Highlights
-
•
A cell cycle analysis workflow was developed for radiosensitizer discovery.
-
•
Nuclear size imaging was employed in a primary screen to recognize G2/M stall.
-
•
29 potential radiosensitizers were identified from screening 1430 FDA approved drugs.
-
•
Proflavine hemisulfate, an approved antimicrobial, was identified as a radiosensitizer.
Keywords: High-content imaging, Proflavine hemisulfate, Cell cycle analysis, X-ray, Radiation therapy
Abstract
Radiation is a powerful tool used to control tumor growth and induce an immune response; however, it is limited by damage to surrounding tissue and adverse effects such skin irritation. Breast cancer patients in particular may endure radiation dermatitis, and potentially lymphedema, after a course of radiotherapy. Radio-sensitizing small molecule drugs may enable lower effective doses of both radiation and chemotherapy to minimize toxicity to healthy tissue. In this study, we identified a novel high-throughput method for screening radiosensitizers by image analysis of nuclear size and cell cycle. In vitro assays were performed on cancer cells lines to assess combined therapeutic and radiation effects. In vivo, radiation in combination with proflavine hemisulfate led to enhanced efficacy demonstrated by improved tumor volume control in mice bearing syngeneic breast tumors. This study provides a proof of concept for utilizing G2/M stall as a predictor of radiosensitization and is the first report of a flavin acting as an X-ray radiation enhancer in a breast cancer mouse model.
Introduction
Approximately 40% of cancer patients undergo radiotherapy (RT) over the course of curative treatment [1]. While effective, RT has limitations such as normal tissue toxicity and radioresistance stemming from hypoxia, altered cell cycle, immune evasion, increased population of cancer stem-like cells, and enhanced DNA damage response [2]. The synergistic combination of RT with chemotherapeutics overcomes some of the limitations of RT alone. Small molecule drugs have been found to sensitize RT by limiting the DNA damage response, reprogramming the cell-cycle, increasing DNA damage, preventing replication, and normalizing the vasculature among other strategies [3]. Despite the frequent application of chemoradiotherapy, few clinically-approved agents reliably sensitize cells to radiation. While effective, current drugs used in conjunction with RT (camptothecin analogs, cisplatin, irinotecan, and PARP inhibitors) are plagued by a variety of limitations. For example, camptothecin is hampered by poor PK properties and drug delivery issues. In addition, radiation contributes to tumor hypoxia leading to chemo- and radioresistance, which is exacerbated with the use of platins. These complications highlight the need for discovery of novel radiosensitizers.
Previously, several radio-sensitizing drugs, including PARP and checkpoint kinase inhibitors, have been identified via high-throughput screening (HTS). Prior reports utilizing HTS have largely focus on cell viability or repair pathway modification as an outcome. For example, Li et al. developed a radioresistant cell line and identified radiosensitizers that influenced viability in a treatment resistant environment [4]. Others have similarly evaluated viability in a hypoxic environment [5]. Several radiosensitizer screens have resulted in clinical trials examining new indications for previously approved drugs. Drug repurposing efforts are potentially cost effective and may leverage prior knowledge of the drug's pharmacokinetic/pharmacodynamic and safety profiles. For instance, Goglia et al. developed a robust screen examining inhibition of DNA damage repair proteins that lead to a new clinical trial of mibefradil as a radiosensitizer [6].
In this study, we developed an image-based HTS method for G2/M arrest to predict radio-sensitizing activity of small molecules. A library of FDA approved drugs was tested against breast and cervical cancer cells followed by high-content imaging to assess nuclear size as a primary readout and cell cycle phase as a secondary readout (Fig. 1, see Supplemental Methods). The screen successfully confirmed established G2/M inhibiting radiosensitizers as well as several novel hits. The activity of one of these candidates, proflavine hemisulfate (PFS), was assessed both in vitro and in vivo.
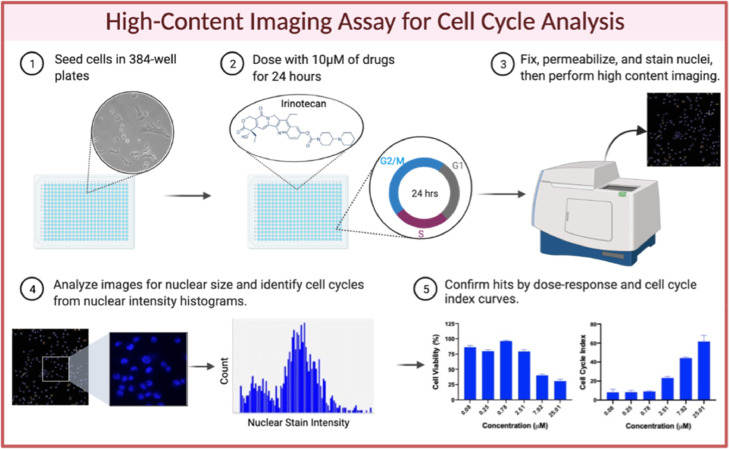
Fig. 1.
