Abstract
Background
Response to targeted therapy varies between patients for largely unknown reasons. Here, we developed and applied an integrative platform using mass spectrometry imaging (MSI), phosphoproteomics, and multiplexed tissue imaging for mapping drug distribution, target engagement, and adaptive response to gain insights into heterogeneous response to therapy.
Methods
Patient-derived xenograft (PDX) lines of glioblastoma were treated with adavosertib, a Wee1 inhibitor, and tissue drug distribution was measured with MALDI-MSI. Phosphoproteomics was measured in the same tumors to identify biomarkers of drug target engagement and cellular adaptive response. Multiplexed tissue imaging was performed on sister sections to evaluate spatial co-localization of drug and cellular response. The integrated platform was then applied on clinical specimens from glioblastoma patients enrolled in the phase 1 clinical trial.
Results
PDX tumors exposed to different doses of adavosertib revealed intra- and inter-tumoral heterogeneity of drug distribution and integration of the heterogeneous drug distribution with phosphoproteomics and multiplexed tissue imaging revealed new markers of molecular response to adavosertib. Analysis of paired clinical specimens from patients enrolled in the phase 1 clinical trial informed the translational potential of the identified biomarkers in studying patient’s response to adavosertib.
Conclusions
The multimodal platform identified a signature of drug efficacy and patient-specific adaptive responses applicable to preclinical and clinical drug development. The information generated by the approach may inform mechanisms of success and failure in future early phase clinical trials, providing information for optimizing clinical trial design and guiding future application into clinical practice.
Keywords: drug distribution, drug response, mass spectrometry imaging, phosphoproteomics, t-CyCIF
Key Points.
Heterogeneous distribution of drug and response help to understand drug resistance.
DDR markers found by phosphoproteomics can be used to map cellular response by multiplexed tissue imaging.
Multimodal platform for development and implementation of targeted therapeutics and identification of biomarkers to evaluate in clinical practice.
Importance of the Study.
We have developed a multimodal platform to measure drug distribution, drug target engagement, and adaptive cellular response in the same tumor. Using this comprehensive discovery-oriented approach, we identified markers of drug target engagement relevant to DNA damage response pathways. Additionally, altered tyrosine phosphorylation signaling pathways were identified as putative mechanisms of cellular adaptation that may play a role in drug resistance to adavosertib. Combining mass spectrometry imaging, phosphoproteomics and multiplexed tissue imaging provide a unique platform to characterize the heterogeneous response to therapy common in cancer models and early phase clinical trials, potentially accelerating drug development and identifying biomarkers that can be applied in routine clinical settings. This platform can be used to study other therapies and solid tumor types and may identify the mechanisms that limit the efficacy of drugs in early phase clinical trials.
The clinical and preclinical studies that are typically conducted to assess endpoints such as progression-free survival or overall survival often suffer from a lack of adequate molecular characterization. When in vivo studies in mouse models of cancer and in patients yield unfavorable results, unanswered questions abound as to whether the drug was appropriately delivered and distributed throughout the tumor, whether the drug successfully inhibited the target, and whether tumor growth continued due to inherent or adaptive therapeutic resistance. In addition, genetic and phenotypic heterogeneity, both across patients or animals and within individual tumors, can affect response to therapy for most tumor types.1,2
Adavosertib is a Wee1 tyrosine kinase (TK) inhibitor that has emerged as a potential adjuvant treatment for high-grade brain tumors such as glioblastoma (GBM) which often overexpress Wee1.3 Wee1 is a negative regulator of entry into mitosis and when inhibited, an early G2 to M phase transition is induced that leads to mitotic catastrophe. Therapeutic resistance eventually develops through unidentified systems-level adaptive response.4 Treatment with adavosertib leads to DNA damage in S-phase cells, as indicated by increased phosphorylation of H2AX.5 Although changes in DNA damage response (DDR) associated markers such as γH2AX and cell cycle markers such as phosphoCDK1/2 have been measured in response to adavosertib treatment,6,7 the overall response to adavosertib-mediated DNA damage is incompletely understood. Other than downregulation of CDK1 Y15 phosphorylation, there is a lack of proven drug efficacy markers for adavosertib. Additional markers may be identified using new approaches for tissue profiling and algorithms that facilitate the integration and analysis of the multiple datasets acquired by these new technologies.
Methods have recently emerged that allow a more comprehensive profiling of the cellular states of tissues including phosphoproteomics, mass spectrometry imaging (MSI), and multiplexed tissue imaging. Phosphoproteomic analysis can simultaneously reveal the quantitative phosphorylation state of thousands of proteins. Characterization of the DDR by measuring the levels of phosphorylation at phosphoserine and phosphothreonine sites in putative ATM/ATR substrates can provide information on drug action,8 and phosphotyrosine levels can provide information on activation of cellular signaling pathways that have been associated with adaptive response and resistance.9–12 Direct mapping and quantifying the spatial distribution of drugs, lipids, or metabolites in tissue by MSI13,14 without the need for molecular labeling provides insight into the pharmacologic mechanisms underlying cellular responses to therapy.15 Genetic diversity is one of the main drivers of GBM heterogeneity,16 and invasiveness of tumor cells17 affect drug distribution.17,18 Single-cell studies have revealed the key role of the tumor microenvironment including immune cells—principally myeloid-lineage cells—and neuronal cells demonstrating cell state plasticity.16,19 Tissue cyclic immunofluorescence (t-CyCIF) is a highly multiplexed antibody-based imaging method that can characterize cell states and tumor architecture and thereby provide insights into the response of tumors to treatment at single-cell and subcellular resolution.20,21 The analysis of samples assayed with different technologies has created a major analytical challenge that requires algorithms that account for multiple experimental and biological factors and that permit cross-modality spatial integration (eg, Harmony).22
To identify key features of response to adavosertib, we developed a platform to study preclinical GBM PDX (patient-derived xenograft) models and clinical trial (“on-treatment”) GBM biopsies. This platform integrates data acquired from different analytical technologies: we use t-CyCIF to measure markers of DNA damage, cell cycle, and apoptosis markers (among others) that were in part selected based on phosphoproteomic analysis of GBM PDX before and after treatment with adavosertib; the results were mapped and quantified at single-cell level (in GBM PDX and then in human GBM resection samples) and correlated with drug distribution measured by MSI. Through this work, we hope to demonstrate an approach for multi-dataset integration that should ultimately benefit clinical practice by reducing complex measurements down to key features (nominated and validated across multiple methods) with the goal of implementation in clinical workflows using widely available and standard methods (eg, IHC).
Materials and Methods
Abridged versions of methods are included below. Please refer to the Supplementary Information for detailed materials and methods.
The clinical trial specimens were obtained from 6 different patients with recurrent GBM enrolled on the surgical arm of a phase I study of adavosertib (MK-1775 or AZD1775) led by the Adult Brain Tumor Consortium (ABTC), approved by the human investigations committee at Dana-Farber/Harvard Cancer Center, and registered on clinicaltrials.gov (NCT01849146). Informed written consent was obtained from each participant. The accepting criteria included participants with recurrent GBM that required surgery for tumor removal. After treatment with 200 mg or 425 mg of adavosertib daily for 5 days, tumors were surgically resected. Tumors were flash-frozen and stored at −80°C.
