Abstract
The accurate and speedy detection of COVID-19 is essential to avert the fast propagation of the virus, alleviate lockdown constraints and diminish the burden on health organizations. Currently, the methods used to diagnose COVID-19 have several limitations, thus new techniques need to be investigated to improve the diagnosis and overcome these limitations. Taking into consideration the great benefits of electrocardiogram (ECG) applications, this paper proposes a new pipeline called ECG-BiCoNet to investigate the potential of using ECG data for diagnosing COVID-19. ECG-BiCoNet employs five deep learning models of distinct structural design. ECG-BiCoNet extracts two levels of features from two different layers of each deep learning technique. Features mined from higher layers are fused using discrete wavelet transform and then integrated with lower-layers features. Afterward, a feature selection approach is utilized. Finally, an ensemble classification system is built to merge predictions of three machine learning classifiers. ECG-BiCoNet accomplishes two classification categories, binary and multiclass. The results of ECG-BiCoNet present a promising COVID-19 performance with an accuracy of 98.8% and 91.73% for binary and multiclass classification categories. These results verify that ECG data may be used to diagnose COVID-19 which can help clinicians in the automatic diagnosis and overcome limitations of manual diagnosis.
Keywords: COVID-19, ECG trace Image, Deep learning, Convolutional neural networks (CNN), Discrete wavelet transform (DWT)
1. Introduction
By the end of December 2019, the new coronavirus was originally registered in Wuhan, China. It has been called COVID- 19 by the World Health Organization (WHO) in 2020 [1]. Later on March 11, the WHO announced COVID-19 as a pandemic [2]. Following the epidemic, the rates of infections have increased tremendously and achieved 190 million infected patients and almost 4 million patients died worldwide by the end of July 2021. This dangerous virus has affected more than 200 countries all over the world [3]. With the fast and huge growing number of COVID-19 infections, several counties faced novel challenges that they haven't experienced before. For instance, the health organizations of such countries were about to collapse due to the excess influx of COVID-19 patients which requires more beds. Besides, these health infrastructures suffered from the lack of crucial medical equipment and the hazard of doctors and nurses infection which disable them from performing their duties. Therefore, there is a vital need for accurate and fast methods to diagnose COVID-19 cases to decrease the death rates and prevent the burden on the healthcare systems.
Nowadays, the current standard method declared by the WHO to diagnose COVID-19 is reverse-transcription polymerase chain reaction (RT-PCR) testing [4]. Even though these tests are the gold standard because of their acceptable sensitivity rates, they struggle with several drawbacks. They need a prolonged time to achieve a result, solid laboratory settings, qualified persons to perform the tests [5], and they might produce a negative result although the patients have confirmed COVID-19 [6]. Several other tests and approaches that are capable of accomplishing faster and more accurate results are yet under examination. One of these approaches utilized for the early diagnosis of COVID-19 is medical imaging involving X-ray and computed tomography (CT) imaging modalities. Chest X-ray and CT images inhibit significant information that can help in the diagnostic procedure of lung-related diseases. Many research articles have confirmed that there are variations found in the chest radiographic images acquired before appearing of COVID-19 symptoms. Moreover, several studies verified that X-ray and CT images can diagnose COVID-19 more accurately than the PCR tests [7,8]. However, these techniques require an expert radiologist to diagnose because of the similarity of COVID-19 patterns with other types of pneumonia. Moreover, manual inspection is time-consuming, and therefore automated diagnostic systems are mandatory to reduce the time made by health professionals and produce a more accurate diagnosis.
With the advent of artificial intelligence (AI) approaches including machine and deep learning, the analysis of medical images is easier and faster. These techniques are widely used to produce accurate results in many related medical problems such as the heart [9,10], brain [[11], [12],13], intestine [14], breast [15,16], eye disease [17]. DL methods are currently used extensively along with chest radiography images to facilitate the diagnosis process of COVID-19 and overcome the limitations of manual diagnosis. Numerous studies [[18], [19], [20], [21], [22], [23], [24]] employed deep learning with X-ray images to diagnose COVID-19 images. Other studies [[25], [26], [27], [28], [29], [30], [31]] utilized deep learning techniques to identify COVID-19 from CT images. Despite, the great success of utilizing deep learning methods along with radiographic images to diagnose COVID-19, these imaging modalities have several limitations involving portability or mobility, being expensive, huge radiation exposure, and the need for skilled technicians for the acquisition and analysis procedures [32]. Thus, novel approaches are required to facilitate the diagnosis of COVID-19 while the pandemic endures.
The novel coronavirus mainly affects the respiratory system, it also influences other human vital organs, particularly the cardiovascular system [33]. The literature has shown several forms of cardiovascular variations in patients with COVID-19 such as QRST anomalies, arrhythmias [34], ST-segment modifications [35], PR interval changes [36], and conduction conditions [37]. These changes could be noticed in the electrocardiogram (ECG) of COVID-19 cases. Those Cardiac alterations [38,39] have encouraged the examination of ECG as a diagnostic tool for COVID-19 diagnosis. Taking into consideration the great benefits of ECG usage such as portability, availability, ease of use, inexpensive, harmlessness, real-time examination, the automatic diagnosis of COVID-19 from ECG can be of substantial importance besides PCR tests and chest X-ray or CT scans. Therefore, this paper investigates the feasibility of using ECG data for diagnosing COVID-19 by proposing a pipeline called ECG-Bi-CoNet based on AI techniques. The traditional way to analyze ECG data using AI methods is to extract hand-crafted features and use them to train machine learning classifiers. However, these approaches regularly involve obtaining a sufficient trade-off concerning computation efficiency and accuracy and might be prone to error [40,41]. The most recent way to analyze ECG is deep learning which can automatically obtain significant features from data and avoid the limitations of the hand-crafted approaches [[42], [43], [44]]. Thus, in this study, the proposed pipeline is based on five deep learning techniques to analyze ECG data and determine the possibility of using ECG data as a diagnostic tool to diagnose COVID-19.
2. Related works
Several studies have utilized AI techniques to detect cardiac problems from ECG data. Some of these studies [[45], [46], [47]] employed 1-D ECG signals to classify the anomalies. Other research articles [[48], [49], [50], [51], [52]] transformed ECG signals to 2-D images using numerous methods such as wavelet transform and short-term frequency transform to detect abnormalities. Although the previous methods achieved significant success using public ECG signals-based datasets, it would be difficult to apply them in the real-world medical environment. This is because most of the current methods depend on time series ECG signals. Nevertheless, in real clinical scenarios, this is not constantly the common case, as the ECG data is acquired and stored as images in actual practice [53]. Unlike the digital ECG data which consists of several clean and fine-detached lead signals, the image-based ECG acquired in real practice is vague. Moreover, in an ECG image, there exists some overlap among waveforms from diverse leads and solid surrounding secondary axes that adds difficulty in accurately extracting useful features. Also, the huge decrease in the sampling rate from around hundreds of hertz in ECG digital signals to tens of hertz or less in ECG images leads to great loss in information which correspondingly influence the performance of AI techniques (either using the handcrafted or deep learning methods). One possible solution to solve this problem is to convert the image into a digital signal [54], however, this transformation has high computational load and the quality of the transformed signal is constrained [55]. This is because the noise produced as a result of this conversion clearly affects the performance of the machine learning classifiers and deep learning techniques. To diagnose the cardiac condition from ECG data, the variations among almost all sorts of cardiac disorders are often small, those tiny differences are the main elements for the abnormality detection. Even with the great learning capacities of the deep learning approaches, they cannot identify the discriminative portions accurately due to the noise generated in the transformed ECG signals.
All the previously mentioned challenges, hinder digital ECG signals from being utilized in practical real-world clinical environments which acquire ECG as images. For these reasons, some studies utilized ECG trace images to detect several cardiac abnormalities, however, the number of these studies is limited. Mohamed et al. [56] employed discrete wavelet transform (DWT) to extract meaningful features from ECG trace images using the “Haar” mother wavelet. The authors used artificial neural networks to discriminate between normal and abnormal ECG patterns reaching an accuracy of 99%. The authors of [57] used ECG trace images to classify four types of cardiac diseases. They employed the MobileNet v2-deep learning technique for the classification and reached an accuracy of 98%. Du et al. [58] presented an end-to-end fine-grained framework to detect multiple cardiac disorders from ECG trace image data. The framework found out the prospective distinguishing portions and adaptively combined them. Afterward, the authors utilized a recurrent neural network and achieved a detection sensitivity and precision of 83.59% and 90.42% respectively. Hoa et al. [59] introduced a framework to diagnose myocardial infractions from ECG trace images. The framework is composed of a multi-branch shallow artificial neural network that used 12-lead ECG. The authors obtained an accuracy of 94.73%. The authors of [60] utilized five types of handcrafted feature extraction methods along with five machine classifiers to diagnose two types of arrhythmia from ECG trace images. The highest accuracy of 96% was obtained using local binary patterns and artificial neural networks. On the other hand, Xie et al. [61] utilized ECG trace images to predict stroke using the DenseNet deep learning technique attaining an accuracy of 85.82%.
