Abstract
Fibroblast growth factors (FGFs) comprise a large family of multifunctional, heparin-binding polypeptides that show diverse patterns of interaction with a family of receptors (FGFR1 to -4) that are subject to alternative splicing. FGFR binding specificity is an essential mechanism in the regulation of FGF signaling and is achieved through primary sequence differences among FGFs and FGFRs and through usage of two alternative exons, IIIc and IIIb, for the second half of immunoglobulin-like domain 3 (D3) in FGFRs. While FGF4 binds and activates the IIIc splice forms of FGFR1 to -3 at comparable levels, it shows little activity towards the IIIb splice forms of FGFR1 to -3 as well as towards FGFR4. To begin to explore the structural determinants for this differential affinity, we determined the crystal structure of FGF4 at a 1.8-Å resolution. FGF4 adopts a β-trefoil fold similar to other FGFs. To identify potential receptor and heparin binding sites in FGF4, a ternary FGF4-FGFR1-heparin model was constructed by superimposing the FGF4 structure onto FGF2 in the FGF2-FGFR1-heparin structure. Mutation of several key residues in FGF4, observed to interact with FGFR1 or with heparin in the model, produced ligands with reduced receptor binding and concomitant low mitogenic potential. Based on the modeling and mutational data, we propose that FGF4, like FGF2, but unlike FGF1, engages the βC′-βE loop in D3 and thus can differentiate between the IIIc and IIIb splice isoforms of FGFRs for binding. Moreover, we show that FGF4 needs to interact with both the 2-O- and 6-O-sulfates in heparin to exert its optimal biological activity.
The fibroblast growth factor (FGF) family consists of 22 polypeptides (FGF1 to-22) with diverse biological activities (16, 21, 41). FGFs modulate proliferation and differentiation of a variety of cells of mesenchymal and neuroectodermal origin (1). FGFs play critical roles during embryonic processes such as mesoderm induction, postimplantation blastocyst development, and limb and lung development (7, 40). Increased FGF signaling leads to a variety of human skeletal disorders, including dwarfism and craniosynostosis syndromes (15, 19, 39). In adult organisms, FGFs are thought to be involved in physiological angiogenesis and wound healing as well as in pathological angiogenesis, such as in tumor neovascularization and diabetic retinopathy (1).
The diverse effects of FGFs are mediated by four receptor tyrosine kinases, FGFR1 to -4, which are composed of an extracellular ligand binding portion consisting of three immunoglobulin (Ig)-like domains (D1 to -3), a single transmembrane helix, and a cytoplasmic portion with protein tyrosine kinase activity. Ligand binding and specificity reside in D2, D3, and the short D2-D3 linker (29, 30, 34).
Receptor dimerization is a prerequisite for FGF signaling and requires heparin or heparan sulfate proteoglycans (HSPGs) (22, 31). The recent crystal structure of a ternary FGF2-FGFR1-heparin complex has provided a mechanistic view of the process by which heparin aids FGFs to induce FGFR dimerization (32). According to the proposed “two-end” model, heparin interacts via its nonreducing end with the heparin binding sites of FGF and FGFR and promotes the formation of a ternary 1:1:1 FGF-FGFR-heparin complex. A second ternary 1:1:1 FGF-FGFR-heparin complex is then recruited to the first ternary complex via interactions of FGFR, FGF, and heparin in one ternary complex with the FGFR in the adjoining ternary complex. A fundamentally different model has emerged from the recent crystal structure of a dimeric FGF1-FGFR2-heparin ternary complex (27). In this structure, a single heparin molecule links two FGF ligands into a dimer that bridges between two receptor chains. The asymmetric heparin binding involves contacts with both FGF molecules, but only one receptor chain. There is essentially no protein-protein interface between the two 1:1 FGF-FGFR complexes in the dimer.
With the exception of FGF1, which is the universal ligand for all FGFRs, most FGFs exhibit specific, albeit promiscuous, patterns of receptor binding affinity (23). Comparison of the crystal structures of FGF1-FGFR1, FGF2-FGFR1, and FGF2-FGFR2 complexes defined a general binding interface for FGF-FGFR complexes that involves contacts made by FGF with D2 and with the D2-D3 linker (30). It was also shown that specificity is achieved through interactions of the FGF N-terminal (immediately preceding the FGF's β-trefoil core domain) and central regions with FGFR D3. These structures have also provided a molecular basis for how alternative splicing in FGFR modulates specificity. In both FGF2-FGFR1 and FGF2-FGFR2 structures, FGF2 makes specific contacts with the βC′-βE loop in D3, which is subject to alternative splicing. Consequently, FGF2 discriminates between the IIIc and IIIb variants of FGFRs. In contrast, FGF1 does not interact with the βC′-βE loop and therefore can bind all FGFRs irrespective of alternative splicing in D3 (30).
FGF4 shares about 30% sequence identity with the prototypical members of the FGF family, FGF1 and FGF2 (4). FGF4, unlike FGF1 and FGF2, has a classical signal peptide and thus is efficiently secreted from cells (2). Most receptor binding studies indicate that FGF4 binds and activates the IIIc splice forms of FGFR1 to -3 to comparable levels, but it shows little activity towards the IIIb splice forms of FGFR1 to -3 as well as towards FGFR4 (23, 36). As for FGF1 and FGF2, heparin greatly augments the biological activity of FGF4 on cells lacking endogenous cell surface HSPG (14). However, employing selectively O-desulfated heparins, Guimond et al. (8) showed that both 2-O- and 6-O-desulfated heparin were able to support the mitogenic activity of FGF4, while neither of these heparins could support the biological activity of FGF1 and FGF2. It has been suggested the sulfation motifs in heparin required for FGF4 activity may differ from those required for the actions of FGF1 and FGF2 (8, 9).
To explore the structural determinants of FGF4 involved in receptor and heparin binding, we determined the crystal structure of FGF4 at a 1.8-Å resolution. As anticipated, FGF4 adopts a β-trefoil fold similar to those of other FGFs. Superimposition of FGF4 structure onto FGF2 bound to FGFR1 and heparin allowed us to identify potential receptor and heparin binding sites in FGF4. Mutation of several key FGF4 residues, observed to interact with FGFR1 in the model, produced ligands with reduced receptor binding and extremely low mitogenic potential. Significantly, the observed interactions between FGF4 and FGFR1 D3 provide a potential basis for preferential affinity of FGF4 towards IIIc splice variants of FGFR1 to -3. Moreover, the presented modeling studies along with mutational data suggest a two-step model for FGF-FGFR binding that involves initial formation of a crucial FGF-D2 interface stabilized by heparin binding followed by secondary FGF-D3 interactions.
MATERIALS AND METHODS
Protein expression and purification of FGF4.
DNA fragments generated by PCR of N-terminally truncated human FGF4 cDNA (encoding residues Gly-79 to Leu-206 [Gly79-Leu206]) were subcloned into the pET-15b bacterial expression vector by using NcoI and XhoI cloning sites. The resulting construct (FGF4-pET15b) was transformed into Escherichia coli strain BL21 (DE3) bacteria, and FGF4 expression was induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside for 5 h. The bacteria were then centrifuged and subsequently lysed in a 25 mM Na/K phosphate buffer (pH 7.5) containing 300 mM NaCl with a French cell press. The N-terminally truncated FGF4 (Gly79-Leu206) was found primarily in the insoluble fraction and was extracted in 25 mM Na/K phosphate buffer (pH 7.5) containing 1 M NaCl at 4°C overnight. Following centrifugation, soluble FGF4 was diluted five times with 25 mM Na/K phosphate buffer (pH 7.5) and loaded onto a Source S column (Pharmacia). Bound FGF4 was eluted by a linear gradient of NaCl to 1 M in a 25 mM Na/K phosphate buffer (pH 7.5). Matrix-assisted laser desorption ionization mass spectrometry of the purified FGF4 gave a molecular mass of 14,244 Da (calculated mass, 14,409 Da). The mass difference was due to the cleavage of initiation methionine of FGF4 upon expression in E. coli and a point mutation (Ser-182→Gly) that resulted from PCR. This mutation had no effect on FGF4 biological activity (data not shown).