The animal studies were performed by Mayo Clinic and approved by the Mayo Institutional Animal Care and Use Committee. Three PDX lines were used: GBM12, GBM22, and GBM84 (PDX National Resource, Mayo Clinic). Athymic nude mice with xenograft implants were treated with adavosertib (Selleck) suspension in 0.5% methocel in water for 9 doses BID PO. Two hours after the last treatment, tumors were resected and flash-frozen.
Frozen tumors were split for MALDI-MSI (matrix-assisted laser desorption/ionization mass spectrometry imaging), phosphoproteomics, and t-CyCIF analysis. Cryo-sections with 10-μm thickness of frozen specimens were mounted either onto indium tin oxide coated glass slides for MALDI-MSI or onto regular microscopy glass slides for H&E staining.
MALDI-MSI analysis was performed on a 9.4 Tesla SolariX XR Fourier transform ion cyclotron resonance mass spectrometer with a resolution of 100 µm for flank tumors and 30 µm for clinical specimens. Phosphoproteomics was performed on frozen tumors as described previously23 with few modifications detailed in the Supplementary Information. Partial least-squares regression (PLSR) analysis and phosphorylation site identification details are described in the Supplementary Information. t-CyCIF was validated in fresh-frozen (FF) samples compared to FFPE (formalin-fixed paraffin-embedded) using a GBM22 PDX line and performed as described in the Supplementary Information. Drug distribution data from MALDI-MSI for adavosertib were integrated with t-CyCIF markers using Harmony pipeline described in Korsunsky et al22 to identify drug distribution associated with spatial DDR.
Results
Integrated Multimodal Platform to Study Drug Distribution and Molecular Response to Drug
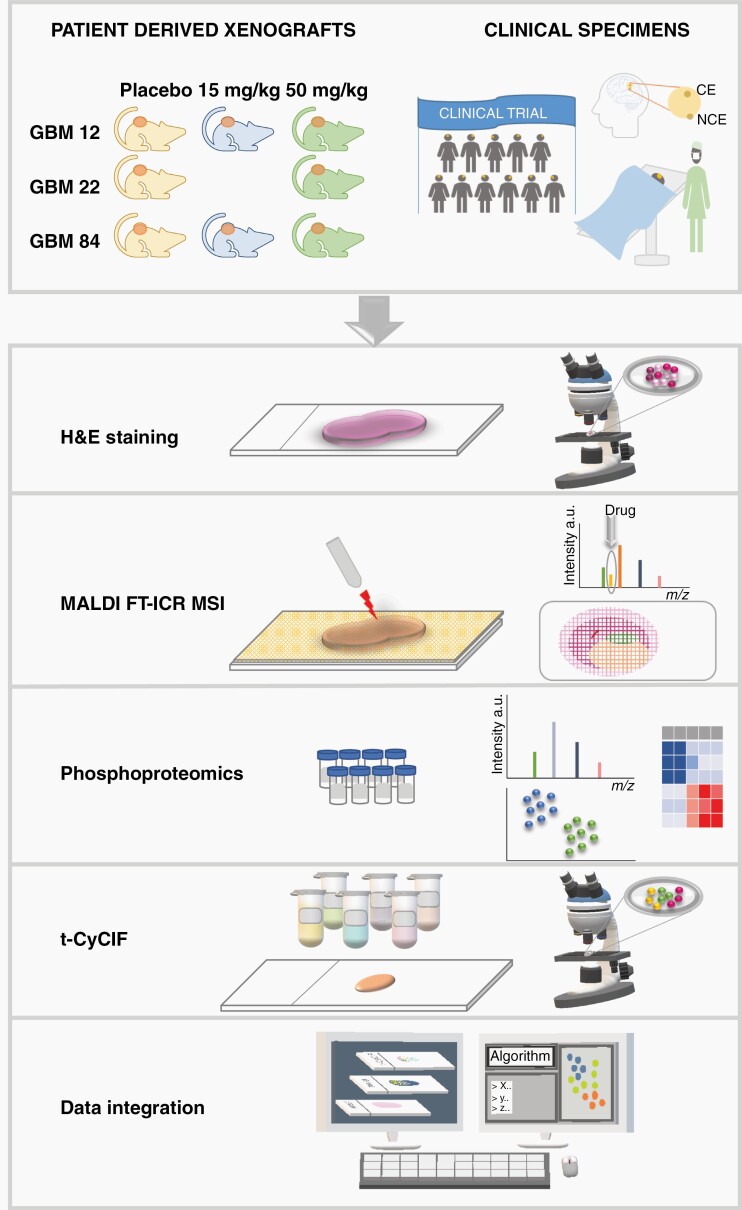
We developed an integrated multimodal platform (Figure 1) to assess drug distribution, drug action, and tumor cell response to the Wee1 kinase inhibitor adavosertib in GBM, a therapeutic that is being assessed in a multisite phase 1 clinical trial. To evaluate the performance of the platform while gaining insight into the in vivo distribution and response to adavosertib, we first selected 3 different PDX lines of GBM (GBM12, GBM22, and GBM84) and generated flank tumors for each in mice. Consecutive tissue sections of tumors before and after treatment were analyzed by histopathology and matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry imaging (MALDI FT-ICR MSI) was used to map the distribution of adavosertib and biometabolites. Pieces of the same PDX tumors were analyzed by phosphoproteomics to characterize the response to drug target engagement (focused on ATM/ATR substrate motifs: phosphoserine/threonine followed by glutamine [pSQ/pTQ])8 and the cellular adaptive response (phosphotyrosine [pTyr]). Finally, t-CyCIF was performed on tissue sections immediately adjacent to those used for MALDI-MSI, thereby allowing us to evaluate the spatial co-localization of drug and the resulting tissue response.
Fig. 1.
Summary of the experimental design and platform used for assessing drug distribution and response in patient-derived xenograft (PDX) models and clinical trial specimens resected from contrast-enhancing (CE) and non–contrast-enhancing (NCE) regions.
MALDI-MSI Reveals Heterogeneous Distribution of Adavosertib in Flank Tumors
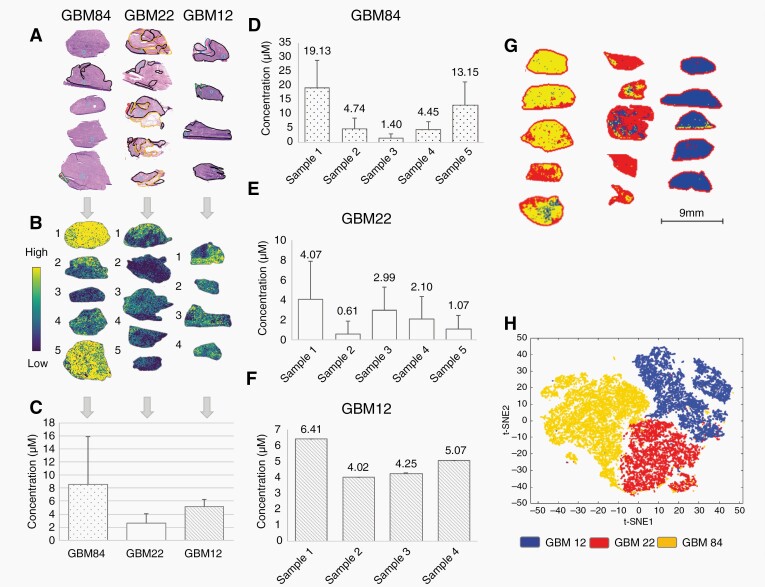
We observed a heterogeneous distribution of adavosertib in tissue sections mapped by MALDI FT-ICR MSI, with no direct spatial correlation of the drug with vasculature in pixel-by-pixel comparisons of drug (according to P values) and heme (a cofactor of hemoglobin) which is a well-validated marker of vasculature24,25 (Figure 2a–c, Supplementary Figure S1). This finding indicates that the drug enters the tumor parenchyma. Substantial heterogeneity between tumors was highlighted in GBM84 where the concentrations of adavosertib varied between 1–19 µM (Figure 2d) for samples in the 50 mg/kg dosing group. The coefficient of variation for drug concentration for GBM22 and GBM84 was above 60% and 20.9% for GBM12 (Figure 2c–f). Spatial distribution of the drug was evaluated relative to corresponding histopathology and showed significantly (P < .001 by ANOVA) lower intensities of adavosertib in necrotic regions annotated by a neuropathologist (Supplementary Figure S2). We also noted baseline metabolic differences between PDX lines independent of treatment (3 distinct clusters—Figure 2g; t-distributed stochastic neighbor embedding26 in Figure 2h).