The success of the previously discussed methods has inspired an inspection of the potential of using ECG trace images along with AI techniques specifically deep learning methods for COVID-19 diagnosis. Thanks for the public dataset [62] that has recently been released by a research group offering a chance to accomplish the recommended goal. The dataset contains images of normal, cardiac disorders, and COVID-19 patients. To our knowledge, until now three research articles have employed this dataset to investigate the possibility of using ECG trace images to diagnose COVID-19. Anwar and Zakir [63] used this dataset to determine the effect of using several augmentation techniques on the COVID-19 classification performance of the EfficientNet deep learning technique. The authors concluded that the augmentation techniques can enhance the performance to some extent. However, the extensive use of the augmentation approaches can degrade the performance. The highest accuracy of 81.8% was attained. On the other hand, the authors of [64] compared the performance of six deep learning techniques to diagnose COVID-19 as binary and multiclass classification problems. Conversely, Ozdemir et al. [65] presented a new approach based on hexaxial feature and Gray-Level Co-Occurrence Matrix (GLCM) methods to obtain significant features and create hexaxial mapping pictures. These produced pictures were then supplied into a deep learning technique to detect COVID-19 as a binary classification problem achieving an accuracy of 96.2%.
The methods mentioned earlier have some drawbacks. The method proposed by Ref. [63] has relatively low performance and cannot be considered a reliable system. The framework proposed in Ref. [64] used a very small number of features for testing which could lead to bias. The method presented in Ref. [65] was only tested on a binary classification problem. Besides, the process of hexaxial feature mapping used in Ref. [65] is extremely sensitive to the ECG trace images resolution and affects the features extracted using the GLCM method. The techniques used in Refs. [63,64,66] employed individual deep learning networks to either perform the classification or feature extraction. However, the studies [[67], [68], [69]] verified that integrating deep features of multiple deep learning techniques is proficient in boosting classification performance. Also, ensemble classification can improve the accuracy of classification.
This paper investigates the potential of utilizing ECG trace images for COVID-19 diagnosis. It introduces an automated diagnostic tool termed ECG-BiCoNet relying on five deep learning methods. Initially, it extracts deep features from two different layers of each deep learning technique. Next, it fuses the features obtained from the higher layers of the five deep learning approaches using the discrete wavelet transform (DWT) to reduce their huge size. Afterward, it combines the features extracted from the lower layers of the five deep learning approaches and then merges them with the higher-layers features. Then, it applies a feature selection approach to reduce the dimension of features after fusion. Finally, it employs an ensemble learning algorithm to boost the classification performance. ECG-BiCoNet accomplishes two classification categories for the automatic diagnosis of COVID-19 from ECG trace images. The first category is binary classification, and its main target is to distinguish between COVID-19 and normal patients (with normal ECG). The second category is a multiclass classification problem that differentiates among normal, COVID-19, and other cardiac findings.
The crucial contributions of the pipeline are:
-
•
A new harmless, cost-effective, sensitive, and fast pipeline called ECG-BiCoNet is introduced as a substitution to the current diagnostic tools to help in the automatic detection of COVID-19.
-
•
The new approach depends on a 2-D ECG trace image to diagnose COVID-19 which is a novel method to perform diagnosis.
-
•
ECG-BiCoNet employs five deep networks of distinct architecture not only one structure.
-
•
Due to the variations in these architectures, ECG-BiCoNet obtains two levels of deep features from two different layers of each deep learning network.
-
•
Features extracted from former layers are of huge dimensions, therefore they are mined via DWT to reduce their dimension, and then merged in a concatenated way with features of latter layers.
-
•
Investigating which level of feature has a greater impact on the performance.
-
•
Exploring if integrating bi-levels of features improve performance especially after feature selection and ensemble classification
3. Material and methods
3.1. Deep learning algorithms
Deep learning algorithms are the latest branch of machine learning approaches. They have several structures involving recurrent neural networks, restricted Boltzmann machines, beliefs networks, auto-encoders, and finally convolutional neural networks (CNNs). Each one of them is employed for a specific type of data. Among these deep learning architectures, the CNNs are the widely used construction appropriate for images [70]. In health informatics, CNNs are the frequently utilized architecture particularly employed in classification or diagnosis processes from medical images [71]. Thus, in this study five CNNs of different structures are employed to implement the proposed diagnostic tool ECG-BiCoNet. These constructions include ResNet-50, Inception V3, Xception, InceptionResNet, and DenseNet-201.
3.2. ECG COVID-19 trace image dataset
This article utilized a new freely available dataset [62] involving ECG trace images of COVID-19 and other cardiac complications. To the best of our knowledge, this is the earliest and only dataset made public for the ECG of the novel coronavirus. The dataset has 1937 ECG images of different classes. These images were investigated by specialists who categorized them to 859 images of normal patients with no heart abnormalities, 250 images of patients with COVID-19, 548 images with abnormal heartbeats, 300 images of patients with a current or previous myocardial infarction (MI).
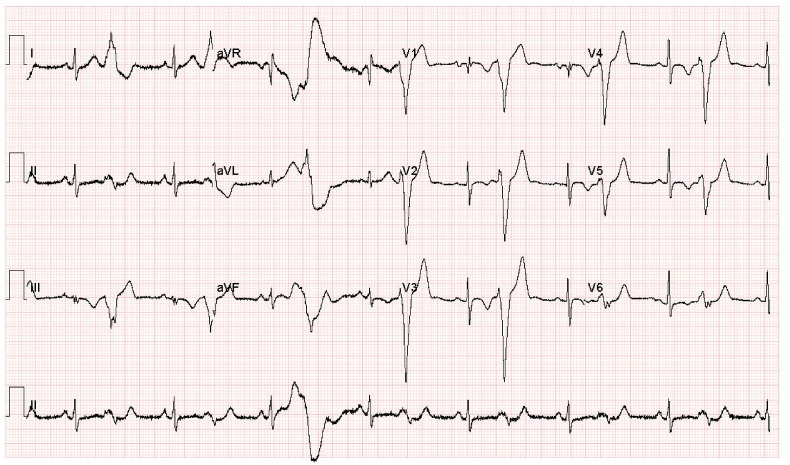
The dataset contains ECG waveforms from a 12-lead system gathered with a 500 Hz sampling frequency using EDAN SE-3 series 3-channel electrocardiograph. For the binary classification category (COVID-19 versus Normal ECG), 250 normal and 250 COVID-19 ECG images are employed. In the multi-class classification categories, 250 normal, 250 COVID-19, and 250 cardiac disorders ECG images are utilized. The reason for choosing this number is to balance the dataset and avoid the imbalance effect that may influence the classification performance. The patients' records that have been used are mentioned in the supplementary material. The images of cardiac disorders refer to cases with current or previous myocardial infarction (MI) as well as abnormal heartbeats for patients recovered from COVID-19 or MI and still have signs of shortness of breath or respiratory sickness. These images include Premature ventricular contractions (PVC) and T wave abnormalities. All cardiac findings regarding variations in ECG due to COVID found in the literature have been noticed on the whole ECG COVID-19 data of this study. A sample of an ECG trace image for a COVID-19 patient is displayed in Fig. 1 .
Fig. 1.
A sample of an ECG trace image for a COVID-19 patient.
3.3. Proposed ECG-BiCoNet
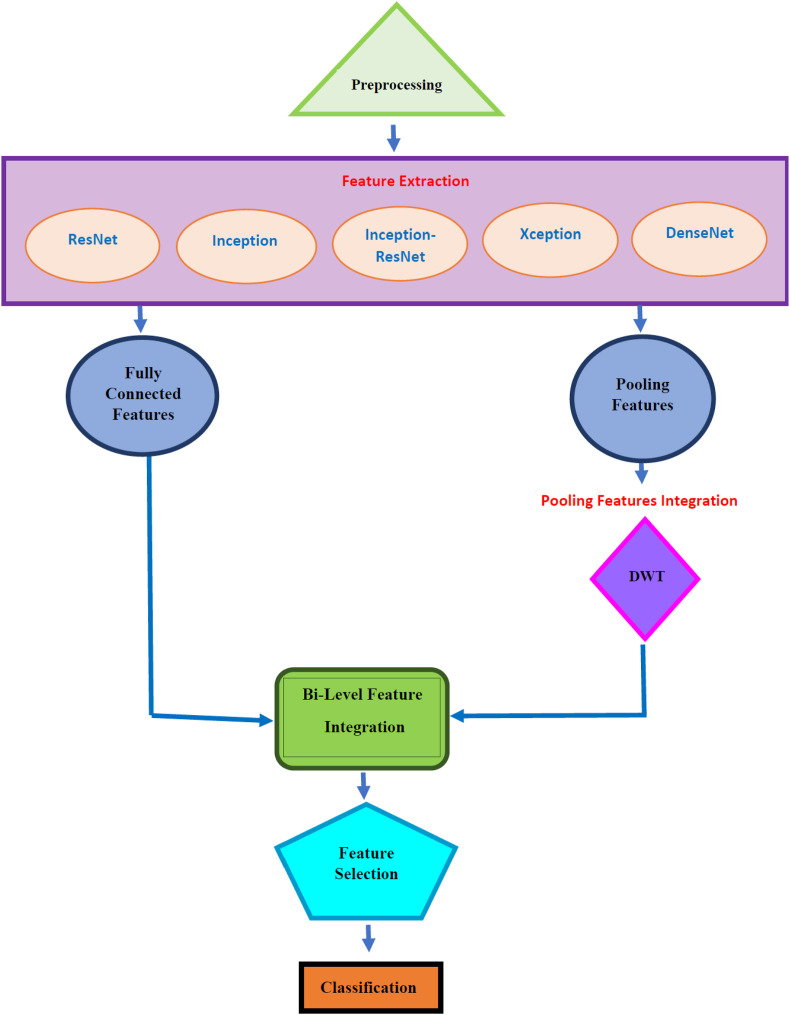
The main aim of this study is to investigate the potential of using ECG data for diagnosing COVID-19 using deep learning techniques. The paper proposes a pipeline namely ECG-BiCoNet to examine the possibility of achieving high diagnostic performance using ECG data and using it as a diagnostic tool. ECG-BiCoNet employs ECG trace images to allow it to be used in real clinical practice. It consists of four stages, preprocessing, feature extraction and integration, feature selection, and classification. In the preprocessing stage, the size of ECG trace images is altered, and the images are then augmented. Next, the five CNNs are trained, and then deep features are extracted from different layers of each CNN including pooling and fully connected layers. The pooling layer features are fused using DWT and subsequentially integrated with fully connected features in the feature extraction and integration stage. Afterward, the size of the integrated features is decreased in the feature selection stage. Finally, in the classification stage, two classifications schemes are constructed. In the first scheme, three individual classifiers are used to diagnose COVID-19 from ECG trace images in both binary and multiclass classification categories. The later scheme involves the construction of multiple classifier systems (MCS) based on voting ensemble classification. The block diagram of ECG-BiCoNet is shown in Fig. 2 .
