Abstract
The levels of amyloid peptides in the brain are regulated by a clearance pathway from neurons to the blood–brain barrier. The first step is thought to involve diffusion from the plasma membrane to the interstitium. However, amyloid peptides are hydrophobic and avidly intercalate within membranes. The ABC transporter P-glycoprotein is implicated in the clearance of amyloid peptides across the blood–brain, but its role at neurons is undetermined. We here propose that P-glycoprotein mediates ’exit’ of amyloid peptides from neurons. Indeed, amyloid peptides have physicochemical similarities to substrates of P-glycoprotein, but their larger size represents a conundrum. This review probes the plausibility of a mechanism for amyloid peptide transport by P-glycoprotein exploiting evolving biochemical and structural models.
Keywords: ABCB1, Alzheimer’s disease, amyloid peptides, blood–brain barrier, hydrophobic peptides, MDR1, membrane transport, Pgp
Why is Pgp implicated in amyloid peptide clearance?
ABBA’s legendary hit song Mamma Mia has inspired people worldwide to dance for several decades. The two legends starring in this perspectives article, namely Amyloid-β (Aβ)and P-glycoprotein (Pgp, MDR1 and ABCB1), have led scientists in their respective research fields on a ‘merry dance’. Aβ, a key peptide in the pathogenesis of Alzheimer’s disease (AD), is small, sticky, neurotoxic and implicated in memory loss [1]. Pgp, a key ABC transporter at the blood–brain barrier, is highly expressed, extremely potent and a master in handling a variety of substrates protecting the brain from toxic compounds. The potential connection between amyloid peptides and the transporter, however, remained unnoticed for decades.
In 2001, Lam et al. [2] published a landmark research article proposing the unthinkable: ‘the ABC transporter known as MDR1 is an Aβ efflux pump’. Using Pgp-overexpressing HEK293 cells and CHO cell membrane vesicles, they provided evidence that Pgp is involved in the transport of Aβ. This was followed by a well-designed in vivo study by Cirrito et al. [3], demonstrating that knocking out Pgp in a murine AD model elevated brain Aβ levels by 40% and prolonged Aβ brain clearance by 2.2-fold. Using freshly isolated mouse brain capillaries, we found that fluorescent-labelled human Aβ accumulated rapidly in the capillary lumen, indicating active transport from bath to vascular space [4,5]. This accumulation was reduced by the Pgp inhibitors PSC833 (valspodar), ivermectin and cyclosporin A, whereas inhibitors of the breast cancer resistance protein (BCRP, ABCG2) and the multidrug resistance protein (MRP1, ABCC1) were without effect. In 2016, Yuede et al. [6] showed in an AD mouse model that inhibiting Pgp resulted in threefold increased brain Aβ levels and 2.8-fold longer Aβ clearance compared with control mice. Other groups have confirmed these findings, supporting the conclusion that Pgp functions to transport Aβ and facilitates Aβ clearance across brain capillary endothelium [7–9]. Moreover, data from human studies indicate that Pgp and Aβ expression levels are inversely correlated and that loss of blood–brain barrier Pgp is associated with extensive deposition of Aβ in the brains of AD patients [10–17]. Together, these studies provide considerable evidence pointing towards a role for Pgp in the export of Aβ peptides across the blood–brain barrier.
One aspect that has been completely overlooked is the potential role of Pgp in neurons, where the Aβ peptides are generated. Two studies have reported neuronal expression of Pgp in the context of schizophrenia [18,19]; however, its function in neurons and how this relates to Aβ export have never been investigated. The mechanism of Aβ peptide export from neurons is generally presumed to be driven by diffusion. However, the peptides are hydrophobic (in particular, the longer species) and membrane-anchored, opening the door for the involvement of a transporter-mediated process that is performed by Pgp in neurons, in addition to its role at the blood–brain barrier. Once released from neurons, Aβ associates with ApoE and the ApoE-Aβ complex serves as a shuttle between neurons and the endothelial cells [20]. In addition to Pgp, the low-density lipoprotein receptor-related protein 1 (LRP1) has been implicated in receptor-mediated Aβ clearance, whereas the receptor for advanced glycation end products (RAGE) has been shown to mediate receptor-mediated transcytosis of Aβ peptides into the brain (for more information, see [20,21]). Together, a complex picture emerges that likely also involves more transport proteins including other ABC transporters and, as a whole, provides exciting opportunities for new discoveries and therapeutic targets.
While direct data showing that Pgp transports Aβ are missing, the indirect evidence suggesting a role for Pgp in Aβ transport is overwhelming. Despite the existing evidence, the concept that Pgp may transport Aβ peptides remains hard to envisage for most ABC researchers due to the size of these peptides when compared to most established Pgp substrates. In this perspective article, we provide evidence that this indeed is a real possibility.
Do Aβ peptides need a transporter to leave neuronal or endothelial cells?
