Abstract
Rpb8p, a subunit common to the three yeast RNA polymerases, is conserved among eukaryotes and absent from noneukaryotes. Defective mutants were found at an invariant GGLLM motif and at two other highly conserved amino acids. With one exception, they are clustered on the Rpb8p structure. They all impair a two-hybrid interaction with a fragment conserved in the largest subunits of RNA polymerases I (Rpa190p), II (Rpb1p), and III (Rpc160p). This fragment corresponds to the pore 1 module of the RNA polymerase II crystal structure and bears a highly conserved motif (P.I.KP..LW.GKQ) facing the GGLLM motif of Rpb8p. An RNA polymerase I mutant (rpa190-G728D) at the invariant glycyl of P.I.KP..LW.GKQ provokes a temperature-sensitive defect. Increasing the gene dosage of another common subunit, Rpb6p, suppresses this phenotype. It also suppresses a conditional growth defect observed when replacing Rpb8p by its human counterpart. Hence, Rpb6p and Rpb8p functionally interact in vivo. These two subunits are spatially separated by the pore 1 module and may also be possibly connected by the disorganized N half of Rpb6p, not included in the present structure data. Human Rpb6p is phosphorylated at its N-terminal Ser2, but an alanyl replacement at this position still complements an rpb6-Δ null allele. A two-hybrid interaction also occurs between Rpb8p and the product of orphan gene YGR089w. A ygr089-Δ null mutant has no detectable growth defect but aggravates the conditional growth defect of rpb8 mutants, suggesting that the interaction with Rpb8p may be physiologically relevant.
Three related RNA polymerases (Pol's) are required to transcribe the nuclear genome of all eukaryotes investigated so far. In yeast (Saccharomyces cerevisiae), they are made of 14 (Pol I), 12 (Pol II), and 17 (Pol III) polypeptides. Ten of these subunits form a conserved core structure containing the two large polypeptides (corresponding to the bacterial β' and β) and eight small subunits with molecular masses ranging between 8 and 25 kDa (38). The Pol II core enzyme is competent for the transcription of nonspecific DNA templates (12). In the cases of Pol I (30) and Pol III (5, 37), it has been demonstrated that additional enzyme-specific subunits are required for promoter-dependent transcription. The backbone structure of the yeast core Pol II (9) reveals a remarkable similarity to the bacterial core structure (43). Both enzymes contain a large DNA channel, leading to an internal catalytic pocket containing one or possibly two catalytic Mg2+ ions. A secondary channel beneath the active site probably allows nucleotide triphosphates to gain access to the catalytic site. This catalytic architecture is primarily determined by the folding of the two largest subunits.
The role of the small polypeptides present in the eukaryotic core structure is still poorly characterized. Four of them form an α-like structure (9, 22). Like the corresponding bacterial α2 homodimer, this structure is located at the rear of the bacterial enzyme and has been implicated in protein assembly but does not appear to directly contribute to the active site (16, 20, 22, 25, 32). It contains two small polypeptides (Rpb10p and Rpc10p) common to the three nuclear enzymes (7) and two subunits shared by Pol I and III (Rpc40p, Rpc19p) which have strong homologs in Pol II (Rpb3p, Rpb11p). It has been known for a long time that Pol's I, II, and III share three other small polypeptides, Rpb5p, Rpb6p, and Rpb8p (6, 39, 42). The first two polypeptides are strongly conserved from archaea to eukaryotes (23), and Rpb6p is also distantly related to the bacterial ω subunit (28). The present study deals with Rpb8p, a 16.5-kDa protein that has no homology in archaeal or bacterial genomes and is thus a uniquely eukaryotic component of the transcription machinery.
MATERIALS AND METHODS
Genetic material.
