Abstract
Drug delivery in a safe manner is a major challenge in the drug development process. Growth factor receptors (GFRs) are known to have profound roles in the growth and progression of cancerous cells making these receptors a therapeutic target in the effective treatment of cancer. This work focused on exploring bioactive compounds that can target GFRs using in silico method. In this study, 50 bioactive compounds from different plant sources were screened as anticancer agent against GFRs using drug likeness parameters of Lipinski’s rule of five. The molecular docking was performed between phytochemicals and GFRs. Ligands with acceptable drug likeness and binding energy comparable to the standard drugs were further screened to determine their pharmacokinetic activities. This study showed phytochemicals with the binding energy comparable with the standard drugs (Dovitinib and Gefitinib), while ADME, bioactivity score, and bioavailability radar analysis gave further insight on these compounds as potent anticancer agents.
Keywords: GFR, Drug delivery, ADMET, Molecular docking, Cancer
Introduction
Multiple alterations in the gene expression lead to imbalance in cellular proliferation resulting in abnormal growth of cells called cancer. Over the years, cancer has been a persistent public health problem recorded as the major cause of death in both developed and developing countries worldwide [1, 2]. Research shows that an estimated number of 23.6 million new cases of cancer are likely to be recorded per year by 2030 [3]. Cancerous cells have special ability of survival, immortality, self-sufficiency of growth signals, unlimited replication, gene instability, evading programmed cell death, continuous angiogenesis, and diverse mutations [4]. Growth factor receptors (GFRs) are proteins that have profound roles in tumor growth of cell, metastasis, angiogenesis, cell survival, cell death, cell migration, differentiation, organogenesis, neovascularization, and chemoresistance, activated by binding with their ligands (growth factors) [5–7]. These receptors highly involved in cancer progression include epidermal growth factor receptors (EGFRs), insulin-like growth factor receptor (IGFR), vascular endothelial growth factor receptor (VEGFR), transforming growth factor-beta receptor (TGF-βR), fibroblast growth factor receptor (FGFR), and platelet-derived growth factor receptor (PDGFR). In some cancer, these receptors are often overexpressed leading to uncontrolled proliferation and differentiation [8], and their amplification activates both inherent and acquired resistance to cancer treatments. Overexpression of HER-2 gene was found in breast, bladder, lung, and glial carcinomas; HER-3 gene in aggressive metastatic breast, ovarian, lung, gastric, invasive urothelial bladder, and endometrial carcinomas; VEGFR1-3genes found in large number of tumors like bladder, colon, breast, lung, brain, prostate, gastric, kidney, thyroid, and ovarian cancer cells; FGFR-2 gene in gastric, breast, endometrial, and lung carcinomas [6, 9]. The role of these receptors in cancer progression has made them a necessary therapeutic target for effective cancer therapy.
Presently, the treatment of cancer involves the use of chemotherapy, radiotherapy, immunotherapy, surgery, and anticancer drugs, all of which have been proven to be less effective due to disadvantages of recurrence, drug resistance, effect on non-targeted cell, and other various side effects and toxicity that accompanies them [2, 4].
Studies have shown that phytochemicals are powerful anticancer agents, with benefits greater than synthetic compounds, ranging from less toxicity, ease of extraction, to large abundance [10]. About 35% of cancer cases can be addressed by establishing a proper dieting which largely involves the consumption of plant-derived foods such as vegetables, fruits, and whole grains that contains carotenoids, flavonoids, and phenolics [11–13]. Phytochemicals possess various mechanisms to shut down or slow down cancer growth and progression by reducing oxidative stress, suppressing proliferation of cells, causing programmed cell death, preventing angiogenic process, and cell cycle arrest [2, 11]. Studies showed that not less than 60 phytochemicals are currently in the pipeline as potential anticancer agents [14–16]. Developing anticancer drug requires blocking one or more proteins or pathways involved in cancer development. In this present study, 50 phytochemicals were evaluated against EGFRs (HER-2 and HER-3), VEGFR (VEGFR-2 and VEGFR-3), and FGFR (FGFR-2) to determine potential lead agents for the development of anticancer drugs.
Materials and Method
Selection and Preparation of Ligands
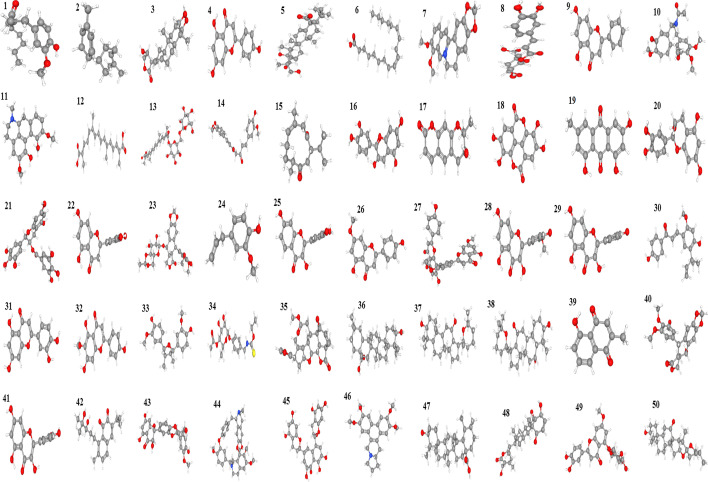
Fifty bioactive compounds from different plant sources collated from public database and published research papers were downloaded from https://pubchem.ncbi.nlm.nih.gov in SDF format (Fig. 1). Also, two standard drugs Dovitinib and Gefitinibwith Pubmed IDCID_135398510 and CID_123631 respectively were downloaded in 3D Sdf format for comparism with the phytochemicals. Using the software open babel (http://openbabel.org/wiki/Main_Page), these ligands were changed to PDB format which makes them suitable for docking analysis. This was followed by setting the torsion requirements for proper binding using Autodock 4.2.6 parameters.
