Abstract
Background
SARS-CoV-2 infection can lead to the abnormal induction of cytokines and a dysregulated hyperinflammatory state that is implicated in disease severity and risk of death. There are several molecules present in blood associated with immune cellular response, inflammation, and oxidative stress that could be used as severity markers in respiratory viral infections such as COVID-19. However, there is a lack of clinical studies evaluating the role of oxidative stress-related molecules including glial fibrillary acidic protein (GFAP), the receptor for advanced glycation end products (RAGE), high mobility group box-1 protein (HMGB1) and cyclo-oxygenase-2 (COX-2) in COVID-19 pathogenesis.
Aim
To evaluate the role of oxidative stress-related molecules in COVID-19.
Method
An observational study with 93 Brazilian participants from September 2020 to April 2021, comprising 23 patients with COVID-19 admitted to intensive care unit (ICU), 19 outpatients with COVID-19 with mild to moderate symptoms, 17 individuals reporting a COVID-19 history, and 34 healthy controls. Blood samples were taken from all participants and western blot assay was used to determine the RAGE, HMGB1, GFAP, and COX-2 immunocontent.
Results
We found that GFAP levels were higher in patients with severe or critical COVID-19 compared to outpatients (p = 0.030) and controls (p < 0.001). A significant increase in immunocontents of RAGE (p < 0.001) and HMGB1 (p < 0.001) were also found among patients admitted to the ICU compared to healthy controls, as well as an overexpression of the inducible COX-2 (p < 0.001). In addition, we found a moderate to strong correlation between RAGE, GFAP and HMGB1 proteins.
Conclusion
SARS-CoV-2 infection induces the upregulation of GFAP, RAGE, HMGB1, and COX-2 in patients with the most severe forms of COVID-19.
Keywords: COVID-19, RAGE, GFAP, COX-2, HMGB1, Neurological dysfunction
1. Introduction
Coronavirus disease 2019 (COVID-19) is defined primarily as a respiratory disease caused by a single-stranded RNA virus known as SARS-CoV-2. This novel betacoronavirus has a genome size of approximately 29.9 kb [1] packed in an envelope with four associated structural (S, E, M, and N) and sixteen non-structural proteins (nsp1 − 16) [2]. There is evidence that the SARS-CoV-2 S protein is essential for infectivity [3]. The binding between the SARS-CoV-2 S protein and the human angiotensin-converting enzyme 2 (hACE2) [4], which is expressed particularly in the kidneys, heart, and respiratory and gastrointestinal tract tissues can lead to the abnormal induction of cytokines and a dysregulated hyperinflammatory state that is implicated in disease severity and risk of death [5], [6].
Growing evidence has shown that hACE2 can also be expressed in brain capillary endothelial cells, neurons, astrocytes, oligodendrocytes, and the olfactory bulb [7]. In addition to the possibility of direct viral invasion, it has been proposed that SARS-CoV-2 infection can affect the nervous system through cerebrovascular changes, immune-mediated injury, and peripheral organ dysfunction [8], [9]. In a study carried out in Wuhan, China, with hospitalized COVID-19 patients, a prevalence of 36.4% of nervous system manifestations was found, mainly dizziness, headache, and chemosensory disturbances. Moreover, neurological symptoms were significantly more common in severe infections and included acute cerebrovascular disease, impaired consciousness, and skeletal muscle injury [10].
Although studies have suggested a possible direct viral invasion of SARS-CoV-2 into hepatocytes [11], cardiomyocytes [12], alveolar epithelial cells [13] and the nervous system [14], most clinical manifestations of COVID-19 are associated with systemic inflammation and oxidative stress. It has been demonstrated that reduction in ACE2 bioavailability after SARS-CoV-2 binding allows angiotensin II to be available to interact with AT1R, which mediates signals for activate nicotinamide adenine dinucleotide phosphate (NADPH) oxidase leading to oxidative stress and inflammatory responses [15]. Therefore, several molecules present in the blood that are associated with immune cellular response, inflammation, and oxidative stress could be used as severity markers in COVID-19 patients. There is evidence that the increased levels of IL-6, C-reactive protein, ferritin, and procalcitonin; increased erythrocyte sedimentation rate; decreased CD4 and CD8 count cells; changes in biochemical indices (albumin, blood urea nitrogen, creatinine, creative kinase, hypersensitive cardiac troponin I, and lactate dehydrogenase); and coagulation abnormalities, including prolonged prothrombin time, increased d-dimer and thrombocytopenia, are important predictors for mortality in patients with COVID-19 [6]. However, there is a lack of clinical studies evaluating the role of oxidative stress-related molecules including glial fibrillary acidic protein (GFAP), receptor for advanced glycation end products (RAGE), high mobility group box-1 protein (HMGB1) and cyclo-oxygenase-2 (COX-2) in COVID-19 pathogenesis.
GFAP is a cytoskeleton glial protein expressed in astrocytes that regulates the morphology and function of these cells in the central nervous system (CNS). The plasmatic concentration of GFAP is typically not detectable in healthy individuals [16], but increased GFAP levels are considered a nonspecific marker of CNS pathology [17]. Recent studies have shown increased levels of GFAP in patients with COVID-19 and discussed the potential role of this biomarker in SARS-CoV-2 neuropathogenesis [18], [19], [20].
RAGE is a member of the immunoglobulin superfamily of receptors and can be present in two forms: membrane-bound RAGE (mRAGE) and soluble RAGE (sRAGE). This receptor is expressed in a wide array of cell types including microglia, neurons, endothelial vascular cells, cardiomyocytes and alveolar pneumocytes [21], [22], [23], and is involved in inflammatory responses, oxidative stress, and cellular dysfunction in several conditions [24], [25]. It was found that RAGE can play an important role as a receptor target for HMGB1, a nonhistone chromosomal protein that mediates late activation of the innate immune response to infection, including the release of chemotactic factors, the upregulation of the COX-2/PGE2 axis, and the production of proinflammatory cytokines [26], [27], [28]. In addition, there is evidence that some respiratory viruses can lead to oxidative stress due to the sequestration of the secretory pathway for the manufacture of viral glycoproteins on infected cells [29], and thus mediate HMGB1 secretion, amplifying the inflammatory response and disease severity [30], [31].
This clinical study evaluated the role of RAGE, HMGB1, GFAP (and breakdown products) and COX-2 in patients with COVID-19.