Diagrams on the mechanism of plaque formation in AD depict the Aβ peptides leaving their neuronal site of synthesis and into the interstitium by a process of diffusion [21]. It is also well accepted that Aβ peptides have a propensity to self-associate into increasingly more complex aggregates or oligomers and this process is driven by their inherent hydrophobicity. The incompatibility between a peptide with high hydrophobicity and a predilection to diffuse out of a membrane surely invokes the old English proverb of ‘having your cake and eating it too’. The hydrophobicity of the Aβ peptides is a clear physicochemical property (as discussed below), which suggests that intercalation within a bilayer represents the lowest free energy state; particularly in comparison with the aqueous sea of the interstitium! Moreover, Aβ peptides do not leave the brain following an arduous ‘diffusive swim’ to the blood-brain barrier since this step involves their packing within apolipoprotein E (ApoE)-containing particles [22]. Aβ peptides also do not just diffuse across the blood–brain barrier to clear themselves from the brain. We argue that extraction of Aβ peptidβs from their site of synthesis in neurons and Aβ clearance across the blood–brain barrier are unlikely to occur by simple diffusion.
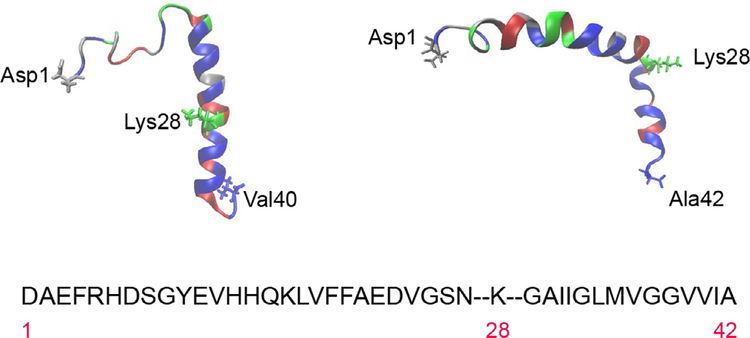
The ‘antidiffusive’ stance is predicated on the physical and chemical properties of Aβ peptides. Figure 1 shows the NMR-derived structures for human Aβ40 (PDB: 1BA4), solved in SDS micelles [23], and human Aβ42 (PDB: 1iYT), solved in aqueous trifluoroethanol [24]. SDS micelles simulate a water-membrane medium, in which Aβ40 is unstructured between residues 1 and 14, with an α-helical conformation from residues 15 to 36 (C terminus) and a kink or hinge at 25–27. Obtaining the structure of Aβ42 eluded many groups for a considerable time, primarily due to the peptide’s hydrophobicity [24], and was only achieved in an organic solvent (hexafluoroisopropanol) that mimicked the lipid bilayer environment. The structure shows two helical regions encompassing residues 8–25 (N terminus) and 28–38 (C terminus), connected by a regular type I β-turn and a relatively rigid ‘elbow’ at the Gln27-Lys28 junction. Residues 28–42 originate from the transmembrane domain of the amyloid precursor protein APP, and the depiction in Fig. 1 reveals the presence of several nonpolar, or hydrophobic, residues and multiple small amino acids (Gly and Ala), frequently associated with transmembrane helices. The only charged residue is lysine 28, found at the N terminus of the short helix. There are numerous investigations reporting the propensity of Aβ peptides to avidly and rapidly intercalate within bilayers, both natural and synthetic, where they aggregate and frequently assemble into pores that conduct metal ions [25–28]. Crescenzi et al. [24] also argued that, from a sequence and structural perspective, the Aβ42 peptide resembled the fusion domain of influenza haemagglutinin that penetrates membranes during viral infection.
Fig. 1.
Amino acid sequence and structure of the Aβ40 and Aβ42 peptides. The single letter code has been used for the sequence, which is divided to highlight lysine 28 for illustrative purposes. Residues have been colour-coded by type for Aβ40 (top left) and Aβ42 (top right). Hydrophobic amino acids are in blue, polar uncharged in red, acidic in silver and basic in green. Structures were obtained from PDB1iyt (Aβ42) and PDB1ba4 (Aβ40).
Using model DMPC liposomes, Ji et al. [29] demonstrated that Aβ peptide interaction with bilayers has a dependence on cholesterol content. At low cholesterol content, Aβ adsorbed to the liposome surface, presumably via electrostatic interactions from the long helix and it was only when the cholesterol content was raised that the short helix penetrated into the bilayer. Intercalation of Aβ peptides into the lipid milieu has profound effects on biophysical properties of membranes including reduced fluidity, altered molecular order, modified lysis tension, bilayer thinning and enhanced conductivity (for reviews, see ref. [30]).
A recent review highlights the intriguing link between APP processing and specific membrane domains [31]. In particular, APP, the secretases and Aβ peptides colocalise within cholesterol- and sphingolipid-rich microdomains, also known as lipid rafts. The physiological purpose of rafts is to ensure containment of interacting proteins within a defined locale, which provides an ideal platform for the complex pathways associated with APP processing.
The preference of Aβ peptides for cholesterol-containing membranes might not simply be a compositional one. Lipid rafts are characterised by their low fluidity, and the inclusion of cholesterol into phospholipid membranes can induce rigid, or highly ordered, domains. Moreover, it has also been shown that other physical properties of bilayers including the torsional effects of leaflet curvature can affect Aβ peptide insertion and aggregation within the lipid milieu [32]. Physical properties of bilayers can also modify the structure adopted by Aβ peptides; for example, the α-helical conformation is preferred in ordered membrane environments in vitro [33–35]. These investigations used circular dichroism and NMR approaches to demonstrate that the β-sheet conformation, which is thought to accelerate aggregation, has a preference for disordered micelle environments and for bilayers with increased negative surface charge.
