Abstract
Background
Gliomas comprise the most common type of primary brain tumor, are highly invasive, and often fatal. IDH-mutated gliomas are particularly challenging to image and there is currently no clinically accepted method for identifying the extent of tumor burden in these neoplasms. This uncertainty poses a challenge to clinicians who must balance the need to treat the tumor while sparing healthy brain from iatrogenic damage. The purpose of this study was to investigate the feasibility of using resting-state blood oxygen level-dependent (BOLD) functional magnetic resonance imaging (fMRI) to detect glioma-related asynchrony in vascular dynamics for distinguishing tumor from healthy brain.
Methods
Twenty-four stereotactically localized biopsies were obtained during open surgical resection from ten treatment-naïve patients with IDH-mutated gliomas who received standard-of-care preoperative imaging as well as echo-planar resting-state BOLD fMRI. Signal intensity for BOLD asynchrony and standard-of-care imaging was compared to cell counts of total cellularity (H&E), tumor density (IDH1 & Sox2), cellular proliferation (Ki67), and neuronal density (NeuN), for each corresponding sample.
Results
BOLD asynchrony was directly related to total cellularity (H&E, P = 4 × 10–5), tumor density (IDH1, P = 4 × 10–5; Sox2, P = 3 × 10–5), cellular proliferation (Ki67, P = .002), and inversely related to neuronal density (NeuN, P = 1 × 10–4).
Conclusions
Asynchrony in vascular dynamics, as measured by resting-state BOLD fMRI, correlates with tumor burden and provides a radiographic delineation of tumor boundaries in IDH-mutated gliomas.
Keywords: BOLD asynchrony, IDH-mutated glioma, infiltration, resting-state fMRI, tumor burden
Key Point.
BOLD asynchrony quantitates tumor burden in IDH-mutated gliomas and elucidates infiltrating tumor undetectable on standard-of-care imaging.
Importance of the Study.
IDH-mutated gliomas are invasive tumors of the central nervous system whose boundaries of infiltration are difficult to ascertain using standard-of-care imaging. In our current study, we show that asynchrony in vascular dynamics measured using resting-state BOLD fMRI allows us to quantify the degree of tumor burden and infiltration in IDH-mutated gliomas. Our study suggests that BOLD asynchrony may allow clinicians to personalize the treatment of brain cancer patients by providing objective coordinates for biopsy collection, boundaries for extent of resection, as well as targets for radiation field mapping that are specific to each patient.
Current standard-of-care imaging for gliomas involves a number of structural magnetic resonance imaging (MRI) modalities that can elucidate gross anatomical abnormalities and reveal areas of blood-brain-barrier breakdown with the use of gadolinium-based contrast. In IDH wild-type gliomas, standard of care involves maximal surgical resection of the contrast-enhancing portion of the tumor, which has been associated with improved survival.1,2 However, in IDH-mutated gliomas, which are typically lower in grade and non-contrast-enhancing, no such radiographic guidelines exist.3 The inability to visualize the spatial extent of tumor burden poses a challenge to clinicians who must balance the need to treat the tumor while protecting healthy brain from iatrogenic damage.4,5 Surgically ambiguous margins impede preoperative planning and intraoperative decision-making.3,5 Furthermore, post-resection uncertainty in tumor boundaries hampers accurate radiation field mapping and has led to significant variability across institutions in their guidelines for field placement.6
To address shortcomings in structural imaging, a variety of metabolic and functional imaging techniques have been employed to better characterize gliomas. Positron emission tomography (PET) has been used to identify metabolically active regions of gliomas, and magnetic resonance spectroscopy has been used to detect the presence of clinically relevant metabolites including D-2-hydroxyglutarate, which is upregulated in IDH-mutated tumors.7–12 However, both techniques have limited spatial resolution, and PET is further hampered by the low metabolic activity of IDH-mutated gliomas, which can reduce clinical utility.8–10 Perfusion weighted imaging (PWI) has also been used to characterize gliomas according to tumor grade and predict disease progression prior to contrast enhancement.13,14 However, in IDH-mutated gliomas, perfusion features are often indistinguishable from that of normal brain, which limits the ability of PWI to delineate tumor boundaries.15,16
Another method for visualizing vascular function is blood oxygen level-dependent (BOLD) functional MRI (fMRI). BOLD fMRI measures hemodynamic fluctuations and has traditionally been used to preoperatively map eloquent cortex using task-evoked signals.17 On resting-state BOLD fMRI, infiltrating glioma has been shown to induce alterations in neurovascular coupling18 and results in locally decreased cerebrovascular reactivity,19 which fundamentally alters the BOLD signal. Building upon these findings, we have recently developed a method that measures asynchrony in BOLD dynamics between tumor and normal cardiovascular fluctuations in healthy brain, and use this measure, termed BOLD asynchrony (BA), to quantify glioma-related vascular dysregulation.20
In our previous work, we have shown that BOLD asynchrony extends beyond tumor visible on standard-of-care imaging and that the extent of BOLD asynchrony detectable in the peritumoral zone accurately predicts IDH mutation status.21 Furthermore, histologic analysis of MRI localized biopsies taken from patients with IDH wild-type gliomas has previously demonstrated that BOLD asynchrony is proportional to tumor burden.22 This suggests that BOLD asynchrony may serve as a viable biomarker for detecting infiltrating non-enhancing tumor in IDH wild-type gliomas. However, it is not known whether BOLD asynchrony can quantify tumor burden in IDH-mutated gliomas, which are typically non-enhancing and have boundaries that can be more difficult to discern using standard-of-care imaging. If BOLD asynchrony is similarly correlated to tumor burden in IDH-mutated gliomas, then BOLD fMRI could be used to generate targets for biopsy acquisition as well as margins for surgical resection and radiation field mapping.