Schematic of High-Throughput Screen. 4T1 and HeLa cells were plated in 384-well plates (1), then dosed with 10 µM of drug per well for 24 h (2). Cells were then fixed, permeabilized, and nuclear stained prior to high content imaging (3). Average nuclear size was derived from the images, and drugs considered hits were 1 SD from the mean for each plate. Histograms were then generated from the images for nuclear size hits, and subsequently cell cycle indexes were derived. Hits were 3 SD away from the mean (4). Select hits for nuclear size and G2/M stall by cell cycle analysis were validated by dose response and cell cycle index (5).
Materials and methods
Materials
The FDA-approved drug library was purchased from Selleckchem (Houston, TX, USA). Crystal violet dye, RPMI 1640 medium, and propidium iodide were purchased from Fisher Scientific (Hampton, NH, USA). Proflavine hemisulfate hydrate was purchased from TCI Chemicals (Tokyo, Japan). All cell lines were purchased from ATCC (Manassas, VA, USA) and maintained in recommended medium supplemented with 10% (v/v) fetal bovine serum (FBS, Hyclone) and 1% (v/v) penicillin/streptomycin.
Flow cytometry cell cycle analysis
4T1 cells were seeded at 100,000 cells per well, MDA-MB-231 cells and MCF7 cells at 300,000 per well, and HCC1937 at 450,000 per well in 6-well plates and allowed to settle overnight. Cells were then treated in duplicate with proflavine hemisulfate (PFS) at 2.5 mM, irinotecan (10 µM) or nocodazole (40 ng/mL) as a positive control, and media alone. 24 h later, the media was aspirated and the cells were trypsinized. The cells were resuspended in 1 mL media, then centrifuged at 300xG and 4 °C for 5 mins. The pellet was resuspended in 1 mL cold PBS, then spun once more. The PBS was aspirated down to 50 mL, which was used to resuspend the pellet. 1 mL 70% EtOH stored at 4 °C was added to the cells dropwise while vortexing and fixed at 4 °C for 2 h. Cells were then spun down at 500xG for 5 mins, then washed with PBS once prior to adding the staining cocktail. In each sample, the cocktail was composed of 0.5 µL Triton-X100, 10 µL RNAase A, 5 µL of propidium iodide (PI) at 2 mg/mL, and 485 µL molecular grade water. The stain was incubated at 37 °C for 1 hr before running on a BD Fortessa Flow Cytometer.
In vitro cytotoxicity
To determine the efficacy of PFS, 4T1 cells were seeded at 4000 cells per well in a 96-well plate and allowed to settle overnight. Cells were then treated with PFS at concentrations ranging from 0.03 µM to 16 µM (Fig. 1D and S1) and incubated at 37 °C. After 48 h, cells were washed once with PBS, then viability was determined by Alamar Blue as previously detailed [7].
Clonogenic assay
The radiosensitizing ability of PFS was assayed by seeding 1 × 106 4T1 cells into T25 flasks, which were allowed to settle overnight and subsequently treated for 24 h with PFS (2.5 µM) and media alone. The assay was carried out as previously described, with the exception of plating numbers which ranged from 300 cells/well for no treatment to 12,000 cells/well for 6 Gy PFS, and a proliferation time of 11 days [7].
Immunofluorescent γH2AX DNA damage assay
To evaluate DNA double strand breaks (DSBs), 4T1 cells were plated at 70,000 cells/well in two gelatin-coated 4-well chamber slides and allowed to settle for 24 h. The media was then replaced with media containing PFS (0.5 µM) or control media. Cells were incubated with these treatments (Fig. 4) for 24 h at which point one of the plates was irradiated with 4 Gy (CellRad). The remainder of the assay was carried out as previously described [7].
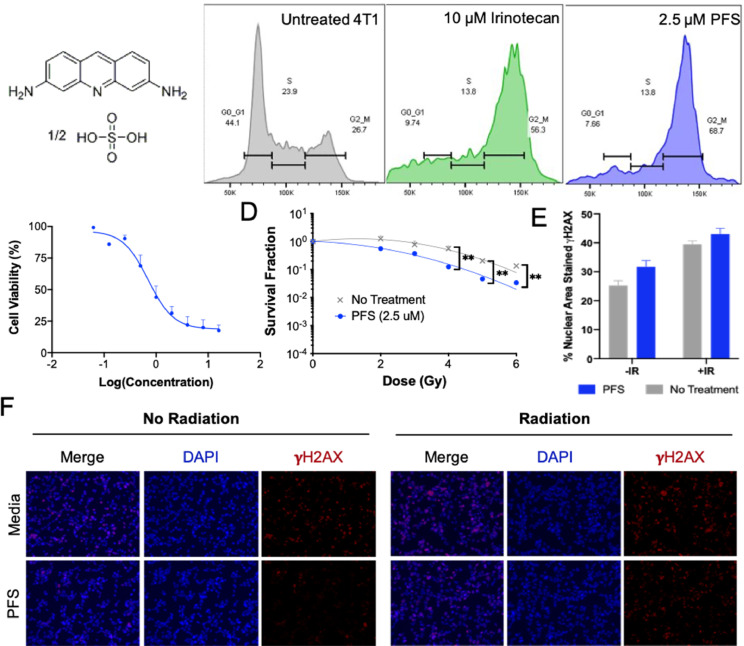
Fig. 4.
