluation of endosomal disruption has been limited to low-throughput imaging methods requiring specialized microscopy modalities (16, 21). We used a genetically encoded endosomal disruption sensor based on the natural clustering of Gal8 molecules at damaged endosomal membranes to detect NP-induced endosomal disruption quantified at the level of intracellular events within single cells. Simultaneously, cellular internalization of NPs could be tracked by delivering nucleic acids labeled with a different fluorophore. We hypothesized that this dual NP uptake and endosomal disruption assay could provide useful information on structure-function relationships when used to screen several NP gene delivery systems.
We used two series of PBAE quadpolymers to validate this screening platform. PBAEs are cationic, biodegradable polymers that have been shown to be highly effective at in vitro delivery of plasmid DNA (38), siRNA (39), mRNA (32), and protein cargos (40). The highly modular nature of these polymers facilitates combinatorial library synthesis via Michael addition of small-molecule precursors, making it possible to systematically vary polymer backbone or end-group characteristics to directly probe the effects of incremental differential polymer structural changes on downstream nucleic acid delivery efficacy. The PBAE quadpolymer is the majority component of all our NP delivery formulations, including systemically administered in vivo formulations, which have 10% PEG-lipid incorporated as a second component, without the presence of other lipids or cholesterol. This approach differs significantly from many previously studied lipid-based NP systems, in which the NP formulation was changed primarily by varying the ratios of incorporated lipids (41) or the structure of the ionizable lipid in an NP system consisting of multiple lipid components (11).
Two polymer series in which polymer backbone hydrophobicity was modulated by varying the content of lipophilic side-chain monomers were synthesized to probe the effect of polymer backbone structure on cellular interactions of polymeric NPs. Traditional metrics of predicting NP function, such as polymer nucleic acid binding affinity, endosomal pH buffering potential, NP hydrodynamic diameter, and zeta potential, generally correlated poorly with functional delivery efficacy of multiple nucleic acid cargos, highlighting the need for new metrics for rapid and meaningful NP screening. The dual NP uptake and endosomal disruption assay presented here showed significant correlations with transfection efficacy for all nucleic acid cargos tested. NP uptake correlated positively with transfection (global r = 0.55, P < 0.001). Endosomal disruption correlated negatively with transfection for these PBAE NPs (that each had greater endosomal disruption capacity than that achieved by the commercial gene delivery materials) (r = −0.57, P < 0.0001). The negative correlation with endosomal disruption is unexpected but may be attributed to the formation of polymer-only NPs that do not contain nucleic acid cargo. Amphiphilic PBAEs like the ones presented in this study have been reported to form polymer-only micellar NPs without any nucleic acid (42). Thus, PBAEs that are effective at endosomal disruption, but not efficient at leading to transfection, may be forming large populations of polymer-only NPs empty of nucleic acid cargo. When these polymer-only NPs are internalized by cells, they could enable endosomal disruption, resulting in high Gal8 counts but low transfection. When this dual NP uptake/Gal8 endosomal disruption assay was applied to commercial gene delivery materials such as Lipofectamine 3000, branched and linear PEI, and PLL, endosomal disruption as indicated by Gal8 puncta count was significantly lower for all of these commercial materials than the PBAE NPs, which, for the most part, also resulted in lower transfection efficacy compared to PBAE NPs. Transfection of these positive control materials correlated positively with endosomal disruption for all cargo types (global r = 0.68, P = 0.02). Together, our data show that a threshold for endosomal disruption, as defined by the amount achieved by the most effective commercial transfection reagent Lipofectamine 3000 (≥2 Gal8 puncta per cell in B16-F10 cells), must be reached in order for gene delivery to efficiently occur. PBAE NPs generally enabled endosomal disruption levels significantly above this threshold in the B16-F10 cells evaluated here and resulted in generally high transfection levels, while commercial materials such as linear PEI and PLL enabled endosomal disruption levels below this threshold and consequently showed negligible transfection levels. The lack of high transfection of PBAE NPs across the board indicates that delivery obstacles further downstream (such as intracellular trafficking or cargo release) may pose significant delivery challenges for some of these materials.
