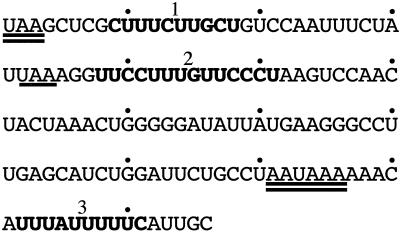Abstract
Human globins are encoded by mRNAs exhibiting high stabilities in transcriptionally silenced erythrocyte progenitors. Unlike α-globin mRNA, whose stability is enhanced by assembly of a specific messenger RNP (mRNP) α complex on its 3′ untranslated region (UTR), neither the structure(s) nor the mechanism(s) that effects the high-level stability of human β-globin mRNA has been identified. The present work describes an mRNP complex assembling on the 3′ UTR of the β-globin mRNA that exhibits many of the properties of the stability-enhancing α complex. The β-globin mRNP complex is shown to contain one or more factors homologous to αCP, a 39-kDa RNA-binding protein that is integral to α-complex assembly. Sequence analysis implicates a specific 14-nucleotide pyrimidine-rich track within its 3′ UTR as the site of β-globin mRNP assembly. The importance of this track to mRNA stability is subsequently verified in vivo using mice expressing human β-globin transgenes that contain informative mutations in this region. In combination, the in vitro and in vivo analyses indicate that the high stabilities of the α- and β-globin mRNAs are maintained through related mRNP complexes that may share a common regulatory pathway.
Eukaryotic mRNAs display half-lives (t1/2s) that range from as short as several minutes (74) to as long as several days (1, 52). Although short-lived mRNAs are typically present in low abundance, their steady-state levels rapidly adjust to reflect fluctuations in gene transcriptional activity. In contrast, long-lived mRNAs may accumulate to high levels that are relatively slow in responding to changes in gene transcription. It is not surprising that mRNAs encoding cytokines, proto-oncogenes, and factors that regulate gene transcription, cell growth, and cell cycling are generally short-lived (6, 49), while mRNAs encoding structural proteins (e.g., collagens) (24, 41) or highly abundant functional products (e.g., crystallins and globins) (4, 36, 52, 66) display longer t1/2s. Although the stabilities of most mRNAs are likely to be constitutive, some—including mRNAs encoding the transferrin receptor (34), histones (5), tubulin (15), and interleukin-2 (11, 12)—display dynamic stabilities that vary in response to changing cellular requirements or environmental conditions (reviewed in reference 49).
The t1/2s of individual mRNAs reflect the combined effects of general and specific determinants of mRNA stability. Two nearly invariant features of eukaryotic mRNAs—the m7G(5′)ppp(5′)N cap (21, 60) and 3′ poly(A) tail (7, 37, 73)—are believed to provide a basal level of stability to all mRNAs by preventing their degradation by cytoplasmic exonucleases. The mRNA-stabilizing properties of the poly(A) tail have been linked both to its poly(A)-binding protein-binding function (7, 50) and to its capacity to stimulate active translation (46; reviewed in references 49 and 57). In addition to these general determinants, a number of well-defined cis elements appear to mediate the stabilities of specific mRNAs. These structurally diverse elements include linear A+U-rich (59) and C+U-rich (72) motifs, as well as an array of stem-and-loop structures (34, 48, 58). mRNA decay rates are modified, either positively or negatively, by a specific trans-acting factor(s) that targets these sites. With few exceptions (30, 61, 74), cis elements are positioned within the 3′ UTR, where their functional interactions are not subject to steric disruption by actively translating ribosomes.
The cis elements required for full α-globin mRNA stability have been particularly well defined. Early observations that α-globin mRNAs containing naturally occurring antitermination mutations failed to accumulate in posttranscriptional reticulocytes, despite their normal levels in transcriptionally active erythroid progenitors, led to the hypothesis that one or more stability-enhancing elements might be positioned within the α 3′ UTR (38). Analyses of α-globin mRNAs containing informative mutations in both cultured cells (71, 72) and in animal models (45, 56) confirmed the importance of the 3′ UTR to α-globin mRNA stability. The capacity of this region to function autonomously was demonstrated in transgenic mice where the stability of human ζ-globin (hζ-globin) mRNA nearly doubled when its 3′ UTR was replaced by an α 3′ UTR (56). Crucial mRNA-stabilizing activity was subsequently mapped to a 3′ UTR region containing a 16-nucleotide (nt) sequence comprised entirely of cytosine and uridine residues (termed the α pyrimidine-rich element or PRE) (33, 67). Although site-specific PRE mutations destabilize α-globin mRNAs expressed in cultured cells (67, 71, 72), their effects on mRNAs expressed in whole-animal models have never been established.
The identification of sequences specifying high α-globin mRNA stability has facilitated a detailed characterization of the mechanism through which this property is effected. When incubated in cytoplasmic extracts, α 3′ UTRs assemble a messenger RNP (mRNP) α complex that is characterized by its mobility and sensitivity to competition by specific homodeoxyribopolymers (28, 67). Mutations within the α PRE have parallel effects on α complex assembly in vitro and on α-globin mRNA stability in cultured cells, linking the mRNP complex to its anticipated molecular function (33, 67). The number and identity of trans-acting factors comprising the complete α complex is still debated (13, 32, 68), although there is general agreement that a ubiquitous 39-kDa RNA-binding factor, αCP, is a core participant (31, 33, 42, 67). Using an RNA titration recruitment assay and density sedimentation analysis, Chkheidze and colleagues have concluded that the α complex is a binary structure comprising the α 3′ UTR and αCP (13). In contrast, Kiledjian et al. have identified AUF-1 (75) as a component of the α complex using a yeast two-hybrid method (32). The same group subsequently demonstrated that assembly of a stable high-order structure comprising αCP and poly(A)-binding protein preserved α-globin mRNA stability by blocking access of an erythroid cell-specific endoribonuclease to its target site within the α PRE (69, 70). Hence, there has been considerable progress in identifying both the cis- and trans-acting elements that are crucial to α-globin mRNA stability, as well as the mechanism through which they function.
Unlike the α-globin mRNA, neither the cis elements nor the trans factors that specify the high stability of β-globin mRNA are known. Estimates from theoretical models (3), 3T3 cells (30), mouse erythroleukemia (MEL) cells (1, 35), cultured mouse spleen cells and reticulocytes (4), human reticulocytes (52), and human bone marrow (51) suggest a t1/2 for β-globin mRNA in the range of 16 to 20 h. Although several hundred naturally occurring mutations are known to affect β-globin gene expression, few offer clues as to the position of a specific mRNA stability-enhancing region or its likely mechanism. Single-nucleotide conversions affecting pre-mRNA splicing or resulting in premature translational termination can accelerate β-globin mRNA degradation, but through pathways that are unlikely to affect a specific mRNA-stabilizing element (reviewed in references 8, 20, and 43). Likewise, frameshift mutations in the terminal coding region that permit ribosomes to read through the proximal one-third of the β 3′ UTR have relatively little impact on overall β-globin expression (9, 18, 19, 54), suggesting that crucial stability elements are downstream of this region. It has also been suggested that β-globin mRNA stability might result from spatially distinct but functionally redundant cis elements (54). Hence, there is a need to define both the structure and function of specific β-globin mRNA stability elements.
The present work investigates the structural basis for the high stability of β-globin mRNA. In vitro conditions are established that promote the assembly of a specific β-globin mRNP complex, whose structural composition and functional properties are compared to those of the stability-enhancing mRNP α complex. This analysis indicates that the β mRNP complex contains an αCP-like factor(s) and implicates a 14-nt PRE within the β-globin 3′ UTR where it is likely to act. The functional importance of this PRE is subsequently verified in terminally differentiating erythroid progenitors in intact mice carrying hβ-globin transgenes with informative mutations in this region. Our results indicate that structurally related mRNPs assemble on PREs within the α- and β-globin 3′ UTRs and that the β PRE is crucial to the high stability of hβ-globin mRNA. The data also encourage speculation that the stabilities of the hα- and hβ-globin mRNAs might be coregulated through a related mechanism.
