Abstract
The plant-specific VQ gene family participates in diverse physiological processes but little information is available on their role in leaf senescence. Here, we show that the VQ motif-containing proteins, Arabidopsis SIGMA FACTOR BINDING PROTEIN1 (SIB1) and SIB2 are negative regulators of abscisic acid (ABA)-mediated leaf senescence. Loss of SIB1 and SIB2 function resulted in increased sensitivity of ABA-induced leaf senescence. In contrast, overexpression of SIB1 significantly delayed this process. Moreover, biochemical studies revealed that SIBs interact with WRKY75 transcription factor. Loss of WRKY75 function decreased sensitivity to ABA-induced leaf senescence, while overexpression of WRKY75 significantly accelerated this process. Chromatin immunoprecipitation assays revealed that WRKY75 directly binds to the promoters of GOLDEN 2-LIKE1(GLK1) and GLK2, to repress their expression. SIBs repress the transcriptional function of WRKY75 and negatively regulate ABA-induced leaf senescence in a WRKY75-dependent manner. In contrast, WRKY75 positively modulates ABA-mediated leaf senescence in a GLK-dependent manner. In addition, SIBs inhibit WRKY75 function in ABA-mediated seed germination. These results demonstrate that SIBs can form a complex with WRKY75 to regulate ABA-mediated leaf senescence and seed germination.
Keywords: Abscisic acid, GOLDEN 2-LIKE1/2, leaf senescence, seed germination, SIGMA FACTOR BINDING PROTEIN, WRKY75
SIBs function as repressors of WRKY75 and regulate expression of GLKs in ABA-mediated leaf senescence and seed germination.
Introduction
Leaf senescence constitutes the last stage of plant development and is an evolutionarily selected developmental process that is controlled by a highly regulated genetic network. Numerous studies have demonstrated that leaf senescence is critical for plant growth and also increases reproductive success and fitness, because it enables the relocation of mobilizable nutrient and energy from aging leaves to reproducing seeds (Lim et al., 2007). Thus, plant senescence represents an important adaptive mechanism that plants use to increase their survival and fitness in their given ecological niches. Senescence is initiated in an age-dependent manner and is also triggered by environmental signals and various phytohormones, including abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA), ethylene (ET), gibberellin (GA) and brassinosteroids, but inhibited by cytokinins and auxin (Lim et al.,2007; Chen et al., 2017).
ABA is known as an important regulator that mediates both plant growth and response to diverse environmental stresses (Fujii and Zhu, 2009). It coordinates a sophisticated gene regulatory network to enable plants to respond properly to both developmental and environmental signals (Hubbard et al., 2010; Chen et al., 2020). Three protein classes have been identified as major components that form a central signaling module to govern ABA signal transduction (Hubbard et al., 2010). These components are composed of the membrane-localized receptors PYRABACTIN RESISTANCE (PYR)/PYR1-LIKE (PYL)/REGULATORY COMPONENT OF ABSCISIC ACID RECEPTOR (RCAR), protein phosphatases type 2Cs (PP2Cs) and SNF1-related kinases 2 (SnRK2s). Under normal conditions, PP2Cs interact with, and dephosphorylate SnRK2s, causing their reduction of catalytic activities (Umezawa et al., 2009; Vladet al., 2009; Dupeux et al., 2011). A raise in ABA concentrations results in PYR/PYL/RCAR receptor-mediated repression of PP2C activity, leading to the activation of SnRK2s, which then phosphorylate downstream transcription factors (TFs) and ultimately activate ABA signaling (Umezawa et al., 2009; Vlad et al., 2009; Dupeux et al., 2011). Interestingly, one recent study revealed that the GARP family of transcription factors GOLDEN 2-LIKE1 (GLK1) and GLK2 can form a transcription module with WRKY40 to suppress ABSCISIC ACID INSENSITIVE 5 (ABI5) expression upon activation via the ABA signaling components PYL/PYRs-PP2Cs-SnRKs, to finally modulate the ABA response (Ahmad et al., 2019). Although numerous studies have demonstrated that ABA plays an important role in diverse physiological processes, such as leaf senescence and seed germination, the underlying mechanisms involved remain to be further investigated.
Recently, a class of plant-specific transcriptional regulators with a short conserved VQ motif (FxxhVQxhTG) was identified and designated as VQ proteins (Jing and Lin, 2015; Yuan et al., 2021). Several studies have demonstrated that the VQ motif has a great impact on the actions or functions of VQ proteins. For instance, the VQ motif has been demonstrated to be essential for both protein–protein interactions and protein sub-cellular localization, and modification of the VQ motif can also change the transcriptional activities of VQ proteins (Jing and Lin, 2015; Jiang and Yu, 2016; Yuan et al., 2021). As a family of transcriptional regulators, VQ proteins often work in concert with their interacting partners to fine-tune the complex regulatory networks that mediate plant growth and stress responses. For example, the structurally related VQ proteins, SIGMA FACTOR BINDING PROTEIN1 (SIB1) and SIB2, function as transcriptional activators of WRKY33 to modulate plant defense against Botrytis cinerea (Lai et al., 2011). In contrast, VQ9 functions with WRKY8 to modulate the plant salt stress response (Hu et al., 2013). The VQ protein IKU1 (VQ14) interacts with MINI3 (WRKY10) to regulate endosperm development and seed size (Wang et al., 2010). Besides WRKY transcription factors, VQ proteins also interact with their other interacting partners, such as PHYTOCHROME INTERACTING FACTOR 4 (PIF4), ABI5, mitogen-activated protein kinases (MAPKs), and RING-type E3 ubiquitin ligase, to coordinate diverse physiological processes (Li et al., 2014; Pecher et al., 2014; Pan et al., 2018; Ali et al., 2019). Although several VQ proteins have been functionally characterized, the biological roles of specific VQ proteins under given conditions are largely unknown. Until now, there has been no report about their involvement in leaf senescence, and thus it is worthwhile to investigate their biological significance in this process.
As a class of important interacting partners of VQ proteins, the WRKY transcription factor family has been shown to form integral parts of the complex signaling networks to regulate both plant growth and stress responses (Rushton et al., 2010; Chen et al., 2012;2017; Guo et al., 2017; Zhang et al., 2018). Recent studies have provided evidence to show that WRKY proteins often interact with important proteins of various phytohormone signaling pathways to regulate diverse physiological processes. For example, WRKY57 can form a complex with both repressors of the JA and auxin signaling pathways, including JASMONATE ZIM-DOMAIN4/8 (JAZ4/8) and the AUX/IAA protein IAA29, and thus function as a node of convergence for JA- and auxin-mediated signaling pathways in JA-induced leaf senescence (Jiang et al., 2014). WRKY45 physically associates with a repressor of the GA signaling pathway, such as the DELLA protein RGA-LIKE1 (RGL1), to positively regulate leaf senescence (Chen et al., 2017). In addition, WRKY12/13 and WKKY75 also participate in GA-mediated floral initiation via interaction with DELLAs (Li et al., 2016; Zhang et al., 2018). Furthermore, WRKY proteins also act as critical components of various phytohormone-mediated signaling pathways to mediate various plant processes. For instance, the structurally related WRKY proteins, including WRKY18/40/60, directly bind to the promoter of several ABA-responsive genes, such as ABI4 and ABI5, to regulate ABA-mediated seed germination(Shang et al., 2010).Similarly, WRKY75 can directly associate with the promoters of SA INDUCTION-DEFICIENT2 (SID2) and ETHYLENE RESPONSIVE FACTOR 59 (ORA59), to regulate SA-promoted leaf senescence. and JA-mediated plant defense to necrotrophic fungal pathogens, respectively (Guo et al., 2017; Chen et al., 2021). However, it is still unclear whether WRKY proteins can function together with their interacting partners, such as VQ proteins, to participate in modulation of ABA-mediated leaf senescence or seed germination.
In this study, in order to explore the possible roles of VQ-WRKY complexes in ABA-mediated leaf senescence or seed germination, we used both molecular and genetic approaches to investigate the physiological effects between SIBs and WRKY75 in these processes. Our results indicated that SIBs function as repressors of WRKY75, and regulate expression of GLKs in ABA-mediated leaf senescence and seed germination.