Previous studies have shown that the structure of PBAE polymer end groups can significantly alter the transfection efficacy of the backbone polymer and impart biomaterial-mediated selectivity in transfection of certain cell types (25–27). We synthesized a polymer series with a common backbone but varying end-group structure and evaluated mRNA delivery efficacy on three cell lines. The endosomal disruption levels of these polymers had positive correlations with transfection efficacy, which were strongest in more difficult-to-transfect cell lines as indicated by Spearman’s coefficients (r) that are closer to 1: r = 0.93 for difficult-to-transfect RAW 264.7 cells but r = 0.47 for easier-to-transfect B16-F10 cells. Differences observed in transfection efficacy were not attributable to polymers’ pH buffering capabilities, which varied with backbone structure but were generally unaffected by end-group structure. Even in the 7-90,c12-63, X% alkyl side-chain polymer series, in which the effective pKa decreased with increasing hydrophobic Sc12 content in the polymer backbone, the correlation between pH buffering and transfection efficacy was poor. This is in contrast to an observation recently reported by our group with hyperbranched PBAEs, where increasing polymer branching by incorporation of a triacrylate monomer in the backbone increased both effective pKa and transfection (38), suggesting that different classes of PBAE polymer structures can enable endosomal escape via differing mechanisms. In the case of the linear lipophilic PBAE quadpolymers, the endosomal disruption mechanism may rely on the lipophilicity of the polymers, causing them to associate with and directly interact with the endosomal membrane, where the charged polymer end groups may cause transient pore formation that leads to NP leakage out of damaged endosomes, similar to that observed with lipid materials (16, 37), rather than acidic buffering-induced complete endosomal rupture as proposed by the proton sponge hypothesis (21). NP uptake of the end-modified linear PBAEs did not correlate significantly with mRNA transfection efficacy (r = 0.11, P = 0.73), although a significant positive correlation was observed when PBAE NPs carrying each of the three nucleic acid cargos were analyzed globally (global r = 0.55, P < 0.001). Collectively, our data suggest that endosomal escape is the primary barrier in mRNA delivery to more difficult-to-transfect cells and that the differential gene delivery efficacy mediated by polymer end groups is largely due to their ability to facilitate endosomal disruption.
Last, we validated these PBAE NPs for in vivo mRNA expression following tail vein injection into mice. NPs formulated by simple mixing of mRNA and polymer in aqueous buffer yielded significantly lower transfection, particularly in the liver, than similar formulations with 10% PEG-lipid dialyzed into the NPs. Using dialyzed PEG-coated formulations, we saw that in vivo mRNA expression levels correlated strongly with in vitro transfection efficacy in B16-F10 cells. Specifically, top performers from in vitro screens such as 7-90,c12-63, 80%-Sc12 NPs resulted in high in vivo mRNA expression, while formulations that enabled low in vitro transfection such as 7-90,c12-7, 50%-Sc12 (E7) NPs resulted in minimal in vivo expression. No significant correlation was found between in vivo expression and NP uptake or endosomal disruption from in vitro screening. This was not unexpected as many obstacles need to be overcome before successful transfection can occur, making delivery mechanisms several steps upstream of transfection unreliable indicators of in vivo gene delivery. Together, these data show that in vitro screening of these polymeric materials has a predictive capacity that is rare in large library screens of lipid materials (43). Increasing polymer backbone hydrophobicity increased whole-body mRNA expression in general, following trends that we observed in vitro, and which could also be due, in part, to improved incorporation of PEG-lipid in hydrophobic formulations that could lead to more stable NPs in the blood (33). Similar to differential transfection of various cell types in vitro, polymer end-group variation also led to tuning of organ tropism in vivo. Unlike most lipid NP formulations that have been demonstrated to predominantly target liver hepatocytes (44, 45), the four top performing NP formulations from the in vitro mRNA transfection screens in the end-group variation polymer series exhibited alternative patterns of expression in nonliver organs, with preferential transfection in the lungs and/or spleen. Particularly, high expression was seen in the lungs for most formulations, which is consistent with previous reports by Kaczmarek et al. (32) using related PBAE lipid-polymer NP formulations for mRNA delivery. Within each organ, multiple cell types were transfected, including endothelial cells, B cells, and macrophages, all of which have distinct clinical relevance. The lipophilic side chains of the polymers enabled the PEG-lipid DMG-PEG2k to be easily incorporated into NP formulations via dialysis, which increased in vivo expression by an order of magnitude compared to NPs without PEG-lipid coating despite slightly lowering in vitro transfection. Cheng et al. (46) recently reported that incorporation of selective organ targeting molecules at defined ratios enabled highly targeted mRNA expression in select organs and that these molecules maintained their organ targeting capabilities across multiple lipid NP platforms. This suggests intriguing future directions where an innate organ tropism of PBAE NP formulations could perhaps be combined with other technologies to enhance selective organ targeting and potentially cell type–specific targeting.
In summary, we have reported a high-content high-throughput quantitative imaging assay capable of simultaneously quantifying NP uptake and endosomal disruption. This assay is robust, has higher predictive capacity for in vitro mRNA delivery efficacy compared to conventionally used metrics of polymer or NP properties, and can be performed with ~100 NP formulations in a few hours. Assay validation using PBAE NPs elucidated structure-function relationships through incremental changes in both the polymer backbone and end groups for these highly modular polymers. Moreover, we showed that this assay is generally applicable across all major nucleic acid types, several different cell lines, and multiple gene delivery systems. The NP screening platform presented here can be a useful tool for high-throughput identification of promising candidates for gene delivery and further elucidation of structure/function relationships for the delivery of DNA, siRNA, and mRNA. Lead nanomaterials composed of PBAE quadpolymers demonstrated safe and effective delivery of mRNA in vivo, including organ-targeted expression based on polymer structure. PEGylated PBAE NPs enabled significant exogenous mRNA expression differentially to the liver, lung, and spleen. Critically, nanomaterial formulations identified as lead candidates in vitro also performed well for in vivo mRNA delivery following systemic intravenous injection. Such a broadly applicable screening method provides a new metric for nanomaterial characterization, which is important for directly comparing and contextualizing the myriad NP systems that have been reported in the burgeoning field of intracellular gene delivery. With further study, the PBAE-based materials investigated here may be promising for mRNA delivery to promote human health.