Fig. 1:
3D Structures of 50 phytochemicals used in this study. (1) 6-Shogaol, (2) α-Zingiberene, (3) Andrographolide, (4) Apigenin, (5) Bassic acid, (6) Linoleic acid, (7) Berberine, (8) Chlorogenic acid, (9) Chrysin, (10) Colchicine, (11) Corydine, (12) Crocetin, (13) Crocin 3, (14) Curcumin, (15) Curdione, (16) Cyanidin, (17) Decursinol, (18) Ellagic acid, (19) Emodin, (20) Epicatechin, (21) Epigallocatechin, (22) Eriodyctiol, (23) Etoposide, (24) Eugenol, (25) Fisetin, (26) Genkwanin, (27) Ginkgetin, (28) Isorhamnetin, (29) Kaempferol, (30) Licochalcone A, (31) Luteolin, (32) Hispidulin, (33) Nectandrin B, (34) Niaziminin, (35) Nimbolide, (36) Oleanolic acid, (37) Panaxadiol, (38) Panaxatriol, (39) Plumbagin, (40) Podophyllotoxin, (41) Quercetin, (42) Salvicine, (43) Silibinin, (44) Tetrandrine, (45) Theaflavin, (46) Tylophorine, (47) Ursolic acid, (48) Withaferin A, (49) Yuanhuanin, (50) Hecogenin
Proteins Preparation
3D structures of human endothelial receptor (HER-1 and HER-2), vascular endothelial growth factor receptor (VEGFR-2 and VEGFR-3), and fibroblast growth receptor (FGFR-2) were downloaded from RCSB protein data bank (http://www.rcsb.org/pdb/home/home.do) with PDB ID 3PP0 for HER-2, 6OP9 for HER-3, 3VHE for VEGFR-2, 4BSJ for VEGFR-3, and 2PVF for FGFR-2. Using Autodock 4.2.6, removal of water molecules and addition of polar hydrogen and Kollman’s charges were done, and these proteins were saved in PDBQT format.
Docking Analysis
After the proteins and ligands preparation, molecular docking was executed using Autodock 4.2.6. For binding to take place, x, y, and z dimensions were set at 60 × 60 × 60 with a resolution of 0.500 Å and grid box centered to obtain favorable docking conformations. The grid file was saved as (.gpf) file and run autogrid. Docking calculation was then carried out using the Lamarckian genetic algorithm with the default at 10 runs. Dock file was saved as dpf and after running autodock, final docked results were obtained in (.dlg) file which showed information such as binding residues, binding energy, and inhibition constant. The structures showing interaction between ligands and proteins were viewed using Discovery Studio.
Rule of Five (RO5)
Lipinski’s RO5 is used for evaluation of drug likeness of a compound, a necessary step in drug discovery which helps to determine if a certain compound is likely to be orally active. In this study, ligands were screened for the RO5 using the Supercomputing facility for bioinformatics and computational biology (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) [17, 18]. Bioactive compounds with binding energy comparable to standard drugs were subjected to this analysis.
In Silico ADME Analysis
Pharmokinetics parameters such as Absorption, Distribution, Metabolism, and Excretion were evaluated in the ligands using SwissADME (http://www.swissadme.ch/index.php). The essence of this assay is to provide insights that will be useful in drug research and development process.
Bioactivity Score and Bioavailabilty Radar
The bioactivity score of ligands was determined employing online Molinspiration software (http://www.molinspiration.com/). This was done by applying canonical SMILES of ligands obtained from PubChem. Properties analyzed include G-protein coupled receptors (GPCR), enzyme inhibitors (EI), kinase inhibitors (KI), nuclear receptors ligands (NRL), and ion channel modulators (ICM). The bioavailability radar ligands were determined by using Swiss ADME (http://www.swissadme.ch/index.php) which instantly shows if a compound is orally bioavailable.
Results and Discussion
Docking Analysis
Due to the roles growth factor receptors play in the development and progression of cancer, it is necessary to develop suitable drug candidates that can effectively inhibit growth factors at the sites of receptors with little or no effects. Binding affinity between ligands and receptors is determined by the binding energy, the lower the energy, the higher the binding affinity [9]. For this purpose, 50 bioactive compounds (Fig. 1) were screened against ERRB2/HER-2, ERRB3/HER-3, VEGFR-2, VEGFR-3, and FGFR-2. Commonly used anticancer drugs (Dovitinib and Gefitinib) that have activities against the target receptors were docked against the proteins and their binding energies used as comparison with the binding energies of the selected. After docking, the binding energy(kcal/mol), number of hydrogen bond, inhibitory constant (μM/nM), and amino acids involved in hydrogen bonding were noted.
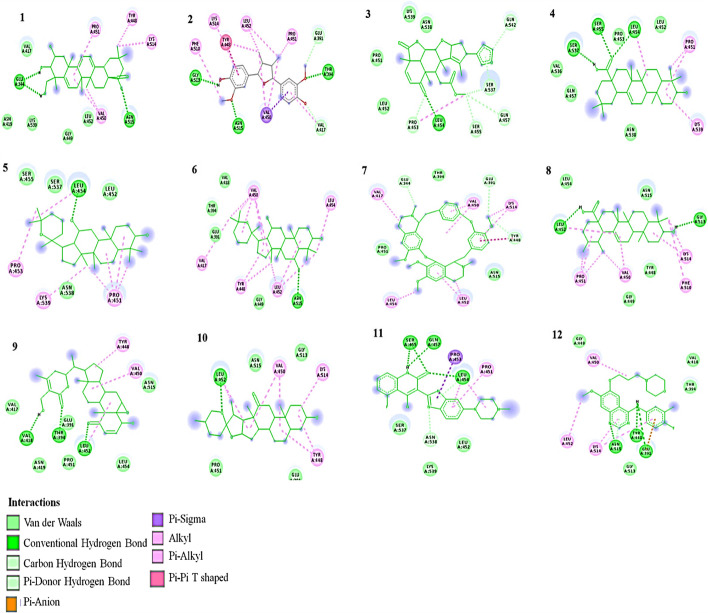
Out of the 50 phytochemicals, 14 have binding energy comparable to the standard drug for HER-2 (Table 1 and Fig. 2). Dovitinib and Gefitinib have binding energies of − 6.03 kcal/mol and − 6.59 kcal/mol, respectively, with amino acid involved in hydrogen bond as LYS1326 and GLN1329 for Dovitinib and LEU1291 for Gefitinib.
Table 1.