In summary, the observations detailed in this section reveal a complex interaction between Aβ peptides and biological membranes. The Aβ peptides are formed within the membrane of neuronal cells and avidly associate with bilayer components. Such avidity of association is a clear indication that, from an energetic standpoint, Aβ peptides are unlikely to diffuse into the incompatible environment of the interstitium. Clearly, the peptides must leave neuronal membranes in order to enter the clearance pathway to exit the brain and under pathological conditions form aggregates, fibrils and plaques in the interstitium. This conundrum is at the crux of our hypothesis that leaving the neuronal membrane requires a transporter-mediated step.
Is the transport of Aβ peptides plausible at a molecular level?
There is considerable biological evidence implicating Pgp in the transport of Aβ peptides across the blood-brain barrier, as introduced earlier in this article. We also provided intuitive justification for a role in the transport of Aβ peptides out of the neuronal plasma membrane to the waiting arms of ApoE-containing lipoproteins.
Yet, there remains scepticism, or a cautious concern, regarding the ability of Pgp to transport Aβ peptides [36]. The scepticism from this citation is largely based on an inability to reproduce the earlier observation that Aβ peptides stimulate the ATPase activity of Pgp [2]; an assay oft-used to identify compounds as substrates or inhibitors of the transporter. The relatively modest extent of stimulation (1.5-fold) reported by Lam et al. [2] may simply suggest a low-affinity substrate.
Do the Aβ peptides share properties with known substrates of Pgp? In one respect, the answer is yes, based on the peptide hydrophobicity and the presence of cationic nitrogen group. However, at ~ 4 kD, the peptides are considerably larger than typical substrates such as vinblastine (Mw = 811) and paclitaxel (Mw = 854). Perhaps the largest documented substrate of Pgp is the fluorescent cyclosporine A derivative, BODIPY-FL-cyclosporine, which weighs in at 1.5 kDa [37].
There are a number of structures, and structural models available for Pgp and the inward-facing (high affinity for substrate) conformations all display a large central cavity that functions as a binding site for some drugs and/or the internal conduit for transport. A key issue to resolve is whether the Aβ peptides are able to fit into this transport nook. To test this hypothesis, we have undertaken molecular docking using the 4m1m structural model of Pgp [38,39]. For nonstructural biologists, it is important to note that any solved protein structure is a snapshot for one of several conformations adopted by the protein in the translocation sequence. The open, inward-facing Pgp conformation was chosen since it represents the initial state that is capable of high-affinity substrate binding.
It is also important to consider that the Aβ peptides vary in conformation under different conditions and are intrinsically unstructured in vivo [40]. Replica exchange molecular dynamics studies suggested that Aβ40 and Aβ42 assume multiple discrete conformations, comprising α-helix or β-sheet conformers, and the structural states transition rapidly. In water, the peptide collapses into a compact series of loops, strands and turns without alpha-helical or beta-sheet structure, but helical structures are more common in a lipidic environment (reviewed in ref. [41]).
Our docking strategy used the 1ba4 and 1iyt structures of Aβ40 and Aβ42, respectively, since these are perhaps the two most accurate NMR structures among many conformers. The 4m1m-corrected PDB structure of mouse Pgp (Mdr1a; [42]) was used for docking since this version has been validated as the most appropriate representation [38]. We used the docking software program CLUSPRO 2.0 ([43]), and PYMOL (Molecular Graphics System, v2.4.0 Schrodinger) to render the figures. The protein–protein docking server CLUSPRO 2.0 performs three computational steps: (a) rigid body docking; (b) RMSD-based clustering of the 1000 lowest energy structures; and (c) removal of steric clashes by energy minimisation. We set up separate docking jobs with both Aβ peptides and Pgp, and these were set up and run as replicates (n = 4) to reveal consistencies or deviations in the docked molecules. For each job, we examined the best 10 output docking modes. Outputs for the four replicate runs were very similar for both peptides docked to Pgp (see Supplementary File for docking method and parameters).
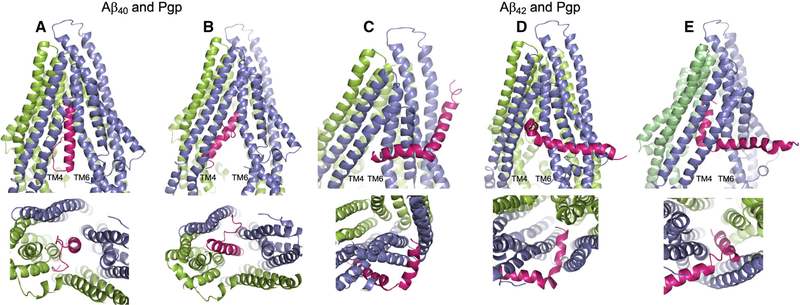
Aβ40 docked to Pgp in a number of partially to fully docked associations, the majority within the central cavity and several ‘clinging’ to the annular face of TM helices. Figure 2 depicts Aβ40 wholly within the inward-facing Pgp cavity, but in two different orientations: the first showing the peptide vertically within the Pgp cavity and with the nonpolar, helical C-terminal end uppermost (Fig. 2A); and the second showing the peptide at an angle to the vertical, still wholly within the Pgp cavity, and with the polar N-terminal random coil uppermost (Fig. 2B). This second docking mode is shown only to illustrate that the reverse docking is possible in silico, but not in vivo.