MATERIALS AND METHODS
Materials
Bisphenol A glycerolate (1 glycerol/phenol) diacrylate (B7; CAS 4687949), 4-(2-aminoethyl)morpholine (S90; CAS 2038-031), octylamine (Sc8; CAS 111-86-4), 1-decylamine (Sc10; CAS 2016-57-1), oleylamine (Sc18; CAS 112-90-3), 1,3-diaminopropane (E1; CAS 109-76-2), tetraethylenepentamine (E31; CAS 1112-57-2), N,N-diethyldiethylenetriamine (E58; CAS 24426-16-2), tris(2-aminoethyl)amine (E32; CAS 4097-89-6), 2-(3-aminopropylamino)ethanol (E6; CAS 4461-39-6), 4,7,10-trioxa-1,13-tridecanediamine (E27; CAS 4246-51-9), and 1-(2-aminoethyl)piperazine (E39; CAS 140-31-8) were purchased from Sigma-Aldrich (St. Louis, MO). 1-Dodecylamine (Sc12; CAS 124-22-1) and 1-(3-aminopropyl)-4-methylpiperazine (E7; CAS 4572-031) were purchased from Alfa Aesar (Tewksbury, MA). Tetradecylamine (Sc14; CAS 2016-42-4) and hexadecylamine (Sc16; CAS 143-27-1) were purchased from Acros Organics (Pittsburgh, PA). Diethylentriamine (E63; CAS 111-40-0) was purchased from EMD Millipore (Burlington, MA). 3,3′-Iminobis(N,N-dimethylpropylamine) (E56; CAS 6711484) was purchased from Santa Cruz Biotechnology (Dallas, TX). 1,4-Bis(3-aminopropyl)piperazine (E65; CAS 7209-38-3) was purchased from MP Biomedicals (Solon, OH). Plasmid eGFP-N1(Addgene 2491) was purchased from Elim Biopharmaceuticals (Hayward, CA) and amplified by Aldevron (Fargo, ND). Cy5-labeled plasmid DNA was synthesized following a method reported by Wilson et al. (47). 5-Methoxyuridine–modified CleanCap eGFP mRNA (L-7201), fLuc mRNA (L-7202), and Cy5-labeled mRNA (L-7702) were purchased from TriLink Biotechnologies (San Diego, CA). Negative control siRNA (1027281) was purchased from Qiagen (Germantown, MD). GFP siRNA targeting the sequence 5′-GCAAGCTGACCCTGAAGTTC-3′ (P-002048-01) was purchased from Dharmacon (Lafayette, CO). Cy5-labeled siRNA (SIC005) was purchased from Sigma-Aldrich (St. Louis, MO). Plasmid DNA encoding a Gal8 fluorescent fusion protein was a gift from the laboratory of C. Duvall and cloned into a PiggyBac transposon vector (PB-mRuby3-Gal8, Addgene #150815) for stable integration into mammalian chromosomal DNA.
Polymer synthesis
Polymers were synthesized using previously reported protocols (38). Briefly, diacrylate monomer B7 and side-chain monomers (S90 and combinations of ScX monomers) were dissolved at 600 mg/ml in dimethylformamide and reacted with stirring for 48 hours at 90°C to allow polymerization via stepwise Michael addition reactions. Monomers were reacted at an overall vinyl:amine ratio of 2.3 to allow acrylate-terminated polymers to form. Polymers were end-capped by further reaction with primary amine-containing E monomers at room temperature for 2 hours [polymer (200 mg/ml) and 0.3 M E monomer in tetrahydrofuran (THF)] and purified by two diethyl ether washes. Diethyl ether was decanted and dried thoroughly under vacuum, and polymers were dissolved in dimethyl sulfoxide at 100 mg/ml and stored at −20°C with desiccant in single-use aliquots.
Polymer characterization
Polymer molecular weight was characterized using gel permeation chromatography (GPC) against linear polystyrene standards (Waters, Milford, MA). Polymers were dissolved in butylated hydroxytoluene (BHT)-stabilized THF and filtered through 0.2-μm polytetrafluoroethylene (PTFE) filters before GPC measurements. Predicted polymer LogP values were calculated using the online cheminformatics software molinspiration.com.
Polymer buffering capacity and determination of effective pKa
pH titrations were performed using a SevenEasy pH meter (Mettler Toledo, Columbus, OH) as previously described (38). Briefly, 10 mg of polymer was dissolved in 10 ml of 100 mM NaCl acidified with HCl and titrated from pH 3.0 to pH 11.0 via stepwise addition of 100 mM NaOH. To calculate the effective pKa of the polymer in the physiologically relevant pH range (pH 5 to 8), normalized buffering capacity was calculated from titration data as Δ(−OH)/Δ(pH) for each titration point. Effective pKa was defined as the pH point corresponding to the maximum normalized buffering capacity.