MATERIALS AND METHODS
Electrophoretic mobility shift assay (EMSA). (i) Preparation of cytoplasmic extracts.
S-100 extracts were prepared from cultured MEL cells as previously described (56). Briefly, ∼2 × 108 cells in the log phase of growth were washed in excess phosphate-buffered saline (PBS), pelleted, and lysed in 3.5 ml of buffer A (10 mM KCl, 1.5 mM MgCl2, 10 mM Tris, pH 7.4, 0.5 mM dithiothreitol [DTT], 200 ng of pepstatin/ml, 200 ng of leupeptin/ml, and 10 μg of aprotinin/ml) by successive passages through 23- and 25-gauge needles. The crude extract was clarified at 4°C by a 10-min spin at 2,000 × g (Sorvall RC-5B; DuPont Instruments) followed by vacuum ultracentrifugation for 60 min in an SW51Ti rotor at 32,500 rpm (Beckman, Columbia, Md.). The supernatant was amended with 0.1 volume of glycerol and stored in aliquots at −80°C. A protein yield of ∼4 mg/ml was typical of this method (bicinchoninic acid kit; Pierce, Rockford, Ill.).
(ii) Generation of mRNAs.
The construction of a cDNA template for the in vitro transcription of hα mRNA 3′ UTRs has been previously described (53). A cDNA template for in vitro transcription of hβ 3′ UTR mRNA was generated by thermal amplification of the cloned gene (54) by Taq polymerase (New England BioLabs, Beverly, Mass.) using forward (5′ ATACgATTTAggTgACACTATAg AACTCgCTCgCTTTCTTgCTgTCC 3′) and reverse (5′ gCAATgAAAATAAATgTTTTTTATTA 3′) oligomers under standard reaction conditions. The forward oligomer contains an SP6 promoter (doubly underlined) and its consensus transcription initiation sequence (singly underlined). Amplified cDNAs were purified over G50 spin columns (Boehringer Mannheim, Indianapolis, Ind.) (56). In vitro transcriptions were carried out in the presence of [α-32P]CTP (400 Ci/mmol, 10 mCi/ml; Amersham, Arlington Heights, Ill.) using a Maxiscript SP6 kit under conditions recommended by the manufacturer (Ambion, Austin, Tex.). An aliquot of each reaction was resolved on a 6% acrylamide–8 M urea gel to assess probe quality (56). Competitor oligomers (Integrated DNA Technologies, Coralville, Iowa) were resuspended at 100 nmol/μl.
(iii) EMSA analyses.
As described previously (56), reaction mixtures (15 μl) assembled from 4 μl of S-100 extract and a ∼50,000-cpm probe in 1× reaction buffer (150 mM KCl, 1.5 mM MgCl2, 10 mM Tris, pH 7.4, 0.5 mM DTT, 200 ng of pepstatin/ml, 200 ng of leupeptin/ml, and 10 μg of aprotinin/ml) were incubated at room temperature for 30 min, supplemented with RNase A (1 μl, 250 μg/ml) and RNase T1 (1 μl, 2,000 U/ml), and incubated for an additional 10 min. Reactions were resolved on a nondenaturing 6.5% polyacrylamide gel (56).
Generation of transgenic mice.
All animal experimentation in this study fully complied with protocols approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania School of Medicine. All of the transgenes utilized in this work derive from a previously described transgene encoding wild-type human β-globin (hβWT) mRNA (27). This pSP72-based plasmid contains a 6.5 kb micro-locus control region (μLCR) within an engineered SstI polylinker site (27, 39, 63) and a 4.1-kb HpaI-XbaI fragment of human genomic DNA encompassing the full-length β-globin gene and its 5′ promoter and 3′ enhancer elements (65) within a unique ClaI site. The cloning strategy preserves the native order and orientation of the μLCR and hβ-globin fragments. EcoRI-EcoNI fragments of the β-globin gene (encompassing exon 3 coding and 3′ UTR sequences and 692 bp of the contiguous 3′ flanking region) that contained the readthrough (βRT), deletion (βDEL), or substitution (βSUB) mutations were ligated into the corresponding site of the βWT transgene. A fragment containing the βRT mutation was prepared from an EcoRI-EcoNI digest of the previously constructed pβB,Af plasmid (54). DNA fragments containing the βDEL and βSUB mutations were generated by splice-overlap-extension PCR (53, 56). To prepare the βDEL fragment, a 166-bp DNA fragment encompassing the exon 3 EcoRI site and the desired deletion was generated by PCR amplification of the β-globin sequence using forward (Fi 5′ AACGTGCTGGTCTGTGTGCT 3′) and reverse (Del-R: 5′ GTAGTTGGACTTCCTTTAATAGAAATTGGACAGC 3′) oligomers. A second 822-bp DNA fragment encompassing the mutation and the EcoNI site was generated from the β-globin gene using forward (Del-F: 5′ CTATTAAAGGAAGTCCAACTACTAAACTGGGGG 3′) and reverse (R: 5′ AGAATGGGACTTCCATTTGG 3′) oligomers. Fifty-microliter reactions containing 20 ng of DNA template, 100 nmol of each oligomer, and 2 mM Mg2SO4 were amplified with Vent polymerase as recommended by the manufacturer (New England BioLabs). A standard amplification program was utilized (cycle 1, 95°C for 3 min, 58°C for 15 s, and 73°C for 45 s; cycles 2 to 30, 92°C for 1 min, 58°C for 15 s, and 73°C for 45 s; and cycle 31, 73°C for 45 s). Aliquots of each of the initial reactions were combined and reamplified using the two flanking oligomers (F and R) but with the extension time prolonged to 60 s. The 980-bp product was digested with EcoRI and EcoNI and was ligated into the cognate site of the βWT transgene plasmid. The βSUB transgene was generated using a similar protocol but with two different internal oligomers (Sub-F, 5′ GAAAAGAAGGAAAgAAGTCCAACTACTAAACTGGGGG 3′; Sub-R, 5′ CTTTCCTTCTTTTCCCTTTAATAGAAATTGGACAGC 3′). The fidelity of the method was verified by sequencing through the coding, 3′ UTR, and proximal 3′ flanking region. Each transgene was subsequently released as a 10.6-kb SalI-EcoRV fragment, purified, and provided to the Transgenic and Chimeric Mouse Facility at the University of Pennsylvania for pronuclear injection (27, 45, 56). Positive founders identified by sequential dot blot and Southern analysis were used to generate FI mice with germ line transgene integration (27, 45, 56). Five independent βWT lines have been previously described (27). Although 12 βRT founders were generated by multiple pronuclear injections performed over the course of more than a year, only a single line could be established that expressed the βRT mRNA (see Results). Three and two independent βDEL and βSUB lines, respectively, were generated.
Recovery and purification of mouse mRNA. (i) Preparation of animals.
Sexually mature male and female mice were pretreated with three intraperitoneal doses of acetyl-2-phenylhydrazine (40 μg/g of body weight; Sigma) on days 0.0, 0.5, and 1.0 and were subsequently sacrificed on day 4.5. Bone marrow and blood were separately collected in PBS-heparin (20 U/ml) as previously described (39, 45, 56).
(ii) RNA purification.
Tissues were lysed in a 5 M guanidine isothiocyanate solution (50 mM Tris [pH 7.4], 10 mM EDTA, 700 mM β-mercaptoethanol, and 1% sarcosyl) and were centrifuged through a 5.7 M CsCl cushion at 42,000 rpm for 16 h in a TL-55 rotor (Beckman). RNAs were resuspended in 10 mM Tris, pH 7.4, 1 mM EDTA, and 1% sodium dodecyl sulfate, extracted twice with phenol-chloroform-isoamyl alcohol, and precipitated. The concentration of the resuspended mRNAs was established spectrophotometrically by densitometry at a wavelength of 260 nm (39, 45, 56).