Crystallization and data collection.
Crystals of FGF4 were grown by vapor diffusion at 20°C by the hanging drop method. Two microliters of protein solution (2 mg/ml in 25 mM HEPES-NaOH buffer [pH 7.5] containing 150 mM NaCl) was mixed with an equal volume of the crystallization buffer (30% polyethylene glycol 8000, 0.2 M ammonium sulfate). FGF4 crystals belong to the orthorhombic space group P212121 with unit cell dimensions of a = 40.37 Å, b = 53.3 Å, and c = 56.23 Å. There is one molecule of FGF4 in the asymmetric unit with a solvent content of ∼43%. Diffraction data were collected from a flash-frozen (in a dry nitrogen stream with mother liquor containing 10% glycerol as cryoprotectant) crystal on an R-Axis IV image plate detector at Beamline X4-A at the National Synchrotron Light Source, Brookhaven National Laboratory, Long Island, N.Y. The data were processed with DENZO and SCALEPACK (25).
Structure determination and refinement.
A molecular replacement solution was found for one copy of FGF4 in the asymmetric unit by using the program AmoRe (20) and the structure of FGF2 (Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, Rutgers, the State University of New Jersey, entry 2FGF) (42) as the search model. Simulated annealing and positional/B-factor refinement were performed with CNS (3). Bulk solvent and anisotropic B-factor corrections were applied. Model building into 2Fo-Fc and Fo-Fc electron density maps was performed with the program O (10). The atomic model contains residues 79 to 206 of FGF4, 3 sulfate ions, and 96 water molecules. The average B-factors are 10.5 Å2 for the FGF4 molecule, 40.5 Å2 for the sulfate ions, and 17.5 Å2 for the water molecules.
Coordinates have been deposited in the RCSB Protein Data Bank under identification code 1IJT.
Production of the mutant FGF4 proteins.
Alanine substitutions were introduced into the N-terminally truncated FGF4 (Gly79-Leu206) by PCR site-directed mutagenesis (Quik Change; Stratagene) with the FGF4-pET15b expression plasmid (described above) as the template and the following mutant oligonucleotides as primers: Y87A (5′-AAGCGGCTGCGGCGGCTCGCATGCAACGTGGGCATCGGC-3′), F129A (5′-GCGTGGTGAGCATCGCCGGCGTGGCCAGCCGG-3′), F151A (5′-CTATGGCTCGCCCTTCGCGACCGATGAGTGCACGTTC-3′), E159A (5′-GATGAGTGCACGTTCAAGGCCATTCTCCTTCCCAAC-3′), Y166A (5′-CTCCTTCCCAACAACGCGAACGCGTACGAGTCC-3′), L203A (5′-CCATGAAGGTCACCCACTTCGCCCCTAGGCTGTGACCC-3′), R205A (5′-CCCACTTCCTCCCCGCGCTGTGACCTTCCAGAGG-3′), N89A, (5′-CGGCGGCTCTACTGCGCCGTGGGCATCGGCTTC-3′), K198A (5′-GTGTCGCCCACCATGGCGGTCACCCACTTCCTC-3′), K183A/K188A (5′-GCCCTGAGCGCGAATGGGAAGACCGCGAAGGGGAAC-3′), R103A (5′-GCGCTCCCCGACGGCGCCATCGGCGGCGCGCAC-3′), and K144A (5′-ATGAGCAGCAAGGGCGCGCTCTATGGCTCGCCC-3′).
The presence of the mutations was confirmed by sequencing. Mutant FGF4-pET15b plasmids were transformed into E. coli strain BL21 (DE3). Expression of the FGF4 proteins was induced as described above. Following centrifugation, cells expressing wild-type and various mutant FGF4 proteins were suspended in a 50 mM HEPES-NaOH buffer (pH 7.4) containing 1 M NaCl and protease inhibitors (phenylmethylsulfonyl fluoride [100 μg/ml], aprotinin [2 μg/ml]) and disrupted by sonication. Lysates were left at 4°C overnight in order to salt-extract the FGF4 proteins from particulate fractions. Following centrifugation, supernatants containing soluble FGF4 proteins were diluted four times with 50 mM HEPES-NaOH buffer (pH 7.4) and loaded onto heparin-Sepharose columns. After washing the columns with 50 mM HEPES-NaOH (pH 7.4) buffer containing 250 mM NaCl, the FGF4 proteins were eluted by a 50 mM HEPES buffer (pH 7.4) containing 1.5 M NaCl. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (15% polyacrylamide), and the purity of the FGF4 proteins was assessed by staining with Coomassie blue R-250.
DNA synthesis assay.
NIH 3T3 cells were seeded at a density of 3 × 104/well in 24-well plates in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum. The following day, the medium was replaced with DMEM containing only 0.5% calf serum, and the cells were allowed to reach quiescence for 48 h. Thereafter, serial dilutions of wild-type full-length FGF4 (Ala31-Leu206) or N-terminally truncated wild-type or mutant FGF4 (Gly79-Leu206) were added for 18 h. Cells were then labeled with 1 μCi of [3H]thymidine for 6 h, washed with Tris-HCl-buffered saline (pH 7.5), and lysed with 0.5 M NaOH. The lysates were then neutralized with 0.5 M HCl, and the radioactivity incorporated into the acid-precipitable material was measured with a β-counter (LKB, Pharmacia). Each assay was performed in triplicate.
Receptor binding assay.
N-terminally truncated FGF4 (Gly79-Leu206) was radioiodinated by the chloramine-T method by a previously described protocol (2). Labeled FGF4 was separated from free iodine over a Sephadex G-25 column, which was previously equilibrated in phosphate-buffered saline containing 1% bovine serum albumin. CHO cells overexpressing FGFR2 (14) were seeded at 106 cells/well in six-well plates in DMEM containing 10% fetal calf serum. The following day, the medium was removed, and the cells were allowed to bind labeled FGF4 (specific activity, 2.5 × 104 cpm/ng) in DMEM containing 25 mM HEPES-NaOH (pH 7.4), 15% gelatin, 10 μg of heparin per ml, and increasing concentrations of the wild-type or mutant FGF4 proteins for 2 h at 4°C. Cells were then washed several times with ice-cold Tris-HCl-buffered saline (pH 7.5), and the receptor-bound radiolabeled FGF4 was released by using a 50 mM sodium acetate buffer (pH 4.0) containing 2 M NaCl. Radioactivity was measured with a γ-counter (LKB-Pharmacia). Binding assays were done in duplicate.
RESULTS AND DISCUSSION
Structure determination.
The mature, secreted form of human FGF4 spans amino acids Ala-31 to Leu-206 (2). Based on the crystal structure of FGF2 (5, 42, 45), the β-trefoil core of FGF4 is expected to start at Leu-83 (Pro-29 in FGF2). Recent crystal structures of three different FGF-FGFR complexes have revealed that the residues immediately preceding the β-trefoil core in FGF1 and FGF2 are involved in FGFR binding (29, 30, 33). These residues correspond to amino acids Gly-79 to Arg-82 of FGF4 (Fig. 1B). Thus, to maximize the likelihood of obtaining diffracting crystals without jeopardizing the biological activity of FGF4, we decided to crystallize an N-terminally truncated FGF4 containing residues Gly-79 to Leu-206 (Gly79-Leu206). Truncated FGF4 was expressed in E. coli and purified to homogeneity (see Materials and Methods). The mitogenic activity of the truncated FGF4 on NIH 3T3 cells was only slightly lower than that of the mature FGF4, indicating that the truncated FGF4 contains the majority of receptor binding sites (not shown). Crystallization trials with FGF4 produced orthorhombic crystals with 1 molecule per asymmetric unit. The crystal structure of FGF4 was solved by molecular replacement (see Materials and Methods) and refined to a 1.8-Å resolution with an R-value of 19.4% (free R-value of 20.7%). The atomic model for FGF4 consists of 1 FGF4 molecule (residues 79 to 206), 3 sulfate ions, and 96 water molecules. Data collection and refinement statistics are given in Table 1.