Common yeast media, growth conditions, and genetic techniques were used as described elsewhere (15, 34, 36). Yeast-peptone-dextrose (YPD) is a standard rich medium, synthetic complete (SC) is a synthetic medium supplemented with amino acids, adenine, and uracil, and SC-U is the corresponding medium without uracil. Strains and plasmids are listed and summarily described in Tables 1 and 2. Gene symbols and their synonyms are listed in the Yeast Protein Data website (http://www.proteome.com/databases/index.html). The corresponding protein symbols are followed by a p (Rpb8p), and the product of the YGR089w open reading frame was thus noted Ygr089p. The human homologue of Rpb6p and Rpb8p were referred to as Hs6 and Hs8.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| YGVS043 | MATaura3-52 his3-Δ200 leu2-Δ1 lys2-Δ201 ade2 trp1-Δ63 rpb8-Δ1::LYS2/pSL103(CEN URA3 RPB8) | 34 |
| YGVS045 | MATaura3-52 his3-Δ200 leu2-Δ1 lys2-Δ201 ade2 trp1-Δ63 rpb8-Δ1::LYS2/pGEN-Hs8(2μm TRP1 Hs8) | 34 |
| YJFB026 | MATaura3-52 his3-Δ200 leu2-Δ201 lys2-Δ201 ade2 trp1-Δ63 rpb8-Δ1::LYS2/pGEN-Hs8(2μm TR P1 Hs8)/pCM189-RPB6(CEN URA3 RPB6) | This study |
| YJFB025 | MATaura3-52 his3-Δ200 leu2-Δ1 lys2-Δ201 ade2 trp1-Δ63 rpb8-Δ1::LYS2/pGEN-rpb8-L122D(2μm TRP1 rpb8-L122D) | This study |
| YJB027 | MATaura3-52 his3-Δ200 leu2-Δ1 lys2-Δ201 ade2 trp1-Δ63 rpb8-Δ1::LYS2/pGEN-Hs8(2μm TRP1 Hs8)/pCM189-Hs6(CEN URA3 Hs6) | This study |
| YGVS003 | MATαCAN1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 rpb6-Δ::LEU2/pGEN-Hs6(2μm TRP1 Hs6) | 34 |
| YGVS004 | MATαCAN1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 rpb6-Δ::LEU2/pGEN-Hs6-S2A(2μm TRP1 Hs6-S2A) | This study |
| YPR2 | MATα ade2-101 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 ygr089w-Δ::URA3 | This study |
| YPR3 | MATaade2-101 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 ygr089w-Δ::URA3 | This study |
| NOY265 | MATα rpa190-3 trp1-Δ1 his4-Δ401 leu2-3,112 ura3-52 CAN1-100 | 41 |
| JAY444 | MATα CAN1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 rpb6::LEU2/pRPO26(CEN URA3 RPB6) | 1 |
| YJB021 | MATaura3-52 his3-Δ200 leu2-Δ1 lys2-801 ade2-1 trp1-Δ63 rpb8-Δ1::LYS2/pGEN-RPB8-G99K(2μm TRP1 rpb8-G99K) | This study |
| YPR6 | MATα ura3-52 his3-Δ200 leu2-Δ1 lys2-801 ade2-1 trp1-Δ63 ygr089-Δ::URA3 rpb8-Δ1::LYS2/pGFN-Hs8(2μm TRP1 RPB8Hs) | This study |
| YPR8 | MATα ura3-52 his3-Δ200 leu2-Δ1 lys2-201 ade2 trp1-Δ63 ygr089-Δ::URA3 rpb8-Δ1::LYS2/pGEN-M28(2μm TRP1 rpb8-G99K) | This study |
TABLE 2.
Plasmids
| Plasmids | Featuresa | Reference or source |
|---|---|---|
| pACT2Δ-RPA190e | 2μm LEU2 pADH1::GAL4(768-881)::RPA190(663-805)::tADH1 | 13 |
| pACT2Δ-RPB1e | 2μm LEU2 pADH1::GAL4(768-881)::RPB1(516-639)::tADH1 | 13 |
| pACT2Δ-RPC160e | 2μm LEU2 pADH1::GAL4(768-881)::RPC160(547-697)::tADH1 | 13 |
| pACT2Δ-YGR089w | 2μm LEU2 pADH1::GAL4(768-881)::YGR089w(497-769)::tADH1 | This study |
| pACT2Δ-NUP82 | 2μm LEU2 pADH1::GAL4(768-881)::NUP82(457-566)::tADH1 | This study |
| Yip5-ygr089w-Δ::URA3 | 2μm URA3 ygr089w-Δ::URA3 | This study |
| pGBT9-RPB8 | 2μm TRP1 pADH1::GAL4(1-147)::RPB8::tADH1 | This study |
| pGBT9-rpb8-xxx | 2μm TRP1 pADH1::GAL4(1-147)::rpb8-xxx::tADH1 | Mutant clones of pGBT9-RPB8 |
| pGEN-RPB8 | 2μm TRP1 pPGK1::RPB8::tCYC1 | 34 |
| pGEN-rpb8-xxx | 2μm TRP1 pPGK1::rpb8-xxx::tCYC1 | Mutant clones of pGEN-RPB8 |
| pGEN-Hs8 | 2μm TRP1 pPGK1::Hs8::tCYC1 | 34 |
| pGBT9-Hs8 | 2μm TRP1 pADH1::GAL4(1-147)::Hs8::tADH1 | This study (cloning of H. sapiens RPB8 in pGBT9) |
| pGBT9-Pb8 | 2μm TRP1 pADH1::GAL4(1-147)::Pb8::tADH1 | This study (cloning of S. pombe RPB8 in pGBT9) |
| pGBT9-RPB8Δ21 | 2μm TRP1 pADH1::GAL4(1-147)::RPB8-Δ21::tADH1 | PCR amplification from pDBSc8Δ21 (40) and cloning in pGBT9 |
| pGEN-Hs6 | 2μm TRP1 pPGK1::Hs6::tCYC1 | 34 |
| pGEN-Hs6S2A | 2μm TRP1 pPGK1::Hs6-S2A::tCYC1 | This study (site-directed mutagenesis of pGEN-Hs6) |
| pFL44-RPB6 | 2μm URA3 RPB6 | This study (1,540-bp BamHI-XhoI fragment cloned into pFL44L [2]) |
| pJFB003 | CEN4 ARS1 URA3 pTet07 RPB6::tCYC1 | This study (coding sequence of RPB6 cloned in pCM189 [17]) |
| pCM189-Hs6 | CEN4 ARS1 URA3 pTet07::Hs6 Tet-VP16::tCYC1 | This study (coding sequence of Hs6 cloned in pCM189 [17]) |
The Amp E. coli marker is present on all plasmids.