Evaluation of proflavine as a radiosensitizer in vitro. (A) Chemical structure of proflavine hemisulfate. (B) Cell cycle analysis of proflavine by flow cytometry, validating it as a hit for G2/M arrest. Irinotecan was used as a positive control for G2/M stall. (C) Cell viability of 4T1 cells dosed with proflavine. (D) Clonogenic assay showing radiositizing capabilities of PFS at 2.5 µM. **p-value < 0.01 (E) Quantification of γH2AX assay images in (F). Radiation dose was 4 Gy.
Apoptosis assay
To analyze the effect of PFS and irradiation on apoptosis and necrosis, 150,000 cells were plated in 6-well plates. After settling overnight, cells were dosed with PFS (1 µM) or media in duplicate. After 24 h, one of the plates was irradiated with 4 Gy. 2 h later, cells were washed, trypsinized, and resuspended in media. An apoptosis kit from Miltenyi Biotec was used to stain the cells prior to flow cytometry (BD Fortessa).
ATP release assay
4T1 cells were plated at 10,000 cells/well in a 96-well plate and allowed to settle for 24 h. After settling, the media was replaced with 50 µL media containing PFS (1 µM and 5 µM), mitoxantrone (1 µM and 5 µM) as a positive control, or media alone. 50 µL of Promega Extracellular ATP Detection Reagent (Promega CS3030A01) was added to all the wells and placed on a plate shaker for 30 secs before detecting luminescence (Tecan). The plate was returned to the incubator until the next timepoint (0–50 h).
Calreticulin assay
4T1 cells were plated at 30,000 cells/well in an 8-well chamber slide and allowed to settle overnight. The media was then replaced with 250 µL of media containing PFS (5 µM) or media/DMSO alone. After 6 h of incubation, media was removed, and wells were washed 1x with PBS. Anti-CRT antibody (ab196159) diluted by 500 in 1% BSA was incubated for 1 hr. Wells were washed with PBS, then wheat germ agglutinin (WGA, AF555 conjugated) was incubated at 5 µg/mL for 1 min to visualize the membrane. Hoescht containing mountant was added to the slide and cells were imaged at 63x (Zeiss Airy Confocal Microscope).
In vivo studies
All animal studies were approved by and conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Oregon Health and Sciences University. Ten days after tumor inoculation (see supplemental methods), 4T1 tumor bearing mice were randomized into 4 groups: PBS, PBS + IR, PFS, PFS + IR. The mice were injected intratumorally with 50 µL of PBS or PFS (10 µM) on days 0 and 1. The average tumor volume on day 0 (80 mm3) was used to calculate the 10 µM dose of PFS, assuming the drug diffuses throughout the tumor. On days 1 and 2, the mice in the IR groups received 2 gray (Gy) radiation (total of 4 Gy). Radiation was delivered (CellRad, Faxitron, 130 kV, 5 mA, 0.5 mm aluminum filter, ∼1.2 Gy/min) selectively to tumors by covering mice with half-moon cutout lead shields (Precision X-Ray, North Branford, CT). Mouse body weight and average tumor diameter (1/2 × length × width2) were recorded 2–3 times per week. On the day that the first mouse from reached an endpoint (either cavitated ulcerations, weight loss at 20% of starting weight, or any diameter at 2 cm), the study was ended.
Statistical analysis
All data are expressed as mean ± SEM. Statistical differences and significance were evaluated using one-way ANOVA, two-way ANOVA, or t-test in the Graph Pad Prism 8 software pack. p < 0.05 was considered statistically significant and represented by *.
Results and discussion
The G2/M phase is recognized as the most radiosensitive stage of the cell cycle, followed by G1, and the late S as the least sensitive [8,9]. Synchronization in the G2/M phase has been shown to sensitize cells to radiation. While this is a well-established concept, G2/M arrest has not yet been exploited thoroughly for screening of radiosensitizers. The classic method for assessing G2/M arrest is cell cycle analysis by flow cytometry. In this procedure, a nuclear dye such as DAPI, Hoescht, or PI can be used to stain DNA content stoichiometrically, which allows for the binning of cells into phases of the cell cycle based on nuclear intensity. Flow cytometry is often used for cell cycle analysis (by plotting nuclear stain intensity histograms) due to its sensitivity, repeatability, and ability to eliminate doublets. However, it is cumbersome for screening numerous drugs and requires trypsinization of cells for each drug tested, which alters the architecture of adherent cells [10]. Alternatively, several recent studies have employed high-content imaging to analyze cell cycle for HTS [10], [11], [12]. Herein we utilized both a novel primary readout based on nuclear size and an established secondary readout based on analyzing cell cycle content after dosing 4T1 murine breast cancer cells and HeLa cervical cancer cells with a library of 1430 FDA approved small molecules. HeLa cells are often employed in chemotherapeutic screens and served to validate our method of screening in breast cancer cells. Based on previous reports, a dose of 10 µM was selected and incubated with cells for 24 h.