Methods
Patient Selection
We prospectively collected MRI localized biopsies from ten treatment-naïve patients undergoing open surgical resection of pathology-proven IDH-mutated glioma at Columbia University Medical Center (CUMC). All patients included in the study provided consent as part of a CUMC institutional review board-approved study protocol. The mean patient age at the time of surgery was 43 years (range: 30–64) with 4 men and 6 women. Additional diagnostic information regarding each patient’s tumor is shown in Supplementary Table 1.
Image Acquisition
All patients received standard-of-care preoperative imaging including T1-weighted pre-contrast, T1-weighted post-contrast, T2-weighted, T2-weighted fluid-attenuated inversion recovery (FLAIR), diffusion-weighted imaging with apparent diffusion coefficient (ADC), and dynamic susceptibility contrast PWI, as well as echo-planar resting-state BOLD fMRI. Imaging parameters are listed in Supplementary Table 2. Contrast-enhanced images were acquired with intravenous gadobenate dimegulmine (Gd) with a weight-based dose of 0.2 mL per kilogram of body weight. The time between intravenous injection and contrast-enhanced T1-weighted imaging was 5 minutes. BOLD images were obtained before contrast administration.
Image Processing of Structural Data
All structural images were skull stripped and co-registered to each patient’s T1-weighted post-contrast sequence using a linear transformation with 6 degrees of freedom in FSL (FMRIB Software Library).23 To standardize voxel intensity values of T1-weighted pre-contrast, T1-weighted post-contrast, T2-weighted, and T2-FLAIR images among patients who underwent structural imaging with different scanning parameters, we implemented a histogram-matching algorithm.24 Histogram matching was performed between the contralesional, radiologically normal hemisphere (the target) and the equivalent hemisphere of a radiologically normal patient (the template), and then applied to the whole image. For each target image, the histogram of voxel intensities was first converted to a cumulative distribution function (CDF). Then the CDF was scaled and offset using a two-parameter linear model, such that the correlation between the image CDF and template CDF was maximal:
| (1) |
where corr represents Pearson correlation.
Mask Generation
Masks for visible tumor and T2-FLAIR hyperintensity were hand-drawn by a board-certified neuroradiologist (C.I.H.) on high-resolution structural images. In this cohort of IDH-mutated, non-enhancing tumors, the visible tumor was defined as T1 hypointensity. To create control masks, each patient’s T1-weighted post-contrast image was registered to Montreal Neurologic Institute 152 (MNI-152) space using affine registration with 12 degrees of freedom in FSL. Then an MNI-152 mask of the contralesional hemisphere was transformed to the patient’s functional space using nearest-neighbor interpolation. Any portion of the visible tumor or T2-FLAIR hyperintensity that extended into the contralesional hemisphere was removed from the control mask.
Image Processing of Perfusion Data
Each perfusion scan was rigid-body motion-corrected, slice time corrected, spatially smoothed (full width at half maximum = 3 mm), and high-pass filtered (50 seconds) using FEAT in FSL. Multiple linear regression was performed using a four-parameter gamma model of the form:
| (2) |
where tmax is time of maximum response, a is amplitude of the response, b is the shape parameter of the gamma function, and t = k to n seconds where k is the bolus arrival time and n is the total acquisition time in seconds. For each voxel’s time series, the free parameters were tmax, a, b, and k. The fitted response was converted to a percent signal change by normalizing to the mean of the baseline i.e. the first 15 seconds post-infusion. Cerebral blood volume (CBV) was computed by integrating the fitted response; relative CBV (rCBV) was computed by dividing the CBV estimate for each voxel by the mean CBV response in the contralesional white matter mask.
Image Processing of BOLD Data
Processing was performed as described in Chow et al20 using FSL and MATLAB (MathWorks). In brief, each functional image was motion-corrected, slice time corrected, and skull stripped. To minimize the effect of head motion on BOLD signal intensity, the motion correction parameters (12 motion regressors consisting of three translations, three rotations, and the first derivative of each) were regressed out of the data. MELODIC-FIX in FSL was then used to remove independent components related to scanner noise and head motion.25
For each patient, the BOLD images were co-registered to the T1-weighted post-contrast image and the tumor mask and the contralesional control mask were transformed to functional space to extract the respective BOLD time series. The two time series were used as regressors in a multiple linear regression model. Two Z-statistic images were generated to describe how synchronous each voxel was with the tumor mask (Ztumor) and contralesional control mask (Zcontrol). These two images were subtracted (Ztumor–Zcontrol) to create a map of BOLD asynchrony. Applying this method on a voxel-by-voxel basis to the whole brain resulted in a broad range of BOLD asynchrony values with most ranging from Z ≈ –10 to +10, where voxels with values closer to -10 having temporal signatures closely resembling that of the global mean signal from the contralesional hemisphere and voxels with values closer to +10 having temporal signatures closely resembling that of visible tumor. Since all biopsies were collected from the diseased hemisphere within the normal surgical plan, the distribution of corresponding BOLD asynchrony values was positively skewed towards greater degrees of BOLD asynchrony.