The ygr089w-Δ::URA3 null mutant was constructed by deleting an internal 810-bp HindIII fragment of YGR089w and replacing it with a 1,170-bp URA3+ fragment. The corresponding SalI-ClaI fragment was introduced into a YPH499 × YPH500 diploid strain by transformation. Integrative transformants generated by homologous recombination at the YGR089w locus were selected on SC-U and confirmed by PCR amplification from primers upstream and downstream of YGR089w. The mutant haploid strains YPR2 and YPR3 were obtained by meiotic sporulation. pCM189-Hs6 was constructed by cloning the PCR-amplified human RPB6 cDNA (Hs6) in pCM189 (17). Site-directed mutagenesis of pGEN-RPB8 and pGBT9-RPB8 was done by PCR amplification with Pfu DNA polymerase (Stratagene), using complementary primers containing the desired mutation. PCR products were digested with DpnI to eliminate the methylated wild-type template and propagated in XL1-Blue Escherichia coli cells. Mutant clones were identified by DNA sequencing and checked for the absence of additional mutations in the RPB8 coding sequence. Their growth phenotype was tested by genetic transformation in strain YGVS043, using a plasmid shuffle assay (34).
Two-hybrid tests were as described before (13). Starting from a random library of genomic fragments (14), 29 clones were isolated from 1.2 × 107 transformants by a double selection based on the activation of the HIS3 and LacZ reporter genes. Five clones were duplicate isolates due to some redundancy in the DNA library, four corresponded to noncoding regions, three were out-of-frame fusions, and three were FAS3 fragments that were isolated in several other screens of the same library. The 15 remaining clones resulted from in-frame fusions in eight distinct genes. Dose-dependent suppressors of the temperature-sensitive growth defect of strain YGVS045 were isolated from a yeast genomic library as previously described (36).
Cell fractionation.
Log-phase cells grown in 250 ml of YPD to an optical density at 600 nm (OD600) of about 0.6 were harvested by centrifugation, washed twice with spheroplasting buffer (50 mM Tris-HCl [pH 7.5], 1.2 M sorbitol, 10 mM NaN3, and 40 mM β-mercaptoethanol), and resuspended in a final volume of buffer adjusted to 1 ml for 40 OD600 units. Zymolyase 100T (ICN Biomedicals) was added at 300 μg/ml. Cells were incubated at 37°C for 30 to 45 min, and spheroplast formation was monitored under a microscope. Cells were washed twice in the same buffer, resuspended in 1 volume of ice-cold lysis buffer (20 mM HEPES-KOH, pH 7.4, 100 mM K-acetate, 5 mM Mg-acetate, 1 mM EDTA, 1 mM DTT) with protease inhibitors (Boehringer Mannheim) and 1 mM phenylmethylsulfonyl fluoride. They were gently crushed in a cold Potter, centrifuged for 3 min at 1,000 × g to remove unbroken cells, and centrifuged again for 15 min at 13,000 × g. The pellet (P13) contained nucleus and some endoplasmic reticulum fragments. The supernatant was purified by ultracentrifugation at 100,000 × g for 1 h, yielding an S100 fraction that contained essentially cytosolic proteins. Twenty to 100 μg of proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was blotted to nitrocellulose membranes (Hybond C-super) and probed with antibodies using the ECL detection system (Amersham Pharmacia Biotech).
RESULTS
Rpb8p interacts in vivo with Rpb6p.
