Abstract
The mechanism of cargo coupling to kinesin motor proteins is a fundamental issue in organelle transport along microtubules. Kinectin has been postulated to function as a membrane anchor protein that attaches various organelles to the prototype motor protein kinesin. To verify the biological relevance of kinectin in vivo, the murine kinectin gene was disrupted by homologous recombination. Unexpectedly, kinectin-deficient mice were viable and fertile, and no gross abnormalities were observed up to 1 year of age. The assembly of the endoplasmic reticulum was essentially unaffected in kinectin-deficient cells. Mitochondria appeared to be correctly distributed throughout the cytoplasm along the microtubules. Furthermore, the stationary distribution and the bidirectional movement of lysosomes did not depend on kinectin. Kinectin-deficient phagocytes internalized and cleared bacteria, indicating that phagosome trafficking and maturation are functional without kinectin. Thus, these data unequivocally indicate that kinectin is not essential for trafficking of lysosomes, phagosomes, and mitochondria in vivo.
Microtubule (MT)-dependent membrane transport is essential for a variety of cellular processes (for a review, see references 22 and 46). MT-dependent intracellular transport processes are mediated by motor proteins of the kinesin and of the cytoplasmic dynein superfamilies. These molecules act as mechanochemical adenosin triphosphatases, converting chemically bound energy into directed motile force along the MT tracks. The majority of the kinesin family members are anterograde motors, conveying their cargoes specifically toward the plus ends of the MT. Some kinesin family members and cytoplasmic dynein function as retrograde motors, mediating cargo transport toward the MT minus ends (reviewed in references 21 and 54). Since the first description of kinesin (6, 56), approximately 30 members of murine kinesin superfamily proteins (KIFs) have been identified (21, 34, 59). Genetic inactivation of various KIFs and of the cytoplasmic dynein heavy chain (cDHC) in the mouse and in Drosophila melanogaster resulted in embryonic or perinatal lethality, thus proving the vital importance of these motor proteins (14, 18, 38, 44, 51, 52, 60). A conventional kinesin consists of two kinesin heavy chains (KHCs) and two kinesin light chains (KLCs). The KHC binds directly to the MT via its globular N-terminal head (23, 45). The C-terminal regions of the KHC associate with the KLCs and are thought to interact with the cargoes (50). Recently, the KLCs have been implicated in the regulation of motor activity. Up to now, it has not been clarified, whether KLCs are directly involved in cargo binding (9, 12, 49).
While many KIFs have been identified and much has been learned about kinesin functions (reviewed by Goldstein and Philip [16] Hirokawa et al. [22]), there is only scarce information available concerning the receptors anchoring the various KIFs to their cognate cargoes. Kinectin (KNT), the first proposed candidate anchor protein for kinesin tails on vesicle membranes, was purified by affinity column chromatography (53). The KHC binding domain of KNT was mapped to a short C-terminal KNT region. Correspondingly, the KNT interaction domain in kinesin was defined to reside in the KHC tail (39). KNT associates with membranes via its N terminus (13, 61). It is an integral membrane protein of ca. 160 kDa, located at the cytoplasmic face of membranous vesicles (53, 61). KNT can form heterodimers with a shorter 120-kDa isoform, presumably by interaction of α-helical coiled coils that encompass the C-terminal two-thirds of the molecule (27). However, the function of the 120-kDa isoform is unclear. Antibody inhibition studies suggested a participation of KNT in MT-dependent vesicle motility. An anti-KNT antibody raised against a native epitope in the KNT cytoplasmic domain was shown to efficiently block plus-end-and; to a lesser extent, also minus-end-directed vesicle motility in an in vitro trafficking assay (28). The same antibody was also reported to reduce kinesin binding to motile vesicles (28). Furthermore, attempts to suppress KNT function by peptide inhibition approaches suggested a requirement for KNT in phagosome and lysosome motility. Here, peptides derived from the central KNT stalk domain but not from the KNT C terminus were described to block bidirectional movement of phagosomes in vitro (3). In this study, the reported content of KNT was elevated on late phagosomes of higher motility. In a different study, overexpression of the KNT-kinesin binding domain was shown to disrupt anterograde lysosome transport (39). However, it remains controversial whether KNT is an essential anchor for kinesin on cargo membranes (21).
To analyze the biological relevance of KNT in cells and in animals, the murine knt gene was disrupted by homologous recombination in embryonic stem (ES) cells. KNT knockout mice were viable, reproduced normally, and did not develop any obvious abnormalities for up to 1 year of age. KNT-null mutant cells showed a normal assembly of endoplasmic reticulum (ER) and cis-Golgi, and the stationary intracellular distribution of mitochondria and lysosomes was unaffected. The lack of KNT did not abrogate lysosome motility. Peritoneal exudate cells (PEC) obtained from knt-deficient mice were capable of phagocytosing and eliminating Escherichia coli bacteria, indicating that phagosome trafficking and maturation occur in the absence of KNT.
MATERIALS AND METHODS
Targeting vector and structure of the genomic knt locus.
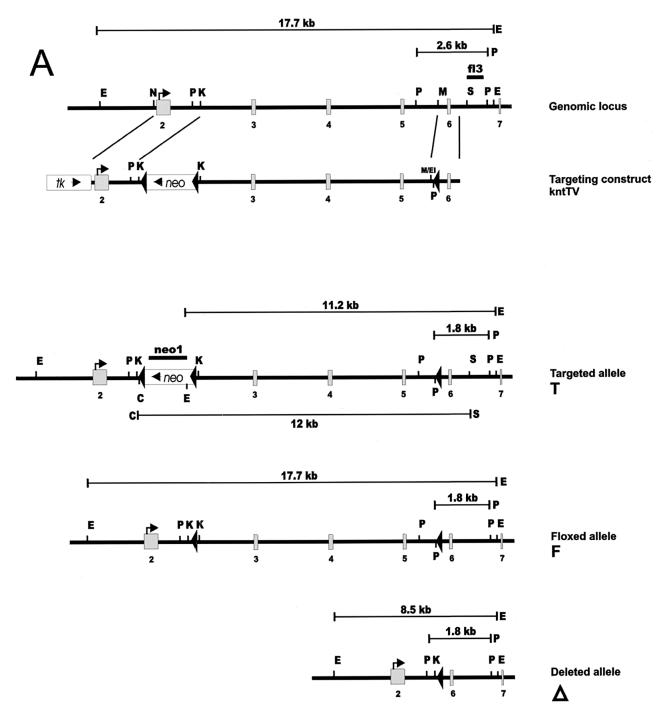
A partial human cDNA (the kind gift of M. Krönke) was used to screen a mouse 129Sv/J genomic library in λDASH II. A 14.5-kb genomic clone hybridizing to the 5′ region of the cDNA was isolated, mapped, and sequenced. It contained exons 2 to 6 and in part exon 7, corresponding to nucleotide (nt) −12 to nt 1163 of the published murine cDNA (GenBank accession no. L43326). The sequenced exons were identical to the murine knt coding sequence. The ATG start site was located at nt 19 in exon 2. The exon borders were as follows: exon 2 (nt −12 to 548), exon 3 (nt 549 to 686), exon 4 (nt 687 to 860), exon 5 (nt 861 to 991), exon 6 (nt 992 to 1108), and exon 7 (nt 1109 to >1163). To insert loxP signals into intron 2 and intron 5 of the knt locus, a targeting vector was constructed as follows (Fig. 1A). A synthetic loxP motif and a PstI and a BglII site were added by PCR to a 0.7-kb genomic DNA fragment located directly 3′ to a unique MfeI site of the genomic clone. This 0.7-kb fragment was inserted into a modified pBluescript vector (Stratagene) using PstI and BamHI to generate the plasmid kntSA. A 1.7-kb fragment containing exon 2 was excised from the genomic clone with NcoI and KpnI and inserted into pGEM7 (Promega) cut with SmaI and KpnI. The 1.7-kb fragment was recovered from this plasmid by digestion with EcoRI and ClaI and ligated to kntSA cut with EcoRI and ClaI to create kntSAL1. The genomic region containing exon 3 to exon 5 was inserted into kntSAL1 cut with KpnI and EcoRI as a 9.2-kb KpnI/MfeI fragment, yielding kntLS. The targeting vector kntTV was completed by inserting a PGK-neor gene flanked by loxP motifs into the KpnI site and the HSV-tk cassette into the SalI site of kntLS, respectively. The orientations of loxP motifs were verified by restriction analysis and sequencing.
