Abstract
We examined the selective pressure for, and the impact of, p53 inactivation during epithelial tumor evolution in a transgenic brain tumor model. In TgT121 mice, cell-specific inactivation of the pRb pathway in brain choroid plexus epithelium initiates tumorigenesis and induces p53-dependent apoptosis. We previously showed that p53 deficiency accelerates tumor growth due to diminished apoptosis. Here we show that in a p53+/− background, slow-growing dysplastic tissue undergoes clonal progression to solid angiogenic tumors in all animals. p53 is inactivated in all progressed tumors, with loss of the wild-type allele occurring in 90% of tumors. Moreover, similar progression occurs in 38% of TgT121p53+/+ mice, also with loss of at least one p53 allele and inactivation of p53. Thus, the selective pressure for p53 inactivation, likely based on its apoptotic function, is high. Yet, in all cases, p53 inactivation correlates with progression beyond apoptosis reduction, from dysplasia to solid vascularized tumors. Hence, p53 suppresses tumor progression in this tissue by multiple mechanisms. Previous studies of fibroblasts and hematopoietic cells show that p53 deficiency can be associated with chromosomal instability, a mechanism that may drive tumor progression. To determine whether genomic gains or losses are present in tumors that progress in the absence of p53, we performed comparative genomic hybridization analysis. Surprisingly, the only detectable chromosomal imbalance was partial or complete loss of chromosome 11, which harbors the p53 gene and is thus the selected event. Flow cytometry confirmed that the majority of tumor cells were diploid. These studies indicate that loss of p53 function is frequent under natural selective pressures and furthermore that p53 loss can facilitate epithelial tumor progression by a mechanism in addition to apoptosis reduction and distinct from chromosomal instability.
The p53 gene is mutated in at least 50% of human cancers, including most tumor types (27, 35, 43). Although many possible mechanisms for p53 tumor suppression have been defined for cultured cells, no mechanism has been fully established in vivo. In cultured cells, p53 is activated in response to a variety of cellular stress signals including DNA damage, aberrant proliferation, hypoxia, and nucleotide deprivation (36, 50, 56). Activation of p53 leads to either growth arrest or apoptosis, a decision that appears to depend on the cell type and specific stimulus (2, 22, 36, 65, 67). Thus, the in vivo signals for p53 tumor suppression and the mechanism by which p53 inactivation contributes to tumorigenesis are likely to vary depending on the cell type and environment. Based on the known functions of p53, there are several mechanisms by which p53 inactivation could contribute to cancer. For example, p53 regulates a G1 checkpoint in fibroblasts in response to DNA damage, a response that requires transcriptional activation of the p53 target gene p21 (7, 14). In addition, p53 can arrest cells in G2, possibly via transcriptional activation of 14-3-3 ς (25) and/or transcriptional repression of cdc2 and cyclin B1 (61). Importantly, Chk1 and Cds1/Chk2 kinases involved in replication and DNA damage-induced G2 arrest have recently been shown to phosphorylate p53 (9, 26, 58). The identification of a Cds1/Chk2 mutation in a Li-Fraumeni syndrome family that lacks p53 mutation supports the idea that Cds1/Chk2 and p53 function in a pathway for tumor suppression (3). Finally, p53 prevents DNA endoreduplication in fibroblasts exposed to mitotic spindle inhibitors (15) and has been implicated in centrosome regulation (20). In the absence of p53, cultured fibroblasts can be readily selected for gene amplification, indicating a tendency for genetic instability upon p53 deficiency (38, 73). These observations led to the “guardian of the genome” hypothesis for p53 tumor suppression (32), which suggests that p53 inactivation may contribute to tumorigenesis by facilitating the propagation of genetically defective cells as a result of checkpoint loss (23). Secondary mutations or chromosomal changes that provide the cell a selective advantage would thus facilitate tumorigenesis. Indeed, hematopoietic tissues from p53-deficient mice contain a high percentage of aneuploid cells (6, 21), the mice are predisposed to thymic lymphoma (17, 30), and lymphomas are aneuploid (37, 64). However, the direct contribution of p53-deficiency-induced genetic instability to tumorigenesis has not been demonstrated in vivo.
Another mechanism by which p53 can suppress tumorigenesis involves the apoptotic response to aberrant proliferation induced by oncogene expression or pRb inactivation. Many studies have demonstrated that this response is p53 dependent (13, 28, 39, 41, 42, 47, 53, 68), indicating that developing tumors could activate p53, thus establishing a selective pressure for p53 inactivation leading to tumor cell survival. We previously demonstrated such a role for p53 in vivo using a transgenic mouse model that undergoes epithelial cell tumorigenesis in response to pRb pathway inactivation (60). In the choroid plexus (CP) epithelium, p53 inactivation alone is inconsequential, as is the case for most mouse epithelial cells (17, 30). However, cell-specific inactivation of the pRb pathway induces aberrant proliferation and dysplastic growth, resulting in p53-dependent apoptosis (see Fig. 1). A similar response occurs in the lens (46) and retina (28) and in several cell types during development (41, 42). In the brain tumor model, germ line p53 deficiency leads to significant acceleration of dysplastic tissue growth due to an 85% reduction in apoptosis (60). Importantly, most human tumors harbor a defect in the pRb pathway (70); thus, p53 and Rb pathway mutations frequently coexist during the course of tumorigenesis. Hence, it is important to understand the dynamic interplay between these aberrations during tumor development. The dependency of tumor cell apoptosis on p53 function suggests that selective pressure for inactivation of the p53 pathway should be high in a developing tumor. Such direct selection for p53 inactivation is unlikely to result from a role in genome maintenance alone. However, once a p53-deficient cell is selected based on survival, its propagation in the absence of p53-dependent checkpoints could result in genetic aberrations that may accelerate tumor progression. By this scenario, p53 inactivation would contribute to tumorigenesis by multiple mechanisms. In the present report, we test this hypothesis by exploring the evolution of developing CP tumors in p53 heterozygous and wild-type backgrounds. We assess the natural selective pressure for p53 inactivation and the contribution of p53 inactivation to tumor progression. In particular, we explore the possibility that chromosomal instability drives tumor progression in tumors that evolve to a p53-deficient state. The contribution of p53 inactivation to tumor progression during the natural evolution of epithelial tumors lacking the Rb pathway has not previously been explored.
FIG. 1.
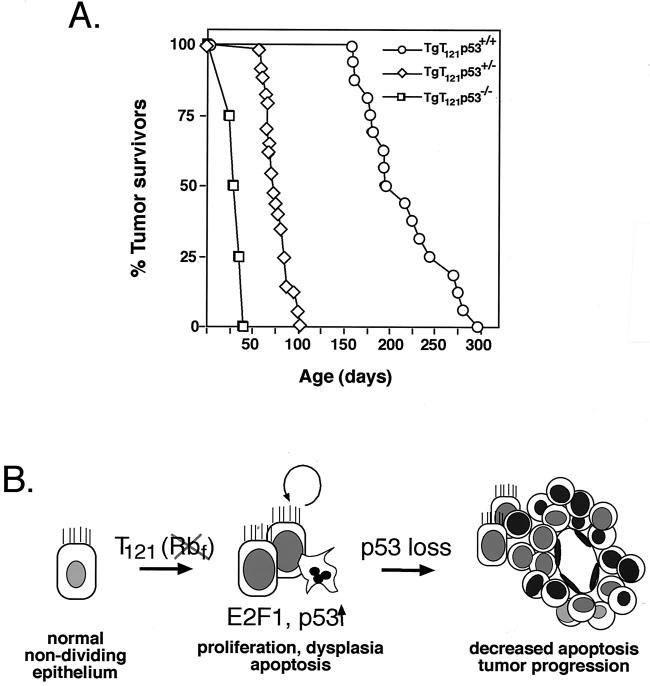
A pathway to tumor development. (A) Time of survival from brain tumors depends on p53 status. Mice were sacrificed when they showed severely bulged crania and decreased activity. Histological examination confirmed the presence of CP masses in all mice. (Data are revised from those presented in reference 60.) (B) The diagram depicts previously elucidated steps in T121-induced development of CP tumors. T121 induces proliferation of normally nondividing epithelial cells by inactivating the pRb family proteins pRb, p107, and p130. p53-dependent apoptosis is then activated by a process requiring E2F1 (48). As shown in the present report, in a p53 heterozygous background, the selective pressure for p53 inactivation is 100%, resulting in the focal development of solid vascularized tumors.
MATERIALS AND METHODS
Mice.
Generation, screening, and characterization of TgT121 transgenic mice (B6D2) were described previously (54, 60). These mice harbor the T121 mutant T-antigen gene under the control of the lymphotropic papovavirus transcriptional signals, resulting in uniformly high levels of expression in the CP (10). The T121 transgene encodes the first 121 amino acids of simian virus 40 (SV40) T antigen and is capable of binding to the pRb family proteins but not to p53. TgT121p53+/− and TgT121p53−/− mice were generated by crossing TgT121+/+ or TgT121+/− mice with p53−/− mice (C57BL6/J; Jackson Laboratories) (30). Genotypes were identified by PCR analysis of tail DNA as described previously (60). Mice were monitored regularly for the outward signs of a brain tumor, which consist of cranial bulging and decreased activity. Mice were then sacrificed, and tumors were either frozen at −80°C for nucleic acid analysis or fixed in 10% formalin as described previously (54).
Histology and immunohistochemistry.
