Abstract
We show that the Mre11 complex associates with E2F family members via the Nbs1 N terminus. This association and Nbs1 phosphorylation are correlated with S-phase checkpoint proficiency, whereas neither is sufficient individually for checkpoint activation. The Nbs1 E2F interaction occurred near the Epstein-Barr virus origin of replication as well as near a chromosomal replication origin in the c-myc promoter region and was restricted to S-phase cells. The Mre11 complex colocalized with PCNA at replication forks throughout S phase, both prior to and coincident with the appearance of nascent DNA. These data suggest that the Mre11 complex suppresses genomic instability through its influence on both the regulation and progression of DNA replication.
The Mre11 complex (composed of Mre11, Rad50, and Nbs1) and the ataxia-telangiectasia mutated (ATM) protein kinase are required to activate a DNA damage-induced S-phase checkpoint in mammalian cells (46). Mutations in the ATM, MRE11, or NBS1 gene (from patients with ataxia-telangiectasia [A-T], ataxia-telangiectasia-like disorder [A-TLD], or Nijmegen breakage syndrome [NBS], respectively) abrogate this checkpoint (12, 52, 58, 66). Mutant cells fail to repress the firing of DNA replication origins in the presence of ionizing radiation (IR)-induced DNA damage, a phenomenon termed radioresistant DNA synthesis (RDS) (28, 42). Hence, the Mre11 complex can act as a negative regulator of DNA replication origins in response to DNA damage.
The Mre11 complex is also important for recombinational DNA repair, as established by genetic analyses with Saccharomyces cerevisiae (21). Both the conservation of Mre11 and Rad50 and in vitro studies of the human Mre11 complex strongly suggest that the human Mre11 complex also functions in DNA recombination (43, 44, 63). DNA recombination and DNA replication functions are intrinsically linked; thus, Mre11 complex recombination functions are implicated in S-phase progression in addition to its role in S-phase regulation. In vertebrates, null mutants of the Mre11 complex are inviable (33, 68, 73), and DT40 cells depleted of Mre11 die with chromosome damage indicative of failure to resolve double-strand breaks arising during DNA replication (69). This suggests that the complex's recombination functions are required for DNA replication in a manner analogous to that of Rad51 (45, 69). In Rad51-deficient cells, spontaneous chromosomal breakage during DNA replication leads to cell death (32, 54, 56, 64). It is not clear whether the Mre11 complex's influence on the S-phase checkpoint is related to its DNA recombination functions.
The Nbs1 protein is an important link between the Mre11 complex and the ATM-controlled S-phase checkpoint. ATM phosphorylates Nbs1 (20, 31, 67, 72), and this event is required for checkpoint activation (31, 72). Its role in cell cycle regulation is consistent with the fact that Nbs1 contains a forkhead-associated (FHA) domain and a BRCA1 C-terminal (BRCT) domain (66), each of which is found in a number of proteins that effect DNA damage-dependent checkpoint functions (4, 10, 22, 57, 59).
We identified the E2F1 transcription factor in a screen for proteins that interacted with the Nbs1 N-terminal region and established evidence that this interaction occurs on chromatin near a defined DNA replication origin. The interaction between E2F1 and Nbs1 was abrogated or significantly reduced in NBS and A-TLD cells, respectively. Further, we found the Mre11 complex undergoes dramatic relocalization during DNA replication in a manner analogous to that seen in damaged cells (35, 37, 38). The data presented in this study suggest that the Mre11 complex directly influences S-phase progression both near replication origins via its interaction with E2F1 and at replication forks.
MATERIALS AND METHODS
Cells.
Normal lymphoblastoid cells (721) were obtained from B. Sugden. Raji 525-7 cells were a gift from D. Eick and were grown in RPMI–10% calf serum–200 μg of hygromycin per ml. E14 embryonic stem cells were propagated as described previously (47). All other cell lines have been described previously (12, 58). Raji cells were synchronized by incubation in the presence of 2 mM thymidine for 14 h, released into drug-free medium for 11 h, and incubated in the presence of 1 μg of aphidicolin/ml for 14 h. Cells were then released into drug-free medium and harvested.
Immunological reagents.
Nbs1 (#16) and Mre11 (#59) antisera were described previously (12, 58). E2F1 C20, E2F1 KH95, E2F2 C20, E2F3 C18, E2F3 N20, E2F4 C20, retinoblastoma (Rb) 1F8, Ets1/2 C275, and promyelocytic leukemia protein PG-M3 antibodies were obtained from Santa Cruz Biotechnology. E2F1 mixed monoclonal antibody (KH20+KH95) was purchased from Upstate Biotechnology. Anti-PCNA PC10 was from Oncogene Research Products. E2F1 mixed monoclonal antibody (SQ41+SQ71) was from Neomarkers. Secondary antibodies for immunofluorescence were obtained from Jackson Immunoresearch Laboratories. Murine Nbs1 rabbit polyclonal antiserum 93 was raised against amino acids (aa) 1 to 645 of murine Nbs1. E2F1 rabbit polyclonal antiserum 70 was raised against aa 282 to 416.
Immunoprecipitation and phosphatase assays.
For coimmunoprecipitation assays, cells were lysed as described previously (58). Cleared lysates were immunoprecipitated with preimmune serum, Nbs1 or Mre11 antiserum, or 5 μg of antibodies against E2F1, E2F2, E2F3, E2F4, or Rb. Immunoprecipitates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose using standard methods (2). Immunoblotting was carried out as described previously (12, 58). To control for artifactual coimmunoprecipitation resulting from contaminating DNA in the extracts, immunoprecipitations were carried out in the presence of 300 μg of ethidium bromide/ml to disrupt protein-DNA interactions, as described previously (25).
For phosphorylation assays, Nbs1 immunoprecipitations and phosphatase assays were carried out, as described previously (31), using 50 U of λ phosphatase (NEB) or an equivalent volume of storage buffer for mock-treated samples.
Immunofluorescence assays.
Cells were plated on glass coverslips 24 h before each experiment. Before fixation, in situ cell fractionation was performed as described previously (39). The cells were then fixed in modified Streck tissue fixative for 30 min at room temperature (RT) and permeabilized for 15 min at RT as described previously (38).
Cells were blocked with 10% fetal calf serum (FCS) in phosphate-buffered saline (PBS) and then stained with primary antibody diluted in PBS with 5% FCS for 1 h at RT, followed by staining with secondary antibody for 30 min. 4′,6′-Diamidino-2-phenylindole (DAPI) counterstain was used at a final concentration of 0.05 μg/ml in the last wash. Controls with preimmune serum or secondary antibody alone were negative.