Fig. 2.
The block diagram of ECG-BiCoNet.
3.3.1. Preprocessing
In this stage, the size of the ECG trace images is modified to be equal to the size of the input layers of each deep learning architecture as described in Table 1 . This table illustrates the sizes of the input layers of the five deep learning architectures and the name of the layers where features were obtained and the mined features and their sizes. Afterward, these images are augmented using several techniques to enlarge the number of images available in the dataset and overcome overfitting that might occur in the training process due to small datasets. These augmentation approaches incorporate flipping in x and y directions, translation (−30,30), scaling (0.9,1.1), and shearing (0,45) in x and y directions.
Table 1.
The sizes of the input layers of the five deep learning architectures and the name of the layers where features were obtained and the mined features sizes.
| CNN Architecture | Size of Input | Feature Extraction Layer Name | Feature Extraction Layer Description | Size of Features |
|---|---|---|---|---|
| ResNet-50 | 224x224x3 | “avg_pool” | Last average pooling layer | 2048 |
| “new_fc” | Last fully connected layer | Binary | ||
| 2 | ||||
| Multiclass | ||||
| 3 | ||||
| DenseNet-201 | 224x224x3 | “avg_pool” | Last average pooling layer | 1920 |
| “new_fc” | Last fully connected layer | Binary | ||
| 2 | ||||
| Multiclass | ||||
| 3 | ||||
| Inception-V3 | 229x229x3 | “avg_pool" | Last average pooling layer | 2048 |
| “new_fc” | Last fully connected layer | Binary | ||
| 2 | ||||
| Multiclass | ||||
| 3 | ||||
| Xception | 229x229x3 | “avg_pool" | Last average pooling layer | 2048 |
| “new_fc” | Last fully connected layer | Binary | ||
| 2 | ||||
| Multiclass | ||||
| 3 | ||||
| Inception-ResNet | 229x229x3 | “avg_pool” | Last average pooling layer | 1536 |
| “new_fc” | Last fully connected layer | Binary | ||
| 2 | ||||
| Multiclass | ||||
| 3 |
3.3.2. Feature extraction and integration
During the training procedure of the CNNs, a few problems might occur such as overfitting and convergence. These problems necessitate the modification of some of the CNN parameters to confirm that all of its layers are learned with similar rates. Transfer learning (TL) provides a solution to these problems by reusing CNNs that were formerly trained on large datasets for similar classification issues but with smaller datasets containing a few numbers of images [72] (such as the one used in this study). The reason for using pre-trained CNNs is that they have been trained on image attributes from a massive number of distinct images and thus boost the performance of classification when used to similar problems [73]. Thus, this stage involves the use of five pre-trained CNNs with TL including ResNet-50, Inception V3, Xception, InceptionResNet, and DenseNet-201 after modifying some parameters which will be explained later in the experimental setup section. TL is also utilized to adjust the output layer's size of the CNNs in order to be suitable to perform binary and multiclass classification categories of COVID-19 diagnosis. Afterward, the five CNNs are trained with the COVID-19 dataset. Next, deep features are extracted using TL from two different layers of each CNN including the last average pooling layer and the last fully connected layer. The CNN is composed of several layers. Each group of layers accomplishes a few sole roles on the input images fed to it. The first layer is the input layer that embraces the raw images. Then, the convolutional layers execute a convolution process among regions of the image and a filter bank and calculate the output volume. Afterward, are the pooling layers that reduce the size of the output of the previous convolutional layer to lower the computation load. Finally, are the fully-connected layers that receive inputs from the preceding layer and generate the class scores [74]. The CNN learns the elementary patterns of the input image in the initial layers and learns the significant features of the input image in the later deeper layers, leading to a more precise image classification [75,76]. Therefore, in this study, the deeper layers of the CNN including the last pooling and last fully connected layers were employed. The size of each deep feature set extracted from each CNN is discussed in Table 1.
Due to the deviations among the performance of various deep learning techniques, feature integration is employed to integrate all deep learning architecture's benefits. It can be noticed from Table 1 that the sizes of deep features obtained from the average pooling layers are large. Therefore, they are integrated using DWT which can be used to reduce the dimension of features. DWT is capable of decreasing the large dimension of data and explaining the time-frequency illustration from it [77]. It employs a cluster of orthogonal wavelet functions to explore data. It operates by entering the 1-D data to low and high pass filters and then downsampling the generated output to reduce its size. The resultant is two groups of coefficients called approximation and detail coefficients. Multilevel DWT decomposition can be performed by further passing the approximation coefficients through multiple low and high pass filters. The number of decomposition levels employed in this paper is 3. The Haar mother wavelet is used for the analysis. The frequency range of the third approximation level coefficients is 0–2.3 KHz. The approximation coefficients of the third DWT level produced from all deep features extracted from the average pooling layer of the five CNNs are integrated. Finally, these integrated reduced features are combined with the features extracted from the last fully connected layer of the five CNNs. The “Haar” wavelets were chosen in this study as they are efficient and are commonly used in several studies analyzing ECG data [56,[78], [79], [80], [81], [82]]. The number of decomposition levels is 3 as it showed excellent performance in various studies analyzing ECG data [[83], [84], [85], [86]]. The approximation coefficients are used as the authors of the study [87] concluded that the approximation coefficients extracted from medical signals have demonstrated superior performance compared to details coefficients.
3.3.3. Feature selection
To accomplish additional reduction to the dimension of features after the integration step, a feature selection approach is necessary. Feature selection (FS) is an essential process commonly employed in automated diagnostic tools to diminish the feature space and eliminate redundant and irrelevant variables [[88], [89], [90]]. They are also utilized to decrease the classification model complexity and avoid overfitting. Thus, a FS approach is used in this stage involving the symmetrical uncertainty (SU) method [91]. SU is a technique that computes the relevance among variables and the class value. It calculates the redundancy among features by measuring the entropy (En) of two features and the information gain between these two variables. SU can be defined using the following equations.
| (1) |
| (2) |
where En(X) and En(Y) are defined as the entropy of variables X and Y, and is the information gain of X once examining variable Y.
3.3.4. Classification
The classification stage is done either using three individual classifiers including LDA, SVM, and RF classifiers or by creating an MCS based on an ensemble voting algorithm. The classification process is accomplished using five scenarios for both the binary and multiclass classification categories. The first four scenarios employ individual classifiers to perform classification. The first scenario involves the fusion of the deep features extracted from the fully connected layers of the five CNNs. The second scenario presents the use of combined features obtained from the last average pooling layers of the five CNNs after passing through DWT. The third scenario includes the integration of the bi-level of features obtained from the previous two scenarios. The fourth scenario represents the classification after the FS approach. Finally, the last scenario compromises the classification after FS and using the MCS. For testing the performance of the classification models, 10-fold cross-validation is used. In 10-fold cross-validation, the dataset is split into 10 folds. For every iteration, 9 folds are used for training the model whereas the 10th fold is used for testing. The performance of the 10 testing folds of all iterations is averaged. For binary class, 500 images were used (where 250 images belong to the normal class and 250 images belong to the COVID-19 class). This means that in this case (25 normal + 25 COVID-19 images) are used for the testing process during each iteration of the 10-fold cross-validation process. For the multiclass classification task, 750 images are utilized including 250 normal, 250 COVID-19, and 250 cardiac disorders ECG images. This means that in this case (25 normal + 25 COVID-19 + 25 Cardiac disorders images) are used for the testing process during each iteration of the 10-fold cross-validation procedure.