Biopsy Acquisition
A total of twenty-four localized biopsies were obtained from the ten patients included in the study and were collected by one of four board-certified neurosurgeons (S.A.S., G.M.M., M.B.S., J.N.B.). All samples were taken within the planned surgical trajectory and obtained before surgical debulking, use of mannitol, cerebrospinal fluid diversion, or hyperventilation to minimize the effects of brain shift and deformation. Frameless stereotactic guidance was provided using the T1-weighted post-contrast sequence uploaded to a neuro-navigation interface (Brainlab Curve). Biopsy location was recorded by screen captures of the neuro-navigation interface and co-registered to the MRI using a semi-automated MATLAB screen capture registration tool, allowing for the downstream determination of the Cartesian coordinates of each biopsy with spatial accuracy to within 3 mm as described in Chang et al.26
Histologic Analysis
Samples were formalin-fixed and paraffin-embedded for histologic analysis. Five-micrometer sections were stained with H&E (hematoxylin and eosin), IDH1 (a mutated enzyme specific for tumor cells in this cohort), Sox2 (a transcription factor overexpressed in malignant glioma), Ki67 (a nuclear protein necessary for cell division), and NeuN (a nuclear protein expressed in neurons). Cell counts for total cellularity, tumor density, cellular proliferation, and neuronal density were calculated using an automated cell-counting algorithm and visually inspected by a board-certified neuropathologist (P.C.) to ensure accuracy.26 Briefly, H&E-stained sections were digitized using a Leica scn400 system (Leica Biosystems, Buffalo Grove, Illinois), with hematoxylin-stained nuclei counted in iteratively processed high-power field (HPFs), defined by a 2.25 × 2.25 mm square (900 × 900 pixels). Total cellularity was counted as the mean number of hematoxylin-stained cells across all HPFs that contained at least 1 nucleus. Each immunoperoxidase-stained slide (IDH1, Sox2, Ki67, NeuN) was digitized using the same setup as above and identification of positively stained cells was established with user-determined RGB (3-channel) thresholding during visual inspection of 9 randomly generated HPFs. Cell counts of tumor density, cellular proliferation, and neuronal density were counted as the mean number of immunoperoxidase-stained cells across all HPFs that contained at least 1 positively stained nucleus.
Statistical Analysis of Histologic and Radiologic Correlations
The MRI intensity of each biopsy was taken from single voxels identified from intraoperative stereotactic registration to MRI using the surgical Brainlab software. A single voxel rather than a cube of voxels centered on the biopsy was chosen because it provided the strongest relationship between histological features and MRI for standard-of-care imaging (Supplementary Figure 1).
All statistical tests were performed using the Statistics Toolbox in MATLAB and statistical significance was set at P < .05. We used linear regression to model the relationship between histological features (total cellularity, tumor density, cellular proliferation, and neuronal density) and MR imaging (T1, T2, FLAIR, ADC, rCBV, and BA). The relationship between BOLD asynchrony and histological features was exponential, thus for H&E, IDH1, Sox2, and Ki67 we performed regression using an exponential model of the form,
| (3) |
where i represents the BA intensity, k is a scaling constant, λ is the rate of change, and H is the histological count. For NeuN we used an exponential model of the form,
| (4) |
Tumor Burden Model
The parameter estimates from the tumor density (IDH1 or Sox2) versus BA relationship were used in conjunction with a distance function to estimate the voxel-wise tumor burden across the entire brain. The distance of each biopsy to the nearest voxel in the visible tumor mask was computed and the distances of all biopsies in the cohort were used to fit to an exponential decay based on previous work.22
| (5) |
This exponential function was normalized to 1 and the value of the exponential at each voxel was multiplied by the tumor burden estimated from BA to produce a distance-adjusted tumor burden estimate.
Results
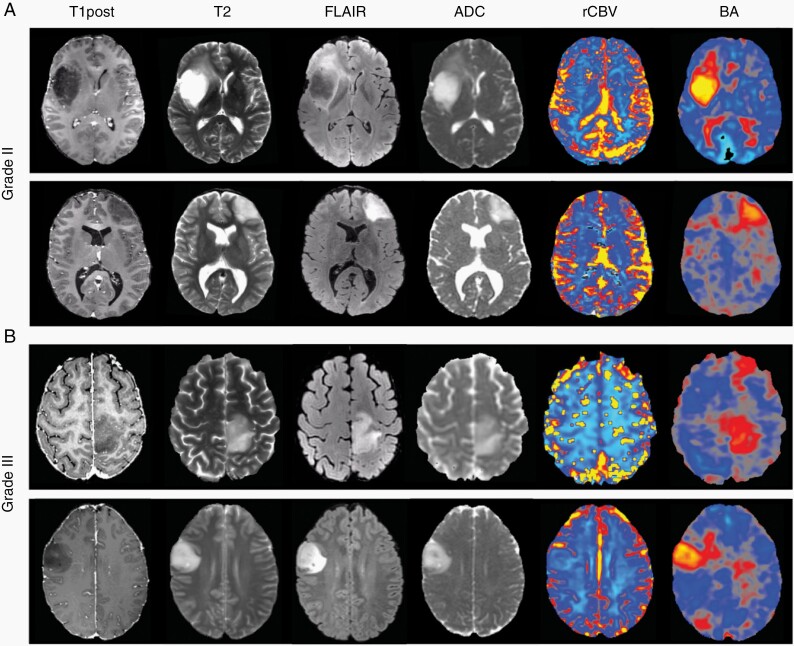
Representative T1 post-contrast, T2, FLAIR, ADC, as well as maps of rCBV and BOLD asynchrony (BA) are shown from two patients with grade II IDH-mutated gliomas (Figure 1A) and two patients with grade III IDH-mutated gliomas (Figure 1B). In all cases, abnormalities in rCBV were difficult to discern in the tumor-infiltrated hemisphere when compared to the contralesional hemisphere. By contrast, the maps of BA were clearly demarcated in both grade II and III IDH-mutated gliomas, highlighting regions of vascular dysregulation that extended beyond structural abnormalities seen on standard-of-care imaging.