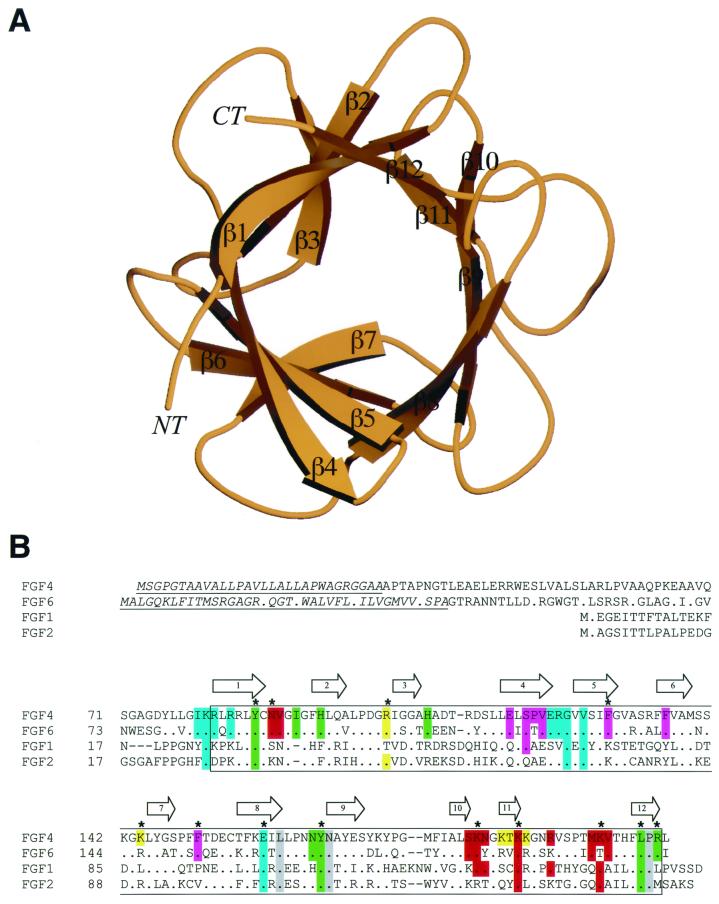
FIG. 1.
Structure and sequence alignment of FGF4. (A) Ribbon diagram of FGF4. Secondary structure assignments were obtained with the program PROCHECK (12). The β-strands of FGF4 are labeled according to the conventional strand nomenclature for FGF1 and FGF2 (6). NT and CT denote the amino and carboxy termini, respectively. This figure was created with the programs Molscript (11) and Raster3D (17). (B) Structure-based sequence alignment of FGFs. Sequence alignment was performed with CLUSTALW (34). All of the FGFs used in this alignment are human. The locations and lengths of the β-strands and α-helices are shown on the top. The signal sequences of FGF4 and FGF6 are indicated by italics and underlining. The box demarcates the boundaries of the β-trefoil core. A period indicates sequence identity to FGF4. A dash represents a gap introduced to optimize the alignment. FGF4 residues are colored with respect to the region on FGFR1 with which they interact: residues that interact with D2 are green, residues that interact with the linker region are gray, and residues that interact with D3 are cyan. FGF4 residues that interact with the βC′-βE loop in D3 of FGFR1 are purple. In red are FGF4 residues that constitute the conventional low- and high-affinity heparin binding sites. In addition, FGF4 residues that localize to the periphery of the high-affinity heparin-binding site and could potentially interact with heparin are yellow. A star indicates FGFR and heparin binding residues that were tested by site-directed mutagenesis.
TABLE 1.
Summary of crystallographic analysis
| Type of statistic | Result |
|---|---|
| Data collection | |
| Resolution (Å) | 25.0–1.8 |
| No. of reflections (total/unique) | 40,562/11,590 |
| Completeness (%) | 99.6 (99.5)b |
| Rsym (%)a | 5.1 (10.9)b |
| Signal (<I/ςI>) | 16.5 |
| Refinementc | |
| Resolution (Å) | 25.0–1.8 |
| No. of reflections | 11,488 |
| Rcryst/Rfree (%)d | 19.4/20.7 |
| rms deviations | |
| Bonds (Å) | 0.005 |
| Angles (°) | 1.31 |
| B-factors (Å2)e | 1.00 |
Rsym = 100 × ΣhklΣi|li(hkl) − <l(hkl)>|/ΣhklΣi li(hkl).
Value in parentheses is for the highest-resolution shell: 1.86 − 1.8 Å.
Atomic model: 994 protein atoms, 3 SO42− ions, and 96 water molecules.
Rcryst/free = 100 × Σhkl ∥Fo(hkl)|−|Fc(hkl)∥ / Σhkl|Fo(hkl)|, where Fo (>0ς) and Fc are the observed and calculated structure factors, respectively. Five percent of the reflections were used for calculation of Rfree.
B-factors for bonded protein atoms.
Description of the structure.
As anticipated by sequence similarity (36), FGF4 adopts a β-trefoil fold (Fig. 1A). Superimposition of the Cα traces located within the β-trefoil core of FGF4 with those of FGF1 (45) and FGF2 (42) gives root-mean-square (rms) deviations of only 0.86 Å (122 common Cα atoms) and 0.76 Å (123 common Cα atoms), respectively. Within the β-trefoil core, the major differences between the FGF4 structure and the structures of FGF1 and FGF2 are the conformations of the β1-β2, β3-β4, and β9-β10 loops. These loops vary in both length and sequence among the various members of the FGF family (Fig. 1B). In FGF4, the β1-β2 loop is one residue longer than the corresponding loops of FGF1 and FGF2. In contrast, the β3-β4 loop in FGF4 is shorter by one residue than the corresponding loops in FGF1 and FGF2. Like FGF2, the β9-β10 loop in FGF4 is shorter by two residues than in FGF1 (Fig. 1B).
As with the structures of free FGF1 and FGF2 (45), a sulfate ion is bound to the predicted high-affinity heparin binding site of FGF4. In addition, two other sulfate ions are coordinated by FGF4 residues, whose corresponding residues in FGF1 and FGF2 have not been observed to bind sulfate ions (see Fig. 5).
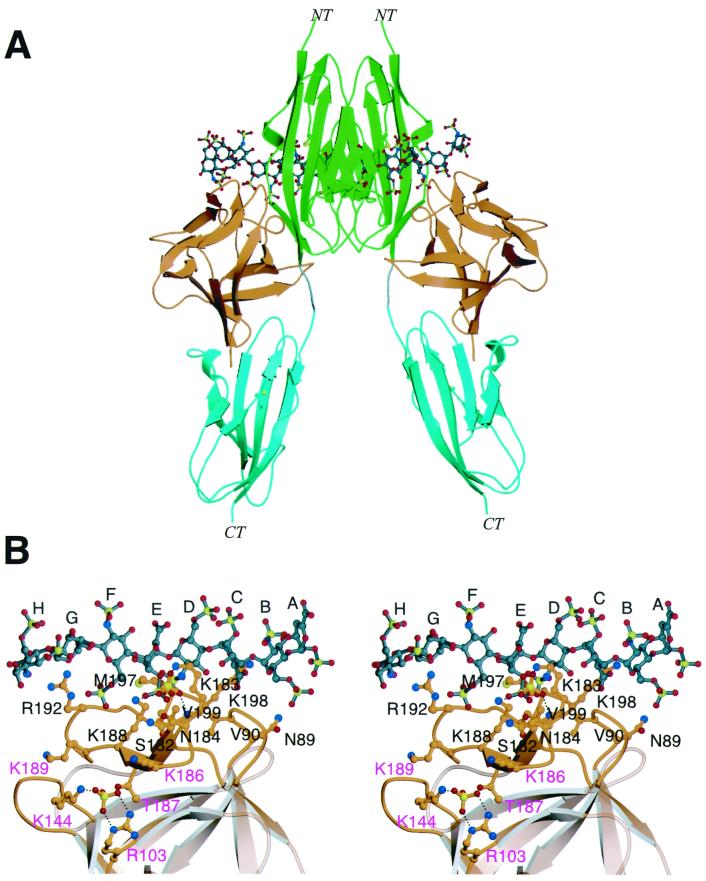
FIG. 5.