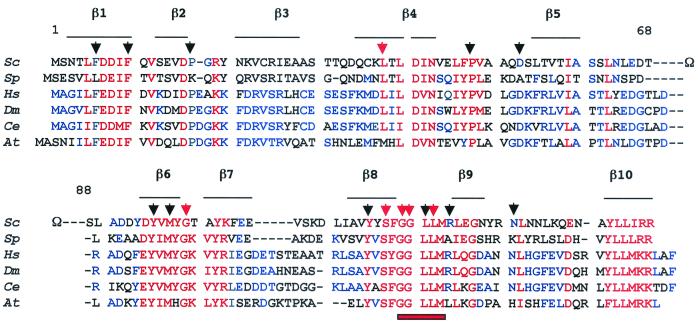
Figure 1 documents the sequence conservation of Rpb8p, based on the inspection of current databanks. This subunit is present in fungus (S. cerevisiae, Schizosaccharomyces pombe), plant (Arabidopsis thaliana), and animal (Drosophila melanogaster, Caenorhabditis elegans, Homo sapiens) genomes. Rpb8p is likely to be shared by all three RNA polymerases in each of these organisms, since the corresponding gene is unique on their genome. Figure 1 shows that Rpb8p contains about 40 highly conserved or invariant positions (i.e., less than one-third of the protein) shared among all the Rpb8p-like subunits identified so far. A disorganized Ω loop (positions 68 to 88) is totally dispensable in vivo (40) and is absent from animal or plant Rpb8p. The spatial structure of the yeast subunit was determined in solution by nuclear magnetic resonance (NMR) (21) and, more recently, by high-resolution crystal data from the core of Pol II (8). These two structures are in fairly good agreement but are not entirely superimposable. The most highly conserved regions on the amino acid sequence correspond to the β1-, β4-, β6-, and β10-strands and to an invariant GGLLM signature between β8 and β9.
FIG. 1.
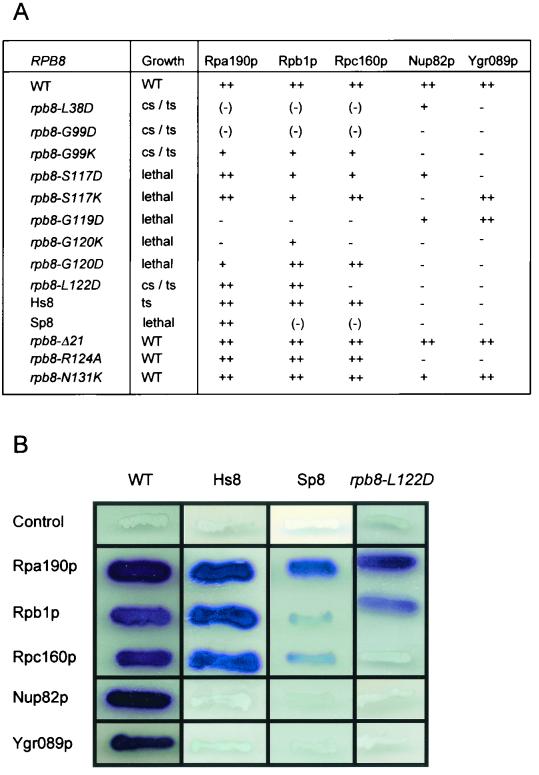
Sequence conservation and site-directed mutagenesis of Rbp8p. Sc, S. cerevisiae; Sp, S. pombe; Hs, H. sapiens; Dm, D. melanogaster; Ce, C. elegans; At, A. thaliana. Shown are invariant or highly conserved positions (red) and positions conserved in at least three sequences (blue). The positions of the β-strands identified on the spatial structure (8, 21) are indicated by horizontal lines. A GGLLM signature motif present in Rpb8p-like sequences is noted by a red bar. The 17 amino acids mutated in this study are indicated by triangles; black triangles denote mutations with no growth phenotype (F6L, F6K, F10D, P17K, P48V, D53A, Y95F, Y95R, Y95E, M97C, M97D, M97K, Y115F, Y115R, Y115E, L121I, L121N, L121K, L121D, R124A, N131K) (the rpb8-Δ21 deletion of the Ω-loop is also silent; 40), and red triangles indicate conditional (L38D, G99D, G99K, L122D) and lethal (S117D, S117K, G119D, G120K, G120D) mutations.