RNase protection assay (RPA). (i) Generation of antisense probes.
A previously constructed cDNA template containing a contiguous region of intron 1 and exon 2 from the mouse α-globin (mα-globin) gene (39) encodes a 235-nt antisense RNA that protects a 179-nt fragment of the fully processed mα-globin mRNA. A cDNA template encoding an antisense hβ-globin mRNA probe was generated from a cDNA fragment containing contiguous regions of intron 1 and exon 2 using forward (5′ GATCCCCGGGTACCCTGATAGGCACTGACTCTCT 3′) and reverse (5′ TTGCATGCCTGCAGCAGCTTGTCACAGTGCAGCT 3′) oligomers in a standard thermal amplification reaction. The 261-bp PstI-KpnI fragment of the PCR-amplified product was inserted into the cognate polylinker site of pSP72, and the sequence was confirmed by automated sequencing. DNAs linearized with EcoRI were transcribed in vitro using SP6 polymerase to generate a 287-nt probe protecting a 199-nt fragment of hβ exon 2. 32P-labeled mα- and hβ-globin probes were transcribed in vitro as described above.
(ii) RPA method.
Purified bone marrow (∼1,500 ng) and peripheral blood (∼100 ng) RNAs were desiccated and resuspended in 20 μl of Berk buffer {80% formamide, 40 mM PIPES [piperazine-N,N′-bis(2-ethane sulfonic acid)] [pH 6.4], 400 mM NaCl, and 1 mM EDTA} supplemented with hβ and mα probes, heat denatured for 10 min at 80°C, and incubated overnight at 52°C. The samples were digested at room temperature for 25 min in 200 μl of buffer (10 mM Tris [pH 7.5], 300 mM NaCl, 5 mM EDTA, 20 mg of RNase A/ml, and 1 mg of RNase T1/ml), and the reaction terminated by addition of 17 μl of stop solution (8% sodium dodecyl sulfate and 2 mg of proteinase K/ml) and incubating at 37°C for an additional 20 min. The samples were extracted with phenol-chloroform-isoamyl alcohol, precipitated with ethyl alcohol, resolved on a denaturing polyacrylamide-urea gel, and quantitated by PhosphorImager analysis (Molecular Dynamics). RNA stability is defined as [(hβ)/(mα)]PB/[(hβ)/(mα)]BM, where PB and BM indicate peripheral blood and bone marrow, respectively.
Northwestern analysis.
EMSA reaction mixtures were resolved on nondenaturing polyacrylamide gels as described above and were then transferred at 4°C to a nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.) in transfer buffer (25 mM Tris [pH 7.4], 190 mM glycine, and 20% methanol) at 30 V using a Hoefer Transphor Tank apparatus (Amersham Pharmacia Biotech, San Francisco, Calif.). Membranes were rinsed in PBS and then gently shaken for 2 h at room temperature in Northwestern buffer (10 mM Tris [pH 7.4], 50 mM NaCl, 1 mM EDTA, and 1× Denhardt's solution) augmented with 1 mM DTT. Membranes were subsequently incubated for 2 h at room temperature in Northwestern buffer augmented with 5 μg of heparin/ml, 10 μg of tRNA/ml, and ∼20,000 cpm of 32P-labeled RNA probe/ml and were then washed in Northwestern buffer and exposed to autoradiography. Reactions containing no 32P-labeled RNA were run in parallel as controls.
Supershift assay.
EMSA reactions supplemented with 1 μl of affinity-purified rabbit antiserum recognizing two independent αCP isoforms (αCP-1 and -2; kind gift of S. Liebhaber [13]) were resolved on nondenaturing polyacrylamide gels as described above. Reactions incubated in the absence of antiserum, either with or without homodeoxyribopoly(C) as competitor, were run in parallel as controls.
RESULTS
The β-globin 3′ UTR assembles an mRNP complex that comigrates with the authentic α complex.
Although the long t1/2 of β-globin mRNA (1, 35, 40, 51, 52), as well as its close phylogenetic relationship to α-globin mRNA (22, 23), suggested the possibility that the β-globin 3′ UTR might contain a conserved α-like PRE, a direct structural comparison failed to reveal any significant sequence homologies (not shown). Consequently, an alternate approach was employed to directly determine whether the β 3′ UTR could assemble an mRNP complex with attributes similar to those of the stability-enhancing α complex (67). Specifically, these characteristics include similar migration on a nondenaturing polyacrylamide gel (28), a high sensitivity to competition by homodeoxyribopoly(C) but not by other homodeoxyribopolymers (28, 67), and a sensitivity to competition by PREs from other mRNAs (28). RNA EMSAs indicate that the migration of in vitro-transcribed 32P-labeled β 3′ UTRs is substantially retarded when they are preincubated in cytoplasmic extract prepared from MEL cells (Fig. 1, lanes 4 and 6). In contrast, 32P-labeled β 3′ UTRs incubated with proteinase K-treated extracts migrate normally (data not shown). These results indicate that the β 3′ UTR assembles with trans-acting factors into an mRNP complex that we have termed the β complex. Importantly, the α and β complexes comigrate on nondenaturing polyacrylamide gels (Fig. 1, lanes 3 and 6). Identical mRNP complexes readily assemble on heat-denatured 32P-labeled α or β 3′ UTRs, suggesting that the participating cis elements are thermodynamically stable and kinetically accessible (data not shown). In contrast to the α complex, the β complex appears to be relatively intolerant of variations in buffer pH and salt concentration, possibly reflecting differences in the affinities of shared trans-acting factors for structurally dissimilar α and β cis elements (data not shown).
FIG. 1.
hα- and hβ-globin mRNA 3′ UTRs assemble comigrating mRNP complexes in vitro. An EMSA was performed by incubating 32P-labeled α and β 3′ UTR probes in MEL cell cytoplasmic (S-100) extract and resolving the RNase-resistant mRNP products on a nondenaturing 6.5% polyacrylamide gel. Extract-free reactions assembled in the absence (lanes 1 and 4) or presence (lanes 2 and 5) of added RNase were run in parallel as controls. The composition of each reaction is indicated at the top of the autoradiograph, and the migration of the mRNP complexes is shown on the left.
The α and β complexes display similar competition profiles.
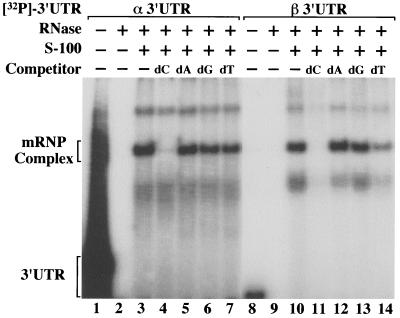
To further establish its similarity to the α complex, the efficiency of β complex assembly was tested in the presence of specific homodeoxyribopolymers (Fig. 2). The β complex is efficiently competed by unlabeled homodeoxyribopoly(C) but not by homodeoxyribopoly(A) or -(G) (Fig. 2, lanes 10 to 13). This competition pattern reproduces the effect of these polymers on α complex assembly (Fig. 2, lanes 3 to 6) (67). In contrast to the α complex, β complex assembly appears to be modestly sensitive to competition by homodeoxyribopoly(T) (Fig. 2, compare lanes 7 and 14), a characteristic that might predict a high uridine content for the β cis element.
FIG. 2.