Nucleic acid binding assays
RiboGreen nucleic acid binding dye (Invitrogen, Carlsbad, CA) was mixed with nucleic acids in 25 mM magnesium acetate buffer (MgAc2, pH 5.0) at a final nucleic acid concentration of 5 μg/ml (siRNA), 2.5 μg/ml (mRNA), or 1 μg/ml (pDNA) and a final 1:2000 RiboGreen dilution. Polymers were dissolved and serially diluted to a range of concentrations in MgAc2, and 25 μl of polymer solution was mixed with 75 μl of nucleic acid/RiboGreen solution per well in 96-well black bottom assay plates. The solutions were incubated at 37°C for 20 min before fluorescence readings were taken on a Biotek Synergy 2 fluorescence multiplate reader (BioTek, Winooski, VT). To characterize nucleic acid binding affinity, the polymer IC50 of binding (polymer concentration at which 50% of RiboGreen fluorescence is quenched by RiboGreen displacement from polymer binding to nucleic acids) was calculated by plotting percent fluorescence quenching as a function of polymer concentration and fitting a sigmoidal curve to the data. Polymer IC50 of binding varies inversely with binding affinity; lower IC50 values indicate higher binding affinity.
NP formulation and characterization
For in vitro studies, NPs were formulated in 25 mM magnesium acetate buffer (MgAc2, pH 5) and added directly to cells without the addition of PEG-lipids or dialysis. Polymers and nucleic acids (plasmid DNA, mRNA, or siRNA) were dissolved separately in 25 mM MgAc2 at concentrations of 0.83 ng/μl for nucleic acids and 50 ng/μl for polymers and mixed together via pipetting at a 1:1 volume ratio. NPs were allowed to self-assemble for 10 min at room temperature; the polymer–to–nucleic acid ratio was 60 by weight (60, w/w) for all experiments.
NP hydrodynamic diameter was measured via dynamic light scattering using Malvern Zetasizer Pro with universal dip cell (Malvern Panalytical, Malvern, UK). Samples were prepared in 25 mM MgAc2 and diluted 1:6 in 150 mM PBS to determine NP characteristics in neutral isotonic buffer. Zeta potential was measured by electrophoretic light scattering on the same instrument. TEM images were captured using a Philips CM120 transmission electron microscope (Philips Research, Cambridge, MA). NP samples (30 μl) were allowed to coat 400–square mesh carbon-coated TEM grids for 20 min. Grids were then rinsed with ultrapure water and allowed to fully dry before imaging.
Cell culture and cell line preparation
B16-F10 murine melanoma and RAW 264.7 murine macrophage cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. GFPd2+ B16-F10 cells used in siRNA knockdown experiments were established previously (40) and cultured using the same medium. NIH/3T3 murine fibroblasts were cultured in DMEM supplemented with 10% bovine calf serum and 1% penicillin/streptomycin. Cells were induced to constitutively express the Gal8-mRuby fusion fluorescent protein construct using the PiggyBac transposon/transposase system. The PiggyBac transposon plasmid carrying the Gal8-mRuby gene was created using restriction enzyme cloning and is available on Addgene (plasmid #150815). The transposase expression plasmid (PB200A-1) was purchased from System Biosciences (Palo Alto, CA). The transposon plasmid was cotransfected with the PiggyBac transposase plasmid using PBAE NPs as described below. mRuby+ cells were isolated using at least two rounds of fluorescence-assisted cell sorting using the Sony SH800 Cell Sorter (Sony Biotechnology, San Jose, CA) to generate stably expressing cell lines.
Transfection
Cells were plated at 10,000 cells per well in 100 μl of complete medium in CytoOne 96-well plates (USA Scientific, Ocala, FL) and allowed to adhere overnight. NPs were formulated following the in vitro transfection formulation described above; 20 μl of NP solution was added to 100 μl of fresh complete medium, and NP medium mixture (120 μl per well) was used to replace the culture medium. For all in vitro transfections, NPs were formulated at 60 (w/w) delivering 50 ng of nucleic acids per well. For cellular uptake experiments, 20% of the total nucleic acid drugs were replaced with Cy5-labeled nucleic acids before mixing with polymers. NPs were incubated with cells at 37°C for the appropriate duration, depending on assay conditions (6 hours for dual uptake/Gal8 assay, 24 hours for mRNA and siRNA transfections, and 48 hours for DNA transfections).