Fig. 1.
Pre-operative scans of pathology-proven IDH-mutated grade II (A) and grade III (B) gliomas displaying both structural MRI (T1 post-contrast, T2, FLAIR, and ADC) in the first four columns and functional MRI (rCBV and BA) in the last two columns. There were no obvious rCBV abnormalities near the tumor. In comparison, well-demarcated areas of vascular dysregulation were captured using BA.
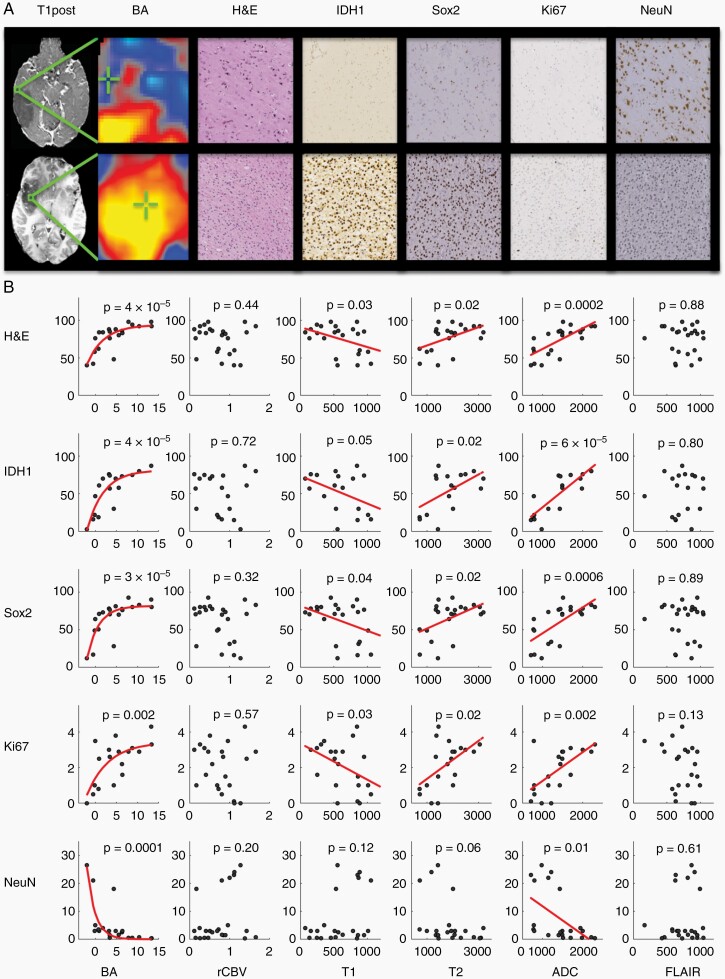
Biopsies within and around the tumor were acquired with a broad range of BA (Z = –3 to 13) and were compared to the underlying histologic characteristics within individual tissue samples. Figure 2A shows an example biopsy with low BA (Z = –3) demonstrating low cellularity (H&E), low tumor density (IDH and Sox2), low cellular proliferation (Ki67), and high neuronal density (NeuN). By contrast, a biopsy with high BA (Z = 10.5) demonstrates high cellularity, high tumor density, high cellular proliferation, and low neuronal density. To summarize the histological-radiographic relationships, we performed a regression of each histological feature against each MR imaging modality (Figure 2B). BA showed a significant relationship to total cellularity (H&E, P = 4 × 10–5), tumor density (IDH1, P = 4 × 10–5; Sox2, P = 3 × 10–5) and cellular proliferation (Ki67, P = .002). This analysis also demonstrated a significant inverse relationship between BA and neuronal density (NeuN, P = 1 x 10–4). rCBV showed no significant relationship with any of the histological markers.
Fig. 2.
(A) Two representative biopsies are displayed from radiographically distinct areas with low BA (Z = –3) in the top row and high BA (Z = 10.5) in the bottom row. The top row depicts low cellularity (H&E), low tumor density (IDH1 and Sox2), low cellular proliferation (Ki67), and high neuronal density (NeuN) whereas the bottom row depicts high cellularity, high tumor density, high cellular proliferation, and low neuronal density with the same markers. (B) Red lines represent significant regression fits between histologic and radiographic data. BA and ADC were significantly predictive of all histological features. In contrast, rCBV and FLAIR showed no significant relationship to histology.
Repeating this analysis with standard-of-care structural imaging showed that T1 pre-contrast signal intensity was inversely related to cellularity (H&E, P = .03), tumor density (IDH1, P = .05; Sox2, P = .04), and cellular proliferation (Ki67, P = .03). T2 signal intensity was directly related to cellularity (H&E, P = .02), tumor density (IDH1, P = .02; Sox2, P = .02), and cellular proliferation (Ki67, P = 0.02). ADC signal intensity was directly related to cellularity (H&E, P = 2 × 10–4), tumor density (IDH1, P = 6 × 10–5; Sox2, P = 6 × 10–4), cellular proliferation (Ki67, P = .002), and inversely related to neuronal density (NeuN, P = .01). Interestingly, the predictive value of FLAIR, though previously proposed to be a proxy for non-contrast-enhancing tumor,27 was not significant for any histological features. Table 1 shows a direct comparison of the proportion of histological variance explained by each imaging modality: BA explained the largest proportion of variance whereas rCBV and FLAIR explained the least.