Heparin binding sites in FGF4. (A) A dimeric FGF4-FGFR1-heparin model was created by superimposition of the Cα traces of two FGF4 structures onto the Cα traces of the two FGF2 molecules in the FGF2-FGFR1-heparin ternary structure. NT and CT denote the amino and carboxy termini, respectively. The coloring for FGF4 and FGFR1 is as presented in Fig. 2. The heparin oligosaccharides are rendered in the ball-and-stick format. The atom coloring for oxygens and nitrogens is as presented in Fig. 2. In addition, sulfur atoms are yellow, and the carbon atoms of oligosaccharides are gray. (B) FGF4 residues that localize to the heparin binding surface of FGF4 in the context of the ternary FGF4-FGFR1-heparin structure are mapped onto the ribbon diagram of FGF4. FGF4 residues that localize to the peripheries of the high-affinity heparin binding site and could potentially bind heparin are labeled in purple letters. A sulfate ion is bound to the conventional high-affinity heparin binding site. Another sulfate ion is bound in the additional potential heparin binding site. The atom coloring is as presented in panel A. Dotted lines represent hydrogen bonds. The sugar rings of heparin are labeled A through H, starting at the nonreducing end of the oligosaccharide. This figure was created with Molscript and Raster3D.
Receptor binding sites and specificity.
To identify potential receptor binding sites in FGF4, we constructed an FGF4-FGFR1 model by superimposing the FGF4 structure onto the FGF2 structure bound to the ligand binding portion of FGFR1 consisting of Ig-like domains 2 (D2) and 3 (D3) (Fig. 2A). Careful inspection of the FGF4-FGFR1 interface showed that the majority of the interactions between FGF4 and FGFR1 in the FGF4-FGFR1 model could be accommodated by minor adjustments of FGF4 side-chain rotamers. Three loop regions, the β1-β2 and β8-β9 loops (within the β-trefoil core) and the N terminus (outside the β-trefoil core), sterically clash with the receptor (Fig. 2A). In the present crystal structure, these loop regions are involved in crystal lattice contacts, implying that the present conformations of these loops are dictated by the lattice contacts. However, upon FGFR binding, these regions are expected to undergo changes in backbone conformation to allow an engagement with FGFR1 to occur.
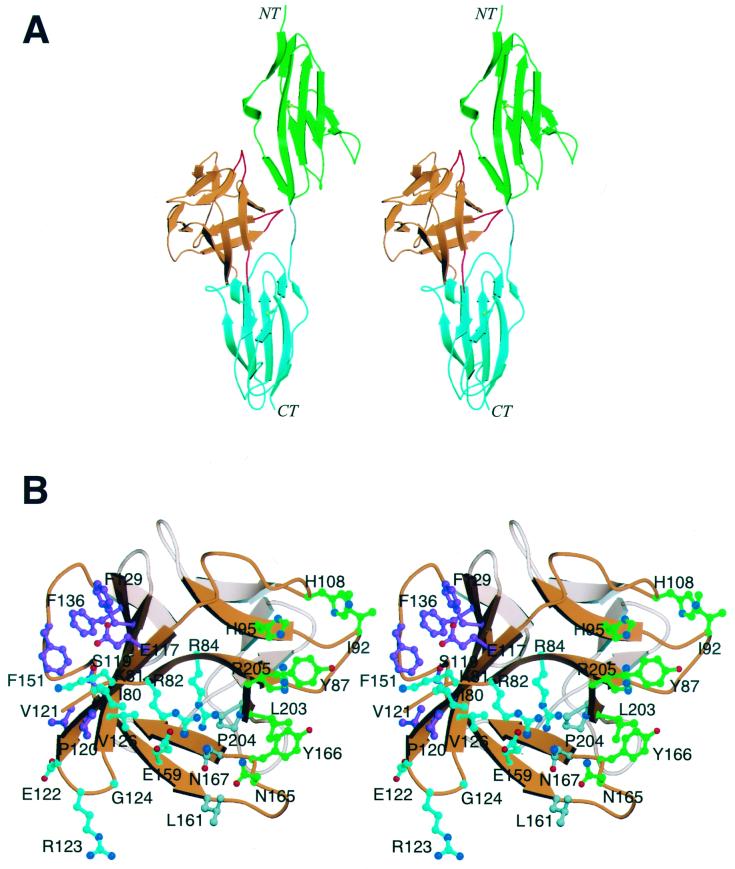
FIG. 2.
Mapping of receptor binding sites in FGF4. (A) A model of the FGF4-FGFR1 structure was generated by superimposition of the Cα traces within the β-trefoil of the FGF4 structure onto the corresponding Cα traces of FGF2 in the FGF2-FGFR1 structure. Color coding is as follows: FGF4 is orange, D2 is green, D3 is cyan, and the linker region is gray. The FGF4 loop regions that clash with FGFR1 are red. NT and CT denote the amino and carboxy termini, respectively. (B) Stereo view of the receptor binding sites on FGF4. FGF4 residues are considered to be in the FGF4-FGFR1 interface if their side-chain or main-chain interatomic distance to FGFR1 is less than or equal to 3.8 Å. FGF4 residues are colored with respect to the FGFR1 regions with which they interact. FGF4 residues that interact with D2 are green, residues that interact with the linker region are gray, and residues that interact with D3 are cyan. FGF4 residues that interact with the βC′-βE loop in D3 of FGFR are purple. Oxygen and nitrogen atoms are red and blue, respectively. This figure was created by using Molscript and Raster3D.
At the FGF4-D2 interface, three highly conserved solvent-exposed FGF4 residues, Tyr-87, Tyr-166, and Leu-203, are predicted to be packed against a highly conserved hydrophobic surface consisting of Ala-167, Pro-169, and Val-248 at the bottom of D2 in FGFR1 (Fig. 2B). Significant differences between FGF4 and FGF2 at the FGF-D2 interface are the substitutions of Phe-40 and Met-151 in FGF2 with His-95 and Arg-205 in FGF4 (Fig. 1B). These substitutions may indicate a weaker hydrophobic FGF4-D2 interface compared to the FGF2-D2 interface. At the FGF4-linker interface, Asn-167, which is also highly conserved among FGFs (Fig. 1B), is expected to make hydrogen bonds with the FGFR-invariant arginine (Arg-250 in FGFR1) in the D2-D3 linker region.
To provide experimental support for the described interactions between FGF4 and FGFR1 at the FGF4-D2 interface, we mutated Tyr-87, Tyr-166, Leu-203, and Arg-205 individually to alanine in the N-terminally truncated FGF4 construct (Gly79-Leu206). Mutant FGF4 proteins were expressed in E. coli, purified to near homogeneity (as described in Materials and Methods), and assayed for the ability to induce DNA synthesis in NIH 3T3 fibroblasts. As shown in Fig. 4 and Table 2, the Y87A, Y166A and L203A mutant FGF4 proteins were severely compromised in their ability to induce DNA synthesis, while the R205A mutant showed a more modest decrease in the induction of DNA synthesis. Thus, these data confirm the observed interactions between FGF4 and D2 in the FGF4-FGFR1 model. Indeed, the corresponding four residues in FGF2 had been shown previously to also be important for biological activity (33).
FIG. 4.