To analyze tumor morphology and development, mouse brains were cut into halves, fixed in 10% formalin, embedded in paraffin, and sectioned for 10 successive layers at 50-μm intervals. A 5-μm section from each layer was stained with hematoxylin and eosin for morphological analysis (60). To measure the tumor size, the slide with the largest tumor cross section was chosen for each sample, and the tumor area was measured using the PAXit image capture and analysis system (MIS, Inc.). The tumor volume was then calculated with the assumption that the tumor has the same volume as a sphere with an equivalent cross-sectional area.
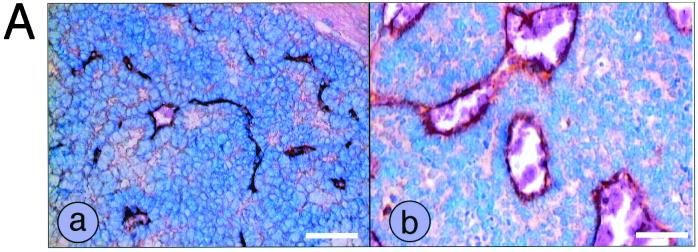
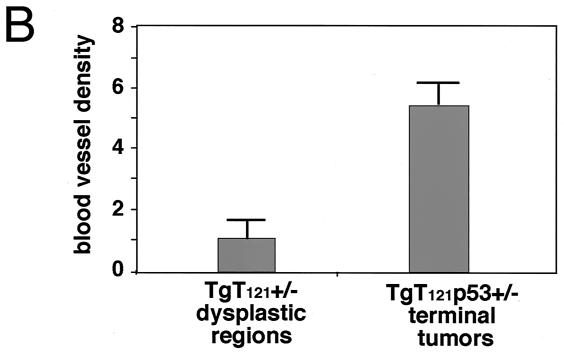
For CD31 immunostaining, sections were treated with 1 mg of trypsin/ml in phosphate-buffered saline (PBS) for 10 min at 37°C and then blocked in 5% normal rabbit serum (NRS) in PBS for 1 h at room temperature (RT). The sections were incubated with anti-CD31 antibody (1:50 in PBS containing 5% NRS; Pharmingen) overnight at 4°C. After three washes in PBS, slides were incubated with biotin-conjugated secondary antibody (1:100 in PBS containing 2% NRS) for 30 min at RT. Slides were washed twice with TS buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.1% Tween 20) and incubated with avidin-biotin-peroxidase–alkaline phosphatase reagent (Vector Laboratories) for 30 min. After two washes with TS buffer, slides were incubated with alkaline phosphate substrate (Vector Laboratories) for 10 min at RT, counterstained with methyl green, and dehydrated in two changes of xylene. The blood vessel pattern of dysplastic CP and tumors was evaluated by quantifying CD31-positive vessels. For each brain sample, the numbers of blood vessels larger than 200 μm2 were counted in fields with abundant blood vessels. Dysplastic CP and terminal tumors from seven T121p53+/− mice each were evaluated; the averages and the standard deviations of their blood vessel counts are represented in Fig. 3.
FIG. 3.
Tumor progression is associated with angiogenesis. Dysplastic CP (a) and tumor (b) sections from T121p53+/− mice were immunostained using an antibody specific for CD31 to detect blood vessels (A). Each white bar represents 50 μm. Sections from brains of seven T121p53+/− mice at 5 to 7 weeks were analyzed. On the average, tumors contained larger vessels than did dysplastic tissue. Since even normal CP contains numerous capillaries owing to its function as a blood-cerebrospinal fluid barrier, vessels larger than 200 μm2 were counted for a quantitative comparison of the dysplastic and tumor vasculatures (Materials and Methods). By this analysis, tumors contained a significant increase in such vessels (B).
Loss-of-heterozygosity (LOH) analysis.
To determine the status of the wild-type p53 allele in terminal TgT121p53+/− tumors, semiquantitative PCR analysis was performed. The wild-type p53 allele was amplified with primers directed against exon 6 (X6.5; 5′-ACAGCGTGGTGGTACCTTAT-3′) and exon 7 (X7; 5′-TATACTCAGAGCCGGCCT-3′), while the p53 null allele was amplified with primers directed against the neomycin gene contained within the targeted locus (neo18; 5′-CTATCAGGACATAGCGTTGG-3′) and p53 exon 7. The 25-μl PCR mixture contained 0.2 mM concentrations of each of the four deoxynucleoside triphosphates, 2 μCi of [α-32P]dCTP (3,000 Ci/mmol), 0.8 μM X6.5 and neo18, 1.6 μM X7, 0.5 U of Taq polymerase (Boehringer Mannheim), and 100 ng of DNA template. PCR was performed for 25 cycles using the following conditions: 1 min at 94°C, 2 min at 60°C, and 2 min at 72°C. PCR products were resolved on an 8% polyacrylamide gel and quantified using a PhosphorImager (Molecular Dynamics). In standardization assays, tail DNA from a p53+/− mouse and a p53−/− mouse were mixed at ratios ranging from 1:2 to 1:50. The ratio of the product intensity to the template concentration was graphed and resulted in a linear relationship. The ratio of wild-type allele intensity in the control lane was normalized to 1.00. With the assumption that the tumor was not likely to include more than 30% nontumor cells, an arbitrary 0.33 intensity ratio threshold was established for determining LOH. The normalized ratio threshold is 0.20. If the ratio of the p53 wild-type signal to p53 null signal was greater than 0.20, the p53 wild-type allele was considered to be retained; a ratio of less than 0.20 indicated loss. Two independent PCR assays were performed for each sample.
To determine the status of the wild-type p53 allele in terminal TgT121p53+/+ tumors, quantitative real-time PCR analysis was performed. The conditions for analysis of the p53 locus were provided by Lynda Chin and colleagues (Harvard University). The primers for the p53 allele were 5′-ATGGCCATCTACAAGAAGTCACAG-3′ and 5′-ATCGGAGCAGCGCTCATG-3′. The sequence of the p53 probe was 5′-ACATGACGGAGGTCGTGAGACGCTG-3′. The primers for the internal control β-actin gene were 5′-AAGAGCTATGAGCTGCCTGA-3′ and 5′-ACGGATGTCAACGTCACACT-3′. The sequence of the β-actin probe was 5′-CACTATTGGCAACGAGCGGTTCCG-3′. Each 25-μl reaction mixture contained 50 ng of DNA template, 18 nM p53 primers, 80 nM β-actin primers, 8 nM probe, and 12.5 μl of TaqMan Universal PCR Master Mix (Applied Biosystems) containing AmpliTaq Gold polymerase, deoxynucleoside triphosphates, and PCR buffer. The cycling conditions were 50°C for 2 min and 95°C for 10 min for 1 cycle and 95°C for 15 s and 60°C for 1 min for 40 cycles. The reactions were performed using an ABI 7700 Sequence Detection system (Applied Biosystems), and the data were analyzed using Sequence Detector 1.7 (Applied Biosystems) and standard protocols (http://www.appliedbiosystems.com). The copy number of each sample was determined by calculating ΔΔCt based on the formula ΔΔCt = [sample Ct(p53) − sample Ct(β-actin)] − [p53+/+ control Ct(p53) − p53+/+ control Ct(β-actin)], where Ct is the number of cycles required to reach a threshold based on linear amplification. Analyses of standard samples (L. Chin, Harvard University, personal communication) indicate that copy numbers of 2, 1, and 0 are indicated by 2−ΔCtn values of >0.6, 0.15 to 0.6, and <0.15, respectively. Standard samples analyzed along with experimental samples confirmed the accuracy of these assignments.
In situ RNA hybridization.
In situ hybridization was carried out as described previously (48). For template preparation, pBS-KSp21 was digested with EcoRI and BamHI. The antisense probe was generated by T7 transcription of an EcoRI-linearized template, and the sense probe was produced by T3 transcription of a BamHI-linearized template. Probes were labeled with [α-35S]UTP (5 × 104 cpm/μl) and hybridized to slides at 50°C overnight. Autoradiography was performed at 4°C for 3 days. Sections were counterstained with 0.2% toluidine blue and observed and photographed using dark-field microscopy.
CGH.
Comparative genomic hybridization (CGH) was performed essentially as described elsewhere (16, 31). Tumor and normal tail DNA were labeled by nick translation using fluorescein-12-dUTP (NEN) and Alex-568-5-dUTP (Molecular Probes), respectively. The optimum probe size was about 600 bp. Labeled tumor and normal DNA (1 μg each) were coprecipitated and dissolved in 10 μl of hybridization solution to obtain a final composition of 50% formamide, 10% dextran sulfate, and 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate [pH 7.0]). The mixture was heated to 70°C for 15 min to denature the DNA and was incubated at 37°C for 10 min. Normal mouse metaphase chromosomes prepared from mouse embryonic fibroblasts were denatured at 70°C in 70% formamide–2× SSC for 4 min and dehydrated through graded ethanols. The hybridization mixture was added to the slides, coverslips were sealed with rubber cement, and the slides were incubated at 37°C for 3 days. The slides were washed in formamide and SSC as previously described (31) and stained with 0.1 μM 4′,6′-diamino-2-phenylindole (DAPI). Digital images of each fluorochrome were obtained using a fluorescence microscope and a charge-coupled device camera. The profiles of different fluorescence intensities and corresponding chromosomal losses and gains were analyzed using Vysis software (Applied Images Inc.). The normalized average ratio of green to red (tumor/normal ratio) is 1.0. The threshold of variability using normal samples was 0.8 to 1.2.