Fig. 2.
Docking of the Aβ40 and Aβ42 peptides to Pgp. The server CLUSPRO 2.0 was used to dock amyloid peptides Aβ40 (PDB: 1BA4) and Aβ42 (PDB: 1iYT) to Pgp (PDB: 4m1m). (A–E) Are side (upper) and bottom (lower) views of either peptide (hot pink) docked to Pgp (slate blue and chartreuse green). Pgp is represented with the domain swapped TM helices 4 and 5 (blue) shown at the left of each upper panel; and the opposite domain swapped TMs 10 and 11 (green) at the back right of each side view panel. (A) Aβ40 docked to Pgp with the nonpolar, helical end of the peptide uppermost in the Pgp cavity; and (B) Aβ40 docked to Pgp with the polar, random coil end of the peptide uppermost in the Pgp cavity. (C–E) Aβ42 docked to Pgp with the peptide at different stages of docking with Pgp. In each of the three docked modes, Aβ42 engages Pgp in the same orientation. (C) The short, nonpolar, C-terminal helix at the Pgp ‘gate’ between TM4 and TM6; (D) the Aβ42 helix within the Pgp cavity and at right angles to the Pgp TM helices; and (E) the helix wholly within the Pgp cavity and parallel to the Pgp TM helices. The figure was rendered with PYMOL (Molecular Graphics System, v2.4.0 Schrodinger). Standard settings were used, and no additional features such as attraction or repulsion were used. Scores for all docking modes were almost identical.
Aβ42 docking to Pgp was more complex, and a significant number of output modes (4/10) depicted the peptide, as shown in Fig. 2C–E, in three possibly progressive frames, all with the nonpolar C-terminal helix as the leading end of the peptide. The shorter of the two helices (residues 28–38) was poised at the cavity/gate between TMs 4 and 6 (Fig. 2C), then just within the cavity with the helix at right angles to the Pgp TM helices (Fig. 2D), and lastly with the short helix fully within the cavity and repositioned parallel to the Pgp TM helices (Fig. 2E), and seemingly interacting with the top portion of TM12. In these frames, the longer, trailing helix was fully outside or with the first one or two residues just inside the gap between TMs 4 and 6. A small number of output modes were associated with peptide docking within the annular region of the protein and may represent initial binding prior to entry into the cavity (not shown). However, the number of output dockings with Aβ42 partly at or within the TM4–6 gate was consistently observed as the major configuration. The long trailing helix might well enter the Pgp cavity fully if the two-helix conformation changed to a helix–coil – as seen with Aβ40 – which is plausible given that Aβ peptides are known to transition between different conformations depending of the nature of their environment [39], as discussed above.
There is considerable biochemical evidence to support the initial binding of Pgp substrates directly from the bilayer milieu [44,45] and the presence of a binding site at the protein annular region [46–49]. Similarly, there is also evidence for substrate binding exclusively within the central cavity of Pgp [50–52]. The likely localisation of Aβ peptides, following cleavage from APP, is in the outer leaflet of the neuronal membrane. Does this have consequences for interaction with Pgp? There is no consensus as to the precise access point of substrates from the lipid milieu; for example, this may occur from a specific hemi-leaflet or both. The Aβ peptides contain several charged amino acids, which would suggest they are most likely to approach the binding site, gate or central cavity from the outer leaflet. Access via the inner leaflet would be problematic given the energetic cost to flip between leaflets. Elucidating such issues such as this will require experimental validation. In summary, the docking data shown in Fig. 2 indicate that the two amyloid peptides can interact with Pgp. That the interaction occurs at distinct sites may propose distinct mechanisms for their subsequent translocation across the membrane.
Does a potential transport mechanism involve gates or zippers?
In the mid-1960s, Jardetzky published a simple mechanistic model for membrane transporters [53]. Our understanding of transporters from both a biochemical and a structural perspective has advanced significantly in the intervening years, yet a significant proportion of the mechanism proposed by Jardetzky remains valid. Considerable biochemical work in the period 1990-2005 extended the principles of this simple model to generate the mechanism used by Pgp to mediate substrate transport [51,54–58]. Substrates enter the central cavity and bind to one of the multiple pharmacological sites, which triggers the nucleotide binding domains to dimerise and ensure tight ATP binding. The binding energy triggers an outward orientation of the central cavity, the substrate is released, and energy from ATP hydrolysis resets the transporter for a subsequent cycle. This sequence of events has been termed the ‘alternating access model’ [59,60]. The more recent advent of structural models in multiple conformational states for Pgp, and other ABC proteins, has largely confirmed the biochemical data [50,61–64]. One caveat of this model remains how substrates enter the cavity and precisely where the binding sites are localised. A considerable amount of early evidence revealed that substrates clearly ‘accessed’ Pgp from the lipid milieu [44,45]. Subsequently, cross-linking and mutagenesis data provided convincing evidence for an initial interaction between drugs and Pgp at the lipid annulus [47,65]. This initial binding has become referred to as a binding gate that provides entry to the central cavity or translocation conduit [46,47,66].