2. Materials and methods
2.1. Study design
This observational study enrolled 93 individuals from Sergipe state, Northeast Brazil, from September 2020 up to April 2021. This group comprised 23 patients with COVID-19 admitted to intensive care unit (ICU), 19 outpatients with COVID-19 with mild to moderate symptoms, 17 individuals reporting a COVID-19 history, and 34 healthy controls. All individuals were tested using real-time reverse-transcriptase polymerase chain reaction (RT-PCR) from a nasopharyngeal swab or by serology because of the limited availability of laboratory resources and molecular tests in a resource poor setting. Blood was collected from all individuals to test for oxidative stress-related molecules using western blot assay. Demographic data (age and sex) were collected from all patients. Participants with negative serology without a history of positive RT-PCR for SARS-CoV-2 were included in the control group. All participants received written information about the study and were asked to give written informed consent for participation. This study was approved by the institutional review board of the Federal University of Sergipe (CAAE: 34240620.7.0000.5546).
2.2. Plasma sample preparation
Blood samples were collected with 4 mL EDTA tubes and processed by centrifugation at 2500 g for 15 min at room temperature. Plasma aliquots were frozen at − 80 °C and the samples were thawed only once during the processing. To perform the analysis, total plasma proteins were measured by Bradford assay [32]. The levels of RAGE, HMGB1, GFAP, and COX-2 were measured in plasma samples as described below.
2.3. Western blot assay
Western blot assay was used to determine the RAGE, HMGB1, GFAP, and COX-2 immunocontent. Plasma aliquots from patients and controls were prepared by addition of Laemmli buffer 4x and heated at 95 °C for 5 min. First, measured plasma proteins (total proteins) allowed the equivalent amount of proteins required for each sample to be determined (40 μg of proteins). Samples were separated by SDS-PAGE and transferred to nitrocellulose membranes (BioRad) for 1 h at 25 V in a transfer buffer (48 mM Tris, 39 mM glycine, 20% methanol and 1.3 mM SDS). The nitrocellulose membranes were washed for 5 min in Tris–buffered saline with 0.1% Tween-20 (T-TBS; 0.5 M NaCl, 20 mM Tris, 0.1% Tween-20, pH 7.5), followed by 1 h incubation in blocking solution (T-TBS plus 5% defatted dried milk). The blots were incubated overnight at 4 °C in blocking solution containing the following antibodies at a dilution of 1:1000: anti-COX2 [(clone D5H5) Cell Signaling Technology], anti-RAGE [Cell Signaling Technology, cs 4679], anti-HMGB1 [(D3E5) Cell Signaling Technology, cs 6893] and anti-GFAP [(GA5) Cell Signaling Technology, cs 3670]. Then, the blots were washed three times for 15 min with T-TBS and incubated for 2 h in blocking solution containing peroxidase conjugated anti-rabbit or anti-mouse IgG diluted 1:4000. Finally, the blots were washed twice again for 15 min with T-TBS and twice for 15 min with TBS, and developed using a chemiluminescence substrate. The densitometric band quantification was performed by ImageJ 1.5 k software (National Institute of Health, USA). Normalization of western blots was performed using Ponceau S. Results were expressed as arbitrary units.
2.4. Statistical analysis
Data were expressed as median and quartiles (Q1 and Q3). Differences in biomarker levels between groups were determined using the Kruskal-Wallis test followed by Dunn's post hoc test for multiple comparisons. Correlations between biomarkers were tested using Spearman's correlation coefficients (rho). The strength of the correlation was considered very weak if the rho was 0–0.19; weak, 0.2–0.39; moderate, 0.4–0.59; strong, 0.6–0.79; and very strong, 0.8–1.0. Two-sided p-values less than 0.05 were considered statistically significant. Analyses were performed using Prism 8.0 software (GraphPad Prism Software Inc., San Diego, CA, USA).
3. Results
3.1. Sample characteristics
The mean age of participants was 44.6 ± 16.4 years (minimum 11, maximum 100), and most of them were female (55.9%; 52/93). All patients with COVID-19 admitted to the ICU had severe or critical COVID-19, whereas all outpatients had mild to moderate symptoms and most individuals with COVID-19 history were asymptomatic
3.2. Immunocontent of GFAP in plasma
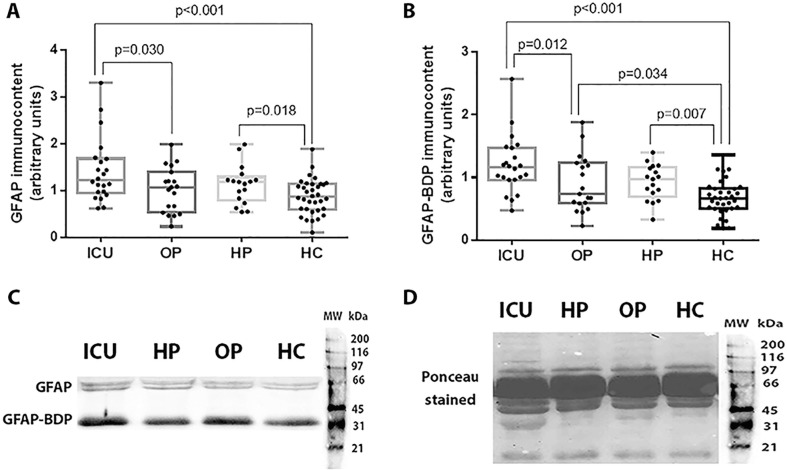
Fig. 1 summarizes the levels of GFAP and GFAP breakdown products (GFAP-BDP) among COVID-19 patients admitted to the ICU, outpatients, individuals with COVID-19 history and controls. We found that GFAP and GFAP-BDP levels were higher in patients with severe or critical COVID-19 compared to the outpatients (GFAP: 1.22 [0.97 – 1.66] vs. 1.07 [0.60 – 1.30]; p = 0.030 / GFAP-BDP: 1.16 [0.97 – 1.43] vs. 0.74 [0.59 – 1.21]; p = 0.012) and controls (GFAP: 1.22 [0.97 – 1.66] vs. 0.86 [0.61 – 1.13]; p < 0.001 / GFAP-BDP: 1.16 [0.97 – 1.43] vs. 0.67 [0.51 – 0.82]; p < 0.001). Individuals with COVID-19 history had also higher levels of GFAP (p = 0.018) and GFAP-BDP (p = 0.007) than controls. In addition, our analysis showed that GFAP-BDP was higher in outpatients with COVID-19 compared to the healthy control group (p = 0.034).
Fig. 1.
GFAP (A) and GFAP-BDP (B) levels among COVID-19 patients admitted to the ICU (ICU), outpatients (OP), patients with COVID-19 history (HP), and healthy controls (HC). Representative blots and Ponceau S stain are shown in Fig. 1C and 1D, respectively. Statistically significant differences were determined by Kruskal-Wallis test followed by Dunn's post hoc test for multiple comparisons.