For bromodeoxyuridine (BrdU) labeling, cells were labeled with 10 μM BrdU for 4 h and then were washed and processed for immunofluorescent staining as described above. After the secondary antibody incubation, samples were fixed again in 4% paraformaldehyde for 30 min at RT and then were incubated with 50 mM glycine for 10 min at RT. DNA was denatured with 4 N HCl plus 0.1% Triton X-100 for 10 min at RT, and then samples were extensively washed in PBS, followed by a 50 mM glycine wash. Cells were incubated for 1 h at RT with fluorescein isothiocyanate-conjugated monoclonal BrdU antibody (Becton Dickinson) diluted at 0.8 μg/ml in PBS plus 5% FCS plus 0.3% Triton X-100.
Images were captured with a charge-coupled device camera (Princeton Instruments), and gray scale images were processed using IP Labs (Scanalytics) and Photoshop 5.5 (Adobe) software.
Yeast two-hybrid assays.
Two-hybrid interaction screening was performed as described previously (12), using Nbs1 expressed as a fusion to the GAL4 DNA binding domain and a human B-lymphoblastoid cDNA library. Derivatives of E2F1 and Nbs1 were cloned by standard methods (2) as GAL4 DNA binding or activation domain fusions. Interaction testing was performed on both histidine (with 3 mM 3-aminotriazole) and adenine selection, using three independent clones for each interaction.
Chromatin immunoprecipitations.
Cells were formaldehyde cross-linked essentially as described previously for HeLa cells (7), except that cells were swelled in 5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 8.0), 85 mM KCl, 0.5% IGEPAL CA-330, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 100 ng of leupeptin/ml, and 100 ng of aprotinin/ml instead of reticulocyte standard buffer. Chromatin was sheared to an average size of approximately 1,000 bp. Each immunoprecipitation reaction contained 1 μg of antibody. One μg of rabbit anti-mouse antibody (ICN) was added to E2F1 (KH20+KH95) antibody reactions for the last hour of immunoprecipitation. Antibody-protein-DNA complexes were isolated by immunoprecipitation with preblocked protein A-positive Staphylococcus aureus cells. Following extensive washes, bound DNA fragments were eluted and analyzed by PCR.
PCR analysis and Southern blotting.
Immunoprecipitates were dissolved in 30 μl of water (except for input samples, which were dissolved in 1,000 μl of water). Each reaction mixture contained 2 μl of immunoprecipitated DNA, 1× Taq reaction buffer, 1.5 mM MgCl2, 50 ng of each primer, 1.7 U of Taq polymerase (Promega), 200 μM each deoxynucleoside triphosphate, and 1 M betaine in a final reaction volume of 20 μl. PCR mixtures were amplified for 1 cycle of 95°C for 5 min, annealing temperature of the primers for 5 min, and 72°C for 3 min and either 28 (episomal) or 33 (endogenous) cycles of 95°C for 1 min, annealing temperature of the primers for 2 min, and 72°C for 1.5 min, followed by incubation at 72°C for 7 min. PCR products were electrophoresed through a 1.5% agarose gel and visualized with ethidium bromide. Linearity of PCR conditions was tested in reaction mixtures containing 0.1, 1, 2, or 4 μl of immunoprecipitated DNA and was analyzed by Southern blotting (6). The PCR primers (University of Wisconsin Biotechnology Center) used were the following: oriP 8099, 5′-CGCTCAGGCGCAAGTGTGTGTA-3′; oriP 8512, 5′-GGCAGGGACCAAGACAGGTGAA-3′; Myc 2411, 5′-GGCTTCTCAGAGGCTTGGCGGC-3′; Myc 2857, 5′-0-3′; MycDE2F, 5′-GGCTTCTCAGAGGCTTGAATTC-3′.
RESULTS
Nbs1 phosphorylation is not sufficient for activation of the S-phase checkpoint.
NBS lymphoblastoid cells homozygous for the common nbs1 657del5 allele (66) express an aberrant Nbs1 protein species, Nbs1p70, that lacks the N-terminal 221 aa but remains associated with the Mre11 complex (36). NBS lymphoblastoid cells exhibited RDS, as shown previously for untransformed lymphocytes (36, 60, 61). Hence, the FHA and BRCT domains missing from the N terminus in Nbs1p70 are likely to influence the S-phase regulatory function of the Mre11 complex.
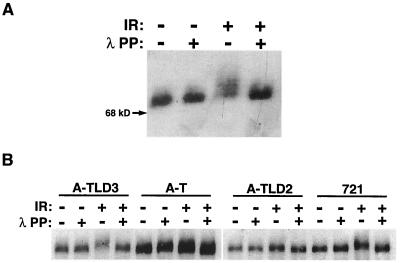
ATM phosphorylates Nbs1 in response to IR, and this event is necessary for activation of the S-phase checkpoint (20, 31, 67, 72). We asked whether Nbs1p70, which contains the primary IR-induced phosphorylation site, serine 343, was phosphorylated in response to IR. Nbs1p70 was phosphorylated after IR (Fig. 1A). In addition, IR-induced Nbs1 phosphorylation was observed in A-TLD3 cells but was nearly absent in A-TLD2 cells (Fig. 1B). Since each of these cells exhibits RDS, phosphorylation of either Nbs1p70 or full-length Nbs1 (present in A-TLD cells) is not sufficient for activation of the DNA damage-induced S-phase checkpoint.
FIG. 1.
IR-induced Nbs1 phosphorylation is compromised in A-TLD2 but not in A-TLD3 or NBS cells. (A) Nbs1p70 is phosphorylated after IR. NBS cells were irradiated with 20 Gy (+) or were mock treated (−) and harvested at 45 min post-IR. Nbs1 immunoprecipitations and lambda phosphatase treatments (λ PP) were performed, resolved by SDS-PAGE, and immunoblotted with Nbs1 antiserum. The migration of a 68-kDa molecular size marker protein is indicated. (B) Lymphoblasts from two different A-TLD patients (A-TLD2 and A-TLD3), A-T lymphoblasts (AT3LA), and a normal control (721) were mock treated (−) or irradiated (+) with 20 Gy and harvested for analysis at 45 min after treatment. Nbs1 immunoprecipitations were performed, and each immunoprecipitation reaction mixture was either mock treated (−) or treated with lambda phosphatase (+) as described for panel A.
Nbs1 associates with E2F1.
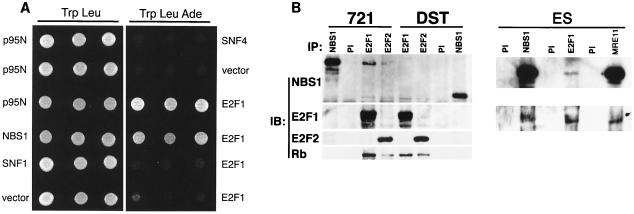
S-phase checkpoint failure in Nbs1p70-containing cells, notwithstanding IR-induced phosphorylation, suggested that the Nbs1 N-terminal region is required for checkpoint activation. We undertook a yeast two-hybrid screen to identify protein interactions mediated by the FHA and BRCT domains in the Nbs1 N terminus. Among 35 positive interactors representing 5 distinct genes, 2 encoded nearly full-length human E2F1 (aa 27 to 437). The interaction with E2F1 occurred via the N terminus of Nbs1 (Fig. 2A, p95N) (data not shown).
