Abstract
We have reported that the papillomavirus E2 protein binds the nuclear factor AMF1 (also called G-protein pathway suppressor 2 or GPS2) and that their interaction is necessary for transcriptional activation by E2. It has also been shown that AMF1 can influence the activity of cellular transcription factors. These observations led us to test whether AMF1 regulates the functions of p53, a critical transcriptional activator that integrates stress signals and regulates cell cycle and programmed cell death. We report that AMF1 associates with p53 in vivo and in vitro and facilitates the p53 response by augmenting p53-dependent transcription. Overexpression of AMF1 in U2OS cells increases basal level p21WAF1/CIP1 expression and causes a G1 arrest. U2OS cells stably overexpressing AMF1 show increased apoptosis upon exposure to UV irradiation. These data demonstrate that AMF1 modulates p53 activities.
The p53 tumor suppressor is a common target for genetic alteration in human tumors and is one of the most frequently altered genes in cancer cells (38, 43). Under normal circumstances, the p53 protein remains in a latent state. Following DNA damage or other forms of cellular stress, the p53 protein rapidly accumulates and becomes activated through posttranslational mechanisms. Wild-type p53 limits cellular proliferation by inducing either a transient G1 arrest or apoptosis, depending on the cellular context (for a review, see reference 41). Differential regulation of p53 transactivation has been observed between cells undergoing growth arrest and apoptosis. Transactivation is required for the efficient execution of p53-mediated growth arrest, yet its role in apoptosis is equivocal (19, 70).
The growth arrest response mediated by p53 relies on its ability to act as a sequence-specific DNA-binding transcription factor (38, 73). A variety of downstream target genes that influence cell growth, including p21WAF1/CIP1, Bax, Mdm2, cyclin G, and Gadd45, have been identified (for a review, see references 38 and 43). The p53-mediated G1 arrest is believed to occur primarily through induction of p21WAF1/CIP1, a general inhibitor of cyclin-dependent kinases whose functions are required for cell cycle progression (18, 28, 75). In contrast, although increases of p21WAF1/CIP1 have been reported to be associated with apoptosis in some cases (51, 68), p21WAF1/CIP1 activity appears to be dispensable for apoptosis (2, 9). The human Mdm2 protein binds to p53 and inhibits its transactivation function by targeting p53 for rapid degradation (29, 40). Transcription of the Mdm2 gene itself is activated by p53, representing a feedback loop to control p53 activity (74).
The human p53 protein consists of 393 amino acids (aa) and contains four major functional domains for a review, see reference 49). At the N terminus is a transcriptional activation domain (aa 1 to 50) which recruits the basal transcriptional machinery, including the TATA box binding protein (TBP) and TBP-associated factors (47, 67). The central region of p53 is the sequence-specific DNA-binding domain (DBD) (aa 102 to 292). It is within this central domain that 80 to 90% of the tumor mutations are found (12). The central region also functions as protein binding domain interacting with simian virus 40 large T antigen (34) and the cellular proteins 53BP1 and 53BP2 (20, 33). The C-terminal portion contains an oligomerization domain (aa 323 to 356) and a regulatory domain (aa 360 to 393). It is well established that p53 forms tetramers via the oligomerization domain (39). Tetramerization appears to be required for efficient transactivation in vivo and for p53-mediated suppression of growth of carcinoma cell lines (55). The extreme C-terminal regulatory domain acts as a negative regulator of p53 sequence-specific DNA binding (1, 60). The activation of p53 in cultured mammalian cells has been correlated with phosphorylation (38), acetylation (59), glycosylation (61), and proteolytic removal (52) of the C-terminal domain. These modifications are thought to activate p53 by causing a conformational change of the protein, regulated by an allosteric effect.
Although it is widely recognized that transcriptional activation plays a pivotal role in mediating the p53 response, relatively limited information is available on the mechanisms through which p53 stimulates transcription. Several cellular proteins have been shown to play roles in p53 transactivation. The human TBP-associated factor TAFII31, a component of TFIID, binds to p53 via residues in the amino-terminal domain that are essential for transcription. Antibodies directed against TAFII31 inhibit p53-activated transcription in vitro (47). Members of the p300 (also known as CREB-binding protein) transcriptional coactivator family were shown to interact with p53 and stimulate its sequence-specific DNA binding (23). Interaction with p300 modulates p53-mediated activation of the p21WAF1/CIP1 and Bax promoters, as well as p53-induced cell cycle arrest and apoptosis (24, 44). Furthermore, p300-binding proteins, such as PCAF and JMY, were also shown to facilitate the p53 response by augmenting p53-dependent transcription and apoptosis (45, 59, 63).
We report here the properties of a novel cellular protein, AMF1 (also called G-protein pathway suppressor 2 or GPS2), and its stimulatory interaction with p53. AMF1 is a human nuclear protein of 327 aa with a predicted molecular size of 37 kDa. It was initially identified as one of two human suppressors of lethal G-protein subunit-activating mutations in the pheromone response pathway of the yeast Saccharomyces cerevisiae and was named GPS2 (64). Meanwhile, this protein was found to interact with the papillomavirus E2 protein activation domain and to be required for the stimulation of E2 transcriptional activity; therefore, it was named AMF1 (for activation-domain modulating factor 1) (8). The human papillomavirus 16 E2 protein was reported to interact with p53 and induce apoptosis (17, 48, 72). Recently, we demonstrated that AMF1 interacts with the transcriptional adapter p300, and both AMF1 and p300 additively increased E2 transactivation (54), suggesting that AMF1 could act as a transcription cofactor. GPS2 also interacts with the human T-cell leukemia virus type 1 (HTLV-1) Tax protein (14, 35) and influences the transcriptional activities of Tax and c-Jun (35, 64). In this study, we investigated the physical and functional relationship between AMF1 and p53.
MATERIALS AND METHODS
Cell lines, transfection, and cell treatments.
The osteosarcoma cell lines U2OS (ATCC HTB96) and Saos-2 (ATCC HTB85) were grown and maintained in Dulbecco's modified Eagle medium (GIBCO/BRL) supplemented with 10% fetal bovine serum and penicillin-streptomycin. For transient transfection, cells seeded onto 60-mm-diameter petri dishes or 24-well plates were transfected with the indicated plasmids at a confluence of about 70%, using the calcium phosphate method. To establish U2OS cell lines stably expressing six-histidine-tagged AMF1 (6H-AMF1) or six-histidine-tagged β-galactosidase (6H–β-Gal), pcDNA3.1/6H:AMF1 or pcDNA3.1/6H:LacZ (54) was transfected into U2OS cells, which were maintained in medium containing G418 (0.5 mg/ml; GIBCO/BRL) for 2 weeks. Representative clones were selected for high-level expression of 6H-AMF1 (these clones were named U2OS/AMF1 cells) or 6H–β-Gal (these clones were named U2OS/β-Gal cells) by Western blotting and used for studies described in Results.
Where indicated, etoposide (13) was added into culture medium (final concentration, 10 μM) at 70% cell confluence. At different time points, drug-containing medium was removed, cells were washed with phosphate-buffered saline (PBS), and harvested. For UV irradiation, medium was removed from the 70% confluent cell culture before exposing cells in a UV Stratalinker (model 1800; Stratagene) and added back after the irradiation. Cells were harvested at different time points after the treatments.
Transcriptional activation.
Transactivation assays with p53 were performed as described (8, 54). Wild-type human p53 was cloned in the pCMV-Neo-Bam plasmid (6), resulting in plasmid pC53SN. The PG13-luciferase reporter was constructed by subcloning the HindIII fragment from the PG-CAT reporter, which contains 13 repeats of a p53 consensus sequence (19, 37) into the pGL2 reporter (Promega). The pWAF1-luciferase reporter consists of a 2.4-kb WAF1 genomic reporter region in pBluescript KS(+) (19). The MDM2-luciferase reporter was constructed by subcloning the MDM2 p53 response element from Cosx1Cat (74) into pGL2. The IGF-BP3 Box B-luciferase reporter consists of the cytomegalovirus promoter and the Box B response element of IGF-BP3 in pUHC13-3 (58), and the BAX-luciferase reporter is the human Bax response element in pGL3 (Promega) (58). All transfections included pSV-β-gal, which was used to standardize p53 activation values for transfection efficiency. Vector plasmid pCG (66) was added to samples, when necessary, to bring the cytomegalovirus promoter-containing DNA to an equal amount. At 32 h after transfection, the luciferase activities in cell lysates were measured with the luciferase assay system (Promega) and presented as the increase in activation over reporter alone.
Protein expression and purification.