3.3. Immunocontent of RAGE and HMGB1 in plasma
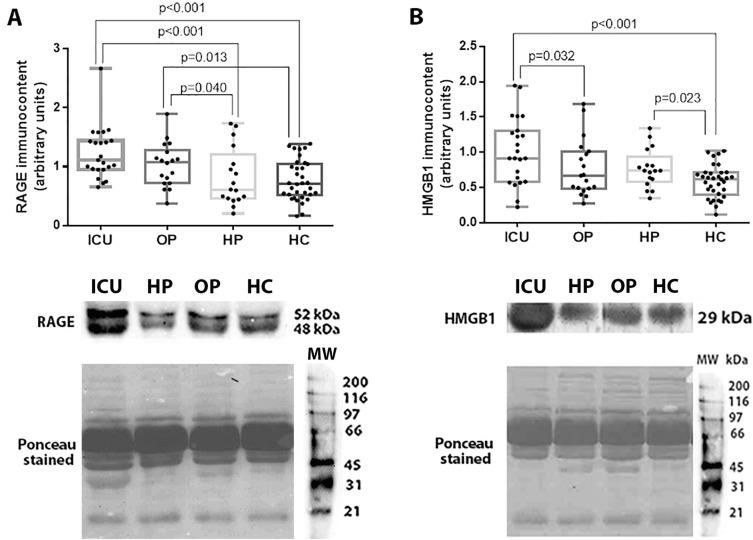
A significant increase in immunocontents of RAGE were found among patients admitted to the ICU (1.11 [0.96 – 1.43]) compared to those with a COVID-19 history (0.61 [0.46 – 1.05]; p < 0.001) and healthy controls (0.71 [0.53 – 1.03]; p < 0.001). Outpatients with COVID-19 also had higher levels of RAGE than individuals with a COVID-19 history (p = 0.040) and controls (p = 0.013) (Fig. 2 A).
Fig. 2.
RAGE (A) and HMGB1 (B) levels among COVID-19 patients admitted to the ICU (ICU), outpatients (OP), patients with COVID-19 history (HC), and healthy controls (HC). Representative blots and Ponceau S stain for RAGE and HMGB1 are shown in Fig. 2C and 2D, respectively. Statistically significant differences were determined by Kruskal-Wallis test followed by Dunn's post hoc test for multiple comparisons.
HMGB1 levels were higher among patients in ICU group (0.91 [0.61 – 1.29]) compared to outpatients with COVID-19 (0.67 [0.49 – 0.99]; p = 0.032) and controls (0.62 [0.42 – 0.71]; p < 0.001). In addition, we found differences between HMGB1 levels between patients with COVID-19 history (higher levels) and healthy controls (p = 0.023) (Fig. 2B). RAGE and HMGB1 levels, representative blots and Ponceau S stain for each study group are shown in Fig. 2.
3.4. Immunocontent of COX-2
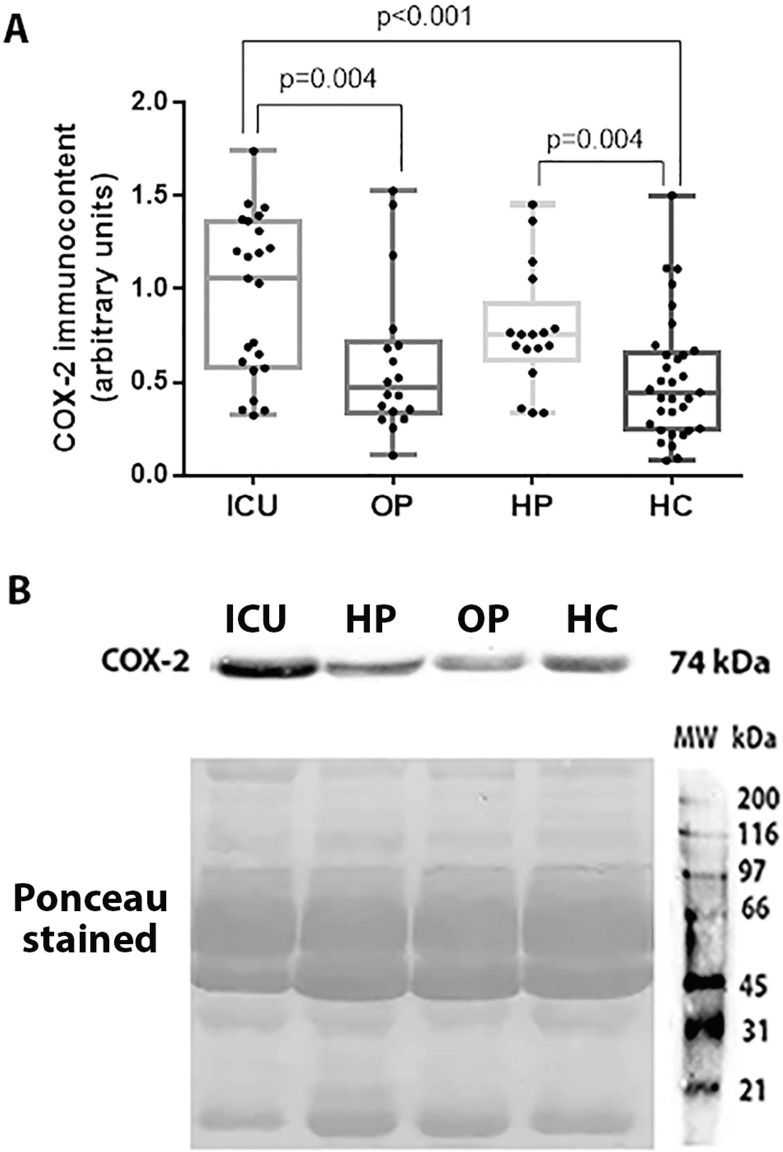
The present study demonstrated that the inducible COX-2 is overexpressed in patients with severe or critical SARS-CoV-2 infection (1.04 [0.60 – 1.33]) compared to patients with mild to moderate COVID-19 (0.47 [0.35 – 0.69]; p = 0.004) and healthy controls (0.45 [0.25 – 0.65]; p < 0.001). Individuals with COVID-19 history had also higher levels of COX-2 than controls (p = 0.004) (Fig. 3 ).
Fig. 3.
COX-2 immunocontent in the ICU group (ICU), outpatients (OP), patients with COVID-19 history (HP), and healthy controls (HC) (A). Representative blots and Ponceau S stained are shown in Figure B. Statistically significant differences were determined by Kruskal-Wallis test followed by Dunn's post hoc test for multiple comparisons.