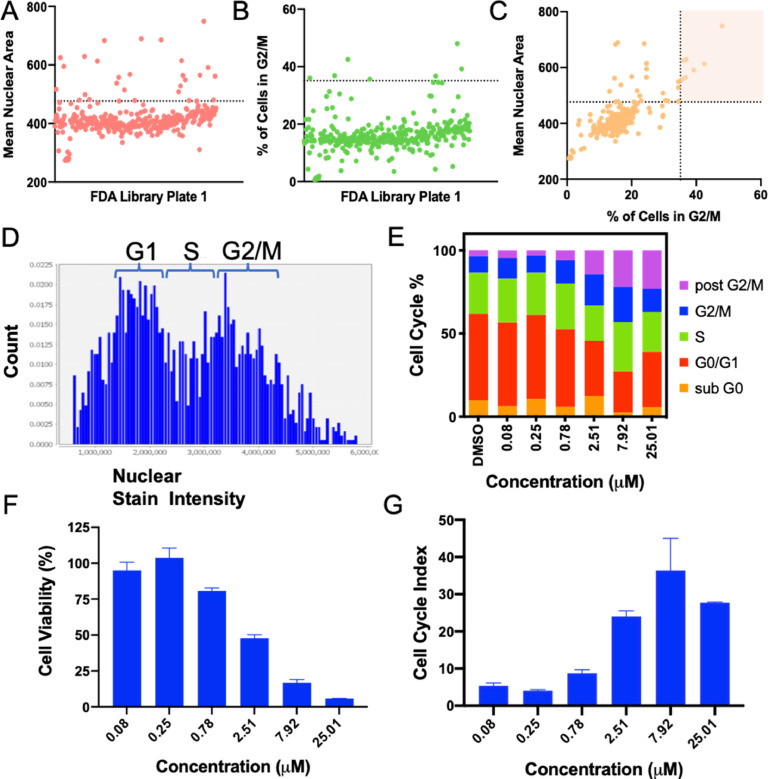
After staining with Hoescht, high-content imaging was performed and outlines of cell nuclei were identified using Cellomics (Thermofisher), from which nuclear area was also calculated. G2/M phase has twice the number of chromosomes causing an enlargement in nuclei. We therefore hypothesized that an increased average nuclear size could be used to identify G2/M arresting drugs. Previously, Ferro et al. had similarly plotted nuclear size vs. nuclear intensity to bin cell clusters into phases [10]. In addition to G2/M arrest, nuclear size can increase due to cellular stresses such as that caused by reactive oxygen species, senescence, and necrosis. However, senescence occurs at a later time (> 24 h) so it was not expected to be a factor in this screen. To reduce false positives due to cellular stress and necrosis, nuclear size was used as a primary endpoint with an approximate cutoff of 1 standard deviation from the mean nuclear size per plate of drugs (Fig. 2A), and further narrowed with classical image-based cell cycle analysis from identification of nuclear intensity, with the 4 N population representing G2/M (Fig. 2B). As seen in Fig. 2C, there was a loose correlation between nuclear area and percent of cells in G2/M, with the false positives for nuclear size eliminated in the upper left quadrant. After identifying hits, histograms were manually reviewed to confirm cell cycle stall (Fig. 2D). Binning by nuclear intensity was based off the histograms for the positive control, irinotecan—a known radiosensitizer and mitotic inhibitor—and the negative control, DMSO. Select hits were further confirmed by evaluating the cell cycle of hits at a range of concentrations (Fig. 2E), as well as the dose-response (Fig. 2F) and cell cycle index (CCI, Fig. 2G). CCI indicates overall deviation of the drug induced cell cycle compared to DMSO controls. Other reported screens have employed a CCI of 10 to recognize cell cycle arrestors [11]. Fig. 2D–G show the response to a novel radiosensitizer from the screen, proflavine hemisulfate, which had a CCI above 10.
Fig. 2.
4T1 Screening Results and Validation. (A) Mean nuclear area from well images was used as a primary analysis for G2/M stall, with hits more than 1 SD from the mean (above dotted line). 1 of 5 plates of the drug library is shown. (B) Nuclear intensity histograms were generated from the same images and used to further narrow in on G2/M arrest hits (above dotted line, indicating 3 SD). (C) Mean nuclear area and nuclear intensity showed a loose correlation with false positives for nuclear size in the upper left quadrant, and hits for both parameters in the highlighted quadrant. After identifying hits, they were validated by dosing at a range of concentrations with an automated printer and generating histograms (D), cell cycle profiles (E), cell viability (F), and cell cycle index (G). *D–G are from G2/M hit proflavine hemisulfate.