In recent years, structural and biochemical data on two prokaryotic ABC proteins that transport large substrates have proffered variations to this alternating access mechanism and will have implications for Aβ peptides. The first is Pglk from Campylobacter jejuni, which is believed to mediate the flipping of a lipid-linked oligosaccharide (LLO) between leaflets of the inner membrane [67]. LLO is subsequently used to glycosylate proteins. Structural data suggest that the pyrophosphate moiety of LLO is recognised by a series of positively charged residues on Pglk, located at the cytosolic surface of the inner membrane leaflet [67,68]. This highly selective initial binding was termed ‘substrate hunting’ since it facilitates the interaction of LLO from a vast sea of lipids in the membrane. The complete LLO macromolecule is thought to be too large to fit within the confines of the Pglk central cavity, which precludes a conventional transport mechanism. It is believed that the lipid polyprenyl tail of LLO is dragged through the lipid milieu at the Pglk annulus, while the oligosaccharide passes through the central conduit. To account for the large size of the oligosaccharide, the authors suggested that Pglk remains in an outward open configuration throughout the translocation process since the alternating switch described above would be constricted by substrate. One may depict this transport process as a zipper mechanism that operates along the gate of Pglk.
The Wzm/Wzt protein also transports a large substrate that is not compatible with the simple alternating access model, as discussed in a structural investigation [69]. The substrate is polysaccharide (O-antigen), which is ultimately attached to lipid A on the outer membrane of Gram-negative bacteria in order to evade the host immune system. The O-antigen is transported as an undecaprenyl-phosphate derivative across the inner membrane. The postulated transport mechanism is initiated following binding of the phosphorylated isoprenoid component at a cytosolic gate. This group enters the cavity and is spontaneously flipped to the periplasmic face of Wzm/Wzt. This movement drags part of the polysaccharide segment into the cavity – the entire molecule is too large to fit. Translocation is achieved by multiple successive rounds of ATP hydrolysis to pull the saccharide through in a processive manner that may be likened to a ratchet mechanism. Once the polysaccharide is translocated, the isoprenoid anchor dissociates from Wzm/Wzt. The ratchet mechanism also requires an extended opening time for the central cavity, rather than the rapid orientation switch implicit within the alternating access mechanism.
The models certainly provide multiple possible mechanisms to transport this cargo, and our docking observations reveal that interaction between Pgp and the Aβ peptides is possible. What are the implications of these distinct mechanisms for the transport of Aβ peptides by Pgp? Transport of Aβ40 according to the alternating access model may require an extended opening time to ensure the long helix is fully translocated. Certainly, the initial interaction of Aβ42 utilises a binding gate access; but does the long helix enter the cavity? If the long helix remains localised at the annulus, then an alternating access mechanism might ‘pinch’ the peptide. Aspects of the ratchet and zipper models provide features that may be desirable to transport Aβ peptides. It is important to note that there is no current precedent for a ratchet- or zipper-like mechanism for Pgp. Our intention is to present ideas to formulate a mechanism for Aβ peptides and to demonstrate that their transport by Pgp is indeed conceivable from a biochemical perspective. The key next step is to develop experimental systems to directly demonstrate transport and elucidate a molecular mechanism. Only ‘by taking a chance’ to provide this information will we determine whether the immortal lyric ‘my, my how can I resist you’ applies to Pgp and Aβ peptides.
Supplementary Material
Acknowledgements
The project was supported by grant number 2R01AG039621 from the National Institute on Aging (to AMSH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging or the National Institutes of Health.
Abbreviations
- ABC
ATP-binding cassette
- AD
Alzheimer’s disease
- ApoE
apolipoprotein E
- Aβ
Amyloid beta peptide
- BCRP
breast cancer resistance protein
- LLO
lipid-linked oligosaccharide
- MRP1
multidrug resistance protein
- Pgp
P-glycoprotein.
Footnotes
Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Supplementary File.