Binding parameters between ligands and target protein HER-2
| Phytochemicals | Binding energy (Kcal/mol) | Inhibition constant (µM) | Number of hydrogen bond | Amino acids involved in hydrogen bonding |
|---|---|---|---|---|
| Andrographolide | − 6.77 | 10.87 | 3 | ASP 950, ASP 871, ILE 872 |
| Bassic acid | − 7.19 | 5.37 | 2 | ASP 769, ARG 985 |
| Crocetin | − 6.73 | 11.76 | 2 | ARG 968, ASP 950 |
| Curdione | − 6.72 | 11.89 | 1 | ILE 714 |
| Nimbolide | − 8.11 | 1.13 | 3 | ARG 968, LYS 765, LYS 954 |
| Oleanolic acid | − 7.84 | 1.78 | 1 | ASP 871 |
| Panaxadiol | − 8.73 | 0.395 | 2 | LYS 765, LEU 870 |
| Panaxatriol | − 8.41 | 0.685 | 2 | LEU 870, LYS 765 |
| Ursolic acid | − 8.05 | 1.25 | 1 | ASP 871 |
| Withaferin A | − 9.55 | 0.998 | 1 | ASP 950 |
| Hecogenin | − 7.93 | 1.54 | 2 | ASP 950 |
| Dovitinib (Standard drug) | − 7.45 | 3.48 | 3 | GLU 971, SER 963, ARG 966 |
| Gefitinib (Standard drug) | − 6.58 | 15.08 | 1 | LYS 765 |
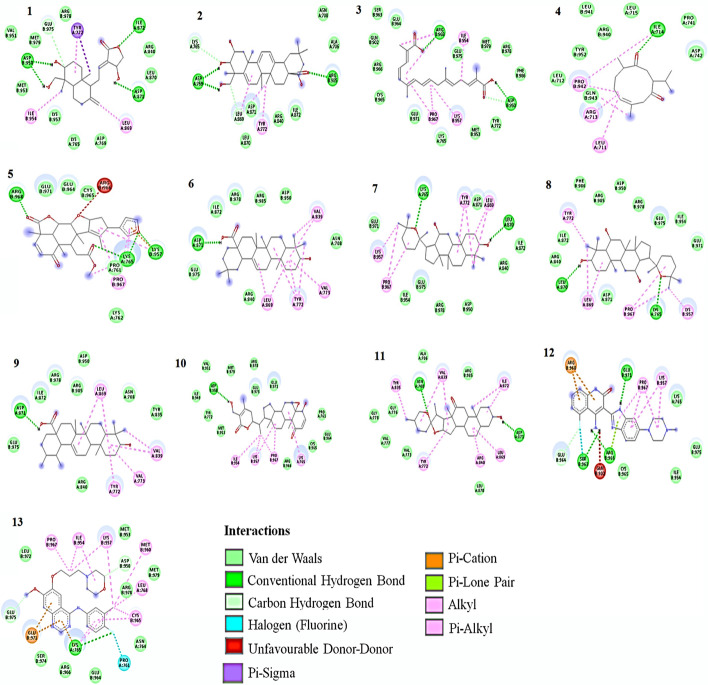
Fig. 2.
2D Interaction of ligands with HER-2. (1)Andrographolide, (2) Bassic acid, (3) Crocetin, (4) Curdione, (5) Nimbolide, (6) Oleanolic acid, (7) Panaxadiol, (8) Panaxatriol, (9) Ursolic acid, (10) Withaferin A, (11) Hecogenin, (12) Dovitinib, (13) Gefitinib
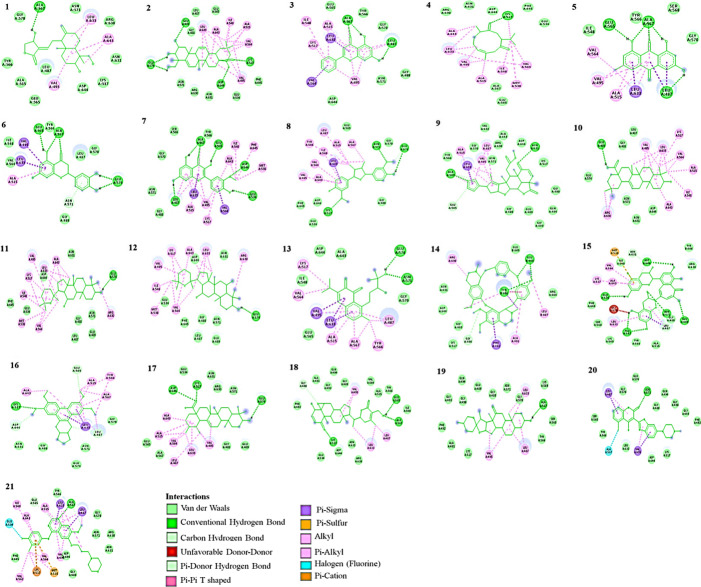
Nineteen phytochemicals for HER-3 as shown in Table 2 and Fig. 3 have Dovitinib and Gefitinib with binding energies of − 8.77 kcal/mol and − 6.89 kcal/mol, with amino acid involved in hydrogen bond as LEU771 for Dovitinib and CYS721 and LEU771 for Gefitinib.
Table 2.