FIG. 1.
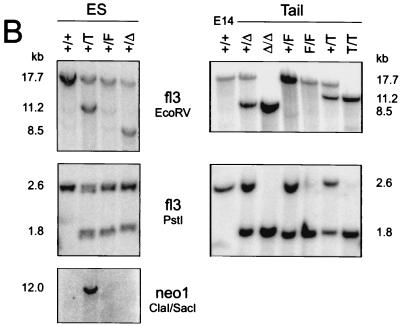
Construction of the KNT targeting vector and generation of mutant mice. (A) Schematic representation of the genomic knt locus, the targeting construct (kntTV) and the different mutated alleles. Homologous recombination of kntTV introduces a loxP-flanked neomycin resistance gene (neo) into intron 2 and a single loxP motif into intron 5 of the knt genomic locus. Cre-mediated recombination at the targeted locus (T) produces either a floxed knt allele (F) or a deleted knt allele (Δ) by excision of the neomycin resistance gene or by deletion of the neomycin resistance gene together with exons 3 to 5, respectively. Exons are depicted as shaded boxes; loxP motifs are indicated as filled triangles. Transcriptional directions of neomycin resistance and tk (open boxes) are indicated by arrows. fl3, 3′ external probe; neo1, neo probe; E, EcoRV; P, PstI; C, ClaI; S, SacI; N, NcoI; K, KpnI; M, MfeI; M/EI, MfeI/EcoRI. (B) Southern blot analysis of genomic DNA from targeted ES cell clones before and after in vitro Cre recombination (left) and from mouse tail biopsies (right). A 1.8-kb restriction fragment after PstI digestion is diagnostic for the mutated alleles in contrast to a 2.6-kb fragment generated from the wild-type allele. Note that the deleted allele is cut at an endogenous PstI site and not at the PstI site introduced with the distal loxP motif. EcoRV digestion generates fragments of 8.5 kb (Δ), 11.2 kb (T), and 17.7 kb (wt and F) in size. Southern detection of the PstI and the EcoRV digests was performed with the external probe fl3. Hybridization with neo1 after ClaI/SacI double digestion yields one 12-kb fragment for the targeted allele (T), indicating a single integration event. E14, genomic DNA from E14.1 ES cells.
Targeting of the knt locus and generation of KNT mutant mice.
For conditional targeting approaches, cointegration of a third, distal loxP motif, together with a floxed marker gene, into the target locus is crucial. Since there is no selection pressure on integration of the distal loxP motif, cointegration can be very inefficient, especially if the distance to the floxed marker cassette is long (1). To enable direct screening for correct recombination of the single loxP site, the targeting vector kntTV was linearized near the single loxP motif with NotI and transfected into embryonic day 14.1 (E14.1) ES cells. ES cell transfection, culture and selection was done as described previously (41). 288 G418- and ganciclovir-resistant ES cell colonies were picked and screened for integration of the single loxP motif by PCR using the external primer knt-S1 (5′-GAACAGATCCACATCACATCC-3′) and the internal primer knt-LF (5′-CCAATTCCTGCAGAGATCTATAAC-3′), which binds to genomic, polylinker, and loxP sequences and is therefore specific for the single loxP motif. Of 29 PCR positive clones, 11 were analyzed by Southern blotting of genomic DNA. The correct recombination event could be verified for 10 of 11 clones by blotting ES cell DNA after digestion with EcoRV or PstI and hybridization with the 3′ flanking probe fl3. Specifically, the PstI digest was indicative for cointegration of the single loxP site, whereas the EcoRV digest was used to analyze the correct localization of the floxed neomycin resistance gene. Single-copy integration was verified by Southern probing a ClaI/SacI double digest of the ES cell DNA with a part of the neomycin resistance cassette. ES cell clones carrying a floxed (F) knt allele were generated by deleting the neomycin resistance gene from the original targeted (T) knt locus in vitro. After transient transfection of 20 μg of supercoiled pIC-Cre plasmid (17), G418-sensitive ES cell clones were selected and characterized by PCR and Southern analysis for the different deletion events. The frequencies of Cre-mediated deletion yielding the floxed and the deleted alleles were 6 and 2%, respectively. Chimeric mice were generated from knt+/T and knt+/F ES cells by injection of C57BL/6 blastocysts and CD1 morula aggregation. Chimeric mice obtained from knt+/T ES cell lines were crossed to the Cre-transgenic deleter strain (47), resulting in deletion of the neomycin resistance gene and exons 3 to 5 of the knt gene (knt+/Δ). Germ line transmission of the targeted and the floxed allele, as well as successful deletion, were identified by Southern blot analysis. Mice were housed in an animal facility with barrier conditions.
Construction of pkntegfpN20.
A full-length murine knt cDNA was isolated from a brain endothelioma cell library by screening with a human knt cDNA and then ligated into an EcoRI/SmaI-cut pEGFP-N1 vector (Clontech) to create pkntegfpN20. The knt cDNA contained the alternatively spliced “inserted sequences” 1 and 4, but not sequences 2, 3, and 5, as confirmed by sequencing (30).
RNA and protein analysis.
Total RNA was extracted as described elsewhere (42). For Northern blotting, 20 μg of RNA was separated on a formaldehyde agarose gel, transferred onto Genescreen Plus nylon membrane (DuPont), and hybridized with the following 32P-labeled murine cDNA probes: knt3′, knt5′ (encompassing the murine cDNA from nt 1250 to 4180 and from nt −43 to 1950, respectively) and glctrp (1.6-kb fragment of the murine facilitated glucose transporter, GenBank accession no. M22998) as a loading control. Reverse transcription-PCR (RT-PCR) analysis was performed with the primers knt#p12 (5′-GAGTCAACTGCTTACAGCGC-3′), knt#p13 (5′-CCCCTTATTCATGCAACTAC-3′), knt#p68 (5′-CCTTGAAGAGCAGGTCATC-3′), knt#p69 (5′-AAGGAGAACTGAGTGGTC-3′), and knt#p70 (5′-GATCTTTCATTCTTGCTA-3′) located in exons 6, 4, 2, 6, and 7, respectively. With the indicated combinations of primers, wild-type and deleted alleles give rise to products of the following sizes: knt#p68 + knt#p12 → 716 and 273 bp, knt#69 + knt#70 → 165 and 165 bp, and knt#p13 + knt#p12 → 400 bp and no product (due to deletion of exon 4).
For protein analysis, organs were homogenized in a lysis buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, and the protease inhibitors (all Roche) bestatin (20 μg/ml), pepstatin (20 μg/ml), aprotinin (30 μg/ml), leupeptin (10 μg/ml), antipain (20 μg/ml), and phenylmethylsulfonyl fluoride (100 μM). The lysates were separated on a sodium dodecyl sulfate–7% polyacrylamide gel and blotted on ProtranBA nitrocellulose membrane (Schleicher & Schuell). Membranes were blocked with 5% skim milk in TBST (50 mM Tris-HCl, pH 7.6; 150 mM NaCl; 0.1% Tween 20), and Western detection was performed with a rabbit anti-human KNT polyclonal serum (RS-TUM; 1:5,000), followed by incubation with goat anti-rabbit horseradish peroxidase (HRP)-conjugated antibody and detection by the ECL Kit (Amersham). RS-TUM was raised against the central part of the human KNT protein (amino acids 373 to 923) (13). It is not directed against parts of the KNT protein that are encoded by exons 3 to 5 (amino acids 177 to 324), which are deleted in the knt (Δ) allele.
Phagocytosis assays.