Expression of p53, E1, and AU1-AMF1 in Sf9 cells was described previously (54). Sequences encoding the N-terminal 103, 160, and 250 aa of AMF1 were derived from AMF1-expressing vector pDB327 (open reading frame of AMF1 cloned in vector pCG) (8) through PCR and inserted into pcDNA3 (Invitrogen) as BamHI-XbaI fragments, creating pcDNA-AMF1(1–103), pcDNA-AMF1(1–160), and pcDNA-AMF1(1–250). These constructs, along with vectors expressing 6H-AMF1 or hemagglutinin (HA)-AMF1 (8), were used for in vitro translation and cell transfection. The glutathione S-transferase (GST)-AMF1 fusion proteins were produced in Escherichia coli and purified as described elsewhere (8). Wild-type p53 and deletion mutants used in in vitro translation assays were cloned in SP65 (Promega). The p53 deletion mutants consist of the amino acids specified in their names. The six-histidine-tagged p53 (6H-p53) was synthesized in E. coli and purified as previously described (54).
Protein association assays.
Coprecipitation of p53 with 6H-AMF1 or 6H–β-Gal from modified U2OS cells (U2OS/AMF1 and U2OS/β-Gal) was carried out as described previously (54). Briefly, cells were lysed in a buffer containing 50 mM Tris (pH 8), 100 mM NaCl, 20 mM NaF, 10 mM KH2PO4, 1% Triton X-100, 0.1 mM dithiothreitol (DTT), 10% glycerol, and protease inhibitors phenylmethylsulfonyl fluoride (PMSF), leupeptin, and pepstatin A. The cell extracts were adjusted to 40 mM imidazole. Nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen) were added in the presence or absence of 10 mM EDTA. Bound proteins were eluted with cell lysis buffer plus 250 mM imidazole. Concentrated eluates were resolved by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) and subjected to Western blotting analysis using anti-p53 monoclonal antibody (MAb) DO-1 (Santa Cruz). Coprecipitation of 6H-AMF1 and endogenous AMF1 was performed by the same procedure except that the elutes were analyzed by Western blotting with anti-AMF1 serum.
The protocol for coimmunoprecipitation of p53 and AMF1 from Sf9 insect cell lysates was the same as before (54). When in vitro self-association of AMF1 or binding of mutants AMF1 to p53 was tested, AMF1 proteins were prepared by in vitro translation and labeled with [35S]methionine; different forms of AMF1 were mixed, or added into Sf9 cell lysate containing baculovirus-expressed p53. The mixture was diluted 1:1 in a buffer containing 50 mM Tris (pH 8.0), 100 mM KCl, 0.1 mM EDTA, 2 mM DTT, 0.2% NP-40, 0.1% nonfat milk, 2.5% glycerol, PMSF (100 μg/ml), leupeptin (0.5 μg/ml), and pepstatin A (1 μg/ml), before addition of antibody and protein A-Sepharose beads (Pharmacia). The reactions were incubated at 4°C for 3 h with shaking. Beads were washed three times in 1 ml of LSAB buffer (100 mM Tris [pH 8], 200 mM NaCl, 0.5% NP-40, 2 mM DTT, PMSF [100 μg/ml]). Proteins remaining on beads were resolved by SDS-PAGE on a 15% polyacrylamide gel and analyzed with a Bio-Rad GS-250 molecular imager.
The procedure for GST pull-down experiments was described (8). Briefly, GST-AMF1 fusion proteins immobilized on glutathione-Sepharose were normalized by SDS–15% PAGE followed by Coomassie blue staining. Binding reactions were assembled by mixing GST-AMF1 proteins with purified 6H-p53 or 10 μl of 35S-labeled protein, prepared by in vitro translation, in 250 μl of LSAB buffer plus 5 mM DTT. After incubation at 4°C for 3 h with shaking, the complexes were washed four times with LSAB and twice with 100 mM Tris (pH 8)–500 mM NaCl–1% NP-40. Bound 6H-p53 was analyzed by Western blotting, and radioactive bound proteins were analyzed as described above.
Yeast two-hybrid assay.
LexA-AMF1 fusion vectors were derived from YEplac181GLexA:AMF3X (8). These constructs along with YEplac112-VP16:AMF1, YEplac112-AMF1, or YEplac112-VP16 (8) were transformed into the yeast S. cerevisiae DBY1 (7) containing the LexA reporter pSH18-34 (26). Transformants were selected at 30°C on yeast minimal medium plates with glucose and replica plated on galactose-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) medium (57). Color formation for each construct was monitored after 24, 48, and 72 h of incubation at 30°C.
Reverse transcription (RT)-PCR analysis.
U2OS/AMF1 and U2OS/β-Gal cells were cultured in 60-mm-diameter dishes and treated with etoposide for 0, 4, 8, or 24 h. Total RNA was isolated using TRIZOL reagent (GIBCO/BRL), and 5 μg of total RNA was reverse transcribed with oligo(dT) by using the Thermoscript RT-PCR system for first-strand cDNA synthesis (GIBCO/BRL). Five percent of the cDNA product was subjected to PCR with two different pairs of primers: p21 oligonucleotides, 5′-CTCTCATGCTCCAGGTGGCTC-3′ and 5′-CCATAGCCTCTACTGCCACCATCT-3′; GAPDH oligonucleotides, 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′. PCR was carried out with the Expand high-fidelity PCR system (Roche Molecular Biochemicals) and run for 15 cycles; the products were loaded on a 1.5% agarose gel.
Flow cytometric analysis.
After treatment with etoposide or UV irradiation as described above, cells were harvested and processed (25). Briefly, at the indicated time intervals, total populations of cells, including floating and adherent, were collected. The cells were fixed in 95% cold ethanol and stored at −20°C for a minimum of 7 h. Subsequently, the fixed cells were washed with PBS, resuspended in PBS, treated with RNase A (20 μg/ml), and incubated at 37°C for 30 min. Propidium iodide (PI) (100 μg/ml) was added and incubated at room temperature, in the dark, for 10 min. Samples were then left on ice, and DNA content was analyzed in a cell sorter (FACSCalibur; Becton Dickinson) measuring PI fluorescence intensity. Cell cycle analysis was carried out using ModFit LT software (Verity Software House, Inc.). Analysis of cells for their sub-G1 DNA content was performed with CellQuest software (Becton-Dickinson).
RESULTS
Establishment of an AMF1-transformed cell line.
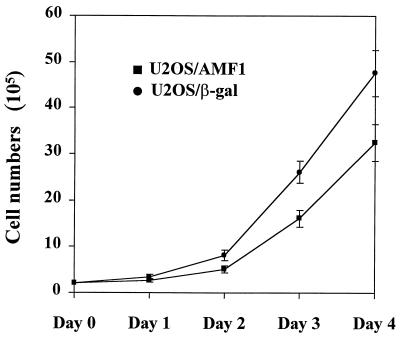
Previously we showed that AMF1 interacts with p300 and papillomavirus E2 proteins as a necessary cofactor of E2-dependent transcription (8, 54). To identify cellular proteins in complex with AMF1, we sought to establish cell lines that would express a high level of His-tagged protein. Human U2OS osteosarcoma cells were transfected with plasmid pcDNA3.1/6H:AMF1 or plasmid pcDNA3.1/6H:LacZ as a control. After G418 selection, two cell lines were established that stably express 6H-AMF1 or 6H–β-Gal, called U2OS/AMF1 or U2OS/β-Gal. Morphologically, U2OS/AMF1 cells at low density exhibited altered morphology and became more spindle shaped (data not shown). In contrast, the U2OS/β-Gal cells remain flat and are morphologically indistinguishable from parental U2OS cells. The growth rate of U2OS/AMF1 is ∼30% slower than that of U2OS/β-Gal (Fig. 1). These observations suggest that AMF1 may impact upon cell cycle control.
FIG. 1.
Stable overexpression of AMF1 decreases growth rate of U2OS cells. A total of 2 × 105 U2OS/AMF1 or U2OS/β-Gal cells were cultured in Dulbecco's modified Eagle medium (GIBCO/BRL) supplemented with 10% fetal bovine serum. Cells from three pairs of U2OS/AMF1 and U2OS/β-Gal dishes were counted at each time point of 24, 48, 72, and 96 h, and values were plotted. Error bars indicate the variation of cell numbers from three identical dishes. The procedures were repeated three times with similar results.