Binding parameters between ligands and target protein HER-3
| Phytochemicals | Binding energy (Kcal/mol) | Inhibition constant (µM) | Number of hydrogen bond | Amino acids involved in hydrogen bonding |
|---|---|---|---|---|
| Andrographolide | 8.07 | 1.21 | 4 | LYS 723, LEU 771, GLY 774, ASP 833 |
| Bassic acid | − 8.14 | 1.08 | 2 | PRO 772, ASN 820 |
| Crocetin | − 7.81 | 1.90 | 2 | GLN 769, LYS 853 |
| Curdione | − 7.26 | 4.76 | 0 | - |
| Etoposide | − 7.51 | 3.12 | 4 | LEU 771, LYS 723, PHE 701, ASN 815 |
| Genkwanin | − 7.00 | 7.37 | 3 | LEU 771, LYS 723, ASP 833 |
| Licochalcone A | − 7.04 | 6.90 | 2 | LYS 723, VAL 753 |
| Nectandrin B | − 7.23 | 4.98 | 2 | ASN 820, PHE 834 |
| Nimbolide | − 7.83 | 1.81 | 3 | PHE 701, LYS 723, LYS 853 |
| Oleanolic acid | − 9.86 | 0.059 | 3 | THR 768, LYS 723, ARG 819 |
| Panaxadiol | − 10.04 | 0.043 | 0 | –– |
| Panaxatriol | − 8.91 | 0.296 | 1 | ASP 833 |
| Salvicine | − 8.83 | 0.337 | 2 | ASP 833, GLN 769 |
| Tetrandine | − 7.34 | 4.18 | 1 | ARG 781 |
| Theaflavin | − 7.90 | 1.62 | 5 | LEU 771, GLN 769, CYS 721, THR 768, ASP 833 |
| Tylophorine | − 7.55 | 2.94 | 2 | CYS 721, LYS 723 |
| Ursolic acid | − 9.72 | 0.075 | 1 | ARG 819 |
| Withaferin A | − 11.69 | 0.0027 | 3 | VAL 753, ASP 833, LEU 771 |
| Hecogenin | − 10.50 | 0.020 | 3 | CYS 721, ASP 833, ASP 778 |
| Dovitinib (Standard drug) | 8.77 | 0.373 | 1 | LEU 771 |
| Gefitinib (Standard drug) | − 6.89 | 8.83 | 2 | CYS 721, LEU 771 |
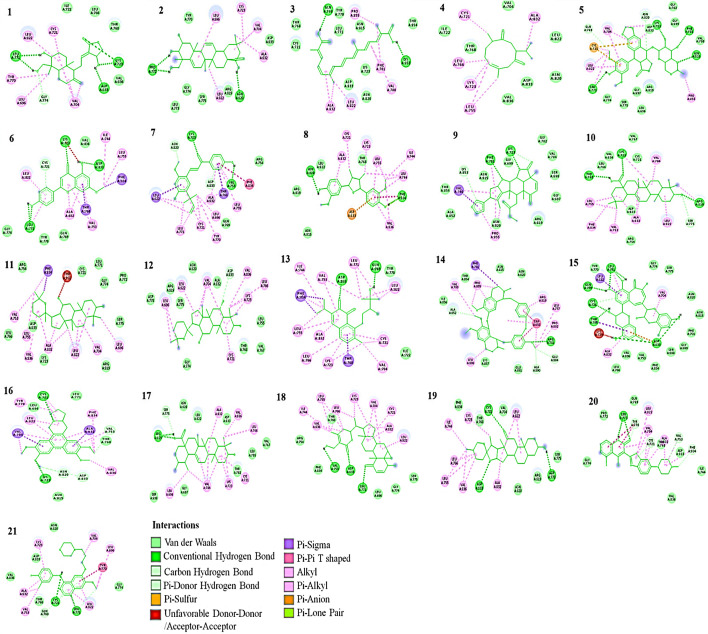
Fig. 3.
2D Interaction of ligands with HER-3. (1) Andrographolide, (2) Bassic acid, (3) Crocetin, (4) Curdione, (5) Etoposide, (6) Genkwanin, (7) Licochalcone A, (8) Nectandrin B, (9) Nimbolide, (10) Oleanolic acid, (11) Panaxadiol, (12) Panaxatriol, (13) Salvicine, (14) Tetrandrine, (15) Theaflavin, (16) Tylophorine, (17) Ursolic acid, (18) Withaferin A, (19) Hecogenin, (20) Dovitinib, (21) Gefitinib
Six phytochemicals for VEGFR-2 as shown in Table 3 and Fig. 4 have − 8.84 kcal/mol and − 8.70 kcal/mol binding energies for Dovitinib and Gefitinib respectively and amino acids GLU917 for Dovitinib and ASP1046 and CYS919 for Gefitinib.
Table 3.
Binding parameters between ligands and target protein VEGFR-2
| Phytochemicals | Binding energy (Kcal/mol) | Inhibition constant (µM) | Number of hydrogen bond | Amino acids involved in hydrogen bonding |
|---|---|---|---|---|
| Oleanolic acid | − 9.05 | 0.231 | 2 | ARG 1027, 1LE 1044 |
| Panaxadiol | − 9.51 | 0.106 | 3 | ARG 1027, HIS 1026, ILE 1044 |
| Panaxatriol | − 9.13 | 0.202 | 3 | ARG 1027, HIS 1026, 1LE 1044 |
| Ursolic acid | − 9.11 | 0.210 | 3 | ASP 814, ARG 1027, ILE 1044 |
| Withaferin A | − 9.53 | 0.103 | 2 | GLU 885, ILE 1025 |
| Hecogenin | − 9.82 | 0.314 | 2 | ARG 1027, GLU 818 |
| Dovitinib (Standard drug) | − 8.84 | 0.332 | 1 | GLU 917 |
| Gefitinib (Standard drug) | − 8.70 | 0.423 | 2 | ASP 1046, CYS 919 |
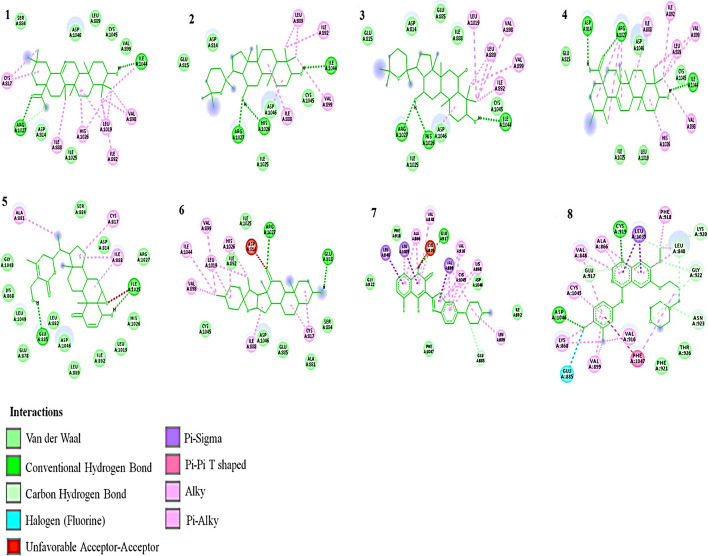
Fig. 4.