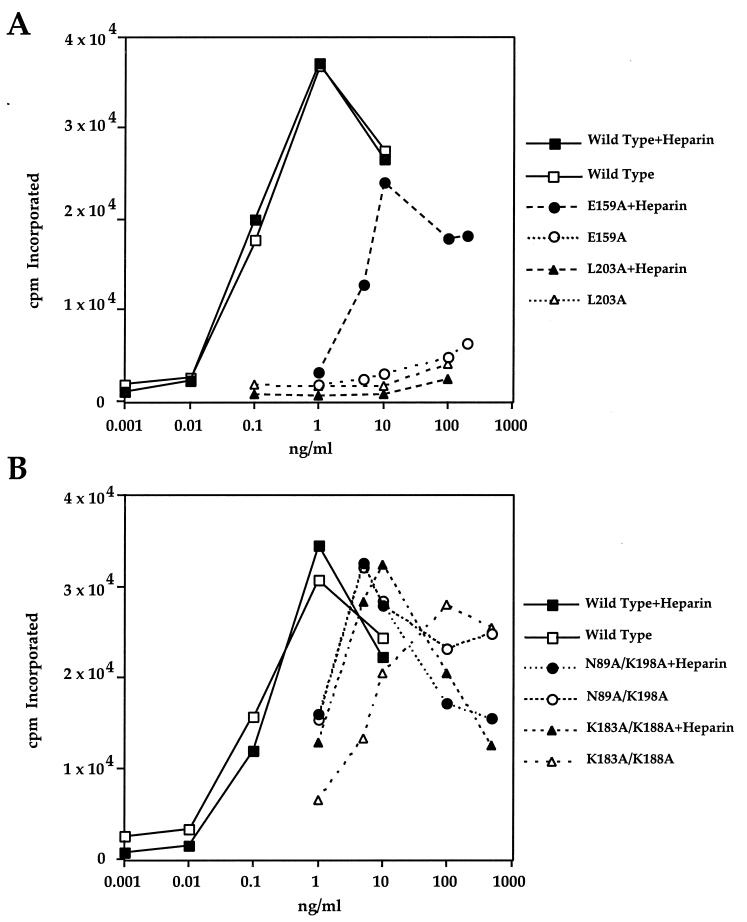
Differential effect of heparin on stimulation of DNA synthesis in NIH 3T3 cells by some FGF4 mutants. Thymidine uptake in NIH 3T3 cells in response to increasing concentrations of wild-type FGF4 and FGF4 mutants in the presence and absence of exogenous heparin was determined as described in Materials and Methods. (A) A representative experiment with the E159A and L203A mutants. (B) A representative experiment with the K183A/K188A and N89A/K198A double mutants. These results and those obtained with other mutants are summarized in Table 2, where we calculate the 50% effective dose—the concentration of mutant necessary to achieve 50% of maximum DNA synthesis produced by the wild-type FGF4.
TABLE 2.
Summary of DNA synthesis activity and receptor binding affinity of mutant FGF4
| Protein (Gly79-Leu206) | DNA synthesis (ED50 for mutant/ED50 for wild type)a
|
Receptor binding (IC50 for mutant/IC50 for wild type)b (+ heparin) | |
|---|---|---|---|
| −Heparin | +Heparin | ||
| Wild type | 1 | 1 | 1 |
| Y87A | >500 | 500 | >100 |
| F129A | >500 | >500 | >100 |
| F151A | 1,000 | 4 | 2.5 |
| E159A | >1,000 | 80 | 5.0 |
| Y166A | 500 | >500 | >100 |
| L203A | >500 | >500 | >100 |
| R205A | 25 | 2.5 | 2.5 |
| N89A/K198A | 10 | 10 | NDc |
| K183A/K188A | 70 | 13 | ND |
| R103A/K144A | 1 | 1 | ND |
ED50, 50% effective dose, defined as the dose of FGF4 necessary to reach 50% of maximum DNA synthesis obtained with the wild-type FGF4.
IC50, 50% inhibitory concentration, defined as the concentration of FGF4 required to compete 50% of binding of labeled wild-type FGF4 to FGFR2.
ND, not determined.
Interactions between FGF4 and D3 occur at the upper part of D3 and involve mainly the βB′-βC, βC′-βE, and βF-βG loops in D3. At the interface between FGF4 and the βB′-βC loop of D3, Glu-159 of FGF4 (an FGF-invariant residue) (Fig. 1B) is expected to make a hydrogen bond with Gln-284 (an FGFR-invariant residue). This prediction is supported by a 1,000-fold reduction in the ability of the E159A mutant FGF4 to induce DNA synthesis in living cells (Fig. 4 and Table 2).
In contrast to the interface between FGF4 and the βB′-βC loop, interactions between FGF4 and the βC′-βE and βF-βG loops are variable. Significantly, FGF4, like FGF1, has a serine (Ser-119) at the position homologous to Gln-65 of FGF2 (Fig. 1B). We have previously shown that Gln-65 of FGF2 makes two hydrogen bonds with Asp-320/Asp-321 in the βC′-βE loop of FGFR1/FGFR2, and as a result, the βC′-βE loop is ordered in both FGF2-FGFR1 and FGF2-FGFR2 structures (29, 30). In contrast, in the FGF1-FGFR1 structure, the βC′-βE loop is disordered because Ser-62 of FGF1 cannot interact with Asp-320 of FGFR1 in the βC′-βE loop (30). Thus, by analogy, we would predict that Ser-119 of FGF4 will also not make hydrogen bonds with Asp-320 located in the βC′-βE loop. However, based on the present model, the side chain of Glu-117 in FGF4 could make hydrogen bonds with Lys-321 located in the βC′-βE loop of FGFR1. This interaction may potentially compensate for the inability of FGF4 to engage Asp-320 and lead to an ordered βC′-βE loop. Moreover, in the FGF4-FGFR1 model, several solvent-exposed hydrophobic residues in FGF4 (Val-121, Phe-129, Phe-136, and Phe-151) are in the vicinity of the βC′-βE loop of FGFR1 and could engage in hydrophobic contacts with Val-316 in the βC′-βE loop of FGFR1. These hydrophobic interactions would further contribute to the ordering of the βC′-βE loop in an FGF4-FGFR1 structure. DNA synthesis assays performed with mutant FGF4 molecules support the aforementioned hypothesis. Substitutions of Phe-129 and Phe-151 with alanine in FGF4 reduced the ability of FGF4 to induce thymidine incorporation in NIH 3T3 cells about a thousand-fold (Table 2). The residue corresponding to Phe-151 in FGF2 has also been shown to be important for FGFR binding (44).
Thus, based on our FGF4-FGFR1 model, we propose that FGF4 like FGF2 may engage the βC′-βE loop of FGFR1. Consequently, sequence variations in the βC′-βE loop resulting from alternative splicing should affect FGF4-FGFR binding affinity. A sequence comparison of FGFRs at the βC′-βE loop region demonstrates that Lys-321 is conserved only in the IIIc isoforms of FGFR1 to -3, providing a potential explanation for reduced affinity of FGF4 towards the IIIb splice variants of FGFR1 to -3 and FGFR4. However, definite proof of this hypothesis will necessitate determination of the crystal structure of the FGF4-FGFR1 complex.
Binding of the FGF4 mutants to FGFR2.
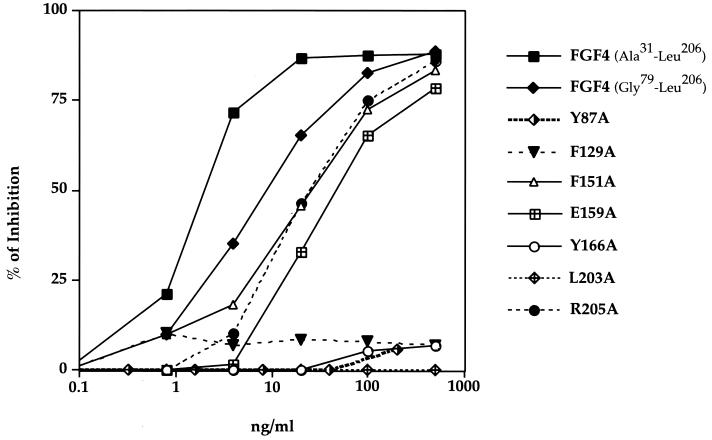
We tested the mutant FGF4 proteins in a receptor binding assay to confirm that the diminished capacity of the mutant FGF4 proteins to induce DNA synthesis is a result of the reduced ability of the mutant FGF4 to interact with FGFR. CHO cells overexpressing FGFR2 (14) were allowed to bind radiolabeled N-terminally truncated FGF4 (Gly79-Leu206) in the presence of increasing concentrations of unlabeled full-length (Ala31-Leu206), N-terminally truncated wild-type, or various N-terminally truncated mutant FGF4 proteins (Fig. 3). The N-terminally truncated wild-type FGF4 bound FGFR2 with only a slightly lower affinity than full-length FGF4 (Ala31-Leu206), indicating that the majority of receptor binding sites are contained within the Gly79-Leu206 construct (Fig. 3). Substitutions of Tyr-87, Tyr-166, and Leu-203 with alanine severely reduced the affinity of FGF4 towards FGFR2 (Fig. 3 and Table 2), emphasizing the importance of the hydrophobic FGF4-D2 interface in providing FGF4-FGFR affinity. In contrast, the R205A mutant FGF4 showed only a slight reduction in FGFR2 binding affinity. The relative decrease in binding affinity of these mutants towards FGFR2 is consistent with the results of the DNA synthesis assay, thus implying that the impaired ability of these mutants to induce DNA synthesis is a consequence of loss of binding affinity to FGFR2.