For transfections using commercially available reagents, Lipofectamine 3000 (Thermo Fisher Scientific) was used as instructed by the manufacturer. 25-kDa branched polyethylenimine (BPEI), 2.5-kDa linear polyethylenimine (LPEI), and 15-kDa PLL were used at the highest concentrations that did not cause significant cytotoxicity [15 (w/w) for BPEI, 60 (w/w) for LPEI, and 30 (w/w) for PLL]. PEI NPs were formulated in 150 mM NaCl solution, and PLL NPs were formulated in 10 M Hepes buffer (pH 7); all formulations delivered 50 ng of nucleic acids to match the dose delivered by PBAE NPs.
Transfection efficacy was evaluated via flow cytometry using a BD Accuri C6 flow cytometer (BD Biosciences, East Rutherford, NJ). For plasmid DNA and mRNA transfections, the expression of a GFP reporter gene was quantified by normalizing the geometric mean fluorescence intensity of each NP treatment to that of the formulation achieving maximum expression. Cells previously engineered to constitutively express GFP (48) were used for siRNA knockdown transfections, and the percentage of cells positively expressing GFP when gated against untreated cells in wells treated with siRNA targeting GFP was normalized against that of wells treated with noncoding control siRNA.
Dual NP uptake and Gal8 endosomal disruption assay
NPs of matching formulation as those used for transfection experiments were used to deliver nucleic acid cargo containing 20% Cy5-labeled nucleic acids to enable visualization of NP uptake. NPs were incubated with Gal8-mRuby+ cells for 6 hours (assay time point optimized in fig. S1), at which point NPs and cell culture medium were removed and cells were washed with PBS and fixed with 10% formalin for 10 min at room temperature. The formalin was then removed, cells were washed with PBS, and Hoechst nuclear stain (1:5000 in PBS) was applied for 10 min. NP uptake and Gal8-mRuby endosomal escape were then quantified by high-content imaging analysis of Cy5 and mRuby puncta per cell, respectively, using a CellInsight CX7 LZR high-content imager (Thermo Fisher Scientific) with HCS Studio analysis software.
NP formulation for in vivo studies
All animal work was done in adherence of the policies and guidelines of the Johns Hopkins University Animal Care and Use Committee. NPs for in vivo mRNA delivery were formulated at 30 (w/w). mRNA was dissolved in MgAc2, while polymer and the PEG-lipid DMG-PEG2k (10% by mass) were dissolved in 100% ethanol. The mRNA and polymer–PEG-lipid solutions were mixed via pipetting at 1:1 volume ratio, and NPs were allowed to self-assemble at room temperature for 10 min. NPs were then dialyzed against cold PBS at 4°C for 75 min using Spectra/Por Float-A-Lyzer G2 dialysis devices (Repligen, Waltham, MA) with 50-kDa molecular weight cutoff. NP volume after dialysis was adjusted with PBS for a final mRNA concentration of 0.1 mg/ml. NPs were administered to animals via 100-μl tail vein injections for a final dose of 10 μg of mRNA per animal.
To investigate the effects of PEGylation and dialysis on in vivo mRNA expression, NPs with no PEG-lipid and no dialysis were formulated in 25 mM MgAc2 at the same final mRNA concentration and weight-to-weight ratio as above. Sucrose solution (500 mg/ml) was used to bring the mixture to isotonicity.
fLuc mRNA in vivo bioluminescence
NPs encapsulating fLuc mRNA were formulated as described above and administered to 6- to 7-week-old male BALB/c mice via lateral tail vein injection. Whole-body bioluminescence was assessed 24 hours after injection. d-Luciferin potassium salt solution (25 mg/ml in PBS; Cayman Chemical Company, Ann Arbor, MI) was administered to mice via 150-μl intraperitoneal injection, and mice were imaged using the IVIS Spectrum Imager (Perkin Elmer, Waltham, MA) 10 min later. The same animals were euthanized immediately after whole-body imaging via cervical dislocation, and select organs were extracted, submerged in d-luciferin solution (250 μg/ml), and imaged with IVIS.
Cre mRNA delivery to Ai9 mice
NPs encapsulating Cre mRNA were formulated with DMG-PEG2k and dialyzed in PBS as described above. NPs were administered to 6-week-old male Ai9 mice via tail vein injection, and tdTomato expression following Cre-Lox recombination was allowed to accumulate for 3 days, at which point animals were euthanized via cervical dislocation. Select organs were extracted and dissociated by a 1-hour incubation in collagenase (2 mg/ml) at 37°C followed by mechanical pressing through a 70-μm cell strainer. Cells were pelleted by centrifugation, the supernatant was removed, and red blood cells in the cell pellet were lysed by incubating in ACK (ammonium-chloride-potassium) lysing buffer (Quality Biological, Gaithersburg, MD) for 1 min at room temperature. Cells were diluted in PBS, passed through a 100-μm cell strainer, pelleted by centrifugation, and resuspended in fluorescence-activated cell sorting (FACS) buffer (2% FBS in PBS with 0.02% sodium azide). Surface staining of cells with fluorescent antibodies was then performed using the antibodies and dilutions listed in table S1 in FACS buffer for 30 min at 4°C, at which time cells were washed twice and resuspended in FACS buffer for further analysis. FACS experiments were performed using an Attune NxT flow cytometer (Thermo Fisher Scientific) and analyzed using FlowJo software (FlowJo, Ashland, OR). Gating strategies to identify cell populations are provided in fig. S13.