2D Interaction of ligands with VEGFR-2. (1) Oleanolic acid, (2) Panaxadiol, (3) Panaxatriol, (4) Ursolic acid, (5) Withaferin A, (6) Hecogenin, (7) Dovitinib, (8) Gefitinib
Ten phytochemicals for VEGFR-3 as seen in Table 4 and Fig. 5 have binding energies of − 6.07 kcal/mol and − 5.94 kcal/mol for Dovitinib and Gefitinib, respectively, having amino acid SER455, GLN457, and LEU454 for Dovitinib and ASN515, TYR448, and GLU391 for Gefitinib.
Table 4.
Binding parameters between ligands and target protein VEGFR-3
| Phytochemicals | Binding energy (Kcal/mol) | Inhibition constant (µM) | Number of hydrogen bond | Amino acids involved in hydrogen bonding |
|---|---|---|---|---|
| Bassic acid | − 6.14 | 31.80 | 2 | GLU 344, ASN 515 |
| Nectandrin B | − 6.08 | 34.69 | 3 | GLY 513, ASN 515, THR 394 |
| Nimbolide | − 6.58 | 15.08 | 1 | LEU 454 |
| Oleanolic acid | − 7.59 | 2.75 | 3 | SER 537, SER 455, LEU 454 |
| Panaxadiol | − 7.11 | 6.10 | 1 | LEU 454 |
| Panaxatriol | − 6.79 | 10.49 | 1 | ASN 515 |
| Tetrandine | − 6.44 | 19.13 | ||
| Ursolic acid | − 7.42 | 3.65 | 2 | LEU 452, GLY 513 |
| Withaferin A | − 7.27 | 4.72 | 3 | LEU 452, VAL 418, THR 394 |
| Hecogenin | − 7.45 | 3.44 | 1 | LEU 452 |
| Dovitinib (Standard drug) | − 6.07 | 35.59 | 3 | SER 455, GLN 457, LEU 454 |
| Gefitinib (Standard drug) | − 5.94 | 44.14 | 3 | ASN 515, TYR 448, GLU 391 |
Fig. 5.
2D Interaction of ligands with VEGFR-3. (1) Bassic acid, (2) Nectandrin B, (3) Nimbolide, (4) Oleanolic acid, (5) Panaxadiol, (6) Panaxatriol, (7) Tetrandine, (8) Ursolic acid, (9) Withaferin A, (10) Hecogenin, (11) Dovitinib, (12) Gefitinib
Lastly, 19 phytochemicals have binding energy comparable to standard drug for FGFR-2 (Table 5 and Fig. 6); Dovitinib has an energy of − 6.72 kcal/mol with amino acid involved in hydrogen bond as ASN571, while Gefitinib has a binding energy of − 7.84 kcal/mol and amino acid ALA567. From these results, we selected 13 phytochemicals with the lowest binding energy comparable to standard drugs found in at least three of the target proteins; these were used for further in silico studies.
Table 5.
Binding parameters between ligands and target protein FGFR-2
| Phytochemicals | Binding energy (Kcal/mol) | Inhibition constant (µM) | Number of hydrogen bond | Amino acids involved in hydrogen bonding |
|---|---|---|---|---|
| Andrographolide | − 7.42 | 3.62 | 1 | ALA 567 |
| Bassic acid | − 7.64 | 2.51 | 4 | GLU 574, GLU 489, ASP 644, LYS 517 |
| Chrysin | − 6.82 | 10.06 | 2 | ALA 567, LEU 487 |
| Curdione | − 7.27 | 4.69 | 1 | LYS 517 |
| Emodin | − 7.37 | 3.94 | 3 | GLU 565, ALA 567, LEU 487 |
| Eriodyctiol | − 6.72 | 11.96 | 3 | GLU 565, ALA 567, GLU 574 |
| Isorhamnetin | − 6.87 | 9.26 | 5 | LEU 487, ALA 567, GLU 565, ASP 644, GLU 534 |
| Nectandrin B | − 7.50 | 3.20 | 3 | ASN 571, GLU 574, LYS 517 |
| Nimbolide | − 8.17 | 1.03 | 3 | ALA 567, ASN 631, GLU 565 |
| Oleanolic acid | − 8.77 | 372.58 nM | 1 | GLU 489 |
| Panaxadiol | − 9.02 | 244.6 nM | 1 | GLU 574 |
| Panaxatriol | − 8.61 | 486.38 nM | 1 | GLU 574 |
| Salvicine | − 7.86 | 1.73 | 2 | GLU 574, ASN 571 |
| Tetrandine | − 7.69 | 2.32 | 2 | GLU 604, TYR 606 |
| Theaflavin | − 6.90 | 8.71 | 5 | ASP 644, GLU 534, ALA 567, ASN 571, GLU 489 |
| Tylophorine | − 7.61 | 2.66 | 1 | LYS 517 |
| Ursolic acid | − 8.61 | 490.22 nM | 3 | ASP 644, LYS 517, GLU 574 |
| Withaferin A | − 8.63 | 469.67 nM | 3 | GLU 565, ALA 567, LYS 517 |
| Hecogenin | − 8.87 | 314.06 nM | 1 | ALA 567 |
| Dovitinib (Standard drug) | − 6.72 | 11.80 | 1 | ASN 571 |
| Gefitinib (Standard drug) | − 7.84 | 1.80 | 1 | ALA 567 |
Fig. 6.
2D Interaction of ligands with FGFR-2. (1)Andrographolide, (2) Bassic acid, (3) Chrysin, (4) Curdione, (5) Emodin, (6) Eriodyctiol, (7) Isorhamnetin, (8) Nectandrin B, (9) Nimbolide, (10) Oleanolic acid, (11) Panaxadiol, (12) Panaxatriol, (13) Salvicine, (14) Tetrandrine, (15) Theaflavin, (16) Tylophorine, (17) Ursolic acid, (18) Withaferin A, (19) Hecogenin, (20) Dovitinib, (21) Gefitinib
Pharmacokinetic and Drug Likeness Screening of Phytochemicals
Drug properties of the 13 selected phytochemicals were screened by Lipinski rule of five and ADME, which was followed by determining the bioavailability radar and bioactive score of the ligands. Lipinski’s rule of five checked parameters like molecular weight, hydrogen donor, hydrogen acceptor, lipophilicity, and molar refractivity [17, 18]. Andrograholide, Bassic acid, Curdione, Nectandrin B, Nimbolide, salvicine, Withaferin A, and Hecogenin satisfied all the five criteria of Lipinski; Tetrandine did not pass the criteria while the others have deviation in at most one of the criteria which is still acceptable (Table 6).