Assembly of α- and β-globin mRNP complexes is inhibited by homodeoxyribopoly(C) but not by other homodeoxyribopolymers. EMSA reactions were performed on 32P-labeled α and β 3′ UTRs supplemented with 100 μg of unlabeled competitor homodeoxyribopolymer (dC, dA, dG, and dT). Control lanes demonstrate migration of RNase-undigested 3′ UTRs (lanes 1 and 8) and RNase-digested 3′ UTRs (lanes 2 and 9) in extract-free reactions. The components of each reaction are indicated at the top of the autoradiograph, and the positions of the α and β mRNP complexes are shown on the left.
The α and β mRNP complexes cross-compete for labeled mRNAs.
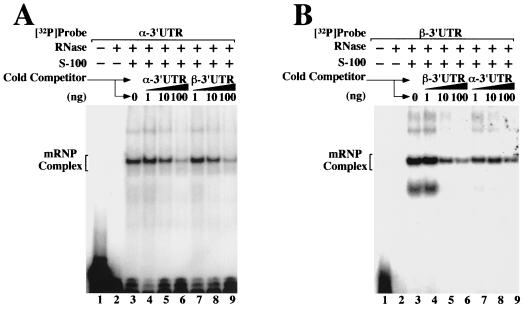
The structural identity of the α and β complexes was further tested using standard cross-competition experiments. 32P-labeled α 3′ UTRs were incubated with cytoplasmic S-100 extracts in the presence of known quantities of unlabeled α- or β-globin 3′ UTRs (Fig. 3A, lanes 4 to 6 and 7 to 9, respectively). For completeness, the converse experiment—competing 32P-labeled β 3′ UTRs with unlabeled α and β 3′ UTRs—was also performed (Fig. 3B). In both cases, the 32P-labeled probes were efficiently competed by increasing levels of either of the two unlabeled competitor RNAs. The outcome of the experiment was not affected by the order in which the cytoplasmic extract, 32P-labeled 3′ UTRs, or unlabeled competitor 3′ UTRs were added to the reaction (data not shown). Confirming experiments indicated that the α 3′ UTRs might be marginally better competitors than the β 3′ UTRs, consistent with the hypothesis that trans-acting components bind to the two 3′ UTRs with different affinities. Nevertheless, the observation that the α and β 3′ UTRs assemble identically migrating, cross-competing mRNP complexes that are effectively competed by homodeoxyribopoly(C) suggests that the β complex shares structural characteristics, and possibly functional properties, with the stability-enhancing α complex.
FIG. 3.
Binding specificity of trans-acting factors for α and β 3′ UTRs. EMSA analyses were performed using in vitro-transcribed 32P-labeled α-globin 3′ UTRs (A) and β-globin 3′ UTRs (B) incubated in MEL cell S-100 extract. Reactions were supplemented with defined quantities of unlabeled α- or β-mRNA 3′ UTR competitor. The composition of each reaction mixture is indicated above the autoradiograph, and the positions of the α and β complexes are shown on the left.
Fully assembled α and β mRNP complexes efficiently bind both α and β 3′ UTRs.
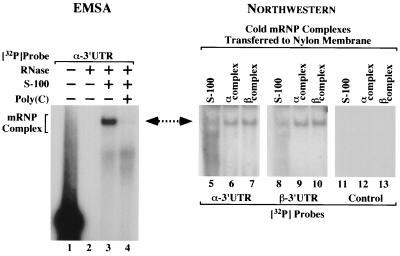
An independent method, Northwestern analysis, was used to confirm the structural similarities between the α and β complexes suggested by the preceding experiments. Unlabeled α and β complexes were resolved in triplicate on the same gel and transferred to nitrocellulose, and the renatured protein complexes were probed with 32P-labeled α or β 3′ UTRs (Fig. 4). On the same gel a 32P-labeled α complex was resolved as a migration marker (Fig. 4, lane 3). 32P-labeled α and β 3′ UTR probes were observed to bind to lanes containing (unlabeled) α and β complexes at positions corresponding to the control α complex (Fig. 4, compare lane 3 to lanes 6, 7, 9, and 10). Control lanes containing cytoplasmic extract alone (Fig. 4, lanes 5 and 8) bound the 32P-labeled 3′ UTR probes at a similar position but with severalfold-lower efficiency, suggesting that mRNP complexes, believed to be assembled on murine globin mRNAs, persist at low levels within the S-100 extract. A control blot probed with a 32P-labeled RNA corresponding to the pSP72 polylinker sequence did not display the mRNP complex band (Fig. 4, lanes 11 to 13). These results indicate a high relative specificity of the α and β complexes for both the α and β 3′ UTRs and support the hypothesis that the two mRNP complexes are structurally related.
FIG. 4.
Fully assembled α and β complexes exchange α- and β-globin 3′ UTRs. In vitro-transcribed α- and β-globin 3′ UTRs were incubated in MEL cell S-100 extract and then resolved on a single polyacrylamide gel. (Left) A reaction utilizing 32P-labeled α 3′ UTR indicates the migration of the α complex (lane 3). Undigested and fully digested probes, as well as a homodeoxyribopoly(C)-competed reaction, were included as controls (lanes 1, 2, and 4). The composition of each reaction is indicated above the autoradiograph, and the position of the α complex is shown on the left. (Right) On the same gel, reactions utilizing α 3′ UTRs (lanes 6, 9, and 12) and β 3′ UTRs (lanes 7, 10, and 13) were resolved in triplicate. Control lanes 5, 8, and 11 contained S-100 extract alone. The complexes were transferred to a nylon membrane and were subsequently probed with either 32P-labeled α 3′ UTR, 32P-labeled β 3′ UTR, or an unrelated [32P]mRNA control (indicated at bottom), and autoradiographs were exposed. The composition of each reaction is indicated above the autoradiograph. A double-headed arrow emphasizes comigration of the α complex and the membrane-bound mRNP complexes that exchange 32P-labeled α and β 3′ UTRs.
Antibodies to αCP recognize elements of the β complex.
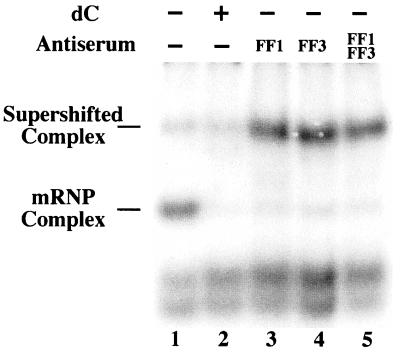
To directly assess whether elements of the α and β complexes were structurally related, supershift analysis of the β complex was carried out using two independently generated rabbit antisera recognizing the core component of the α complex. Affinity-purified antiserum against two different αCP isoforms (αCP-1 and αCP-2) were utilized, as both isoforms are known to participate in the mRNP α complex (13, 42). Reactions utilizing the antiserum, both alone and in combination, displayed a reduction in the intensity of the β complex and the reciprocal appearance of a more slowly migrating band (Fig. 5). It is likely that the new band comprises antibody-bound β complex, although we cannot rule out the possibility that it represents a secondary mRNP complex that assembles as a result of antibody sequestration of αCP. Both possibilities directly implicate αCP or an αCP-like factor as a participant in both the α and β complexes and suggest a structural basis for the functional similarities that they exhibit.
FIG. 5.
Anti-αCP antibodies bind the mRNP β complex. 32P-labeled β 3′ UTR was incubated in MEL cell S-100 extract in the absence (lanes 1 and 2) or presence (lanes 3 to 5) of anti-αCP antibodies, and the reactions were resolved on a nondenaturing polyacrylamide gel. FF1 and FF3 denote rabbit antisera raised against αCP-1 and αCP-2 isoforms, respectively. The reaction shown in lane 2 was supplemented with homodeoxyribopoly(C) to verify the position of the β complex. The composition of each reaction is indicated at the top of the gel, and the positions of the native and supershifted β complexes are given on the left.
PREs are present in the 3′ UTR of hβ-globin mRNA.