3.5. Correlations between biomarkers levels
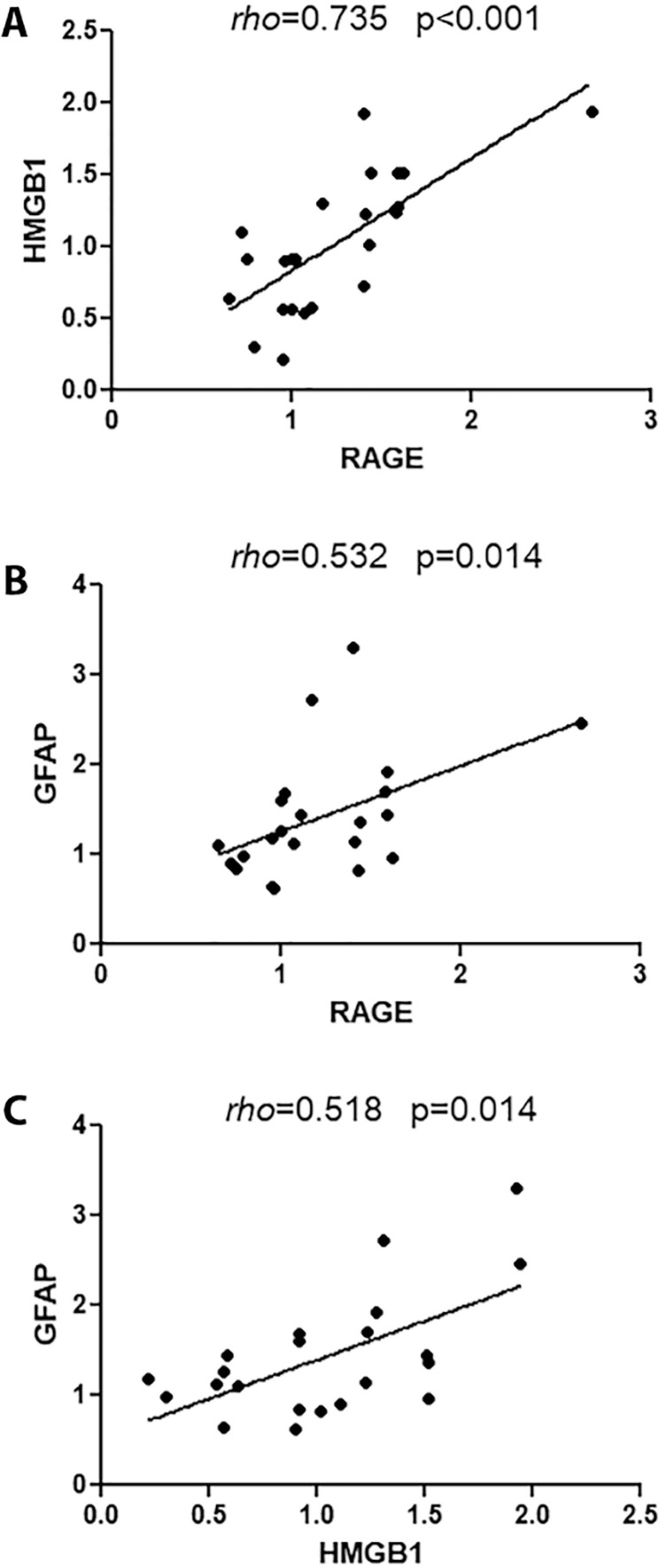
We explored the relationship between (1) RAGE and HMGB1; (2) RAGE and GFAP; (3) RAGE and COX-2; (4) HMGB1 and GFAP; and (5) HMGB1 and COX-2 using the samples from patients admitted to the ICU. There was a strong positive correlation between RAGE and HMGB1 (p < 0.001; rho = 0.735) (Fig. 4 A) and a moderate positive correlation between RAGE and GFAP (p = 0.014; rho = 0.532) (Fig. 4B). In addition, we found a moderate positive correlation between HMGB1 and GFAP (p = 0.014; rho = 0.518) (Fig. 4C). No correlation was found between RAGE and GFAP-BDP (p = 0.434), RAGE and COX-2 (p = 0.141), HMGB1 and GFAP-BDP (p = 0.360), and HMGB1 and COX-2 (p = 0.375).
Fig. 4.
Correlations between RAGE and HMGB1 (A), RAGE and GFAP (B), and HMGB1 and GFAP (C) in patients admitted to the ICU.
4. Discussion
There is evidence that SARS-CoV-2 infection can increase the risk of cardiac, renal, gastrointestinal, and endocrine complications [33], [34]. Moreover, patients with COVID-19 can develop a broad spectrum of neurological symptoms including ischemic stroke, meningoencephalitis, encephalopathy, acute necrotizing encephalopathy, Guillain-Barré syndrome, and acute disseminated encephalomyelitis [14], [35], [36], [37]. A wide range of biomolecules associated with the inflammatory and immune cell response, which are used as predictors of organ failure and severity in other diseases, are also being studied in patients with COVID-19 [38], [39]. It has been found that several inflammatory and oxidative mediators are misregulated in COVID-19 and are associated with multiorgan damage and poor disease prognosis [40], [41].
This study evaluated the role of the inflammatory and oxidative biomolecules GFAP, RAGE, HMGB1 and COX-2 in the COVID-19 pathophysiology. Our findings showed that the levels of these biomarkers were higher in patients with the most severe forms of COVID-19. In addition, we found a moderate to strong correlation between RAGE, GFAP and HMGB1 proteins.
The results obtained from the analysis of GFAP and GFAP-DBP levels may suggest that patients with SARS-CoV-2 infection are at increased risk of astrocytic damage and neural dysfunction, especially among those with severe or critical COVID-19. These findings have also been suggested in other studies [19], [42], [43] and reinforce the capacity for neural damage caused by the infection. There is growing evidence of the role of astrocytes in the brain response to SARS-CoV-2-triggered neuroinflammation. It has been found that these cells might be targets of SARS-CoV-2 [44] and play a key role in the control of the cytokine microenvironment and brain function in COVID-19 [45], [46]. Therefore, elevated serum levels of GFAP may be useful for diagnosis and prognosis of COVID-19 and brain dysfunction.
Furthermore, the disintegration of astrocytes, detected by the GFAP upregulation, can stimulate the overexpression of other inflammatory mediators involved in neurological damage. Necroptotic astrocytes release the HMGB1 protein and stimulate the HMGB1-RAGE axis [47], [48] that can induce inflammatory events leading to organ dysfunctions, including the brain. The binding of HMGB1 to RAGE promotes the transcription of inflammatory factors including the NFκB and MAPK pathways [49], and its sustained activation causes cell death by stimulating the production of reactive oxygen species [50]. It has been found that the HMGB1-RAGE axis plays a significant role in a wide spectrum of pathological responses such as Alzheimer's disease, brain injury-induced pulmonary dysfunction, chronic inflammatory conditions, cancer, and sepsis [51], [52], [53], [54], [55].