Table 6.
Lipinski’s rule of 5
| Phytochemicals | Mass | Hydrogen bond donor | Hydrogen bond acceptor | LOGP | Molar refractivity |
|---|---|---|---|---|---|
| Andrographolide | 350 | 3 | 5 | 1.96 | 93.5 |
| Bassic acid | 486 | 4 | 5 | 4.305 | 135.46 |
| Curdione | 236 | 0 | 2 | 3.55 | 69.73 |
| Nectandrin B | 344 | 2 | 5 | 4.19 | 94.21 |
| Nimbolide | 466 | 0 | 7 | 3.74 | 119.39 |
| Oleanolic acid | 456 | 2 | 3 | 7.233 | 132.68 |
| Panaxadiol | 460 | 2 | 3 | 6.74 | 133.81 |
| Panaxatriol | 476 | 3 | 4 | 5.71 | 135.20 |
| Salvicine | 330 | 2 | 4 | 2.86 | 93.96 |
| Tetrandine | 622 | 0 | 8 | 7.16 | 177.68 |
| Ursolic acid | 456 | 2 | 3 | 7.09 | 132.61 |
| Withaferin A | 470 | 2 | 6 | 3.35 | 124.46 |
| Hecogenin | 430 | 1 | 4 | 4.92 | 118.11 |
| Dovitinib (Standard drug) | 392 | 4 | 6 | 2.23 | 111.83 |
| Gefitinib (Standard drug) | 446.5 | 1 | 7 | 4.28 | 118.15 |
The phytochemicals were screened for ADME properties calculated from SwissADME, a free web tool which predicts and evaluates pharmacokinetics and drug likeness of molecules and built on several models [19]. In silico pharmacokinetics of the ligands as shown in Table 7 reveals that the drug likeness and pharmacokinetics of some of the phytochemicals are comparable to the standard drugs. The estimated solubility (ESOL) showed the phytochemicals curdione, andrographolide, and salvicine to be more soluble than the standard drugs Dovitinib and Gefitinib with lipophilicity comparable to Dovitinib but less than Gefitinib. Lipophilicity expressed as iLogP affects the absorption of drug, the lower the iLOGP value, the higher the absorption, and vice versa. Gastrointestinal absorption (GIA) and blood–brain barrier (BBB) permeation are predicted by BOILED model which utilizes the polarity and lipophilicity of compounds [20]. GIA was high for all the ligands except oleanolic acid and ursolic acid. In line with this, the bioavailability score of all the ligands falls in the normal range except that of ursolic and oleanolic acids. Major therapeutic agents are substrates to p-glycoprotein, which in most cases have the potential to reduce absorption, permeability, oral bioavailability, and retention time of drugs [21, 22]. P-glycoproteins are overly expressed in cancer cells, a major barrier in cancer treatment that causes drug efflux and makes chemotherapy quite ineffective [22, 23]. Hence, ligands that are non-substrate of p-gp are most preferable for cancer treatment.
Table 7.
In silico pharmacokinetics of ligands using Swiss ADME
| Phytochemicals | ESOL (Log S) | GIA | BBB permeant | P-gp substrate | CYP3A4 inhibitor | CYP1A2 inhibitor | (iLOGP) | Bioavailabilty score |
|---|---|---|---|---|---|---|---|---|
| Andrographolide | − 3.18 (S) | High | No | Yes | No | No | 2.45 | 0.55 |
| Bassic acid | − 5.91 (MS) | High | No | Yes | No | No | 3.32 | 0.56 |
| Curdione | − 2.91 (S) | High | Yes | No | No | No | 2.80 | 0.55 |
| Nectandrin B | − 4.42 (MS) | High | Yes | Yes | Yes | No | 2.75 | 0.55 |
| Nimbolide | − 3.94 (S) | High | No | Yes | No | No | 3.51 | 0.55 |
| Oleanolic acid | − 7.32 (PS) | Low | No | No | No | No | 3.92 | 0.85 |
| Panaxadiol | − 6.96 (PS) | High | No | No | No | No | 4.61 | 0.55 |
| Panaxatriol | − 6.22 (PS) | High | No | Yes | No | No | 4.24 | 0.55 |
| Salvicine | − 3.58 (S) | High | Yes | No | No | No | 2.93 | 0.55 |
| Ursolic acid | − 7.23 (PS) | Low | No | No | No | No | 4.01 | 0.85 |
| Withaferin A | − 4.97 (MS) | High | No | Yes | No | No | 3.39 | 0.55 |
| Hecogenin | − 5.55 (MS) | High | Yes | Yes | No | No | 4.02 | 0.55 |
| Dovitinib (Standard drug) | − 3.66 (S) | High | No | Yes | No | Yes | 2.26 | 0.55 |
| Gefitinib (Standard drug) | − 5.05 (MS) | High | Yes | No | Yes | No | 4.04 | 0.55 |
As shown in Table 7, most of the phytochemicals are non-inhibitors of CYP3A4 and CYP1A2, members of drug-metabolizing enzymes cytochrome P450, an enzyme that possesses important role in drug metabolism. The interaction of cytochrome 450 isoenzymes with drug could result in either rapid metabolism when the drug is a substrate of any CYP causing induction or accumulation of the drug when the drug is an inhibitor that causes inhibition, which in both cases are undesirable [24]. Therefore in silico analysis in predicting the interaction of compounds or drugs with CYP isoenzymes is important in drug development process.