Fig. 2.
Distribution and quantification of adavosertib (m/z 501.2706) and clustering of untreated tumors. (a) Annotated H&E staining of the flank tumors. Necrotic regions are delineated in yellow, mainly dense tumor in black, hemorrhagic tumor in red, and infiltrated skeletal muscle in green. (b) Spatial distribution of adavosertib in each of the tissue sections. (c) Average concentration of adavosertib measured in each GBM line with coefficient of variation of 65.0% for GBM22, 85.6% for GBM84, and 20.9% for GBM12. (d) Variation of the concentration of adavosertib in GBM84. (e) Variation of the concentration of adavosertib in GBM22. (f) Variation of the concentration of adavosertib in GBM12. The animal dosing was 50 mg/kg in all cases. (g) Clustering of the 3 control GBM models using Segmentation by bisecting k-means—3 clusters. (h) t-distributed stochastic neighbor embedding (t-SNE) of the 3 control PDX lines of GBM. Abbreviations: GBM, glioblastoma; PDX, patient-derived xenograft.
Potential underlying mechanisms of heterogeneous drug distribution were assessed by comparing the distribution of ions with high spatial variance to the corresponding histopathology (Supplementary Figure S1). For instance, in GBM22 adavosertib levels did not correlate with regions of hemorrhage or the vasculature (marked by heme) suggesting that the drug penetrated into the tumor parenchyma and distributed beyond the blood vessels. Selected ceramides such as cer(d34:1) were significantly higher in necrotic regions according to ANOVA, especially in GBM22 which displayed larger necrotic regions (Supplementary Figure S2), while ATP distribution was significantly higher in non-necrotic regions (as would be expected for viable and proliferating tumor). Treated samples showed an inverse distribution of adavosertib and the identified ceramides, coinciding with the lower amounts of drug found in necrotic tissue (Supplementary Figure S2), thus suggesting that intragroup variations of drug levels were in part affected by the presence of necrotic regions.
Adavosertib Leads to Dose-Dependent Phosphorylation Changes in DDR Network
Inhibition of Wee1 by adavosertib should result in increased DNA damage which can be monitored by measuring substrate phosphorylation in each PDX tumor line. Phospho-SQ/TQ-containing peptides were enriched by immunoprecipitation with a phospho-motif-specific antibody and quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using isobaric tags and multiplexed analysis. Adavosertib treatment resulted in a significant increase in phosphorylation of 19%, 60%, and 22% of sites across multiple DDR associated pathways in GBM12, GBM22, and GBM84 (50 mg/kg group), respectively (Supplementary Figure S3a), with substantial heterogeneity in response noted for each GBM line (Figure 3a, Supplementary Figure S3b and c, Supplementary Tables S1 and S2).
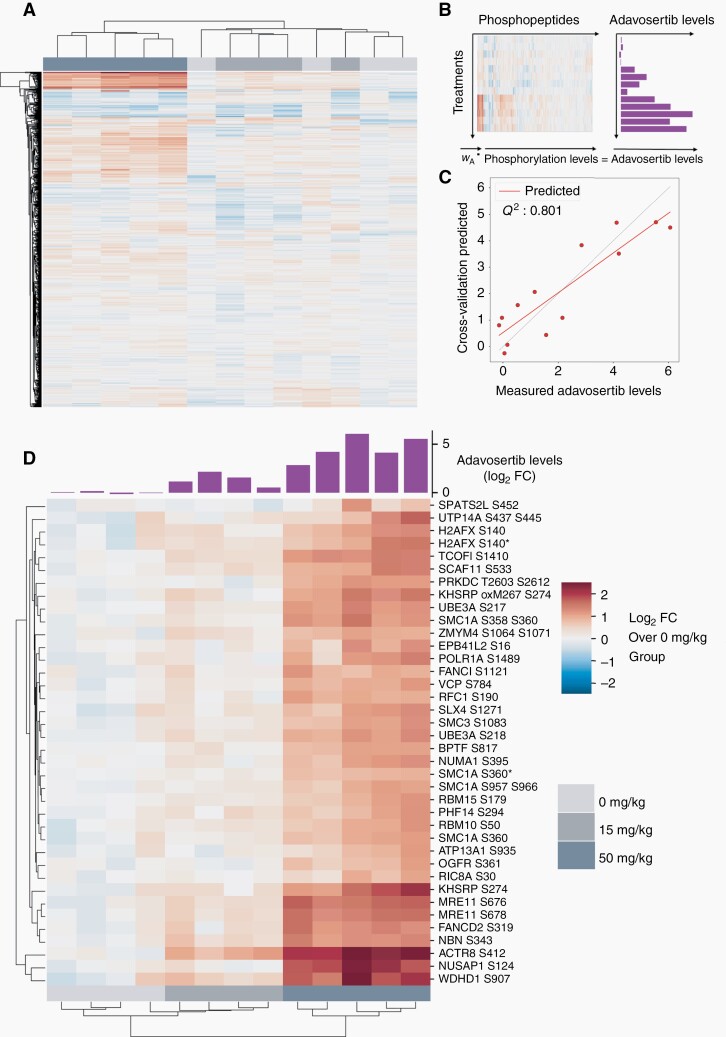
Fig. 3.
Identification and quantification of DDR markers after treatment with adavosertib. (a) HCA of phosphorylation levels of pSQ/pTQ-containing peptides in GBM84 PDX tumors treated with different doses. (b) Construction of the PLSR model. Quantified levels of phosphopeptides were regressed against adavosertib levels measured by MALDI-MSI. (c) Correlation plot of measured and predicted adavosertib levels from a cross-validated model built for GBM84. Q2 is the coefficient of determination (R2 of prediction from cross-validated model). (d) HCA of peptides that contribute most (variable importance in projection (VIP) score > 2) to the model built for GBM84. A barplot with measured drug levels is overlaid on top of the clusters. Measured drug levels and phosphorylation levels are shown as log2-fold change over the average of vehicle-treated tumors. Miscleaved peptides are indicated by *. Abbreviations: DDR, DNA damage response; GBM, glioblastoma; HCA, hierarchical clustering analysis; MALDI-MSI, matrix-assisted laser desorption/ionization mass spectrometry imaging; PDX, patient-derived xenograft.