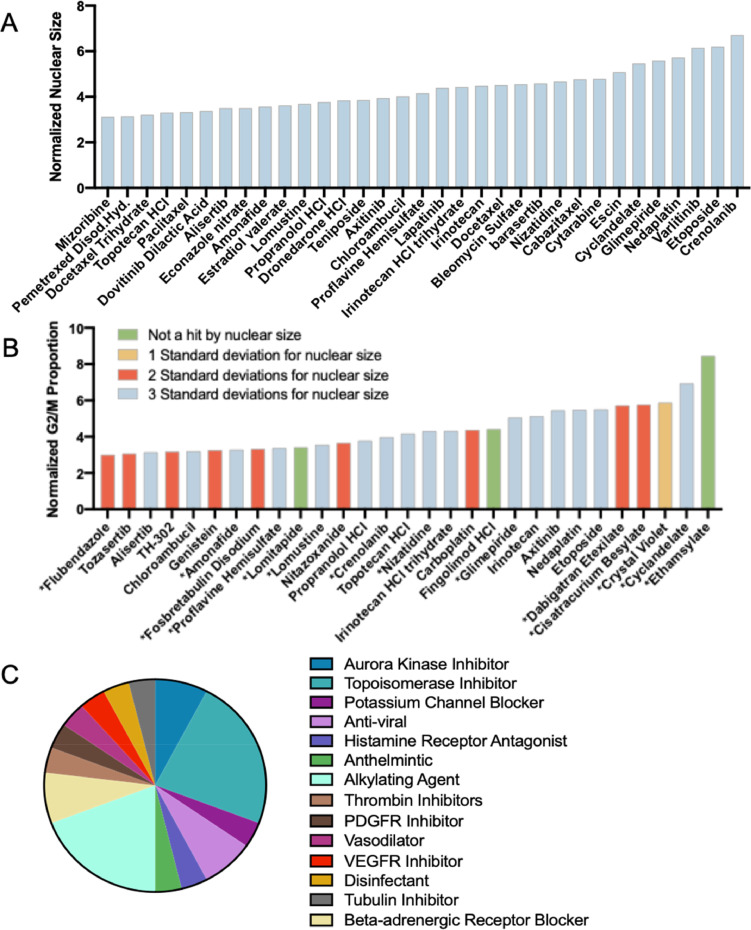
The screen resulted in 122 hits for nuclear size, or an 8.5% hit rate, and 29 hits from the secondary cell cycle analysis, 26 of which were validated by nuclear size giving a final hit rate of 1.8%. Furthermore, the strongest hits for the primary and secondary readouts largely overlap (Fig. 3A,B, blue bars). Many of the hits, including etoposide, carboplatin and nedaplatin, and genistein are known G2/M arrestors and radiosensitizers validating the method. In addition, the screen identified G2/M arrestors that other cell cycle assays had identified in HeLa cells, such as chloroambucil and etoposide (Fig. S2) [11]. Amongst the hits, there were 14 different FDA indications (Fig. 3C). Using strictly standardized mean difference (SSMD) as the parameter for evaluating the assay quality, the primary screen for 4T1 resulted in an average SSMD of 9.22 +/- 2.34 and 9.65 +/- 1.62 for HeLa, and the secondary screen had an SSMD of 6.94 +/- 1.10 and 4.64 +/- 1.36 for 4T1 and HeLa respectively. For very strong controls, an SSMD of greater than 5 is considered excellent and 3–5 considered good [13].
Fig. 3.
Hits from the Screen. (A) Strong hits (3 standard deviations from the mean) for nuclear size increase in 4T1 cells. (B) Strong hits (3 standard deviations from mean) for G2/M proportion from nuclear intensity histograms in 4T1. Hits that were also strong for nuclear size are identified in blue, with weaker hits for nuclear size in red and yellow. *drugs represent novel G2/M arrestors. (C) Varying indications of the hits from both A and B, suggesting new potential classes of interest for radiosensitzer repurposing.
Proflavine hemisulfate was identified as a strong candidate radiosensitizer in 4T1s based on nuclear size and cell cycle analysis. PFS is classified as an acriflavine derivative commonly applied as a topical disinfectant bacteriostatic agent [14]. PFS intercalates in DNA strands, thereby altering DNA synthesis and leading to accumulation of mutations. Small ligands, like proflavine, that are often polycyclic, aromatic, and planar can intercalate in DNA by inserting themselves between two adjacent base pairs in the DNA strand, both unwinding the helical twist and disrupting DNA repair (Fig. 4A) [15]. While proflavine has been proposed as a chemotherapy as early 1922 [16], it has only been FDA approved for topical use preventing gram-positive bacterial infections and is in clinical trials as a diagnostic for Barrett's Esophagus, cervical cancer, and colonic polyps due to its fluorescent properties. The development of proflavine as a chemotherapy has been limited by its classification as a mutagen, not unlike cisplatin. Here we report on proflavine as having radiosensitizing capabilities at low doses.
PFS was validated as a G2/M arrestor by flow cytometry in the murine 4T1 cell line (Figs. 4B, S3) as well as human triple negative breast cancer cell lines, HCC1937 and MDA-MB-231 (Fig. S4). However, with a 24 h incubation at 2.5 μM PFS arrested ER+PR+ MCF-7 cells in G1 instead (Fig. S4G–I). After validating cell cycle arrest, PFS was evaluated for radiosensitizing capabilities by the clonogenic assay and the γH2AX puncti formation assay (Fig. 4D–F). The clonogenic assay is considered the gold standard of radiosensitivity assays and evaluates radiobiological death via clonogenicity. Above 4 Gy PFS was able to significantly decrease the survival fraction compared to radiation alone. In analyzing DNA damage, we found that PFS can enhance the number of DSBs caused by radiation, but only minimally and with lower staining intensity (Fig. 4E, F). Wang et al. observed a lack of evidence of DNA damage signaling (including γH2AX) for acridine derivatives, and instead report activation of p53 transcriptional responses that trigger cell cycle arrest and apoptosis [17]. Previous reports found that flavins produce ROS independently or when activated by UV radiation, which may be responsible for the radiosensitivity observed via the clonogenic assay with only modest increase in DSBs [18,19].