Flow cytometry.
Brain tumor cells were isolated and stained as described (52). Since normal CP is difficult to cleanly dissect in sufficient quantity, normal spleen cells were used as a diploid control. Glass slides and 70-μm-pore-size cell strainers (Becton Dickinson) were used to dissociate tumor and spleen cells. The cells were washed with PBS, fixed in 70% ethanol, and stained with 25 μg of propidium iodide/ml containing 0.1 mg of RNase A/ml. Flow cytometry was carried out using a FACScan cytometer (Becton Dickinson), and data were analyzed with Cyclops software (Cytomation).
RESULTS
High rates of tumor progression in TgT121p53+/− and TgT121p53+/+ mice.
In TgT121 transgenic mice, pRb and related proteins p107 and p130 (collectively referred to here as pRbf) are inactivated in the CP by tissue-specific expression of the truncated SV40 T antigen, T121 (see Materials and Methods). Disruption of the pRb pathway in CP induces slow-growing dysplastic masses, resulting in death of the mice at an average of 26 weeks (Fig. 1A) (60). Upon pRbf inactivation, CP cells are induced to proliferate aberrantly (54), resulting in the induction of p53-dependent apoptosis (Fig. 1B) (60). In a p53−/− background, growth of the CP masses is accelerated approximately sevenfold (50% survival time = 4 weeks [Fig. 1A]) due to an 85% reduction in cell death, although masses remain morphologically dysplastic (60). We previously showed that TgT121p53+/− mice survive to an intermediate age (50% survival time = 11 weeks [Fig. 1]) and show morphological signs of clonal tumor progression (Fig. 1B) (60). As expected, apoptosis levels were significantly reduced in progressed tumors. This could occur by direct p53 inactivation or by inactivation of other genes in the relevant p53 pathway (if a linear pathway is involved) or in parallel pathways with similar function. To determine the genetic basis for tumor progression and the selective pressure for and the contribution of p53 inactivation, we carried out a comprehensive developmental analysis of tumor evolution.
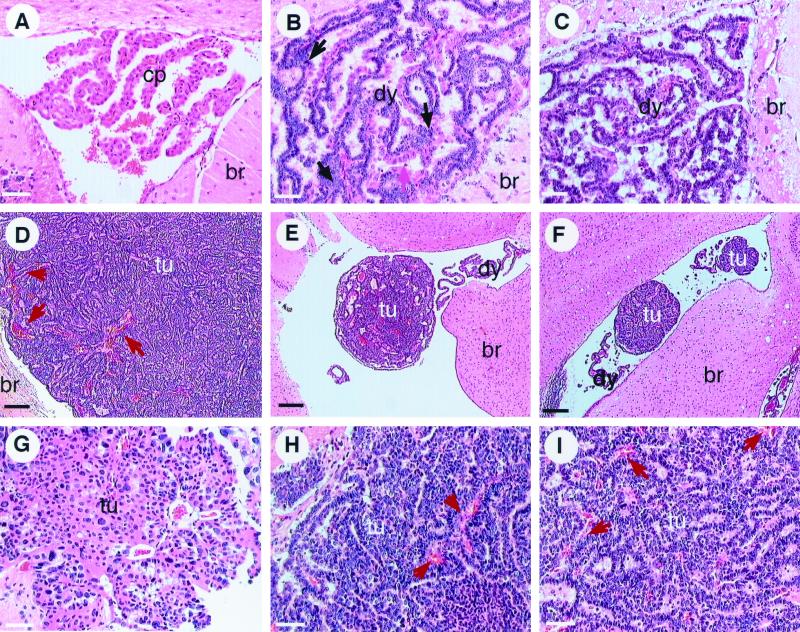
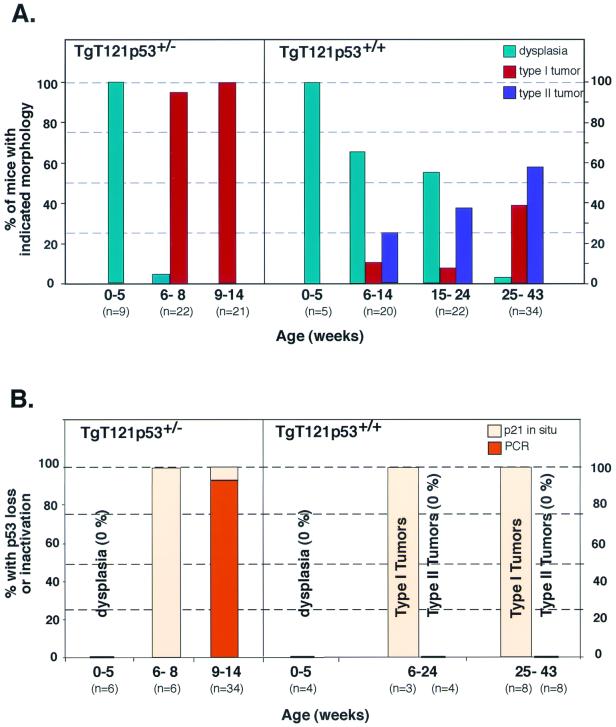
To more fully characterize tumor progression in TgT121p53+/− and TgT121p53+/+ mice, the frequency, timing, and morphological characteristics of tumor progression were analyzed. Step sections were examined at 50-μm intervals for each brain. In 42 of 43 TgT121p53+/− mice analyzed beyond 6 weeks of age, the CP showed evidence of focal progression from dysplasia to solid tumors (Fig. 2; see also Fig. 4). Normal CP appears as frond-like papillary structures extending from the walls of lateral ventricles and the roofs of the third and fourth ventricles. Each frond consists of a single uniform layer of differentiated epithelial cells (Fig. 2A). TgT121p53+/− mice younger than 6 weeks all showed only dysplastic changes in CP (Fig. 2B). The nuclei in these lesions were crowded and elongated with a high nuclear/cytoplasmic ratio, and the chromatin was dense and coarse. The single-layer structure was disrupted with multilayer regions (Fig. 2B, arrows), but the overall frond-like structure of the CP was maintained. No morphologically distinct tumor foci were observed prior to 6 weeks of age. In contrast, TgT121p53+/− mice sacrificed at 6 to 8 weeks of age showed emergence of focal tumors in addition to the preexisting dysplasia. The foci were characterized by nodules of solid tumor arising from the papillary, dysplastic CP cells (Fig. 2E). Tumors were relatively small at this stage; 16 of 22 tumors were smaller than 10 mm3, with an average volume of 1.1 ± 2.0 mm3.
FIG. 2.
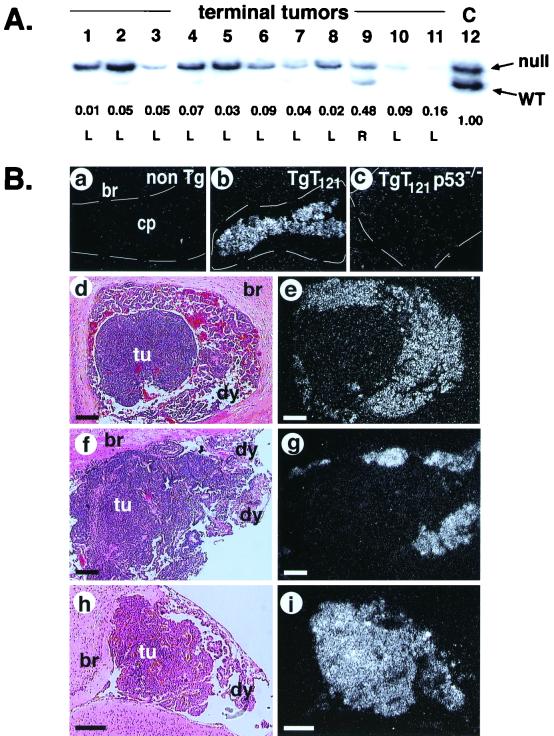
Tumor progression in T121p53+/− and T121p53+/+ mice. Normal CP of a 3-month-old nontransgenic mouse (br, brain tissue). Cells contain regularly sized and shaped nuclei with significant cytoplasm in a single layer forming a papillary architecture (A). Due to T121 expression, the CP of young T121p53+/− (3 weeks) (B) and T121p53+/+ (5 weeks) (C) mice appears dysplastic (dy) with nuclei of aberrant sizes and a high nuclear/cytoplasmic ratio in most cells. The overall papillary organization of the tissue is retained; however, multilayered regions are present. The black arrow indicates dysplastic cells, and the pink arrow indicates normal cells. Focal solid masses indicative of tumor (tu) progression are detectable by 6 to 8 weeks in all T121p53+/− mice (7-week-old mouse shown in panel E) and at various times beyond 7 weeks in some T121p53+/+ mice (7-week-old mouse shown in panel F). These focal tumors arise among the background of dysplastic CP. Solid vascularized tumors (designated type I) grow rapidly and become life threatening by 12 weeks in all T121p53+/− mice (12-week-old mouse shown in panels D and H) and at 25 to 43 weeks in 38% of T121p53+/+ mice (38-week-old mouse shown in panel I). The red arrows in these figures indicate blood vessels. Tumors of distinct morphology (type II) also arise in 59% of T121p53+/+ mice (38-week-old mouse shown in panel G). Loosely organized cells within these masses often contain copious cytoplasm and grossly enlarged nuclei. These tumors appear to grow slowly, since they do not reach the large sizes observed for type I tumors. All slides were stained with hematoxylin and eosin. Each white bar represents 50 μm, and each black bar represents 200 μm.