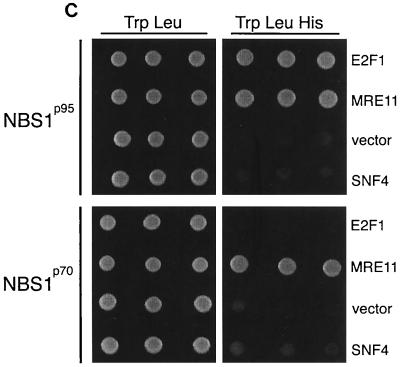
FIG. 2.
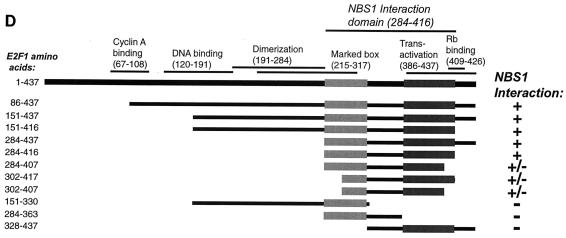
Nbs1 associates with E2F1. (A) The N terminus of Nbs1 associates with E2F1. The N terminus of Nbs1 (p95N; aa 1 to 180) was tested for association with E2F1 (aa 284 to 416), empty vector, or a nonspecific control (SNF1) by yeast two-hybrid testing. Positive (Nbs1) and negative (vector and SNF4) controls for E2F1 association are shown. Growth on Trp-Leu-Ade and the Trp-Leu control plates is shown; results on Trp-Leu-His plates were identical. (B) Coimmunoprecipitation of Nbs1 with E2F1 in wild-type but not NBS cells. Lysates were prepared from wild-type (721) or NBS (DST) lymphoblasts, and immunoprecipitations (IP) were performed with E2F1 or E2F2 antibodies or with Nbs1 or preimmune (PI) antiserum. The immune complexes were resolved by SDS-PAGE and serially immunoblotted (IB) with Nbs1 antiserum, E2F1, E2F2, and Rb antibodies. Reciprocal IPs from murine embyronic stem (ES) cell extracts were carried out with Nbs1 antiserum 93, E2F1 antiserum 70, and Mre11 antiserum 59 (58) immunoblotted with Nbs1 93 and KH95. (C) Nbs1p70 associates with Mre11 but not E2F1. GAL4 DNA binding domain fusions of full-length Nbs1 or Nbs1p70 were tested for interaction with GAL4 activation domain fusions of E2F1 (aa 284 to 416), Mre11, empty vector, or a nonspecific control (SNF4). Growth on Trp-Leu-His and the Trp-Leu control plates is shown; results on Trp-Leu-Ade plates were identical. (D) Deletion analysis of the E2F1-Nbs1 interaction domain. Various fragments of E2F1 were cloned as fusions to the GAL4 activation domain and were tested for the ability to associate with full-length Nbs1 fused to the GAL4 DNA binding domain. The portions of each E2F1 fusion protein are shown as bars under the diagram depicting the locations of known motifs and interaction domains in E2F1 (55). A positive interaction (+) was defined as growth on both His and Ade test plates, and a weak interaction (+/−) was scored as growth on His plates only. The shaded boxes represent minimally defined regions of E2F1 necessary for Nbs1 association.
Antibodies to E2F1 and E2F2 immunoprecipitated Nbs1 and Mre11 from normal human lymphoblast cell lysates (Fig. 2B and data not shown) as well as from Raji cells and murine embryonic stem cells. E2F1 and E2F2 antibodies failed to coimmunoprecipitate Nbs1p70 from NBS lymphoblasts (Fig. 2B), and Nbs1p70 did not associate with E2F1 in yeast two-hybrid assays (Fig. 2C). Thus, the FHA and BRCT domains missing from Nbs1p70 are required for Nbs1's association with E2F1 but not Mre11.
Deletion analyses indicated that the E2F1 Nbs1 interaction domain (Fig. 2D) encompassed portions of the marked box (aa 284 to 320) (30) and the transactivation and Rb-binding domains (aa 363 to 416) (55). Neither of these regions of E2F1 alone was sufficient for interaction with Nbs1 (Fig. 2D). Although Rb immunoprecipitated with E2F1 and Nbs1 (Fig. 2B), this assay could not distinguish whether the Mre11 complex-E2F1 association is exclusive of E2F1 association with Rb. Since the Mre11 complex is much more abundant than E2F1, it is expected that most Mre11 complex members are not associated with E2F. Nonetheless, E2F1 is present in Nbs1 and Mre11 immunoprecipitates from murine embryonic stem cell extracts (Fig. 2B).
E2F1 targets the Mre11 complex to E2F sites.
Recent findings for Drosophila melanogaster suggest that dE2F is important for the proper localization of the origin recognition complex (ORC) complex within the chorion gene cluster during embryogenesis. This effect does not require dE2F transcriptional activity, suggesting that dE2F acts directly at or near origins of replication (51).
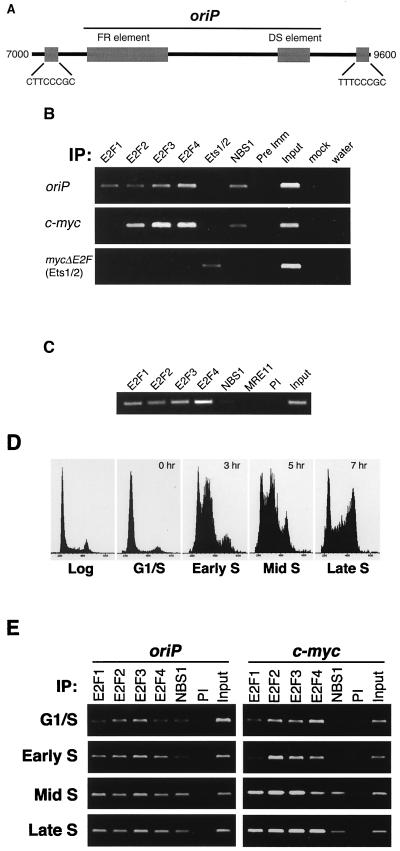
The Epstein-Barr virus (EBV) latent origin of replication, oriP, is regulated similarly to chromosomal origins during normal growth as well as after DNA damage (1, 13, 26, 71). Therefore, we examined oriP as a potential site of Nbs1-E2F interaction. Two E2F binding sites were found within 400 bp of oriP (Fig. 3A), as determined by DNA sequencing and mobility shift assays (data not shown). Chromatin immunoprecipitations were prepared from formaldehyde-cross-linked, logarithmically growing cells to test whether Nbs1 and E2F1 bound E2F sites near oriP and at chromosomal loci linked to replication origin activity. Nbs1 (and Mre11) and E2F family members bound near EBV oriP in Raji cells containing an oriP episome (Fig. 3B and data not shown). Nbs1, presumably via E2F2, also bound at chromosomal E2F sites in the vicinity of the c-myc promoter, a region which has been shown to contain replication origin activity (62) (Fig. 3B).