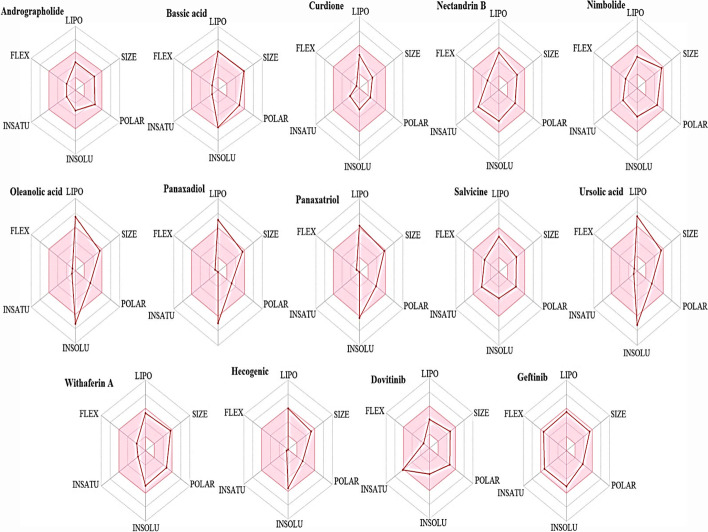
Bioavailability Radar
Bioavailability radar provides a rapid assessment of the drug likeness of a compound. As seen in Fig. 6, the pink area shows the optimal range of each parameter; when considering the parameters of a phytocompound, the radar plot of the compound has to fall in the pink area in order to be considered drug-like; hence, the ligands are either predicted to be orally bioavailable or not orally bioavailable via the radar plot. Flexibility (FLEX) and polarity (polar) are two essential properties that determine the bioavailability of compounds. Flexibility is determined by rotatable bonds; compounds with rotatable bonds > 10 are predicted to have low oral bioavailability while polarity as determined by topological polar surface implies that compound with TPSA > 20 Å2 < 130 Å2 have high oral bioavailability [24]. Seven of the phytochemicals (Andrographolide, Curdione, Nectandrin B, Nimbolide, Salvicine, Withaferin A, and Hecogenic) under study are noted to satisfy the radar plot criteria and hence can be suggested to be orally bioavailable (Fig. 7).
Fig. 7.
Radar plots of ligands
Bioactivity Score
Bioactivity score is used to calculate the drugability properties of ligands such as GPCR, ICM, KI, NRL, PI, and EI. Molinspiration online server was used to predict the scores of the ligands. Scores greater than 0.00 denote high activity, scores ranging from − 0.5 to − 0.00 show moderate activity, and scores lower than − 0.5 imply inactivity [25]. The phytochemicals under study showed a good score of high to moderate activity except for Hecogenic that showed inactivity in KI with a score of − 0.57 as shown in Table 8. For GPCR and NRL, Andrographolide showed the highest score of 0.32 and 0.94, respectively; salivicine showed the highest score of 0.50 for ICM; Withaferin A had the highest score of 0.94 for EI; and Andrographolide and Salivicine both showed a high score of 0.26 for PI. Comparing this to the standard drugs, Dovitinib has good scores for all the properties while Gefitinib showed inactivity in some of the properties as seen in Table 8. Good bioactivity scores revealed the potentials of these bioactives as potent therapeutic agents, the higher the scores, the better the activity.
Table 8.
Bioactivity score of compounds using Molinspiration.com
| Compound | GPCR | ICM | KI | NRL | PI | EI |
|---|---|---|---|---|---|---|
| Andrographolide | 0.32 | 0.17 | − 0.01 | 0.94 | 0.26 | 0.81 |
| Bassic acid | 0.23 | − 0.28 | − 0.35 | 0.79 | 0.16 | 0.66 |
| Curdione | − 0.26 | − 0.01 | − 1.04 | − 0.05 | − 0.040 | − 0.33 |
| Nectandrin B | 0.08 | − 0.16 | − 0.16 | 0.20 | − 0.14 | 0.10 |
| Nimbolide | 0.22 | 0.22 | − 0.36 | 0.32 | 0.04 | 0.36 |
| Oleanolic acid | 0.26 | − 0.06 | − 0.40 | 0.77 | 0.15 | 0.65 |
| Panaxadiol | 0.16 | 0.15 | − 0.26 | 0.54 | 0.19 | 0.66 |
| Panaxatriol | 0.19 | 0.16 | − 0.27 | 0.55 | 0.19 | 0.73 |
| Salvicine | 0.10 | 0.50 | − 0.06 | 0.30 | 0.26 | 0.61 |
| Ursolic acid | 0.28 | − 0.03 | − 0.50 | 0.89 | 0.23 | 0.69 |
| Withaferin A | 0.07 | 0.14 | − 0.49 | 0.76 | 0.15 | 0.94 |
| Hecogenin | 0.05 | 0.04 | − 0.57 | 0.47 | 0.08 | 0.61 |
| Dovitinib (Standard drug) | 0.23 | 0.10 | 0.84 | − 0.01 | − 0.06 | 0.27 |
| Gefitinib (Standard drug) | − 0.18 | − 0.54 | − 0.07 | − 0.62 | − 0.67 | − 0.26 |
Conclusion
In silico study is carried out to explore the potentials of various phytochemicals to inhibit cancer growth and progression through modulation of growth factor receptors. Based on our analysis, seven phytochemicals (Andrographolide, Curdione, Nectandrin B, Nimbolide, Salvicine, Withaferin A, and Hecogenic) possessed drug likeness and pharmacokinetics activities that are comparable with the standard drugs (Dovitinib and Gefitinib). This reveals that bioactive compounds have the ability to bind with GFRs causing inhibition of growth factors which in turn hinders cancer cell proliferation. Further investigation is needed to establish the pharmacodynamics and kinetic properties of these phytochemicals; also, mechanism of action of these phytochemicals as nanoparticle carrier of anticancer drug for effective cancer treatment can be found.
Acknowledgements
The authors are thankful to B.S. Abdur Rahman Institute of Science & Technology, Chennai, for providing research facilities in school of life sciences.
Author Contribution
SH conceived and designed research. ELM performed experiments. All authors wrote the manuscript. All authors read and approved the manuscript.
Funding
The authors also gratefully acknowledge ASEAN University Network (AUN)/Southeast Asia Engineering Education Development Network (SEED)/Japan International Cooperation Agency (JICA) SPRAC (SN042/MI.KU/2020) and Ministry of Science and Technology, Department of Science and Technology (DST/SATYAM/COVID-19/2020/213 (G)).
Data availability
Data will be available on request.
Code Availability
Not applicable.