Given that drug distribution and DDR phosphorylation data were both heterogeneous, we assessed the response profile relative to the average amount of drug present in a given tumor. We built a PLSR model for GBM84 (Figure 3b) to identify phosphorylation sites that are highly correlated with drug levels in each tumor. The PLSR model built for GBM84 was predictive of drug levels when assessed by leave-one-out cross-validation resulting in the coefficient of determination (Q2) between the measured drug levels and cross-validation predicted drug levels of 0.80 (Figure 3c). The heatmap resulting from hierarchical clustering analysis (HCA) of the phosphorylation sites that were contributing most to the model (Figure 3d) demonstrated a strong positive correlation between drug amount and phosphorylation response. Sites in the phosphorylation signature included proteins that are commonly associated with DDR pathways such as MRE11 and NBN (double-stranded break repair), SMC1A and SMC3 (structural maintenance of chromosomes), and NUMA1 (mitotic apparatus). We also observed upregulation of the canonical DNA damage marker γH2AX (H2AFX pS140), although other sites were more responsive to drug levels. Phosphorylation sites most predictive of drug levels in this model were found in WDHD1 (WD repeat and HMG box DNA-binding protein 1), also known as AND-1, a protein required for homologous recombination-based repair, and in UBE3A (ubiquitin-protein ligase E3A), a protein involved in p53 stability and apoptosis. Separate PLSR models for GBM12 and GBM22 were also predictive of drug levels (Q2 = 0.80 and 0.87, respectively), with a similar set of phosphorylation sites associated with drug levels (Supplementary Figure S4). Taken together, adavosertib led to increased phosphorylation of numerous putative ATM/ATR substrates in each of the PDX lines suggesting that the drug is inhibiting Wee1 resulting in increased DNA damage. Additionally, the level of DDR was directly correlated with the amount of drug distributed in the tumors.
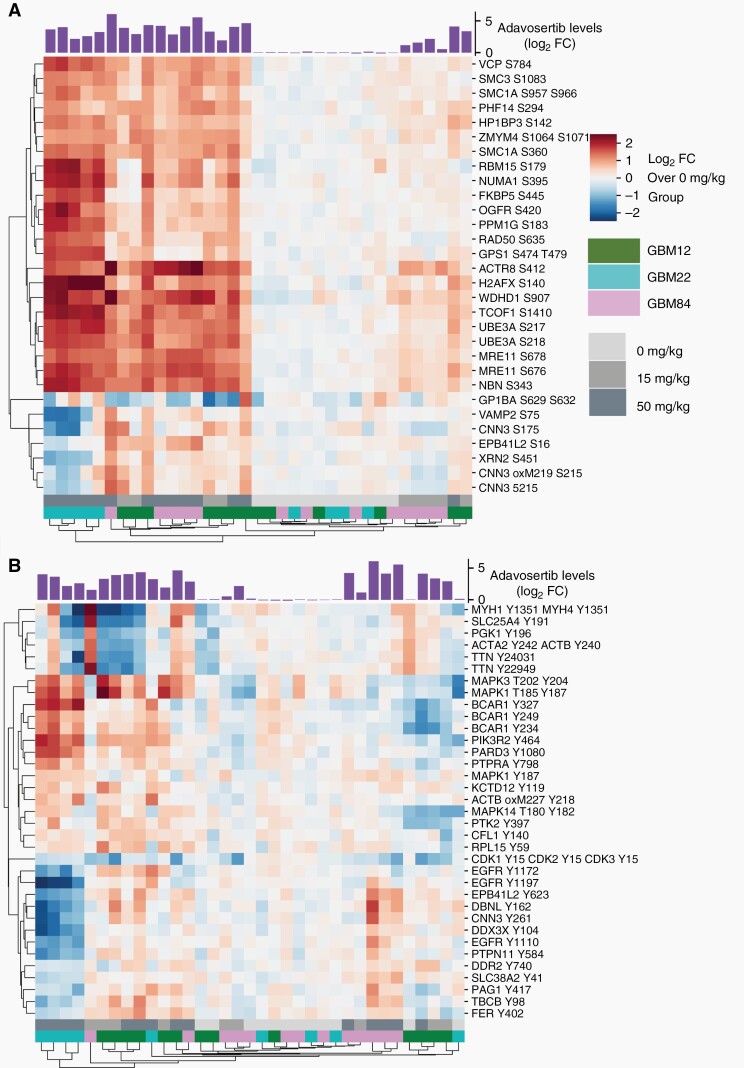
Considering the overlap in predictive phosphorylation sites across the 3 PDX lines, we built a predictive model encompassing data from all 3 PDX lines, yielding an overlap of 272 unique pSQ/pTQ-containing peptides and identifying a common phosphorylation signature of drug action. The PLSR “cross-model” was predictive of adavosertib levels across all tumors (Q2 of 0.83) (Supplementary Figure S5a) using leave-one-out cross-validation. Predictive values (Q2 between 0.44 and 0.65) were also obtained when 1 of the 3 GBM lines was used as test set while other 2 lines were used as training set (Supplementary Figure S5b–d), indicating that this phosphorylation signature may be extendable to other GBM PDXs. Phosphorylation sites that were most positively correlated with drug level consisted of well-characterized proteins such as MRE11, NBN, RAD50, and H2AX (Figure 4a). Several sites on VAMP2, XRN2, and CNN3 were differentially phosphorylated in GBM22, where they were weakly negatively correlated with drug level, suggesting that there were some differences between the PDX lines. Taken together, the results suggest that heterogeneous drug distribution leads to drug exposure-dependent changes in the DDR network. Moreover, across PDXs from different patients, a common set of phosphorylation sites were identified. The phosphorylation data are summarized in Supplementary Table S1.
Fig. 4.
Quantification of DNA damage and adaptive response to adavosertib across PDXs of GBM12, GBM22, and GBM84. (a) HCA heatmap showing the pSQ/TQ-containing peptides with variable importance in projection (VIP) score > 1.4 for a PLSR model built with pSQ/pTQ data from all 3 lines. (b) HCA heatmap of pY-containing peptides with VIP score > 1 for a PLSR model built with phosphotyrosine data from all 3 PDX lines. A barplot with measured drug levels is overlaid on top of the clusters. Measured drug levels and phosphorylation levels are shown as log2-fold change over the average of vehicle-treated tumors. Abbreviations: GBM, glioblastoma; PDX, patient-derived xenograft; PLSR, partial least-squares regression.