FIG. 3.
Nbs1-E2F1 localizes to EBV oriP. (A) Location of oriP E2F target sites. A segment of the EBV genome, nt 7000 to 9600 (70), including oriP, is depicted. The FR and DS elements (large gray boxes) of oriP are located at nt 7421 to 8043 and nt 8891 to 9131, respectively. The two E2F elements (small gray boxes with their corresponding sequences) are located at nt 7051 to 7058 and 9418 to 9425, respectively. Primers for chromatin immunoprecipitation are at the 3′ end of the FR element at nt 8099 and 8412. (B) Nbs1 localization to E2F target sites in log phase cells. Chromatin immunoprecipitations (IP) were performed on log phase Raji cells carrying an episomal plasmid containing a mutant E2F site in the c-myc promoter (mycDE2F), allowing Ets1/2 binding. Primers near E2F sites in oriP and the promoters of c-myc and mycDE2F were used to amplify DNA fragments from the indicated immunoprecipitations. Input, 0.1% of the total isolated chromatin; mock, no-antibody control; Pre Imm, Nbs1 preimmune antiserum (#16). PCRs were performed in the linear range (see Materials and Methods). (C) The Mre11 complex fails to bind to E2F sites in NBS cells. Chromatin immunoprecipitations were performed as described in the legend to panel B with the indicated antibodies from formaldehyde-cross-linked NBS lymphoblasts, and PCR was per- formed with c-myc-specific primers to detect bound DNA. PI, preimmune. (D) Cell cycle profiles of synchronized Raji cells used for the experiment depicted in panel E. Cells were synchronized at the G1/S border and released into S phase. Cells were harvested for propidium iodide staining and for chromatin immunoprecipitations (shown in panel E). The cells were harvested at 0 h (G1/S phase), 3 h (early S phase), 5 h (mid-S phase), and 7 h (late S phase) after release from synchrony. A representative log phase profile is shown for comparison. (E) Nbs1 localization to E2F sites in synchronized cells. Raji cells were synchronized at the G1/S border, released into S phase, and harvested at various times after release (given in the legend to panel D). Chromatin immunoprecipitations using the indicated antibodies were performed, and PCR using primers specific to the indicated loci was used to amplify those DNA fragments in the immunoprecipitates.
We tested whether Nbs1-E2F1 binding to origin-proximal E2F sites was altered during S phase. Chromatin immunoprecipitations were performed on synchronized cells harvested at time points during S-phase progression (Fig. 3D) and were examined for E2F site occupancy. Nbs1 and E2F1 binding at the c-myc E2F site as well as at the episomal oriP site increased markedly as cells progressed from G1/S phase to mid-S phase (Fig. 3E). In contrast, E2F3 and E2F4 occupancy did not appear to be modulated during S-phase progression. These data suggest the E2F1-Nbs1 interaction at the origin-proximal E2F site is enhanced when replication origins are active.
Neither Nbs1 nor E2F bound to an episomal c-myc promoter in which the E2F site was mutated to allow Ets1/2 binding (mycDE2F; Fig. 3B), nor were they found at a genomic locus (Fra1) lacking E2F binding sites (data not shown). In NBS cells, neither Nbs1p70 nor Mre11 bound to E2F sites, whereas E2F1 binding was unaffected (Fig. 3C). Thus, Nbs1 binding to E2F sites was dependent on E2F1.
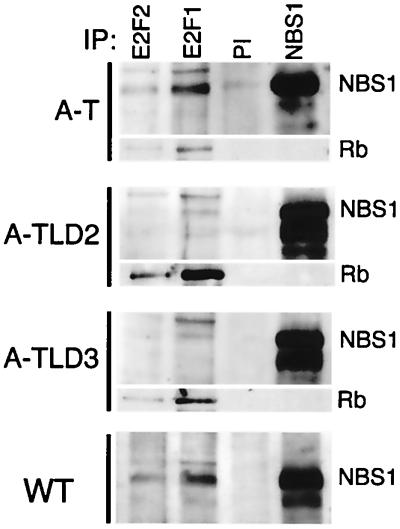
A-TLD but not A-T cells are deficient in Nbs1-E2F1 association.
To determine whether lack of Nbs1-E2F1 interaction was correlated with abrogation of the S-phase checkpoint, we carried out immunoprecipitations from A-TLD and A-T cells. The Mre11 complex-E2F1 association was reduced in both A-TLD2 (R633X Mre11 mutant [58]) and A-TLD3 cells (N117S Mre11 mutant) but remained intact in A-T cells (Fig. 4). Rb coimmunoprecipitated with E2F1 in both types of A-TLD cells (Fig. 4), demonstrating that the defective Nbs1-E2F1 association was intrinsic to the mutant Mre11 complex. Thus, disruption of the Mre11 complex-E2F1 association is common to Mre11 complex mutant cells that lack the S-phase checkpoint. Reduced levels of Nbs1-E2F complexes in A-TLD cells may reflect that Nbs1 is less abundant and is primarily cytoplasmic in these mutants (58). This interaction remains intact in A-T cells, consistent with the hypothesis that both Nbs1 phosphorylation and E2F interaction are necessary for S-phase checkpoint activation.
FIG. 4.
Nbs1-E2F1 association is maintained in A-T cells but is reduced in A-TLD cells. Immunoprecipitations (IP) were performed as described in the legend to Fig. 2B from the indicated cell lines and were immunoblotted serially with Nbs1 and Rb antibodies. WT, wild type; PI, preimmune.
The Mre11 complex localizes with replication sites.