4. Experimental setup
4.1. The setting of the CNN parameters
Some parameters of the five CNNs are altered involving the mini-batch size, validation frequency, the initial learning rate, and the number of epochs. For the multiclass and binary classification categories, the learning rate is 3 x 10 −4, the number of epochs is 30, and the mini-batch size is 4. The validation frequency is 87 for the binary class, while it is equal to 131 for the multi-class. Stochastic gradient descent with momentum procedure is implemented for the training process. The experiments of ECG-BiCoNet are accomplished using Matlab 2020 a and Weka Data Mining Tool [92].
4.2. Evaluation measures
The measures used to evaluate the capacity of the proposed ECG-BiCoNet are the F1-measure, accuracy, sensitivity, precision, and specificity calculated using equations (3), (4), (5), (6), (7).
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
Where; TN is the true negative corresponding to the non-COVID-19 images correctly classified. TP is the true positive equivalent to the COVID-19 images correctly diagnosed. FN is the false negative the number of the COVID-19 images incorrectly classified as non-COVID-19, and FP is the false positive representing the number of the non-COVID-19 images incorrectly classified as COVID-19.
5. Results
5.1. Scenario I, II, and III classification results
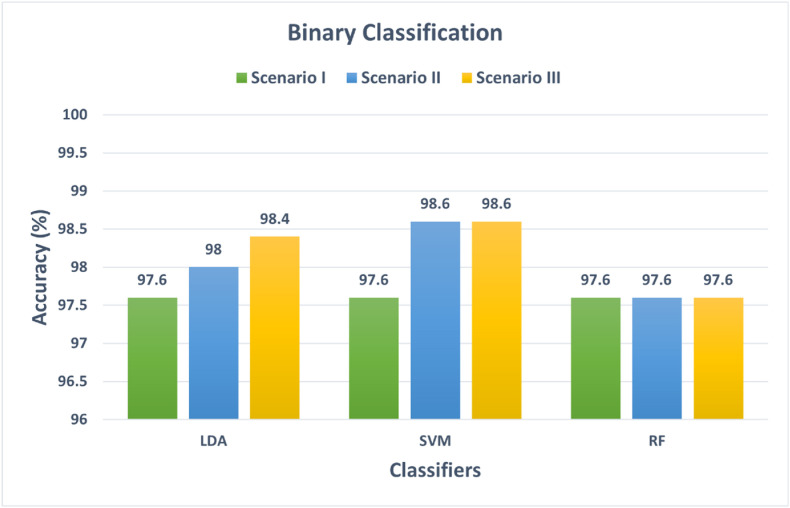
This section discusses the results of the first three classification scenarios. These three scenarios use three individual classifiers to perform the classification process. Scenario I corresponds to the use of the fused fully connected features extracted from the five CNNs. Scenario II represents the classification using the integrated pooling features of the five CNNs fused by DWT. Scenario III represents the classification using the combination of scenario I and II features. Fig. 3 shows the binary classification accuracies for scenarios I, II, and III. For scenario I, the accuracies achieved using the LDA, SVM, RF classifiers trained with the combined fully connected features are 97.6%, 97.6%, and 97.6%. Whereas for scenario II, the fused pooling features achieved higher accuracies of 98%, 98.6% for the LDA and SVM classifiers, and the same accuracy of 97.6% for the RF classifier. While for scenario III, the fusion of both pooling and fully connected features has increased the accuracy of the LDA classifier to 98.4%, but the same accuracy is attained for both the SVM and RF classifier. However, the improvement in accuracies in scenario III is clearer in the multiclass classification category shown in Fig. 4 . The performance metrics such as sensitivity, specificity, precision, and F1 score are shown in Table 2 .
Fig. 3.
Binary classification accuracy for scenarios I, II, and III.
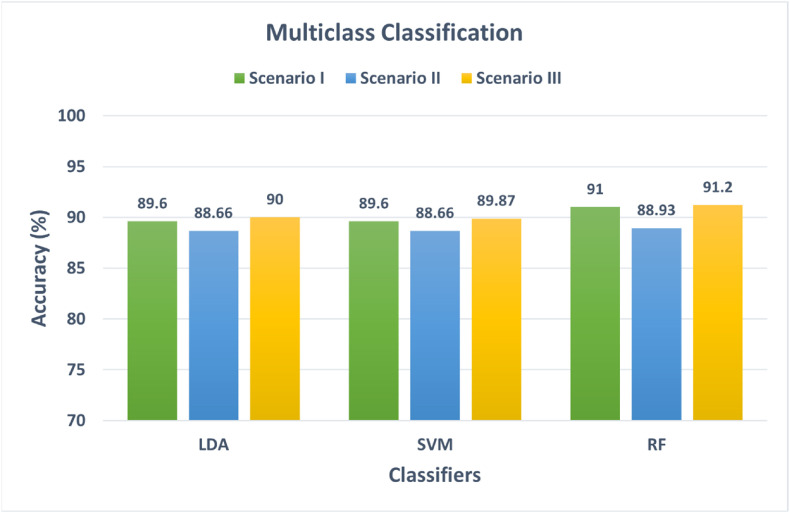
Fig. 4.
Multiclass classification accuracy for scenarios I, II, and III.
Table 2.
The performance metrics of the binary classification task achieved using the LDA, SVM, and RF classifier for scenarios I, II, and III.
| Models | Sensitivity | Specificity | Precision | F1 score |
|---|---|---|---|---|
| Scenario I | ||||
| LDA | 0.976 | 0.986 | 0.977 | 0.976 |
| SVM | 0.976 | 0.986 | 0.977 | 0.976 |
| RF | 0.976 | 0.986 | 0.977 | 0.976 |
| Scenario II | ||||
| LDA | 0.98 | 0.98 | 0.981 | 0.98 |
| SVM | 0.986 | 0.996 | 0.986 | 0.986 |
| RF | 0.976 | 0.986 | 0.977 | 0.976 |
| Scenario III | ||||
| LDA | 0.984 | 0.984 | 0.984 | 0.984 |
| SVM | 0.986 | 0.986 | 0.986 | 0.986 |
| RF | 0.976 | 0.977 | 0.976 | 0.976 |
In the case of the multiclass classification category, it is obvious from Fig. 4 that the combined fully connected features have higher accuracies than that of the integrated pooling features. This is because the accuracies attained in scenario II are 89.6%, 89.6%, and 91% which are greater than the 88.66%, 88.6%, and 88.93% of scenario I using the LDA, SVM, and RF classifiers respectively. The fusion of both fully connected and pooling features in scenario III has enhanced the accuracy compared to scenario I and II to reach 90%, 89.87%, and 91.2% for LDA, SVM, and RF classifiers respectively. This enhancement shows that combining bilevels of deep features can boost the performance of the classifiers. The performance metrics attained for the multiclass classification task are shown in Table 3 .
Table 3.
The performance metrics of the multiclass classification task achieved using the LDA, SVM, and RF classifier for scenarios I, II, and III.
| Models | Sensitivity | Specificity | Precision | F1 score |
|---|---|---|---|---|
| Scenario I | ||||
| LDA | 0.896 | 0.948 | 0.899 | 0.897 |
| SVM | 0.896 | 0.948 | 0.898 | 0.897 |
| RF | 0.911 | 0.955 | 0.913 | 0.911 |
| Scenario II | ||||
| LDA | 0.887 | 0.943 | 0.889 | 0.888 |
| SVM | 0.887 | 0.943 | 0.889 | 0.888 |
| RF | 0.889 | 0.945 | 0.892 | 0.890 |
| Scenario III | ||||
| LDA | 0.9 | 0.95 | 0.902 | 0.901 |
| SVM | 0.899 | 0.949 | 0.901 | 0.9 |
| RF | 0.903 | 0.951 | 0.905 | 0.904 |
5.2. Scenario IV classification results
This section shows the classification results of the SU feature selection procedure used to reduce the dimension of integrated features of scenario III (fully connected + pooling features). In the binary classification category, the results of the SVM classifier after the SU feature selection approach are shown in Table 4 . The SVM is chosen as it achieved the highest accuracy for the binary classification. Table 2 confirms that the SU feature selection method has successfully lowered the number of features to 204 while attaining a higher accuracy of 98.8% using the SU method compared to the 98.6% using the whole pooling and fully connected features (310 features of scenario I incase of binary classification category). Moreover, the sensitivity (0.988), specificity (0.988), precision (0.988), and F1-score (0.988) attained using the SU are higher than that achieved using the scenario III features which are all equal to 0.986.
Table 4.
The binary classification results that are obtained using the SVM classifier after the two feature selection approaches.
| Method | No of features | Accuracy (%) | Sensitivity | Specificity | Precision | F 1-Score |
|---|---|---|---|---|---|---|
| Before Feature Selection | ||||||
| Pooling + Fully Connected Features (scenario III) | 310 | 98.6 | 0.986 | 0.986 | 0.986 | 0.986 |
| After Feature Selection | ||||||
| SU | 204 | 98.8 | 0.988 | 0.988 | 0.988 | 0.988 |
For the multiclass classification category, the results obtained after the feature selection process are displayed in Table 5 . The results included in Table 5 prove that the SU feature selection method has successfully reduced the number of features to 223 while achieving an accuracy of 91.47%, a sensitivity of 0.915, specificity of 0.957, precision of 0.917, and F1-score of 0.915 which are higher than the accuracy (90%), sensitivity (0.9), specificity (0.95), precision (0.902), and F1 score (0.901) achieved using scenario III features before feature selection (315 features).