FIG. 4.
Frequency and timing of tumor progression (A) and p53 inactivation (B) in T121p53+/− and T121p53+/+ mice. (A) Before 6 weeks of age, the CP of all T121p53+/− mice was dysplastic with no evidence of tumor progression. From 6 to 8 weeks, 95% of T121p53+/− mice developed small focal tumors. From 9 to 14 weeks, these solid tumors rapidly increased in size (up to >100 mm3) and became life threatening. Tumor progression occurred later and with a lower frequency in T121p53+/+ mice. Only dysplasia was observed in most mice younger than 15 weeks. Focal development of two distinct morphologies could first be observed in mice at 6 to 14 weeks. Ten percent of focal tumors were morphologically indistinguishable from those developing in T121p53+/− mice (type I tumors), while 25% of tumors appeared distinct (type II [Fig. 2]). Beyond 25 weeks, 38% of mice had developed type I tumors and 59% had developed type II tumors. (B) Inactivation of p53 was detected by in situ detection of p21 transcripts and/or by gene loss using PCR analysis as indicated (Fig. 5). More than 90% of T121p53+/− terminal tumors showed loss of the wild-type p53 allele. Terminal tumors that did not demonstrate p53 LOH as well as early focal tumors (6 to 8 weeks) showed functional loss of p53 based on loss of p21 expression. One-hundred-percent correlation of p53 inactivation with tumor progression indicates that p53 inactivation is likely the facilitating event. In T121p53+/+ mice, all type I tumors, but none of the type II tumors, showed loss of p53 function. All three terminal type I tumors further analyzed by quantitative PCR showed a corresponding loss of at least one p53 allele, while the three type II tumors analyzed retained both copies (real-time PCR data are presented in Table 2 and are not depicted in the histogram).
From 9 to 14 weeks, all TgT121p53+/− tumors became life threatening. At this stage, tumor morphology was similar to the previous stage, but the tumors were much larger (Table 1). The large tumors possessed an extensive vasculature (Fig. 2D and H, e.g., arrows), and necrotic foci were sometimes present (data not shown). CD31 staining of blood vessels showed that tumors contained approximately five times the density of enlarged vessels of that of dysplastic CP (Fig. 3). Thus, tumor progression was associated with promotion of angiogenesis. In some regions, tumor cells displayed a perivascular orientation, and thus, the tumors resembled poorly differentiated papillary adenocarcinomas (Fig. 2D and H). In some terminally ill mice, tumors spread into the subarachnoid space surrounding the brain, but invasion into adjacent brain parenchyma was rare (2 of 21 mice). In summary, these studies showed that the probability of tumor progression in mice with a single functional p53 allele was 100%. Furthermore, the event(s) responsible for tumor progression appeared to occur within a predictable time, with the earliest morphological signs of progression detectable at 6 to 8 postnatal weeks (Fig. 4).
TABLE 1.
Summary of tumor sizesa
| Genotype | Tumor type | No. of tumors | Tumor vol (mm3)
|
|
|---|---|---|---|---|
| Range | Avg | |||
| T121p53+/− | I | 14 | 26.2–268.1b | >95.8 ± 70.4b |
| T121p53+/+ | I | 5 | 10.1–122.4 | 42.6 ± 46.4 |
| T121p53+/+ | II (a)c | 12 | 0.4–5.0 | 2.1 ± 1.4 |
| T121p53+/+ | II (b)c | 3 | 24.1–56.7 | 40.7 ± 16.3 |
Tumor volumes were measured based on area measurements in brain sections from terminally ill mice (see Materials and Methods).
Many of the largest T121p53+/− tumors from the same brains were dissected for DNA analysis and thus not measured.
A subset of tumors with type II morphology was heterogeneous. These tumors (type IIb) grew to larger sizes than those consisting of only type II morphology (type IIa) and also contained regions of densely packed cells.
Tumor progression also occurred with a high frequency in TgT121p53+/+ mice, although the timing was much less predictable (Fig. 4), consistent with the requirement for a minimum of two stochastic genetic lesions to inactivate p53. Emerging tumors could be classified into two distinct morphological types (arbitrarily referred to as types I and II). Type I tumors were identical to those arising in TgT121p53+/− mice (Fig. 2, compare panels E and F and panels H and I). Focal tumors with this morphology were observed in 10% of mice 6 to 24 weeks old; at 25 to 43 weeks, 38% of mice harbored type I tumors (Fig. 4). While morphologically identical to TgT121p53+/− terminal tumors, these tumors were often smaller (Table 1), possibly owing to the older ages at which they developed. For example, life-threatening hydrocephalus, which was not present in TgT121p53+/− mice, developed in 31% of the TgT121p53+/+ mice with type I tumors.
Type II tumors were less compact and were characterized by considerable nuclear polymorphism. Type II cells had a lower nuclear/cytoplasmic ratio than did type I cells; the cytoplasm was copious, and nuclei were often grossly enlarged (Fig. 2G). Apoptotic cells were also evident (data not shown). While apoptosis in type I tumors had dropped to 3.6% ± 0.2%, compared to 12.6% ± 2.8% in dysplastic tissue, apoptosis in type II tumors was at an intermediate level (6.8% ± 1.8%). Type II tumors were generally much smaller than type I tumors, although a subset of tumors containing type II morphology grew to larger sizes (Table 1). These larger tumors were heterogeneous and contained regions of densely packed cells intermixed with typical type II morphology. Type II foci were first observed in 9-week-old mice and were present in 25% of mice at 15 to 24 weeks and 56% of mice at 25 to 43 weeks. Hydrocephalus developed in 70% of mice with type II tumors and likely accounts for the life-threatening condition despite the relatively smaller tumor size. In general, TgT121p53+/+ mice developed either type I or type II tumors; of the 33 mice analyzed with tumors, only 2 harbored both tumor types. Of the terminally ill mice, 41% had type I tumors and 59% had type II tumors.
p53 is inactivated in all type I tumors.
To determine whether p53 inactivation was associated with tumor progression and to assess the selective pressure for p53 inactivation, we examined TgT121p53+/− tumors for loss of the wild-type p53 allele. Semiquantitative PCR was used to detect p53 wild-type and null alleles (Materials and Methods). DNA isolated from terminal TgT121p53+/− tumors was compared with tail DNA from the same mice. Due to the presence of stroma and vasculature in the tumor samples, a reduction of two-thirds or more in the wild-type signal relative to the null signal was considered a loss. A representative result is shown in Fig. 5A. Of 34 TgT121p53+/− tumors analyzed, 32 showed convincing loss of the wild-type allele (Fig. 4B). All tumors retained the null allele, indicating that gene loss was not due to random genetic instability. Thus, the selective pressure for p53 loss during tumor progression was extremely high, with 94% of tumors having specifically lost the functional p53 allele.
FIG. 5.
p53 is inactivated in all T121p53+/− and T121p53−/− type I tumors. (A) Genomic DNAs from TgT121p53+/− terminal tumors (lanes 1 to 11) and TgT121p53+/− tail DNA (“C,” lane 12) were analyzed by semiquantitative PCR to detect null (upper band) and wild-type (WT; lower band) p53 alleles (see Materials and Methods). Samples shown are representative of 34 tumors analyzed. The signal from the wild-type p53 allele is selectively reduced in all but one tumor (lane 9) relative to the upper band (the p53 null allele). The normalized intensity ratios of wild-type to null alleles are listed under corresponding lanes. L (loss) and R (retention) were determined by using the arbitrary threshold (see Materials and Methods). A total of 32 of 34 tumors analyzed in this fashion had lost the wild-type p53 allele (Fig. 4B). (B) Functional assessment of p53 by in situ hybridization analysis of p21 transcript levels. p21 expression in CP is an indicator of p53 function. p21 transcripts are undetectable in nontransgenic CP (a). p21 is induced upon activation of p53 by T121 (b). p21 expression is fully p53 dependent since it is absent in TgT121p53−/− CP (c). Functional assessment of p53 in TgT121p53+/− (d) or TgT121p53+/+ (f) CP tissue shows that p21 expression (p53 activity) is absent in type I tumor nodules but present in surrounding dysplastic CP (e and g). Type II tumors (h) retain p21 expression (i). Subpanels d, f, and h show hematoxylin and eosin staining as viewed by bright-field microscopy. Subpanels e, g, and i show in situ hybridization with an antisense p21 RNA probe viewed by dark-field microscopy. No signal was detectable using the sense probe (data not shown). Each bar represents 200 μm. The frequency and timing of p53 inactivation for all samples analyzed are graphed in Fig. 4. non Tg, nontransgenic; br, brain tissue; tu, tumor; dy, dysplasia.