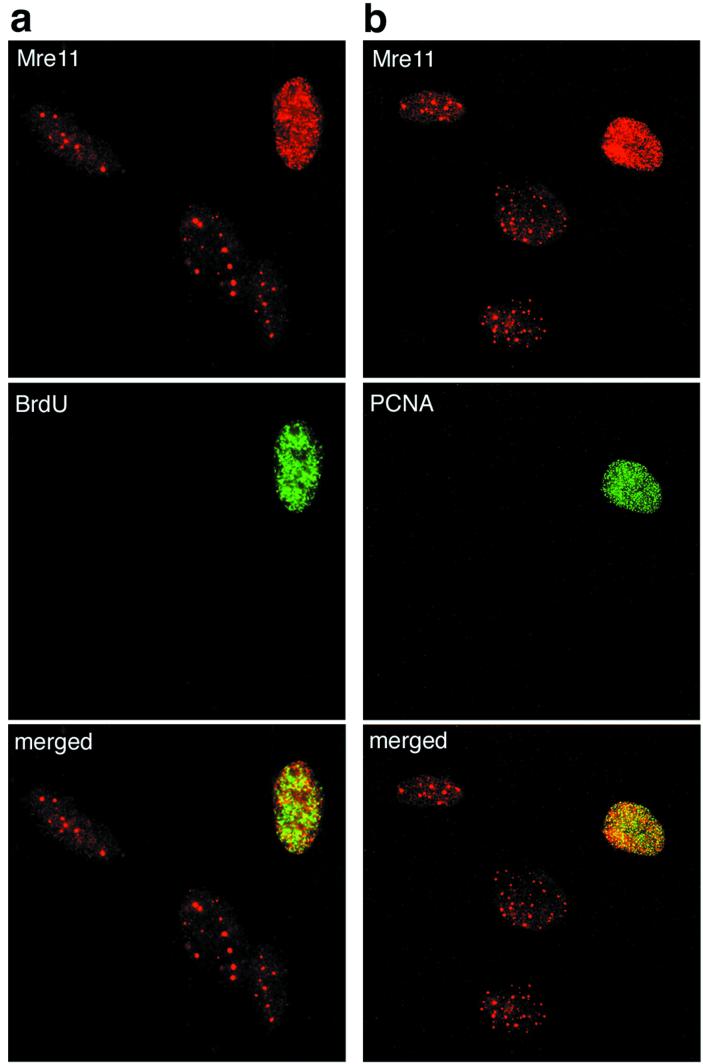
The phenotypic features of mammalian and yeast Mre11 complex mutants suggest that the complex plays an important role in the regulation of DNA replication in response to DNA damage. We adapted an in situ fractionation technique to assess whether the cytologic behavior of the Mre11 complex during cell cycle progression reflected its S-phase-dependent association with origin-proximal E2F sites (24, 37). We found that 10 to 20% of normally growing cells exhibited Mre11 and Nbs1 foci similar to those observed in irradiated cells with the same in situ fractionation conditions (37) (Fig. 5 and data not shown). To determine whether the focus-positive cells were in S phase, normal human fibroblasts were pulsed with BrdU and stained for both BrdU and Mre11. Nuclei with Mre11 foci were BrdU positive (Fig. 5a). PCNA positivity following extraction was also used to identify S-phase cells (8). As with BrdU incorporation, Mre11 foci were only seen in PCNA-positive cells (Fig. 5b). These data indicated that the Mre11 complex was associated with sites of DNA replication under normal (i.e., nonstressed) growth conditions.
FIG. 5.
The speckled Mre11 focus pattern is found in nonirradiated S-phase cells. (a) Speckled Mre11 foci are contained in BrdU-positive cells and overlap with BrdU incorporation. Nonirradiated 37Lu human fibroblasts were doubly stained for Mre11 (top panel, red signal) and BrdU incorporation (middle panel, green signal), and the images were merged (bottom panel). Overlap between the two signals appears yellow. The three nuclei that do not stain with BrdU correspond to those containing promyelocytic leukemia protein-associated Mre11 foci (37). (b) Speckled Mre11 foci are contained in PCNA-positive S-phase cells. Cells were doubly labeled for Mre11 (top panel, red signal) and PCNA (middle panel, green signal), and the images were merged (bottom, yellow signal). Overlap between the two signals appears yellow.
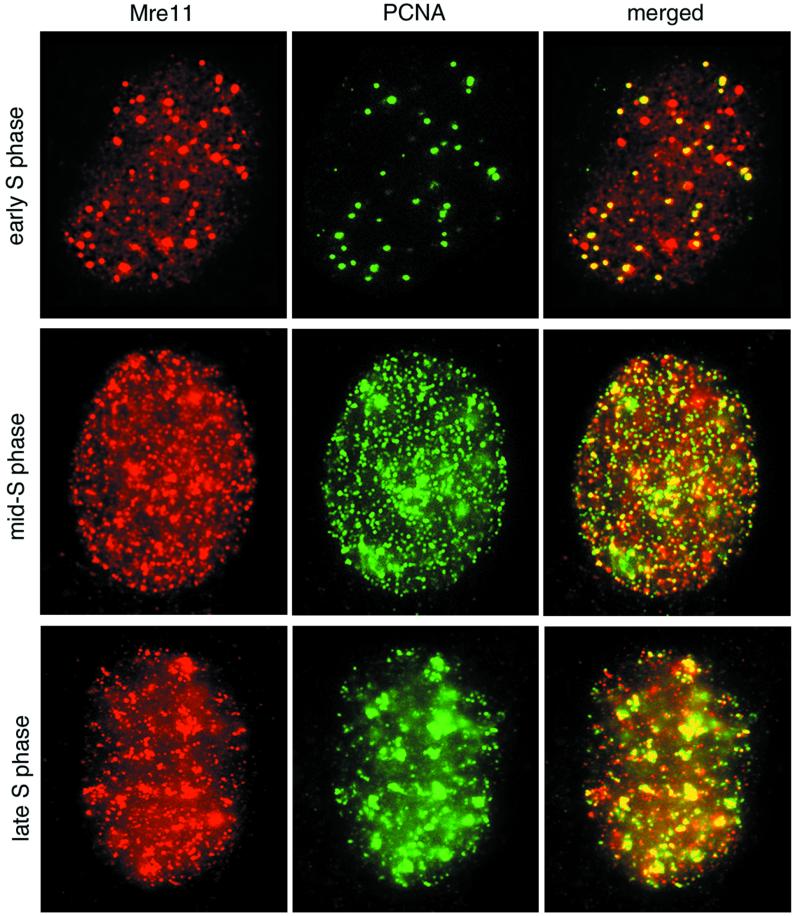
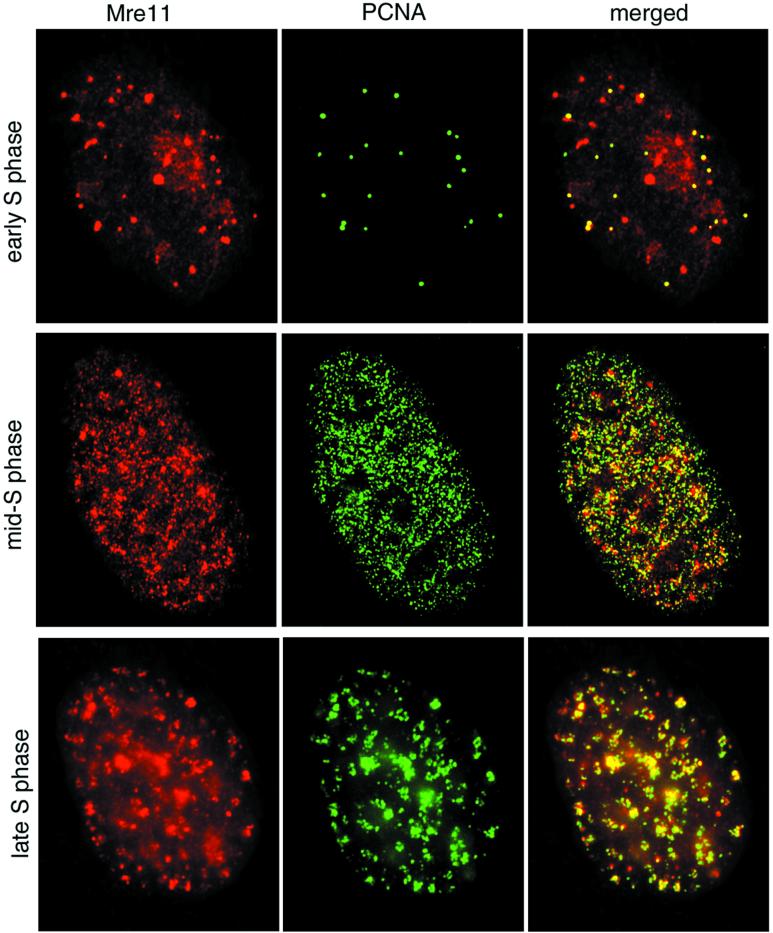
PCNA-containing DNA replication foci undergo characteristic changes as replication progresses (8, 24). We examined the temporal and spatial relationship between Mre11 and PCNA foci during S-phase progression. At the G1/S transition, PCNA is localized at a few discrete sites prior to the appearance of nascent DNA (24). These early PCNA foci colocalized with many Mre11 foci, consistent with chromatin immunoprecipitation data placing the complex at sites adjacent to replication origins (Fig. 6, top row). In early- to mid-S phase, Mre11 and PCNA foci were distributed throughout the nucleus and were substantially colocalized (Fig. 6, middle row). In late S phase, PCNA staining is localized at heterochromatic regions containing late replicating DNA (24). The majority of these late replication clusters contained both PCNA and Mre11 (Fig. 6, bottom row). Thus, colocalization of Mre11 was observed with normal DNA replication patterns throughout S phase.