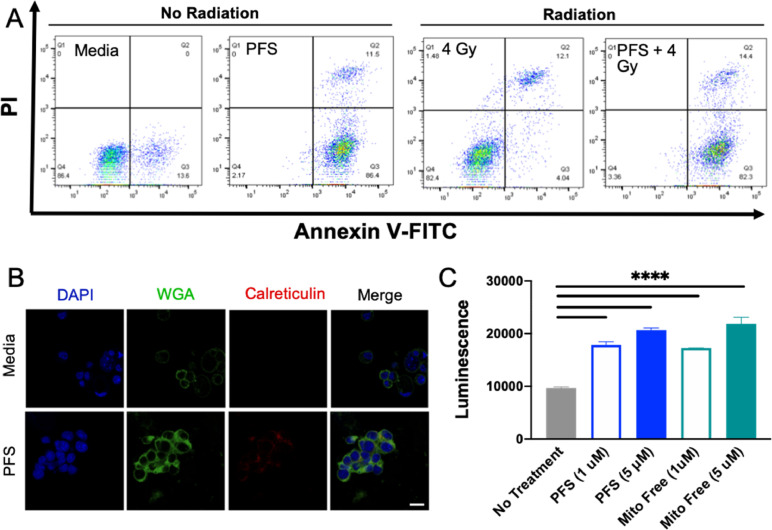
Recently, the ability of chemotherapy to induce an immune response, rather than a tolerogenic or suppressive response has become an area of critical investigation. The induction of immunogenic cell death (ICD) is one mode of death stimulated by a local antitumor immune response causing increased immune cell infiltration in tumor stroma. Some chemotherapeutics induce ICD, such as oxaliplatin and mitoxantrone. Herein we assessed the ability of PFS to induce ICD. Previous reports indicated that proflavine contributes to apoptosis and autophagy [20]. Here, PFS was evaluated by a PI/annexin V assay for contribution to early and late apoptosis/necrosis with radiation. Proflavine alone and with radiation caused late apoptotic/necrotic cell death along with early apoptosis (Fig. 5A). Apoptosis was previously considered tolerogenic, however with ICD markers is considered immunogenic. The expression of damage-associated molecular patterns (DAMPs), such as calreticulin (CRT) can be evaluated at early timepoints, and the release of ATP at later timepoints. Cells were treated with the drug combination and stained for membrane CRT expression (Fig. 5B). Co-localization of the cell membrane stain, WGA, with the CRT stain indicated translocation. PFS caused substantial translocation of CRT to the external leaflet of the cell membrane as seen in Fig. 5B, as well as significant ATP release, comparable to mitoxantrone as a positive control.
Fig. 5.
Immunogenic Cell Death Induction. (A) Apoptosis and necrosis assay showing increase of apoptosis and necrosis with proflavine. (B) Visualization of immunogenic cell death by surface exposure of calreticulin at 6 hr. (C) 50 hr timepoint of ATP release from untreated 4T1, PFS, and mitoxantrone (mito) as a positive control. ****p-value < 0.0001.
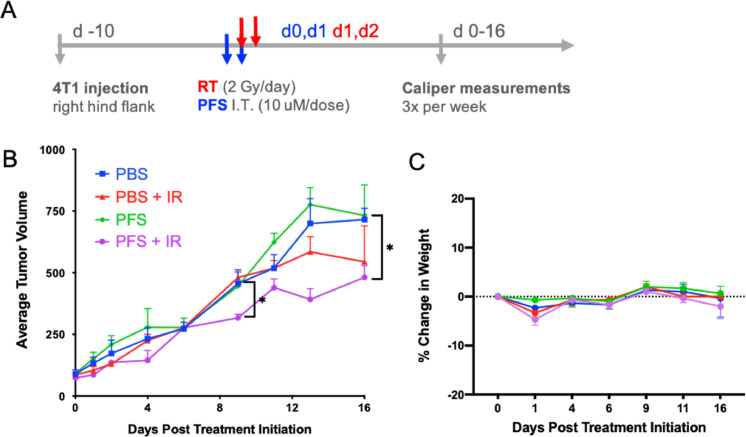
Lastly, the in vivo efficacy of PFS as a radiosensitizer was assessed in a syngeneic 4T1 breast cancer mouse model evaluating tumor volume. Mice were split into four treatment arms: PBS only, PBS + IR, PFS, and PFS + IR. When tumors reached an average size of 80 mm3, the first dose of PFS (10 μM) was delivered intratumorally as a priming dose to stall cells in G2/M (Fig. 6A). A second dose of PFS was delivered on the following day with concomitant radiation (2 Gy). A second fraction (2 Gy) was delivered on the third day to conclude treatment. As seen in Fig. 6B, one round of treatment was sufficient to cause separation of the PFS + IR arm of the study from the untreated and PFS or IR alone arms at day 9 of the study. Notably, PFS was only effective at controlling tumor volume when combined with radiation. However, by day 16 the IR and PFS + IR arms were starting to have similar tumor volume indicating the animal study could have benefited from a second round of PFS dosing. The treatment regimen did not show toxicity by the lack of 20% fluctuation in mouse body weight (Fig. 6C).