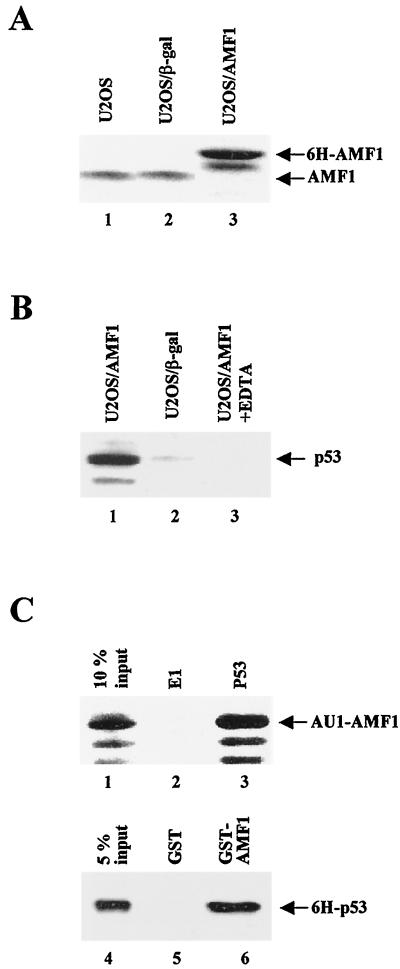
To compare the expression of AMF1 in different U2OS cell lines, we performed Western blotting using cell lysates from U2OS, U2OS/β-Gal, and U2OS/AMF1 lines. Shown in Fig. 2A is a quantitative analysis of AMF1 in these cells. In U2OS/AMF1 cells, only the 6H-AMF1 species was detected, with the endogenous AMF1 protein being below the detection level of the assay (Fig. 2A, lane 3), implying that expression of endogenous AMF1 is suppressed. Endogenous AMF1 was detected in the same amount of extract from the parental U2OS (lane 1) and in U2OS/β-Gal (lane 2) cells; however, these AMF1 levels are lower than those in U2OS/AMF1 cells (compare lanes 1 and 2 to lane 3). U2OS/AMF1 cells express approximately two- to threefold higher levels of AMF1 protein. RT-PCR analysis confirmed that U2OS/AMF1 cells contain higher levels of AMF1 mRNA (data not shown). Papillomavirus E2 activated transcription of an E2-dependent reporter efficiently in U2OS/AMF1 cells (data not shown), indicating that the 6H-AMF1 protein is able to carry out the function of endogenous AMF1.
FIG. 2.
In vivo and in vitro complex formation of AMF1 and p53. (A) AMF1 expression in human U2OS cell lines. Equal amounts of total cellular protein extracts from parental U2OS (lane 1), U2OS/β-Gal (lane2), and U2OS/AMF1 (lane 3) cells were subjected to Western blotting with polyclonal rabbit serum against AMF1. (B) U2OS/AMF1 or U2OS/β-Gal cells were treated with 10 μM etoposide for 8 h, extracts were prepared and incubated with Ni-NTA resin in the presence (lane 3) or absence (lanes 1 and 2) of 10 mM EDTA. After extensive washing, six-histidine-tagged proteins were eluted and concentrated. Copurification of p53 with 6H-AMF1 (lane 1) or 6H–β-Gal (lane 2) was probed by Western blotting with MAb DO-1. (C) In vitro binding of AMF1 to p53. (Upper panel) Sf9 cells were infected by recombinant baculoviruses expressing AU1-AMF1, p53, or bovine papillomavirus E1 proteins, harvested 40 h postinfection, and lysed as described previously (54). Cell extract with AU1-AMF1 was incubated with extract containing E1 (lane 2) or p53 (lane 3). P53 was immunoprecipitated with MAb pAb421 and protein A-conjugated Sepharose beads. After washing, the immunoprecipitates were analyzed by Western blotting with polyclonal antibody against AMF1. Input cell extract (10%) containing AU1-AMF1 was loaded in lane 1. (Lower panel) GST (lane 5) or GST-AMF1 fusion (lane 6) was incubated with 6H-p53 and washed extensively. Bound proteins were resolved by SDS-PAGE followed by Western blotting with MAb DO-1 against p53. Input 6H-p53 (5%) was loaded in lane 4.
AMF1 and p53 bind to each other in vivo and in vitro.
Given that p300 interacts with both AMF1 and p53 (5, 54), we asked whether AMF1 also interacts with p53. To obtain complexes of AMF1 and p53, the U2OS/AMF1 and U2OS/β-Gal cells were treated with etoposide, a topoisomerase inhibitor reported to cause crisis in cells and increase p53 levels (4, 42). The 6H-AMF1 and 6H–β-Gal in cell extracts were bound to Ni-NTA resin, washed extensively, and eluted with imidazole. Eluates were subjected to SDS-PAGE and blotting for p53. As shown in Fig. 2B, p53 copurified with the 6H-AMF1 protein from U2OS/AMF1 cell extract (lane 1), but not with the 6H–β-Gal protein from U2OS/β-Gal cell extract (lane 2). No p53 was detected when binding was performed in the presence of 10 mM EDTA (lane 3), which blocks binding of the six-histidine-tagged proteins to the Ni-NTA resin (31). These results demonstrated that AMF1 and p53 exist in a complex in vivo.
The association of AMF1 and p53 was also confirmed by in vitro binding assays. First, AMF1 and p53 were produced in Sf9 cells using recombinant baculoviruses. Cell lysates containing p53 or papillomavirus E1 (as a negative control) were combined with lysate containing AMF1. Immunoprecipitation with anti-p53 antibody pulled down AMF1 from the lysate with p53 (Fig. 2C, lane 3), but not from the lysate with BPV-1 E1 (Fig. 2C, lane 2), indicating that the AMF1-p53 interaction is specific. Second, in a GST pull-down assay, about 10% of the input 6H-p53 was bound to GST-AMF1 (Fig. 2C, lane 6), while none was retained by GST alone (Fig. 2C, lane 5), demonstrating that AMF1-p53 interaction is direct.
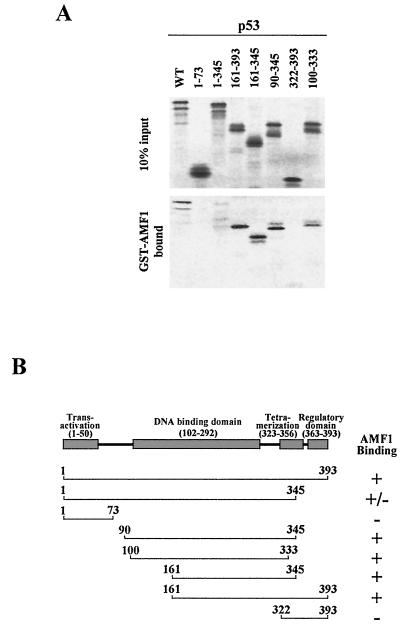
The region in p53 responsible for the interaction with AMF1 was identified using a panel of truncated p53 polypeptides made by in vitro translation. In this binding assay, wild-type and mutant p53 proteins were mixed with GST-AMF1. As shown in Fig. 3, 35S-labeled p53 (wild type) bound to AMF1, consistent with the results demonstrated in Fig. 2. p53 is composed of several subdomains that mediate distinct functions (for a review, see reference 49) (Fig. 3B). We suspected that the AMF1-binding site may be within the transactivation domain (aa 1 to 50), as is the AMF1-binding site in the papillomavirus E2 protein (8). However, no binding was detected with a peptide containing aa 1 to 73 of p53, suggesting that AMF1 does not interact with the transactivation domain. Similarly, the regulatory domain (aa 363 to 393) showed no binding to AMF1. On the other hand, fragments covering aa 90 to 345, aa 100 to 333, aa 161 to 345, and aa 161 to 393 strongly bound to AMF1, defining the AMF1-binding site on p53 from aa 161 to 333, where it overlaps the DNA binding domain (aa 102 to 292). Whether or not AMF1 interaction impacts DNA binding activity of p53 is unclear.
FIG. 3.
Mapping the AMF1 binding domain on p53. (A) Coprecipitation of wild-type and deletion mutant p53 with GST-AMF1 fusion protein. GST-AMF1 was expressed in E. coli and affinity purified with glutathione-Sepharose. Wild-type and mutant p53 were prepared by in vitro translation. (Upper panel) Input p53 proteins (10%); (lower panel) p53 proteins bound on GST-AMF1. (B) Schematic representation of wild-type p53 (49) and results for AMF1 binding assays. Numbers represent amino acid residues.