Adavosertib Leads to PDX Line-Specific Adaptive Response
Although tyrosine phosphorylation (pTyr) only represents 0.1%–1% of total protein phosphorylation, it can provide a readout of the adaptive cell response to drug exposure.9,27 Some common mechanisms of drug resistance result in dysregulation of receptor TK networks, and the canonical central regulator pathways including PI3K and ERK pathways. The activities of these pathways are regulated by pTyr, so we captured the adaptive cellular response to adavosertib through quantification of pTyr. As with the pSQ/pTQ data, pTyr levels demonstrated a high degree of inter-tumor heterogeneity within treatment groups. PLSR models were generated for each line by regressing pTyr data against the average adavosertib level measured for that tumor. Tyrosine phosphorylation was associated with drug levels, although less compared to the pSQ/pTQ data, as demonstrated by Q2 values of 0.56, 0.46, and 0.72 for GBM12, GBM22, and GBM84, respectively (Supplementary Figure S6a–c). The adaptive response across the entire set of tumors was investigated by building a PLSR “cross-model” of the association between pTyr and adavosertib levels. Excluding an entire PDX line from the training set resulted in very poor Q2 values of 0.28, −9.55, and −0.26 when predicting for GBM12, GBM22, and GBM84, respectively, although leave-one-tumor-out cross-validation yielded a Q2 value of 0.75 (Supplementary Figure S6d–g), suggesting differences in pTyr signaling among the PDX lines in response to adavosertib. HCA of the most correlated sites provided 3 clusters, including a MAPK and p130Cas (BCAR1) cluster, an EGFR/Src family kinase (SFK) cluster, and a cytoskeletal cluster featuring sites on actin, myosin, and titin (Figure 4b). However, unlike the pSQ/pTQ data, these clusters were differentially regulated in each line. For instance, the EGFR/SFK cluster was positively correlated with drug levels in GBM84, yet negatively correlated with drug levels in GBM22. Similarly, the MAPK/p130Cas cluster demonstrated positive correlation with drug levels in GBM22 and GBM12, yet was either unchanged or slightly decreased with increasing adavosertib in GBM84, suggesting probable bypass adaptive resistance mechanisms in GBM12 and GBM22 (Supplementary Figure S6h and i). Together, the pSQ/pTQ and pTyr data provide unprecedented systems-level characterization of tumor response to adavosertib, including well-characterized and largely conserved activation of DNA damage repair pathways and previously uncharacterized activation of TK networks that are tumor-specific. Further characterization of these pathways and their potential effects on cellular phenotypes may help to provide insight into patient-specific response and resistance mechanisms.
t-CyCIF Shows Differential Cellular Response to Drug at High Spatial Resolution
While we had characterized the association of drug level and tumor response at the bulk tissue level using phosphoproteomics, MSI quantification in tissue sections revealed that adavosertib distribution was spatially heterogeneous within each tumor. To characterize the spatial heterogeneity of the tumor response to adavosertib on a cellular level for each tumor, we mapped cell states using t-CyCIF in the same specimens that we had analyzed by MSI and phosphoproteomics. We selected antibodies against DDR phosphorylation sites that were present in the predictive phosphorylation signature (such as SMC1_S957, MRE11_S676, NBN_S343, and H2AX_S139) along with markers of cell proliferation and cell cycle phase (such as Ki67, PCNA, CDT1, Geminin, Histone H3_S10, and Cyclin A1).
We validated the t-CyCIF antibody staining on FFPE tissue samples and compared the results with FF tissue. t-CyCIF is typically performed on FFPE tissue but drug distribution is assessed by MSI from FF tissue, so direct comparison of t-CyCIF with MSI results required validation of the t-CyCIF method in FF tissue. Single cells were segmented, and the fluorescence signal was calculated on a per cell basis. We calculated the percentage of positive cells identified per marker in both the FFPE and FF sections and found a correlation value of R = .8986, P = .004 indicating that the antibodies performed similarly in FF and FFPE samples (Supplementary Figure S7).
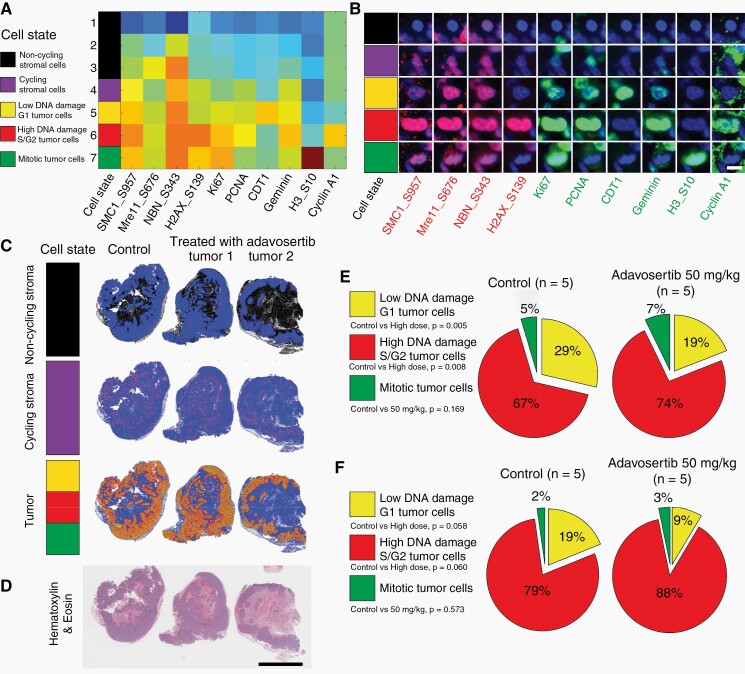
In the FF tissues, we used clustering analysis and binned cells into 5 different cell state categories (non-cycling stromal cells, cycling stromal cells, low DNA damage G1 tumor cells, high DNA damage S/G2 tumor cells, and mitotic tumor cells) depending on the co-expression of different markers (Figure 5a, Supplementary Table S3). Cells with high Geminin, high PCNA, low CDT1, and high phosphorylation of DDR proteins were classified as “high DNA damage S/G2 tumor cells.” Representative t-CyCIF images of individual cells in each of the 5 cell states are shown in Figure 5b; maps showing the spatial distribution of these cell states (relative to their corresponding histopathology) provided spatial information for the evaluation of cycling vs non-cycling stroma, along with different tumor cell states (Figure 5c and d). t-CyCIF performed on adjacent sections to MSI and histology sections from each GBM PDX tumor highlighted a shift in tumor cell state following adavosertib treatment, with an increase in high DNA damage S/G2 cells and a corresponding decrease in low DNA damage G1 tumor cells in adavosertib-treated GBM22 and GBM84 tumors relative to their untreated controls (Figure 5e and f, Supplementary Table S3) supporting target engagement and the expected effects on DNA damage and the cell cycle.
Fig. 5.
Effects of adavosertib treatment in GBM-derived lines characterized by t-CyCIF technique. (a) Heatmap of each cell state (clusters) using k-means clustering from all cells in FFPE section from GBM22 derived PDX line batch 1 (control, n = 1; adavosertib high dose, 50 mg/kg, n = 2) using 4 DNA damage related markers (SMC1_S957, Mre11_S676, NBN_S343, and H2AX_S139) and 6 cell cycle related markers (Ki67, PCNA, CDT1, Geminin, H3_S10, and Cyclin A1) (Heatmap: red—high expression; blue—low expression). All cells were clustered into the following cell states: non-cycling stromal cells (black, Ki67-negative/PCNA-negative, cluster 1–3); Cycling stromal cells (purple, Ki67 low/PCNA low/DNA damage low, cluster 4); Low DNA damage G1 tumor cells (yellow, CDT1 high/Ki67 low/PCNA low/DNA damage low, cluster 5); High DNA damage S/G2 tumor cells (red, Geminin high/PCNA high/CDT1 low/DNA damage high, cluster 6); Mitotic tumor cells (green, pH3 high/Ki67 high/PCNA low, cluster 7). (b) Representative cells for each cell state in the FFPE section from GBM22-derived PDX line batch 1 (scale bar: 10 µm). (c) Dot plots showed the distribution of each cell state (non-cycling stroma, cycling stroma and tumor) in FFPE section from GBM22-derived PDX line batch 1 (control, n = 1; adavosertib high dose, 50 mg/kg, n = 2). (d) The H&E image from adjacent FFPE section showed the tumor region (scale bar: 5000 µm). (e) The percentage of tumor cells of each cell cycle phase to all tumor cells was calculated and presented as a pie chart for frozen sections from GBM22 derived PDX line batch 2 (control, n = 5; adavosertib high dose, 50 mg/kg, n = 5). (f) GBM84 derived PDX line (control, n = 5; adavosertib high dose, 50 mg/kg, n = 5). Detailed information for individual PDX is listed in Supplementary Table S4. Abbreviations: GBM, glioblastoma; FFPE, formalin-fixed paraffin-embedded; PDX, patient-derived xenograft, FFPE, PDX.