Table 1.
Proportion of Variance Explained (R2) for BA and Standard-of-Care Imaging for Predicting Histological Counts of Sample Cellularity (H&E), Tumor Density (IDH1 and Sox2), Cellular Proliferation (Ki67), and Neuronal Density (NeuN) Using the Models Illustrated in Figure 2B
| Proportion of Variance Explained | |||||
|---|---|---|---|---|---|
| H&E | IDH1 | Sox2 | Ki67 | NeuN | |
| BA | 0.61 | 0.67 | 0.61 | 0.43 | 0.57 |
| ADC | 0.50 | 0.66 | 0.42 | 0.36 | 0.22 |
| T2 | 0.46 | 0.54 | 0.38 | 0.28 | 0.18 |
| T1 | 0.29 | 0.28 | 0.21 | 0.17 | 0.05 |
| FLAIR | 0.01 | 0.00 | 0.01 | 0.10 | 0.04 |
| rCBV | 0.01 | 0.00 | 0.02 | 0.01 | 0.02 |
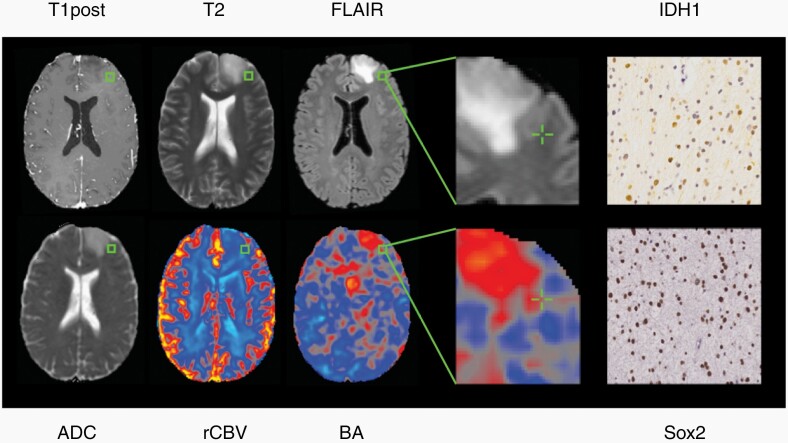
Because of the widespread use of FLAIR to demarcate tumor boundaries, we evaluated whether FLAIR could still be used as a binary marker for the presence or absence of tumor, despite its lack of correlation to tumor burden. A t-test comparing FLAIR normo-intense versus hyperintense biopsies for detecting tumor showed no significant differences between the two groups for any of the histological measures (P > .05). We then assessed whether BA can detect tumor burden outside of FLAIR hyperintense regions using 10 of the 24 biopsies that had normal FLAIR intensities. Figure 3 shows an example biopsy with high tumor burden in which standard-of-care imaging was radiographically normal, yet BA was abnormally high. A regression analysis between the histological features and each MR imaging modality for this subset of biopsies confirmed that BA can detect tumors outside of conventional tumor boundaries (Supplementary Figure 2).
Fig. 3.
A representative biopsy acquired from radiographically normal brain on standard-of-care imaging but with high BA and tumor burden as measured by IDH1 and Sox2.
Although BA has the strongest relationship to histological features, it is possible that by combining imaging modalities one could further improve the prediction of tumor burden. We, therefore, performed a stepwise regression using BA in the first step and each of the standard-of-care imaging modalities in the second to predict histological features. None of the standard-of-care imaging modalities could explain a significant proportion of the histological variance after first regressing out BA (P > .05). In contrast, when we performed a stepwise regression with standard-of-care imaging in the first step, we found that BA still explained a significant proportion of the variance in most cases (Table 2). This demonstrates that BA provides significantly more predictive power than standard-of-care imaging for elucidating underlying histology.
Table 2.
Stepwise Regression Revealed That BA Has Significantly More Predictive Power for all Histological Features Compared to T1, rCBV, and FLAIR and More Predictive Power for Some Histological Features Compared to ADC and T2
| BA Significance After Removing | H&E | IDH1 | Sox2 | Ki67 | NeuN | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| R 2 | P-Value | R 2 | P-Value | R 2 | P-Value | R 2 | P-Value | R 2 | P-Value | |
| ADC | 0.10 | .2331 | 0.12 | .2528 | 0.16 | .1186 | 0.08 | .2851 | 0.26 | .0432 |
| T2 | 0.17 | .1082 | 0.25 | .0801 | 0.25 | .0487 | 0.10 | .2362 | 0.33 | .0194 |
| T1 | 0.41 | .0075 | 0.57 | .0027 | 0.46 | .0037 | 0.30 | .0294 | 0.49 | .0024 |
| rCBV | 0.54 | .0011 | 0.65 | .0008 | 0.54 | .0012 | 0.38 | .0104 | 0.50 | .0023 |
| FLAIR | 0.63 | .0002 | 0.67 | .0006 | 0.64 | .0002 | 0.56 | .0009 | 0.63 | .0003 |
All the P-values in this table refer to the BOLD significance after removing the effect of a standard of care imaging modality.