Table 5.
The results of the multiclass classification category that are achieved using the LDA classifier after the two feature selection approaches.
| Method | No of features | Accuracy (%) | Sensitivity | Specificity | Precision | F 1-Score |
|---|---|---|---|---|---|---|
| Before Feature Selection | ||||||
| Pooling + Fully Connected Features (scenario III) | 315 | 90 | 0.9 | 0.95 | 0.902 | 0.901 |
| After Feature Selection | ||||||
| SU | 223 | 91.467 | 0.915 | 0.957 | 0.917 | 0.915 |
5.3. Scenario V classification results
This section shows the classification results of the MCS which combines the predictions of LDA, SVM, and RF classifiers using the voting ensemble technique. These classifiers were trained with scenario IV features (integrated pooling and fully connected features after feature selection). The results of the MCS for the binary and multiclass classification categories are shown in Fig. 5 . It is clear from Fig. 5 that for the binary classification category, the MCS has improved the accuracy attained using the LDA (98%) and RF (97.6%) to reach 98.8% which is similar to that obtained using the SVM classifier. On the other hand, for the multiclass category, the MCS has enhanced the accuracy of the three classifiers to reach 91.73% which is higher than the 90.8% of the RF classifier, 90 SVM classifier, and 91.467% of the LDA classifier. These results indicate the MCS can enhance performance. The performance metrics attained for the binary and multiclass classification tasks using the LDA, SVM, and RF classifiers are shown in Table 6 . The classification accuracy for each class of the multiclass classification task is shown in Table 7 . Table 7 indicates that the classification accuracies of the cardiac abnormalities (89.2%) and normal classes (91.7%) achieved using ensemble classification are higher than that of individual classifiers. Furthermore, the accuracy of the COVID-19 (95.2%) attained using the ensemble is similar to that of the individual classifiers.
Fig. 5.
The MCS results for binary and multiclass classification categories.
Table 6.
The performance metrics of the binary and multiclass classification tasks achieved using the LDA, SVM, RF, and ensemble classifiers for scenarios V.
| Models | Sensitivity | Specificity | Precision | F1 score |
|---|---|---|---|---|
| Binary Class Task | ||||
| LDA | 0.98 | 0.98 | 0.981 | 0.98 |
| SVM | 0.988 | 0.988 | 0.988 | 0.988 |
| RF | 0.976 | 0.977 | 0.976 | 0.976 |
| Voting | 0.988 | 0.988 | 0.988 | 0.988 |
| Multiclass Task | ||||
| LDA | 0.915 | 0.957 | 0.917 | 0.915 |
| SVM | 0.9 | 0.95 | 0.903 | 0.901 |
| RF | 0.908 | 0.954 | 0.910 | 0.909 |
| Voting | 0.917 | 0.959 | 0.919 | 0.918 |
Table 7.
The accuracy of each class of the multiclass classification tasks achieved using the LDA, SVM, RF, and ensemble classifiers for scenarios V.
| Class | LDA | SVM | RF | Voting |
|---|---|---|---|---|
| Abnormalities | 88.8 | 88 | 87.6 | 89.2 |
| COVID-19 | 95.2 | 95.2 | 95.2 | 95.2 |
| Normal | 90.4 | 86.8 | 88.5 | 91.7 |
5.4. Comparison with related studies
The results of ECG-BiCoNet are compared with relevant studies and illustrated in Table 8 . The table proves that ECG-BiCoNet has a competing performance compared to other related studies for both the binary and multiclass classification categories. This is because, for the binary class category, ECG-BiCoNet has an accuracy of 98.8%, a sensitivity of 0.988, specificity of 0.988, and precision of 0.988 which are higher than that obtained by Ref. [64], and [65] methods. Besides, for the multiclass classification category, an accuracy of 91.73%, a sensitivity of 0.917, specificity of 0.959, and precision of 0.919 are obtained using ECG-BiCoNet which are greater than achieved by Ref. [64], and [63] methods. This outstanding performance verifies the capability of the proposed ECG-BiCoNet. It also shows the possible use of ECG trace images to diagnose COVID-19. It might be considered as a new diagnostic tool to be employed in real clinical practice which may help clinicians in the accurate diagnosis of COVID-19, and diminish the likelihood of misdiagnosis during manual diagnosis and its challenges.
Table 8.
A comparison between ECG-BiCoNEt and other relevant studies.
| Binary classification | |||||
|---|---|---|---|---|---|
| Reference | Method | Sensitivity (%) | Precision (%) | Specificity (%) | Accuracy (%) |
| [65] | hexaxial feature mapping + GLCM + CNN | 0.984 | 0.943 | 0.94 | 96.2 |
| [64] | ResNet-18 | 0.986 | 0.985 | 0.96 | 98.62 |
| Proposed ECG-BiCoNet |
Pooling features after DWT + Fully connected Features + SU + MCS |
0.988 |
0.988 |
0.988 |
98.80 |
| Multiclass Classification | |||||
| Sensitivity (%) |
Precision (%) |
Specificity (%) |
Accuracy (%) |
||
| [63] | EfficientNet | 0.758 | 0.808 | – | 81.8 |
| [64] | MobileNet | 0.908 | 0.913 | 0.928 | 90.79 |
| Proposed ECG-BiCoNet | Pooling features after DWT + Fully connected Features + SU + MCS | 0.917 | 0.919 | 0.959 | 91.73 |
6. Discussion
The literature has indicated numerous types of cardiovascular alteration in ECG data of patients with COVID-19 such as QRST anomalies, arrhythmias, and ST-segment modifications. On the other hand, some studies urged that the novel coronavirus may not be the main cause of these cardiovascular abnormalities, however, it must be highlighted that it could uncover the inherent conditions or degrade them [93]. All findings in the literature have been noticed on the whole ECG data of this study. The key aim of this study is to differentiate the ECG data of COVID-19 cases from numerous types of ECGs without COVID-19 (having other cardiac findings). Consequently, a new tool has been proposed based on 2D ECG trace images acquired from 12 leads.
This paper investigated the potential of using ECG data for COVID-19 diagnosis. To our knowledge, only a few papers explored the use of ECG data for diagnosing COVID-19. With the latest advancement of AI techniques especially deep learning, the analysis of medical images has become simpler and quicker. This study provided a new method based on deep learning techniques for diagnosing Covid-19 using ECG trace images. It proposed a pipeline called ECG-BiCoNet based on five pre-trained deep learning models of different architectures for feature extraction. It extracted features from two different deep layers including the last pooling and last fully connected layers. Features obtained from former layers are large compared to the later layers and therefore mined via DWT to decrease their dimension. The paper first explored if the few features mined from the later deeper layers (last fully connected layers) are enough to produce an accurate diagnosis of COVID-19 compared to the former features pulled out from the pooling layers. Then, it examined the impact of fusing features of these two layers on diagnostic accuracy. Due to this fusion, features dimension increase, and therefore their dimension was reduced using the SU feature selection method. Three machine learning classifiers including SVM, LDA, and RB were used for the classification process. Finally, the paper constructed an MCS to study the influence of ensemble classification using the voting method on diagnostic accuracy. Five classification scenarios were performed for binary and multiclass classification to search for the best scenario which enhances the performance of the proposed pipeline. The initial four scenarios utilized individual classifiers where the fifth used ensemble classifiers. Scenario I implied the combination of deep features obtained from the fully connected layers of the five CNNs. Scenario II introduced the fused deep features mined from the average pooling layers of the five CNNs after going through DWT. Scenario III involved the incorporation of the bi-level of features extracted from the earlier two scenarios. Scenario IV represents the classification after the FS approach. Finally, scenario V represented the classification after FS and the MCS.
A comparison was performed to compare the results of the five scenarios. Fig. 3, Fig. 4 compared the accuracies of the first three scenarios. It can be observed from Fig. 3 that for binary classification categories, scenario II features which have higher dimensions than features of scenario I have improved the accuracy by 0.4% for LDA and 1% for SVM compared to scenario I while maintaining the same accuracy using RF classifier. Moreover, scenario III improved the accuracy of LDA by 0.8% and SVM by 1% and achieved the same accuracy for the RF classifier compared to scenario I. These results showed that this improvement in accuracy but also has increased the computation of the training models. Therefore, there should be a tradeoff between accuracy and computation cost. On the other hand, it can be noticed from Fig. 4 that for multiclass, scenario II has slightly lower performance than scenario I, and the difference between the accuracy attained in scenarios I and II is 0.2–0.4% and scenario II and III is 1.21–2.23% for the three classifiers. This shows that scenario III has an improvement compared to scenario II and a slight enhancement compared to scenario I at the expense of computation load.