PEC were obtained by peritoneal lavage from mice challenged with thioglycolate (Sigma) for 4 days. Then, 5 × 105 PEC were incubated with 2.5 × 107 yellow-green fluorescent FluoSpheres (2 μm; Molecular Probes F-8827) for 5 to 240 min at 37°C. As a negative control cells were left on ice for 60 min. PEC were separated from unincorporated beads by density centrifugation through fetal calf serum (FCS) (360 relative centrifugal force [RCF], 10 min), washed, and fixed in 1% paraformaldehyde. Cytospins were assayed by confocal microscopy. For analysis of phagosome function, PEC were challenged with bacteria. Overnight cultures of Escherichia coli DH5α transformed with GFPmut2 (expressing a GFP mutant suitable for fluorescence-activated cell sorting detection [8], The kind gift of H. Häcker) and of untransformed controls were washed three times with phosphate-buffered saline (PBS) and adjusted to an optical density at 600 nm of 1. A volume of 2.5 ml of the bacterial suspensions was added to 106 PEC in 10 ml of Dulbecco modified Eagle medium, supplemented with 10% fetal calf serum, and 0.1% 2-mercaptoethanol (all from Gibco), and 100 U of penicillin per ml, 100 U of streptomycin per ml, and 2 nM l-glutamine. After 1 h of incubation, the PEC were washed and centrifuged at 240 REF three times to remove free bacteria. After a further 1, 4, and 17 h, PEC were washed with PBS, fixed in 2% formalin, and analyzed by flow cytometry.
Lysosome assay.
Lysosome motility was analyzed according to the method of Heuser (20). In brief, embryonic fibroblasts (EF) were removed from the CO2 incubator, washed three times for 15 min with Ringer's solution (R; 155 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 2 mM NaH2PO4, 10 mM HEPES buffer [pH 7.2], 10 mM Glucose, 0.5 mg of bovine serum albumin [BSA] per ml), stained for 5 min with 330 nM Mitotracker in R, washed briefly, and incubated without CO2 at 37°C for 30 min. Cells were then transferred to NH4Cl R (BR; 30 mM NH4Cl in R), acetate R (AR; 80 mM NaCl, 70 mM sodium acetate, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 2 mM NaH2PO4, 10 mM HEPES buffer [pH 6.9], 10 mM glucose), or R alone for 60 min. Subsequently, cells were fixed with 2% paraformaldehyde–0.1% glutaraldehyde in PBS for 10 min at ambient temperature and then processed for immunofluorescence. For the rebound experiments, cells were incubated in BR for 30 min, washed three times in R, and incubated for 30 min in AR before fixation. For quantification, lysosome localization was scored for one of three possibilities: clustered, distributed, or peripheral. One hundred cells per sample were analyzed microscopically without knowing the treatment and genotype of the cell samples.
Immunocytochemistry.
EF were obtained from embryos of the different genotypes as described previously (42). EF were grown overnight on glass slides, fixed with 2% paraformaldehyde–0.1% glutaraldehyde in PBS for 10 min at 37°C, permeabilized with 1% saponin in PBS for 10 min at 37°C, and blocked with 5% skim milk powder in PBS for 30 min at room temperature, followed by 1-h incubations at room temperature with primary and secondary antibodies diluted with 1% skim milk in PBS. Each incubation step was followed by brief washing steps. Mouse anti-α tubulin (1:200, clone DM 1A; Sigma), mouse anti-gm130 (1:400, clone 35: Transduction Laboratories), rabbit anti-KNT (1:100; RS-TUM), or rat anti-LAMP-1 fluorescein isothiocyanate (FITC; 1:200, clone 1D4B; PharMingen) were used as primary antibodies and detected with Alexa488- or Alexa546-labeled goat anti-mouse IgG; immunoglobulin G (1:200; Molecular Probes) or HRP-conjugated goat anti-rabbit IgG (1:1,000; Bio-Rad) secondary antibodies. HRP development was performed as described earlier (42). Cells were observed through a Leica DMRBE microscope or a Zeiss 410 confocal microscope. For labeling of the ER, cells were fixed with ice-cold ethanol (95%)–acetic acid (5%) for 1.5 min, blocked for 30 min with 8% BSA in PBS, and incubated with rabbit anticalreticulin (1:100; Upstate) and Alexa546-conjugated goat anti-rabbit IgG (1:200; Molecular Probes) antibody in 1% BSA-PBS.
RESULTS
Generation of kntΔ/Δ and kntF/F mice.
The targeted disruption of various KIFs resulted in embryonic or perinatal lethality (38, 51, 52, 60). KNT was postulated to be a major kinesin-binding protein contained in motile vesicles (53). Thus, a targeted knt-null mutation could be assumed to be fatal. Therefore, a conditional targeting strategy (17) based on the Cre/loxP recombination system was employed to inactivate the knt gene. The targeting vector was constructed in a way that a loxP flanked neomycin resistance gene and a third, distal, loxP site were inserted into intron 2 and intron 5 of a genomic knt fragment, respectively (Fig. 1A). The herpes simplex virus thymidine kinase (tk) gene was included to enable negative selection against random integrants. Linearization of the targeting construct close to the neomycin resistance cassette used for positive selection may result in a high proportion of clones with correct recombination of the floxed neomycin resistance marker but without cointegration of the distal loxP motif (1, 29). (T. Plitz and K. Pfeffer, unpublished data). Therefore, the targeting vector was linearized at the 3′ end near the distal loxP motif and transfected into E14.1 ES cells (41). Transfectants were selected with G418 and ganciclovir, and double-resistant clones were screened for correct integration of the distal loxP motif by PCR (see Materials and Methods). Correct homologous recombination encompassing all three loxP motifs could be confirmed by genomic Southern blot analysis in 10 of 11 ES cell clones tested (Fig. 1B. data not shown). To obtain ES cell clones carrying a floxed (F) knt allele, the floxed neomycin resistance gene was removed from the targeted (T) locus by transient Cre transfection. Chimeric mice were generated from ES cell clones harboring the targeted or the floxed knt allele. Chimeras harboring the targeted knt allele were bred with transgenic “deleter” mice expressing Cre recombinase in early development (47). This cross gave rise to mice heterozygous for the deleted knt (Δ) allele. Germ line transmission of the targeted (T), the floxed (F), and the deleted (Δ) alleles was proven by Southern blot analysis (Fig. 1B).
The genetic inactivation of murine cDHC and all KIFS analyzed to date (see Introduction) shows the crucial involvement of these motor proteins during ontogeny. To study the possible role of KNT in murine development, mice heterozygous for the deleted allele (knt+/Δ), as well as knt+/F control mice, were intercrossed to produce homozygous offspring. Both crosses gave rise to litters of normal size, with wild-type, heterozygous (knt+/Δ or knt+/F), and homozygous (kntΔ/Δ or kntF/F) offspring present at a frequency consistent with Mendelian inheritance (data not given). kntΔ/Δ and kntF/F animals appeared healthy and reproduced normally. Gross pathological examination of mice up to 1 year of age did not reveal abnormalities.
The deleted knt allele is a null allele.