Recent studies have suggested the activation of HMGB1-RAGE cascade in COVID-19 pathogenesis [56], [57] and an association with disease severity [21], [58]. In this respect, our results showed an overexpression of HMGB1 and RAGE proteins in the plasma of patients with COVID-19, confirming that this effect is a common finding in the most severe forms of the disease. In addition, the misregulation of RAGE signaling has been reported as an important factor in sustaining the pathological states in the most common comorbidities of COVID-19 [59] . Therefore, the assessment of the HMGB1-RAGE axis as a predictor of COVID-19 severity may be important in individuals with pre-existing medical conditions.
The cyclooxygenase family has also been found to play an important role in COVID-19 [60]. The inducible COX‐2 enzyme has a central role in viral infections and regulates the expression levels of many serum proteins, including proinflammatory cytokines that directly influence the physiological regulation [61]. There is evidence that COX-2 overexpression has been described in patients who have died of H5N1 infection [62], suggesting that this enzyme is associated with poor clinical outcomes in viral infections. Moreover, the production of COX‐2 metabolites can lead to coagulopathy and hyperinflammation [63], common findings in patients with severe COVID-19. Recently, it was found that SARS-CoV-2 induced COX-2 upregulation in diverse human cell culture and mouse systems [64], which may be implicated in regulating lung inflammation and disease severity as observed in the present study. In addition, the upregulation of COX-2 could increase the mortality and morbidity rate in COVID-19 patients. The high levels of COX-2 cause a depletion in the endogenous antiviral compound arachidonic acid, making individuals more susceptible to COVID-19 [65].
Finally, the hyperinflammation induced by SARS-CoV-2 infection can potentiate the generation of oxygen reactive species leading to mitochondrial damage, apoptosis, expression of inflammatory cytokines, vascular endothelial changes, disruption of cellular repair mechanisms, and disease progression [66], [67]. In addition, oxidative stress genes were found to be upregulated in the whole blood of COVID-19 patients [68]. Therefore, the imbalance in homeostasis redox contributes to the pathophysiology of COVID-19 [69] and is an important prognostic factor of the disease.
Although the study was the first to assess all these biomarkers together as predictors of severity for COVID-19, it has some limitations which should be noted. First, we used a convenience sample which may increase the risk of selection bias. Second, due to the nature of the study, patients were not followed and outcomes such as death were not evaluated.
5. Conclusion
SARS-CoV-2 infection induces the upregulation of GFAP, RAGE, HMGB1, and COX-2 in patients with the most severe forms of COVID-19 and can be associated with organ dysfunctions including neural structures. The levels of these biomolecules can reflect inflammatory, oxidative, and neurological alterations in patients with SARS-CoV-2 infection and guide future patient-targeted therapies in clinical practice.
CRediT authorship contribution statement
FabiollaRocha Santos Passos: Conceptualization, Resources, Data curation, Formal analysis, Software, Investigation, Methodology, Writing-original-draft. LuanaHeimfarth: Conceptualization, Resources, Data curation, Formal analysis, Software, Investigation, Methodology, Writing-original-draft. BrendaSouza Monteiroa: Data curation, Formal analysis, Software. CristianeBani Corrêa: Data curation, Formal analysis, Software. TatianaRodrigues de Moura: Data curation, Formal analysis, Software. AdrianoAntunes de Souza Araújo: Data curation, Formal analysis, Supervision, Validation, Visualization, Writing-review-editing. PauloRicardo Martins-Filho: Supervision, Validation, Visualization, Writing-review-editing. LucindoLucindo JoséJosé Quintans: Conceptualization, Resources, Funding acquisition, Project administration. Jullyanade Souza Siqueira Quintans: Conceptualization, Resources, Data curation, Formal analysis, Software, Investigation, Methodology, Writing-original-draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from FAPESE, FAPITEC-SE, CNPq and CAPES, all agencies from Brazil. The Federal University of Sergipe has been one of the leading institutions in the fight against COVID-19 in the state of Sergipe. This study is part of the EpiSERGIPE project, which has helped to guide the response to COVID-19 in our state. We dedicate this article to all frontline healthcare professionals who have died or are on the frontline against COVID-19.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2021.108502.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
References
- 1.Godeau D., Petit A., Richard I., Roquelaure Y., Descatha A. Return-to-work, disabilities and occupational health in the age of COVID-19. Scand. J. Work Environ. Health. 2021;47(5):408–409. doi: 10.5271/sjweh.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M.-Y., Zhao R., Gao L.-J., Gao X.-F., Wang D.-P., Cao J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front Cell Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q.i., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 4.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martins-Filho P.R., Tavares C.S.S., Santos V.S. Factors associated with mortality in patients with COVID-19. A quantitative evidence synthesis of clinical and laboratory data. Eur. J. Intern. Med. 2020;76:97–99. doi: 10.1016/j.ejim.2020.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen R., Wang K., Yu J., Howard D., French L., Chen Z., Wen C., Xu Z. The Spatial and Cell-Type Distribution of SARS-CoV-2 Receptor ACE2 in the Human and Mouse Brains. Front Neurol. 2020;11 doi: 10.3389/fneur.2020.573095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed M.U., Hanif M., Ali M.J., Haider M.A., Kherani D., Memon G.M., Karim A.H., Sattar A. Neurological Manifestations of COVID-19 (SARS-CoV-2): A Review. Front Neurol. 2020;11:518. doi: 10.3389/fneur.2020.