Table 2, Table 3 compared the results of scenarios III and IV (using the SU feature selection method). Scenario IV was performed to lower the computation of the training models by decreasing the dimension of the input features (scenario III features). Scenario IV achieved a slight improvement over the other two scenarios for both the binary and multiclass classification categories. For the binary class task, Table 2 proved that the dimension of features has been reduced with an extra slight improvement in accuracy of 0.2%. On the other hand, for the multiclass task, Table 3 showed that the FS has successfully lowered the size of scenario III features which correspondingly reduced the computation with an additional enhancement in the accuracy of 0.467%. Finally, Fig. 5 compared the accuracies of the ensemble classification (scenario V) with the individual classification of scenario IV (after feature selection). The figure indicated that the accuracy improved by 0.8–1.2% for LDA and RF classifiers but maintained the same best accuracy for the SVM classifier. For the multiclass task, the ensemble classification provided an improvement of only 0.3% compared to the LDA best classifier for this task, whereas for the SVM and RF classifiers, it has a better enhancement of 1.73% and 0.93%. The results of scenario V showed an increase in the computation overhead compared to the scenario I with a slight improvement in accuracy. It is worth mentioning that the few features of scenario I may be enough for the two tasks at hand compared to the higher computation obtained using scenario V and this greater computation can be considered the main limitation of this study.
All leads of ECG images of every patient were employed in constructing the classification models in order to involve the information of the whole ECG channels. This is because some cardiac abnormalities occur more clearly in some channels than others. No proof was demonstrated that the anomalies that occurred due to COVID-19 could be split on a channel base. For the binary classification task, the results of scenario III showed an accuracy of 98.6%, sensitivity of 98.6%, and specificity of 98.6% using the SVM classifier. These results show that the proposed pipeline correctly diagnosed 98.6% of the COVID-19 ECG images as COVID-19, and 98.6% of normal ECG images as normal. These results verify the robustness of the proposed tool and confirm the cardiac variations found in ECG images due to COVID-19 can be significantly distinguished from normal ECG images with no cardiac findings. On the other hand, for the multiclass task, scenario III achieved an accuracy of 91.2%, a sensitivity of 90.3%, a specificity of 95.1% which indicate that the proposed tool has successfully identified COVID-19 patients (as the specificity is 95.1%), but it has a lower ability in identifying normal and other cardiac abnormalities. In other words, the proposed model accurately diagnosed 95.1% of the COVID-19 ECG images as COVID-19 but misdiagnosed 9.7% of normal and ECG images other cardiac abnormalities.
These previous results were then enhanced in scenarios IV and V as the accuracy, sensitivity, and specificity reached 98.8%, 98.8%, and 98.8% for the binary classification task (scenario V), and the accuracy, sensitivity, and specificity reached 91.73%, 91.7%, and 95.9% for the multiclass classification task (scenario V). The results of the binary task confirm the great capacity of the proposed tool to discriminate among normal and COVID-19 ECG as the proposed pipeline misdiagnosed only 1.2% and 1.2% of the normal and COVID-19 ECG images. This proves that there are obvious differences between normal ECG and COVID-19 ECG which has cardiovascular variations. Furthermore, for the multiclass task, the accuracies achieved for each class of ECG are 95.2% for the COVID-19 ECG, 89.2% for the ECG with other cardiac findings, and 91.7% for the normal ECG. These accuracies show that the tool can accurately identify COVID-19 ECG compared to ECG with other abnormalities and normal cases. The proposed pipeline misdiagnosed 8.3% of the normal cases and 10.8% of the ECG images with other cardiac abnormalities. The similarity among several cardiac variations that occur in the ECG with other cardiac abnormalities (not COVID-19) degrades the proposed tool capacity to accurately recognize these cardiac abnormalities. The ability of the introduced pipeline to classify COVID-19 ECGs could be evidence of the occurrence of COVID-19-induced cardiovascular variations.
7. Conclusion
This paper examined the possibility of using ECG data to diagnose COVID-19. It proposed a novel pipeline called ECG-BiCoNet for the automatic diagnosis of COVID-19 from ECG trace image data based on an integration of deep learning techniques. The pipeline has two classification categories, binary and multiclass. The binary class is employed to differentiate between COVID-19 and normal patients, while the multiclass is to distinguish normal, COVID-19, and other cardiac findings. ECG-BiCoNet extracted two levels of deep features (including pooling and fully connected layers) from five CNNs. It then reduced the dimension of pooling features using the discrete wavelet transform and then merge it with fully connected features. Afterward, it investigated if integrating these features influences diagnostic accuracy. It then examined if feature selection can reduce the number of features while enhancing the performance of classification. Finally, it created a multiple classifier system (MCS) to study the possibility of a further enhancement in the accuracy. The results obtained indicated that feature integration had improved the accuracy. Furthermore, the feature selection approach had additional improvement in classification performance. Besides, further enhancement was achieved using the MCS, especially for the multiclass classification category. For the binary classification task, the results of scenario III showed an accuracy of 98.6%, sensitivity of 98.8%, (accuracy for normal class), and specificity of 98.8% (accuracy of COVID-19 class) using the SVM classifier which verifies the robustness of the proposed tool and confirms the cardiac variations found in ECG images due to COVID-19 can be significantly distinguished from normal ECG images with no cardiac findings. Furthermore, for the multiclass task, the accuracy achieved for the COVID-19 class is 95.2%, for other cardiac abnormalities is 89.2%, and for the normal class is 91.7%. This means that the proposed pipelined has correctly diagnosed 98.8% and 95.2% of the COVID-19 ECG images as COVID-19 for the binary and multiclass tasks respectively. It only misdiagnosed 1.2% of the COVID-19 images as normal for the binary class task and 4.8% of the COVID-19 ECG images as either normal or other cardiac abnormalities for the multiclass task. These results prove that the proposed pipeline is capable of accurately identifying COVID-19 ECG. The ability of the introduced pipeline to classify COVID-19 ECGs could be evidence of the occurrence of COVID-19-induced cardiovascular variations. These results verified that the proposed ECG-BiCoNet has attained promising performance for the automatic diagnosis of COVID-19 from ECG trace image data. Moreover, the ability of ECG-BiCoNet to distinguish between COVID-19 and other cardiac findings can prove that ECG data can be used to diagnose COVID-19. The proposed method can be employed to avoid the limitations of PCR tests and imaging modalities. It can be considered a simpler, harmless, quicker, cheaper, more mobile, and more sensitive method compared to PCR tests and imaging modalities. Therefore, it can help doctors in the automatic diagnosis of COVID-19 and avoid limitations of manual diagnosis. It can also lower the infection and the hospital load by avoiding excessive hospital visits. This study did not employ techniques that handle the class imbalance problem. Upcoming work will consider techniques that handle class imbalance problems such as Synthetic Minority Oversampling Technique (SMOTE). It is worth declaring that the few features extracted from the fully connected layers might be sufficient for the binary and multiclass tasks compared to the greater computation attained using the fused features of both fully connected and pooling layers which are then reduced and used to train the MCS. Although, these fused features enhanced the performance, however, it increased the computation. This larger computation is believed to be the key limitation of this study which will be addressed in future work. Moreover, future work may focus on testing the effectiveness of the proposed method in real clinical practice. Other deep learning approaches can be investigated.
Declaration of competing interest
The authors declare that they have no conflict of interests.
Acknowledgement
The author declares that she did not receive any source of funding.