FIG. 3.
Comparison of the binding affinities of various FGF4 mutants towards FGFR2. The capacity of the various FGF4 mutants to compete with binding of the N-terminally truncated wild-type FGF4 to FGFR2 binding was measured as described in Materials and Methods. The data are expressed as percent inhibition of wild-type FGF4 binding to FGFR2 by the indicated amount of unlabeled mutant FGF4. The results presented in this figure are also summarized in Table 2, where we have calculated for each mutant a 50% inhibitory concentration, or the concentration of mutant FGF4 necessary to compete off 50% of wild-type FGF4.
Alanine substitutions of FGF4 residues predicted to interact with D3 also reduced the binding affinity of FGF4 for FGFR2. The F129A mutant showed a large decrease (more than 100-fold) in receptor binding affinity, which paralleled the severe impairment of this mutant in induction of DNA synthesis (Fig. 3 and Table 2). In contrast, the F151A and E159A mutants were only slightly affected (2.5- and 5-fold, respectively) in FGFR2 binding (Fig. 3 and Table 2), yet these mutants were severely compromised in induction of DNA synthesis (Table 2). This was particularly unexpected for the E159A mutant, because Glu-159 is highly conserved among FGFs (Fig. 1B) and the corresponding glutamic acid in FGF2 (Glu-96) was shown to be critical for binding of FGF2 to FGFR1 (43).
We reasoned that this discrepancy between receptor binding and DNA synthesis data for the F151A and E159A mutants may be due to a difference between the experimental conditions used for the receptor binding and DNA synthesis assays. Since NIH 3T3 cells naturally express cell surface HSPG in abundance, they do not require exogenous heparin to respond fully to FGF4. Therefore, we did not include exogenous heparin in the DNA synthesis assays. In contrast, the receptor binding assays were performed in the presence of exogenous heparin in order to exclude binding of FGF to the abundantly expressed cell surface HSPG. In the absence of heparin, binding to these low-affinity but very abundant receptors cannot easily be distinguished from binding to FGFR. Although methods to differentiate between FGF2 binding to HSPGs and FGFR binding have been described (18), our attempts to perform meaningful binding experiments with FGF4 in the absence of heparin were not successful.
Since heparin stabilizes FGF-FGFR interactions, it was possible that the presence of exogenous heparin in the receptor binding assay could have partially reversed the reduced ability of the F151A and E159A mutants to interact with FGFR. To test this possibility, we repeated the DNA synthesis assays in the presence of exogenous soluble heparin. While, as expected, heparin had no effect on the mitogenic ability of wild-type FGF4, heparin dramatically enhanced the capacity of the F151A and E159A mutants to induce DNA synthesis (Fig. 4A and Table 2). In contrast, addition of heparin had no effect on the Y87A, F129A, Y166A, and L203A mutants and enhanced the ability of R205A to induce DNA synthesis only by about 10-fold (Fig. 4A and Table 2).
Analysis of the location of the various FGF4 mutations in the ternary FGF4-FGFR1-heparin model provides a potential explanation for the differential ability of heparin to rescue only some of the mutants. Both the F151A and E159A mutations, which display the greatest potentiation upon addition of heparin, affect FGF4 interaction with FGFR D3 (Fig. 1B). In contrast, with the exception of the F129A mutant, all of the mutations that are not rescued by heparin affect FGF4's interaction with FGFR D2. Upon binding of FGF to FGFR, a continuous negatively charged surface is formed by the heparin binding sites of FGF and FGFR D2, to which heparin binds (29). Simultaneous binding of the same heparin polymer to both FGF and FGFR will clearly increase apparent FGF-FGFR affinity (32). Mutations affecting the FGF-D2 interface will hamper a productive juxtaposition of the heparin binding sites of FGF and FGFR to form a continuous heparin binding surface, and thus heparin will not reverse the deleterious effects of these mutations. In contrast, mutations affecting the FGF-D3 interface will not interfere with the formation of a productive heparin binding surface by FGF and FGFR D2, and heparin can enhance FGF-FGFR affinity by interacting with both FGF and FGFR D2.
The mutagenesis data suggest that interactions of FGF with FGFR D2 provide the primary FGF-FGFR binding affinity. Indeed, Wang et al. have shown that several FGFs can bind to the isolated D2 domain of FGFR1 in vitro in the presence of heparin (38). It is therefore likely that FGFR binds FGF first via D2. Heparin then stabilizes the FGF-D2 interaction and facilitates formation of an FGF-D3 interface.
Heparin binding sites.
Recent biochemical and structural data demonstrate that FGF in the absence of heparin can form an initial low-affinity complex with FGFR (26). In the presence of heparin, the low-affinity complexes become stabilized, which in turn leads to stable 2:2 FGF-FGFR signaling complexes. The recent crystal structure of a dimeric 2:2:2 FGF2-FGFR1-heparin complex provides a molecular basis for how heparin enhances FGF-FGFR affinity and promotes dimerization (32). Within each ternary 1:1:1 FGF-FGFR-heparin complex, heparin makes numerous contacts with the heparin binding residues of FGF and FGFR, thereby increasing the affinity of FGF towards FGFRs. In addition, heparin also interacts with the heparin binding residues in D2 of the adjoining FGFR, thereby augmenting the weak interactions of FGF and FGFR in one ternary complex with the FGFR in the adjoining ternary complex. Since FGFs differ in the primary sequences of heparin binding sites, each FGF may require different heparin motifs (sulfation pattern and/or length) in order to exert its optimal biological activities (6, 32).
To evaluate the potential heparin binding sites of FGF4, a dimeric FGF4-FGFR-heparin model was generated by superimposing two copies of the FGF4 structure onto the two copies of FGF2 in the dimeric FGF2-FGFR1-heparin ternary complex (Fig. 5A). FGF4 residues corresponding to the heparin binding residues of FGF2 along with other FGF4 surface residues that could bind heparin were mapped onto the ribbon diagram of FGF4 (Fig. 5B). With the exception of Lys-188 (Lys-134 in FGF2) and Lys-198 (Lys-144 in FGF2), the remainder of the heparin binding residues differ between FGF4 and FGF2 (Fig. 1B). These differences are likely to determine the optimal sulfation motifs in heparin that are required to support FGF4 or FGF2 biological activities. Interestingly, Asn-36 and Gln-143, two critical heparin binding residues in FGF2, are substituted by hydrophobic residues Val-90 and Met-197 in FGF4 (Fig. 5B and Fig. 1B). Therefore, these residues are unable to make hydrogen bonds with the hydroxyl group and N-sulfate group of ring D of heparin. Moreover, in the model, the side chain of Val-199 of FGF4 (Ala-145 in FGF2) clashes with the N-sulfate of ring D (Fig. 5B). These observations indicate that FGF4 may not require N-sulfate on ring D for heparin binding. Two other significant differences between FGF2 and FGF4 are the substitutions of Lys-35 and Lys-128 of FGF2 with Asn-89 and Ser-182, respectively, in FGF4 (Fig. 1B). Based on the present model, Asn-89 and Ser-182 would better engage the 6-O-sulfate of ring B and the 2-O-sulfate group of ring E (Fig. 5B).