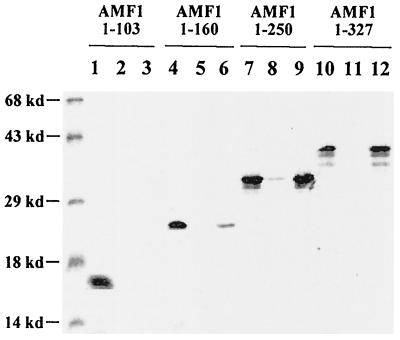
We also determined the region in AMF1 that is responsible for p53 binding. A group of C-terminal deletion mutants, as well as wild-type AMF1, were in vitro translated and mixed with Sf9 cell lysate containing p53. Immunoprecipitation with anti-p53 antibody efficiently pulled down wild-type AMF1 (lanes 10, 11, and 12 in Fig. 4) and a mutant containing aa 1 to 250 (lanes 7, 8, and 9). aa 1 to 160 weakly bound to p53 (lanes 4, 5, and 6). No binding was detected to AMF1(1–103) (Fig. 4, lanes 1, 2, and 3). We conclude from these binding experiments that the region on AMF1 responsible for binding to p53 is localized to its aa 103 to 250.
FIG. 4.
N terminus of AMF1 interacts with p53. Six-histidine-tagged wild-type and mutant AMF1 proteins were prepared by in vitro translation. These proteins (named by the number of amino acids contained) were incubated with Sf9 cell extract containing either papillomavirus E1 (lanes 2, 5, 8, and 11) or p53 (lanes 3, 6, 9, and 12), in the presence of MAb pAb421 against p53. Immunocomplexes were precipitated with protein A-Sepharose. After washing, AMF1 proteins remaining on the beads were run on an SDS–15% polyacrylamide gel together with 10% volume of each input (lanes 1, 4, 7, 10). The gel was analyzed with a Bio-Rad GS-250 molecular imager.
AMF1 stimulates p53-dependent transcription.
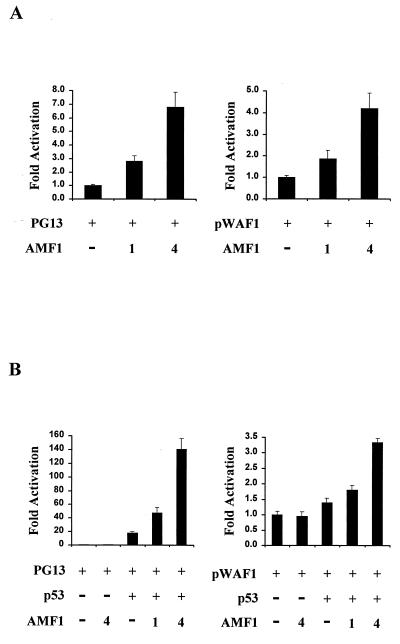
As p300 regulates p53 activity and since AMF1 and p300 interact (5, 24, 54), we examined whether AMF1 is involved in the p53 transcriptional activation. In a transient-transfection experiment, the synthetic p53-dependent luciferase reporter PG13, which contains multiple p53 binding sites (19, 37), was transfected into human U2OS cells with or without AMF1 expression vector. Transcription was efficiently induced by the endogenous p53 expressed in U2OS cells. As shown in the left panel of Fig. 5A, exogenous expression of AMF1 stimulated transactivation by p53. A titratable increase in p53-dependent transcription was apparent as the level of AMF1 was increased. Typically, cotransfection of 4 μg of vector DNA expressing AMF1 increased transactivation approximately sixfold.
FIG. 5.
AMF1 stimulates transcriptional activation by p53. (A) Transient transactivation in U2OS cells. A total of 500 ng of p53-dependent luciferase reporters PG13 or pWAF1-luciferase (19) was cotransfected into U2OS cells with 0, 1, or 4 μg of AMF1-expressing vector pDB327. At 32 h after transfection, luciferase activities were measured and are presented as the increase in activation over that of the reporter alone. Each sample was analyzed in triplicate, and standard deviations are shown (error bars). (B) Transient transactivation in Saos-2 cells. The same procedure as that for panel A was used, except that 50 ng of vector pC53SN was cotransfected to provide wild-type p53 activity in the cell.
It is established that p53 activates gene expression of p21WAF1/CIP1, Mdm2, IGF-BP3, and Bax under certain circumstances (38). To evaluate the effect of AMF1 on expression of these genes, we used luciferase reporters containing p53-responsive elements from gene p21WAF1/CIP1, Mdm2, IGF-BP3, or Bax. These reporter constructs were transfected into U2OS cells with or without AMF1, as performed with the PG13 reporter. Luciferase assays showed that AMF1 stimulated transcription from each reporter construct, although transactivation levels varied. With 4 μg of AMF1-expressing DNA, luciferase gene expression from the p21WAF1/CIP1 promoter was increased approximately fourfold (right panel of Fig. 5A), and expression from Mdm2, IGF-BP3, and Bax promoters increased approximately six-, approximately five-, and approximately twofold, respectively (data not shown).
We also tested the effect of AMF1 on exogenously expressed p53 by using p53-null Saos-2 cells. Low-level luciferase activities were detected from these cells when reporter alone was transfected (Fig. 5B). Cotransfection of AMF1 with the reporter constructs had little effect in Saos-2 cells (Fig. 5B). Cotransfection of 50 ng of p53-expressing vector with the PG13 reporter increased transactivation approximately 20-fold. Addition of 4 μg of AMF1-expressing vector further increased luciferase activity sevenfold (left panel of Fig. 5B), mimicking the results in U2OS cells. Transactivation from other reporters was not as efficient as that from PG13. For example, the pWAF1 luciferase reporter was stimulated by p53 plus AMF1 1.5- to 3-fold (right panel of Fig. 5B). The differences could be explained by the multiple p53 binding sites on reporter PG13. Taken together, these results show that AMF1 cooperates with both endogenously and exogenously expressed p53 and enhances p53-dependent transcription. These enhancements are not cell type specific.
Oligomerization of AMF1 is necessary for p53 transactivation.
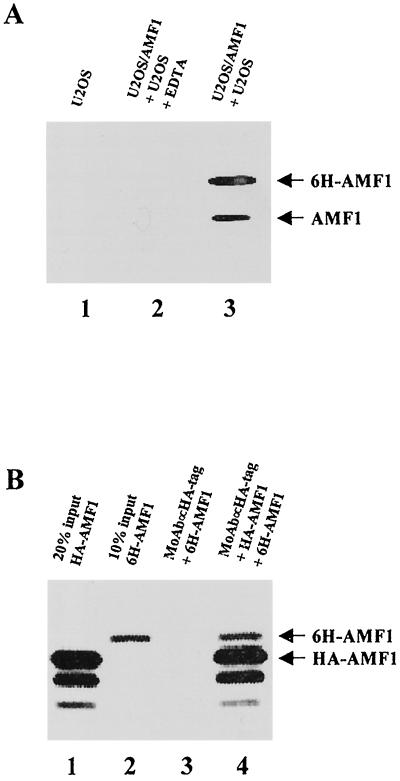
While performing binding experiments with a mixture of U2OS and U2OS/AMF1 cell extracts, we noticed that Ni-NTA beads bound two AMF1 species; one is 6H-AMF1, and the other is endogenous AMF1 (Fig. 6A, lane 3). We initially suspected that the endogenous AMF1 was nonspecifically pulled down by the Ni-NTA beads. Therefore, we tested U2OS cell extract alone for binding but found that no AMF1 was bound to the beads (Fig. 6A, lane 1), suggesting that AMF1 was precipitated via binding to 6H-AMF1. In addition, the presence of EDTA in the binding reactions with both U2OS and U2OS/AMF1 extracts totally removed both AMF1 bands from the precipitates (Fig. 6A, lane 2), further demonstrating that the AMF1–6H-AMF1 complex was bound to the Ni-NTA beads through the histidine motif on 6H-AMF1. These results suggest that AMF1 exists as oligomers in vivo.
FIG. 6.
Self-association of AMF1 in vivo and in vitro. (A) Coprecipitation of endogenous AMF1 with 6H-AMF1 from U2OS cells. Cell extracts of parental U2OS and U2OS/AMF1 were mixed and incubated with Ni-NTA resin in the presence (lane 2) or absence (lane 3) of 10 mM EDTA. Parental U2OS cell extract alone was also incubated with Ni-NTA resin without EDTA as a control (lane 1). After washing, proteins bound on the resin were eluted and concentrated. AMF1 proteins were detected by Western blotting. (B) In vitro association of HA-AMF1 with 6H-AMF1. Both proteins were prepared by in vitro translation and labeled with [35S]methionine. 6H-AMF1 was incubated with anti-HA MAb 12CA5 (Boehringer Mannheim) and protein A-Sepharose beads with (lane 4) or without (lane 3) HA-AMF1. Protein complexes were resolved on an SDS–15% polyacrylamide gel and analyzed with a Bio-Rad GS-250 molecular imager. Input HA-AMF1 (20%) (lane 1) and input 6H-AMF1 (10%) (lane 2) are shown.