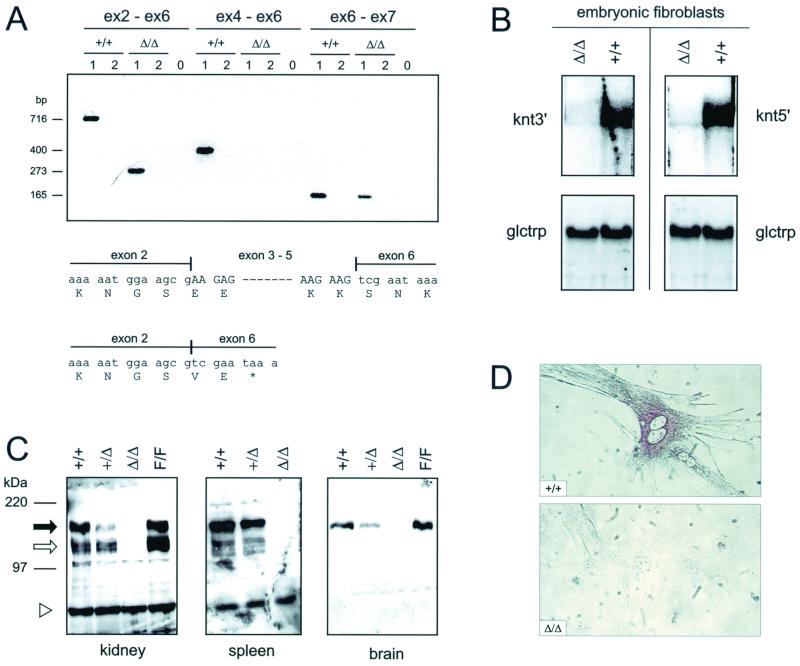
To verify that the targeting resulted in a knt null mutation, KNT expression was analyzed in kntΔ/Δ mice. Cre-mediated deletion between the outer loxP sites results in excision of a genomic region encompassing exons 3 to 5 of the knt gene. The deletion leads to direct splicing from exon 2 to exon 6, thus generating a frameshift mutation that results in a premature termination codon (PTC). Therefore, putatively, a truncated protein consisting of the N-terminal 178 amino acids of KNT might be translated. However, such a putative truncated KNT protein would lack the interaction domain with kinesin (39). Using RT-PCR, the engineered splicing of exon 2 directly to exon 6 in the knt transcripts could be identified in kntΔ/Δ EF, but not in wild-type EF controls (Fig. 2A). Sequencing of these RT-PCR products was employed to verify the expected exon 2 to exon 6 join in the kntΔ/Δ transcripts (Fig. 2A). In general, a mutation generating a PTC in an upstream exon leads to rapid degradation of the mRNA by a mechanism known as nonsense-mediated decay (reviewed by Hentze and Kulozik [19]). Consistently, by using knt cDNA probes that cover the entire coding sequence, knt mRNA could not be detected in kntΔ/Δ EF by Northern blot analysis. Both cDNA probes, however, hybridized to the knt transcripts contained in wild-type RNA (Fig. 2B). As predicted from these results, no KNT protein was detected in kntΔ/Δ mice, as verified by Western blot analysis of tissue homogenates from different organs (Fig. 2C) or immunolabeling of KNT in EF (Fig. 2D). However, KNT was readily detectable in protein extracts from kntF/F mice, indicating that the insertion of loxP sites into introns of the knt locus did not affect KNT biosynthesis. These data prove that KNT was successfully inactivated in kntΔ/Δ mice.
FIG. 2.
The deleted knt allele is a null allele. (A) RT-PCR analysis of knt RNA obtained from kntΔ/Δ and knt+/+ EF. PCR with primers located in exon 2 and exon 6 (ex2-ex6, left) yields amplification products of 273 bp (kntΔ/Δ) and 716 bp (knt+/+), indicative of splicing of exon 2 directly to exon 6 at the kntΔ/Δ mRNA. This results in a frameshift mutation and generates a PTC. That exon 2 to exon 6 join in the kntΔ/Δ transcripts was verified by DNA sequencing. PCR encompassing exons 4 to 6 (ex4-ex6, middle) yields a 400-bp product in the wild type. No amplification is obtained on the kntΔ/Δ template due to the engineered deletion of exons 3 to 5. Splicing of exon 6 to exon 7 occurs in transcripts from the deleted and the wild-type knt alleles (ex6-ex7, right), yielding products of 165 bp. For primer sequences, see Material and Methods. Lanes 1, with RT; lanes 2, without RT; lanes 0, negative controls (no template). (B) Northern blot analysis, verifying the absence of stable knt transcripts in kntΔ/Δ mice. The same RNA preparations as used in panel A were hybridized to knt5′ and knt3′, two cDNA probes encompassing the entire coding sequence of the knt transcript. The blots were stripped and rehybridized to glctrp (mouse facilitated glucose transporter) as a loading control. kntΔ/Δ transcripts are rapidly degraded, most probably due to nonsense mediated decay. (C) Western blot analysis showing the absence of KNT protein form kntΔ/Δ mice. Organ homogenates of kidney, spleen, and brain tissues obtained from mice of the indicated genotypes were probed with a polyclonal anti-KNT serum (RS-TUM) directed against the central part of the KNT protein. Black arrow, 160-kDa full-length KNT; open arrow, 120-kDa form of KNT (27, 32); open triangle, irrelevant protein. (D) Immunolabeling of EF with RS-TUM shows the absence of KNT from kntΔ/Δ EF. Magnification, ×400.
Subcellular localization of KNT.
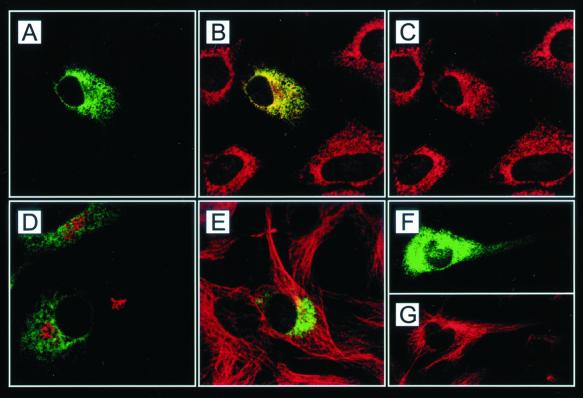
To establish the subcellular compartmentalization of KNT, fibroblasts were transiently transfected with pkntegfpN20, encoding a murine N-terminal KNT-eGFP fusion protein. The resulting expression of green fluorescent protein (GFP)-tagged KNT enables the detection of cellular compartments associated with KNT. Confocal microscopy of pkntegfpN20-expressing cells revealed a reticular, perinuclear staining pattern (Fig. 3A), corresponding to previous reports using anti-KNT antibodies (13, 53). This result establishes a correct localization of the exogenously expressed eGFP fusion protein. Calreticulin and gm130 can be used as ER and cis-Golgi markers, respectively (40) (35). In the transfected cells, KNT-eGFP almost exclusively colocalized with the ER and not with the Golgi compartment (Fig. 3B and D). Virtually no colocalization of KNT-eGFP was detectable with microtubules (Fig. 3E) identified by labeling with anti-α-tubulin antibody (4). KNT-eGFP was restricted to the perinuclear region in almost all inspected cells. Pronounced KNT-eGFP staining in the cell periphery, implying the association of KNT with anteriogradely transported organelles, could not be detected. Only in very few heavily overexpressing cells, KNT-eGFP was found as a punctate staining pattern in the periphery (Fig. 3F and G). This label may also colocalize with ER structures, since the ER is known to extend to the cell periphery (31). These data confirm that KNT is almost exclusively targeted to the ER.
FIG. 3.
Subcellular localization of KNT analyzed by confocal microscopy. Double labeling of KNT-eGFP (A, D, E, and F; green)-transfected 3T3 cells with anticalreticulin (B and C; red), anti-gm130 (D; red) and anti-α-tubulin (E and G; red) antibody. The merged image (B) shows the colocalization of KNT-eGFP (A) with the ER (C). KNT-eGFP does not colocalize with cis-Golgi cisternae (D) and microtubules (E). (F and G) In heavily overexpressing cells, KNT-eGFP can be detected in the periphery. Magnification, ×1,000.
KNT deficiency does not alter the stationary localization of membrane organelles.
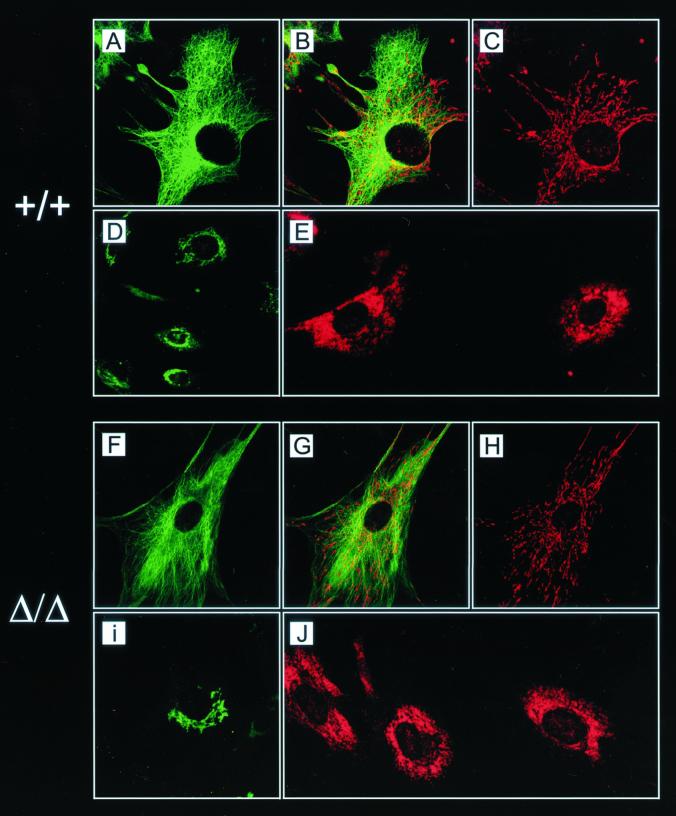
Since KNT was postulated to be a kinesin-binding protein that is required for kinesin-based vesicle motility (28), kntΔ/Δ EF were analyzed for a possible transport defect of membrane organelles. Labeling of the ER with an anti-calreticulin antibody resulted in a perinuclear reticular staining pattern in kntΔ/Δ and knt+/+ cells (Fig. 4J and E). The ER structures seemed equally large and spread out in cells of both genotypes, indicating that the assembly of the ER is independent from KNT. This is intriguing since KNT predominantly localizes to the ER (Fig. 3B). In the null mutant EF the perinuclear, tubular staining pattern of the cis-Golgi apparatus with an anti-gm130 antibody was indistinguishable from the gm130 staining pattern in wild-type cells (Fig. 4I and D).