In light of the above observations, the β-globin 3′ UTR was reexamined to identify pyrimidine-rich tracks containing any combination of cytosine and/or uridine residues (Fig. 6). Although the C+U content of the α and β 3′ UTRs is similar (62 and 58%, respectively), nearly half of the 76 pyrimidines within the β 3′ UTR are clustered into three tracks that we have designated β PRE-1, -2, and -3. Of these three elements, we hypothesized that the longest, β PRE-2, was likeliest to play a functional role in β-globin mRNA stability. In contrast to the α PRE, β PRE-2 has a higher proportion of uridine residues (57 versus 31%) (28) and is interrupted by a single-nucleotide purine G. These features are consistent with the heterogeneity in both overall length and cytosine/uridine ratio that characterize PREs assembling α complexes on α globin (67), tyrosine hydroxylase (16), 15-lipoxygenase (47), and α1(I)-collagen (62) mRNAs. β-Globin genes containing antitermination frameshift mutations permitting ribosomes to read through β PRE-1 to a UAA termination codon 10 codons into the 3′ UTR are expressed at normal or near-normal levels (9, 18, 19, 54), suggesting that the structural integrity of β PRE-1 is not required for high β-globin mRNA stability. Likewise, β PRE-3 is a less attractive candidate than β PRE-2 as a stabilizing cis element because of its short length (10 nt), cytosine deficiency (a single residue), and positioning 3′ to the AAUAAA polyadenylation hexanucleotide. These structural considerations predicted that β PRE-2 would be the likeliest of the three C+U-rich tracks to effect high β-globin mRNA stability.
FIG. 6.
The β-globin 3′ UTR contains three extended PREs. The β-globin 3′ UTR is displayed with the native UAA termination codon and AAUAAA polyadenylation signal doubly underlined. β-Globin mRNAs carrying naturally occurring +2 frameshift antitermination mutations terminate translation at an in-frame UAA 10 codons into the 3′ UTR (underlined; see text). Pyrimidine-rich tracks designated β PRE-1, -2, and -3 are boldfaced and labeled.
Generation of mice containing hβ-globin transgenes with antitermination mutations.
The preceding structural considerations predicted that the β-globin 3′ UTR, and specifically β PRE-2, might play a functional role in β-globin mRNA stability. To investigate this possibility, we generated a series of transgenic mouse lines that expressed hβ-globin mRNAs containing informative site-specific mutations. Previous work using transiently transfected MEL cells demonstrated that full-length hβ-globin mRNAs can be destabilized by a pair of site-specific mutations (Fig. 7A) (54). We generated transgenic mice containing this βRT gene (Fig. 7A) linked in its native orientation to a β μLCR (27, 63). Only a single transgenic line expressing the βRT mRNA could be established from 12 βRT founders, despite repeated pronuclear injections using independently prepared DNAs, reflecting the possible toxicity of extended hβ-globin chains to mouse erythroid progenitors (reviewed in reference 64). The tandem mutations permit ribosomes to read 36 codons past the native UAA into the 3′ UTR where they sterically disrupt functional high-order mRNA structures and mRNP interactions (Fig. 7B). In addition to the βRT transgenics, five independent βWT transgenic lines were generated from the cloned hβ-globin gene for use as controls (27) (Fig. 7A). Where possible, age- and sex-matched mice were used for all subsequent studies.
FIG. 7.
Construction of transgenes encoding βWT and βRT β-globin mRNAs. (A) Transgene structures. The βWT and βRT transgenes comprise the full-length hβ-globin gene, including native promoter (P) and 3′ enhancer (E) elements, linked in their native orientation to a β μLCR (LCR) (63). The two transgenes are identical except for two tandem mutations (asterisks) in exon 3 of the βRT transgene (54). Tick marks indicate the positions of the native translation initiation and termination codons. (B) Structures of βWT and βRT mRNAs. The 5′-cap (●), poly(A) tail (An), and native translation initiation and termination codons (tick marks) are indicated. Asterisks denote the positions of tandem mutations in the βRT mRNA. The region of each mRNA that is actively translated is indicated by an arrow.
An antitermination mutation destabilizes hβ-globin mRNA.
The stabilities of the βWT and βRT mRNAs were assessed using a previously described method designed specifically for this purpose (38, 45, 56). The assay tests the decline in transgenic mRNA levels, relative to the levels of endogenous mα-globin mRNA, in the transcriptionally silent interval between the marrow and peripheral reticulocyte stages of terminal erythroid differentiation. One advantage of this method is that it does not require the use of global transcriptional inhibitors, such as actinomycin D or 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), that may produce anomalous results (26) and can paradoxically stabilize some mRNAs (61).
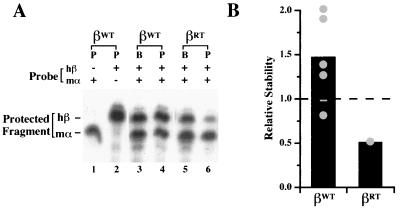
The levels of βWT and βRT mRNAs in paired marrow and reticulocyte samples were determined by an RPA in which 32P-labeled antisense hβ and mα probes protected defined fragments of the transgenic hβ- and endogenous mα-globin mRNAs. Two or more mice from each independent transgenic line were each analyzed at least twice using this method. A representative two-probe RPA demonstrates that the stability of βWT mRNA is nearly threefold greater than that of the βRT mRNA (Fig. 8A; for each mouse compare any change in the level of hβ mRNA between the marrow and reticulocyte samples to the change in mα mRNA levels between these two tissues). Analyses of mice from all transgenic lines indicate that the average stability of βWT mRNA was reduced nearly threefold by the introduction of the tandem readthrough mutations (Fig. 8B). This result confirms in a whole-animal model the destabilizing effect of readthrough mutations on β-globin mRNA initially established in transiently transfected MEL cells (54), thus indicating the existence of a stability determinant in the β-globin 3′ UTR.
FIG. 8.
Translation antitermination mutations reduce the stability of hβ-globin mRNA in intact erythroid cells. (A) Representative two-probe RPA. RNA prepared from bone marrow progenitors (B) and peripheral blood reticulocytes (P) obtained from representative mice containing the βWT transgene (lanes 3 and 4) or the βRT transgene (lanes 5 and 6) was analyzed using 32P-labeled probes complementary to hβ-globin (hβ) and mα-globin (mα) mRNAs. The migration of the protected mα and hβ probe fragments is indicated by control reactions utilizing either of the probes alone (lanes 1 and 2). The composition of each reaction and the migration of the protected fragments are shown above and to the left of the autoradiograph, respectively. Control lanes indicating functional probe excess, performed for every experiment, were cropped from the figure to maintain clarity. (B) Stabilities of βWT and βRT mRNAs in multiple transgenic lines. The average stabilities of βWT and βRT mRNAs in each of five and one independent line, respectively, are plotted (●). For each line, a minimum of two mice were studied on at least two separate occasions using the two-probe (hβ and mα) RPA. The stability of each mRNA, averaged across all lines, is indicated as a bar; the stability of mα-globin mRNA, defined as 1.0, is denoted by a dashed line.
β-Globin mRNA is destabilized by replacement or deletion of β PRE-2.