To confirm that other cellular proteins do not mediate the AMF1 self-interaction, we performed in vitro binding assays. In these experiments, 6H-AMF1 and HA-AMF1 were prepared by in vitro translation. A mixture of both proteins was immunoprecipitated with anti-HA antibody. As expected, 6H-AMF1 was pulled down when HA-AMF1 was present (lane 4 of Fig. 6B) but not when it was absent (lane 3). Based on these observations, we postulate that oligomerization is an intrinsic property of AMF1 and probably represents the functional form of the protein.
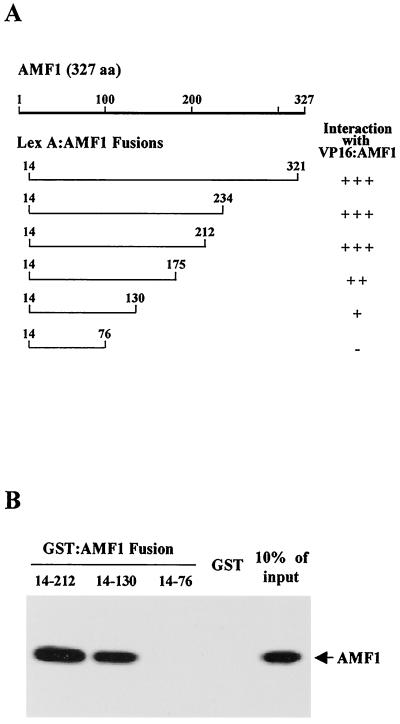
The oligomerization domain of AMF1 was also identified by using yeast two-hybrid and in vitro protein association assays. A series of truncated AMF1 mutants fused to the LexA DBD were examined for interaction with VP16-AMF1 by using a lexA operator-lacZ reporter (Fig. 7A). The hybrid LexA-AMF1(14–76) did not cooperate with VP16-AMF1 to activate the LexA-dependent reporter. Among the constructs positively cooperating with VP16-AMF1, interaction of LexA-AMF1(14–130) with VP16-AMF1 suggested that the N-terminal 130 aa of AMF1 is sufficient for mediating self-interaction; however, aa 130 to 212 may facilitate the oligomerization process (Fig. 7A). Furthermore, binding of full-length AMF1 to selected AMF1 truncations was confirmed by using GST fusions. GST-AMF1(14–212) and GST-AMF1(14–130) specifically retained in vitro-translated AMF1, but not by using GST-AMF1(14–76) and GST alone (Fig. 7B). These results are in agreement with the two-hybrid analyses.
FIG. 7.
Mapping the oligomerization domain on AMF1. (A) Interaction of VP16-AMF1 with LexA-AMF1 fusion proteins. Galactose-inducible expression vectors containing the LexA-AMF1 deletion fusions were transformed along with VP16-AMF1, AMF1, or the VP16 AD into DBY1 containing a lexA operator-lacZ reporter. Colonies were transferred to galactose-X-Gal plates and color formation was monitored. All constructs except LexA-AMF1(14–76) activate transcription of pSH18-34. A plus sign indicates earlier and more intense color formation in the presence of VP16-AMF1 than in the presence of the VP16 AD or AMF1. (B) In vitro binding of deletion mutant AMF1 and wild-type AMF1. Three of the AMF1 deletion mutants, AMF1(14–212), AMF1(14–130), and AMF1(14–76), were subcloned into vector pGEX2T (Pharmacia), and expressed in E. coli BL21::DE3(pLysS). Purified GST and GST-AMF1 fusion proteins were incubated with 35S-labeled in vitro-translated full-length AMF1. Bound AMF1 was resolved by SDS–15% PAGE and analyzed with a Bio-Rad GS-250 molecular imager. The right side lane shows 10% of input AMF1.
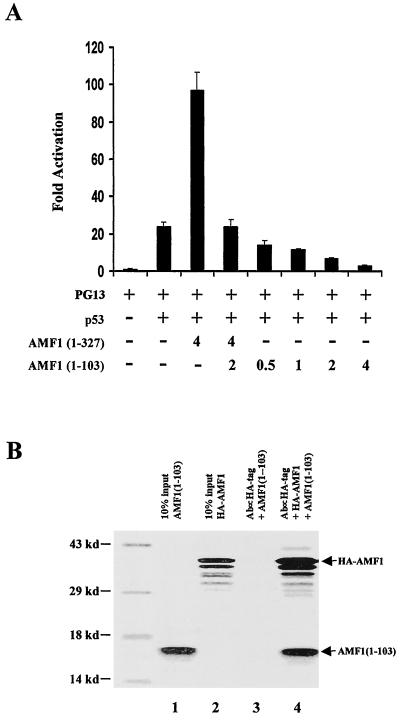
If oligomerization is requested for AMF1 activation of p53-dependent transcription, then an AMF1 mutant that binds the wild type but forms nonfunctional complexes might be valuable in addressing the significance of AMF1 in p53-dependent transcription. For this purpose, we used the AMF1 mutant containing the N-terminal 103 aa (Fig. 4). Binding of AMF1(1–103) to wild-type AMF1 was confirmed by in vitro binding assay (Fig. 8B). Anti-HA antibody could specifically precipitate AMF1(1–103) in the presence (lane 4) but not absence (lane 3) of HA-AMF1. Plasmids expressing wild-type or mutant AMF1 were cotransfected into Saos-2 cells with p53-expressing vector and a p53-dependent luciferase reporter, PG13. As shown in Fig. 8A, wild-type AMF1 increased luciferase expression about fourfold, consistent with previous results. Interestingly, cotransfection of 2 μg of plasmid DNA expressing AMF1(1–103) decreased the luciferase level to that without transfection of wild-type AMF1. Furthermore, AMF1(1–103) decreased p53 transactivation in a dose-dependent fashion (Fig. 8A), demonstrating that AMF1(1–103) can neutralize endogenous AMF1 in Saos-2 cells as well. These results indicate that AMF1 is required for maximal p53-dependent transcription. It is unlikely that AMF1(1–103) interferes with p53 directly, because no interaction was detected between these two proteins (Fig. 4). Binding of AMF1(1–103) to wild-type AMF1 and interfering with its functional interaction with p53 could cause the inhibitory effect of AMF1(1–103) in the transactivation assay. These experiments imply that oligomeric forms of wild-type AMF1 are necessary for stimulation of p53-dependent transcription.
FIG. 8.
Expression of AMF1(1–103) interferes with wild-type AMF1 function in vivo. (A) AMF1(1–103) inhibits p53 transcriptional activation in Saos-2 cells. A total of 500 ng of p53-dependent luciferase reporter PG13 was cotransfected into Saos-2 cells with or without vectors expressing p53, AMF1(1–327) (wild type), and AMF1(1–103) as indicated. A total of 100 ng of pC53SN plasmid was used. Total DNA for each transfection was made up to 6.6 μg by using vector plasmid pCG. Luciferase activities were measured at 32 h after transfection and are presented as the increase in activation over reporter alone. Each sample was analyzed in triplicate, and standard deviations are shown (error bars). (B) In vitro binding of AMF1(1–103) to HA-AMF1. Both proteins were prepared by in vitro translation. Experimental procedures were the same as in Fig. 5B. Each input (10%) is shown in the first two lanes. Lanes 3 and 4 demonstrate that AMF1(1–103) can be coimmunoprecipitated by HA-AMF1 but not the anti-HA MAb.
Overexpression of AMF1 increases basal level expression of p21WAF1/CIP1 and promotes G1 arrest.