FIG. 6.
Mre11 complex and PCNA colocalization during S-phase progression in normal cells. 37Lu fibroblasts were doubly labeled with Mre11 (red) and PCNA (green) as for Fig. 5b, and the images were merged. Overlap between the two signals in merged images appears yellow. Top row, early S phase; middle row, early- to mid-S phase; bottom row, mid- to late-S phase. Stages were determined by the method of Kill et al. (24).
We tested whether Mre11 complex localization to replication sites during normal S phase was ATM dependent. A-T fibroblasts were double labeled for Mre11 and PCNA. The pattern of Mre11 (and Nbs1; data not shown) and PCNA staining in S-phase A-T cells was indistinguishable from that of normal cells (Fig. 7). Thus, as in cells treated with IR (37), ATM is not required for relocalization of the Mre11 complex in S phase.
FIG. 7.
Mre11 colocalization with PCNA in A-T fibroblasts. Cells and images were prepared as described in the legend to Fig. 6. Top row, early S phase; middle row, early- to mid-S phase; bottom row, mid- to late-S phase. Stages were determined by the method of Kill et al. (24).
DISCUSSION
In this study, we demonstrated that the Mre11 complex physically associates with E2F protein family members via the N terminus of Nbs1. We established evidence that the Nbs1-E2F interaction occurs near an origin of DNA replication and that interaction at those sites is essentially restricted to S-phase cells, suggesting that the interaction coincides with origin activity. Abrogation of the interaction is correlated with S-phase checkpoint deficiency—the failure to suppress origin firing in response to DNA damage—irrespective of ATM-mediated phosphorylation of Nbs1, suggesting that this interaction may facilitate both positive and negative influences on DNA replication. Mre11 complex proteins were also associated with sites of DNA replication removed from the replication origin. We observed colocalization with PCNA at sites of DNA synthesis throughout S phase. Together, these data suggest that the Mre11 complex is important in both the regulation and completion of DNA replication.
Activation of the S-phase checkpoint suppresses the firing of replication origins, whereas the progression of established replication forks is essentially unimpeded (27, 28, 42, 50). The targets of E2F transcriptional control include genes that encode enzymes required for DNA synthesis; these enzymes are presumably required for replication fork progression (16). Thus, it is unlikely that the Nbs1-E2F1 interaction influences the S-phase checkpoint through altering E2F1-dependent transcriptional regulation. Supporting this interpretation, we found that gamma irradiation of murine embryonic fibroblasts or human lymphoblastoid cells transfected with an E2F1-dependent luciferase reporter did not alter luciferase expression (data not shown).
Chromatin immunoprecipitation demonstrated E2F-Nbs1 occupancy of E2F sites in close proximity (within 500 nucleotides [nt]) to replication origins at both oriP and c-myc (62). These data suggest that a subset of E2F sites may be associated with replication origins in human cells. The E2F-Nbs1 interaction at sites adjacent to replication origins was largely restricted to cells in S phase (Fig. 3D and E). Therefore, we hypothesize that the influence of the Mre11 complex and E2F1 on origins of DNA replication is direct. Precedent for this interpretation comes from D. melanogaster. During Drosophila embyrogenesis, endoreduplication of the chorion gene cluster in follicle cells leads to amplification of the chorion genes (11, 41). The switch from normal replication to endoreduplication is associated with relocalization of the ORC complex at replication origins within the chorion gene cluster (51). dE2F mutants impair ORC complex localization to those sites. The effect is seen in a dE2F mutant with reduced DNA binding activity that retains transactivation and Rb binding functions. Conversely, ORC localization is normal in a dE2F mutant that retains DNA binding capacity but lacks transactivation and Rb binding activity (51). Finally, evidence for direct physical interaction between dE2F and the ORC complex has been established (5), consistent with a similar role for human E2F1. These data support a model wherein the sequence-specific DNA binding of E2F1 directs the Mre11 complex to origins of replication where its functions are relevant to origin function. Such recruitment of Mre11 complex functions is also observed at mammalian telomeres via the complex's interaction with the telomere protection protein, TRF2 (74). Interestingly, the presumptive TRF2 binding sequence, TTAGGG, is found at two sites in oriP and one site in the myc promoter region. Since chromatin immunoprecipitation of oriP with Nbs1 antiserum depends on E2F binding (Fig. 3B), TRF2-Mre11 complex interaction does not occur at those sites.