A sulfate ion (provided by the crystallization buffer) is coordinated at the predicted high-affinity heparin binding site of FGF4 by Lys-183 and Lys-188 (Fig. 5B). These two lysines are expected to bind the 2-O-sulfate group of ring E of heparin. In fact, the sulfate ion in the FGF4 structure nearly colocalizes with the 2-O-sulfate group of ring E in the FGF4-heparin model (Fig. 5B). To provide experimental support for the modeled FGF4-heparin interactions, we generated mutant FGF4 proteins in which FGF4 residues predicted to coordinate the 2-O-sulfate of ring E (K183 and K188) or the 6-O-sulfate of ring B (N89 and K198) are substituted with alanines. Both the doubly mutated K183A/K188A and N89A/K198A FGF4 proteins showed diminished ability to induce DNA synthesis in NIH 3T3 cells (Fig. 4B and Table 2). Thus, as for FGF2, both the 6-O-sulfate of ring B and the 2-O-sulfate group of ring E of heparin may play important roles in promoting heparin-dependent FGF-FGFR interaction and dimerization. Our data are also consistent with the finding that a high content in 6-O-sulfate groups in heparin is required for specific interaction with FGF4 (9).
Another sulfate ion is coordinated by the side chains of Arg-103 and Lys-144 in the crystal structure of FGF4 (Fig. 5B). Since bound sulfate ions in the crystal structures of free FGFs often indicate potential heparin binding sites in FGFs, we also generated a doubly mutated R103/K144 FGF4 protein. This mutant FGF4 protein induced DNA synthesis in NIH 3T3 cells to a level comparable to that of the wild-type FGF4 (Table 2), suggesting that Arg-103 and Lys-144 most likely do not participate in heparin binding.
We also tested whether exogenous heparin can compensate for the reduced ability of the K183A/K188A and N89A/K198A mutant FGF4 proteins in the DNA synthesis assay. As shown in Fig. 4B, exogenously added heparin significantly enhanced the ability of the K183A/K188A mutant FGF4 to induce DNA synthesis, but had no effect on the N89A/K198A mutant. A possible explanation for the differential effect of heparin on the activity of these two mutants lies in the heterogeneous nature of commercial heparin. It is known that heparin is a mixture of oligosaccharides of different lengths and sulfate contents, generated by the polymerization of repeating disaccharide units consisting of d-glucosamine (GlcN) and l-iduronic acid (IdoA). During biosynthesis, heparin is sulfated by the sequential actions of three different sulfotransferases: an N-sulfotransferase, a 2-O-sulfotransferase, and a 6-O-sulfotransferase (13). In general, these reactions proceed in the order indicated, but often fail to go to completion, resulting in tremendous chemical heterogeneity in sulfation patterns in heparin. The observation that addition of exogenous heparin partially rescues only the K183A/K188A mutant (predicted to coordinate 6-O-sulfate), but not the N89A/K198A mutant (predicted to coordinate 2-O-sulfate), is probably due to the fact that the heparin subfraction containing 2-O-sulfate is more abundant than the subfraction containing 6-O-sulfate.
The heterogeneity in sulfation pattern is even more profound in heparan sulfate moieties of cell surface HSPGs, which are thought to cooperate with FGFs to induce FGFR dimerization and activation. The requirement for a specific sulfation motif in heparan sulfate for optimal FGF4 action may be a mechanism to fine tune FGF4-FGFR interactions and to restrict FGF4 signaling to a specific set of cells in a specific tissue during various stages of embryonic development, in which spatial and temporal regulation by FGF is critically required.
Implications for the general mode of FGF-FGFR binding.
It is important to note that some of the data presented in this report are not consistent with the model of FGF1-FGFR2 binding described in the recently published crystal structure of a ternary FGF1-FGFR2-heparin complex (27). In this structure, the FGFR-invariant Pro-253 located in the D2-D3 linker is found in a cis configuration, while in all previously reported binary FGF-FGFR structures, Pro-253 is found only in a trans configuration (29, 30, 34). Consequently, relative to its position in all binary FGF-FGFR structures, the receptor D3 in the ternary FGF1-FGFR2-heparin structure is swiveled around the linker region by more than 160°. This creates a completely different set of interactions at the FGF-D3 interface. Pellegrini et al. (27) propose that this D3 rotation is caused by a heparin-mediated trans-to-cis isomerization of Pro-253 in the D2-D3 linker region, but our mutagenesis data do not support this hypothesis. Based on this ternary FGF1-FGFR2-heparin structure (27), neither F129 nor F151 in FGF4 is predicted to make any contacts with D3. Thus, the drastically reduced mitogenic capacity of the F129A and F151A mutants is in disagreement with the mode of FGF-FGFR binding described by these authors. We believe that the cis isomerization of Pro-253 observed in the FGF1-FGFR2-heparin structure (27) is probably the result of partial refolding of FGFR2.
In the preceding sections, we proposed a sequential model of FGF-FGFR binding in which interaction of FGF with the FGFR D2 domain provides the primary FGF-FGFR binding surface and heparin facilitates the formation of an FGF-D3 interface by stabilizing the FGF-D2 interaction. This hypothesis could explain the exclusively heparin-dependent binding of FGF1 to an in vitro-refolded FGFR2 described by Pellegrini et al. (27). As discussed above, it is likely that the FGFR2 used by these authors was not properly refolded, and consequently D3 is in a different position from the one observed in the previously reported FGF-FGFR crystal structures. Despite the lack of sufficient contact between FGF1 and FGFR2 D3, the FGF1-FGFR2 complex could still be captured in the presence of heparin, as evident from the crystal structure (27).
In conclusion, the data presented in this report show that FGF4 adopts a typical β-trefoil fold similar to other FGFs (24, 28, 43). A ternary FGF4-FGFR1-heparin model constructed by superimposing FGF4 onto FGF2 in the FGF2-FGFR1-heparin structure assisted the identification of several key residues in FGF4 involved in receptor and heparin binding. Substitution of several of these residues with alanine produced FGF4 molecules with reduced receptor binding and mitogenic potential, which could, in some cases, be partially reversed by excess soluble heparin. Significantly, the modeling and mutagenesis data show that FGF4 interacts with the βC′-βE loop in FGFR D3 and provide a molecular basis for why FGF4, like FGF2, but unlike FGF1, can discriminate between the IIIc and IIIb splice variants of FGFRs for binding. These studies should help understanding of the molecular basis for specific FGF-FGFR interactions and could contribute to the design of novel FGF molecules with increased or altered binding specificity.
ACKNOWLEDGMENTS
M.M. acknowledges S. R. Hubbard for advice in structure determination and C. Ogata for assistance at Beamline X4-A at the Brookhaven National Synchrotron Light Source, a Department of Energy facility. Beamline X4-A is supported by the Howard Hughes Medical Institute. We also thank J. Schlessinger, S. R. Hubbard, A. Mansukhani, and B. K. Yeh for critically reading the manuscript.
This work was supported by National Institutes of Health grants DE13686 (to M.M.) and CA42568 (to C.B) and by a grant from Collateral Therapeutics, Inc. (to C.B).
P.B., A.I., and A.N.P. contributed equally to this work.