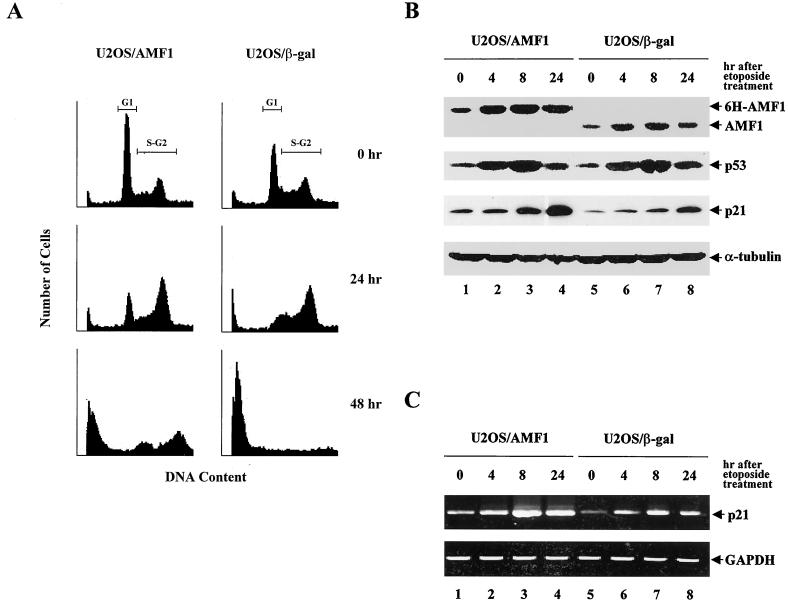
U2OS/AMF1 cells express higher levels of AMF1 than parental U2OS and U2OS/β-Gal cells (Fig. 2A). The U2OS/AMF1 cells also display distinct morphology (data not shown) and growth rate (Fig. 1). S. cerevisiae cells overexpressing GPS2 and STE4 are greatly enlarged and elongated, indicating a direct or indirect effect of GPS2 (AMF1) on G1 cyclin control (64). To examine the role of AMF1 in mammalian cell cycle control, we analyzed the U2OS/AMF1 cells and U2OS/β-Gal cells, as a control, by flow cytometry. Cultured cells were stained with PI (25) and sorted by measuring PI fluorescence intensity. We found that in comparison with U2OS/β-Gal cells, a greater proportion of U2OS/AMF1 cells was in G1 (Fig. 9A, top panel), suggesting that overexpression of AMF1 caused a cell cycle arrest. This may explain the lower growth rate of U2OS/AMF1 cells.
FIG. 9.
Overexpression of AMF1 affects cellular responses under etoposide treatment. (A) Overexpression of AMF1 in U2OS holds more cells in G1 phase and inhibits etoposide-induced S-G2 arrest and apoptosis. U2OS/AMF1 or U2OS/β-Gal cells were treated with 10 μM etoposide for 0, 24, and 48 h. Cells were harvested, processed, and subjected to flow cytometric analysis. Cell cycle stages are represented by the cellular DNA content, which was analyzed by PI staining and fluorescence-activated cell sorting. Boundaries for G1 and S-G2 phases are labeled on the top pair of graphs. (B) Western blotting analysis of AMF1, p53, and p21WAF1/CIP1 in U2OS cells at 0, 4, 8, and 24 h after addition of 10 μM etoposide into culture medium. Protein concentrations in cell extracts were determined, and equal amounts were loaded in each lane as judged by the level of α-tubulin. Intensity of bands at 0 h shows the baseline level of each protein. (C) RT-PCR analysis of p21WAF1/CIP1 transcripts in U2OS cells at 0, 4, 8, and 24 h after addition of 10 μM etoposide into culture medium. The 370-bp product was amplified using p21 oligonucleotides. As a control, the cDNA encoding GAPDH was amplified with specific oligonucleotides.
It is intuitive to propose that the G1 arrest in U2OS/AMF1 cells is caused by AMF1-enhanced activity of p53. The p53-mediated G1 arrest is believed to occur primarily through induction of p21WAF1/CIP1 (18, 28, 75). We therefore measured the basal level expression of p21WAF1/CIP1 and p53 in both U2OS/AMF1 and U2OS/β-Gal cells by Western blotting. As shown in Fig. 9B, without treatment, U2OS/AMF1 cells express higher level 6H-AMF1 than U2OS/β-Gal cells (0 h, AMF1 panel), consistent with the results reported in Fig. 2. Using equal amounts of cellular protein (as visualized by the level of α-tubulin), about twofold more p21WAF1/CIP1 protein was detected in U2OS/AMF1 cells (panels p21WAF1/CIP1 in Fig. 9B, comparing 0 h in both U2OS/AMF1 and U2OS/β-Gal cells). Experimentally, p21WAF1/CIP1 could be easily detected in U2OS/AMF1 cells, while it was sometimes undetectable under the same conditions in U2OS/β-Gal cells. RT-PCR analysis also confirmed that the level of p21WAF1/CIP1 transcript is twofold higher in U2OS/AMF1 cells than that in U2OS/β-Gal cells (Fig. 9C). Importantly, expression of p53 appeared to be the same in both cells (panel p53 in Fig. 9B, comparing protein levels at 0 h). These results imply that overexpression of AMF1 in U2OS cells increases the endogenous level of p53-responsive gene, such as p21WAF1/CIP1, but does not affect p53 protein levels.
Following exposure to DNA-damaging agents, intracellular p53 levels rise dramatically and cells undergo either cell cycle arrest, predominantly in G1 (36), or apoptosis (15, 46). Obviously, the G1 arrest in U2OS/AMF1 cells differs in that p53 expression is not changed. It was of interest to examine the responses from U2OS/AMF1 and U2OS/β-Gal cells after treatment with a DNA-damaging agent. For this reason, both cell types were incubated with etoposide, a chemotherapeutic agent that complexes with topoisomerase II and enhances cleavage of DNA (10, 27). Previous studies demonstrated that etoposide blocks cell cycle in S and G2 (13, 32). After 24 h of etoposide treatment, the U2OS/β-Gal cells were primarily in S and G2, consistent with other reports (13, 32), whereas a significant proportion of the U2OS/AMF1 cells remained in G1 (Fig. 9A, middle panel). With 10 μM etoposide treatment for 48 h, the sub-G1 population of U2OS/AMF1 cells was approximately 30% less than that of U2OS/β-Gal cells (Fig. 9A, bottom panel), suggesting that AMF1 delays apoptosis or cell death. This is not surprising, because previous reports showed that increasing expression of p21WAF1/CIP1 induces cell cycle arrest instead of apoptosis (9, 16, 70).
We monitored AMF1, p53, and p21WAF1/CIP1 protein levels by Western blotting. Cell lysates were prepared from both U2OS/AMF1 and U2OS/β-Gal cells at 0, 4, 8, or 24 h after etoposide treatment. Baseline protein levels are represented at 0 h. To our surprise, AMF1 expression increased and peaked at 4 to 8 h after etoposide treatment. The mechanism for this change is unknown. Similar to other reports (4, 42), p53 levels increased dramatically following etoposide treatment. In our experiments, p53 levels were highest after 4 to 8 h of treatment (p53 panel in Fig. 9B). Correspondingly, p21WAF1/CIP1 protein expression increased to a maximum at 24 h (p21 panel in Fig. 9B), similar to the observations reported for X-ray treatment (2), while the mRNA level of p21WAF1/CIP1 reaches a peak at 8 h (p21 panel in Fig. 9C). Both protein and mRNA levels of p21WAF1/CIP1 are higher in U2OS/AMF1 cells than in U2OS/β-Gal cells along the course of etoposide treatment. It is worthwhile to mention that although ectopic expression of AMF1 was able to arrest U2OS cells in G1, ultimately cells underwent apoptosis (Fig. 9A, 48 h), suggesting that AMF1 delays but does not block apoptosis.
AMF1 sensitizes U2OS cells to UV irradiation-induced apoptosis.
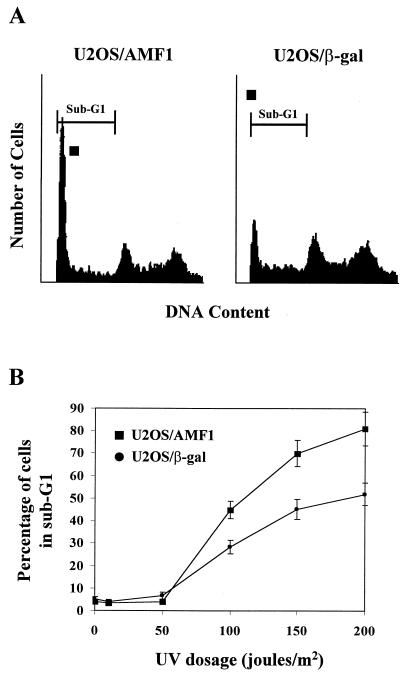
Given that a major biological function of p53 is to induce apoptosis when cells are genetically compromised, we set out to determine the physiological consequence of AMF1 overexpression in an apoptotic reaction. Both U2OS/AMF1 and U2OS/β-Gal cells were exposed to various doses of UV irradiation, previously reported to induce p53-dependent apoptosis in U2OS cells (2). Cells were incubated for 48 h after exposure, followed by flow cytometry analysis. As shown in Fig. 10A, exposure to UV irradiation at 100 J/m2 resulted in substantial apoptosis as detected by the presence of cells in the phase of sub-G1 (most U2OS cells were in G1 and S-G2 phases without treatment [Fig. 9A, 0 h]). Surprisingly, however, significantly more U2OS/AMF1 cells (average of 43%) underwent apoptosis compared to control U2OS/β-Gal cells (average of 28%) at 48 h (Fig. 10A). This suggests that overexpression of AMF1 in UV-irradiated cells enhances p53-dependent apoptosis. The cytotoxic effect of UV irradiation was dose dependent (Fig. 10B), as reported (2). Increased levels of apoptosis were observed with treatment with UV irradiation at 50 to 200 J/m2. Interestingly, the sensitivity of U2OS/AMF1 to apoptosis was also increased at higher UV dosages (Fig. 10B). Thus, in a similar fashion to the effects on transcription and cell growth arrest, in some cases AMF1 can augment the p53 activity involved in apoptosis.