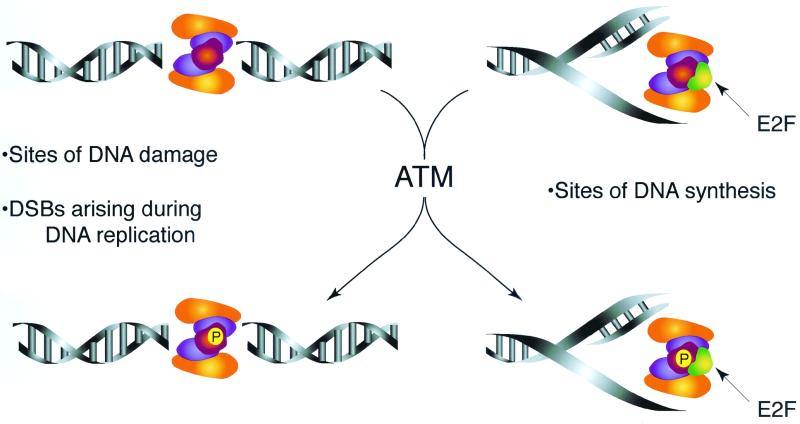
Previous studies established that the complex's function in S-phase checkpoint activation required phosphorylation of Nbs1 by ATM (31, 72). In this study, we found that phosphorylation of Nbs1p70 and Nbs1 occurred in gamma-irradiated NBS and A-TLD3 cells, respectively, indicating that this event is not sufficient for checkpoint activation. We demonstrated that NBS and A-TLD cells were deficient in Nbs1-E2F1 association. Hence, Nbs1-E2F interaction and Nbs1 phosphorylation may both be required for suppression of origin firing in response to DNA damage. The interaction of E2F1-Nbs1 on chromatin or in protein extracts was not affected by DNA damage (data not shown). This suggests that ATM acts on the complex at the sites of DNA replication and presumably alters its activity. This view is consistent with previous data that suggest ATM phosphorylates Nbs1 when it is bound to DNA damage (37) (Fig. 8).
FIG. 8.
Functional interaction of ATM and the Mre11 complex. The Mre11 complex relocalization in response to DNA damage (37) and during DNA replication is independent of ATM. We therefore infer that the Mre11 complex is situated at spontaneously arising DNA breaks or proximal to sites of DNA replication as part of its function in normally growing cells. We propose that upon the induction of DNA damage, ATM is activated and acts on the Mre11 complex engaged at those structures. This implies that the complex's role in S-phase checkpoint activation is partially dependent upon ATM effects on the complex's function at those sites. It is conceivable that ATM modification may also enhance Mre11 complex DNA repair functions, as has been suggested for Tel1-dependent modification of the S. cerevisiae Mre11 complex (65).
The nature of the change in activity imparted by Nbs1 phosphorylation and the function(s) of the Mre11 complex at sites of DNA replication remain to be established. Previous analyses have not implicated these proteins in origin firing or in the assembly of the ORC complex (3, 17). The Mre11 complex may mediate the same functions proximate to origins and at replication forks, as suggested for the minichromosome maintenance proteins (17). Like the homologous SbcCD complex (14), the Mre11 complex may process DNA secondary structures that arise at replication forks. Its presence at origins may reflect the processing of secondary structures forming at sites of localized helical distortions during the initiation of DNA synthesis.
An alternative possibility is suggested by the behavior of S. cerevisiae Mre11 complex mutants in the initiation of meiotic recombination. The formation of nuclease hypersensitive sites in meiotic chromatin prior to the initiation of meiotic recombination is altered in Mre11 mutants (19, 40). The Mre11 complex could affect DNA replication origins by influencing analogous changes in local chromatin architecture (3), with specificity for replication origins conferred by E2F1. Modulation of chromatin structure at the origin may also be governed by E2F1. E2F-Rb and related complexes have been shown to recruit histone modification proteins that alter local chromatin structure during transcriptional activation (18, 48). A similar role for dE2F in modulation of chromatin structure at replication origins has been proposed (51).
The colocalization of Mre11 complex proteins with PCNA throughout S phase indicates that the complex also functions at established replication forks. Recombination and replication are intimately linked (49). Homologous recombination requires both leading and lagging strand replication proteins (23). Conversely, DNA recombination is required to reestablish collapsed replication forks (15, 34, 53), and DNA recombination intermediates are formed during DNA replication in normally growing S. cerevisiae cells (75).
The complex's role in DNA recombination (9, 21) may reflect that human Mre11 complex proteins colocalized with PCNA are functioning in the resolution of damaged or stalled replication forks. This interpretation is consistent with the functions of the bacterial homologue of the Mre11 complex, SbcCD, which has been proposed to degrade secondary structures on the lagging strand template and thereby induces recombination between sister chromatids. This mechanism would stabilize DNA sequences, such as palindromes, that are prone to form aberrant DNA structures (14, 29). It is likely that the spontaneous genomic instability observed in Mre11 complex mutant cells stems from reduced efficiency in this function and that this contributes to the disease phenotypes observed in NBS and A-TLD patients. The loss of Mre11 complex-E2F interaction must also be considered as a contributing factor in the diverse pathology associated with those syndromes.
ACKNOWLEDGMENTS
We thank D. Eick for the gift of Raji 525-7 cells, T. de Lange for helpful discussion and comments, and Allen Edmonds for critical insight.
This work was supported by the Milwaukee Foundation (J.H.J.P.), the National Institutes of Health (GM59413 to J.H.J.P., CA09681 to J.W.), the Department of Energy (J.H.J.P.), and the Public Health Service (CA45240 to P.J.F.).
Footnotes
Report 3572 from the University of Wisconsin—Madison Laboratory of Genetics.