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heneka M.T., Golenbock D., Latz E., Morgan D., Brown R. Immediate and long-term consequences of COVID-19 infections for the development of neurological disease. Alzheimers Res. Ther. 2020;12:69. doi: 10.1186/s13195-020-00640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., Miao X., Li Y., Hu B. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2019;77(2020):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Yijin, Liu Shuhong, Liu Hongyang, Li Wei, Lin Fang, Jiang Lina, Li Xi, Xu Pengfei, Zhang Lixin, Zhao Lihua, Cao Yun, Kang Jiarui, Yang Jianfa, Li Ling, Liu Xiaoyan, Li Yan, Nie Ruifang, Mu Jinsong, Lu Fengmin, Zhao Shousong, Lu Jiangyang, Zhao Jingmin. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J. Hepatol. 2020;73(4):807–816. doi: 10.1016/j.jhep.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A., Garcia G., Arumugaswami V., Svendsen C.N. Human iPSC-Derived Cardiomyocytes are Susceptible to SARS-CoV-2 Infection. BioRxiv. 2020 doi: 10.1101/2020.04.21.051912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Huilan, Zhou Peng, Wei Yanqiu, Yue Huihui, Wang Yi, Hu Ming, Zhang Shu, Cao Tanze, Yang Chengqing, Li Ming, Guo Guangyun, Chen Xianxiang, Chen Ying, Lei Mei, Liu Huiguo, Zhao Jianping, Peng Peng, Wang Cong-Yi, Du Ronghui. Histopathologic Changes and SARS-CoV-2 Immunostaining in the Lung of a Patient With COVID-19. Ann. Intern. Med. 2020;172(9):629–632. doi: 10.7326/M20-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Araújo N.M., Ferreira L.C., Dantas D.P., Silva D.S., Dos Santos C.A., Cipolotti R., Martins-Filho P.R. First Report of SARS-CoV-2 Detection in Cerebrospinal Fluid in a Child With Guillain-Barré Syndrome. Pediatr. Infect Dis. J. 2021;40:e274–e276. doi: 10.1097/INF.0000000000003146. [DOI] [PubMed] [Google Scholar]
- 15.Beltrán-García J., Osca-Verdegal R., Pallardó F.V., Ferreres J., Rodríguez M., Mulet S., Sanchis-Gomar F., Carbonell N., García-Giménez J.L. Oxidative Stress and Inflammation in COVID-19-Associated Sepsis: The Potential Role of Anti-Oxidant Therapy in Avoiding Disease Progression. Antioxidants (Basel). 2020;9:E936. doi: 10.3390/antiox9100936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Missler U., Wiesmann M., Wittmann G., Magerkurth O., Hagenström H. Measurement of Glial Fibrillary Acidic Protein in Human Blood: Analytical Method and Preliminary Clinical Results. Clin. Chem. 1999;45:138–141. doi: 10.1093/clinchem/45.1.138. [DOI] [PubMed] [Google Scholar]
- 17.Brunkhorst Robert, Pfeilschifter Waltraud, Foerch Christian. Astroglial proteins as diagnostic markers of acute intracerebral hemorrhage-pathophysiological background and clinical findings. Transl Stroke Res. 2010;1(4):246–251. doi: 10.1007/s12975-010-0040-6. [DOI] [PubMed] [Google Scholar]
- 18.Cooper J., Stukas S., Hoiland R.L., Fergusson N.A., Thiara S., Foster D., Mitra A., Stoessl J.A., Panenka W.J., Sekhon M.S., Wellington C.L. Quantification of Neurological Blood-Based Biomarkers in Critically Ill Patients With Coronavirus Disease 2019. Critical Care Explorations. 2020;2 doi: 10.1097/CCE.0000000000000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanberg Nelly, Ashton Nicholas J., Andersson Lars-Magnus, Yilmaz Aylin, Lindh Magnus, Nilsson Staffan, Price Richard W., Blennow Kaj, Zetterberg Henrik, Gisslén Magnus. Neurochemical evidence of astrocytic and neuronal injury commonly found in COVID-19. Neurology. 2020;95(12):e1754–e1759. doi: 10.1212/WNL.0000000000010111. [DOI] [PubMed] [Google Scholar]
- 20.Virhammar Johan, Nääs Anja, Fällmar David, Cunningham Janet L., Klang Andrea, Ashton Nicholas J., Jackmann Sven, Westman Gabriel, Frithiof Robert, Blennow Kaj, Zetterberg Henrik, Kumlien Eva, Rostami Elham. Biomarkers for central nervous system injury in cerebrospinal fluid are elevated in COVID-19 and associated with neurological symptoms and disease severity. Eur. J. Neurol. 2021;28(10):3324–3331. doi: 10.1111/ene.14703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yalcin Kehribar Demet, Cihangiroglu Mustafa, Sehmen Emine, Avci Bahattin, Capraz Aylin, Yildirim Bilgin Ayse, Gunaydin Caner, Ozgen Metin. The receptor for advanced glycation end product (RAGE) pathway in COVID-19., Biomarkers : Biochemical Indicators of Exposure. Response, and Susceptibility to Chemicals. 2021;26(2):114–118. doi: 10.1080/1354750X.2020.1861099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brett J., Schmidt A.M., Yan S.D., Zou Y.S., Weidman E., Pinsky D., Nowygrod R., Neeper M., Przysiecki C., Shaw A. Survey of the distribution of a newly characterized receptor for advanced glycation end products in tissues. Am. J. Pathol. 1993;143:1699–1712. [PMC free article] [PubMed] [Google Scholar]
- 23.Daffu G., del Pozo C.H., O’Shea K.M., Ananthakrishnan R., Ramasamy R., Schmidt A.M. Radical roles for RAGE in the pathogenesis of oxidative stress in cardiovascular diseases and beyond. Int. J. Mol. Sci. 2013;14:19891–19910. doi: 10.3390/ijms141019891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt Ann Marie, Yan Shi Du, Yan Shi Fang, Stern David M. The biology of the receptor for advanced glycation end products and its ligands. Biochim. Biophys. Acta. 2000;1498(2-3):99–111. doi: 10.1016/s0167-4889(00)00087-2. [DOI] [PubMed] [Google Scholar]
- 25.Anzilotti Serenella, Giampà Carmela, Laurenti Daunia, Perrone Lorena, Bernardi Giorgio, Melone Mariarosa A.B., Fusco Francesca R. Immunohistochemical localization of receptor for advanced glycation end (RAGE) products in the R6/2 mouse model of Huntington’s disease. Brain Res. Bull. 2012;87(2-3):350–358. doi: 10.1016/j.brainresbull.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Lian Y.-J., Gong H., Wu T.-Y., Su W.-J., Zhang Y., Yang Y.-Y., Peng W., Zhang T., Zhou J.-R., Jiang C.-L., Wang Y.-X. Ds-HMGB1 and fr-HMGB induce depressive behavior through neuroinflammation in contrast to nonoxid-HMGB1. Brain Behav. Immun. 2017;59:322–332. doi: 10.1016/j.bbi.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Wang Haichao, Bloom Ona, Zhang Minghuang, Vishnubhakat Jaideep M., Ombrellino Michael, Che Jiantu, Frazier Asia, Yang Huan, Ivanova Svetlana, Borovikova Lyudmila, Manogue Kirk R., Faist Eugen, Abraham Edward, Andersson Jan, Andersson Ulf, Molina Patricia E., Abumrad Naji N., Sama Andrew, Tracey Kevin J. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285(5425):248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 28.Leclerc P., Wähämaa H., Idborg H., Jakobsson P.J., Harris H.E., Korotkova M. IL-1β/HMGB1 complexes promote The PGE2 biosynthesis pathway in synovial fibroblasts. Scand. J. Immunol. 2013;77(5):350–360. doi: 10.1111/sji.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Alice C-H, Burr Lucy, McGuckin Michael A. Oxidative and endoplasmic reticulum stress in respiratory disease. Clin. Transl. Immunology. 2018;7(6):e1019. doi: 10.1002/cti2.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Y., Tang D., Kang R. Oxidative stress-mediated HMGB1 biology. Front Physiol. 2015;6:93. doi: 10.3389/fphys.2015.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu W., Lu Y., Yao J., Li Z., Chen Z., Wang G., Jing H., Zhang X., Li M., Peng J., Tian X. Novel role of resveratrol: suppression of high-mobility group protein box 1 nucleocytoplasmic translocation by the upregulation of sirtuin 1 in sepsis-induced liver injury. Shock. 2014;42:440–447. doi: 10.1097/SHK.0000000000000225. [DOI] [PubMed] [Google Scholar]
- 32.Bradford Marion M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 33.Gavriatopoulou Maria, Korompoki Eleni, Fotiou Despina, Ntanasis-Stathopoulos Ioannis, Psaltopoulou Theodora, Kastritis Efstathios, Terpos Evangelos, Dimopoulos Meletios A. Organ-specific manifestations of COVID-19 infection. Clin. Exp. Med. 2020;20(4):493–506. doi: 10.1007/s10238-020-00648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Lu, Zhao Peijun, Tang Dazhong, Zhu Tong, Han Rui, Zhan Chenao, Liu Weiyong, Zeng Hesong, Tao Qian, Xia Liming. Cardiac Involvement in Patients Recovered From COVID-2019 Identified Using Magnetic Resonance Imaging. JACC Cardiovasc Imaging. 2020;13(11):2330–2339. doi: 10.1016/j.jcmg.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helms Julie, Kremer Stéphane, Merdji Hamid, Clere-Jehl Raphaël, Schenck Malika, Kummerlen Christine, Collange Olivier, Boulay Clotilde, Fafi-Kremer Samira, Ohana Mickaël, Anheim Mathieu, Meziani Ferhat. Neurologic Features in Severe SARS-CoV-2 Infection. N. Engl. J. Med. 2020;382(23):2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsons Timothy, Banks Sarah, Bae Chay, Gelber Joel, Alahmadi Hussein, Tichauer Matthew. COVID-19-associated acute disseminated encephalomyelitis (ADEM) J. Neurol. 2020;267(10):2799–2802. doi: 10.1007/s00415-020-09951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toscano Gianpaolo, Palmerini Francesco, Ravaglia Sabrina, Ruiz Luigi, Invernizzi Paolo, Cuzzoni M. Giovanna, Franciotta Diego, Baldanti Fausto, Daturi Rossana, Postorino Paolo, Cavallini Anna, Micieli Giuseppe. Guillain-Barré Syndrome Associated with SARS-CoV-2. N. Engl. J. Med. 2020;382(26):2574–2576. doi: 10.1056/NEJMc2009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kermali Muhammed, Khalsa Raveena Kaur, Pillai Kiran, Ismail Zahra, Harky Amer. The role of biomarkers in diagnosis of COVID-19 - A systematic review. Life Sci. 2020;254 doi: 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henry B.M., de Oliveira M.H.S., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease, (COVID-19): a meta-analysis. Clin. Chem. Lab Med. 2019;58(2020):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 40.Akboga S.A., Gokce A., Hatipoglu M., Beyoglu M.A., Inan K., Sezen A.I., Dal H.C., Akkas Y., Turan S., Kocer B. The relationship between mortality and inflammatory markers and the systemic immune inflammatory index in patients in the intensive care unit with a pneumothorax as a complication of COVID-19 disease. Ir. J. Med. Sci. 2021 doi: 10.1007/s11845-021-02740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Çakırca G., Damar Çakırca T., Üstünel M., Torun A., Koyuncu İ. Thiol level and total oxidant/antioxidant status in patients with COVID-19 infection. Ir. J. Med. Sci. 2021 doi: 10.1007/s11845-021-02743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Lorenzo Rebecca, Loré Nicola I., Finardi Annamaria, Mandelli Alessandra, Cirillo Daniela M., Tresoldi Cristina, Benedetti Francesco, Ciceri Fabio, Rovere-Querini Patrizia, Comi Giancarlo, Filippi Massimo, Manfredi Angelo A., Furlan Roberto. Blood neurofilament light chain and total tau levels at admission predict death in COVID-19 patients. J. Neurol. 2021;268(12):4436–4442. doi: 10.1007/s00415-021-10595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frithiof Robert, Rostami Elham, Kumlien Eva, Virhammar Johan, Fällmar David, Hultström Michael, Lipcsey Miklós, Ashton Nicholas, Blennow Kaj, Zetterberg Henrik, Punga Anna Rostedt. Critical illness polyneuropathy, myopathy and neuronal biomarkers in COVID-19 patients: A prospective study. Clin. Neurophysiol. 2021;132(7):1733–1740. doi: 10.1016/j.clinph.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMahon Courtney L., Staples Hilary, Gazi Michal, Carrion Ricardo, Hsieh Jenny. SARS-CoV-2 targets glial cells in human cortical organoids. Stem Cell Rep. 2021;16(5):1156–1164. doi: 10.1016/j.stemcr.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lavi Ehud, Cong Lin. Type I astrocytes and microglia induce a cytokine response in an encephalitic murine coronavirus infection. Exp. Mol. Pathol. 2020;115:104474. doi: 10.1016/j.yexmp.2020.104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boldrini M., Canoll P.D., Klein R.S. How COVID-19 Affects the Brain. JAMA Psychiatry. 2021;78:682–683. doi: 10.1001/jamapsychiatry.2021.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan H., Zhang K., Shan L., Kuang F., Chen K., Zhu K., Ma H., Ju G., Wang Y.-Z. Reactive astrocytes undergo M1 microglia/macrohpages-induced necroptosis in spinal cord injury. Mol. Neurodegener. 2016;11:14. doi: 10.1186/s13024-016-0081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan H., Tang H.-B., Chen Z., Wang H.-Q., Zhang L., Jiang Y., Li T., Yang C.-F., Wang X.-Y., Li X., Wu S.-X., Zhang G.-L. Inhibiting HMGB1-RAGE axis prevents pro-inflammatory macrophages/microglia polarization and affords neuroprotection after spinal cord injury. J. Neuroinflammation. 