REFERENCES
- 1.Basilico C, Moscatelli D. The FGF family of growth factors and oncogenes. Adv Cancer Res. 1992;59:115–165. doi: 10.1016/s0065-230x(08)60305-x. [DOI] [PubMed] [Google Scholar]
- 2.Bellosta P, Talarico D, Rogers D, Basilico C. Cleavage of K-FGF produces a truncated molecule with increased biological activity and receptor binding affinity. J Cell Biol. 1993;121:705–713. doi: 10.1083/jcb.121.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruenger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nigles M, Pannu N S, Read R J, Rice L M, Simonson T, Warren G L. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 4.Delli Bovi P, Curatola A M, Kern F G, Greco A, Ittmann M, Basilico C. An oncogene isolated by transfection of Kaposi's sarcoma DNA encodes a growth factor that is a member of the FGF family. Cell. 1987;50:729–737. doi: 10.1016/0092-8674(87)90331-x. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson A E, Cousens L S, Weaver L H, Matthews B M. Three-dimensional structure of human basic fibroblast growth factor. Proc Natl Acad Sci USA. 1991;88:3441–3445. doi: 10.1073/pnas.88.8.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faham S, Linhardt R J, Rees D C. Diversity does make a difference: fibroblast growth factor-heparin interactions. Curr Opin Struct Biol. 1998;8:578–586. doi: 10.1016/s0959-440x(98)80147-4. [DOI] [PubMed] [Google Scholar]
- 7.Goldfarb M. Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev. 1996;7:311–325. doi: 10.1016/s1359-6101(96)00039-1. [DOI] [PubMed] [Google Scholar]
- 8.Guimond S, Maccarana M, Olwin B B, Lindahl U, Rapraeger A C. Activating and inhibitory heparin sequences for FGF-2 (basic FGF). Distinct requirements for FGF-1, FGF-2, and FGF-4. J Biol Chem. 1993;268:23906–23914. [PubMed] [Google Scholar]
- 9.Ishihara M. Structural requirements in heparin for binding and activation of FGF-1 and FGF-4 are different from that for FGF-2. Glycobiology. 1994;4:817–824. doi: 10.1093/glycob/4.6.817. [DOI] [PubMed] [Google Scholar]
- 10.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 11.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 12.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 13.Lindahl U, Kusche-Gullberg M, Kjellen L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 14.Mansukhani A, Dell'Era P, Moscatelli D, Kornbluth S, Hanafusa H, Basilico C. Characterization of the murine BEK fibroblast growth factor (FGF) receptor: activation by three members of the FGF family and requirement for heparin. Proc Natl Acad Sci USA. 1992;89:3305–3309. doi: 10.1073/pnas.89.8.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIntosh I, Bellus G A, Jab E W. The pleiotropic effects of fibroblast growth factor receptors in mammalian development. Cell Struct Funct. 2000;25:85–96. doi: 10.1247/csf.25.85. [DOI] [PubMed] [Google Scholar]
- 16.McKeehan W L, Wang F, Kan M. The heparan sulfate-fibroblast growth factor family: diversity of structure and function. Prog Nucleic Acid Res Mol Biol. 1998;59:135–176. doi: 10.1016/s0079-6603(08)61031-4. [DOI] [PubMed] [Google Scholar]
- 17.Merritt E A, Bacon D J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 18.Moscatelli D. Metabolism of receptor-bound and matrix-bound basic fibroblast growth factor by bovine capillary endothelial cells. J Cell Biol. 1988;107:753–759. doi: 10.1083/jcb.107.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naski M C, Ornitz D M. FGF signaling in skeletal development. Front Biosci. 1998;3:D781–D794. doi: 10.2741/a321. [DOI] [PubMed] [Google Scholar]
- 20.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr Sect A. 1994;50:157–163. [Google Scholar]
- 21.Nishimura T, Nakatake Y, Konishi M, Itoh N. Identification of a novel FGF, FGF-21, preferentially expressed in the liver. Biochim Biophys Acta. 2000;1492:203–206. doi: 10.1016/s0167-4781(00)00067-1. [DOI] [PubMed] [Google Scholar]
- 22.Ornitz D M. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. Bioessays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 23.Ornitz D M, Xu J, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 24.Osslund T D, Syed R, Singer E, Hsu E W, Nybo R, Chen B L, Harvey T, Arakawa T, Narhi L O, Chirino A, Morris C F. Correlation between the 1.6 A crystal structure and mutational analysis of keratinocyte growth factor. Protein Sci. 1998;7:1681–1690. doi: 10.1002/pro.5560070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Pantoliano M W, Horlick R A, Springer B A, Van Dyk D E, Tobery T, Wetmore D R, Lear J D, Nahapetian A T, Bradley J D, Sisk W P. Multivalent ligand-receptor binding interactions in the fibroblast growth factor system produce a cooperative growth factor and heparin mechanism for receptor dimerization. Biochemistry. 1994;33:10229–10248. doi: 10.1021/bi00200a003. [DOI] [PubMed] [Google Scholar]
- 27.Pellegrini L, Burke D F, von Delft F, Mulloy B, Blundell T L. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 28.Plotnikov A N, Eliseenkova A V, Ibrahimi O A, Shriver Z, Sasisekharan R, Lemmon M A, Mohammadi M. Crystal structure of fibroblast growth factor 9 (FGF9) reveals regions implicated in dimerization and autoinhibition. J Biol Chem. 2001;276:4322–4329. doi: 10.1074/jbc.M006502200. [DOI] [PubMed] [Google Scholar]
- 29.Plotnikov A N, Schlessinger J, Hubbard S R, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 30.Plotnikov A N, Hubbard S R, Schlessinger J, Mohammadi M. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell. 2000;101:413–424. doi: 10.1016/s0092-8674(00)80851-x. [DOI] [PubMed] [Google Scholar]
- 31.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 32.Schlessinger J, Plotnikov A N, Ibrahimi O A, Eliseenkova A V, Yeh B K, Yayon A, Linhardt R J, Mohammadi M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 33.Springer B A, Pantoliano M W, Barbera F A, Gunyuzlu P L, Thompson L D, Herblin W F, Rosenfeld S A, Book G W. Identification and concerted function of two receptor binding surfaces on basic fibroblast growth factor required for mitognesis. J Biol Chem. 1994;269:26879–26884. [PubMed] [Google Scholar]
- 34.Stauber D J, DiGabriele A D, Hendrickson W A. Structural interactions of fibroblast growth factor receptor with its ligands. Proc Natl Acad Sci USA. 2000;97:49–54. doi: 10.1073/pnas.97.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J D, Higgins D G, Gibson T J. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vainikka S, Partanen J, Bellosta P, Coulier F, Birnbaum D, Basilico C, Jaye M, Alitalo K. Fibroblast growth factor receptor-4 shows novel features in genomic structure, ligand binding and signal transduction. EMBO J. 1993;11:4273–4280. doi: 10.1002/j.1460-2075.1992.tb05526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkataraman G, Raman R, Sasisekharan V, Sasisekharan R. Molecular characteristics of fibroblast growth factor-fibroblast growth factor receptor-heparin-like glycosaminoglycan complex. Proc Natl Acad Sci USA. 1999;96:3658–3663. doi: 10.1073/pnas.96.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F, Lu W, McKeehan K, Mohamedali K, Gabriel J L, Kan M, McKeehan W L. Common and specific determinants for fibroblast growth factors in the ectodomain of the receptor kinase complex. Biochemistry. 1999;38:160–171. doi: 10.1021/bi981758m. [DOI] [PubMed] [Google Scholar]
- 39.Wilkie A O. Craniosynostosis: genes and mechanisms. Hum Mol Genet. 1997;6:1647–1656. doi: 10.1093/hmg/6.10.1647. [DOI] [PubMed] [Google Scholar]
- 40.Xu X, Weinstein M, Li C, Deng C. Fibroblast growth factor receptors (FGFRs) and their roles in limb development. Cell Tissue Res. 1999;296:33–43. doi: 10.1007/s004410051264. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita T, Yoshioka M, Itoh N. Identification of a novel fibroblast growth factor, FGF-23, preferentially expressed in the ventrolateral thalamic nucleus of the brain. Biochem Biophys Res Commun. 2000;277:494–498. doi: 10.1006/bbrc.2000.3696. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J D, Cousens L S, Barr P J, Sprang S R. Three-dimensional structure of human basic fibroblast growth factor, a structural homolog of interleukin 1 beta. Proc Natl Acad Sci USA. 1991;88:3446–3450. doi: 10.1073/pnas.88.8.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H, Ramnarayan K, Anchin J, Miao W Y, Sereno A, Millman L, Zheng J, Balaji V N, Wolff M E. Glu-96 of basic fibroblast growth factor is essential for high affinity receptor binding. Identification by structure-based site-directed mutagenesis. J Biol Chem. 1995;270:21869–21874. doi: 10.1074/jbc.270.37.21869. [DOI] [PubMed] [Google Scholar]
- 44.Zhu H, Ramnarayan K, Menzel P, Miao Y, Zhang J, Mong S. Identification of two new hydrophobic residues on basic fibroblast growth factor important for fibroblast growth factor receptor binding. Protein Eng. 1998;11:937–940. doi: 10.1093/protein/11.10.937. [DOI] [PubMed] [Google Scholar]
- 45.Zhu X, Komiya H, Chirino A, Faham S, Fox G M, Arakawa T, Hsu B T, Rees D C. Three-dimensional structures of acidic and basic fibroblast growth factors. Science. 1991;251:90–93. doi: 10.1126/science.1702556. [DOI] [PubMed] [Google Scholar]