Materials and methods
Plant growth conditions and materials
Plants used in this study were derived from Arabidopsis Col-0 ecotype. Seeds were surface sterilized with 20% bleach for 15 min and sown on half-strength Murashige and Skoog (MS) media for 3 d at 4 °C. Plants were transferred to soil 7 d after germination and were grown in a greenhouse at 22 °C under a 16h light/8h dark photoperiod. N. benthamiana were grown in a green house at 25 °C under a 16 h light/8 h dark photoperiod. The wrky75-1(SALK_101367), wrky75-25, sib1-4(SM_3.30596), sib2-1 (SM_3.16236), WRKY75:YFP-WRKY75:3’-WRKY75, and 35S:WRKY75-L3 have been described in previous studies(Rishmawi et al., 2014; Zhang et al., 2018; Lv et al., 2019).To generate SIB1 overexpression transgenic plants, the full-length cDNA of SIB1 was cloned into the binary vector pOCA30 in the sense orientation behind a CaMV35S promoter. Taq DNA polymerase was purchased from Takara Biotechnology Co. Ltd (Japan), ABA was purchased from Sigma Co. Ltd (USA) and other chemicals were purchased from Shanghai Sangon Biotechnology Co. Ltd (China).
Expression analysis
ABA was dissolved in 90 µl of ethanol, and water was added to obtain a 10mM stock solution. The ABA stock solution was diluted to 100 µM with distilled water and sprayed onto plants. Water was sprayed onto plants as a control. Total RNA was extracted using Trizol reagent (Invitrogen, USA) from leaves of different ages, or leaves treated with or without 100 µM ABA. About 1 μg of DNase-treated RNA was used for complementary DNA (cDNA) synthesis using M-MLV reverse transcriptase (TaKaRa, Japan), followed by PCR on a Roche LightCycler 480 real-time PCR machine using SYBR Premix Ex Taq™ II (Roche, Mannheim, Germany). ACTIN2 (AT3G18780) and UBQ5 (AT3G62250) were used as internal controls in quantitative RT–PCR. Analysis was conducted following the minimum information for publication of quantitative Real-Time PCR experiments guidelines (Bustin et al., 2009). The gene-specific primers used for qRT-PCR are listed in Supplementary Table S1.
Assays of ABA-induced senescence
The fifth and sixth rosette leaves from 4-week-old plants were placed onto Petri dishes filled with distilled water supplemented with or without 100 µM ABA, and then the plates were kept under weak light (20 µmolm-2s-1photosynthetic photon flux density)for 0, 4 or 5 d at 22 °C. Three-week-old plants grown in soil were sprayed with or without 100 µM ABA and placed under weak light (20 µmolm-2s-1 photosynthetic photon flux density) for 3 dat 22 °C.
Chlorophyll of detached leaves was extracted with 80% acetone, according to Lichtenthaler (1987). Cell death rate was detected by Trypan blue staining; the leaves of indicated genotypes treated with or without 100 µM ABA were soaked in 0.05% Trypan blue solution, kept at80 °C for 2 min, and then cleared with chloral hydrate.
Measurement of germination and greening rates
Seeds were stratified for 3 d at 4 °C. Germination was determined based on the appearance of the embryonic axis (i.e. radicle protrusion), as observed under a microscope (Olympus, Japan). Seedling greening was determined based on the appearance of green cotyledons in a seedling. To measure the ABA sensitivity of germination and greening, seeds were plated on half-strength MS supplemented with different concentrations of ABA(0, 0.5, 0.75, or 1µM). Three independent experiments were conducted, and similar results were obtained.
Yeast two-hybrid screening and confirmation
The coding sequences of full-length WRKY75 (from 35S:WRKY75 construct; Zhang et al., 2018 ) and its derivatives were cloned into bait vector pGBKT7 (Clontech, USA), which was transformed into the yeast strain Y2H Gold (Clontech, USA). Two-hybrid screening was performed according to the mating protocol described in the Clontech Matchmaker TM Gold Yeast Two-Hybrid user manual. To confirm protein-protein interactions, the coding sequences of full-length SIB1or SIB2 which were amplified from a senescence-associated cDNA library and their derivatives were fused to the prey vector pGADT7. The primers used for Y2H screening are shown in Supplementary Table S1.
LUC complementation imaging assays
The full-length WRKY75 CDS was cloned intopCAMBIA1300-nLUC, and the full-length SIB1 or SIB2 CDS were cloned into pCAMBIA1300-cLUC. All plasmids were introduced into Agrobacterium tumefaciens strainEHA105,and then infiltration of N. benthamiana leaves was performed as described in Chen et al. (2008). Infected leaves were analyzed 72 h after infiltration under a low-light cooled CCD imaging apparatus (Tanon-5200, China) to acquire the LUC images. Before luminescence detection, the leaves were sprayed with 0.5 mM fluorescein and kept in the dark for 5 min. The primers used for LUC complementation imaging assays (LCI) are shown in Supplementary Table S1.
Pull-down and electrophoretic mobility shift assay (EMSA)
The full-length SIB1 and WRKY75 CDS were cloned intopGEX-4T-1 (Zhang et al., 2018), and the full-length WRKY75 CDS was cloned into pET-28a(+). All plasmids were introduced into Escherichia coli BL21 cells, and Glutathione S-transferase (GST), GST-SIB1, and His-WRKY75 protein expression was induced by 0.5 mM isopropyl-b-thiogalactopyranoside for 24 h at 16 °C. Soluble GST and GST-SIB1 were extracted and immobilized to glutathione beads (Thermo Fisher Scientific, USA). His-WRKY75 fusion protein from E. coli cell lysate was co-incubated with the immobilized GST and GST–SIB1 fusion proteins for 4 h at 4 °C. Proteins were eluted with elution buffer, and then western blot was used to determine the interaction between WRKY75 and SIB1. The purified GST-WRKY75 protein was confirmed by SDS-PAGE and used for EMSA. EMSA was performed using a Chemiluminescent EMSA Kit (Beyotime, China).The probes were synthesized and labeled with biotin at the Beijing Genomics Institute.
GUS staining and determination of YFP fluorescence
GUS staining was performed as described by Chen et al.(2021). Seeds or 10-day-old seedlings of WRKY75p:GUS transgenic plants floated on water or 100 µM ABA were used as materials for GUS staining. Chlorophyll was removed using several changes of 70% (v/v) ethanol, and the tissues were subsequently photographed. Furthermore, roots of WRKY75:YFP-WRKY75:3’-WRKY75 plants were treated with water or ABA, and then YFP fluorescence was observed under a confocal laser scanning microscope (Olympus, Japan; Rishmawi et al., 2014).
Chromatin immunoprecipitation assays
Two-week-old and ABA-pre-treated seedlings of Col-0andMyc-WRKY75 were harvested for chromatin immunoprecipitation(ChIP) assays, as described previously (Saleh et al., 2008).The Myc antibody was used to immunoprecipitate the protein-DNA complex, and the precipitated DNA was purified using a PCR purification kit for qRT–PCR analysis. The ChIP experiments were performed three times. Chromatin precipitated without antibody was used as the negative control, whereas the isolated chromatin before precipitation was used as the input control. ChIP results are presented as a percentage of input DNA. The primers used for qRT–PCR amplification of different promoters are listed in Supplementary Table S1.
Transcriptional activity assays
The native promoter (about 2.6kb)of GLK1 was inserted into pGWB35 to generate a GLK1p:LUC reporter construct using Gateway technology (Invitrogen, USA).The reporter plasmid and the constructs containing 35S:WRKY75, 35S:SIB1 and 35S:SIB2 were transformed into A. tumefaciens strain EHA105. The strains were incubated overnight in Luria-Bertani medium and resuspended in infiltration buffer (10 mM MES, 0.2 mM acetosyringone, and 10 mM MgCl2) at an optical density at OD600=1. Equal amounts of different combined bacterial suspensions were infiltrated into the 5-week-old N. benthamiana leaves using a needleless syringe. After infiltration, the LUC images were acquired under a low-light cooled CCD imaging apparatus (Tanon-5200, China). Experiments were performed with three independent biological replicates, and generated similar results.
Statistical analysis
Statistical analysis between samples were performed by Student’s t-test or analysis of variance (ANOVA). Sample differences were considered to be statistically significant (indicated with * or different letters) if P<0.05.