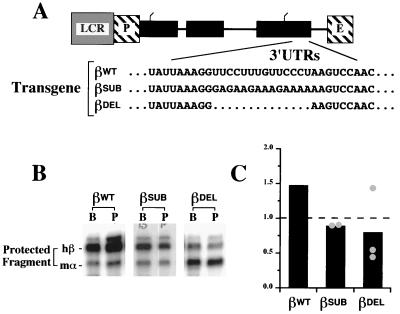
The in vivo effects of antitermination mutations (indicating the importance of the 3′ UTR to β-globin mRNA stability), the in vitro EMSA analyses (demonstrating functional similarity between the mRNP β complex and the stability-enhancing α complex), and results from supershift experiments (indicating participation of αCP in both the α and β complexes) each suggested an important role for β PRE-2 in effecting the high stability of β-globin mRNA. To test the functional properties of this region, we constructed β-globin transgenes in which β PRE-2 was replaced by an equal-length polypurine sequence (βSUB) or deleted in its entirety (βDEL) (Fig. 9A). Independent mouse lines were generated that contained the βSUB (two lines) or βDEL transgenes (three lines), and the presence of the relevant mutation was confirmed by sequencing the appropriate reverse transcriptase PCR product from reticulocyte mRNA (data not shown). The stabilities of the βSUB and βDEL mRNAs were tested in duplicate in at least two mice from each independent line using the two-probe RPA described above. A typical autoradiograph indicates a reduction in the stability of the βWT mRNA when β PRE-2 was either replaced or deleted (Fig. 9B). Data from each of the five independent lines demonstrate that either replacement or deletion of β PRE-2 results in a twofold reduction in β-globin mRNA stability (Fig. 9C). These data indicate the functional importance of β PRE-2 to normal β-globin mRNA stability in terminally differentiating erythroid progenitors.
FIG. 9.
β-Globin mRNAs containing deletion or replacement of PRE-2 are destabilized in vivo. (A) Structures of wild-type and variant β-globin mRNA 3′ UTRs. Transgenes encoding variant β-globin mRNAs were constructed from the βWT transgene by exchanging β PRE-2 for an equal-length purine-rich sequence (βSUB) or deleting it in its entirety (βDEL). The relevant segments of the βWT, βDEL, and βSUB 3′ UTRs have been aligned for comparison. The β μLCR (LCR), promoter (P), and 3′ enhancer elements (E) are indicated. (B) Relative stabilities of βWT, βSUB, and βDEL mRNAs in representative transgenic mice. RNAs recovered from bone marrow (B) and peripheral blood (P) erythroid cells from mice containing the βWT, βSUB, and βDEL transgenes were analyzed by two-probe RPA. The positions of the protected [32P]-labeled mα and hβ RNA fragments are indicated on the left. (C) Stabilities of βSUB and βDEL mRNAs in multiple transgenic lines. The average stabilities of βSUB and βDEL mRNAs in each of two and three independent transgenic lines, respectively, are plotted (●). For each line, a minimum of two mice was studied on at least two separate occasions using two-probe (hβ and mα) RPA. The stability of each mRNA, averaged across all lines, is indicated as a bar; the stability of mα-globin mRNA, defined as 1.0, is denoted by a dashed line. A bar indicating the average stability of transgenic human βWT mRNA is reproduced from Fig. 8 for comparison.
DISCUSSION
The normal expression of human globins is crucially dependent upon the high stability of their encoding mRNAs. Over a period lasting at least 4 days, erythroid progenitor cells in the marrow cease transcription, extrude their nuclei, and migrate into the peripheral circulation as RNA-rich reticulocytes. With their long t1/2s, globin mRNAs survive at high levels in these cells and continue to translate substantial quantities of their cognate globin proteins. Consequently, small changes in globin mRNA stabilities can disproportionately impact the levels of their encoded protein products. Studies of fos-β-globin chimeras (30) and informative β-globin mRNA variants (54) suggest that 3′ UTR determinants may be crucial to normal β-globin mRNA stability. This arrangement would be consistent with the observation that stability-determining elements are commonly positioned in that region, including one that dictates the stability of hα-globin mRNA. The present report confirms that the β-globin 3′ UTR harbors at least one such stability element that maps to a 14-nt PRE. The data also directly demonstrate in a whole-animal model system that site-specific PRE mutations can destabilize globin mRNAs. The structural similarities of the mRNP complexes effecting their long t1/2s suggest that the stabilities of the α- and β-globin mRNAs may be coregulated.
At least three factors have impeded previous attempts to identify specific stability-defining regions of β-globin mRNA. First, unlike α-globin mRNA (14, 17, 25, 44), naturally occurring mutations that destabilize β-globin mRNA by disrupting the function of a specific mRNA stability element are not known to exist. Although β-globin genes have been described that contain single-nucleotide substitutions or small deletions within their 3′ UTRs, these mutations do not appear to affect globin mRNA stability (2, 10, 29; data not shown). β-Globin genes containing frameshift antitermination mutations (permitting ribosomes to read through the initial third of the 3′ UTR) are expressed at near-normal levels (9, 18, 19), suggesting that mRNA stability-defining determinants are positioned further downstream. β-Globin mRNAs containing engineered mutations that permit ribosomes to read further into the 3′ UTR are destabilized both in cultured MEL cells (54), as well as in situ in whole-animal models (Fig. 7 and 8). Replacement or deletion of β PRE-2 within this downstream region reduces β-globin mRNA stability by half (Fig. 9), implicating this element as crucial for normal β-globin mRNA stability. Hence, the importance of the 3′ UTR, as well as specific internal regions, can be demonstrated in mice containing artificial readthrough mutations.
The structural dissimilarity between the α and β 3′ UTRs is a second factor that has misdirected efforts to identify a β-globin mRNA stability element. The expectation that the stabilities of the two mRNAs were similarly regulated was inconsistent with the fact that the β 3′ UTR did not contain a cytosine-rich PRE homologous to the α PRE. It has recently been recognized that α complexes can assemble on PREs sharing minimal homology (16, 47, 62, 67), suggesting that pyrimidine content, and not the cytosine-to-uridine ratio per se, might define functionally important stability determinants. Using this criteria we identified three candidate β PREs that were 10, 14, and 10 nt in length (Fig. 6). The largest, β PRE-2, displays a higher uridine content than the α PRE, possibly explaining both the modest susceptibility of the β complex to competition by poly(T) as well as its sensitivity to small changes in buffer pH and salt concentrations (Fig. 2 and data not shown). This possibility is consistent with previous demonstrations that the efficiency of mRNP complex assembly can be significantly altered by single-nucleotide changes within the α PRE (28, 56). We suggest that the observed heterogeneity in functional PREs may permit distinct RNAs to exhibit different t1/2s while coordinating their relative levels of expression through a common regulatory pathway. Such a mechanism would account for the nearly fourfold difference in the t1/2s of the homologous β- and δ-globin mRNAs (51) and might possibly play a role in balancing the expression of α- and β-globin proteins both in normal and in thalassemic erythroid cells.
Finally, previously stated conclusions that the stabilities of the α- and β-globin mRNAs are independently regulated reference unsuccessful attempts to identify candidate mRNP complexes on the β-globin 3′ UTR (55, 67). In contrast, we readily assembled mRNP complexes on the β-globin 3′ UTR that were nearly indistinguishable from the authentic α complex, with the exception that assembly of the β complex appeared to be more sensitive to competition by homodeoxyribopoly(T) (Fig. 1 to 4). Our success in assembling β complexes may reflect unintended but fortuitous variations in the buffer pH or salt concentrations utilized, which we have noted to be crucial determinants of in vitro β-globin mRNP assembly. In addition, we (and others) have noted that the quality of S-100 extracts may vary among independent preparations, affecting the efficiency of mRNP assembly for reasons that remain unclear (data not shown). Each of these possibilities emphasizes the importance of confirming EMSA findings (Fig. 1 to 5) with alternate, preferably functional, analyses (Fig. 7 to 9).
The functional similarities between the α and β complexes (Fig. 1 to 4) predicted that the two mRNPs might share specific structural features. Antibodies raised against αCP, a critical component of the α complex, bind to β complex epitopes, indicating that an αCP-like factor, perhaps even αCP itself, participates in both the mRNP α and β complexes (Fig. 5). The expanding number of mRNAs that support assembly of αCP-containing mRNPs suggests that these factors may coordinate the expression of multiple genes that are active during the terminal stages of erythroid cell differentiation. The likelihood that the α- and β-globin genes are posttranscriptionally coregulated may also have important implications relating to the pathophysiology of thalassemias characterized by imbalance in the levels of the α- and β-globin mRNAs.