Declarations
Ethics Approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors read and approved for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thun MJ, DeLancey JO, Center MM, Jemal A, Ward EM. The global burden of cancer: Priorities for prevention. Carcinogenesis. 2010;31(1):100–110. doi: 10.1093/carcin/bgp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choudhari AS, Mandave PC, Deshpande M, Ranjekar P, Prakash O. Phytochemicals in cancer treatment: From preclinical studies to clinical practice. Frontiers in pharmacology. 2020;10:1614. doi: 10.3389/fphar.2019.01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., &Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians, 68(6), 394–424. 10.3322/caac.21492 [DOI] [PubMed]
- 4.Abinaya, M., Priya, S., Karunya, J. R., Ranjani, S., &Hemalatha, S. (2021). Screening the efficacy of compounds from ghee to control cancer: An in silico approach. Biointerface research in applied chemistry11(6) 14115–14126. 10.33263/BRIAC116.1411514126
- 5.Lutfiya AS, Priya S, Manzoor MAP, Hemalatha S. Molecular docking and interactions between vascular endothelial growth factor (VEGF) receptors and phytochemicals: An in-silico study. Biocatalysis and Agricultural Biotechnology. 2019;22:101424. doi: 10.1016/j.bcab.2019.101424. [DOI] [Google Scholar]
- 6.Tiash S, Chowdhury EH. Growth factor receptors: promising drug targets in cancer. Journal of Cancer Metastasis and Treatment. 2015;1:190–200. doi: 10.4103/2394-4722.163151. [DOI] [Google Scholar]
- 7.Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley interdisciplinary reviews. Developmental biology. 2015;4(3):215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennasroune A, Gardin A, Aunis D, Crémel G, Hubert P. Tyrosine kinase receptors as attractive targets of cancer therapy. Critical reviews in oncology/hematology. 2004;50(1):23–38. doi: 10.1016/j.critrevonc.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Sarkar B, Ullah MA, Islam SS, Rahman MH, Araf Y. Analysis of plant-derived phytochemicals as anti-cancer agents targeting cyclin dependent kinase-2, human topoisomerase IIa and vascular endothelial growth factor receptor-2. Journal of receptor and signal transduction research. 2021;41(3):217–233. doi: 10.1080/10799893.2020.1805628. [DOI] [PubMed] [Google Scholar]
- 10.Atanasov AG, Waltenberger B, Pferschy-Wenzig EM, Linder T, Wawrosch C, Uhrin P, Temml V, Wang L, Schwaiger S, Heiss EH, Rollinger JM, Schuster D, Breuss JM, Bochkov V, Mihovilovic MD, Kopp B, Bauer R, Dirsch VM, Stuppner H. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnology advances. 2015;33(8):1582–1614. doi: 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qawoogha SS, Shahiwala A. Identification of potential anticancer phytochemicals against colorectal cancer by structure-based docking studies. Journal of receptor and signal transduction research. 2020;40(1):67–76. doi: 10.1080/10799893.2020.1715431. [DOI] [PubMed] [Google Scholar]
- 12.Anand P, Kunnumakkara AB, Sundaram C, Harikumar KB, Tharakan ST, Lai OS, Sung B, Aggarwal BB. Cancer is a preventable disease that requires major lifestyle changes. Pharmaceutical research. 2008;25(9):2097–2116. doi: 10.1007/s11095-008-9661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boeing H, Bechthold A, Bub A, Ellinger S, Haller D, Kroke A, Leschik-Bonnet E, Müller MJ, Oberritter H, Schulze M, Stehle P, Watzl B. Critical review: Vegetables and fruit in the prevention of chronic diseases. European journal of nutrition. 2012;51(6):637–663. doi: 10.1007/s00394-012-0380-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saklani A, Kutty SK. Plant-derived compounds in clinical trials. Drug Discovery Today. 2008;13(3–4):161–171. doi: 10.1016/j.drudis.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Tan G, Gyllenhaal C, Soejarto DD. Biodiversity as a source of anticancer drugs. Current drug targets. 2006;7(3):265–277. doi: 10.2174/138945006776054942. [DOI] [PubMed] [Google Scholar]
- 16.Fischbach MA, Walsh CT. Directing biosynthesis. Science. 2006;314(5799):603–605. doi: 10.1126/science.1132692. [DOI] [PubMed] [Google Scholar]
- 17.Lipinski CA. Lead- and drug-like compounds: The rule-of-five revolution. Drug discovery today. Technologies. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Jayaram, B., Singh, T., Mukherjee, G., Mathur, A., Shekhar, S., &Shekhar, V. (2012). Sanjeevini: A freely accessible web-server for target directed lead molecule discovery. BMC bioinformatics, 13 Suppl 17(Suppl 17), S7. 10.1186/1471-2105-13-S17-S7 [DOI] [PMC free article] [PubMed]
- 19.Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Report. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daina A, Zoete V. A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11(11):1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin JH, Yamazaki M. Role of P-glycoprotein in pharmacokinetics: Clinical implications. Clinical pharmacokinetics. 2003;42(1):59–98. doi: 10.2165/00003088-200342010-00003. [DOI] [PubMed] [Google Scholar]
- 22.Amin ML. P-glycoprotein Inhibition for optimal drug delivery. Drug target insights. 2013;7:27–34. doi: 10.4137/DTI.S12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bansal, T., Jaggi, M., Khar, R. K., &Talegaonkar, S. (2009). Emerging significance of flavonoids as P-glycoprotein inhibitors in cancer chemotherapy. Journal of pharmacy & pharmaceutical sciences : a publication of the Canadian Society for Pharmaceutical Sciences, Societecanadienne des sciences pharmaceutiques, 12(1), 46–78. 10.18433/j3rc77 [DOI] [PubMed]
- 24.Ji D, Xu M, Udenigwe CC, Agyei D. Physicochemical characterisation, molecular docking, and drug-likeness evaluation of hypotensive peptides encrypted in flaxseed proteome. Current Research in Food Science. 2020;3:41–50. doi: 10.1016/j.crfs.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan T, Dixit S, Ahmad R, Raza S, Azad I, Joshi S, Khan AR. Molecular docking, PASS analysis, bioactivity score prediction, synthesis, characterization and biological activity evaluation of a functionalized 2-butanone thiosemicarbazone ligand and its complexes. Journal of chemical biology. 2017;10(3):91–104. doi: 10.1007/s12154-017-0167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available on request.
Not applicable.