Results
SIBs negatively modulate ABA-mediated leaf senescence and seed germination
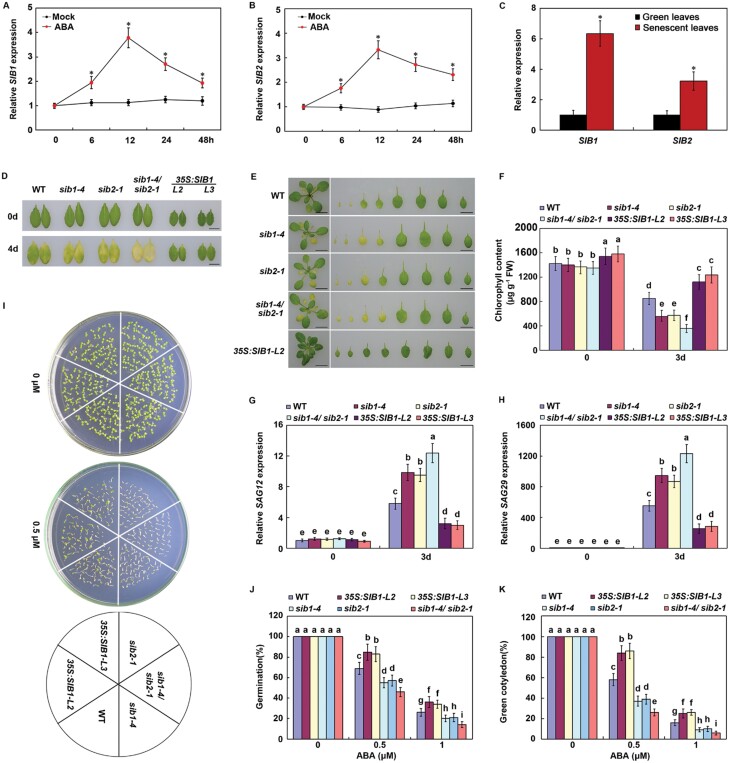
SIB1 and SIB2 were previously found to function as important regulators in JA-mediated defense against necrotrophic pathogens and SA-primed cell death (Lai et al., 2011; Li et al., 2020). To investigate the possible involvement of SIBs in ABA-mediated responses, we first examined the inducibility and temporal kinetics of both SIB1 and SIB2 expression upon ABA treatment. As shown in Fig. 1A, B, expression of both SIB1 and SIB2 was induced by ABA. Further expression analysis showed that SIB1 and SIB2 also have strong expression in senescent leaves (Fig. 1C). Thus, these results imply that SIB1 and SIB2 may play a role in ABA-induced leaf senescence. Next, their single and double mutants were used to investigate their function in ABA-induced leaf senescence (Lai et al., 2011; Lv et al., 2019; Li et al., 2020). Both the detached leaves and the whole plants of the wild type, sibs single and double mutants, or SIB1 overexpression lines (Supplementary Fig. S1), were used for ABA-induced leaf senescence assays. Upon ABA treatment, the sib1 and sib2 single mutants showed more serious yellowing, which is a typical characteristic of leaf senescence, when compared with wildtype (WT; Fig. 1D, E). Interestingly, the sib1/sib2 double mutant further showed more serious yellowing than the single mutants, implying that SIBs may function redundantly in ABA-induced leaf senescence (Fig. 1D, E). Consistent with these findings, chlorophyll content was lower, and expression of representative SENESCENCE ASSOCIATED GENES (SAGs; e.g. SAG12 and SAG29) was stronger in sib mutants than in WT plants (Fig. 1F–H). In contrast, 35S:SIB1 transgenic plants (35S:SIB1-L2 and35S:SIB1-L3) showed remarkably delayed leaf senescence upon ABA treatment, accompanied with higher chlorophyll content, but lower SAG expression, compared with WT (Fig. 1D–H; Supplementary Fig. S1). Together, these observations suggest that SIBs negatively modulate ABA-induced leaf senescence.
Fig. 1.
SIB1/2 negatively modulate ABA-induced leaf senescence and seed germination.(A, B). qRT–PCR analysis of SIB1 and SIB2 transcript levels in 4-week-old WT leaves upon 100 µM ABA treatment. (C) qRT–PCR analysis of SIB1 and SIB2 transcript levels in green and senescent leaves. For A-C, transcript levels of SIB1 and SIB2 in untreated or non-senescent leaves were arbitrarily set to 1. ACTIN2 and UBQ5 were used as internal controls. Error bars represent ±SD from three independent biological replicates. *P<0.05, Student’s t-test compared with mock or green leaves. (D) Senescence phenotypes of the 4-week-old detached leaves of the indicated genotypes treated with or without 100 µM ABA for 4 d. Scale bar =1 cm. (E) Senescence phenotypes of the indicated genotypes treated with or without 100 µM ABA for 4 d. Scale bar =1cm. (F) Chlorophyll content in the indicated genotypes treated with or without 100 µM ABA for 3 d. (G,H) qRT–PCR analysis of SAG12 and SAG29 expression in the leaves of the indicated genotypes treated with or without 100 µM ABA for 3 d. Transcript levels of SAG12 or SAG29 in untreated green leaves were arbitrarily set to 1. ACTIN2and UBQ5 were used as internal controls. (I) Phenotypes of the indicated genotypes grown on half-strength MS medium with 0 or 0.5 µM ABA for 5 d. (J) Germination rates of the indicated genotypes grown on half-strength MS medium with 0, 0.5, or 1 µM ABA for 3 d (K) Cotyledon greening rates of the indicated genotypes grown on half-strength MS medium with 0, 0.5, or 1 µM ABA for 6 d. For F-H, J, and K, error bars represent ±SD from three independent biological replicates. Bars with different letters are significantly different from each other (ANOVA; P<0.05).
Because both SIB1 and SIB2 are induced by ABA, we hypothesized that they may also participate in other ABA-mediated responses, such as seed germination. To confirm our hypothesis, we compared the phenotypes of the wild type, sibs single and double mutants, and SIB1 overexpression lines, in response to ABA during seed germination. As shown in Fig. 1I–K, there were no great differences in phenotypes among WT, sibs single and double mutants, and SIB1 overexpression lines, on half-strength MS medium. Subsequently, we further investigated the phenotype of these seeds after treatment with exogenous ABA. As shown in Fig. 1I–K, the sib1 and sib2 single mutant seeds were more sensitive to ABA compared with WT during seed germination and post-germinative growth. Furthermore, the sib1/sib2 double mutant seeds showed even more sensitivity to ABA compared with their single mutants, implying that SIBs also function redundantly in ABA-mediated seed germination and post-germinative growth (Fig. 1I–K). In contrast, the seeds of 35S:SIB1 transgenic plants showed much higher germination and greening cotyledons than the WT (Fig. 1I–K). Thus, our results support the notion that SIBs function as negative regulators in ABA-mediated seed germination and early seedling growth.
Physical interaction between SIBs and WRKY75
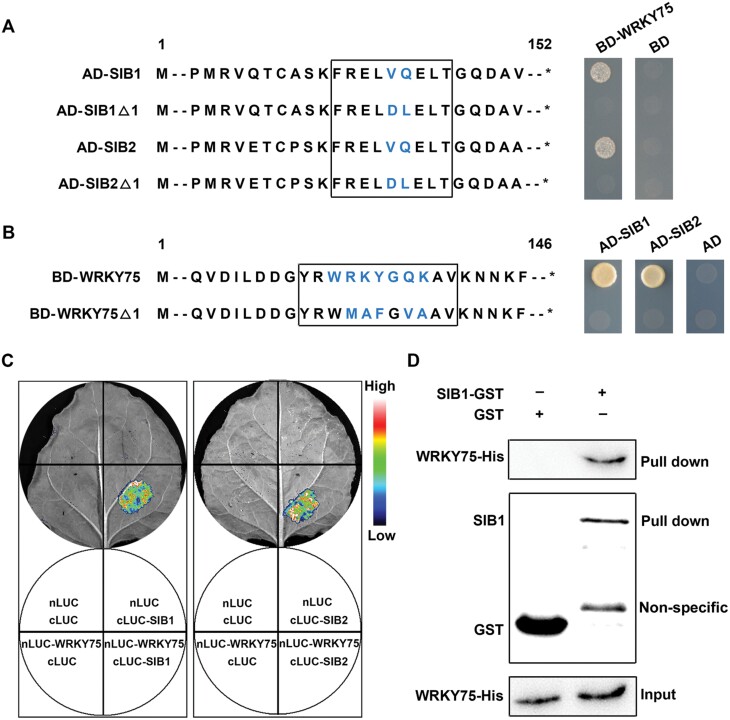
During our screening of potential interaction partners of WRKY75using the yeast two-hybrid system (Y2H), we found that WRKY75 can interact with both SIB1 and SIB2 (Chen et al., 2021). Furthermore, the biological significance of their interaction remains to be determined. Interestingly, WRKY75 has been revealed to participate in several physiological processes through JA, GA, or SA pathways (Guo et al., 2017; Zhang et al., 2018; 2021; Chen et al., 2021). Thus, we speculated that WRKY75 may form a complex with SIBs to co-regulate ABA mediated responses. We further confirmed their interaction using Y2H and luciferase complementation imaging (LCI) assay. As shown in Fig. 2A, B, both SIB1 and SIB2 interact with WRKY75, and the WRKYGQK sequence and VQ-motif are essential for their interaction. For the LCI assay, cLUC-SIBs/nLUC-WRKY75, cLUC/nLUC, cLUC-SIBs/nLUC, and cLUC/nLUC-WRKY75 were co-expressed together in Nicotiana benthamiana leaves at the same time. As shown in Fig. 2C, a LUC signal was only detected when cLUC-SIBs/nLUC-WRKY75 were co-injected into the N.benthamiana leaves. We also confirmed their interactions by performing in vitro pull-down assays. The results of the assay showed that the GST-fused SIB1 was able to retain WRKY75-His, whereas GST alone could not (Fig. 2D). Taken together, these results demonstrate that SIBs physically interact with WRKY75 and they may form a complex to co-regulate ABA-mediated leaf senescence and seed germination.