In vivo NP safety profile
Hepatotoxicity after a single intravenous injection of mRNA NPs was assessed by collecting blood from animals 24 hours and 7 days after treatment. Serum was collected by centrifugation at 1500 relative centrifugal force for 15 min, and AST and ALT activity assays (Sigma-Aldrich, St. Louis, MO) were performed according to the manufacturer’s instructions. Body mass was monitored daily. Untreated animals were used as controls.
Graphical illustrations
Graphical illustrations were created using BioRender (https://biorender.com/).
Statistical analysis
Curve plotting and statistical analysis were performed using Prism 8 (GraphPad, La Jolla, CA). Data are shown as means ± SD for groups of three or more replicates or as individual values with the mean indicated. Unless otherwise stated, the absence of statistical significance markings, where a test was stated to have been performed, signifies no statistical significance. The statistical tests used for each figure are indicated in the figure captions. Statistical significance is denoted as follows: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. n.s., not significant.
Acknowledgments
Funding: We thank the following organizations for financial support: NIH R01CA228133 (J.J.G.), NIH R01EY031097 (J.J.G.), NIH P41EB028239 (J.J.G.), NIH F31CA250319 (Y.R.), NIH R37CA246699 (S.Y.T.), NIH P30EY001765 (Wilmer Core Grant), NSF Graduate Research Fellowship DGE-0707427 (D.R.W.), NSF Graduate Research Fellowship DGE-1232825 (Y.R.), Bloomberg~Kimmel Institute for Cancer Immunotherapy (J.J.G.), Research to Prevent Blindness James and Carole Free Catalyst Award (J.J.G.), and AstraZeneca (J.J.G.).
Author contributions: Conceptualizations: Y.R., D.R.W., S.Y.T., and J.J.G. Methodology: Y.R., D.R.W., S.Y.T., C.A.B., D.J.Z., and J.J.G. Investigation: Y.R., D.R.W., S.Y.T., H.M.Y., D.S., and M.C. Resources and funding acquisition: A.T. and J.J.G. Writing and editing: Y.R., D.R.W., S.Y.T., H.M.Y., D.S., M.C., C.A.B., D.J.Z., A.T., and J.J.G. Supervision and administration: D.J.Z. and J.J.G.
Competing interests: J.J.G., Y.R., S.Y.T., and D.R.W. are inventors on a patent application related to this work filed by Johns Hopkins University (no. C16397, PCT/US2021/052405, filed on 28 September 2021). J.J.G. is a member of the board of directors of VasoRx and is a member of the scientific advisory board of Tidal Therapeutics. The authors declare no other competing interests. Any potential conflicts of interests are managed by the Johns Hopkins Committee on Outside Interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Table S1
REFERENCES AND NOTES
- 1.Karikó K., Muramatsu H., Welsh F. A., Ludwig J., Kato H., Akira S., Weissman D., Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thess A., Grund S., Mui B. L., Hope M. J., Baumhof P., Fotin-Mleczek M., Schlake T., Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 23, 1456–1464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corbett K. S., Edwards D. K., Leist S. R., Abiona O. M., Boyoglu-Barnum S., Gillespie R. A., Himansu S., Schäfer A., Ziwawo C. T., DiPiazza A. T., Dinnon K. H., Elbashir S. M., Shaw C. A., Woods A., Fritch E. J., Martinez D. R., Bock K. W., Minai M., Nagata B. M., Hutchinson G. B., Wu K., Henry C., Bahl K., Garcia-Dominguez D., Ma L. Z., Renzi I., Kong W. P., Schmidt S. D., Wang L., Zhang Y., Phung E., Chang L. A., Loomis R. J., Altaras N. E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M. K., Shi W., Leung K., Yang E. S., West A., Gully K. L., Stevens L. J., Wang N., Wrapp D., Doria-Rose N. A., Stewart-Jones G., Bennett H., Alvarado G. S., Nason M. C., Ruckwardt T. J., McLellan J. S., Denison M. R., Chappell J. D., Moore I. N., Morabito K. M., Mascola J. R., Baric R. S., Carfi A., Graham B. S., SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandal P. K., Ferreira L. M. R., Collins R., Meissner T. B., Boutwell C. L., Friesen M., Vrbanac V., Garrison B. S., Stortchevoi A., Bryder D., Musunuru K., Brand H., Tager A. M., Allen T. M., Talkowski M. E., Rossi D. J., Cowan C. A., Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15, 643–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monroe K. M., Yang Z., Johnson J. R., Geng X., Doitsh G., Krogan N. J., Greene W. C., IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343, 428–432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabnis S., Kumarasinghe E. S., Salerno T., Mihai C., Ketova T., Senn J. J., Lynn A., Bulychev A., McFadyen I., Chan J., Almarsson Ö., Stanton M. G., Benenato K. E., A novel amino lipid series for mRNA delivery: Improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 26, 1509–1519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel A. K., Kaczmarek J. C., Bose S., Kauffman K. J., Mir F., Heartlein M. W., DeRosa F., Langer R., Anderson D. G., Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv. Mater. 31, 1805116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Q., Wei T., Jia Y., Farbiak L., Zhou K., Zhang S., Wei Y., Zhu H., Siegwart D. J., Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv. Mater. 