FIG. 10.
Overexpression of AMF1 sensitizes U2OS cells to UV irradiation-induced apoptosis. (A) U2OS/AMF1 and U2OS/β-Gal cells were exposed to UV irradiation (100 J/m2) at 70% confluence. Cells were harvested after 48 h of incubation, processed, and subjected to flow cytometric analysis. Boundaries for sub-G1, indicative of apoptosis, are marked on top of each graph. (B) Series of UV dosages were tested under the same conditions as in panel A. The percentage of cells with a sub-G1 DNA content is graphed. Data were derived from three separate experiments. Error bars, standard errors.
DISCUSSION
p53 is a transcriptional activator with a sequence-specific DNA binding domain. In response to genotoxic stress, p53 induces cell cycle arrest or apoptosis, presumably to prevent the genome from accumulating mutations (9, 16, 30, 56, 71). A number of p53-responsive genes have been identified (for a review, see references 38 and 43). p53 activity is controlled through several mechanisms (for a review, see references 49, 50, and 69). Cellular proteins such as Mdm2 (53, 76) and ARF (62) have been shown to regulate p53 protein stability. p300 (5, 24) and JMY (63), on the other hand, enhance p53 transcriptional activity. The data presented here demonstrate that AMF1 has a significant effect on the transcriptional activity of p53. The facts that AMF1 stimulates reporter gene expression from p53-dependent promoters and that U2OS/AMF1 cells contain a higher basal level of p21WAF1/CIP1 and are partially G1 arrested imply that AMF1 could be an important modulator in p53 transactivation.
Increasing evidence suggests that coactivators play a significant role in regulating eukaryotic transcription (11). We previously reported that AMF1 activates papillomavirus E2-dependent transcription and directly interacts with both E2 and p300 (8, 54). Complex formation among E2, AMF1, and p300 may function by bringing p300 (with histone acetyltransferase activity) close to initiation sites of transcription and viral DNA replication. Histone acetylation of nearby nucleosomes is thought to enhance access of the transcriptional or replication machinery to DNA (8, 22, 54, 65). Since p300 proteins have been shown to act as coactivators in p53-dependent transcription, participation of AMF1 may further strengthen the connection between p300 and p53. AMF1 did not stimulate a p53-dependent promoter in Soas-2 cells (p53 null, Fig. 5B) or in C33A cells that express mutant p53 (data not shown).
This report shows that there is direct interact between AMF1 and p53. First, AMF1 and p53 coprecipitated in U2OS/AMF1 cells (Fig. 2B); second, AMF1 bound p53 in different in vitro binding assays. We also performed coimmunoprecipitations of AMF1 and p53 from untreated U2OS cells. Using anti-p53 antibody pAb421, native AMF1 was coimmunoprecipitated as detected by Western blotting (data not shown), however, this result was not reproduced in all experiments. This can be explained by the following: (i) the endogenous AMF1 bound to p53 is below detectable amounts or (ii) the AMF1-p53 complex may be transient. In addition, we attempted unsuccessfully to coprecipitate p53 from untreated U2OS cell lysates using polyclonal rabbit antisera against AMF1. It is possible that the antiserum blocks p53 binding. The AMF1 binding domain on p53 was mapped to aa 161 to 333, and the region on AMF1 responsible for binding to p53 was localized to aa 103 to 250. In contrast to what was shown for Mdm2 (21, 29, 40, 43), interaction between AMF1 and p53 does not seem to alter intracellular p53 protein levels before or after etoposide treatment (Fig. 9B). Cotransfection of wild-type p53 with or without AMF1 into Saos-2 cells showed equal levels of p53 (data not shown).
To better understand the mechanisms of how AMF1 functionally interacts with p53, we identified a dominant-negative version of AMF1, AMF1(1–103), which did not bind p53 (Fig. 4) but formed complexes with wild-type AMF1 (Fig. 8B). The observation that AMF1(1–103) can compromise the effects of wild-type AMF1 and inhibit p53-dependent transcriptional activation (Fig. 8A) argue that proper oligomerization of AMF1 is necessary for its activation of p53 transcription. AMF1(1–103) probably forms a nonfunctional complex with wild-type AMF1 in vivo, resulting in a decreased amount of functional oligomers formed between wild-type molecules.
A significant implication of the present study relates to the importance of transcriptional activation in the p53 response. Previous studies have established that increased p53 transactivation can lead to either cell growth arrest or apoptosis, depending on the cell type and DNA-damaging agent used (18, 38, 71). Our studies showed that etoposide treatment of U2OS cells results in the same outcome (growth arrest) as X-ray treatment (2). However, overexpression of AMF1 in U2OS/AMF1 cells delays but does not block apoptosis, probably by raising the p21WAF1/CIP1 protein level. A similar effect was observed with ectopic expression of p21WAF1/CIP1 (2). In contrast, U2OS/AMF1 cells are more sensitive to UV irradiation-mediated apoptosis. It was previously shown that the UV-mediated apoptosis in U2OS cells is p53 dependent (p53-dominant negative U2OS clones barely undergo apoptosis under UV irradiation), but not via a p21WAF1/CIP1-activated pathway, as p21WAF1/CIP1 expression was suppressed upon exposure to UV irradiation (2). Our results confirmed that p21WAF1/CIP1 protein in U2OS/AMF1 cells was undetectable 2 h after UV irradiation (data not shown), indicating that other p53-dependent genes are responsible for the augmented apoptosis in U2OS/AMF1 cells. Ongoing projects will examine the effects of AMF1 on the expression of apoptotic targets of p53.
It is possible for a coactivator to exert variable effects on different classes of transcription activator (11), and in this respect, it will be interesting to determine the specificity of AMF1-mediated coactivation. It was reported that binding of AMF1 to the HTLV-1 oncoprotein Tax can potently suppress its activation of Jun N-terminal protein kinase 1 (JNK1) (35). Interestingly, HTLV-1 Tax oncoprotein also repressed the p53-mediated transactivation through coactivator CREB-binding protein sequestration (3). Like other cell cycle regulators, AMF1 may play multiple roles in different signal transduction pathways. Genetic analysis in yeast indicates that AMF1 functions downstream of STE4-encoded Gβ subunit and upstream of STE20 (35, 64). In NIH 3T3 cells, overexpression of AMF1 can suppress G-protein (RAS)-mediated and mitogen-activated protein kinase-mediated signaling and interfere with JNK1 activity (64). We also found that AMF1 can stimulate the transcription function of a c-Jun AD-GAL4 DBD fusion that is subject to regulation by JNK1 (8). These data imply that AMF1 may play important roles in signal transduction as well as being a coactivator in E2- or p53-dependent transcription. Given the multiple and complex signaling effects of AMF1, it will be of interest to establish a mouse knockout model to further explore its physiological roles.
ACKNOWLEDGMENTS
We thank Karen Vousden for reviewing the manuscript and providing valuable suggestions. We are grateful to Leonard Buckbinder, Wafik El-Deiry, Steve Grossman, Moshe Owen, Kevin Ryan, and Bert Vogelstein for providing reagents. We thank members of Androphy laboratory for many useful discussions.
This work was supported by NIH grants R01 CA58376 and U01 AI38001 to E.J.A. and P30 AI42853 to C.M.