Fig. 2.
SIB1 and SIB2 physically interact with WRKY75. (A) Yeast-two-hybrid assays. The VQ domains of SIB1 and SIB2 are necessary for their interaction with WRKY75. Sequences of full-length and mutated SIB1 or SIB2 were fused to the pGADT7 activation domain (AD, prey), and sequences of full-length WRKY75 were fused to the pGBKT7 binding domain (BD, bait). Interactions were indicated by the ability of yeast cells to grow on selective media lacking Leu, Trp, His, and Ade. The empty pGBKT7 prey vector was used as a negative control. *Represents stop codon. (B) Yeast-two-hybrid assays. The WRKYG domain of WRKY75 is essential for its interaction with SIB1 and SIB2. Sequences of full-length and mutated WRKY75 were fused to the pGBKT7 binding domain (BD, bait), sequences of full-length SIB1 and SIB2 were fused to the pGADT7 activation domain (AD, prey). Interactions were indicated by the ability of yeast cells to grow on selective media lacking Leu, Trp, His, and Ade. The empty pGADT7 prey vector was used as a negative control. (C) LUC complementation imaging (LCI) assay detecting the interaction between WRKY75 and SIB1 or SIB2. Images of N. benthamiana leaves are displayed two days after infiltration. The pseudocolor bar shows the range of luminescence intensity. (D) In vitro pull-down assays. Purified GST or SIB1–GST was incubated with the WRKY75–His protein. GST was used as a negative control.
WRKY75 positively regulates ABA-mediated leaf senescence and seed germination
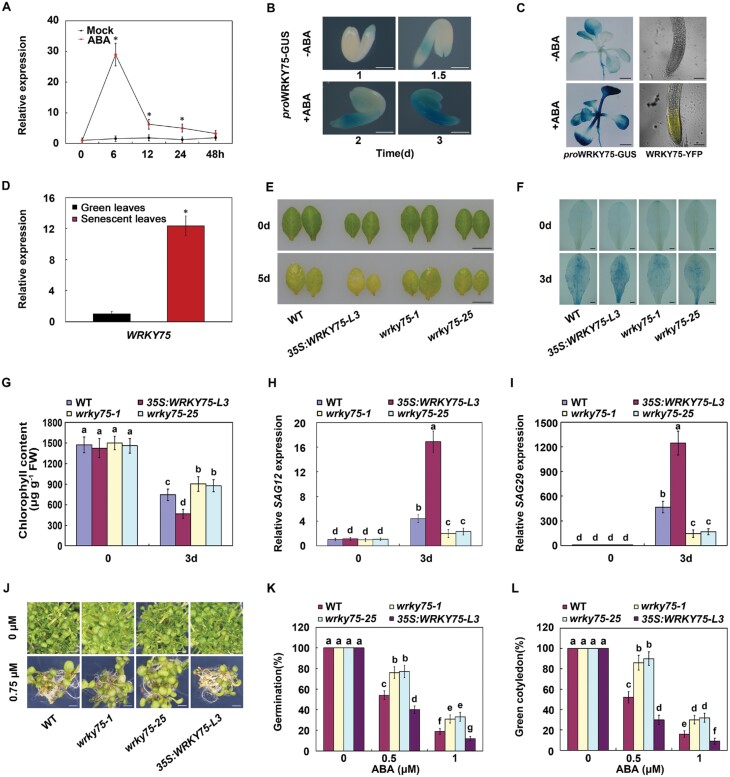
Since SIBs participate in ABA-mediated leaf senescence and seed germination and interact with WRKY75, we speculated that WRKY75 may also participate in ABA responses through interaction with SIBs. To investigate the possible involvement of WRKY75 in ABA-mediated responses, we first determined the induced expression of WRKY75 upon ABA treatment. As shown in Fig. 3A–C, WRKY75 expression was induced by ABA at both mRNA and protein levels. Combined with its strong expression in senescent leaves (Fig. 3D; Guo et al., 2017; Zhang et al., 2021), we deduced that WRKY75 may also play a role in ABA-induced leaf senescence. Therefore, the detached leaves of the WT, wrky75 mutants, or WRKY75 overexpression plants were used for ABA-induced leaf senescence assays. Upon ABA treatment, the wrky75 mutants showed delayed leaf senescence compared with WT plants (Fig. 3E). The mutant plants also displayed decreased cell death, higher chlorophyll content and lower expression of SAGs than WT plants (Fig. 3F–I). In contrast, 35S:WRKY75 transgenic plants showed accelerated leaf senescence upon ABA treatment, accompanied by enhanced cell death and lower chlorophyll content, but higher SAG expression (Fig. 3F–I). Thus, these observations suggest that WRKY75 positively modulates ABA-induced leaf senescence.
Fig. 3.
WRKY75 positively regulates ABA-induced leaf senescence and seed germination. (A) qRT–PCR analysis of WRKY75 transcript levels in 4-week-old WT leaves upon 100 µM ABA treatment . Transcript levels of WRKY75 in untreated leaves were arbitrarily set to 1. ACTIN2 and UBQ5 were used as internal controls. Error bars represent ±SD from three independent biological replicates. *P<0.05, Student’s t-test compared with mock treatment. (B) GUS expression in WRKY75p:GUS transgenic seedlings grown on half-strength MS medium at the indicated times, treated with or without 100 µM ABA. Scale bar =200 µm. (C) Left: GUS staining of 10-d-old WRKY75p:GUS seedlings treated with or without 100 µM ABA for 2 h. Scale bar=1mm; Right: YFP detection of WRKY75 in 5-d-old WRKY75:YFP-WRKY75:3’-WRKY75 seedlings treated with or without ABA for 2 h. Scale bar =100 µm. (D) qRT–PCR analysis of WRKY75 transcript levels in 24-day-old non-senescent and 40-day-old senescent leaves. Transcript levels of WRKY75 in non-senescent leaves were arbitrarily set to 1. ACTIN2and UBQ5 were used as internal controls. Error bars represent ±SD from three independent biological replicates.*P<0.05, Student’s t-test compared with green leaves. (E)Senescence phenotypes of the 4-week-old leaves of the indicated genotypes treated with or without 100 µM ABA for 4 d. Scale bar =1 cm. (F) Trypan blue staining of the indicated genotypes treated with or without ABA for 3 d. Scale bar =2 mm. (G) Chlorophyll content in the indicated genotypes. (H, I) qRT–PCR analysis of SAG12 and SAG29 transcript levels in the indicated genotypes. Transcript levels of WRKY75 in control WT leaves were arbitrarily set to 1. ACTIN2 and UBQ5 were used as internal controls. (J) Phenotypes of the indicated genotypes on half-strength MS medium with 0 or 0.75 µM ABA for 7 d. Scale bar =2 mm. (K) Germination rates of the indicated genotypes on half-strength MS medium with 0, 0.5, or 1 µM ABA treatment for 2 d. (L) Greening rates of the indicated genotypes on half-strength MS medium with 0, 0.5, or 1 µM ABA for 6 d. For G-I, K, and L, error bars represent ±SD from three independent biological replicates. Bars with different letters are significantly different from each other (ANOVA; P<0.05).
Similarly, we also used seeds of WT, wrky75 mutants, and WRKY75 overexpression plants to determine possible participation of WRKY75 in ABA-mediated seed germination. As shown in Fig. 3J–L, there were no great differences among WT, wrky75 mutants, and WRKY75 overexpression plants on half-strength MS medium. We further investigated the phenotype of these seeds after treatment with exogenous ABA. As shown in Fig. 3J–L, the wrky75 mutant seeds were more insensitive to ABA compared with WT during seed germination and post-germinative growth. In contrast, seeds of the 35S:WRKY75 transgenic plants showed much lower germination and greening cotyledons than the WT (Fig. 3J–L). Taken together, our results support the notion that WRKY75 also functions as a positive regulator in ABA-mediated seed germination and post-germinative growth.