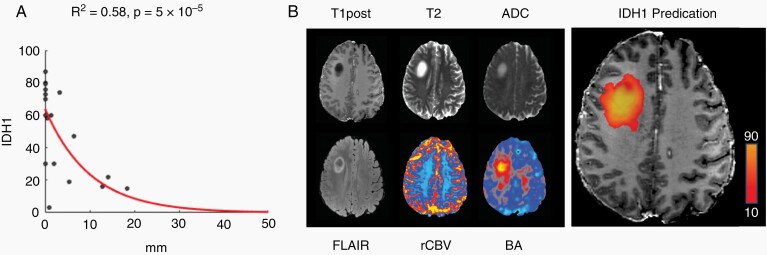
These results suggest that BA can be used to generate a predictive model of tumor burden. We used the IDH1 model parameters (Eq. 3: λ = 4.1, k = 95.5, H0 = –5.3) from the BA regression model in Figure 2B to calculate voxel-wise estimates of tumor burden across the whole brain. To minimize false positives, we modeled the effect of distance on tumor infiltration (Figure 4A) (Eq. 5: λ = 10.1, k = 1, H0 = 0), normalized to 1, then multiplied the value of the exponential at each voxel by the tumor burden estimated from Eq. 3 to produce a distance-adjusted tumor burden estimate. After applying a minimum tumor density threshold of 10 cells per HPF, our model predicted that infiltrative margins extend beyond visible tumor by an average of 4.6 ± 1.4 mm and that 99% of the tumor detected by BA is within 18.4 mm of the tumor visible on standard-of-care imaging (Figure 4B). A similar result was obtained using Sox2 (Supplementary Figure 3).
Fig. 4.
Tumor infiltration decreases exponentially as a function of distance (A) and can be used in conjunction with BA to generate a predictive model of tumor burden (B) that extends beyond what is visible on standard-of-care imaging.
Discussion
The intratumoral heterogeneity and diffusely infiltrative pattern of IDH-mutated gliomas pose significant challenges to using current standard-of-care imaging for elucidating tumor burden and underlying histopathological changes induced by these neoplasms.28 Our results using T1-weighted pre-contrast imaging affirm the conventional notion that areas of T1 hypointensity represent the bulk of visible tumor.29 Interestingly, T2-weighted hyperintensity but not FLAIR hyperintensity was found to be correlated with tumor burden. In our cohort of WHO grade II and III IDH-mutated tumors, this result may be a consequence of T2-FLAIR mismatch where T2 signal appears homogenous but FLAIR appears inhomogeneous and typically displays a penumbra of hyperintensity along the periphery of visible tumor (see the first example in Figure 1A).30 While the mechanisms driving this highly reported imaging finding remain unknown, our results suggest that FLAIR hyperintensity may not be optimal for guiding treatment in this subset of gliomas.
ADC signal intensity has been shown to be inversely correlated with tumor burden in WHO grade IV glioblastomas,31 but in our cohort of grade II and III IDH-mutated gliomas, we demonstrated that ADC was directly correlated to tumor burden. This finding is consistent with Stadlbauer et al32 who used MRI localized biopsies to demonstrate that tumor burden is correlated to ADC in patients with grade II and III gliomas. Similarly, purely radiographic studies have also demonstrated that grade II and III IDH-mutated tumors have a higher mean ADC than their IDH wild-type counterparts.33 The discrepancy in ADC findings between glioblastomas and lower grade gliomas may be related to the slow-growing nature of the latter, which do not achieve the hypercellularity34 that is likely required to reduce ADC. We also found that rCBV did not reveal any obvious abnormalities near the tumor nor was it predictive of any underlying histologic features, a result that may be related to the low angiogenic potential of IDH-mutated tumors.35,36
In contrast to standard-of-care imaging, resting-state BOLD fMRI allowed for the generation of BOLD asynchrony maps that clearly demarcated regions of vascular dysregulation in IDH-mutated gliomas. Histological analysis of MRI localized biopsies confirmed that the vascular dysregulation was caused by local tumor infiltration and not indirectly caused by mass-effect or edema. More importantly, the degree of BOLD asynchrony was found to be directly correlated with tumor burden, a result that is analogous to what we have previously demonstrated in IDH wild-type gliomas.22 Furthermore, our BOLD asynchrony model predicted that the bulk of detectable tumor (>99%) is within 1.84 cm of the margins visible on standard-of-care imaging, which is similar to the 2 cm estimated by Pallud et al.37
The mechanisms by which invading glioma cells perturb cerebrovascular function remain poorly understood but several theories exist. For example, mouse models have shown that glioma infiltration severely changes neurovascular coupling and stimulus-evoked hemodynamic responses.38 In humans, BOLD fMRI has shown glioma-related impairments in vascular reactivity, particularly within gray matter.39–43 Additionally, infiltrating glioma cells may release chemical mediators that directly modulate astrocyte vascular responsiveness or interfere with the function of normal signaling molecules.44 Further studies will be necessary to elucidate whether any of the aforementioned mechanisms are responsible for the BOLD asynchrony signal.
There are, however, several limitations to BOLD asynchrony. Like PWI, BOLD asynchrony has lower spatial resolution than structural MRI and should always be used in tandem with standard-of-care imaging. Furthermore, consideration of anatomical landmarks is necessary to minimize the effect of false positives. Importantly, it is possible that at the margins of infiltration, BOLD asynchrony may underestimate tumor burden either because tumor burden is too low to disrupt vascular function or because there is a delay between infiltration and vascular dysregulation. Therefore, other advanced techniques such as intraoperative fluorescein,45 5-ALA,46 or stimulated Raman histology47 should also be used to augment BOLD asynchrony.
In conclusion, our findings suggest that preoperative maps of BOLD asynchrony may be useful for guiding surgery in patients with IDH-mutated gliomas in light of growing evidence advocating for their aggressive initial resection.27,48 In fact, supra-total resections of low-grade gliomas, which are typically IDH-mutated, have been shown to provide a significant survival benefit to patients, likely due to excision of tumor that is not visible on standard-of-care imaging.49,50 Furthermore, BOLD asynchrony could be useful for guiding tissue sampling for more accurate and timely histological diagnoses as well as for improving radiation field placement, which would maximize dosing of tumor while minimizing damage to healthy brain. Clinical trials will ultimately be needed to further validate the utility of this imaging method.