To determine whether p53 function was also disrupted in the tumors that retained the wild-type p53 allele and to determine whether p53 loss occurred concomitantly with tumor progression rather than subsequently, we assessed p53 activity in situ. As described previously, the p53 target gene p21 is transcriptionally activated in CP upon T121 expression (48). In this system, p21 expression is entirely dependent upon p53 activation, since it is undetectable in TgT121p53−/− CP (48) (Fig. 5B, subpanels a to c). Furthermore, we have shown elsewhere through genetic experiments that p21 is not required for p53-dependent apoptosis in this system (C. Yin et al., unpublished data). Thus, there should be no selective pressure for p21 loss in the absence of p53 loss. In situ hybridization was used to detect p21 transcripts in TgT121p53+/− CP in which both dysplasia and a progressed tumor were present (Fig. 5B, subpanel d). p53 function, as indicated by the presence of p21 transcripts, was clearly absent from the progressed tumor but was present in the surrounding dysplastic regions (Fig. 5B, subpanel e). In addition, five of five terminal tumors that showed p53 LOH had also lost p21 expression, indicating the consistency of this assay in detecting p53 inactivation (Fig. 4B).
The two tumors that had retained the wild-type p53 allele by LOH analysis showed loss of p21 transcripts, indicating that the p53 pathway was functionally inactivated in 100% of progressed tumors (Fig. 4B). To assess the correlation between p53 inactivation and tumor progression, brains with the earliest detectable focal lesions were analyzed. Brains from all six TgT121p53+/− mice analyzed at 6 to 8 weeks old showed clear loss of p53 function in focal tumors with retention of function in dysplastic regions (Fig. 4B). Thus, p53 inactivation was likely causal in the progression of these tumors.
To determine whether type I tumors in TgT121p53+/+ mice showed similar loss of p53 function, we examined 11 tumors at various stages. p21 in situ analysis again showed loss of p53 function in all type I tumors (Fig. 5B, subpanels f and g, and 4B). Consistent with results from TgT121p53+/− tumors, p53 activity was lost at an early stage when tumor foci were very small (data not shown), indicating that loss of p53 is a rate-limiting event in tumor progression. Real-time PCR analysis showed that at least one p53 allele was lost in three of three tumors analyzed (Table 2), suggesting that p53, and not an upstream factor, was the target for inactivation. In contrast to type I tumors, 11 of 11 TgT121p53+/+ type II tumors analyzed at both early and late stages retained p53 activity, and three of these tumors analyzed for gene loss retained both wild-type p53 alleles (Fig. 5B, subpanels h and i; Table 2). These results indicate that the genetic basis for the differences between type I and type II tumors includes the absence and presence, respectively, of p53 activity. Thus, p53 loss is associated with progression to highly vascularized aggressive tumors. These results further emphasize that the selective pressure for p53 inactivation is high. Moreover, since two functional alleles of p53 are present in these mice, the strong correlation between p53 inactivation and type I tumor progression indicates that loss of the p53 pathway, likely of p53 itself, is the major rate-limiting step to tumor progression (see Discussion). Furthermore, since p53 inactivation is associated with tumor progression rather than simply faster-growing dysplastic masses, p53 inactivation must contribute to tumorigenesis by mechanisms in addition to apoptosis modulation.
TABLE 2.
Quantitative analysis of p53 gene loss in TgT121p53+/+ tumorsa
| Sample no. | Tissue | Mouse genotype | 2−ΔΔCtb | No. of assays | No. of wild-type p53 alleles |
|---|---|---|---|---|---|
| 1 | Tail | p53+/+ | 1.0 | 4 | 2 |
| 2 | Tail | p53+/− | 0.33 ± 0.24 | 4 | 1 |
| 3 | Tail | p53+/− | 0.21 ± 0.02 | 2 | 1 |
| 4 | Tumor (I) | TgT121p53+/− | 0.17 ± 0.05 | 4 | 1c |
| 5 | Tumor (I) | TgT121p53+/− | 0.11 ± 0.04 | 2 | 0 |
| 6 | Tumor (I) | TgT121p53+/+ | 0.17 ± 0.01 | 2 | 1d |
| 7 | Tumor (I) | TgT121p53+/+ | 0.48 ± 0.14 | 2 | 1 |
| 8 | Tumor (I) | TgT121p53+/+ | 0.14 ± 0.01 | 2 | 0d |
| 9 | Tumor (II) | TgT121p53+/+ | 0.73 ± 0.02 | 2 | 2 |
| 10 | Tumor (II) | TgT121p53+/+ | 1.21 ± 0.62 | 2 | 2 |
| 11 | Tumor (II) | TgT121p53+/+ | 1.56 ± 0.69 | 3 | 2 |
Real-time PCR was performed to determine the status of the wild-type p53 alleles in TgT121p53+/+ terminal tumors (see Materials and Methods). Among six TgT121p53+/+ tumors, all three type I tumors showed loss of at least one wild-type p53 allele while all three type II tumors retained both wild-type alleles. Two TgT121p53+/− tumors shown to have lost the wild-type p53 allele by allele-specific PCR and CGH were used as controls.
ΔΔCt = [sample Ct(p53) − sample Ct(β-actin)] − [p53+/+ control Ct(p53) − p53+/+ control Ct(β-actin)], where Ct is the number of cycles required to reach a threshold based on linear amplification (see Materials and Methods). Analysis of standard samples indicated that copy numbers of 2, 1, and 0 are indicated by 2−ΔΔCt values of >0.6, between 0.15 and 0.6, and <0.15, respectively.
Although the value obtained here indicates detection of a wild-type allele in this TgT121p53+/− tumor, this is likely due to the contamination of nontumor tissue in the dissection procedure. Previous conventional PCR analysis and CGH all showed that tumor 4 had p53 LOH.
The 2−ΔΔCt value for tumors 6 and 8 that arose in mice with two wild-type alleles is near the low end of the single-copy range. Further analysis will be required to determine with certainty whether one or both wild-type alleles are absent in these tumors. However, there is clearly loss of at least one allele.
Does p53 inactivation during tumor progression cause genetic instability?
A widely held hypothesis is that loss of p53 function results in genomic instability and therefore could contribute to tumor progression by facilitating the propagation of genetic alterations. Although p53-deficient tumors and cell lines often contain genetic aberrations (almost exclusively aneuploidy), whether loss of p53 is a direct cause of genetic instability is unknown (34). If inactivation of p53 function in TgT121p53+/− CP results in genetic instability that facilitates clonal tumor progression, such changes should be detectable in tumors. To determine whether this is the case, we analyzed TgT121p53+/− terminal tumors by CGH. In CGH, fluorescently labeled normal and tumor DNAs are competitively hybridized to normal metaphase chromosomes to assess genome-wide copy number changes. With a resolution of approximately 10 Mb, CGH readily detects aneuploidy as well as partial chromosome gains and losses (19).
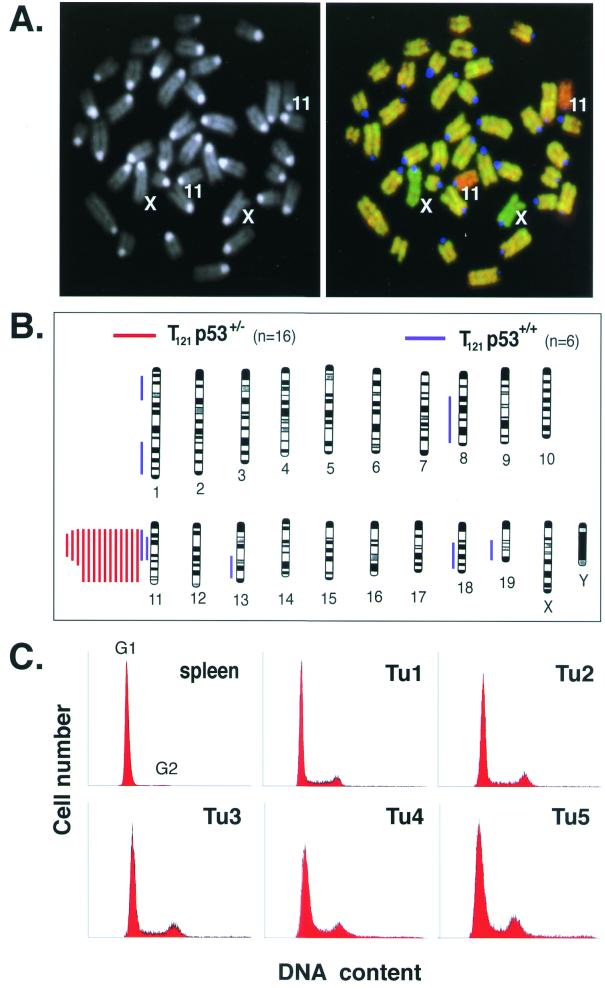
Sixteen TgT121p53+/− terminal tumors were analyzed by CGH (Fig. 6A and B). Normal tail DNA was used as a negative control, while previously karyotyped aneuploid thymic lymphoma DNA was used as a positive control. A sex-mismatched DNA mixture was also used as an internal control for copy number changes. As anticipated based on the p53 LOH results described above, most tumors (14 of 16) showed complete or partial loss of a single copy of chromosome 11 (the location of the p53 gene) (Table 3). Surprisingly, chromosome 11 loss was the only aberration detected in any of the 16 tumors analyzed. We also performed CGH analysis on six terminal TgT121p53+/+ tumors. Four tumors harbored no detectable changes. These tumors were not characterized histologically and could have possessed either type I or type II characteristics. Two tumors showed loss of chromosome 11 and were presumably type I tumors based on the association of p53 inactivation with these tumors as described above. Both tumors also harbored two to four additional changes (Fig. 6B; Table 3). Since type I TgT121p53+/+ tumors appear after long latency likely due to the need to inactivate two wild-type p53 alleles, it is possible that events leading to these changes preceded p53 inactivation in these tumors. This explanation would account for the difference in the level of chromosome changes between TgT121p53+/+ and TgT121p53+/− tumors that have undergone p53 inactivation. Four TgT121p53−/− CP masses were also analyzed and shown to contain no detectable chromosomal losses or gains (Table 3). A binomial statistical test comparing these results to previous CGH studies with p53+/− and p53−/− tumors from other models (16, 64) indicates that the lack of chromosomal instability in TgT121p53−/− CP is statistically significant (P < 0.01). As a positive control for CGH studies, thymic lymphomas known to be aneuploid by karyotype analysis (37) were analyzed by CGH and shown to contain numerous copy number changes (Table 3). Since CGH detects only unbalanced chromosomal gains or losses, it was a formal possibility that tumors were polyploid. To test this possibility, tumor DNAs were analyzed by flow cytometry. The majority of cells in all five TgT121p53+/− tumors analyzed were diploid (Fig. 6C). These results indicate that, contrary to popular hypothesis, p53 inactivation is not sufficient to cause chromosomal instability during tumor progression. Furthermore, the lack of chromosomal instability in these tumors indicates that p53 inactivation contributes to tumor progression by an as yet unidentified mechanism(s).