WRKY75 binds to the promoters of GLK1 and GLK2 to inhibit gene expression
WRKY transcription factors often perform their biological functions by specially binding to the W-box (T/CTGACC/T), present in the promoters of their target genes (Eulgem et al., 2000; Ulker and Somssich, 2004). Our results indicate that WRKY75 may play a role in ABA-mediated leaf senescence and seed germination by modulating the expression of both senescence and seed germination-associated genes in the ABA signaling pathway. Interestingly, several putative W-box elements were found to be present in promoters ofGLK1 and GLK2. Previous studies have demonstrated that GLKs participate in both ABA-mediated leaf senescence and seed germination processes (Rauf et al., 2013; Ahmad et al., 2019). Thus, we speculated that WRKY75 may directly bind the promoters of GLKs to inhibit their expression, and finally modulate ABA responses.
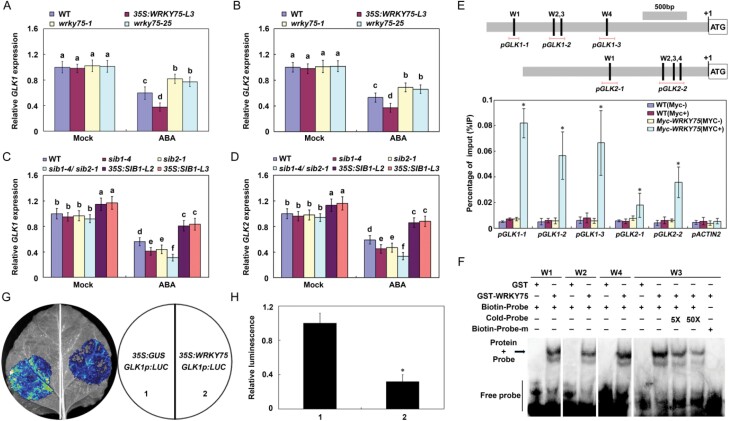
To determine whether GLKs are direct targets of WRKY75, we then compared their expression in WT, wrky75 mutants, and WRKY75 overexpression plants upon ABA treatment. As shown in Fig. 4A, B, the expression of both GLK1 and GLK2 was higher in wrky75 mutants, but was lower in WRKY75 overexpression plants, compared with those in WT. Since SIBs physically interact with WRKY75 and may function together to regulate ABA-mediated leaf senescence and seed germination, we also compared their expression in WT, sib single and double mutants, and SIB1 overexpression lines, upon ABA treatment. Expression of both GLK1 and GLK2 was lower in sib mutants, but was higher in SIB1 overexpression plants compared with those in WT (Fig. 4C, D). Thus, both WRKY75 and SIBs may regulate ABA-mediated responses through GLK1 and GLK2.
Fig. 4.
WRKY75 directly represses the expression of GLK1 and GLK2. (A, B).Transcript levels of GLK1 and GLK2 in 4-week-old WT, WRKY75 mutants and overexpressing plants upon 100µM ABA treatment for 12 h. (C, D)Transcript levels of GLK1 and GLK2 in the WT, sib single or double mutants, and SIB1 overexpressing plants upon 100µM ABA treatment for 12 h. For A-D, error bars represent ±SD from three independent biological replicates. Bars with different letters are significantly different from each other (ANOVA; P<0.05). (E) ChIP-qPCR analysis of the relative binding of WRKY75 to the promoters of GLK1 and GLK2. The promoter structures of both GLK1 and GLK2 and fragment used in the ChIP assay. (Upper) W1, W2, etc. denote each W-box, numbered from left to right with sequence sites relative to the ATG start codon. Red lines indicate the sequences detected by ChIP assays. (Bottom) Real-time RT-PCR results showed that WRKY75 binds to the promoters of GLK1 and GLK2. ChIP assays were performed with chromatin prepared from Myc-WRKY75 plants using an anti-Myc antibody. ChIP results are presented as a percentage of input DNA. Error bars represent ±SD from three independent biological replicates. (F) EMSA of the binding of recombinant WRKY75 proteins to the promoter of GLK1. The oligonucleotides (proGLK1-W1/2/3/4 and proGLK1-W3-m) were used as the probes. Mutated probe indicates a single nucleic acid mutation from TGAC to TAAC.GST-WRKY75, biotin-probe, labeled mutated probe, and unlabeled probe at a 5× and 50× molar excess were present (+) or absent (-) in each reaction. (G,H) Transient transcriptional activity assays in N. benthamiana. A representative leaf image is shown in (G), and the quantification of corresponding relative luminescence intensities was done in (H) by using n=15 independent leaves. Error bars represent ±SD.*P<0.05, Student’s t-test compared with control.
To determine whether GLKs are direct targets of WRKY75, we then conducted in vivo ChIP assays using 35S:Myc-WRKY75 transgenic plants (Zhang et al., 2018). The ChIP-qPCR results revealed that WRKY75 could bind to the promoters of both GLK1 and GLK2 via the W-box sequence (Fig. 4E). Furthermore, we also performed EMSAs with the GST-WRKY75 recombinant protein to determine the in vitro binding of WRKY75 to the GLK1 promoter. As shown in Fig. 4F, WRKY75 could bind all the probes containing W-box sequence (W1, W2, W3, and W4). The binding signals decreased after the addition of unlabeled WT competitors. In contrast, the WRKY75 protein did not bind to the mutant probe carrying a mutated W-box (Fig. 4F). The GST protein alone also did not bind to the GLK1 promoter (Fig. 4F). These data suggest that WRKY75 may directly bind to the promoters of GLKs to modulate ABA responses.
Next, to further confirm the direct regulation of WRKY75 on expression of GLKs, we also performed transient expression assays in N. benthamiana leaves using the GLK1 promoter (2.6 kb) fused with the LUC gene (GLK1p:LUC). Effector plasmids were generated that contained either a WRKY75 or GUS gene driven by the Cauliflower mosaic virus (CaMV) 35S promoter (35S:WRKY75 and 35S:GUS). As shown in Fig. 4G, H, co-expression of the WRKY75 gene with the reporter plasmid resulted in dramatically reduced LUC signals compared with the control. This supports the hypothesis that WRKY75 is a direct transcriptional repressor of GLK1 expression.
ABA-induced leaf senescence delayed by SIBs is WRKY75-dependent
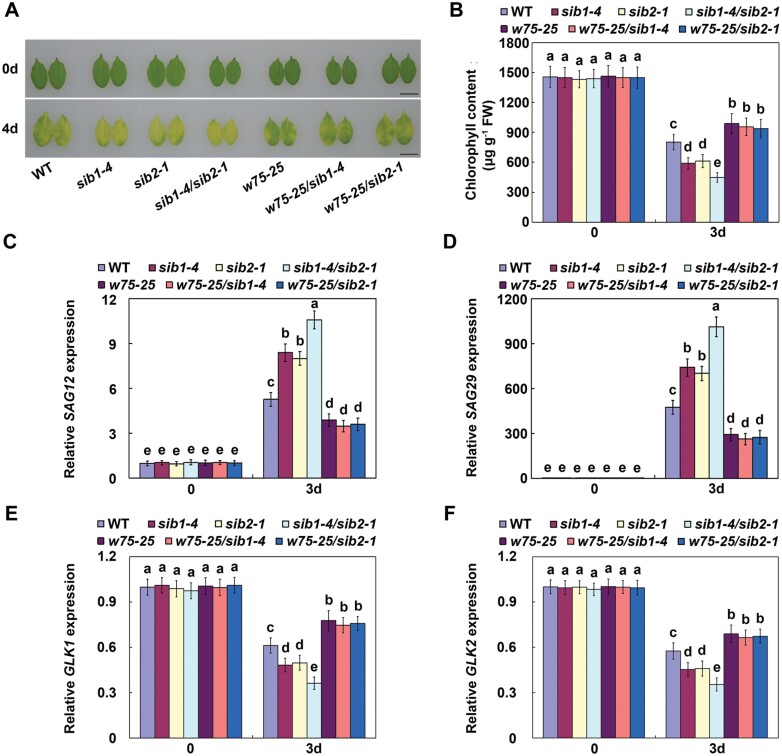
We have shown that SIBs interact with WRKY75, according to both Y2H and LCI assays (Fig. 2), and that SIBs and WRKY75 function in opposite ways in ABA-mediated leaf senescence and seed germination. We therefore deduced that the role of SIBs in ABA responses may be mediated by its interaction with WRKY75. To confirm our speculation, we examined whether the accelerated leaf senescence of sib1 and sib2 mutants is WRKY75-dependent. We then crossed both sib1-4 and sib2-1 mutants with wrky75-25 to produce wrky75-25/sib1-4 and wrky75-25/sib2-1 (Rishmawi et al., 2014). As shown in Fig. 5A, mutation of SIB1 or SIB2 could not accelerate ABA induced leaf senescence of wrky75-25. Consistent with the senescence phenotypes, the chlorophyll content and expression of both SAGs and GLKs were similar between wrky75-25 and wrky75-25/sib1-4 or wrky75-25/sib2-1 (Fig. 5B–F). Thus, ABA-induced leaf senescence delayed by SIBs is WRKY75-dependent.