REFERENCES
- 1.Abarzua P, LoSardo J E, Gubler M L, Spathis R, Lu Y A, Felix A, Neri A. Restoration of the transcription activation function to mutant p53 in human cancer cells. Oncogene. 1996;13:2477–2482. [PubMed] [Google Scholar]
- 2.Allan L A, Fried M. p53-dependent apoptosis or growth arrest induced by different forms of radiation in U2OS cells: p21WAF1/CIP1 repression in UV induced apoptosis. Oncogene. 1999;18:5403–5412. doi: 10.1038/sj.onc.1202931. [DOI] [PubMed] [Google Scholar]
- 3.Ariumi Y, Kaida A, Lin J Y, Hirota M, Masui O, Yamaoka S, Taya Y, Shimotohno K. HTLV-1 tax oncoprotein represses the p53-mediated trans-activation function through coactivator CBP sequestration. Oncogene. 2000;19:1491–1499. doi: 10.1038/sj.onc.1203450. [DOI] [PubMed] [Google Scholar]
- 4.Arriola E L, Lopez A R, Chresta C M. Differential regulation of p21waf-1/cip-1 and Mdm2 by etoposide: etoposide inhibits the p53-Mdm2 autoregulatory feedback loop. Oncogene. 1999;18:1081–1091. doi: 10.1038/sj.onc.1202391. [DOI] [PubMed] [Google Scholar]
- 5.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 6.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 7.Breiding D E, Grossel M J, Androphy E J. Genetic analysis of the bovine papillomavirus E2 transcriptional activation domain. Virology. 1996;221:34–43. doi: 10.1006/viro.1996.0350. [DOI] [PubMed] [Google Scholar]
- 8.Breiding D E, Sverdrup F, Grossel M J, Moscufo N, Boonchai W, Androphy E J. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol Cell Biol. 1997;17:7208–7219. doi: 10.1128/mcb.17.12.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 10.Burden D A, Kingma P S, Froelich-Ammon S J, Bjornsti M-A, Patchan M W, Thompson R B, Osheroff N. Topoisomerase II · etoposide interactions direct the formation of drug-induced enzyme-DNA cleavage complexes. J Biol Chem. 1996;271:29238–29244. doi: 10.1074/jbc.271.46.29238. [DOI] [PubMed] [Google Scholar]
- 11.Chang M, Jaehning J A. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 13.Chow K C, Ross W E. Topoisomerase-specific drug sensitivity in relation to cell cycle progression. Mol Cell Biol. 1987;7:3119–3123. doi: 10.1128/mcb.7.9.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun A C, Zhou Y, Wong C M, Kung H F, Jeang K T, Jin D Y. Coiled-coil motif as a structural basis for the interaction of HTLV type 1 Tax with cellular cofactors. AIDS Res Hum Retrovir. 2000;16:1689–1694. doi: 10.1089/08892220050193155. [DOI] [PubMed] [Google Scholar]
- 15.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hooper M L, Wyllie A H. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 16.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 17.Desaintes C, Goyat S, Garbay S, Yaniv M, Thierry F. Papillomavirus E2 induces p53-independent apoptosis in HeLa cells. Oncogene. 1999;18:4538–4545. doi: 10.1038/sj.onc.1202818. [DOI] [PubMed] [Google Scholar]
- 18.el-Deiry W S, Harper J W, O'Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill. Wang D E Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 19.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 20.Gorina S, Pavletich N P. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 21.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 22.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 24.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 25.Guasch R M, Guerri C, O'Connor J E. Flow cytometric analysis of concanavalin A binding to isolated Golgi fractions from rat liver. Exp Cell Res. 1993;207:136–141. doi: 10.1006/excr.1993.1172. [DOI] [PubMed] [Google Scholar]
- 26.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 27.Hande K R. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur J Cancer. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 28.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 29.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 30.Hermeking H, Lengauer C, Polyak K, He T C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 31.Hoffmann A, Roeder R G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991;19:6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inaba M, Mitsuhashi J, Kawada S, Nakano H. Different modes of cell-killing action between DNA topoisomerase I and II inhibitors revealed by kinetic analysis. Jpn J Cancer Res. 1994;85:187–193. doi: 10.1111/j.1349-7006.1994.tb02081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwabuchi K, Bartel P L, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenkins J R, Chumakov P, Addison C, Sturzbecher H W, Wade-Evans A. Two distinct regions of the murine p53 primary amino acid sequence are implicated in stable complex formation with simian virus 40 T antigen. J Virol. 1988;62:3903–3906. doi: 10.1128/jvi.62.10.3903-3906.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin D Y, Teramoto H, Giam C Z, Chun R F, Gutkind J S, Jeang K T. A human suppressor of c-Jun N-terminal kinase 1 activation by tumor necrosis factor alpha. J Biol Chem. 1997;272:25816–25823. doi: 10.1074/jbc.272.41.25816. [DOI] [PubMed] [Google Scholar]
- 36.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 37.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 38.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 39.Kraiss S, Quaiser A, Oren M, Montenarh M. Oligomerization of oncoprotein p53. J Virol. 1988;62:4737–4744. doi: 10.1128/jvi.62.12.4737-4744.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kubbutat M H, Ludwig R L, Ashcroft M, Vousden K H. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lakin N D, Jackson S P. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 42.Lehar S M, Nacht M, Jacks T, Vater C A, Chittenden T, Guild B C. Identification and cloning of EI24, a gene induced by p53 in etoposide-treated cells. Oncogene. 1996;12:1181–1187. [PubMed] [Google Scholar]
- 43.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 44.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 47.Lu H, Levine A J. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massimi P, Pim D, Bertoli C, Bouvard V, Banks L. Interaction between the HPV-16 E2 transcriptional activator and p53. Oncogene. 1999;18:7748–7754. doi: 10.1038/sj.onc.1203208. [DOI] [PubMed] [Google Scholar]
- 49.May P, May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 50.Meek D W. Mechanisms of switching on p53: a role for covalent modification? Oncogene. 1999;18:7666–7675. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- 51.Mukhopadhyay T, Roth J A. Induction of apoptosis in human lung cancer cells after wild-type p53 activation by methoxyestradiol. Oncogene. 1997;14:379–384. doi: 10.1038/sj.onc.1200835. [DOI] [PubMed] [Google Scholar]
- 52.Okorokov A L, Milner J. Proteolytic cleavage of p53: a model for the activation of p53 in response to DNA damage. Oncol Res. 1997;9:267–273. [PubMed] [Google Scholar]
- 53.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 54.Peng Y C, Breiding D E, Sverdrup F, Richard J, Androphy E J. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J Virol. 2000;74:5872–5879. doi: 10.1128/jvi.74.13.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pietenpol J A, Tokino T, Thiagalingam S, el-Deiry W S, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 57.Prakash S S, Grossman S R, Pepinsky R B, Laimins L A, Androphy E J. Amino acids necessary for DNA contact and dimerization imply novel motifs in the papillomavirus E2 trans-activator. Genes Dev. 1992;6:105–116. doi: 10.1101/gad.6.1.105. [DOI] [PubMed] [Google Scholar]
- 58.Ryan K M, Vousden K H. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol Cell Biol. 1998;18:3692–3698. doi: 10.1128/mcb.18.7.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selivanova G, Ryabchenko L, Jansson E, Iotsova V, Wiman K G. Reactivation of mutant p53 through interaction of a C-terminal peptide with the core domain. Mol Cell Biol. 1999;19:3395–3402. doi: 10.1128/mcb.19.5.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw P, Freeman J, Bovey R, Iggo R. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 62.Sherr C J, Weber J D. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 63.Shikama N, Lee C W, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue N B. A novel cofactor for p300 that regulates the p53 response. Mol Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 64.Spain B H, Bowdish K S, Pacal A R, Staub S F, Koo D, Chang C Y, Xie W, Colicelli J. Two human cDNAs, including a homolog of Arabidopsis FUS6 (COP11), suppress G-protein- and mitogen-activated protein kinase-mediated signal transduction in yeast and mammalian cells. Mol Cell Biol. 1996;16:6698–6706. doi: 10.1128/mcb.16.12.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 67.Thut C J, Chen J L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 68.Vater C A, Bartle L M, Dionne C A, Littlewood T D, Goldmacher V S. Induction of apoptosis by tamoxifen-activation of a p53-estrogen receptor fusion protein expressed in E1A and T24 H-ras transformed p53−/− mouse embryo fibroblasts. Oncogene. 1996;13:739–748. [PubMed] [Google Scholar]
- 69.Vousden K H. p53: death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 70.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 71.Waldman T, Lengauer C, Kinzler K W, Vogelstein B. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature. 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 72.Webster K, Parish J, Pandya M, Stern P L, Clarke A R, Gaston K. The human papillomavirus (HPV) 16 E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. J Biol Chem. 2000;275:87–94. doi: 10.1074/jbc.275.1.87. [DOI] [PubMed] [Google Scholar]
- 73.White E. Life, death, and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 74.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 75.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 76.Zauberman A, Barak Y, Ragimov N, Levy N, Oren M. Sequence-specific DNA binding by p53: identification of target sites and lack of binding to p53-MDM2 complexes. EMBO J. 1993;12:2799–2808. doi: 10.1002/j.1460-2075.1993.tb05941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]