FIG. 6.
Chromosomal stability in p53-deficient tumors. (A) DAPI-stained metaphase chromosomes from normal mouse embryonic fibroblasts are shown, with chromosomes 11 and X indicated (left). A composite digital image from CGH analysis of a representative TgT121p53+/− terminal tumor is shown on the right. Tumor DNA (fluorescein isothiocyanate, green) and normal tail DNA (Alex-568, red) were hybridized to the metaphase spread shown in the left panel. Relative green regions indicate increased copy numbers in the tumor, while red regions indicate decreased copy numbers. The centromeres appear blue because repeated satellite sequences were blocked with unlabeled mouse Cot-1 DNA. This tumor sample shows loss of chromosome 11. Apparent gain of the X chromosome serves as an internal control; tumor DNA was derived from a female mouse, and normal DNA was from a male mouse. (B) Summary of CGH analysis of 14 TgT121p53+/− and 6 TgT121p53+/+ tumors. Each bar to the left of a depicted chromosome represents DNA copy number losses at the corresponding regions in a single tumor; no gains were detected. Signal intensities were consistent with loss of a single copy. Red bars represent TgT121p53+/− tumors, and blue bars represent TgT121p53+/+ tumors. Chromosomes were identified by DAPI banding. Black bands in the chromosome diagrams represent the observed DAPI staining pattern. (C) CP tumor cells from five TgT121 p53+/− mice were analyzed by propidium iodide staining and fluorescence-activated cell sorting analysis for DNA content. More than 90% of cells in all five tumors are diploid.
TABLE 3.
Comparative genomic hybridization analysis of tumor DNAsa
| Tumor type | No. of tumors | Chromosomal change(s) |
|---|---|---|
| TgT121p53+/− CP | 12 | dec11 |
| 1 | dec11 (A2-B4) | |
| 1 | dec11 (A1-B3) | |
| 2 | None | |
| TgT121p53+/+ CP | 1 | dec11 (A1-B4); dec18 (A2-E4) |
| 1 | dec11 (A1-B3); dec1 (A2-B2); dec8 (E3-H5); dec19 (B-D3) | |
| 4 | None | |
| TgT121p53−/− CP | 4 | None |
| TgTΔN TL | 1 | inc4; inc5; inc10; inc15 (D1-F3) |
| 1 | inc4 (A5-E2); inc5; inc10; inc15 (A2-F3) |
Chromosomal changes are indicated as “dec” for DNA copy number decreases and “inc” for increases, followed by the affected mouse chromosome numbers or subchromosomal regions. For TgT121p53+/− CP tumors, where indicated the signal intensity was consistent with the loss of a single copy of chromosome 11. TgTΔN mice develop thymic lymphoma as a result of expressing the SV40 T-antigen mutant dl1135, thus inactivating p53, in thymocytes. TL, thymic lymphoma.
DISCUSSION
High selection for p53 inactivation during tumor evolution.
The data presented here show that there is immense selective pressure for p53 inactivation during the natural evolution of epithelial tumors initiated by inactivation of the pRb pathway. Progression from dysplasia to solid aggressive tumors occurs in 100% of TgT121p53+/− mice, and all of these tumors have inactivated p53. Even more indicative of the high pressure for p53 inactivation was that 38% of TgT121p53+/+ tumors inactivated p53 function. Moreover, p53 was nonfunctional in 100% of type I tumors arising in this background. In TgT121p53+/− tissue, where only a single allele loss is required for p53 inactivation, 95% of the progressed tumors had selectively lost the wild-type p53 allele; the remaining tumors inactivated p53 by some other mechanism. Tumors that did not show p53 gene loss, and all type I tumors from TgT121p53+/+ mice, had lost p53-dependent activation of p21 transcription. Thus, since p21 activation is immediately downstream of p53, no tumors inactivated events downstream of p53. Furthermore, real-time PCR analysis indicated that even p53+/+ tumors had lost at least one p53 allele, suggesting that p53 is the target for inactivation and thus the rate-limiting step in tumor progression. This indicates that there is no single linear pathway upstream or downstream of p53 that is responsible for tumor suppression. Consistent with this observation, loss of p53 is associated with tumor progression, rather than just rapid tissue growth, implying that multiple p53-dependent mechanisms are responsible for overall tumor suppression. While p53-mediated apoptosis significantly slows dysplastic tissue growth and likely accounts for the high selective pressure for p53 inactivation, other p53 functions appear to suppress tumor progression (see model in Fig. 1B).
By CGH analysis, we found that partial or complete loss of a single copy of chromosome 11 occurred with a high frequency to facilitate p53 inactivation in TgT121p53+/− tumors. We considered the possibility that hemizygosity of other tumor suppressors located on this chromosome could contribute to tumor progression. However, two observations indicate that this is not the case. First, some TgT121p53+/− tumors with identical characteristics of progression did not lose chromosome 11 (Tables 2 and 3). Second, we have induced morphologically identical tumors by focal transgenic expression of full-length SV40 T antigen (11, 63). Since p53 is inactivated by T antigen in this case, there is no selective pressure for p53 gene loss in these tumors. Indeed, by CGH these tumors showed no loss of chromosome 11 (data not shown). Thus, p53 inactivation appears to be the critical event causing tumor progression
Together with previous studies of p53 LOH in mouse tumor models (40), the study presented here indicates that the selective pressure for p53 inactivation may vary with the cell type, likely reflecting distinct mechanisms of p53 tumor suppression or the tissue-specific presence of alternate tumor suppression pathways. Pituitary and thyroid tumors of Rb+/−p53+/− mice undergo Rb loss at a high frequency but rarely lose the wild-type p53 allele (71). In addition to the cell type difference between these studies, tumors in the present study were initiated by inactivation of all three pRb-related proteins. Thus, the difference in selective pressure for p53 inactivation could reflect cell type or mechanistic differences. In a study of lymphomas and sarcomas developing in p53+/− mice, the frequency of p53 LOH was 15 to 50%, depending on the age of tumor-bearing mice (64). In that study, p53 expressed in tumors without LOH appeared to be functional in that it was inducible by irradiation and was able to bind DNA specifically. The frequency of p53 LOH in mammary tumors arising in Wnt-1p53+/− mice was also about 50% (16), and in a chemically induced skin tumor model, p53 LOH occurred in 64% of carcinomas (8). A study of salivary and mammary tumors arising in the same MMTV-ras/p53+/− strain of mice offers a clear demonstration of tissue-specific p53 LOH (29). While 10 of 10 salivary tumors lost the wild-type p53 allele, all 7 mammary tumors analyzed retained the locus. Furthermore, by comparing this study of mammary tumors (0% p53 LOH) to the study with Wnt-1 mice (50% p53 LOH), it is clear that the selective pressure for p53 inactivation appears to be influenced by the preceding oncogenic event in addition to the tissue type. Indeed, the frequency of p53 LOH in mammary tumors from Brca1ko/cop53+/− mice was 80% (72). With the exception of the analysis of p53+/− lymphomas and sarcomas, these studies did not address whether p53 function was intact in the tumors that retained a wild-type allele and thus represent a minimum estimate of p53 inactivation.
When evaluating the impact of p53 inactivation on tumor progression, it is important to examine the morphological evolution of the tumor and its correlation with p53 inactivation. As we observed with TgT121p53+/+ mice, tumors may progress to multiple grades within the same genotype. In this model, inactivation of p53 correlated precisely with the development of characteristic carcinomas, while slower-growing tumors of distinct morphology retained p53 function. In most previous studies, the relationship between p53 status and histologic grade was not assessed. However, in the Wnt-1 mammary and MMTV-ras mammary and salivary tumors described above, p53 inactivation also correlated with a higher histologic grade.
In our studies, the perfect correlation between p53 inactivation and aggressive CP tumor progression indicates that p53 inactivation is a highly selected event and may be sufficient for progression. In all other models analyzed thus far, the basis of selective pressure for p53 inactivation is unknown. In dysplastic CP, p53 is clearly required for apoptosis, such that cells that inactivate other tumor suppression pathways but retain p53 would still undergo cell death. Thus, if apoptosis is not an element of p53 tumor suppression in a given cell type, a more random distribution of p53 inactivation may be observed.
p53 inactivation, genomic instability, and tumor progression.