FIG. 4.
Unaltered stationary localization of membrane organelles in EF lacking KNT as shown by immunolabeling of untreated control EF (A to E) and kntΔ/Δ EF (F to J). (A and F) Anti-α-tubulin indirect immunofluorescence staining. (C and H) Mitochondrial labeling with Mitotracker. (B and G) Merged image of mitochondria and microtubules showing the correct colocalization of mitochondria with the microtubules in wild-type and null mutant cells. Anti-gm130 (D and I) and anticalreticulin (E and J) indirect immunofluorescence labeling indicates the unimpaired assembly of cis-Golgi and ER in the absence of KNT. Magnification, ×1,000.
Mitochondria localize to and move along microtubules (2). Anterograde transport of mitochondria is dependent on kinesin since the genetic inactivation of kif5B results in a perinuclear clustering of microtubules (52). To examine the possible involvement of KNT in mitochondrion transport, these organelles were stained with Mitotracker (58). Comparable to knt+/+ control cells, mitochondria in the kntΔ/Δ cells were radially arranged and stretched out along microtubules (Fig. 4A to C and F to H). These data indicate that KNT is not required for the transport of mitochondria along and attachment of these organelles to the cytoskeleton.
KNT is dispensable for anterograde lysosome transport in EF.
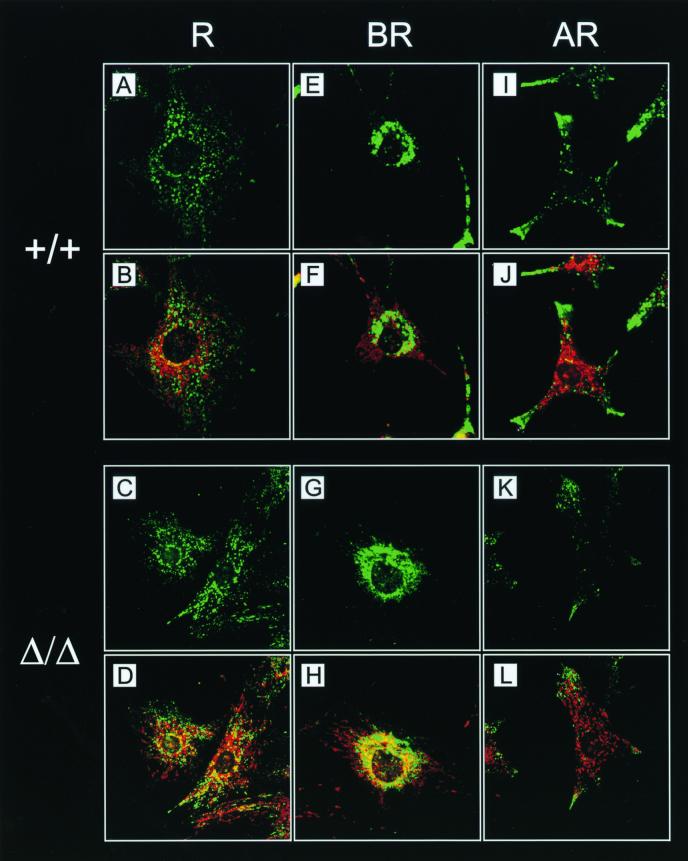
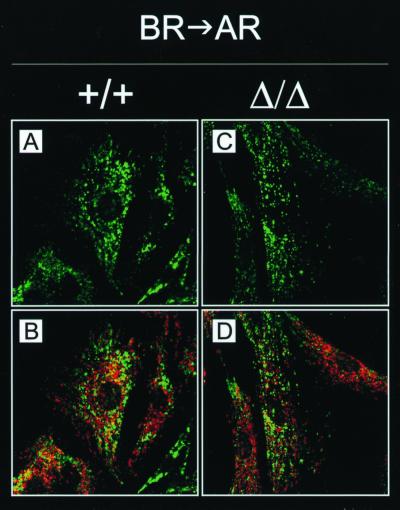
Like mitochondria, lysosomes are attached to and move along microtubules (20). Lysosome localization can be synchronized by pharmacological treatments, thus facilitating the analysis of lysosome transport in vivo (20). Cytoplasmic alkalinization induced by NH4Cl treatment of the cells causes lysosomes to cluster around the nucleus, while cytoplasmic acidification achieved by acetate treatment leads to dispersion of the lysosomes throughout the cytoplasm, eventually resulting in their accumulation at the very edge of the cell. The dependence of anterograde lysosome transport on kinesin has been shown in cells expressing a rigor neuronal kinesin mutant or in cells lacking KIF5B (36, 52). Furthermore, a recent report suggested an involvement of KNT in lysosome trafficking (39). To test lysosome motility in the absence of KNT, lysosomes were localized by labeling with anti-LAMP-1 antibody (7) in kntΔ/Δ, kntF/F, and knt+/+ EF after incubation in ringer (R) solution or in basic R (BR) or acidic R (AR). Staining of mitochondria, which do not redistribute during pH changes (20), was included to monitor the integrity of cells during treatment. In 95.5% of the control EF incubated in R, lysosomes were dispersed throughout the cytoplasm (Fig. 5A and B and Table 1). This stationary distribution of lysosomes was nearly identical in kntΔ/Δ EF. Here, a dispersed lysosome localization was observed in 92% of the cells (Fig. 5C and D and Table 1). Perinuclear clustering of lysosomes upon alkalinization in BR occurred similarly in control and null mutant cells, resulting in clustered lysosomes in more than 85% of both cell types (Fig. 5E to H and Table 1). Acidification in AR induced centrifugal motility of lysosomes in kntΔ/Δ and knt+/+ EF, causing peripheral accumulation of lysosomes in 77 and 88.5% of the cells, respectively (Fig. 5lF to L and Table 1). During these experiments, kntF/F EF were undistinguishable from wild-type control or kntΔ/Δ cells (Table 1). To further analyze anterograde lysosome transport, cells were treated with BR and subsequently transferred to AR. Redistribution of initially clustered lysosomes to the cell periphery could be observed in kntΔ/Δ EF, as well as wild-type control EF (Fig. 6). The fraction of cells with redistributed lysosomes was identical with 97.7% for the null mutant and 97% for the wild-type cells (Table 2). These data prove that kinesin-driven anterograde transport of lysosomes in EF does not depend on KNT.
FIG. 5.
Intact anterograde and retrograde lysosome transport in kntΔ/Δ and knt+/+ EF. (A to L) Lysosomes were directly labeled with FITC-conjugated anti-LAMP-1 antibody in wild-type (A, B, E, F, I, and J) and kntΔ/Δ (C, D, G, H, K, and L) EF. Labeling of mitochondria with Mitotracker (B, D, F, H, J, and L) in red was used to control cellular integrity. (A to D) Stationary localization of lysosomes in EF incubated in R. (E to H) Perinuclear clustering of lysosomes was caused by alkaline treatment (BR) in wild-type and kntΔ/Δ cells. (I to L) In cells of both genotypes, acidification of the culture medium (AR) resulted in transport of lysosomes to the cell periphery. Magnification, ×900.