Supplementary Material
Funding
This study was supported by National Institute of Health/National Institute of Neurological Disorders and Stroke [R01 NS103473].
Conflicts of Interest Statement. None of the authors report any conflicts of interest.
Authorship Statement. P.D.P., P.C., and J.G. conception of study and experimental design. All authors. acquisition of data and performance of research. P.D.P., C.H., P.C., and J.G. analysis and interpretation of data. P.D.P. and J.G. wrote the article. All authors assisted in writing and critically revised the article.
References
- 1. Oszvald A, Güresir E, Setzer M, et al. Glioblastoma therapy in the elderly and the importance of the extent of resection regardless of age. J Neurosurg. 2012;116(2):357–364. [DOI] [PubMed] [Google Scholar]
- 2. Sanai N, Polley MY, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg. 2011;115(1):3–8. [DOI] [PubMed] [Google Scholar]
- 3. Thon N, Tonn JC, Kreth FW. The surgical perspective in precision treatment of diffuse gliomas. Onco Targets Ther. 2019;12:1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Waldman AD, Jackson A, Price SJ, et al. Quantitative imaging biomarkers in neuro-oncology. Nat Rev Clin Oncol. 2009;6(8):445–454. [DOI] [PubMed] [Google Scholar]
- 5. Price SJ, Gillard JH. Imaging biomarkers of brain tumour margin and tumour invasion. Br J Radiol. 2011;84 Spec No 2:S159–S167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kruser TJ, Bosch WR, Badiyan SN, et al. NRG brain tumor specialists consensus guidelines for glioblastoma contouring. J Neurooncol. 2019;143(1):157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kunz M, Thon N, Eigenbrod S, et al. Hot spots in dynamic (18)FET-PET delineate malignant tumor parts within suspected WHO grade II gliomas. Neuro Oncol. 2011;13(3):307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. la Fougère C, Suchorska B, Bartenstein P, Kreth FW, Tonn JC. Molecular imaging of gliomas with PET: opportunities and limitations. Neuro Oncol. 2011;13(8):806–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiang GC, Kovanlikaya I, Choi C, Ramakrishna R, Magge R, Shungu DC. Magnetic resonance spectroscopy, positron emission tomography and radiogenomics-relevance to Glioma. Front Neurol. 2018;9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horská A, Barker PB. Imaging of brain tumors: MR spectroscopy and metabolic imaging. Neuroimaging Clin N Am. 2010;20(3):293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi C, Ganji SK, DeBerardinis RJ, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18(4):624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elkhaled A, Jalbert LE, Phillips JJ, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4(116):116ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Law M, Yang S, Babb JS, et al. Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. AJNR Am J Neuroradiol. 2004;25(5):746–755. [PMC free article] [PubMed] [Google Scholar]
- 14. Danchaivijitr N, Waldman AD, Tozer DJ, et al. Low-grade gliomas: do changes in rCBV measurements at longitudinal perfusion-weighted MR imaging predict malignant transformation? Radiology. 2008;247(1):170–178. [DOI] [PubMed] [Google Scholar]
- 15. Jooma R, Waqas M, Khan I. Diffuse low-grade glioma—changing concepts in diagnosis and management: a review. Asian J Neurosurg. 2019;14(2):356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanai N, Chang S, Berger MS. Low-grade gliomas in adults. J Neurosurg. 2011;115(5):948–965. [DOI] [PubMed] [Google Scholar]
- 17. Silva MA, See AP, Essayed WI, Golby AJ, Tie Y. Challenges and techniques for presurgical brain mapping with functional MRI. Neuroimage Clin. 2018;17:794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Agarwal S, Sair HI, Yahyavi-Firouz-Abadi N, Airan R, Pillai JJ. Neurovascular uncoupling in resting state fMRI demonstrated in patients with primary brain gliomas. J Magn Reson Imaging. 2016;43(3):620–626. [DOI] [PubMed] [Google Scholar]
- 19. Pak RW, Hadjiabadi DH, Senarathna J, et al. Implications of neurovascular uncoupling in functional magnetic resonance imaging (fMRI) of brain tumors. J Cereb Blood Flow Metab. 2017;37(11):3475–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chow DS, Horenstein CI, Canoll P, et al. Glioblastoma induces vascular dysregulation in nonenhancing peritumoral regions in humans. AJR Am J Roentgenol. 2016;206(5):1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Englander ZK, Horenstein CI, Bowden SG, et al. Extent of BOLD vascular dysregulation is greater in diffuse gliomas without isocitrate dehydrogenase 1 R132H mutation. Radiology. 2018;287(3):965–972. [DOI] [PubMed] [Google Scholar]
- 22. Bowden SG, Gill BJA, Englander ZK, et al. Local glioma cells are associated with vascular dysregulation. AJNR Am J Neuroradiol. 2018;39(3):507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62(2):782–790. [DOI] [PubMed] [Google Scholar]
- 24. Shapira D, Avidan S, Hel-Or Y. Multiple histogram matching. Paper presented at: 2013 IEEE International Conference on Image Processing, Melbourne, Australia; 15–18 Sept. 2013.