A common hypothesis for p53 tumor suppression is that p53 prevents genetic instability. Based on this idea, we tested whether CP tumor progression after p53 inactivation was facilitated by the accumulation of chromosomal abnormalities. Aneuploidy is observed in p53−/− hematopoietic cells (6, 21), in p53-deficient Li-Fraumeni syndrome (4) and mouse (24, 62) fibroblasts, and in mouse thymic lymphomas induced by p53 deficiency (37, 64). However, TgT121p53+/− CP tumors were diploid and showed partial or complete loss of a single copy of chromosome 11 as the only detectable chromosomal aberration. The mouse p53 gene is located on chromosome 11, and thus, its loss represents the selected causal event for tumor progression. Hence, in brain epithelium p53 inactivation can contribute to tumor progression by mechanisms other than the induction of chromosomal instability.
Although loss of p53 function has been associated with genetic instability in numerous studies, there has been no direct proof that p53 inactivation is sufficient to cause genetic instability or that such a function facilitates tumorigenesis in the absence of other changes. In some mouse cancer models, tumors that develop in the absence of p53 become aneuploid. For example, CGH analysis of Wnt-1p53+/− terminal mammary tumors showed that tumors with p53 LOH contained more chromosomal changes (4.3 changes per tumor) than did those without p53 LOH (1 change per tumor) (16). A similar observation was made for p53+/− thymic lymphomas and sarcomas, where tumors with p53 LOH contained an average of 5.6 changes per tumor compared to an average of 1 change without p53 LOH (64). Likewise, mammary and salivary tumors induced by MMTV-ras were largely diploid in the presence of p53 but aneuploid in its absence (29). These studies indicate that loss of p53 function in tumors often correlates with chromosomal instability.
Two alternative explanations could account for the apparent differences in the level of genetic instability observed in p53-deficient mouse tumors. One possibility is that these differences reflect true tissue-specific differences in p53 tumor suppression mechanisms. A second possibility is that p53 deficiency is necessary but not sufficient to induce chromosomal instability. Indeed, some studies of cultured p53-deficient cells have indicated that p53 inactivation is not sufficient to drive genetic instability (18, 49). Furthermore, in the study of Wnt-1-induced mammary tumors, karyotype analysis indicated that several p53-deficient tumors had nearly diploid profiles, similar to those of p53 wild-type tumors, indicating that p53 inactivation was insufficient to cause aneuploidy (16). This notion is also supported by the observation that the rapidly dividing cells of p53-deficient embryos are diploid. However, combined deficiencies in Brca1 and p53 induce aneuploidy in these cells (57). Thus, in previous studies, tumors that harbored numerous chromosomal changes may have undergone causal stochastic mutations in addition to p53 loss. In the previous studies, tumors were analyzed after substantial growth, during which such events could have occurred. Although the TgT121p53+/− CP tumors were from terminally ill mice, brain tumors are limited in the extent of their growth due to adverse effects on the host. To determine whether p53 inactivation alone can facilitate genetic instability and to rule out the possibility that secondary changes are responsible, tumors must be analyzed for chromosomal instability at early stages post-p53 loss. It is possible that analysis of the brain tumors in the present study represents such an early assessment of the impact of p53 inactivation.
Our study of p53 deficiency in an evolving tumor showed that genomic instability resulting from p53 loss did not drive epithelial tumor progression. In a recent study, the frequency of epithelial tumors was significantly increased in p53-deficient mice that had been propagated through several generations in the absence of telomerase function (1). Furthermore, these tumors show chromosome instability including end-to-end fusion and aneuploidy. Since such epithelial tumors are not observed in the absence of telomere shortening, these results indicate that p53 deficiency is required to propagate chromosomal instability when driven by other mechanisms but is itself not the driving force. This mechanism also appears to be operative in mouse lymphomas caused by a truncation of Brca2 (33). In that study, p53 inactivation was shown to be required to propagate the Brca2 mutant cells due to severe chromosomal aberrations caused by the Brca2 defect.
If not chromosomal instability, what facilitates CP tumor progression? Although aneuploidy is the most common genetic change observed in p53-deficient cells, it is possible that p53 inactivation results in a type of genetic instability that is undetectable by CGH. Point mutations, small deletions, inversions or amplifications, balanced translocations, and microsatellite instability would all escape detection by CGH. Currently, there is no convincing precedent for the association of p53 inactivation with such changes. Although some studies have suggested that p53 affects base (45) and nucleotide (69) excision repair activities in vitro, the point mutation frequency is normal in p53-deficient cells, tissues, and thymic lymphomas (44, 55). Future experiments to detect the incidence of small deletions-amplifications-inversions in progressing TgT121p53+/− tumors will be required to determine whether p53 inactivation causes any form of genomic instability.
Another possibility is that yet other p53 functions suppress tumor progression. For example, previous studies indicate that p53 can regulate the expression of angiogenesis factors. In cotransfection experiments, p53 can induce transcription of the angiogenesis inhibitor thrombospondin 1 (12) and other potential angiogenesis inhibitors such as maspin (75) and BAI1 (59) through p53-specific DNA binding elements. In addition, the angiogenesis activator vascular endothelial growth factor is induced in p53-deficient cells by an unknown mechanism (5, 51, 66, 74). Since we show here that angiogenesis is a feature of CP tumor progression associated with p53 inactivation, we are currently exploring the possibility that p53 inactivation plays a direct role in modulating the expression of angiogenesis factors, thus facilitating tumor progression.
In summary, p53 appears to suppress tumorigenesis in a single tissue by multiple mechanisms. When tumor growth is suppressed by p53-dependent apoptosis as in the CP tumors, there is a high selective pressure for p53 inactivation. Inactivation of p53 in turn facilitates tumor progression by a mechanism that, surprisingly, does not include the induction of chromosomal instability. These studies underscore the complexity of p53 tumor suppression and the need to understand the mechanisms in the context of natural selective pressures occurring during tumor evolution in vivo.
ACKNOWLEDGMENTS
We thank Lynda Chin and colleagues (Harvard University) for supplying real-time PCR conditions and Hua Wu (T.V. lab) for assistance in developing the assay. Robert Flandermyer (UCSF) and Le Zhang (T.V. lab) provided excellent technical assistance. We acknowledge the UNC Lineberger Comprehensive Cancer Center Histology Core for processing tissues used in this study and the UNC Division of Laboratory Animals for excellent animal care.
This work was supported by NCI grants 1 R01 CA46283 and 5 U01 CA84314.
REFERENCES
- 1.Artandi S E, Chang S, Lee S L, Alson S, Gottlieb G J, Chin L, DePinho R A. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 2.Bates S, Vousden K H. p53 in signaling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:12–19. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 3.Bell D W, Varley J M, Szydlo T E, Kang D H, Wahrer D C, Shannon K E, Lubratovich M, Verselis S J, Isselbacher K J, Fraumeni J F, Birch J M, Li F P, Garber J E, Haber D A. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff F Z, Yim S O, Pathak S, Grant G, Siciliano M J, Giovanella B C, Strong L C, Tainsky M A. Spontaneous abnormalities in normal fibroblasts from patients with Li-Fraumeni cancer syndrome: aneuploidy and immortalization. Cancer Res. 1990;50:7979–7984. [PubMed] [Google Scholar]
- 5.Bouck N, Stellmach V, Hsu S C. How tumors become angiogenic. Adv Cancer Res. 1996;69:135–174. doi: 10.1016/s0065-230x(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 6.Bouffler S D, Kemp C J, Balmain A, Cox R. Spontaneous and ionizing radiation-induced chromosomal abnormalities in p53-deficient mice. Cancer Res. 1995;55:3883–3889. [PubMed] [Google Scholar]
- 7.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 8.Burns P A, Kemp C J, Gannon J V, Lane D P, Bremner R, Balmain A. Loss of heterozygosity and mutational alterations of the p53 gene in the skin tumors of interspecific hybrid mice. Oncogene. 1991;6:2363–2369. [PubMed] [Google Scholar]
- 9.Chehab N H, Malikzay A, Appel M, Halazonetis T D. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Neilson K, Van Dyke T. Lymphotropic papovavirus early region is specifically regulated in transgenic mice and efficiently induces neoplasia. J Virol. 1989;63:2204–2214. doi: 10.1128/jvi.63.5.2204-2214.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Van Dyke T. Uniform cell-autonomous tumorigenesis of the choroid plexus by papovavirus large T antigens. Mol Cell Biol. 1991;11:5968–5976. doi: 10.1128/mcb.11.12.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dameron K M, Volpert O V, Tainsky M A, Bouck N. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 13.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 14.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 15.Di Leonardo A, Khan S H, Linke S P, Greco V, Seidita G, Wahl G M. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 16.Donehower L A, Godley L A, Aldaz C M, Pyle R, Shi Y P, Pinkel D, Gray J, Bradley A, Medina D, Varmus H E. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 1995;9:882–895. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- 17.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A J, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 18.Filatov L, Golubovskaya V, Hurt J C, Byrd L L, Phillips J M, Kaufmann W K. Chromosomal instability is correlated with telomere erosion and inactivation of G2 checkpoint function in human fibroblasts expressing human papillomavirus type 16 E6 oncoprotein. Oncogene. 1998;16:1825–1838. doi: 10.1038/sj.onc.1201711. [DOI] [PubMed] [Google Scholar]