References
- 1.Hardy J and Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297, 353–356. [DOI] [PubMed] [Google Scholar]
- 2.Lam FC, Liu R, Lu P, Shapiro AB, Renoir JM, Sharom FJ and Reiner PB (2001) beta-Amyloid efflux mediated by p-glycoprotein. J Neurochem 76, 1121–1128. [DOI] [PubMed] [Google Scholar]
- 3.Cirrito JR, Deane R, Fagan AM, Spinner ML, Parsadanian M, Finn MB, Jiang H, Prior JL, Sagare A, Bales KR et al. , (2005) P-glycoprotein deficiency at the blood-brain barrier increases amyloid-beta deposition in an Alzheimer disease mouse model. J Clin Invest 115, 3285–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartz AM, Miller DS and Bauer B (2010) Restoring blood-brain barrier P-glycoprotein reduces brain amyloid-beta in a mouse model of Alzheimer’s disease. Mol Pharmacol 77, 715–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartz AM, Zhong Y, Wolf A, LeVine H 3rd, Miller DS and Bauer B (2016) A beta40 reduces P-glycoprotein at the blood-brain barrier through the ubiquitin-proteasome pathway. J Neurosci 36, 1930–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuede CM, Lee H, Restivo JL, Davis TA, Hettinger JC, Wallace CE, Young KL, Hayne MR, Bu G, Li C-Z and et al. , (2016) Rapid in vivo measurement of β-amyloid reveals biphasic clearance kinetics in an Alzheimer’s mouse model. J Exp Med 213, 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruckmann S, Brenn A, Grube M, Niedrig K, Holtfreter S, von Bohlen und Halbach O, Groschup M, Keller M and Vogelgesang S (2017) Lack of P-glycoprotein results in impairment of removal of beta-amyloid and increased intraparenchymal cerebral amyloid angiopathy after active immunization in a transgenic mouse model of Alzheimer’s disease. Curr Alzheimer Res. 14, 656–667. [DOI] [PubMed] [Google Scholar]
- 8.Kuhnke D, Jedlitschky G, Grube M, Krohn M, Jucker M, Mosyagin I, Cascorbi I, Walker LC, Kroemer HK, Warzok RW and et al. , (2007) MDR1-P-glycoprotein (ABCB1) mediates transport of Alzheimer’s amyloid-beta peptides–implications for the mechanisms of Abeta clearance at the blood-brain barrier. Brain Pathol 17, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JD, el Khafagy S, Khanafer K, Takayama S and ElSayed ME (2016) Organization of endothelial cells, pericytes, and astrocytes into a 3D microfluidic in vitro model of the blood-brain barrier. Mol Pharm 13, 895–906. [DOI] [PubMed] [Google Scholar]
- 10.Carrano A, Snkhchyan H, Kooij G, van der Pol S, van Horssen J, Veerhuis R, Hoozemans J, Rozemuller A and de Vries HE (2014) ATP-binding cassette transporters P-glycoprotein and breast cancer related protein are reduced in capillary cerebral amyloid angiopathy. Neurobiol Aging 35, 565–575. [DOI] [PubMed] [Google Scholar]
- 11.Chiu C, Miller MC, Monahan R, Osgood DP, Stopa EG and Silverberg GD (2015) P-glycoprotein expression and amyloid accumulation in human aging and Alzheimer’s disease: preliminary observations. Neurobiol Aging 36, 2475–2482. [DOI] [PubMed] [Google Scholar]
- 12.Deo AK, Borson S, Link JM, Domino K, Eary JF, Ke B, Richards TL, Mankoff DA, Minoshima S, O’Sullivan F et al. , (2014) Activity of P-glycoprotein, a beta-amyloid transporter at the blood-brain barrier, Is compromised in patients with mild Alzheimer disease. J Nucl Med. 55, 1106–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartz AMS, Zhong Y, Shen AN, Abner EL and Bauer B (2018) Preventing P-gp ubiquitination lowers abeta brain levels in an Alzheimer’s disease mouse model. Front Aging Neurosci 10, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeynes B and Provias J (2011) An investigation into the role of P-glycoprotein in Alzheimer’s disease lesion pathogenesis. Neurosci Lett 487, 389–393. [DOI] [PubMed] [Google Scholar]
- 15.Kannan P, Schain M, Kretzschmar WW, Weidner L, Mitsios N, Gulyas B, Blom H, Gottesman MM, Innis RB, Hall MD and et al. , (2017) An automated method measures variability in P-glycoprotein and ABCG2 densities across brain regions and brain matter. J Cereb Blood Flow Metab 37, 2062–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Assema DM, Lubberink M, Bauer M, van der Flier WM, Schuit RC, Windhorst AD, Comans EF, Hoetjes NJ, Tolboom N, Langer O et al. , (2012) Blood-brain barrier P-glycoprotein function in Alzheimer’s disease. Brain 135, 181–189. [DOI] [PubMed] [Google Scholar]
- 17.Wijesuriya HC, Bullock JY, Faull RL, Hladky SB and Barrand MA (2010) ABC efflux transporters in brain vasculature of Alzheimer’s subjects. Brain Res 1358, 228–238. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein HG, Hildebrandt J, Dobrowolny H, Steiner J, Bogerts B and Pahnke J (2016) Morphometric analysis of the cerebral expression of ATP-binding cassette transporter protein ABCB1 in chronic schizophrenia: Circumscribed deficits in the habenula. Schizophr Res 177, 52–58. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein HG, Holzl G, Dobrowolny H, Hildebrandt J, Trubner K, Krohn M, Bogerts B and Pahnke J (2014) Vascular and extravascular distribution of the ATP-binding cassette transporters ABCB1 and ABCC1 in aged human brain and pituitary. Mech Ageing Dev 141–142, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf A, Bauer B and Hartz A (2012) ABC Transporters and the Alzheimer’s disease enigma. Front Psychiatry 3, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chai AB, Leung GKF, Callaghan R and Gelissen IC (2020) P-glycoprotein: a role in the export of amyloid-beta in Alzheimer’s disease? FEBS J 287, 612–625. [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki Y, Zhao N, Caulfield TR, Liu CC and Bu G (2019) Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat Rev Neurol 15, 501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coles M, Bicknell W, Watson AA, Fairlie DP and Craik DJ (1998) Solution structure of amyloid beta-peptide(1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry 37, 11064–11077. [DOI] [PubMed] [Google Scholar]