Drug Distribution and Correlated Response in Clinical Trial Specimens
We used this multimodal platform to study drug distribution and cellular response in human GBM tissue specimens acquired as part of a multisite clinical trial of adavosertib designed to evaluate intratumoral drug distribution in patients with recurrent GBM. Six patients that underwent surgery in 3 clinical sites were randomly assigned to receive a daily dose of either 200 or 425 mg adavosertib without other treatments starting 4 days prior to surgery. Considering the variability in blood brain barrier (BBB) penetration of adavosertib,6,13,28 contrast-enhancing (CE) and non–contrast-enhancing (NCE) areas were both surgically sampled based on preoperative MRI and studied to evaluate the propensity of the drug to distribute into different tumor compartments.
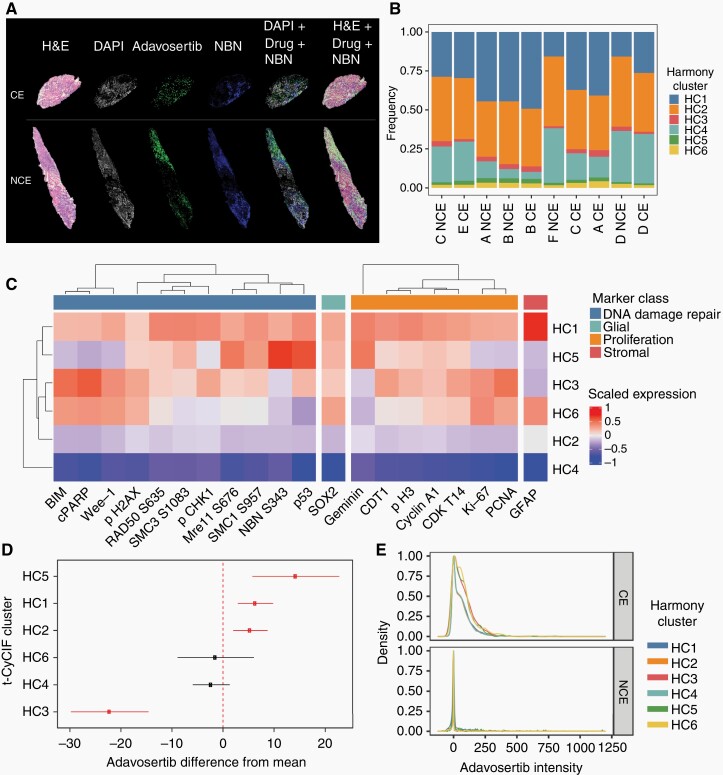
Adjacent tissue sections from each tumor specimen were analyzed by histology, MSI, and t-CyCIF from each patient (Figure 6a), and images were registered using a nonlinear approach. Regions of tumor with high adavosertib levels showed high levels of DDR (Nibrin (NBN) pS343 overlayed with adavosertib levels in Figure 6a), however, initial analysis of drug response in specimens from all patients showed that metabolomic and proteomic analyses as well as t-CyCIF were confounded by collection site (Supplementary Figure S8), likely reflecting the effect of different specimen collection protocols at each site.
Fig. 6.
Multimodal platform application to clinical specimens. (a) Integration of the spatial distribution of tumor cells (histology), drug (MALDI-MSI), and antibody (t-CyCIF) from the CE and NCE specimens from the same patient (case C). From left to right, histological image, DAPI (in gray) used in the corresponding IF cycle where NBN was stained to control for cell loss, drug distribution image acquired by MALDI-MSI (green), NBN distribution acquired by t-CyCIF (blue), nonlinear registration of DAPI, drug, and NBN, and nonlinear registration of the histological image to drug and NBN. (b) Harmony clustering analysis of t-CyCIF images identified 6 joint clusters with similar proportions across 10 distinct images. (c) Differential expression analysis of the t-CyCIF markers shows distinct associations of molecules related to DNA damage repair (blue), glial (green), proliferation (orange), and stromal lineage (red). Colors represent scaled log-fold changes, scaled to z scores for each marker. (d) Integrated analysis of MSI and t-CyCIF associates DDR and proliferation phenotypes with spatial distribution of adavosertib. Spatial distribution of adavosertib is higher (FDR = 0.09) near cluster 5 and lower (FDR = 0.004) near cluster 3 cells. (e) Differential response between CE and NCE regions for patient D based on the t-CyCIF harmony clusters reveal differences between them. Abbreviations: CE, contrast-enhancing; DAPI, diamidino-2-phenylindole; DDR, DNA damage response; FDR, false discovery rate; IF, immunofluorescence; MALDI-MSI, matrix-assisted laser desorption/ionization mass spectrometry imaging; NCE, non–contrast-enhancing.
To study the tissues from the 6 donors spanning 3 collection sites, we integrated the data into a common coordinated space to find overlapping signals of drug distribution and cellular DDR. Batch effect was corrected from the t-CyCIF profiles using Harmony and identified 6 “harmonic” clusters (HC1-6) that were present in all 6 patients (Figure 6b), but with distinct profiles (Figure 6c). HC1 and HC6 both expressed astrocytic marker GFAP and HC1, HC3, HC5, and HC6 expressed SOX2, which is a marker of glial tumor cells. Cluster HC1 was more enriched in DDR markers pSMC1, pSMC3, and RAD50 and proliferation markers pH3 and CDT1. Cluster HC5 had the strongest expression of drug response associated markers pNBN, p53, pMre11, and Geminin. Cluster HC3 likely reflects an apoptotic population, expressing BIM and cPARP. Finally, clusters HC2 and HC4 did not have any distinguishing markers in this panel and may reflect unresponsive stromal cells.
The cellular markers were further correlated to drug distribution, using mixed-effects regression to associate adavosertib levels to t-CyCIF clusters, accounting in this way for the batch effects that were confounding the drug levels. This analysis revealed the highest levels of adavosertib in cluster HC5 (which also showed high DDR markers supporting target engagement) and slightly increased drug levels in cluster HC1 which also showed high DDR markers. The lowest drug levels and highest apoptotic markers (BIM and cleaved PARP) were observed in cluster HC3 (Figure 6c and d). Among the 3 clusters HC1, HC3, and HC5 that showed increased levels of adavosertib, cluster HC5 had the lowest expression of Wee1. Case D was notable for a significant higher adavosertib levels in the CE specimen compared to the NCE specimen (Supplementary Figures S8b and S9a). Consistent with higher drug levels, the Harmony analysis showed higher DDR in the CE specimen compared to the NCE specimen (Figure 6e).