2020;17:295. doi: 10.1186/s12974-020-01973-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie Jianling, Méndez José D., Méndez-Valenzuela Verna, Aguilar-Hernández María Montserrat. Cellular signalling of the receptor for advanced glycation end products (RAGE) Cell Signal. 2013;25(11):2185–2197. doi: 10.1016/j.cellsig.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Vincent Andrea M., Perrone Lorena, Sullivan Kelli A., Backus Carey, Sastry Ann Marie, Lastoskie Christian, Feldman Eva L. Receptor for advanced glycation end products activation injures primary sensory neurons via oxidative stress. Endocrinology. 2007;148(2):548–558. doi: 10.1210/en.2006-0073. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt Ann Marie, Yan Shi Du, Yan Shi Fang, Stern David M. The multiligand receptor RAGE as a progression factor amplifying immune and inflammatory responses. J. Clin. Invest. 2001;108(7):949–955. doi: 10.1172/JCI14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber Daniel J., Allette Yohance M., Wilkes David S., White Fletcher A. The HMGB1-RAGE Inflammatory Pathway: Implications for Brain Injury-Induced Pulmonary Dysfunction. Antioxid Redox Signal. 2015;23(17):1316–1328. doi: 10.1089/ars.2015.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang F., Su X., Huang G., Xin X.-F., Cao E.-H., Shi Y., Song Y. sRAGE alleviates neutrophilic asthma by blocking HMGB1/RAGE signalling in airway dendritic cells. Sci. Rep. 2017;7:14268. doi: 10.1038/s41598-017-14667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paudel Y.N., Angelopoulou E., Piperi C., Othman I., Aamir K., Shaikh M.F. Impact of HMGB1, RAGE, and TLR4 in Alzheimer’s Disease (AD): From Risk Factors to Therapeutic Targeting. Cells. 2020;9 doi: 10.3390/cells9020383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe H., Son M. The Immune Tolerance Role of the HMGB1-RAGE Axis. Cells. 2021;10:564. doi: 10.3390/cells10030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Ruochan, Huang Yan, Quan Jun, Liu Jiao, Wang Haichao, Billiar Timothy R., Lotze Michael T., Zeh Herbert J., Kang Rui, Tang Daolin. HMGB1 as a potential biomarker and therapeutic target for severe COVID-19. Heliyon. 2020;6(12) doi: 10.1016/j.heliyon.2020.e05672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim A., Radujkovic A., Weigand M.A., Merle U. Soluble receptor for advanced glycation end products (sRAGE) as a biomarker of COVID-19 disease severity and indicator of the need for mechanical ventilation ARDS and mortality. Annals of Intensive Care. 2021;11:50. doi: 10.1186/s13613-021-00836-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiappalupi S., Salvadori L., Donato R., Riuzzi F., Sorci G. Hyperactivated RAGE in Comorbidities as a Risk Factor for Severe COVID-19-The Role of RAGE-RAS Crosstalk. Biomolecules. 2021;11:876. doi: 10.3390/biom11060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Egaña-Gorroño L., López-Díez R., Yepuri G., Ramirez L.S., Reverdatto S., Gugger P.F., Shekhtman A., Ramasamy R., Schmidt A.M. Receptor for Advanced Glycation End Products (RAGE) and Mechanisms and Therapeutic Opportunities in Diabetes and Cardiovascular Disease: Insights From Human Subjects and Animal Models. Front Cardiovasc. Med. 2020;7:37. doi: 10.3389/fcvm.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baghaki S., Yalcin C.E., Baghaki H.S., Aydin S.Y., Daghan B., Yavuz E. COX2 inhibition in the treatment of COVID-19: Review of literature to propose repositioning of celecoxib for randomized controlled studies. Int. J. Infect. Dis. 2020;101:29–32. doi: 10.1016/j.ijid.2020.09.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capuano Annalisa, Scavone Cristina, Racagni Giorgio, Scaglione Francesco. Italian Society of Pharmacology, NSAIDs in patients with viral infections, including Covid-19: Victims or perpetrators? Pharmacol. Res. 2020;157 doi: 10.1016/j.phrs.2020.104849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee Suki M.Y., Cheung Chung‐Yan, Nicholls John M., Hui Kenrie P.Y., Leung Connie Y.H., Uiprasertkul Mongkol, Tipoe George L., Lau Yu‐Lung, Poon Leo L.M., Ip Nancy Y., Guan Yi, Peiris J.S. Malik. Hyperinduction of cyclooxygenase-2-mediated proinflammatory cascade: a mechanism for the pathogenesis of avian influenza H5N1 infection. J. Infect Dis. 2008;198(4):525–535. doi: 10.1086/590499. [DOI] [PubMed] [Google Scholar]
- 63.Prasher P., Sharma M., Gunupuru R. Targeting cyclooxygenase enzyme for the adjuvant COVID-19 therapy. Drug Dev. Res. 2021;82:469–473. doi: 10.1002/ddr.21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J.S., Alfajaro M.M., Wei J., Chow R.D., Filler R.B., Eisenbarth S.C., Wilen C.B. Cyclooxgenase-2 is induced by SARS-CoV-2 infection but does not affect viral entry or replication. BioRxiv. 2020 doi: 10.1101/2020.09.24.312769. [DOI] [Google Scholar]
- 65.Hoxha Malvina. What about COVID-19 and arachidonic acid pathway? Eur. J. Clin. Pharmacol. 2020;76(11):1501–1504. doi: 10.1007/s00228-020-02941-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schieber Michael, Chandel Navdeep S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24(10):R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marinescu Ileana, Marinescu Dragoş, Mogoantă Laurenţiu, Efrem Ion Cristian, Stovicek Puiu Olivian. SARS-CoV-2 infection in patients with serious mental illness and possible benefits of prophylaxis with Memantine and Amantadine. Rom. J. Morphol. Embryol. 2021;61(4):1007–1022. doi: 10.47162/RJME.61.4.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saheb Sharif-Askari N., Saheb Sharif-Askari F., Mdkhana B., Hussain Alsayed H.A., Alsafar H., Alrais Z.F., Hamid Q., Halwani R. Upregulation of oxidative stress gene markers during SARS-COV-2 viral infection. Free Radic. Biol. Med. 2021;172:688–698. doi: 10.1016/j.freeradbiomed.2021.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suhail Shanzay, Zajac Jonathan, Fossum Carl, Lowater Harrison, McCracken Cailin, Severson Nathaniel, Laatsch Bethany, Narkiewicz-Jodko Alex, Johnson Benjamin, Liebau Jessica, Bhattacharyya Sudeep, Hati Sanchita. Role of Oxidative Stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) Infection: A Review. Protein J. 2020;39(6):644–656. doi: 10.1007/s10930-020-09935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]