Biography

Omneya Attallah received her B.Sc. and M.Sc. degrees in 2006 and 2009, respectively from the electronics and communications engineering department in the Arab Academy for Science, Technology, and Maritime Transport (AASTMT), Alexandria, Egypt. She received a full scholarship from the AASTMT for her B.Sc. and M.Sc. studies. She was the first in her class in the B.Sc degree. She received her Ph.D. degree in electrical end electronic engineering in 2016 from Aston University, Birmingham, UK. From 2008 to 2011 she was a teaching and research assistant with the department of electronics and communication engineering, AAST. From 2011 to 2015, she was a Ph.D. student at the school of engineering and applied sciences, Aston University, Birmingham, UK. From 2016 to 2020, she was working as an assistant professor teaching and researching at the electronics and communications engineering department in the AAST, Alexandria, Egypt. Since 2020, she has been an associate professor at the same department. She is a reviewer of IEEE access journal, Scientific Reports journals by Nature, and Journal of Ambient Intelligence and Humanized computing by springer and several other reputable journals in Springer, MDPI, and Wiley publishers. She is currently a mentor at Neuromatch Academy. Her current research interests include signal/image processing, biomedical engineering, biomedical informatics, neuroinformatics, pattern recognition, machine/deep learning, and data mining.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105210.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Fan Y., Zhao K., Shi Z.-L., et al. Bat coronaviruses in China. Viruses. 2019;11:210–223. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chauhan S. Comprehensive review of coronavirus disease 2019 (COVID-19) Biomed. J. 2020;43:334–340. doi: 10.1016/j.bj.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abhijit B., et al. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomed. Signal Process Control. 2022;71(Part B):103182. doi: 10.1016/j.bspc.2021.103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong D., Tang Z., Wang S., et al. The role of imaging in the detection and management of COVID-19: a review. IEEE Rev. Biomed. Eng. 2020;14:16–29. doi: 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- 5.Corman V.M., Landt O., Kaiser M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai T., Yang Z., Hou H., et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasawa T., Sato M., Yamaya T., et al. Ultra-high-resolution computed tomography can demonstrate alveolar collapse in novel coronavirus (COVID-19) pneumonia. Jpn. J. Radiol. 2020;38:394–398. doi: 10.1007/s11604-020-00956-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rousan L.A., Elobeid E., Karrar M., et al. Chest x-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020;20:1–9. doi: 10.1186/s12890-020-01286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attallah O., Ma X. Bayesian neural network approach for determining the risk of re-intervention after endovascular aortic aneurysm repair. Proc. IME H J. Eng. Med. 2014;228:857–866. doi: 10.1177/0954411914549980. [DOI] [PubMed] [Google Scholar]
- 10.Karthikesalingam A., Attallah O., Ma X., et al. An artificial neural network stratifies the risks of Reintervention and mortality after endovascular aneurysm repair; a retrospective observational study. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attallah O. MB-AI-His: histopathological diagnosis of pediatric medulloblastoma and its subtypes via AI. Diagnostics. 2021;11:359–384. doi: 10.3390/diagnostics11020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attallah O., Sharkas M.A., Gadelkarim H. Fetal brain abnormality classification from MRI images of different gestational age. Brain Sci. 2019;9:231–252. doi: 10.3390/brainsci9090231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attallah O. CoMB-deep: composite deep learning-based pipeline for classifying childhood medulloblastoma and its classes. Front. Neuroinf. 2021;15 doi: 10.3389/fninf.2021.663592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attallah O., Sharkas M. GASTRO-CADx: a three stages framework for diagnosing gastrointestinal diseases. PeerJ Comput. Sci. 2021;7:e423. doi: 10.7717/peerj-cs.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anwar F., Attallah O., Ghanem N., et al. 2019 International Conference on Advances in the Emerging Computing Technologies (AECT) IEEE; 2020. Automatic breast cancer classification from histopathological images; pp. 1–6. [Google Scholar]
- 16.Ragab D.A., Attallah O., Sharkas M., et al. A framework for breast cancer classification using multi-DCNNs. Comput. Biol. Med. 2021;131 doi: 10.1016/j.compbiomed.2021.104245. [DOI] [PubMed] [Google Scholar]
- 17.Attallah O. DIAROP: automated deep learning-based diagnostic tool for retinopathy of prematurity. Diagnostics. 2021;11:2034. doi: 10.3390/diagnostics11112034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan A.I., Shah J.L., Bhat M.M. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Progr. Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouchicha C., Ammor O., Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos, Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabik S., Gómez-Ríos A., Martín-Rodríguez J.L., et al. COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on Chest X-Ray images. IEEE J. Biomed. Health Inform. 2020;24:3595–3605. doi: 10.1109/JBHI.2020.3037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karhan Z., Fuat A. 2020 Medical Technologies Congress (TIPTEKNO) IEEE; 2020. Covid-19 classification using deep learning in chest X-ray images; pp. 1–4. [Google Scholar]
- 22.Yoo S.H., Geng H., Chiu T.L., et al. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front. Med. 2020;7:427. doi: 10.3389/fmed.2020.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammoudi K., Benhabiles H., Melkemi M., et al. Deep learning on chest x-ray images to detect and evaluate pneumonia cases at the era of covid-19. J. Med. Syst. 2021;45:1–10. doi: 10.1007/s10916-021-01745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altan A., Karasu S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos, Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y., Zheng S., Li L., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE ACM Trans. Comput. Biol. Bioinf. 2021;18:2775–2780. doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alshazly H., Linse C., Barth E., et al. Explainable covid-19 detection using chest ct scans and deep learning. Sensors. 2021;21:455. doi: 10.3390/s21020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Z., Xingming Z., Tao G., et al. Classification of COVID-19 by compressed chest CT image through deep learning on a large patients cohort. Interdiscipl. Sci. Comput. Life Sci. 2021;13:73–82. doi: 10.1007/s12539-020-00408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S., Kang B., Ma J., et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) Eur. Radiol. 2021;31:6096–6104. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attallah O., Ragab D.A., Sharkas M. MULTI-DEEP: a novel CAD system for coronavirus (COVID-19) diagnosis from CT images using multiple convolution neural networks. PeerJ. 2020;8 doi: 10.7717/peerj.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou T., Lu H., Yang Z., et al. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ragab D.A., Attallah O. FUSI-CAD: coronavirus (COVID-19) diagnosis based on the fusion of CNNs and handcrafted features. PeerJ Comput. Sci. 2020;6 doi: 10.7717/peerj-cs.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawlak A., Ręka G., Olszewska A., et al. Methods of assessing body composition and anthropometric measurements–a review of the literature. J. Education Health Sport. 2021;11:18–27. [Google Scholar]
- 33.Soumya R.S., Unni T.G., Raghu K.G. Impact of COVID-19 on the cardiovascular system: a review of available reports. Cardiovasc. Drugs Ther. 2021;35:411–425. doi: 10.1007/s10557-020-07073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denegri A., Pezzuto G., D'Arienzo M., et al. Clinical and electrocardiographic characteristics at admission of COVID-19/SARS-CoV2 pneumonia infection. Int. Emerg. Med. 2021;16:1451–1456. doi: 10.1007/s11739-020-02578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bangalore S., Sharma A., Slotwiner A., et al. ST-segment elevation in patients with Covid-19—a case series. N. Engl. J. Med. 2020;382:2478–2480. doi: 10.1056/NEJMc2009020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pavri B.B., Kloo J., Farzad D., et al. Behavior of the PR interval with increasing heart rate in patients with COVID-19. Heart Rhythm. 2020;17:1434–1438. doi: 10.1016/j.hrthm.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khawaja S.A., Mohan P., Jabbour R., et al. COVID-19 and its impact on the cardiovascular system. Open Heart. 2021;8 doi: 10.1136/openhrt-2020-001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azevedo R.B., Botelho B.G., de Hollanda J.V.G., et al. Covid-19 and the cardiovascular system: a comprehensive review. J. Hum. Hypertens. 2021;35:4–11. doi: 10.1038/s41371-020-0387-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Predabon B., Souza A.Z.M., Bertoldi G.H.S., et al. The electrocardiogram in the differential diagnosis of cardiologic conditions related to the covid-19 pandemic. J. Cardiac Arrhythmias. 2020;33:133–141. [Google Scholar]
- 40.Nanni L., Ghidoni S., Brahnam S. Handcrafted vs. non-handcrafted features for computer vision classification. Pattern Recogn. 2017;71:158–172. [Google Scholar]
- 41.Attallah O., Anwar F., Ghanem N.M., et al. Histo-CADx: duo cascaded fusion stages for breast cancer diagnosis from histopathological images. PeerJ Comput. Sci. 2021;7 doi: 10.7717/peerj-cs.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiranyaz S., Ince T., Gabbouj M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE (Inst. Electr. Electron. Eng.) Trans. Biomed. Eng. 2015;63:664–675. doi: 10.1109/TBME.2015.2468589. [DOI] [PubMed] [Google Scholar]
- 43.Rajpurkar P, Hannun AY, Haghpanahi M, et al. Cardiologist-level Arrhythmia Detection with Convolutional Neural Networks. arXiv preprint arXiv:170701836.