30, 1805308 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Cao J., An D., Galduroz M., Zhuo J., Liang S., Eybye M., Frassetto A., Kuroda E., Funahashi A., Santana J., Mihai C., Benenato K. E., Kumarasinghe E. S., Sabnis S., Salerno T., Coughlan K., Miracco E. J., Levy B., Besin G., Schultz J., Lukacs C., Guey L., Finn P., Furukawa T., Giangrande P. H., Saheki T., Martini P. G. V., mRNA therapy improves metabolic and behavioral abnormalities in a murine model of citrin deficiency. Mol. Ther. 27, 1242–1251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billingsley M. M., Singh N., Ravikumar P., Zhang R., June C. H., Mitchell M. J., Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T cell engineering. Nano Lett. 20, 1578–1589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao L., Li L., Huang Y., Delcassian D., Chahal J., Han J., Shi Y., Sadtler K., Gao W., Lin J., Doloff J. C., Langer R., Anderson D. G., Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 37, 1174–1185 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Liu J., Chang J., Jiang Y., Meng X., Sun T., Mao L., Xu Q., Wang M., Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 31, 1902575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller J. B., Zhang S., Kos P., Xiong H., Zhou K., Perelman S. S., Zhu H., Siegwart D. J., Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. 56, 1059–1063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rui Y., Wilson D. R., Green J. J., Non-viral delivery to enable genome editing. Trends Biotechnol. 37, 281–293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M., Epstein-Barash H., Zhang L., Koteliansky V., Fitzgerald K., Fava E., Bickle M., Kalaidzidis Y., Akinc A., Maier M., Zerial M., Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 31, 638–646 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Tamura A., Oishi M., Nagasaki Y., Enhanced cytoplasmic delivery of siRNA using a stabilized polyion complex based on PEGylated nanogels with a cross-linked polyamine structure. Biomacromolecules 10, 1818–1827 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Akita H., Kogure K., Moriguchi R., Nakamura Y., Higashi T., Nakamura T., Serada S., Fujimoto M., Naka T., Futaki S., Harashima H., Nanoparticles for ex vivo siRNA delivery to dendritic cells for cancer vaccines: Programmed endosomal escape and dissociation. J. Control. Release 143, 311–317 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Kilchrist K. V., Evans B. C., Brophy C. M., Duvall C. L., Mechanism of enhanced cellular uptake and cytosolic retention of MK2 inhibitory peptide nano-polyplexes. Cell. Mol. Bioeng. 9, 368–381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittrup A., Ai A., Liu X., Hamar P., Trifonova R., Charisse K., Manoharan M., Kirchhausen T., Lieberman J., Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 33, 870–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wojnilowicz M., Glab A., Bertucci A., Caruso F., Cavalieri F., Super-resolution imaging of proton sponge-triggered rupture of endosomes and cytosolic release of small interfering RNA. ACS Nano 13, 187–202 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Hadari Y. R., Paz K., Dekel R., Mestrovic T., Accili D., Zick Y., Galectin-8. J. Biol. Chem. 270, 3447–3453 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Thurston T. L. M., Wandel M. P., von Muhlinen N., Foeglein Á., Randow F., Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilchrist K. V., Dimobi S. C., Jackson M. A., Evans B. C., Werfel T. A., Dailing E. A., Bedingfield S. K., Kelly I. B., Duvall C. L., Gal8 visualization of endosome disruption predicts carrier-mediated biologic drug intracellular bioavailability. ACS Nano 13, 1136–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishra B., Wilson D. R., Sripathi S. R., Suprenant M. P., Rui Y., Wahlin K. J., Berlinicke C. A., Green J. J., Zack D. J., A combinatorial library of biodegradable polyesters enables non-viral gene delivery to post-mitotic human stem cell-derived polarized RPE monolayers. Regen. Eng. Transl. Med. 6, 273–285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunshine J. C., Peng D. Y., Green J. J., Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Mol. Pharm. 9, 3375–3383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J., Sunshine J. C., Green J. J., Differential polymer structure tunes mechanism of cellular uptake and transfection routes of poly(β-amino ester) polyplexes in human breast cancer cells. Bioconjug. Chem. 25, 43–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson D. R., Sen R., Sunshine J. C., Pardoll D. M., Green J. J., Kim Y. J., Biodegradable STING agonist nanoparticles for enhanced cancer immunotherapy. Nanomedicine 14, 237–246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzeng S. Y., Patel K. K., Wilson D. R., Meyer R. A., Rhodes K. R., Green J. J., In situ genetic engineering of tumors for long-lasting and systemic immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 117, 4043–4052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A. R. K., Shou Y., Chan B., L K., Tay A., Materials for improving immune cell transfection. Adv. Mater. 