TABLE 1.
Synchronization of lysosome localization by pharmacological treatment
| Treatment medium and genotype | Subcellular localization of lysosomes (avg deviation)a
|
||
|---|---|---|---|
| % Clustered | % Dispersed | % Peripheral | |
| R | |||
| +/+ | 4.5 (3.5) | 95.5 (3.5) | 0 (0) |
| Δ/Δ | 5.8 (3.3) | 92.0 (3.0) | 2.2 (1.8) |
| F/F | 2.5 (2.5) | 88.5 (6.5) | 9.0 (9.0) |
| BR | |||
| +/+ | 85.5 (0.5) | 14.5 (0.5) | 0 (0) |
| Δ/Δ | 85.8 (5.4) | 14.0 (5.0) | 0.2 (0.4) |
| F/F | 82.5 (4.5) | 17.5 (4.5) | 0 (0) |
| AR | |||
| +/+ | 0 (0) | 11.5 (3.5) | 88.5 (3.5) |
| Δ/Δ | 0 (0) | 23.0 (1.0) | 77.0 (1.0) |
| F/F | 0 (0) | 13.5 (9.5) | 86.5 (9.5) |
One hundred cells per sample were scored for the subcellular localization of their lysosomes blindly with respect to the prior treatment and genotype of the cell samples. Percentages were calculated from two independent experiments, and the average deviations are given in parentheses. The Δ/Δ values were calculated from two independent Δ/Δ EF lines; a third Δ/Δ EF line that was included in only one of the experiments showed similar results. For experimental procedures, see Fig. 5 and Materials and Methods.
FIG. 6.
Lysosome redistribution in a pH rebound condition. Double labeling of wild-type (A and B) and null mutant (C and D) EF with FITC-conjugated anti-LAMP-1 antibody (A to D; green) and Mitotracker (B and D) in red. After alkalization in BR for 30 min, cells were transferred to AR. Centrifugal motility was observable in cells of both genotypes. Magnification, ×1,000.
TABLE 2.
Redistribution of lysosomes by AR after BR treatment
| Genotype | Subcellular localization of lysosomes (avg deviation)a
|
|
|---|---|---|
| % Clustered | Redistributed | |
| +/+ | 3.0 (3.0) | 97.0 (3.0) |
| Δ/Δ | 2.3 (1.1) | 97.7 (1.1) |
Cells were treated with BR for 30 min to induce perinuclear clustering of lysosomes. At 30 min after transfer to AR, the percentages of the cells with lysosomes redistributed to the cell periphery or remaining clustered around the nucleus were determined as described in Table 1.
PEC show full phagocytic capacity despite the absence of KNT.
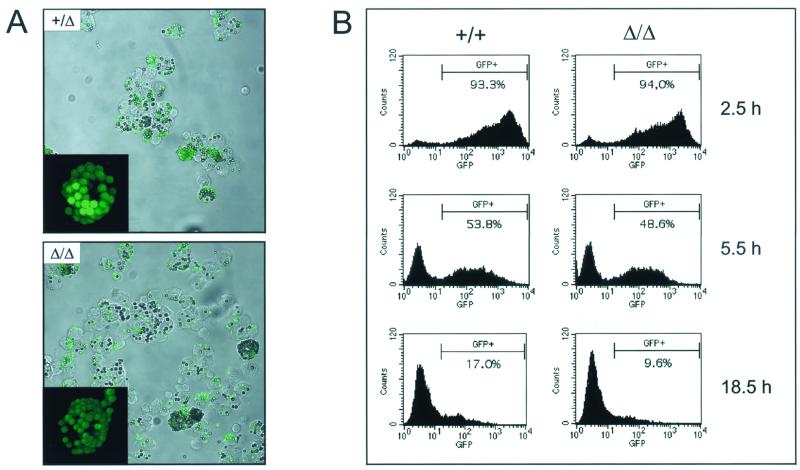
Cells can internalize large particulate matter by phagocytosis. After their formation at the plasma membrane, phagosomes are delivered to the prelysosomal compartment, where they mature to phagolysosomes (43). The extensive trafficking necessary to enable the interaction of phagosomes with lysosomes in the perinuclear area to form the phagolysosome is microtubule dependent (11, 25). The bidirectional movement of phagosomes along microtubules was shown to be mediated by cytoplasmic dynein and kinesin and could be inhibited by KNT peptides (3). Furthermore, KNT was reported localizing on phagosomes and, compared to early phagosomes, elevated KNT protein levels were described on late phagosomes of higher motility (3). To test the contribution of KNT to the maturation and function of phagosomes, PEC obtained from kntΔ/Δ and control mice were analyzed for their phagocytic capacity. In a first set of experiments, PEC were incubated with fluorescent latex microspheres. Both, kntΔ/Δ and knt+/Δ PEC phagocytosed the beads without differrences in kinetics or efficiency (Fig. 7A and data not shown). Especially after longer incubation periods, PEC of both genotypes were loaded with many beads, accumulating inside the cell (Fig. 7A, insets). To provide a more physiological substrate for phagocytosis and to enable the examination of phagosome maturation and function, PEC were challenged with E. coli expressing GFP. The uptake and the elimination of the GFP-labeled bacteria by PEC were monitored by flow cytometry. It could be detected that the bacteria were rapidly taken up, and more than 80% of the PEC were GFP+ after 10 min. After 60 min, more than 98% of the PEC of both genotypes had incorporated bacteria (data not shown). During a following 24-h chase period, the fluorescence of the PEC decreased as they degraded the GFP+ bacteria (Fig. 7B). In this assay system, kntΔ/Δ PEC were undistinguishable from knt+/+ controls with respect to the kinetics and efficiency of phagocytosis and the clearance of E. coli bacteria. These data imply that cellular trafficking enabling the formation of fully functional phagolysosomes is not impaired in the absence of KNT.
FIG. 7.
PEC can phagocytose and degrade bacteria. (A) Cytospins of kntΔ/Δ and knt+/Δ PEC after 1 h of incubation with fluorescent latex beads. Insets show a three-dimensional reassembly of a stack of confocal images of a single PEC after 240 min of incubation. (B) kntΔ/Δ and knt+/+ PEC were challenged with GFP-expressing E. coli. Unincorporated bacteria were removed by centrifugation, and PEC were fixed after the indicated chase periods. For analysis by flow cytometry, PEC were gated by forward and side scatter and examined for green fluorescence. Unlabeled E. coli cells were used as controls (not shown).
DISCUSSION
Kinesin-driven intracellular membrane transport is of vital importance for a large variety of cellular processes, yet how cargoes are coupled to the kinesin motors is still ill defined. KNT has been proposed to attach cargoes to the prototypic motor protein conventional kinesin (16). The present study describes a novel mouse mutant deficient in KNT. Unexpectedly, KNT-deficient mice appear healthy and fertile. Detailed analysis of kntΔ/Δ mice reveals that KNT cannot be considered to function as a general anchor for the kinesin-driven transport of mitochondria, lysosomes, or phagosomes.
At present, there are conflicting reports about the involvement of kinesin in Golgi-to-ER transport. Redistribution of Golgi markers to the ER could be induced by brefeldin A treatment in conventional KHC (kif5B)-null mutant cells (52). In contrast, injection of anti-kinesin antibody into cells or inhibition of KHC expression by antisense oligonucleotide suppression blocked this Golgi-ER redistribution (31, 52). Additionally, there is evidence that establishment and placement of the ER inside the cell are driven by kinesins (10, 55), although these processes might not be dependent on KIF5B (52). Disruption of the knt gene (this study) apparently does not affect ER and cis-Golgi morphology. These data provide evidence that ER assembly does not depend on KNT or, alternatively, that the kinesin motors needed for ER integrity do not bind to KNT.