- 44.Attallah O., Sharkas M.A., Gadelkarim H. Deep learning techniques for automatic detection of embryonic neurodevelopmental disorders. Diagnostics. 2020;10:27–49. doi: 10.3390/diagnostics10010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sannino G., De Pietro G. A deep learning approach for ECG-based heartbeat classification for arrhythmia detection. Future Generat. Comput. Syst. 2018;86:446–455. [Google Scholar]
- 46.Mathews S.M., Kambhamettu C., Barner K.E. A novel application of deep learning for single-lead ECG classification. Comput. Biol. Med. 2018;99:53–62. doi: 10.1016/j.compbiomed.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Ullah A., Tu S., Mehmood R.M., et al. A hybrid deep CNN model for abnormal arrhythmia detection based on cardiac ECG signal. Sensors. 2021;21:951. doi: 10.3390/s21030951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naz M., Shah J.H., Khan M.A., et al. From ECG signals to images: a transformation based approach for deep learning. PeerJ Comput. Sci. 2021;7 doi: 10.7717/peerj-cs.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ullah A., Anwar S.M., Bilal M., et al. Classification of arrhythmia by using deep learning with 2-D ECG spectral image representation. Rem. Sens. 2020;12:1685. [Google Scholar]
- 50.Zhou H., Kan C. Tensor-based ECG anomaly detection toward cardiac monitoring in the internet of health things. Sensors. 2021;21:4173. doi: 10.3390/s21124173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozdemir M.A., Guren O., Cura O.K., et al. 2020 Medical Technologies Congress (TIPTEKNO) IEEE; 2020. Abnormal ecg beat detection based on convolutional neural networks; pp. 1–4. [Google Scholar]
- 52.Eltrass A.S., Tayel M.B., Ammar A.I. A new automated CNN deep learning approach for identification of ECG congestive heart failure and arrhythmia using constant-Q non-stationary Gabor transform. Biomed. Signal Process Control. 2021;65 [Google Scholar]
- 53.Badilini F., Erdem T., Zareba W., et al. ECGscan: a method for digitizing paper ECG printouts. J. Electrocardiol. 2003;36:39. doi: 10.1016/j.jelectrocard.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Mishra S., Khatwani G., Patil R., et al. ECG paper record digitization and diagnosis using deep learning. J. Med. Biol. Eng. 2021;41:422–432. doi: 10.1007/s40846-021-00632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Widman L.E., Freeman G.L. A-to-D conversion from paper records with a desktop scanner and a microcomputer. Comput. Biomed. Res. 1989;22:393–404. doi: 10.1016/0010-4809(89)90033-5. [DOI] [PubMed] [Google Scholar]
- 56.Mohamed B., Issam A., Mohamed A., et al. ECG image classification in real time based on the haar-like features and artificial neural networks. Procedia Comput. Sci. 2015;73:32–39. [Google Scholar]
- 57.Khan A.H., Hussain M., Malik M.K. Cardiac disorder classification by electrocardiogram sensing using deep neural network. Complexity 2021. 2021 [Google Scholar]
- 58.Du N., Cao Q., Yu L., et al. FM-ECG: a fine-grained multi-label framework for ECG image classification. Inf. Sci. 2021;549:164–177. [Google Scholar]
- 59.Hao P., Gao X., Li Z., et al. Multi-branch fusion network for Myocardial infarction screening from 12-lead ECG images. Comput. Methods Progr. Biomed. 2020;184 doi: 10.1016/j.cmpb.2019.105286. [DOI] [PubMed] [Google Scholar]
- 60.Ferreira M.A.A., Gurgel M.V., Marinho L.B., et al. 2019 International Joint Conference on Neural Networks (IJCNN) IEEE; 2019. Evaluation of heart disease diagnosis approach using ECG images; pp. 1–7. [Google Scholar]
- 61.Xie Y., Yang H., Yuan X., et al. Stroke prediction from electrocardiograms by deep neural network. Multimed. Tool. Appl. 2021;80:17291–17297. [Google Scholar]
- 62.Khan A.H., Hussain M., Malik M.K. ECG images dataset of cardiac and COVID-19 patients. Data Brief. 2021;34 doi: 10.1016/j.dib.2021.106762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anwar T., Zakir S. 2021 International Conference on Artificial Intelligence (ICAI) IEEE; 2021. Effect of image augmentation on ECG image classification using deep learning; pp. 182–186. [Google Scholar]
- 64.Rahman T, Akinbi A, Chowdhury ME, et al. COV-ECGNET: COVID-19 Detection Using ECG Trace Images with Deep Convolutional Neural Network. arXiv preprint arXiv:210600436. [DOI] [PMC free article] [PubMed]
- 65.Ozdemir M.A., Ozdemir G.D., Guren O. Classification of COVID-19 electrocardiograms by using hexaxial feature mapping and deep learning. BMC Med. Inf. Decis. Making. 2021;21:1–20. doi: 10.1186/s12911-021-01521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J. Big Data. 2019;6:1–48. doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amin S.U., Alsulaiman M., Muhammad G., et al. Deep Learning for EEG motor imagery classification based on multi-layer CNNs feature fusion. Future Generat. Comput. Syst. 2019;101:542–554. [Google Scholar]
- 68.Xu Q., Wang Z., Wang F., et al. Multi-feature fusion CNNs for Drosophila embryo of interest detection. Phys. Stat. Mech. Appl. 2019;531 [Google Scholar]
- 69.Zhang Q., Li H., Sun Z., et al. Deep feature fusion for Iris and periocular biometrics on mobile devices. IEEE Trans. Inf. Forensics Secur. 2018;13:2897–2912. [Google Scholar]
- 70.Hongtao L., Qinchuan Z. Applications of deep convolutional neural network in computer vision. J. Data Acquis. Process. 2016;31:1–17. [Google Scholar]
- 71.Ravì D., Wong C., Deligianni F., et al. Deep learning for health informatics. IEEE J. Biomed. Health Inform. 2016;21:4–21. doi: 10.1109/JBHI.2016.2636665. [DOI] [PubMed] [Google Scholar]
- 72.Bhuiyan M.N.Q., Shamsujjoha M., Ripon S.H., et al. Big Data Analytics for Intelligent Healthcare Management. Elsevier; 2019. Transfer learning and supervised classifier based prediction model for breast cancer; pp. 59–86. [Google Scholar]
- 73.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009;22:1345–1359. [Google Scholar]
- 74.Liu W., Wang Z., Liu X., et al. A survey of deep neural network architectures and their applications. Neurocomputing. 2017;234:11–26. [Google Scholar]
- 75.Jogin M., Madhulika M.S., Divya G.D., et al. 2018 3rd IEEE International Conference on Recent Trends in Electronics, Information & Communication Technology (RTEICT) IEEE; 2018. Feature extraction using convolution neural networks (CNN) and deep learning; pp. 2319–2323. [Google Scholar]
- 76.Rawat W., Wang Z. Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput. 2017;29:2352–2449. doi: 10.1162/NECO_a_00990. [DOI] [PubMed] [Google Scholar]
- 77.Sundararajan D. John Wiley & Sons; 2016. Discrete Wavelet Transform: a Signal Processing Approach. [Google Scholar]
- 78.Khairuddin A.M., Azir K.K. International Conference on Industrial, Engineering and Other Applications of Applied Intelligent Systems. Springer; 2021. Using the HAAR wavelet transform and K-nearest neighbour algorithm to improve ECG detection and classification of arrhythmia; pp. 310–322. [Google Scholar]
- 79.Li P., Liu M., Zhang X., et al. A low-complexity ECG processing algorithm based on the Haar wavelet transform for portable health-care devices. Sci. China Inf. Sci. 2014;57:1–14. [Google Scholar]
- 80.Vijendra V., Kulkarni M. 2016 International Conference on Emerging Trends in Engineering, Technology and Science (ICETETS) IEEE; 2016. ECG signal filtering using DWT haar wavelets coefficient techniques; pp. 1–6. [Google Scholar]
- 81.Gutierrez A., Hernandez P., Lara M., et al. vol. 25. IEEE; 1998. A QRS detection algorithm based on Haar wavelet; pp. 353–356. (Computers in Cardiology 1998). Cat. No. 98CH36292. [Google Scholar]
- 82.Delgado J.A., Altuve M., Homsi M.N. 2015 20th Symposium on Signal Processing, Images and Computer Vision (STSIVA) IEEE; 2015. Haar wavelet transform and principal component analysis for fetal QRS classification from abdominal maternal ECG recordings; pp. 1–6. [Google Scholar]
- 83.Long Z., Liu G., Dai X. 2010 International Conference on Biomedical Engineering and Computer Science. IEEE; 2010. Extracting emotional features from ECG by using wavelet transform; pp. 1–4. [Google Scholar]
- 84.De Chazal P., Celler B.G., Reilly R.B. Proceedings of the 22nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (Cat. No. 00CH37143) IEEE; 2000. Using wavelet coefficients for the classification of the electrocardiogram; pp. 64–67. [Google Scholar]
- 85.Arumugam M., Sangaiah A.K. Arrhythmia identification and classification using wavelet centered methodology in ECG signals. Concurrency Comput. Pract. Ex. 2020;32 [Google Scholar]
- 86.Castro B., Kogan D., Geva A.B. 21st IEEE Convention of the Electrical and Electronic Engineers in Israel. Proceedings (Cat. No. 00EX377) IEEE; 2000. ECG feature extraction using optimal mother wavelet; pp. 346–350. [Google Scholar]
- 87.Enamamu T., Otebolaku A., Marchang J., et al. Continuous m-Health data authentication using wavelet decomposition for feature extraction. Sensors. 2020;20:5690. doi: 10.3390/s20195690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li J., Liu H. Challenges of feature selection for big data analytics. IEEE Intell. Syst. 2017;32:9–15. [Google Scholar]
- 89.Attallah O., Karthikesalingam A., Holt P.J., et al. Feature selection through validation and un-censoring of endovascular repair survival data for predicting the risk of re-intervention. BMC Med. Inf. Decis. Making. 2017;17:115–133. doi: 10.1186/s12911-017-0508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Attallah O., Karthikesalingam A., Holt P.J., et al. Using multiple classifiers for predicting the risk of endovascular aortic aneurysm repair re-intervention through hybrid feature selection. Proc. IME H J. Eng. Med. 2017;231:1048–1063. doi: 10.1177/0954411917731592. [DOI] [PubMed] [Google Scholar]
- 91.Potharaju S.P., Sreedevi M., Amiripalli S.S. Cognitive Informatics and Soft Computing. Springer; 2019. An ensemble feature selection framework of sonar targets using symmetrical uncertainty and multi-layer perceptron (su-mlp) pp. 247–256. [Google Scholar]
- 92.Hall M., Frank E., Holmes G., et al. The WEKA data mining software: an update. ACM SIGKDD Explor. Newslett. 2009;11:10–18. [Google Scholar]
- 93.Haseeb S., Gul E.E., Çinier G., et al. Value of electrocardiography in coronavirus disease 2019 (COVID-19) J. Electrocardiol. 2020;62:39–45. doi: 10.1016/j.jelectrocard.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.