The data in this report do not rule out the possibility that additional stability elements may exist elsewhere within the β-globin mRNA. β-Globin mRNAs containing tandem readthrough mutations (Fig. 8) appear to be less stable than mRNAs with deletion or purine replacement of β PRE-2 (Fig. 9). This observation suggests that the function of additional (unrecognized) 3′ UTR stability elements located outside the β PRE-2 may be compromised by undiscriminating readthrough ribosomes. The hypothesis that multiple regions contribute to mRNA stability could be tested by combinatorial mutations of one, two, or all three β PREs, although the extensive sequence alterations required for this approach might nonspecifically affect β-globin mRNA stability. The manner in which multiple elements might interact to maintain the high stability of human β-globin mRNAs during terminal differentiation would benefit from fine mapping studies of the β 3′ UTR, focusing initially on the β PRE-2 element identified in the present work.
ACKNOWLEDGMENTS
This work was supported in part by grant HL56038 from the National Heart, Lung and Blood Institute (J. E. R.).
We thank Z. He for expert technical assistance.
REFERENCES
- 1.Aviv H, Voloch Z, Bastos R, Levy S. Biosynthesis and stability of globin mRNA in cultured erythroleukemic Friend cells. Cell. 1976;8:495–503. doi: 10.1016/0092-8674(76)90217-8. [DOI] [PubMed] [Google Scholar]
- 2.Basak AN, Ozer A, Kirdar B, Akar N. A novel 13 bp deletion in the 3′UTR of the β-globin gene causes β-thalassemia in a Turkish patient. Hemoglobin. 1993;17:551–555. doi: 10.3109/03630269309043496. [DOI] [PubMed] [Google Scholar]
- 3.Bastos R, Aviv H. Theoretical analysis of a model for globin messenger RNA accumulation during erythropoiesis. J Mol Biol. 1977;110:205–218. doi: 10.1016/s0022-2836(77)80069-7. [DOI] [PubMed] [Google Scholar]
- 4.Bastos R, Volloch Z, Aviv H. Messenger RNA population analysis during erythroid differentiation: a kinetic approach. J Mol Biol. 1977;110:191–203. doi: 10.1016/s0022-2836(77)80068-5. [DOI] [PubMed] [Google Scholar]
- 5.Baumbach L L, Stein G S, Stein J L. Regulation of human histone gene expression: transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry. 1987;26:6178–6187. doi: 10.1021/bi00393a034. [DOI] [PubMed] [Google Scholar]
- 6.Beelman C, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein P, Peltz S W, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunn H F, Forget B G. Hemoglobin: molecular, genetic, and clinical aspects. Philadelphia, Pa: The W. B. Saunders Co.; 1986. [Google Scholar]
- 9.Bunn H F, Schmidt G J, Haney D N, Dluhy R G. Hemoglobin Cranston, an unstable variant having an elongated beta chain due to nonhomologous crossover between two normal beta chain genes. Proc Natl Acad Sci USA. 1975;72:3609–3613. doi: 10.1073/pnas.72.9.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai S P, Eng B, Francombe W H, Olivieri N F, Kendall A G, Waye J S, Chui D H. Two novel beta-thalassemia mutations in the 5′ and 3′ noncoding regions of the beta-globin gene. Blood. 1992;79:1342–1346. [PubMed] [Google Scholar]
- 11.Chen C Y, Del Gatto-Koncziak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 12.Chen C Y, Gherzy R, Andersen J S, Gaietta G, Jurchott K, Royer H D, Mann M, Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–1258. [PMC free article] [PubMed] [Google Scholar]
- 13.Chkheidze A N, Lyakhov D L, Makeyev A V, Morales J, Kong J, Liebhaber S A. Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein αCP. Mol Cell Biol. 1999;19:4572–4581. doi: 10.1128/mcb.19.7.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clegg J B, Weatherall D J, Contopolou-Griva I, Cartousos K, Poungouras P, Tsevrenis H. Haemoglobin Icaria, a new chain-termination mutant which causes α thalassemia. Nature. 1974;251:245–247. doi: 10.1038/251245a0. [DOI] [PubMed] [Google Scholar]
- 15.Cleveland D W, Lopata M A, Sherline P, Kirschner M W. Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell. 1981;25:537–546. doi: 10.1016/0092-8674(81)90072-6. [DOI] [PubMed] [Google Scholar]
- 16.Czyzyk-Krzeska M F, Beresh J E. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J Biol Chem. 1996;271:3293–3299. doi: 10.1074/jbc.271.6.3293. [DOI] [PubMed] [Google Scholar]
- 17.De Jong W W, Meera Khan P, Bernini L F. Hemoglobin Koya Dora: high frequency of a chain termination mutant. Am J Hum Genet. 1975;27:81–90. [PMC free article] [PubMed] [Google Scholar]
- 18.Delanoe-Garin J, Blouquit Y, Arous N, Kister J, Poyart C, North M L, Bardakdjian J, Lacombe C, Rosa J, Galacteros F. Hemoglobin Saverne: a new variant with elongated beta chains: structural and functional properties. Hemoglobin. 1988;12:337–352. doi: 10.3109/03630268808998034. [DOI] [PubMed] [Google Scholar]
- 19.Flatz G, Kinderlerer J L, Kilmartin J V, Lehmann H. Haemoglobin Tak: a variant with additional residues at the end of the beta-chains. Lancet. 1971;i:732–733. doi: 10.1016/s0140-6736(71)91994-5. [DOI] [PubMed] [Google Scholar]
- 20.Frischmeyer P A, Dietz H C. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 21.Furuichi Y, LaFiandra A, Shatkin A J. 5′-Terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 22.Goodman M. Decoding the pattern of protein evolution. Prog Biophys Mol Biol. 1981;38:105–164. doi: 10.1016/0079-6107(81)90012-2. [DOI] [PubMed] [Google Scholar]
- 23.Goodman M, Weiss M L, Czelusniak J. Molecular evolution above the species level: branching patterns, rates, and mechanisms. Sys Zool. 1982;31:376–399. [Google Scholar]
- 24.Hamalainen L, Oikarinen J, Kivirikko K I. Synthesis and degradation of type I procollagen mRNAs in cultured human skin fibroblasts and the effect of cortisol. J Biol Chem. 1985;260:720–725. [PubMed] [Google Scholar]
- 25.Hanash S M, Winter W P, Rucknagel D L. Synthesis of haemoglobin Wayne in erythroid cells. Nature. 1977;269:717–719. doi: 10.1038/269717a0. [DOI] [PubMed] [Google Scholar]
- 26.Harrold S, Genovese C, Kobrin B, Morrison S L, Milcarek C. A comparison of apparent mRNA half-life using kinetic labeling techniques vs decay following administration of transcriptional inhibitors. Anal Biochem. 1991;198:19–29. doi: 10.1016/0003-2697(91)90500-s. [DOI] [PubMed] [Google Scholar]
- 27.He Z, Lian L, Asakura T, Russell J E. Functional effects of replacing human α and β globins with their embryonic globin homologues in defined haemoglobin heterotetramers. Br J Haematol. 2000;109:882–890. doi: 10.1046/j.1365-2141.2000.02065.x. [DOI] [PubMed] [Google Scholar]
- 28.Holcik M, Liebhaber S A. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis- and trans components. Proc Natl Acad Sci USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jankovic L, Dimovski A J, Kollia P, Karageorga M, Loukopoulos D, Huisman T H J. A C----G mutation at nt position 6 3′ to the terminating codon may be the cause of a silent beta-thalassemia. Int J Hematol. 1991;54:289–293. [PubMed] [Google Scholar]
- 30.Kabnick K S, Housman D E. Determinants that contribute to cytoplasmic stability of human c-fos and β-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988;8:3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiledjian M, Day N, Trifillis P. Purification and RNA binding properties of the polycytidylate-binding proteins αCP1 and αCP2. Methods. 1999;17:84–91. doi: 10.1006/meth.1998.0710. [DOI] [PubMed] [Google Scholar]