Fig. 5.
SIB1/2 negatively modulate ABA-induced leaf senescence in a WRKY75-dependent manner. (A) Senescence phenotypes of the indicated genotypes treated with or without 100 µM ABA for 4 d. Scale bar =1 cm. (B) Chlorophyll content in the indicated genotypes treated with or without ABA for 3 d.(C-F) qRT–PCR analysis of SAG12, SAG29, GLK1, and GLK2 transcript levels in the indicated genotypes treated with or without ABA for 3 d. Transcript levels of SAG12, SAG29,GLK1 and GLK2 in non-treated WT leaves were arbitrarily set to 1. ACTIN2 and UBQ5 were used as internal controls. For B-F, error bars represent ±SD from three independent biological replicates. Bars with different letters are significantly different from each other (ANOVA; P<0.05).
SIB1 and SIB2 are negative interactors of WRKY75
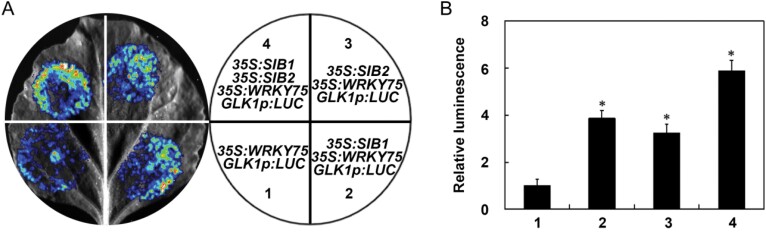
Previous studies have revealed that VQ proteins themselves do not bind DNA directly but form complexes with several types of transcription factors to either promote or suppress their transcriptional activity, and finally exert their regulatory effects (Lai et al., 2011; Li et al., 2014; Jing and Lin, 2015; Lei et al., 2017). Having demonstrated that SIB1 and SIB2 physically and genetically interact with WRKY75, we speculated that they might affect the transcriptional function of WRKY75. To test this possibility, we used the LUC reporter approach to analyze the effects of SIB1 and SIB2 on the transcriptional activity of WRKY75. GLK1p:LUC was again used as a reporter. Effector constructs were generated that contained either a SIB1, SIB2, or WRKY75 gene driven by the CaMV35S promoter (35S:SIB1, 35S:SIB2, and 35S:WRKY75). As shown in Fig. 6, co-expression of SIB1 or SIB2 with WRKY75 resulted in enhanced LUC signals compared with the expression of WRKY75 alone (Fig. 6). More importantly, co-expression of SIB1 and SIB2 with WRKY75 further enhanced the LUC signals compared with the expression of WRKY75 alone, or co-expression of WRKY75 and SIB1 or SIB2. These results support the hypothesis that SIB1 and SIB2 act as negative interactors of WRKY75.
Fig. 6.
SIB1/2 repress the transcriptional activity of WRKY75. Transient transcriptional activity assays in N. benthamiana revealing the repression of GLK1 by WRKY75. A representative leaf image is shown in (A), and the quantification of the corresponding relative luminescence intensities is shown in (B) by using n=15 independent leaves. Error bars represent ±SD.*P<0.05, Student’s t-test compared with control (35S:WRKY75+GLK1p:LUC,1).
WRKY75 promotes both ABA-mediated leaf senescence and seed germination in a GLK-dependent manner
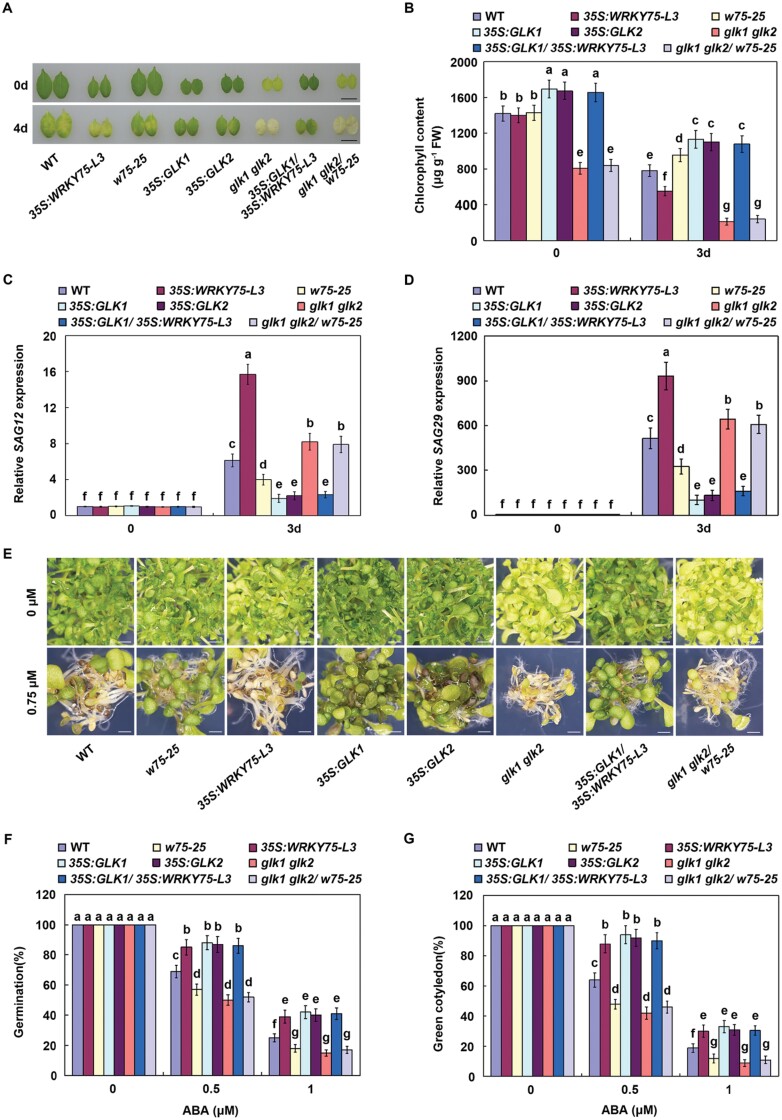
The phenotypic analysis, and biochemical and molecular evidence indicated that WRKY75 positively modulates both ABA-mediated leaf senescence and seed germination through the direct inhibition of GLK expression. To further confirm this conclusion, the genetic relationship between WRKY75 and GLKs was explored. The wrky75-25 mutant was crossed with glk1/glk2 double mutant to produce glk1/glk2/wrky75-25, and 35S:WRKY75-L3 was crossed with 35S:GLK1 to generate 35S:GLK1/35S:WRKY75-L3 (Supplementary Fig. S2). Then both the ABA-mediated leaf senescence and seed germination among these transgenic lines were examined. As shown in Fig. 7A, mutation of WRKY75 could not delay leaf senescence of glk1/glk2, and overexpression of WRKY75 also could not promote leaf senescence of 35S:GLK1. Consistent with these senescence phenotypes, chlorophyll content and SAG expression were similar between glk1/glk2 and glk1/glk2/wrky75-25, or between 35S:GLK1 and 35S:GLK1/35S:WRKY75-L3(Fig. 7B–D). Similarly, mutation of WRKY75 could not promote seed germination and early seedling growth of glk1/glk2, and overexpression of WRKY75 also could not delay seed germination and early seedling growth of 35S:GLK1(Fig. 7E–G). Thus, the genetic analysis indicated that WRKY75 acts upstream of GLKs and functions as a positive regulator of both ABA-mediated leaf senescence and seed germination in a GLK-dependent manner.
Fig. 7.