- 19.Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi O. Genome screening by comparative genomic hybridization. Trends Genet. 1997;13:405–409. doi: 10.1016/s0168-9525(97)01244-4. [DOI] [PubMed] [Google Scholar]
- 20.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Voude G F. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 21.Fukasawa K, Wiener F, Vande Woude G F, Mai S. Genomic instability and apoptosis are frequent in p53 deficient young mice. Oncogene. 1997;15:1295–1302. doi: 10.1038/sj.onc.1201482. [DOI] [PubMed] [Google Scholar]
- 22.Hansen R, Oren M. p53; from inductive signal to cellular effect. Curr Opin Genet Dev. 1997;7:46–51. doi: 10.1016/s0959-437x(97)80108-6. [DOI] [PubMed] [Google Scholar]
- 23.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 24.Harvey M, Sands A T, Weiss R S, Hegi M E, Wiseman R W, Pantazis P, Giovanella B C, Tainsky M A, Bradley A, Donehower L A. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- 25.Hermeking H, Lengauer C, Polyak K, He T C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 26.Hirao A, Kong Y Y, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge S J, Mak T W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 27.Hollstein M, Shomer B, Greenblatt M, Soussi T, Hovig E, Montesano R, Harris C C. Somatic point mutations in the p53 gene of human tumors and cell lines: updated compilation. Nucleic Acids Res. 1996;24:141–146. doi: 10.1093/nar/24.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howes K A, Ransom N, Papermaster D S, Lasudry J G H, Albert D M, Windle J J. Apoptosis or retinoblastoma: alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev. 1994;8:1300–1310. doi: 10.1101/gad.8.11.1300. [DOI] [PubMed] [Google Scholar]
- 29.Hundley J E, Koester S K, Troyer D A, Hilsenbeck S G, Subler M A, Windle J J. Increased tumor proliferation and genomic instability without decreased apoptosis in MMTV-ras mice deficient in p53. Mol Cell Biol. 1997;17:723–731. doi: 10.1128/mcb.17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacks T, Remington L, Williams B, Schmitt E, Halachmi S, Bronson R, Weinberg R. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 31.Kallioniemi A, Kallionemi O, Sudar D, Rutovitz D, Gray J, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 32.Lane D P. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 33.Lee H, Trainer A H, Friedman L S, Thistlethwaite F C, Evans M J, Ponder B A, Venkitaraman A R. Mitotic checkpoint inactivation fosters transformation in cells lacking the breast cancer susceptibility gene, Brca2. Mol Cell. 1999;4:1–10. doi: 10.1016/s1097-2765(00)80182-3. [DOI] [PubMed] [Google Scholar]
- 34.Lengauer C, Kinzler K W, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 35.Levine A, Momand J, Finlay C. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 36.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 37.Liao M-J, Zhang X, Hill R, Gao J, Qumsiyeh M, Nichols W, Van Dyke T. No requirement for V(D)J recombination in p53-deficient thymic lymphoma. Mol Cell Biol. 1998;18:3495–3501. doi: 10.1128/mcb.18.6.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 39.Lowe S, Jacks T, Housman D, Ruley H. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci USA. 1994;91:2026–2030. doi: 10.1073/pnas.91.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lozano G, Liu G. Mouse models dissect the role of p53 in cancer and development. Semin Cancer Biol. 1998;8:337–344. doi: 10.1006/scbi.1998.0096. [DOI] [PubMed] [Google Scholar]
- 41.Macleod K F, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 42.Morgenbesser S, Williams B, Jacks T, DePinho R. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- 43.Nigro J M, Baker S J, Preisinger A C, Jessup J M, Hostetter R, Cleary K, Bigner S H, Davidson N, Baylin S, Devilee P, Glover T, Collins F S, Weston A, Modali R, Harris C C, Vogelstein B. Mutations in the p53 gene occur in diverse human tumor types. Nature. 1989;342:705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 44.Nishino H, Knoll A, Buettner V L, Frisk C S, Maruta Y, Haavik J, Sommer S S. p53 wild-type and p53 nullizygous Big Blue transgenic mice have similar frequencies and patterns of observed mutation in liver, spleen, and brain. Oncogene. 1995;11:263–270. [PubMed] [Google Scholar]
- 45.Offer H, Wolkowicz R, Matas D, Blumenstein S, Livneh Z, Rotter V. Direct involvement of p53 in the base excision repair pathway of the DNA repair machinery. FEBS Lett. 1999;450:197–204. doi: 10.1016/s0014-5793(99)00505-0. [DOI] [PubMed] [Google Scholar]
- 46.Pan H, Griep A E. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 47.Pan H, Griep A E. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes Dev. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 48.Pan H, Yin C, Dyson N, Harlow E, Yamasaki L, Van Dyke T. A key role for E2F1 in p53-dependent apoptosis and cell division within developing tumors. Mol Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- 49.Paulson T G, Almasan A, Brody L L, Wahl G M. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prives C, Hall P A. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 51.Ravi R, Mookerjee B, Bhujwalla Z M, Sutter C H, Artemov D, Zeng Q, Dillehay L E, Madan A, Semenza G L, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson J P, Darzynkiewicz Z, Dean P N, Orfao A, Rabinovitch P S, Stewart C C, Tanke H J, Wheeless L L. Current protocols in cytometry. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 53.Sabbatini P, Lin J, Levine A J, White E. Essential role for p53-mediated transcription in E1A-induced apoptosis. Genes Dev. 1995;9:2184–2192. doi: 10.1101/gad.9.17.2184. [DOI] [PubMed] [Google Scholar]
- 54.Saenz Robles M T, Symonds H, Chen J, Van Dyke T. Induction versus progression of brain tumor development: differential functions for the pRB- and p53-targeting domains of simian virus 40 T antigen. Mol Cell Biol. 1994;14:2686–2698. doi: 10.1128/mcb.14.4.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sands A T, Suraokar M B, Sanchez A, Marth J E, Donehower L A, Bradley A. p53 deficiency does not affect the accumulation of point mutations in a transgene target. Proc Natl Acad Sci USA. 1995;92:8517–8521. doi: 10.1073/pnas.92.18.8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwartz D, Rotter V. p53-dependent cell cycle control: response to genotoxic stress. Semin Cancer Biol. 1998;8:325–336. doi: 10.1006/scbi.1998.0095. [DOI] [PubMed] [Google Scholar]
- 57.Shen S X, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan X Y, Ried T, Deng C X. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- 58.Shieh S Y, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 59.Shiratsuchi T, Nishimori H, Ichise H, Nakamura Y, Tokino T. Cloning and characterization of BA12 and BA13, novel genes homologous to brain-specific angiogenesis inhibitor 1 (BAI1) Cytogenet Cell Genet. 1997;79:103–108. doi: 10.1159/000134693. [DOI] [PubMed] [Google Scholar]
- 60.Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell. 1994;78:703–711. doi: 10.1016/0092-8674(94)90534-7. [DOI] [PubMed] [Google Scholar]
- 61.Taylor W R, Agarwal M L, Agarwal A, Stacey D W, Stark G R. p53 inhibits entry into mitosis when DNA synthesis is blocked. Oncogene. 1999;18:283–295. doi: 10.1038/sj.onc.1202516. [DOI] [PubMed] [Google Scholar]
- 62.Tsukada T, Tomooka Y, Takai S, Ueda Y, Nishikawa S, Yagi T, Tokunaga T, Takeda N, Suda Y, Abe S. Enhanced proliferative potential in culture of cells from p53-deficient mice. Oncogene. 1993;8:3313–3322. [PubMed] [Google Scholar]
- 63.Van Dyke T A, Finlay C, Miller D, Marks J, Lozano G, Levine A J. Relationship between simian virus 40 large tumor antigen expression and tumor formation in transgenic mice. J Virol. 1987;61:2029–2032. doi: 10.1128/jvi.61.6.2029-2032.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venkatachalam S, Shi Y P, Jones S N, Vogel H, Bradley A, Pinkel D, Donehower L A. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vogelstein B, Lane D, Levine A J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 66.Volpert O V, Dameron K M, Bouck N. Sequential development of an angiogenic phenotype by human fibroblasts progressing to tumorigenicity. Oncogene. 1997;14:1495–1502. doi: 10.1038/sj.onc.1200977. [DOI] [PubMed] [Google Scholar]
- 67.Vousden K H. p53: death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 68.Wagner A J, Kokontis J M, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21 waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 69.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G, et al. p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 70.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 71.Williams B O, Remington L, Albert D M, Mukai S, Bronson R T, Jacks T. Cooperative tumourigenic effects of germline mutations in Rb and p53. Nat Genet. 1994;7:480–484. doi: 10.1038/ng0894-480. [DOI] [PubMed] [Google Scholar]
- 72.Xu X, Wagner K U, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng C X. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 73.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L, Yu D, Hu M, Xiong S, Lang A, Ellis L M, Pollock R E. Wild-type p53 suppresses angiogenesis in human leiomyosarcoma and synovial sarcoma by transcriptional suppression of vascular endothelial growth factor expression. Cancer Res. 2000;60:3655–3661. [PubMed] [Google Scholar]
- 75.Zou Z, Gao C, Nagaich A K, Connell T, Saito S, Moul J W, Seth P, Appella E, Srivastava S. p53 regulates the expression of the tumor suppressor gene maspin. J Biol Chem. 2000;275:6051–6054. doi: 10.1074/jbc.275.9.6051. [DOI] [PubMed] [Google Scholar]