- 25. Salimi-Khorshidi G, Douaud G, Beckmann CF, Glasser MF, Griffanti L, Smith SM. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang PD, Malone HR, Bowden SG, et al. A multiparametric model for mapping cellularity in glioblastoma using radiographically localized biopsies. AJNR Am J Neuroradiol. 2017;38(5):890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanai N, Berger MS. Surgical oncology for gliomas: the state of the art. Nat Rev Clin Oncol. 2018;15(2):112–125. [DOI] [PubMed] [Google Scholar]
- 28. Nicholson JG, Fine HA. Diffuse glioma heterogeneity and its therapeutic implications. Cancer Discov. 2021;11(3):575–590. [DOI] [PubMed] [Google Scholar]
- 29. Larsen J, Hoggard N, McKevitt FM. Imaging in low-grade glioma: a guide for neurologists. Pract Neurol. 2018;18(1):27–34. [DOI] [PubMed] [Google Scholar]
- 30. Jain R, Johnson DR, Patel SH, et al. “Real world” use of a highly reliable imaging sign: “T2-FLAIR mismatch” for identification of IDH mutant astrocytomas. Neuro Oncol. 2020;22(7):936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eidel O, Neumann JO, Burth S, et al. Automatic analysis of cellularity in glioblastoma and correlation with ADC using trajectory analysis and automatic nuclei counting. PLoS One. 2016;11(7):e0160250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stadlbauer A, Ganslandt O, Buslei R, et al. Gliomas: histopathologic evaluation of changes in directionality and magnitude of water diffusion at diffusion-tensor MR imaging. Radiology. 2006;240(3):803–810. [DOI] [PubMed] [Google Scholar]
- 33. Thust SC, Hassanein S, Bisdas S, et al. Apparent diffusion coefficient for molecular subtyping of non-gadolinium-enhancing WHO grade II/III glioma: volumetric segmentation versus two-dimensional region of interest analysis. Eur Radiol. 2018;28(9):3779–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cui D, Ren J, Shi J, et al. R132H mutation in IDH1 gene reduces proliferation, cell survival and invasion of human glioma by downregulating Wnt/β-catenin signaling. Int J Biochem Cell Biol. 2016;73:72–81. [DOI] [PubMed] [Google Scholar]
- 35. Reis M, Czupalla CJ, Ziegler N, et al. Endothelial Wnt/β-catenin signaling inhibits glioma angiogenesis and normalizes tumor blood vessels by inducing PDGF-B expression. J Exp Med. 2012;209(9):1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pallud J, Varlet P, Devaux B, et al. Diffuse low-grade oligodendrogliomas extend beyond MRI-defined abnormalities. Neurology. 2010;74(21):1724–1731. [DOI] [PubMed] [Google Scholar]
- 38. Montgomery MK, Kim SH, Dovas A, et al. Glioma-induced alterations in neuronal activity and neurovascular coupling during disease progression. Cell Rep. 2020;31(2):107500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Agarwal S, Sair HI, Pillai JJ. The resting-state functional magnetic resonance imaging regional homogeneity metrics-kendall’s coefficient of concordance-regional homogeneity and coherence-regional homogeneity-are valid indicators of tumor-related neurovascular uncoupling. Brain Connect. 2017;7(4):228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Agarwal S, Lu H, Pillai JJ. Value of frequency domain resting-state functional magnetic resonance imaging metrics amplitude of low-frequency fluctuation and fractional amplitude of low-frequency fluctuation in the assessment of brain tumor-induced neurovascular uncoupling. Brain Connect. 2017;7(6):382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pillai JJ, Zacà D. Comparison of BOLD cerebrovascular reactivity mapping and DSC MR perfusion imaging for prediction of neurovascular uncoupling potential in brain tumors. Technol Cancer Res Treat. 2012;11(4):361–374. [DOI] [PubMed] [Google Scholar]
- 42. Zacà D, Jovicich J, Nadar SR, Voyvodic JT, Pillai JJ. Cerebrovascular reactivity mapping in patients with low grade gliomas undergoing presurgical sensorimotor mapping with BOLD fMRI. J Magn Reson Imaging. 2014;40(2):383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iranmahboob A, Peck KK, Brennan NP, et al. Vascular reactivity maps in patients with gliomas using breath-holding BOLD fMRI. J Neuroimaging. 2016;26(2):232–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Araque A, Parpura V, Sanzgiri RP, Haydon PG. Tripartite synapses: glia, the unacknowledged partner. Trends Neurosci. 1999;22(5):208–215. [DOI] [PubMed] [Google Scholar]
- 45. Bowden SG, Neira JA, Gill BJA, et al. Sodium fluorescein facilitates guided sampling of diagnostic tumor tissue in nonenhancing gliomas. Neurosurgery. 2018;82(5):719–727. [DOI] [PubMed] [Google Scholar]
- 46. Widhalm G, Olson J, Weller J, et al. The value of visible 5-ALA fluorescence and quantitative protoporphyrin IX analysis for improved surgery of suspected low-grade gliomas. J Neurosurg. 2020;133(1):79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hollon TC, Pandian B, Adapa AR, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med. 2020;26(1):52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beiko J, Suki D, Hess KR, et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol. 2014;16(1):81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yordanova YN, Moritz-Gasser S, Duffau H. Awake surgery for WHO Grade II gliomas within “noneloquent” areas in the left dominant hemisphere: toward a “supratotal” resection. Clinical article. J Neurosurg. 2011;115(2):232–239. [DOI] [PubMed] [Google Scholar]
- 50. Duffau H. Long-term outcomes after supratotal resection of diffuse low-grade gliomas: a consecutive series with 11-year follow-up. Acta Neurochir (Wien). 2016;158(1):51–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.