WRKY75 promotes both ABA-induced leaf senescence and seed germination in a GLK-dependent manner. (A) Senescence phenotypes of the indicated genotypes treated with or without 100 µM ABA for 4 d. Scale bar =1 cm. (B) Chlorophyll content in the indicated genotypes treated with or without 100 µM ABA for 3 d. (C, D) qRT–PCR analysis of SAG12 and SAG29 in the indicated genotypes treated with or without 100 µM ABA for 3 d. Transcript levels of SAG12 or SAG29 in non-treated WT leaves were arbitrarily set to 1. ACTIN2 and UBQ5 were used as internal controls. (E) Phenotypes of the indicated genotypes grown on half-strength MS medium with 0 or 0.5 µM ABA for 5 d. Scale bar =2 mm. (F) Germination rates of the indicated genotypes grown on half-strength MS medium with 0, 0.5, or 1 µM ABA for 3 d. (G) Cotyledon greening rates of the indicated genotypes grown on half-strength MS medium with 0, 0.5, or 1 µM ABA for 6 d. For B-D, F and G, error bars represent ±SD from three independent biological replicates. Bars with different letters are significantly different from each other (ANOVA; P<0.05).
Discussion
Although VQ proteins have been identified in several plants, and substantial progress in their function has been obtained in recent years, our knowledge on their functions and mechanisms of action still remains largely unknown. Previous studies have demonstrated that VQ proteins constitute a class of transcription regulators that do not bind DNA directly but form complexes with transcription factors to fine-tune downstream gene transcription (Jing and Lin, 2015; Yuan et al., 2021). Consequently, VQ proteins function as both positive and negative regulators to modulate plant growth and development, such as endosperm growth, seed size, and light morphogenesis, as well as stress responses, including plant immunity and several abiotic stresses (Yuan et al., 2021). Here, we demonstrate that the VQ proteins SIB1 and SIB2 function as repressors of WRKY75 in ABA-mediated leaf senescence and seed germination.
We found that both SIB1 and SIB2 are expressed in senescing leaves and also induced by ABA; both sib1 and sib2 single mutants and their double mutants exhibit accelerated ABA-induced leaf senescence, while SIB1-overexpressing plants show dramatically delayed ABA-induced leaf senescence compared with WT plants (Fig. 1), indicating that both SIB1 and SIB2act as negative regulators of ABA-mediated leaf senescence. These ABA-induced senescence phenotypes were correlated with the altered expression of several senescence-associated genes, such as SAG12 and SAG29 (Fig. 1). Similar to their role in ABA-mediated leaf senescence, SIBs also function redundantly and negatively in ABA-mediated seed germination. As the plant-specific protein family, the VQ family in Arabidopsis consists of 34 members with functional diversity (Yuan et al., 2021). However, until now, there is little report about their involvement in the plant senescence response. Our results thus shed new insights into the importance of VQ proteins, especially in the plant senescence response.
As a class of transcription regulators, VQ proteins often interact with transcription factors to fine-tune the regulatory machinery associated with plant growth and development, and also response to diverse environmental stresses (Jing and Lin, 2015). Previous studies have provided evidence to show that VQ-WRKY interaction represents one important mechanism of action of the VQ proteins. Both SIB1 and SIB2 interact with WRKY33 to positively regulate plant defense against necrotrophic pathogens (Lai et al., 2011). VQ20 can form complexes with both WRKY2 and WRKY34 to positively regulate pollen development (Lei et al., 2017). In contrast, VQ9 physically interacts with WRKY8 to function antagonistically in the regulation of salt stress response (Hu et al., 2013). Thus, VQ proteins can function as both activators and inhibitors of WRKY transcription factors, and they form complexes to fine-tune the complex regulatory networks. In this study, we also identified SIB1 and SIB2 as two interacting partners of WRKY75 using both in vivo and in vitro biochemical analyses. Expression analysis revealed that WRKY75 is strongly induced by ABA at both mRNA and protein levels and is strongly expressed in senescing leaves, indicating that WRKY75 may also be involved in ABA-induced leaf senescence (Fig. 3). In contrast with SIBs, wrky75 mutant plants show delayed ABA-induced leaf senescence, while WRKY75-overexpressing plants exhibit dramatically accelerated ABA-induced leaf senescence, when compared with the WT plants (Fig. 3). Similarly, WRKY75 also has a positive role in ABA-mediated seed germination(Fig. 3). Thus SIBs may form a complex with WRKY75 to modulate ABA-mediated leaf senescence and seed germination.
Plant senescence is precisely controlled by complex senescence-associated transcriptional regulatory networks, in which numerous transcription factors participate. Interestingly, WRKY proteins were identified as the second largest group of transcription factors in the Arabidopsis senescence transcriptome (Guo et al., 2004), and accordingly, several WRKY members have been reported to participate in plant senescence regulation, including WRKY6 (Robatzek and Somssich, 2002), WRKY22 (Zhou et al., 2011), WRKY53 (Miao and Zentgraf, 2007), WRKY54 (Besseau et al., 2012), WRKY57 (Jiang et al., 2014), and WRKY70 (Ulker et al., 2007; Besseau et al., 2012). Recently, WRKY75 was shown to participate in GA-mediated leaf senescence, and also participate in the formation of a tripartite amplification loop together with SA and reactive oxygen species to accelerate leaf senescence (Guo et al., 2017; Zhang et al., 2021). Despite their wide involvement in plant senescence, it is still unclear whether there are certain senescence-associated WRKY members that participate in leaf senescence through the ABA pathway. Here we identified WRKY75 as a critical component in ABA-induced leaf senescence.
Despite their functional diversity, WRKY proteins were found to perform their biological functions by directly binding to the W-box (TTGACC/T) present in their target promoters. Interestingly, we here revealed that WRKY75 can directly target GLKs through the W-boxes in their promoters, and subsequently regulate both ABA-mediated leaf senescence and seed germination (Fig. 4). Because of the contrasting expression pattern of both WRKY75 and GLKs in WRKY75 mutants and overexpression plants, and the reduced LUC signal in transient expression assays (Fig. 4), we deduced that WRKY75 functions as a negative regulator of GLKs. Genetic analysis further revealed that WRKY75 acts upstream of GLKs and participates in ABA-mediated leaf senescence and seed germination in a GLK-dependent manner (Fig. 7). Previously, WRKY75 was also demonstrated to directly activate several senescence-associated genes and SA INDUCTIONDEFICIENT 2 (SID2), but repress CATALASE2 (CAT2) during leaf senescence (Guo et al., 2017; Zhang et al., 2021). Thus, WRKY75 functions as an activator as well as a repressor to fine-tune leaf senescence.
Consistent with the opposite phenotypes in ABA-mediated leaf senescence and seed germination, we also observed opposite expression of GLKs between SIBs and WRKY75 mutants or overexpression plants (Fig. 4). Furthermore, SIBs can repress the transcriptional inhibitory effect of WRKY75 on GLK1 expression and also function by depending on WRKY75 (Figs 5;6). SIBs thus may form a complex with WRKY75 and function to maintain the appropriate ABA signaling level, finally modulating ABA-associated responses. Previous studies have demonstrated that VQs interact with diverse transcription factors to affect their DNA-binding activity or transcriptional activation/inhibition ability. For example, SIBs enhanced the DNA-binding activity of WRKY33, while VQ9 reduced the DNA-binding activity of WRKY8 (Lai et al., 2011; Hu et al., 2013). Both VQ18 and VQ26 repressed the transcriptional activation ability of ABI5, while VQ20 promoted the transcriptional inhibition ability of WRKY2 and WRKY34 (Lei et al., 2017; Pan et al., 2018). Furthermore, SIBs function in B. cinerea defense responses in a WRKY33-dependent manner (Lai et al., 2011). Thus, VQs perform their regulatory roles through interaction with diverse transcription factors, and function synergistically or antagonistically to fine-tune the complex regulatory networks.
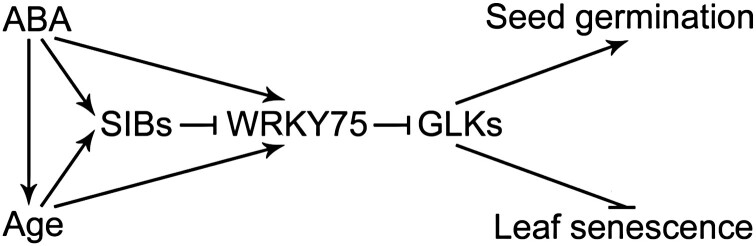
Our results reveal the molecular mechanisms underlying the regulation of ABA-mediated leaf senescence and seed germination by the SIB-WRKY75 complex. They demonstrated that both SIBs and WRKY75 function as novel components of an ABA-mediated regulatory network, and they physically interact and function to fine-tune the ABA-mediated responses. Finally, a working model for the function of SIB-WRKY75 complex in ABA-mediated responses was proposed (Fig. 8). Both SIBs and WRKY75 are up-regulated during leaf senescence and induced by ABA, and the proteins form a complex to regulate GLK expression during ABA-mediated leaf senescence and seed germination. Given that SIBs show slightly altered expression in ABA signaling mutants (e.g. abi4 and abi5;Supplementary Fig. S3), SIB1-overexpressing plants still show ABA-promoted senescence (Fig. 1), and wrky75 mutants and GLK-overexpressing plants also show ABA-dependent senescence (Fig. 2), we speculate that alternative ABA mechanisms may also participate in ABA-mediated leaf senescence. It will be interesting to further investigate how transcription and translation of SIBs or WRKY75 are regulated upon ABA stimuli, and how they form protein complexes during this process. It will also be necessary to determine whether there exist other VQ members that participate in the senescence process. Further systematic analysis of the biological significance of VQ-WRKY interaction will add to our understanding of the involvement of VQ-WRKY complexes in diverse biological processes.