- 24.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D’Ursi AM, Temussi PA and Picone D (2002) Solution structure of the Alzheimer amyloid beta-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur J Biochem 269, 5642–5648. [DOI] [PubMed] [Google Scholar]
- 25.Bode DC, Baker MD and Viles JH (2017) Ion channel formation by amyloid-beta42 oligomers but not amyloid-beta40 in cellular membranes. J Biol Chem 292, 1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koo EH, Lansbury PT and Kelly JW (1999) Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc Natl Acad Sci USA 96, 9989–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngo ST, Derreumaux P and Vu VV (2019) Probable transmembrane amyloid alpha-helix bundles capable of conducting Ca(2+) ions. J Phys Chem B 123, 2645–2653. [DOI] [PubMed] [Google Scholar]
- 28.ärmländer SKTS, Österlund N, Wallin C, Wu J, Luo J, Tiiman A, Jarvet J and Gräslund A (2019) Metal binding to the amyloid-β peptides in the presence of biomembranes: potential mechanisms of cell toxicity. J Biol Inorg Chem 24, 1189–1196. [DOI] [PubMed] [Google Scholar]
- 29.Ji SR, Wu Y and Sui SF (2002) Cholesterol is an important factor affecting the membrane insertion of beta-amyloid peptide (A beta 1–40), which may potentially inhibit the fibril formation. J Biol Chem 277, 6273–6279. [DOI] [PubMed] [Google Scholar]
- 30.Askarova S, Yang X and Lee JC (2011) Impacts of membrane biophysics in Alzheimer’s disease: from amyloid precursor protein processing to abeta Peptide-induced membrane changes. Int J Alzheimers Dis 2011, 134971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabiani C and Antollini SS (2019) Alzheimer’s disease as a membrane disorder: spatial cross-talk among beta-amyloid peptides, nicotinic acetylcholine receptors and lipid rafts. Front Cell Neurosci 13, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terakawa MS, Lin Y, Kinoshita M, Kanemura S, Itoh D, Sugiki T, Okumura M, Ramamoorthy A and Lee YH (2018) Impact of membrane curvature on amyloid aggregation. Biochim Biophys Acta Biomembr 1860, 1741–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bokvist M, Lindström F, Watts A and Gröbner G (2004) Two types of Alzheimer’s b-amyloid (1–40) peptide membrane interactions: aggregation preventing transmembrane anchoring versus accelerated surface fibril formation. J Mol Biol 335, 1039–1049. [DOI] [PubMed] [Google Scholar]
- 34.Liu D, Xu Y, Feng Y, Liu H, Shen X, Chen K, Ma J and Jiang H (2006) Inhibitor discovery targeting the intermediate structure of b-Amyloid peptide on the conformational transition pathway: implications in the aggregation mechanism of b-amyloid peptide. Biochemistry 45, 10963–10972. [DOI] [PubMed] [Google Scholar]
- 35.Mandal P and Pettegrew J (2005) Alzheimer’s disease: soluble oligomeric Abeta(1–40) peptide in membrane mimic environment from solution NMR and circular dichroism studies. Neurochem Res 29, 2267–2272. [DOI] [PubMed] [Google Scholar]
- 36.Bello I and Salerno M (2015) Evidence against a role of P-glycoprotein in the clearance of the Alzheimer’s disease Aβ1–42 peptides. Cell Stress Chaperones 20, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sajid A, Raju N, Lusvarghi S, Vahedi S, Swenson RE and Ambudkar SV (2019) Synthesis and characterization of bodipy-FL-cyclosporine A as a substrate for multidrug resistance-linked P-glycoprotein (ABCB1). Drug Metab Dispos 47, 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carey Hulyer AR, Briggs DA, O’Mara ML, Kerr ID, Harmer JR and Callaghan R (2020) Cross-linking, DEER-spectroscopy and molecular dynamics confirm the inward facing state of P-glycoprotein in a lipid membrane. J Struct Biol 211, 107513. [DOI] [PubMed] [Google Scholar]
- 39.Condic-Jurkic K, Subramanian N, Mark AE and O’Mara ML (2018) The reliability of molecular dynamics simulations of the multidrug transporter P-glycoprotein in a membrane environment. PLoS One 13, e0191882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang M and Teplow DB (2008) Amyloid beta-protein monomer folding: free-energy surfaces reveal alloformspecific differences. J Mol Biol 384, 450–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen GF, Xu TH, Yan Y, Zhou YR, Jiang Y, Melcher K and Xu HE (2017) Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol Sin 38, 1205–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Jaimes KF and Aller SG (2014) Refined structures of mouse P-glycoprotein. Protein Sci 23,34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D and Vajda S (2017) The ClusPro web server for protein-protein docking. Nat Protoc 12, 255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Homolya L, Hollo Z, Germann UA, Pastan I, Gottesman MM and Sarkadi B (1993) Fluorescent cellular indicators are extruded by the multidrug resistance protein. J Biol Chem 268, 21493–21496. [PubMed] [Google Scholar]