Discussion
Here we have developed and implemented a multimodal platform for the analysis of drug distribution, drug efficacy, and tumor cell response, with high spatial resolution. Application of this platform to GBM PDX models treated with adavosertib, a WEE1 inhibitor in clinical testing for a number of different malignancies, provided a biomarker signature for drug efficacy. Components of this signature were then spatially connected to drug distribution in human tumor specimens from a clinical trial for adavosertib in GBM. Together, these data quantify the effects of therapy at a cellular level in tumors from patients in a clinical trial, thus providing insight into mechanisms of action or failure in ongoing clinical trials. This platform can be used in trials to study a range of therapies, including cytotoxic chemotherapies, targeted therapeutics such as kinase inhibitors, and biologicals including antibodies and antibody-drug conjugates. The insights from multimodal studies can then be used to identify biomarkers that may be implemented in routine clinical practice.
Previous studies attributed the heterogeneous distribution of adavosertib in an intracranial GBM22 line to the disruption of the BBB.13 Intriguingly, we observed that PDX tumors grown in the flank of animals also resulted in heterogeneous drug distribution, despite the absence of the BBB. Heterogeneous drug distribution correlated with selected MSI-defined biometabolite distributions within individual tumors, and generally followed distinct histological features such as necrosis, tumor cell density, and hemorrhagic regions, indicating that drug distribution is affected by the biochemical and biophysical properties of the tumor, even in flank tumors. In this context, understanding the spatial distribution of metabolites such as ceramides relative to drug levels in brain tumors can be useful in implementing combination therapy since the acid ceramidase which metabolizes ceramides is involved in the development of resistance to radiotherapy in cases of GBM29 and acid ceramidase inhibitors have been proposed as a means to overcome treatment limitations in GBM.30
Despite the inherent heterogeneity between different PDX tumor lines, PLS-based regression modeling provided a biomarker signature (eg, consensus pSQ/pTQ sites) that was predictive of drug levels in all 3 PDX lines. Although H2AX phosphorylation is widely used as a DNA damage marker,8 other less characterized proteins such as ACTR8 and WDHD1 had a greater increase in phosphorylation on adavosertib exposure, suggesting their utility in measuring DDR. Interestingly, while all 3 PDX lines shared consensus DDR pathways, adaptive response pathways were dissimilar among lines, suggesting that different patient tumors mount different responses to therapy that may engage different resistance mechanisms. The synergistic effect of drug combinations is currently being investigated to reduce tumor growth and improve patient outcomes31,32 and the GBM Adaptive Global Innovative Learning Environment (GBM AGILE)33 counts on a multidisciplinary team of professionals to identify effective therapies and biomarkers worldwide. For immune-based therapies in GBM,34 identification of the adaptive response pathways could lead to optimized personalized combination strategies.
Spatially resolved evaluation of drug levels and DDR, confirmed by statistically significant associations (FDR <10%), allowed mapping of the mechanism of action of adavosertib at cellular resolution. Highest phosphorylation levels for DDR markers in cluster HC5 were consistent with the highest drug levels in same cluster.35 DNA damage associated with drug treatment was highlighted in regions enriched in DNA damage and cell cycle response markers (cluster HC3), consistent with the expected G2/M arrest following adavosertib treatment.36 Finally, adavosertib has been reported to have limited penetration through the BBB,13 and astrocytes play an important role in maintaining the BBB integrity.37 It was observed that regions with high drug intensities presented low levels of the astrocyte marker GFAP, highlighting regions that may have a compromised BBB.
The integrated models provided a biomarker signature of drug efficacy that was applicable across PDX lines and phase 1 clinical trial specimens. Furthermore, the approach allowed us to resolve the intra-tumor heterogeneity of drug distribution and efficacy in human tumors, critical to understanding differential response to therapy amongst patients. The comparison of multiple specimens with different drug levels from each patient provides an opportunity to investigate drug exposure-dependent response directly in humans. Considering the limited dataset, our goal is to encourage further analyses of clinical trials and the use of such platforms to measure drug response. This multimodal platform, while developed and implemented specifically for the study of adavosertib, should facilitate across a range of clinical trials the assessment of drug distribution and drug efficacy, information that has previously been largely inaccessible. The underlying technologies are increasingly accessible at academic clinical centers so in the near term some of the presented analyses could be centralized in these centers for the study of clinical trials and annotated clinical cohorts.38 However, efforts are underway to introduce MALDI-MSI39,40 and a wide range of multiplexed tissue imaging techniques41 into standard clinical diagnostics and to generate phosphoproteomic data from the formalin-fixed surgical resection tissues common in pathology laboratories.42,43 Ultimately, the goal is to identify critical markers of response that can be widely implemented in routine clinical workflows using standard and widely accessible methodologies.
Supplementary Material
Funding
The clinical samples were obtained from a clinical trial sponsored by the Adult Brain Tumor Consortium (ABTC) National Institutes of Health/National Cancer Institute (NIH/NCI) UM1 CA137443 and AstraZeneca (clinicaltrials.gov identifier NCT01849146). This work was funded by NIH U54 CA210180 MIT/Mayo Physical Science Oncology Center for Drug Distribution and Drug Efficacy in Brain Tumors, the MIT Center for Precision Cancer Medicine, NIH U54-CA225088 HMS/Cancer Systems Biology Consortium, and the Dana-Farber Cancer Institute PLGA Fund. N.Y.R.A. receives support from the Ferenc Jolesz National Center for Image Guided Therapy NIH P41-EB-015898 and NIH R01CA201469. E.C.R. receives support from NIH R25 (R25 CA-89017). S.S.B. receives support from NIH Training Grant T32 HL007627. A.R.C. receives funding from the Harvard Program in Therapeutic Science Therapeutics Graduate Program (T32-GM-7306-42). S.A.S. is supported by an NIH T32 (1T32EB025823-01A1).
Conflict of interest statement. N.Y.R.A. is a scientific advisor to BayesianDx and key opinion leader to Bruker Daltonics. E.Q.L. reports royalties from Wolters Kluwer (UpToDate, Inc.) and is a consultant to Prime Oncology.
Authorship statement. Manuscript writing: B.G.C.L., I.N.K., Z.D., E.C.R., N.Y.R.A., A.R.C., F.M.W., and S.S.B. Research design: B.G.C.L., I.N.K., Z.D., P.Y.W., B.A., J.S., S.S. F.M.W., N.Y.R.A., and E.Q.L. Performed research: B.G.C.L., I.N.K., Z.D., M.S.R., B.M.M., A.C.T., D.M.B., and S.A.S. Data analysis contribution: B.G.C.L., I.N.K., Z.D., Y.D., G.G., E.C.R., S.S., W.A., Y.D., G.G., I.K., and S.R. Analytical tools contribution: J.N.A.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE44 partner repository with the dataset identifier PXD018782 and 10.6019/PXD018782. The MALDI-MSI data are available upon request. The processed t-CyCIF montage images were uploaded to Omero.