Fig. 6.
In vivo Efficacy Study. (A) Dosing schedule for the efficacy study. Two doses of PFS were delivered intratumorally and 2 doses of 2 Gy X-ray radiation to subcutaneous tumors. (B) Relative tumor volume curves for mice in efficacy study treated with various therapeutic agents following treatment regimen in (A). (C) Lack of 20% change in body weight indicates safety of the combination.
The promising performance of PFS as a radiosensitizer supports the ability of the screening method to enable discovery of new radiosensitizers. While PFS does not overcome all of the limitations of clinical radiosensitizers mentioned above, it does address some, such as solubility, dependence on DNA repair status, and mode of cell death. Prior to further development of PFS, further studies on long-term safety of PFS are needed due to its status as a mutagen. Of note, cisplatin is likewise classified as a mutagen, causing 0.8 mutations per Mb.[21] However, liposomal formulations help mitigate toxicity of cisplatin, and similarly PFS would likely benefit from a carrier or prodrug formulation.
Conclusion
There are a limited number of clinically available radiosensitizers that can be applied independent of DNA repair status. Various existing screens report on novel radiosensitizers that are dependent on the expression of certain proteins, narrowing the utility and application to various cancers and subtypes. Herein we report on a novel screening method that has the potential to be utilized for various cancer cell lines for radiosensitizer discovery. This screen exploited the simple principle that cells are most sensitive to x-ray radiation in the G2/M phase of the cell cycle. Using 4T1 breast cancer cells, we achieved a high hit rate of 1.8% with an FDA approved library of drugs including 14 novel G2/M cycle arrestors, as well as identified 15 known arrestors. One of the previously undiscovered hits, PFS, was found to be an effective radiosensitizer in vitro and in vivo.
CRediT authorship contribution statement
Madeleine Landry: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization. Dylan Nelson: Conceptualization, Methodology, Validation, Formal analysis, Investigation. Eunseo Choi: Methodology, Investigation, Writing – review & editing. Allison DuRoss: Conceptualization, Methodology, Writing – review & editing. Conroy Sun: Conceptualization, Methodology, Formal analysis, Resources, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
None.
Acknowledgments
This work was supported by the NIH NIGMS as a Maximizing Investigators’ Research Award, 1R35GM119839–01 (C.S.) and Oregon State University College of Pharmacy Start-up Funds. M.R.L. was funded by AFPE as an Herb and Nina Demuth Pre-Doctoral Fellow and currently by the PhRMA Foundation Predoctoral Drug Delivery Fellowship.
The authors thank the High Throughput Screening Core at Oregon State University for their assistance with the screen presented in this study, and Brianna Garcia at the OHSU Flow Cytometry core for assistance with cell cycle analysis.
The schematic in Fig. 1 was made in ©BioRender -biorender.com.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101336.
Contributor Information
Dylan Nelson, Email: Dylan.Nelson@oregonstate.edu.
Conroy Sun, Email: sunc@ohsu.edu.
Appendix. Supplementary materials
References
- 1.Ahmad S.S., Crittenden M.R., Tran P.T., Kluetz P.G., Blumenthal G.M., Bulbeck H., Baird R.D., Williams K.J., Illidge T., Hahn S.M., Lawrence T.S., Spears P.A., Walker A.J., Sharma R.A. Clinical Development of Novel Drug-Radiotherapy Combinations. Clin. Cancer Res. 2019;25:1455. doi: 10.1158/1078-0432.CCR-18-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buckley A.M., Lynam-Lennon N., O’Neill H., O’Sullivan J. Targeting hallmarks of cancer to enhance radiosensitivity in gastrointestinal cancers. Nat. Rev. Gastroenterol. Hepatol. 2020;17:298. doi: 10.1038/s41575-019-0247-2. [DOI] [PubMed] [Google Scholar]
- 3.Sharma R.A., Plummer R., Stock J.K., Greenhalgh T.A., Ataman O., Kelly S., Clay R., Adams R.A., Baird R.D., Billingham L., Brown S.R., Buckland S., Bulbeck H., Chalmers A.J., Clack G., Cranston A.N., Damstrup L., Ferraldeschi R., Forster M.D., Golec J., Hagan R.M., Hall E., Hanauske A.-.R., Harrington K.J., Haswell T., Hawkins M.A., Illidge T., Jones H., Kennedy A.S., McDonald F., Melcher T., O’Connor J.P.B., Pollard J.R., Saunders M.P., Sebag-Montefiore D., Smitt M., Staffurth J., Stratford I.J., Wedge S.R. Clinical development of new drug–radiotherapy combinations. Nat. Rev. Clin. Oncol. 2016;13:627. doi: 10.1038/nrclinonc.2016.79. [DOI] [PubMed] [Google Scholar]