- 45.Raviv Y, Pollard HB, Bruggemann EP, Pastan I and Gottesman MM (1990) Photosensitized labeling of a functional multidrug transporter in living drug-resistant tumor cells. J Biol Chem 265, 3975–3980. [PubMed] [Google Scholar]
- 46.Crowley E and Callaghan R (2010) Multidrug efflux pumps: drug binding – gates or cavity? FEBS J 277, 530–539. [DOI] [PubMed] [Google Scholar]
- 47.Loo TW and Clarke DM (2005) Do drug substrates enter the common drug-binding pocket of P-glycoprotein through "gates"? Biochem Biophys Res Commun 329, 419–422. [DOI] [PubMed] [Google Scholar]
- 48.Ecker GF, Csaszar E, Kopp S, Plagens B, Holzer W, Ernst W and Chiba P (2002) Identification of ligand-binding regions of P-glycoprotein by activated-pharmacophore photoaffinity labeling and matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Mol Pharmacol 61, 637–648. [DOI] [PubMed] [Google Scholar]
- 49.Mittra R, Pavy M, Subramanian N, George AM, O’Mara ML, Kerr ID and Callaghan R (2017) Location of contact residues in pharmacologically distinct drug binding sites on P-glycoprotein. Biochem Pharmacol 123,19–28. [DOI] [PubMed] [Google Scholar]
- 50.Alam A, Kung R, Kowal J, McLeod RA, Tremp N, Broude EV, Roninson IB, Stahlberg H and Locher KP (2018) Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1. Proc Natl Acad Sci USA 115, E1973–E1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loo TW and Clarke DM (2002) Location of the rhodamine-binding site in the human multidrug resistance P-glycoprotein. J Biol Chem 277, 44332–44338. [DOI] [PubMed] [Google Scholar]
- 52.Loo TW and Clarke DM (2015) Mapping the binding site of the inhibitor tariquidar that stabilizes the first transmembrane domain of P-glycoprotein. J Biol Chem 290, 29389–29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jardetzky O (1966) Simple allosteric model for membrane pumps. Nature 211, 969–970. [DOI] [PubMed] [Google Scholar]
- 54.al-Shawi MK and Senior AE (1993) Characterization of the adenosine triphosphatase activity of Chinese hamster P-glycoprotein. J Biol Chem 268, 4197–4206. [PubMed] [Google Scholar]
- 55.Liu R and Sharom FJ (1998) Proximity of the nucleotide binding domains of the P-glycoprotein multidrug transporter to the membrane surface: a resonance energy transfer study. Biochemistry 37, 6503–6512. [DOI] [PubMed] [Google Scholar]
- 56.Martin C, Berridge G, Higgins CF, Mistry P, Charlton P and Callaghan R (2000) Communication between multiple drug binding sites on P-glycoprotein. Mol Pharmacol 58, 624–632. [DOI] [PubMed] [Google Scholar]
- 57.Sauna ZE and Ambudkar SV (2000) Evidence for a requirement for ATP hydrolysis at two distinct steps during a single turnover of the catalytic cycle of human P-glycoprotein. Proc Natl Acad Sci USA 97, 2515–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urbatsch IL, Tyndall GA, Tombline G and Senior AE (2003) P-glycoprotein catalytic mechanism: studies of the ADP-vanadate inhibited state. J Biol Chem 278, 23171–23179. [DOI] [PubMed] [Google Scholar]
- 59.van Wonderen JH, McMahon RM, O’Mara ML, McDevitt CA, Thomson AJ, Kerr ID, MacMillan F and Callaghan R (2014) The central cavity of ABCB1 undergoes alternating access during ATP hydrolysis. FEBS J 281, 2190–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verhalen B, Dastvan R, Thangapandian S, Peskova Y, Koteiche HA, Nakamoto RK, Tajkhorshid E and McHaourab HS (2017) Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature 543, 738–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL and et al. , (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323, 1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dawson RJ and Locher KP (2007) Structure of the multidrug ABC transporter Sav 1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett 581, 935–938. [DOI] [PubMed] [Google Scholar]
- 63.Frank GA, Shukla S, Rao P, Borgnia MJ, Bartesaghi A, Merk A, Mobin A, Esser L, Earl LA, Gottesman MM et al. , (2016) Cryo-EM analysis of the conformational landscape of human P-glycoprotein (ABCB1) during its catalytic cycle. Mol Pharmacol 90, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Locher KP, Lee AT and Rees DC (2002) The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296, 1091–1098. [DOI] [PubMed] [Google Scholar]
- 65.Romsicki Y and Sharom FJ (1999) The membrane lipid environment modulates drug interactions with the P-glycoprotein multidrug transporter. Biochemistry 38, 6887–6896. [DOI] [PubMed] [Google Scholar]
- 66.Pleban K, Kopp S, Csaszar E, Peer M, Hrebicek T, Rizzi A, Ecker GF and Chiba P (2005) P-glycoprotein substrate binding domains are located at the transmembrane domain/transmembrane domain interfaces: a combined photoaffinity labeling-protein homology modeling approach. Mol Pharmacol 67, 365–374. [DOI] [PubMed] [Google Scholar]
- 67.Perez C, Gerber S, Boilevin J, Bucher M, Darbre T, Aebi M, Reymond JL and Locher KP (2015) Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438. [DOI] [PubMed] [Google Scholar]
- 68.Perez C, Mehdipour AR, Hummer G and Locher KP (2019) Structure of outward-facing PglK and molecular dynamics of lipid-linked oligosaccharide recognition and translocation. Structure 27, 669–678.e5. [DOI] [PubMed] [Google Scholar]
- 69.Bi Y, Mann E, Whitfield C and Zimmer J (2018) Architecture of a channel-forming O-antigen polysaccharide ABC transporter. Nature 553, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.