33, 2007421 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Moradian H., Roch T., Lendlein A., Gossen M., mRNA transfection-induced activation of primary human monocytes and macrophages: Dependence on carrier system and nucleotide modification. Sci. Rep. 10, 4181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaczmarek J. C., Kauffman K. J., Fenton O. S., Sadtler K., Patel A. K., Heartlein M. W., DeRosa F., Anderson D. G., Optimization of a degradable polymer–lipid nanoparticle for potent systemic delivery of mrna to the lung endothelium and immune cells. Nano Lett. 18, 6449–6454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eltoukhy A. A., Chen D., Alabi C. A., Langer R., Anderson D. G., Degradable terpolymers with alkyl side chains demonstrate enhanced gene delivery potency and nanoparticle stability. Adv. Mater. 25, 1487–1493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerji A., Wickner P. G., Saff R., Stone C. A. Jr., Robinson L. B., Long A. A., Wolfson A. R., Williams P., Khan D. A., Phillips E., Blumenthal K. G., mRNA vaccines to prevent COVID-19 disease and reported allergic reactions: Current evidence and suggested approach. J. Allergy Clin. Immunol. Pract. 9, 1423–1437 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mui B. L., Tam Y. K., Jayaraman M., Ansell S. M., du X., Tam Y. Y. C., Lin P. J. C., Chen S., Narayanannair J. K., Rajeev K. G., Manoharan M., Akinc A., Maier M. A., Cullis P., Madden T. D., Hope M. J., Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. Nucleic Acids 2, e139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R., Buganim Y., Schroeder A., Langer R., Anderson D. G., Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 31, 653–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehman Z. u., Hoekstra D., Zuhorn I. S., Mechanism of polyplex- and lipoplex-mediated delivery of nucleic acids: Real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano 7, 3767–3777 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Wilson D. R., Rui Y., Siddiq K., Routkevitch D., Green J. J., Differentially branched ester amine quadpolymers with amphiphilic and pH-sensitive properties for efficient plasmid DNA delivery. Mol. Pharm. 16, 655–668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlsson J., Rui Y., Kozielski K. L., Placone A. L., Choi O., Tzeng S. Y., Kim J., Keyes J. J., Bogorad M. I., Gabrielson K., Guerrero-Cazares H., Quiñones-Hinojosa A., Searson P. C., Green J. J., Engineered nanoparticles for systemic siRNA delivery to malignant brain tumours. Nanoscale 11, 20045–20057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rui Y., Rui Y., Wilson D. R., Choi J., Varanasi M., Sanders K., Karlsson J., Lim M., Green J. J., Carboxylated branched poly(β-amino ester) nanoparticles enable robust cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 5, eaay3255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sago C. D., Lokugamage M. P., Paunovska K., Vanover D. A., Monaco C. M., Shah N. N., Gamboa Castro M., Anderson S. E., Rudoltz T. G., Lando G. N., Munnilal Tiwari P., Kirschman J. L., Willett N., Jang Y. C., Santangelo P. J., Bryksin A. V., Dahlman J. E., High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. U.S.A. 115, E9944–E9952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson D. R., Mosenia A., Suprenant M. P., Upadhya R., Routkevitch D., Meyer R. A., Quinones-Hinojosa A., Green J. J., Continuous microfluidic assembly of biodegradable poly(beta-amino ester)/DNA nanoparticles for enhanced gene delivery. J. Biomed. Mater. Res. A 105, 1813–1825 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Paunovska K., Sago C. D., Monaco C. M., Hudson W. H., Castro M. G., Rudoltz T. G., Kalathoor S., Vanover D. A., Santangelo P. J., Ahmed R., Bryksin A. V., Dahlman J. E., A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation. Nano Lett. 18, 2148–2157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akinc A., Maier M. A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., du X., Hope M. J., Madden T. D., Mui B. L., Semple S. C., Tam Y. K., Ciufolini M., Witzigmann D., Kulkarni J. A., van der Meel R., Cullis P. R., The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 14, 1084–1087 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J. B., Karmali P. P., Chivukula P., Verma I. M., Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. U.S.A. 114, E1941–E1950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Q., Wei T., Farbiak L., Johnson L. T., Dilliard S. A., Siegwart D. J., Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson D. R., Routkevitch D., Rui Y., Mosenia A., Wahlin K. J., Quinones-Hinojosa A., Zack D. J., Green J. J., A triple-fluorophore-labeled nucleic acid pH nanosensor to investigate non-viral gene delivery. Mol. Ther. 25, 1697–1709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rui Y., Wilson D. R., Sanders K., Green J. J., Reducible branched ester-amine quadpolymers (rBEAQs) codelivering plasmid DNA and RNA oligonucleotides enable CRISPR/Cas9 genome editing. ACS Appl. Mater. Interfaces 11, 10472–10480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Table S1