Fig. 8.
Model for SIB-WRKY75interaction in ABA-mediated leaf senescence and seed germination. Both SIBs and WRKY75 are up-regulated during leaf senescence and induced by ABA. The ABA pathway generally promotes leaf senescence in Arabidopsis. SIBs interact with WRKY75 to inhibit its transcriptional function. GLKs negatively modulate ABA-mediated leaf senescence but positively modulate ABA-mediated seed germination. WRKY75 directly binds to the promoters of GLKs to repress their expression during ABA-mediated leaf senescence and seed germination.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Confirmation of SIB1 overexpressing plants.
Fig. S2. Confirmation of GLK1, GLK2, and WRKY75 overexpressing plants.
Fig. S3.Identification of SIBs in ABA synthesis or signaling mutants.
Table S1. Primers used in this study.
Acknowledgements
We thank Chanhong Kim, Yijun Qi, Dong-Tao Ren, and Martin Hülskamp for sharing research materials.
Author contributions
LGC and HYZ planned and designed the research; HYZ, LPZ, YRJ, YFJ, LXL, YLC, RLW, and HMZ performed experiments; LGC and HYZ analyzed data; LGC and HYZ wrote the manuscript.
Conflict of interest
The authors declare no competing interests.
Funding
This work was supported by the Yunnan Fundamental Research Projects(2019FA010 and 2019FB029), Natural Science Foundation of China(31671275), and Strategic Leading Science & Technology Programme (XDA24030301) of CAS.
Data availability
Sequence data from this article can be found in the GenBank/EMBL libraries (https://www.ncbi.nlm.nih.gov/) under the following accession numbers: SIB1 (At3G56710), SIB2 (At2G41180), WRKY75 (AT5G13080), GLK1 (AT2G20570), GLK2 (AT5G44190), SAG12 (AT5G45890), SAG29 (AT5G13170), ACTIN2 (AT3G18780), and UBQ5 (AT3G62250).
References
- Ahmad R, Liu Y, Wang TJ, Meng Q, Yin H, Wang X, Wu Y, Nan N, Liu B, Xu ZY. 2019. GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiology 179, 1844–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MRM, Uemura T, Ramadan A, Adachi K, Nemoto K, Nozawa A, Hoshino R, Abe H, Sawasaki T, Arimura GI. 2019. The ring-type E3 ubiquitin ligase JUL1 targets the VQ-motif protein JAV1 to coordinate jasmonate signaling. Plant Physiology 179, 1273–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Li J, Palva ET. 2012. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany 63, 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y. 2020. Abscisic acid dynamics, signaling, and functions in plants. Journal of Integrative Plant Biology 62, 25–54. [DOI] [PubMed] [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D. 2012. The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta 1819, 120–128. [DOI] [PubMed] [Google Scholar]
- Chen L, Xiang S, Chen Y, Li D, Yu D. 2017. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Molecular Plant 10, 1174–1189. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang L, Xiang S, Chen Y, Zhang H, Yu D. 2021. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. Journal of Experimental Botany 72, 1473–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. 2008.Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiology 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Antoni R, Betz K, et al. 2011. Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiology 156, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. 2000. The WRKY superfamily of plant transcription factors. Trends in Plant Science 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. 2009. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceedings of the National Academy of Sciences, USA 106, 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S. 2004. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environment 27, 521–549. [Google Scholar]
- Guo P, Li Z, Huang P, Li B, Fang S, Chu J, Guo H. 2017. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. The Plant Cell 29, 2854–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D. 2013. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. The Plant Journal 74, 730–745. [DOI] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. 2010. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes & Development 24, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D. 2014. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. The Plant Cell 26, 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Yu D. 2016. The WRKY57 transcription factor affects the expression of jasmonate ZIM-domain genes transcriptionally to compromise Botrytis cinerea resistance. Plant Physiology 171, 2771–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Lin R. 2015. The VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiology 169, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z. 2011. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. The Plant Cell 23, 3824–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei R, Li X, Ma Z, Lv Y, Hu Y, Yu D. 2017. Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function. The Plant Journal 91, 962–976. [DOI] [PubMed] [Google Scholar]
- Li Z, Dogra V, Lee KP, Li R, Li M, Li M, Kim C. 2020. N-terminal acetylation stabilizes SIGMA FACTOR BINDING PROTEIN1 involved in salicylic acid-primed cell death. Plant Physiology 183, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jing Y, Li J, Xu G, Lin R. 2014. Arabidopsis VQ MOTIF-CONTAINING PROTEIN29 represses seedling deetiolation by interacting with PHYTOCHROME-INTERACTING FACTOR1. Plant Physiology 164, 2068–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang H, Yu D. 2016. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Molecular Plant 9, 1492–1503. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. 1987. Chlorophylls and carotenoids-pigments of photosynthetic biomembranes. Methods in Enzymology 148, 350–382. [Google Scholar]
- Lim PO, Kim HJ, Nam HG. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136. [DOI] [PubMed] [Google Scholar]
- Lv R, Li Z, Li M, Dogra V, Lv S, Liu R, Lee KP, Kim C. 2019. Uncoupled expression of nuclear and plastid photosynthesis-associated genes contributes to cell death in a lesion mimic mutant. The Plant Cell 31, 210–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. 2007. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. The Plant Cell 19, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Wang H, Hu Y, Yu D. 2018. Arabidopsis VQ18 and VQ26 proteins interact with ABI5 transcription factor to negatively modulate ABA response during seed germination. The Plant Journal 95, 529–544. [DOI] [PubMed] [Google Scholar]
- Pecher P, Eschen-Lippold L, Herklotz S, Kuhle K, Naumann K, Bethke G, Uhrig J, Weyhe M, Scheel D, Lee J. 2014. The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of ‘VQ-motif’-containing proteins to regulate immune responses. New Phytologist 203, 592–606. [DOI] [PubMed] [Google Scholar]
- Rauf M, Arif M, Dortay H, Matallana-Ramírez LP, Waters MT, Gil Nam H, Lim PO, Mueller-Roeber B, Balazadeh S. 2013. ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Reports 14, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishmawi L, Pesch M, Juengst C, Schauss AC, Schrader A, Hülskamp M. 2014. Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiology 165, 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. 2002. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes & Development 16, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. 2010. WRKY transcription factors. Trends in Plant Science 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. 2008. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protocols 3, 1018–1025. [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, et al. 2010. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. The Plant Cell 22, 1909–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B, Shahid Mukhtar M, Somssich IE. 2007. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226, 125–137. [DOI] [PubMed] [Google Scholar]
- Ulker B, Somssich IE. 2004. WRKY transcription factors: from DNA binding towards biological function. Current Opinion in Plant Biology 7, 491–498. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. 2009. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. 2009. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. The Plant Cell 21, 3170–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Garcia D, Zhang H, Feng K, Chaudhury A, Berger F, Peacock WJ, Dennis ES, Luo M. 2010. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. The Plant Journal 63, 670–679. [DOI] [PubMed] [Google Scholar]
- Yuan G, Qian Y, Ren Y, Guan Y, Wu X, Ge C, Ding H. 2021. The role of plant-specific VQ motif-containing proteins: an ever-thickening plot. Plant Physiology and Biochemistry 159, 12–16. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen L, Yu D. 2018. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiology 176, 790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang L, Wu S, Chen Y, Yu D, Chen L. 2021. AtWRKY75 positively regulates age-triggered leaf senescence through gibberellin pathway. Plant Diversity. doi: 10.1016/j.pld.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jiang Y, Yu D. 2011.WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Molecules and Cells 31, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from this article can be found in the GenBank/EMBL libraries (https://www.ncbi.nlm.nih.gov/) under the following accession numbers: SIB1 (At3G56710), SIB2 (At2G41180), WRKY75 (AT5G13080), GLK1 (AT2G20570), GLK2 (AT5G44190), SAG12 (AT5G45890), SAG29 (AT5G13170), ACTIN2 (AT3G18780), and UBQ5 (AT3G62250).