References
- 1. Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saunders NA, Simpson F, Thompson EW, et al. Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO Mol Med. 2012;4(8):675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mir SE, De Witt Hamer PC, Krawczyk PM, et al. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18(3):244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sen T, Tong P, Diao L, et al. Targeting AXL and mTOR pathway overcomes primary and acquired resistance to WEE1 inhibition in small-cell lung cancer. Clin Cancer Res. 2017;23(20):6239–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bukhari AB, Lewis CW, Pearce JJ, Luong D, Chan GK, Gamper AM. Inhibiting Wee1 and ATR kinases produces tumor-selective synthetic lethality and suppresses metastasis. J Clin Invest. 2019;129(3):1329–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanai N, Li J, Boerner J, et al. Phase 0 trial of AZD1775 in first-recurrence glioblastoma patients. Clin Cancer Res. 2018;24(16):3820–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keenan TE, Li T, Vallius T, et al. Clinical efficacy and molecular response correlates of the WEE1 inhibitor adavosertib combined with cisplatin in patients with metastatic triple-negative breast cancer. Clin Cancer Res. 2021;27(4):983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. [DOI] [PubMed] [Google Scholar]
- 9. Emdal KB, Dittmann A, Reddy RJ, et al. Characterization of in vivo resistance to osimertinib and JNJ-61186372, an EGFR/Met bispecific antibody, reveals unique and consensus mechanisms of resistance. Mol Cancer Ther. 2017;16(11):2572–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773(8):1263–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sasaki T, Koivunen J, Ogino A, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71(18):6051–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Amit I, Wides R, Yarden Y. Evolvable signaling networks of receptor tyrosine kinases: relevance of robustness to malignancy and to cancer therapy. Mol Syst Biol. 2007;3:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pokorny JL, Calligaris D, Gupta SK, et al. The efficacy of the Wee1 Inhibitor MK-1775 combined with temozolomide is limited by heterogeneous distribution across the blood-brain barrier in glioblastoma. Clin Cancer Res. 2015;21(8):1916–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Groseclose MR, Castellino S. A mimetic tissue model for the quantification of drug distributions by MALDI imaging mass spectrometry. Anal Chem. 2013;85(21):10099–10106. [DOI] [PubMed] [Google Scholar]
- 15. Calandra E, Posocco B, Crotti S, et al. Cross-validation of a mass spectrometric-based method for the therapeutic drug monitoring of irinotecan: implementation of matrix-assisted laser desorption/ionization mass spectrometry in pharmacokinetic measurements. Anal Bioanal Chem. 2016;408(19):5369–5377. [DOI] [PubMed] [Google Scholar]
- 16. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parker JJ, Canoll P, Niswander L, Kleinschmidt-DeMasters BK, Foshay K, Waziri A. Intratumoral heterogeneity of endogenous tumor cell invasive behavior in human glioblastoma. Sci Rep. 2018;8(1):18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qazi MA, Vora P, Venugopal C, et al. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol. 2017;28(7):1448–1456. [DOI] [PubMed] [Google Scholar]
- 19. Friebel E, Kapolou K, Unger S, et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell. 2020;181(7):1626–1642.e20. [DOI] [PubMed] [Google Scholar]
- 20. Lin JR, Izar B, Wang S, et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife. 2018;7:e31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du Z, Lin JR, Rashid R, et al. Qualifying antibodies for image-based immune profiling and multiplexed tissue imaging. Nat Protoc. 2019;14(10):2900–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dittmann A, Kennedy NJ, Soltero NL, et al. High-fat diet in a mouse insulin-resistant model induces widespread rewiring of the phosphotyrosine signaling network. Mol Syst Biol. 2019;15(8):e8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Randall EC, Lopez BGC, Peng S, et al. Localized metabolomic gradients in patient-derived xenograft models of glioblastoma. Cancer Res. 2020;80(6):1258–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu X, Ide JL, Norton I, et al. Molecular imaging of drug transit through the blood-brain barrier with MALDI mass spectrometry imaging. Sci Rep. 2013;3:2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Der Maaten L, Hinton G.. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 27. Randall EC, Emdal KB, Laramy JK, et al. Integrated mapping of pharmacokinetics and pharmacodynamics in a patient-derived xenograft model of glioblastoma. Nat Commun. 2018;9(1):4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li J, Wu J, Bao X, et al. Quantitative and mechanistic understanding of AZD1775 penetration across human blood-brain barrier in glioblastoma patients using an IVIVE-PBPK modeling approach. Clin Cancer Res. 2017;23(24):7454–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doan NB, Nguyen HS, Al-Gizawiy MM, et al. Acid ceramidase confers radioresistance to glioblastoma cells. Oncol Rep. 2017;38(4):1932–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doan NB, Alhajala H, Al-Gizawiy MM, et al. Acid ceramidase and its inhibitors: a de novo drug target and a new class of drugs for killing glioblastoma cancer stem cells with high efficiency. Oncotarget. 2017;8(68):112662–112674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Graham-Gurysh EG, Murthy AB, Moore KM, Hingtgen SD, Bachelder EM, Ainslie KM. Synergistic drug combinations for a precision medicine approach to interstitial glioblastoma therapy. J Control Release. 2020;323:282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McNeill RS, Canoutas DA, Stuhlmiller TJ, et al. Combination therapy with potent PI3K and MAPK inhibitors overcomes adaptive kinome resistance to single agents in preclinical models of glioblastoma. Neuro Oncol. 2017;19(11):1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alexander BM, Ba S, Berger MS, et al. Adaptive global innovative learning environment for glioblastoma: GBM AGILE. Clin Cancer Res. 2018;24(4):737–743. [DOI] [PubMed] [Google Scholar]
- 34. Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15(7):422–442. [DOI] [PubMed] [Google Scholar]
- 35. Zhu JY, Cuellar RA, Berndt N, et al. Structural basis of wee kinases functionality and inactivation by diverse small molecule inhibitors. J Med Chem. 2017;60(18):7863–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hirai H, Iwasawa Y, Okada M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8(11):2992–3000. [DOI] [PubMed] [Google Scholar]
- 37. Abbott NJ, Rönnbäck L, Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat Rev Neurosci. 2006;7(1):41–53. [DOI] [PubMed] [Google Scholar]
- 38. Rozenblatt-Rosen O, Regev A, Oberdoerffer P, et al. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell. 2020;181(2):236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Basu SS, Agar NYR. Bringing matrix-assisted laser desorption/ionization mass spectrometry imaging to the clinics. Clin Lab Med. 2021;41(2):309–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Basu SS, Regan MS, Randall EC, et al. Rapid MALDI mass spectrometry imaging for surgical pathology. NPJ Precis Oncol. 2019;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bodenmiller B. Multiplexed epitope-based tissue imaging for discovery and healthcare applications. Cell Syst. 2016;2(4):225–238. [DOI] [PubMed] [Google Scholar]
- 42. Kohale IN, Burgenske DM, Mladek AC, et al. Quantitative analysis of tyrosine phosphorylation from FFPE tissues reveals patient-specific signaling networks. Cancer Res. 2021;81(14):3930–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kalocsay M, Maliga Z, Nirmal AJ, et al. Multiplexed proteomics and imaging of resolving and lethal SARS-CoV-2 infection in the lung. bioRxiv. Published online October 15, 2020. doi: 10.1101/2020.10.14.339952 [DOI] [Google Scholar]
- 44. Perez-Riverol Y, Csordas A, Bai J, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE44 partner repository with the dataset identifier PXD018782 and 10.6019/PXD018782. The MALDI-MSI data are available upon request. The processed t-CyCIF montage images were uploaded to Omero.