REFERENCES
- 1.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. 1–3. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 3.Bogan J A, Natale D A, Depamphilis M L. Initiation of eukaryotic DNA replication: conservative or liberal? J Cell Physiol. 2000;184:139–150. doi: 10.1002/1097-4652(200008)184:2<139::AID-JCP1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 5.Bosco G, Du W, Orr-Weaver T L. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat Cell Biol. 2001;3:289–295. doi: 10.1038/35060086. [DOI] [PubMed] [Google Scholar]
- 6.Boyd K E, Farnham P J. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol Cell Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bravo R, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bressan D A, Baxter B K, Petrini J H J. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callebaut I, Mornon J P. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 11.Calvi B R, Lilly M A, Spradling A C. Cell cycle control of chorion gene amplification. Genes Dev. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carney J P, Maser R S, Olivares H, Davis E M, Le Beau M J, Yates III R, Hays L, Morgan W F, Petrini J H J. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 13.Cleaver J E, Rose R, Mitchell D L. Replication of chromosomal and episomal DNA in X-ray-damaged human cells: a cis- or trans-acting mechanism? Radiat Res. 1990;124:294–299. [PubMed] [Google Scholar]
- 14.Connelly J C, Kirkham L A, Leach D R. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 16.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. . (Erratum, 15:5846–5847.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutta A, Bell S P. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 18.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 19.Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatei M, Young D, Cerosaletti K M, Desai-Mehta A, Spring K, Kozlov S, Lavin M F, Gatti R A, Concannon P, Khanna K K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 21.Haber J E. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann K, Bucher P. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 23.Holmes A M, Haber J E. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 24.Kill I R, Bridger J M, Campbell K H, Maldonado-Codina G, Hutchison C J. The timing of the formation and usage of replicase clusters in S-phase nuclei of human diploid fibroblasts. J Cell Sci. 1991;100:869–876. doi: 10.1242/jcs.100.4.869. [DOI] [PubMed] [Google Scholar]
- 25.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb J R, Petit-Frere C, Broughton B C, Lehmann A R, Green M H. Inhibition of DNA replication by ionizing radiation is mediated by a trans-acting factor. Int J Radiat Biol. 1989;56:125–130. doi: 10.1080/09553008914551271. [DOI] [PubMed] [Google Scholar]
- 27.Larner J M, Lee H, Hamlin J L. S phase damage sensing checkpoints in mammalian cells. Cancer Surv. 1997;29:25–45. [PubMed] [Google Scholar]
- 28.Larner J M, Lee H, Little R D, Dijkwel P A, Schildkraut C L, Hamlin J L. Radiation down-regulates replication origin activity throughout the S phase in mammalian cells. Nucleic Acids Res. 1999;27:803–809. doi: 10.1093/nar/27.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leach D R. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 30.Lees J A, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim D-S, Kim S-T, Xu B, Maser R S, Lin J, Petrini J H J, Kastan M B. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 32.Lim D-S, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo G, Yao M S, Bender C F, Mills M, Bladl A R, Bradley A, Petrini J H. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci USA. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marians K J. PriA-directed replication fork restart in Escherichia coli. Trends Biochem Sci. 2000;25:185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 35.Maser R S, Monsen K J, Nelms B E, Petrini J H J. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maser R S, Zinkel R, Petrini J H J. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- 37.Mirzoeva O, Petrini J H J. DNA damage dependent nuclear dynamics of the MRE11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelms B E, Maser R S, MacKay J F, Lagally M G, Petrini J H J. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 39.Nickerson J A, Krockmalnic G, He D C, Penman S. Immunolocalization in three dimensions: immunogold staining of cytoskeletal and nuclear matrix proteins in resinless electron microscopy sections. Proc Natl Acad Sci USA. 1990;87:2259–2263. doi: 10.1073/pnas.87.6.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohta K, Nicolas A, Furuse M, Nabetani A, Ogawa H, Shibata T. Mutations in the MRE11, RAD50, XRS2, and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc Natl Acad Sci USA. 1998;95:646–651. doi: 10.1073/pnas.95.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orr-Weaver T L. Drosophila chorion genes: cracking the eggshell's secrets. Bioessays. 1991;13:97–105. doi: 10.1002/bies.950130302. [DOI] [PubMed] [Google Scholar]
- 42.Painter R B. Radioresistant DNA synthesis: an intrinsic feature of ataxia telangiectasia. Mutat Res. 1981;84:183–190. doi: 10.1016/0027-5107(81)90061-0. [DOI] [PubMed] [Google Scholar]
- 43.Paull T T, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 44.Paull T T, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrini J H. The mammalian Mre11-Rad50-Nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am J Hum Genet. 1999;64:1264–1269. doi: 10.1086/302391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrini J H. The Mre11 complex and ATM: collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 47.Petrini J H J, Xiao Y-H, Weaver D T. DNA ligase I mediates essential functions in mammalian cells. Mol Cell Biol. 1995;15:4304–4308. doi: 10.1128/mcb.15.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson K D, Ait-Si-Ali S, Yokochi T, Wade P A, Jones P L, Wolffe A P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 49.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break.”. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 50.Rowley R, Phillips E N, Schroeder A L. The effects of ionizing radiation on DNA synthesis in eukaryotic cells. Int J Radiat Biol. 1999;75:267–283. doi: 10.1080/095530099140456. [DOI] [PubMed] [Google Scholar]
- 51.Royzman I, Austin R J, Bosco G, Bell S P, Orr-Weaver T L. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 1999;13:827–840. doi: 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 53.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 54.Sharan S K, Morimatsu M, Albrecht U, Lim D S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 55.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. In: Farnham P J, editor. Transcriptional control of cell growth: the E2F gene family. Vol. 208. New York, N.Y: Springer-Verlag; 1996. pp. 1–30. [DOI] [PubMed] [Google Scholar]
- 56.Sonoda E, Sasaki M S, Buerstedde J M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soulier J, Lowndes N F. The BRCT domain of the S. cerevisiae checkpoint protein Rad9 mediates a Rad9-Rad9 interaction after DNA damage. Curr Biol. 1999;9:551–554. doi: 10.1016/s0960-9822(99)80242-5. [DOI] [PubMed] [Google Scholar]
- 58.Stewart G S, Maser R S, Stankovic T, Bressan D A, Kaplan M I, Jaspers N G J, Raams A, Byrd P J, Petrini J H J, Taylor A M R. The DNA double strand break repair gene hMre11, is mutated in individuals with a new ataxia telangiectasia like disorder (ATLD) Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 59.Sun Z, Hsiao J, Fay D S, Stern D F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 60.Taalman R D, Hustinx T W, Weemaes C M, Seemanova E, Schmidt A, Passarge E, Scheres J M. Further delineation of the Nijmegen breakage syndrome. Am J Med Genet. 1989;32:425–431. doi: 10.1002/ajmg.1320320332. [DOI] [PubMed] [Google Scholar]
- 61.Taalman R D, Jaspers N G, Scheres J M, de Wit J, Hustinx T W. Hypersensitivity to ionizing radiation, in vitro, in a new chromosomal breakage disorder, the Nijmegen breakage syndrome. Mutat Res. 1983;112:23–32. doi: 10.1016/0167-8817(83)90021-4. [DOI] [PubMed] [Google Scholar]
- 62.Tao L, Dong Z, Leffak M, Zannis-Hadjopoulos M, Price G. Major DNA replication initiation sites in the c-myc locus in human cells. J Cell Biochem. 2000;78:442–457. doi: 10.1002/1097-4644(20000901)78:3<442::aid-jcb9>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 63.Trujillo K M, Yuan S-S F, Lee E Y-H P, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 64.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Usui, T., H. Ogawa, and J. H. J. Petrini. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell, in press. [DOI] [PubMed]
- 66.Varon R, Vissinga C, Platzer M, Cerosaletti K M, Chrzanowska K H, Saar K, Beckmann G, Seemanova E, Cooper P R, Nowak N J, Stumm M, Weemaes C M, Gatti R A, Wilson R K, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 67.Wu X, Ranganathan V, Weisman D S, Heine W F, Ciccone D N, O'Neill T B, Crick K E, Pierce K A, Lane W S, Rathbun G, Livingston D M, Weaver D T. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 68.Xiao Y, Weaver D T. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamaguchi-Iwai Y, Sonoda E, Sasaki M S, Morrison C, Haraguchi T, Hiraoka Y, Yamashita Y M, Yagi T, Takata M, Price C, Kakazu N, Takeda S. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao S, Weng Y-C, Yuan S-S F, Lin Y-T, Hsu H-C, Lin S-C J, Gerbino E, Song M-H, Zdzienicka M Z, Gatti R A, Shay J W, Ziv Y, Shiloh Y, Lee E Y-H P. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- 73.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 74.Zhu X D, Kuster B, Mann M, Petrini J H, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 75.Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]