- 32.Kiledjian M, DeMaria C T, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol Cell Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiledjian M, Wang X, Liebhaber S A. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klausner R D, Rouault T A, Harford J B. Regulating the fate of mRNA: the control of cellullar iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 35.Krowczynska A, Yenofsky R, Brawerman F. Regulation of messenger RNA stability in mouse erythroleukemia cells. J Mol Biol. 1985;181:231–239. doi: 10.1016/0022-2836(85)90087-7. [DOI] [PubMed] [Google Scholar]
- 36.Li X A, Beebe D C. Messenger RNA stabilization in chicken lens development: a reexamination. Dev Biol. 1991;146:239–241. doi: 10.1016/0012-1606(91)90463-d. [DOI] [PubMed] [Google Scholar]
- 37.Lieberman A P, Pitha P M, Shin M L. Poly(A) removal is the kinase-regulated step in tumor necrosis factor mRNA decay. J Biol Chem. 1992;267:2123–2126. [PubMed] [Google Scholar]
- 38.Liebhaber S A, Kan Y W. Alpha thalassemia caused by an unstable alpha-globin mutant. J Clin Investig. 1983;71:461–466. doi: 10.1172/JCI110790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liebhaber S A, Wang Z, Cash F E, Monks B, Russell J E. Developmental silencing of the embryonic ζ-globin gene: concerted action of the promoter and the 3′-flanking region combined with stage-specific silencing by the transcribed segment. Mol Cell Biol. 1996;16:2637–2646. doi: 10.1128/mcb.16.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lodish H F, Small B. Different lifetimes of reticulocyte messenger RNA. Cell. 1976;7:59–65. doi: 10.1016/0092-8674(76)90255-5. [DOI] [PubMed] [Google Scholar]
- 41.Maatta A, Ekholm E, Penttinen R P. Effect of the 3′-untranslated region on the expression levels and mRNA stability of alpha 1 (I) collagen gene. Biochim Biophys Acta. 1995;1260:294–300. doi: 10.1016/0167-4781(94)00207-j. [DOI] [PubMed] [Google Scholar]
- 42.Makeyev A V, Chkheidze A N, Liebhaber S A. A set of highly conserved RNA-binding proteins, αCP-1 and αCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J Biol Chem. 1999;274:24849–24857. doi: 10.1074/jbc.274.35.24849. [DOI] [PubMed] [Google Scholar]
- 43.Maquat L E. When cells stop making sense: effects of nonsence codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 44.Milner P F, Clegg J B, Weatherall D J. Haemoglobin H disease due to a unique haemoglobin variant with an elongated α-chain. Lancet. 1971;i:729–732. doi: 10.1016/s0140-6736(71)91992-1. [DOI] [PubMed] [Google Scholar]
- 45.Morales J, Russell J E, Liebhaber S A. Destabilization of human alpha globin mRNA by translation anti-termination is controlled during erythroid terminal differentiation and paralleled by phased shortening of the poly(A) tail. J Biol Chem. 1997;272:6607–6613. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- 46.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translation initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostareck-Lederer A, Ostareck D H, Standart N, Thiele B J. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandey N B, Marzluff W F. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ross J, Kobs G, Brewer G, Peltz S W. Properties of the exonuclease activity that degrades H4 histone mRNA. J Biol Chem. 1987;262:9374–9381. [PubMed] [Google Scholar]
- 51.Ross J, Pizarro A. Human beta and delta globin messenger RNAs turn over at different rates. J Mol Biol. 1983;167:607–617. doi: 10.1016/s0022-2836(83)80101-6. [DOI] [PubMed] [Google Scholar]
- 52.Ross J, Sullivan T D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66:1149–1154. [PubMed] [Google Scholar]
- 53.Russell J E, Liebhaber S A. Reversal of lethal α and β thalassemias in mice by expression of human embryonic globins. Blood. 1998;92:3057–3063. [PubMed] [Google Scholar]
- 54.Russell J E, Liebhaber S A. The stability of human beta-globin mRNA is dependent on structural determinants positioned within its 3′ untranslated region. Blood. 1996;87:5314–5323. [PubMed] [Google Scholar]
- 55.Russell J E, Morales J, Liebhaber S A. The role of globin mRNA stability in the control of globin gene expression. Prog Nucleic Acid Res Mol Biol. 1997;57:259–287. doi: 10.1016/s0079-6603(08)60283-4. [DOI] [PubMed] [Google Scholar]
- 56.Russell J E, Morales J, Makeyev A V, Liebhaber S A. Sequence divergence in the 3′ untranslated regions of human ζ- and α-globin mRNAs mediates a difference in their stabilities and contributes to efficient α-to-ζ gene developmental switching. Mol Cell Biol. 1998;18:2173–2183. doi: 10.1128/mcb.18.4.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukayotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 58.Scheper W, Meinsma D, Holthuizen P E, Sussenbach J S. Long-range RNA interaction of two sequence elements required for endonucleolytic cleavage of human insulin-like growth factor II mRNAs. Mol Cell Biol. 1995;15:235–245. doi: 10.1128/mcb.15.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 60.Shimotohno K, Kodama Y, Hashimoto J, Miura K I. Importance of 5′-terminal blocking structure to stabilize mRNA in eukaryotic protein synthesis. Proc Natl Acad Sci USA. 1977;74:2734–2738. doi: 10.1073/pnas.74.7.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shyu A B, Greenberg M E, Belasco J G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 62.Stefanovic B, Hellerbrand C, Holcik M, Briendl M, Liebhaber S A, Brenner D A. Posttranscriptional regulation of collagen α1(I) mRNA in hepatic satellite cells. Mol Cell Biol. 1997;17:5201–5209. doi: 10.1128/mcb.17.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Talbot D, Collis P, Antoniou M, Vidal M, Grosveld F, Greaves D R. A dominant control region from human beta-globin locus conferring integration site independent gene expression. Nature. 1989;33:352–355. doi: 10.1038/338352a0. [DOI] [PubMed] [Google Scholar]
- 64.Thein S L. Structural variants with a β-thaslassemic phenotype. In: Steinberg M A, Forget B G, Higgs D R, Nagel R L, editors. Disorders of hemoglobin: genetics, pathophysiology, and clinical management. Cambridge, England: Cambridge University Press; 2001. pp. 342–355. [Google Scholar]
- 65.Trudel M, Constantini F. A 3′ enhancer contributes to the stage-specific expression of the human beta-globin gene. Genes Dev. 1987;1:954–961. doi: 10.1101/gad.1.9.954. [DOI] [PubMed] [Google Scholar]
- 66.Volloch V, Housman D. Stability of globin mRNA in terminally differentiating murine erythroleukemia cells. Cell. 1981;23:509–514. doi: 10.1016/0092-8674(81)90146-x. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, Kiledjian M, Weiss I M, Liebhaber S A. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability. Mol Cell Biol. 1995;15:1769–1777. doi: 10.1128/mcb.15.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Z, Kiledjian M. Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. EMBO J. 2000;19:295–305. doi: 10.1093/emboj/19.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Z, Kiledjian M. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol. 2000;20:6334–6341. doi: 10.1128/mcb.20.17.6334-6341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weiss I M, Liebhaber S A. Erythroid cell-specific determinants of α-globin mRNA stability. Mol Cell Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiss I M, Liebhaber S A. Erythroid cell-specific mRNA stability elements in the α(2)-globin 3′ nontranslated region. Mol Cell Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson T, Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 74.Wisdom R, Lee W. The protein coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991;5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- 75.Zhang W, Wagner B, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF-1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]