Two murine KIFs, KIF1B and KIF5B, have been implicated in the movement of mitochondria (37) (52). Both KIFs associate with mitochondria. While evidence for an involvement of KIF1B mainly came from in vitro trafficking assays, genetic inactivation experiments clearly demonstrated that transport of mitochondria in intact cells requires KIF5B. In KIF5B-null mutant cells obtained from extraembryonic membranes, mitochondria clustered perinuclearly. Ectopic reconstitution of KIF5B expression in kif5B−/− cells resulted in a dispersion of mitochondria. In contrast, in kntΔ/Δ EF cells, mitochondria were not found around the nucleus but were distributed throughout the cytoplasm and aligned along MT, thus indicating that KNT does not contribute to mitochondrion motility. Provided that there is no synergistic use of motors other than KIF5B to drive anterograde mitochondrion transport in EF, it could be suggested that KNT does not anchor conventional KHC., i.e., KIF5B, to mitochondria.
The bidirectional transport of phagosomes has been reported to depend on KNT (3). Additionally, the increased motility of late phagosomes has been attributed to the accumulation of KNT on phagosomes over time, suggesting a contribution of KNT to phagosome maturation (3). However, kntΔ/Δ PEC are fully able to phagocytose and clear E. coli bacteria, providing evidence for the conclusion that the trafficking and maturation of phagosomes can occur in the absence of KNT. In the study by Blocker et al. (3), peptide inhibition was chosen to suppress KNT function. Fragments of KNT were produced in bacteria, purified, and added to an in vitro system reconstituting phagosome transport. In this assay, two fragments derived from the central part of the KNT protein (encompassing residues 295 to 612 and 568 to 943) blocked the bidirectional movement of phagosomes, whereas a third, C-terminal KNT fragment (residues 924 to 1321) had no effect. One of the central KNT fragments (residues 568 to 943) is recognized by a monoclonal anti-KNT antibody that interferes with the motility of microsome preparations in vitro (3, 28). The conclusion that the central KNT coiled-coil region is indispensable for phagosome transport is contradictory in the light of a recent study characterizing the interacting domains of KNT and kinesin (39). Here, the kinesin-binding domain was mapped to a short region (residues 1188 to 1288) located at the KNT C terminus. The region of the KNT protein that blocked phagosome transport (residues 295 to 612 and 568 to 943) and interacted with the inhibitory anti-KNT antibody (residues 568 to 943) appears not to be involved in kinesin binding. These conflicting results might be due to unspecific peptide inhibition. Since the inhibitory KNT fragments form α-helical coiled coils, a common motif found in many proteins, these peptides may well interfere with cellular processes other than KNT function. The anti-KNT antibody does not necessarily need to bind directly to the KNT-kinesin interaction domain to disrupt vesicle motility. The antibody could interfere sterically with motor binding, provided that KNT is part of or situated near a kinesin receptor complex on the vesicle membrane. The removal of KNT from such a complex might still allow kinesin binding to the organelle, whereas the addition of the antibody to the complex does not.
The anterograde transport of lysosomes depends on kinesins (52), whereas the retrograde movement is mediated by cytoplasmic dynein (18). The overexpression of KNT-kinesin binding domains (KNT residues 1188 to 1288) suggested a function of KNT in anterograde lysosome trafficking (39). However, the requirement of KNT for lysosome transport could not be verified after the genetic inactivation of the knt gene (this study). Comparable to wild-type controls, anterograde lysosome transport does occur in the KNT-null mutant EF. Thus, it might be proposed that KNT does not play a role in lysosome motility or, alternatively, that KNT function is redundant and compensated for by hitherto-unidentified anchor proteins.
In summary, the role of KNT in kinesin-driven motility remains obscure. The functions of KNT, as suggested by various inhibition studies, could not be confirmed in a genetic approach by establishment of a KNT-defective mouse strain. Therefore, it must be determined whether KNT is merely part of a large multiprotein complex anchoring kinesin to the vesicle membrane but not essential for the interaction of the motor with the cargo. The analogy to the dynactin complex regulating cargo binding to the retrograde cytoplasmic dynein motors, as well as the recent reports of two distinct multiprotein anchors on vesicles for KIF13 and KIF17 may argue for this speculation (15, 24, 33, 48). However, the coupling of kinesin motors to vesicles need not be mediated by large multiprotein complexes. Very recently, APP and SYD (also known as JIP) have been described to bind directly to the TPR domain of the KLC (5, 26, 57). These transmembrane proteins could serve as cargo receptors for kinesin in fast axonal transport and post-Golgi transport, respectively. Whether KNT directly interacts with kinesin in vivo but is involved in the transport of a distinct class of vesicles rather than being an universal kinesin anchor remains to be determined. Alternatively, redundant family members might compensate for the KNT deficiency. Thus far, only one murine knt gene is known, and the search for homologues merely yielded one kinectin pseudogene (Plitz and Pfeffer, unpublished). However, the existence of a knt gene family cannot be ruled out. The KNT knockout mouse will provide a valuable tool to further analyze the biological significance of KNT.
ACKNOWLEDGMENTS
The continuous and generous support of H. Wagner is greatly appreciated. We thank M. Krönke for RS-TUM and the human knt cDNA, H. Häcker for GFPmut2, and B. Clausen for pBSlox-PneoloxP. We thank U. Huffstadt and E. Schaller for technical assistance and A. Fütterer, H. Flaswinkel, R. Endres, H. Häcker, and P. Ahmad-Nejad for scientific advice.
This work was supported by the Sonderforschungsbereich 391.
REFERENCES
- 1.Alimzhanov M B, Kuprash D V, Kosco-Vilbois M H, Luz A, Turetskaya R L, Tarakhovsky A, Rajewsky K, Nedospasov S A, Pfeffer K. Abnormal development of secondary lymphoid tissues in lymphotoxin beta-deficient mice. Proc Natl Acad Sci USA. 1997;94:9302–9307. doi: 10.1073/pnas.94.17.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball E H, Singer S J. Mitochondria are associated with microtubules and not with intermediate filaments in cultured fibroblasts. Proc Natl Acad Sci USA. 1982;79:123–126. doi: 10.1073/pnas.79.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blocker A, Severin F F, Burkhardt J K, Bingham J B, Yu H, Olivo J C, Schroer T A, Hyman A A, Griffiths G. Molecular requirements for bi-directional movement of phagosomes along microtubules. J Cell Biol. 1997;137:113–129. doi: 10.1083/jcb.137.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blose S H, Meltzer D I, Feramisco J R. 10-nm filaments are induced to collapse in living cells microinjected with monoclonal and polyclonal antibodies against tubulin. J Cell Biol. 1984;98:847–858. doi: 10.1083/jcb.98.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman A B, Kamal A, Ritchings B W, Philp A V, McGrail M, Gindhart J G, Goldstein L S. Kinesin-dependent axonal transport is mediated by the Sunday driver (SYD) protein. Cell. 2000;103:583–594. doi: 10.1016/s0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- 6.Brady S T. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen J W, Murphy T L, Willingham M C, Pastan I, August J T. Identification of two lysosomal membrane glycoproteins. J Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 9.Coy D L, Hancock W O, Wagenbach M, Howard J. Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 10.Dabora S L, Sheetz M P. The microtubule-dependent formation of a tubulovesicular network with characteristics of the ER from cultured cell extracts. Cell. 1988;54:27–35. doi: 10.1016/0092-8674(88)90176-6. [DOI] [PubMed] [Google Scholar]