- 4.Li Z., Tamari K., Seo Y., Minami K., Takahashi Y., Tatekawa S., Otani K., Suzuki O., Isohashi F., Ogawa K. Dihydroouabain, a novel radiosensitizer for cervical cancer identified by automated high-throughput screening. Radiother. Oncol. 2020;148:21. doi: 10.1016/j.radonc.2020.03.047. [DOI] [PubMed] [Google Scholar]
- 5.Tamari K., Seo Y., Takahashi Y., Otani K., Kawashima A., Suzuki O., Isohashi F., Ogawa K. Ro90-7501 Is Identified As a Radiosensitizer By High Throughput Screening. Int. J. Radiat. Oncol. Biol. Phys. 2017;99:E619. [Google Scholar]
- 6.Goglia A.G., Delsite R., Luz A.N., Shahbazian D., Salem A.F., Sundaram R.K., Chiaravalli J., Hendrikx P.J., Wilshire J.A., Jasin M., Kluger H., Glickman J.F., Powell S.N., Bindra R.S. Identification of Novel Radiosensitizers in a High-Throughput, Cell-Based Screen for DSB Repair Inhibitors. Mol. Cancer Ther. 2015;14:326. doi: 10.1158/1535-7163.MCT-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landry M.R., DuRoss A.N., Neufeld M.J., Hahn L., Sahay G., Luxenhofer R., Sun C. Low Dose Novel PARP-PI3K Inhibition via Nanoformulation Improves Colorectal Cancer Immuno-Radiotherapy. Mater. Today Bio. 2020 doi: 10.1016/j.mtbio.2020.100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall E.J., Giaccia A.J. Radiobiology for the Radiologist. 7th Ed. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2012. Radiobiology for the Radiologist. [Google Scholar]
- 9.Seiwert T.Y., Salama J.K., Vokes E.E. The concurrent chemoradiation paradigm—general principles. Nat. Rev. Clin. Oncol. 2007;4:86. doi: 10.1038/ncponc0714. [DOI] [PubMed] [Google Scholar]
- 10.Ferro A., Mestre T., Carneiro P., Sahumbaiev I., Seruca R., Sanches J.M. Blue intensity matters for cell cycle profiling in fluorescence DAPI-stained images. Lab. Invest. 2017;97:615. doi: 10.1038/labinvest.2017.13. [DOI] [PubMed] [Google Scholar]
- 11.Lo Y.C., Senese S., France B., Gholkar A.A., Damoiseaux R., Torres J.Z. Computational Cell Cycle Profiling of Cancer Cells for Prioritizing FDA-Approved Drugs with Repurposing Potential. Sci. Rep. 2017;7:11261. doi: 10.1038/s41598-017-11508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roukos V., Pegoraro G., Voss T.C., Misteli T. Cell cycle staging of individual cells by fluorescence microscopy. Nat. Protoc. 2015;10:334. doi: 10.1038/nprot.2015.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bray M.A., Carpenter A., et al. In: Assay GuidanceManual. Markossian S., Grossman A., Brimacombe K., Arkin M., Auld D., Austin C.P., et al., editors. Eli Lilly & Company and the National Center for Advancing Translational Sciences; Bethesda (MD): 2004. Advanced Assay Development Guidelines for Image-Based High Content Screening and Analysis. [PubMed] [Google Scholar]
- 14.NCATS Inxight: drugs-PROFLAVINE, 2021.
- 15.Li S., Cooper V.R., Thonhauser T., Lundqvist B.I., Langreth D.C. Stacking Interactions and DNA Intercalation. J. Phys. Chem. B. 2009;113:11166. doi: 10.1021/jp905765c. [DOI] [PubMed] [Google Scholar]
- 16.Browning C.H., Cohen J.B., Gaunt R., Gulbransen R. Relationships between Antiseptic Action and Chemical Constitution with Special Reference to Compounds of the Pyridine, Quinoline, Acridine and Phenazine Series. Proc. R. Soc. Lond. Ser. B. 1922;93:329. Containing Papers of a Biological Character. [Google Scholar]
- 17.Wang W., El-Deiry W. DNA-intercalating drugs activate p53 transcriptional responses without evidence of DNA-damage signaling. Cancer Res. 2007;67:2541. [Google Scholar]
- 18.Fedoseeva I.V., Pyatrikas D.V., Stepanov A.V., Fedyaeva A.V., Varakina N.N., Rusaleva T.M., Borovskii G.B., Rikhvanov E.G. The role of flavin-containing enzymes in mitochondrial membrane hyperpolarization and ROS production in respiring Saccharomyces cerevisiae cells under heat-shock conditions. Sci. Rep. 2017;7:2586. doi: 10.1038/s41598-017-02736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M.Y., Chang C.J., Chen L.Y. Blue light induced reactive oxygen species from flavin mononucleotide and flavin adenine dinucleotide on lethality of HeLa cells. J. Photochem. Photobiol. B Biol. 2017;173:325. doi: 10.1016/j.jphotobiol.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Zhang M.S., Niu F.W., Li K. Proflavin suppresses the growth of human osteosarcoma MG63 cells through apoptosis and autophagy. Oncol. Lett. 2015;10:463. doi: 10.3892/ol.2015.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szikriszt B., Póti A., Pipek O., Krzystanek M., Kanu N., Molnár J., Ribli D., Szeltner Z., Tusnády G., Csabai I., Szallasi Z., Swanton C., Szüts D. A comprehensive survey of the mutagenic impact of common cancer cytotoxics. Genome Biology. 2016;17 doi: 10.1186/s13059-016-0963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.