- 11.Desjardins M, Huber L A, Parton R G, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman D S, Vale R D. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 13.Futterer A, Kruppa G, Kramer B, Lemke H, Kronke M. Molecular cloning and characterization of human kinectin. Mol Biol Cell. 1995;6:161–170. doi: 10.1091/mbc.6.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gepner J, Li M, Ludmann S, Kortas C, Boylan K, Iyadurai S J, McGrail M, Hays T S. Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics. 1996;142:865–878. doi: 10.1093/genetics/142.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill S R, Schroer T A, Szilak I, Stener E R, Sheetz M P, Cleveland D W. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein L S, Philp A V. The road less traveled: emerging principles of kinesin motor utilization. Annu Rev Cell Dev Biol. 1999;15:141–183. doi: 10.1146/annurev.cellbio.15.1.141. [DOI] [PubMed] [Google Scholar]
- 17.Gu H, Zou Y R, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 18.Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol. 1998;141:51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentze M W, Kulozik A E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 20.Heuser J. Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. J Cell Biol. 1989;108:855–864. doi: 10.1083/jcb.108.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 22.Hirokawa N, Noda Y, Okada Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr Opin Cell Biol. 1998;10:60–73. doi: 10.1016/s0955-0674(98)80087-2. [DOI] [PubMed] [Google Scholar]
- 23.Hirokawa N, Pfister K K, Yorifuji H, Wagner M C, Brady S T, Bloom G S. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- 24.Holleran E A, Karki S, Holzbaur E L. The role of the dynactin complex in intracellular motility. Int Rev Cytol. 1998;182:69–109. doi: 10.1016/s0074-7696(08)62168-3. [DOI] [PubMed] [Google Scholar]
- 25.Jahraus A, Storrie B, Griffiths G, Desjardins M. Evidence for retrograde traffic between terminal lysosomes and the prelysosomal/late endosome compartment. J Cell Sci. 1994;107Pt 1:145–157. doi: 10.1242/jcs.107.1.145. [DOI] [PubMed] [Google Scholar]
- 26.Kamal A, Stokin G B, Yang Z, Xia C, Goldstein L S. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron. 2000;28:449–459. doi: 10.1016/s0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 27.Kumar J, Erickson H P, Sheetz M P. Ultrastructural and biochemical properties of the 120-kDa form of chick kinectin. J Biol Chem. 1998;273:31738–31743. doi: 10.1074/jbc.273.48.31738. [DOI] [PubMed] [Google Scholar]
- 28.Kumar J, Yu H, Sheetz M P. Kinectin, an essential anchor for kinesin-driven vesicle motility. Science. 1995;267:1834–1837. doi: 10.1126/science.7892610. [DOI] [PubMed] [Google Scholar]
- 29.Kuprash D V, Alimzhanov M B, Tumanov A V, Anderson A O, Pfeffer K, Nedospasov S A. TNF and lymphotoxin beta cooperate in the maintenance of secondary lymphoid tissue microarchitecture but not in the development of lymph nodes. J Immunol. 1999;163:6575–6580. [PubMed] [Google Scholar]
- 30.Leung E, Print C G, Parry D A, Closey D N, Lockhart P J, Skinner S J, Batchelor D C, Krissansen G W. Cloning of novel kinectin splice variants with alternative C-termini: structure, distribution and evolution of mouse kinectin. Immunol Cell Biol. 1996;74:421–433. doi: 10.1038/icb.1996.72. [DOI] [PubMed] [Google Scholar]
- 31.Lippincott-Schwartz J, Cole N B, Marotta A, Conrad P A, Bloom G S. Kinesin is the motor for microtubule-mediated Golgi-to-ER membrane traffic. J Cell Biol. 1995;128:293–306. doi: 10.1083/jcb.128.3.293. . (Ellatum, 129: 893). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machleidt T, Geller P, Schwandner R, Scherer G, Kronke M. Caspase 7-induced cleavage of kinectin in apoptotic cells. FEBS Lett. 1998;436:51–54. doi: 10.1016/s0014-5793(98)01095-3. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa T, Seton M, Seog D, Ogasawara K, Dohmae N, Takio K, Hirokawa N. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell. 2000;103:569–581. doi: 10.1016/s0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa T, Tanaka Y, Matsuoka E, Kondo S, Okada Y, Noda Y, Kanai Y, Hirokawa N. Identification and classification of 16 new kinesin superfamily (KIF) proteins in mouse genome. Proc Natl Acad Sci USA. 1997;94:9654–9659. doi: 10.1073/pnas.94.18.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis T E, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 38.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. . (Erratum, 99:117, 1999.) [DOI] [PubMed] [Google Scholar]
- 39.Ong L L, Lim A P, Er C P, Kuznetsov S, Yu H. Kinectin-kinesin binding domains and their effects on organelle motility. J Biol Chem. 2000;275:32854–32860. doi: 10.1074/jbc.M005650200. [DOI] [PubMed] [Google Scholar]
- 40.Ostwald T J, MacLennan D H. Isolation of a high affinity calcium-binding protein from sarcoplasmic reticulum. J Biol Chem. 1974;249:974–979. [PubMed] [Google Scholar]
- 41.Pfeffer K, Matsuyama T, Kundig T M, Wakeham A, Kishihara K, Shahinian A, Wiegmann K, Ohashi P S, Kronke M, Mak T W. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 1993;73:457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 42.Plitz T, Huffstadt U, Endres R, Schaller E, Mak T W, Wagner H, Pfeffer K. The resistance against Listeria monocytogenes and the formation of germinal centers depend on a functional death domain of the 55 kDa tumor necrosis factor receptor. Eur J Immunol. 1999;29:581–591. doi: 10.1002/(SICI)1521-4141(199902)29:02<581::AID-IMMU581>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 43.Rabinowitz S, Horstmann H, Gordon S, Griffiths G. Immunocytochemical characterization of the endocytic and phagolysosomal compartments in peritoneal macrophages. J Cell Biol. 1992;116:95–112. doi: 10.1083/jcb.116.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saxton W M, Hicks J, Goldstein L S, Raff E C. Kinesin heavy chain is essen-for viability and neuromuscular functions in Drosophila, but mutants show no defects in mitosis. Cell. 1991;64:1093–1102. doi: 10.1016/0092-8674(91)90264-y. [DOI] [PubMed] [Google Scholar]
- 45.Scholey J M, Heuser J, Yang J T, Goldstein L S. Identification of globular mechanochemical heads of kinesin. Nature. 1989;338:355–357. doi: 10.1038/338355a0. [DOI] [PubMed] [Google Scholar]
- 46.Schroer T A, Sheetz M P. Functions of microtubule-based motors. Annu Rev Physiol. 1991;53:629–652. doi: 10.1146/annurev.ph.53.030191.003213. [DOI] [PubMed] [Google Scholar]
- 47.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Setou M, Nakagawa T, Seog D H, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 49.Skouflas D A, Cole D G, Wedaman K P, Scholey J M. The carboxyl-terminal domain of kinesin heavy chain is important for membrane binding. J Biol Chem. 1994;269:1477–1485. [PubMed] [Google Scholar]
- 50.Stenoien D L, Brady S T. Immunochemical analysis of kinesin light chain function. Mol Biol Cell. 1997;8:675–689. doi: 10.1091/mbc.8.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda S, Yonekawa Y, Tanaka Y, Okada Y, Nonaka S, Hirokawa N. Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J Cell Biol. 1999;145:825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 53.Toyoshima I, Yu H, Steuer E R, Sheetz M P. Kinectin, a major kinesin-binding protein on ER. J Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vale R D, Fletterick R J. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 55.Vale R D, Hotani H. Formation of membrane networks in vitro by kinesin-driven microtubule movement. J Cell Biol. 1988;107:2233–2241. doi: 10.1083/jcb.107.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vale R D, Reese T S, Sheetz M P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verhey K J, Meyer D, Deehan R, Blenis J, Schnapp B J, Rapoport T A, Margolis B. Cargo of kinesin identified as jip scaffolding proteins and associated signaling molecules. J Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitaker J E, Moore P L, Haugland R P, Haugland R P. Dihydrotetramethylrosamine: a long-wavelength, fluorogenic peroxidase substrate evaluated in vitro and in a model phagocyte. Biochem Biophys Res Commun. 1991;175:387–393. doi: 10.1016/0006-291x(91)91576-x. [DOI] [PubMed] [Google Scholar]
- 59.Yang Z, Hanlon D W, Marszalek J R, Goldstein L S. Identification, partial characterization, and genetic mapping of kinesin-like protein genes in mouse. Genomics. 1997;45:123–131. doi: 10.1006/geno.1997.4901. [DOI] [PubMed] [Google Scholar]
- 60.Yonekawa Y, Harada A, Okada Y, Funakoshi T, Kanai Y, Takei Y, Terada S, Noda T, Hirokawa N. Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J Cell Biol. 1998;141:431–441. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu H, Nicchitta C V, Kumar J, Becker M, Toyoshima I, Sheetz M P. Characterization of kinectin, a kinesin-binding protein: primary sequence and N-terminal topogenic signal analysis. Mol Biol Cell. 1995;6:171–183. doi: 10.1091/mbc.6.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]