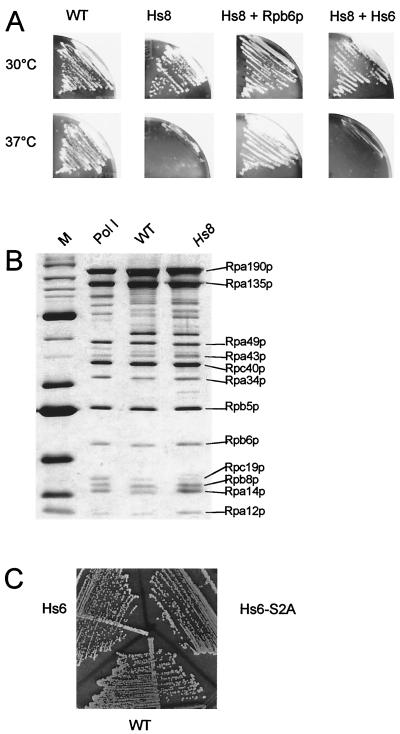
The human subunit (Hs8) differs from S. cerevisiae by 107 of its 150 amino acids, with up to 80 nonconservative changes. Accordingly, replacing the yeast subunit by its human counterpart (strain YGVS045) amounts to a major multisite modification. We have previously shown that complementation between yeast and human RPB8 is functional at 30°C, with a sharp growth defect at 37°C (34) (Fig. 2A). Moreover, the wild-type and hybrid Pol I (containing Hs8 instead of Rpb8p), purified from cells grown at the permissive temperature, have an identical subunit composition (Fig. 2B) with a comparable specific activity on a poly(dA-dT) template, indicating that Hs8 is effectively and stably incorporated in the yeast enzyme under these conditions (data not shown).
FIG. 2.
Genetic interactions between Rpb6p and Rbp8p. (A) Strains YGVS043 (rpb8-Δ::LYS2 with the yeast RPB8 gene [WT]), YGVS045 (rpb8-Δ::LYS2 with the human Hs8 gene [Hs8]), YJB026 (YGVS045 with a centromeric plasmid expressing the yeast RPB6 gene [Hs8 + Rpb6p]), and YJB027 (YGVS045 with a centromeric plasmid expressing the human Hs6 gene [Hs8 + Hs6]) were streaked on YPD and incubated for 3 days at 37°C. Strain genotypes are listed in Table 1. (B) Five micrograms of a highly purified and catalytically active preparation of Pol I and of two purified preparations from YGVS043 (WT) and YGVS045 (Hs8) strains was separated by SDS–8 to 12% PAGE and silver stained. Individual subunits were identified from their apparent molecular weight, in parallel with standard protein markers (lane M) and a purified Pol I sample (15). (C) Strains JAY444 (rpb6-Δ::LEU2 with the yeast RPB6 gene [WT]), YGVS003 (rpb6-Δ::LEU2 with the human Hs6 gene [Hs6]), YGVS004 (rpb6-Δ::LEU2 with the human Hs6 gene and an S2A mutation [Hs6-S2A]) were streaked on YPD and incubated for 3 days at 30°C, showing the same growth pattern (this observation was made at temperatures ranging from 16°C to 37°C). Strain constructions and genotypes are in Table 1.
The temperature-sensitive (ts) growth defect associated with the human Rpb8p subunit (Hs8) provided a simple way to look for physiological partners of Rpb8p by dose-dependent extragenic suppression (36). Of the 12 clones thus obtained, three contained RPB8 itself. The nine others harbored SSD1 (four clones), UBI2 (three clones), UBI4 (one clone), and SMY2 (one clone). UBI2 and UBI4 (encoding ubiquitin) also weakly suppress a temperature-sensitive Pol III mutant, rpc31-236. Ubiquitin targets proteins for degradation by the 26S proteasome, and suppression may therefore be due to the proteolysis of incorrectly folded mutant polypeptides. SMY2 and SSD1 suppress various mutants unrelated to Pol's (24, 36) and may have a general effect on the adaptation of S. cerevisiae at 37°C.
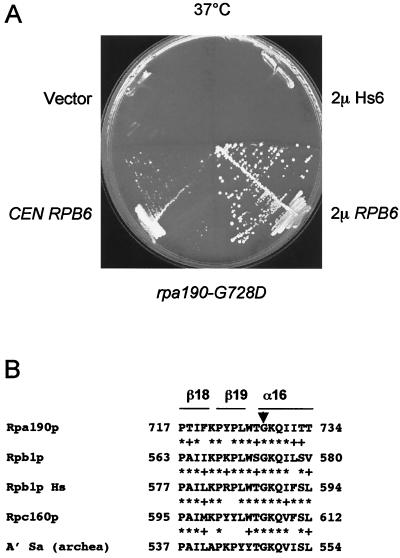
We separately tested all RNA polymerase subunits and found strong suppression by RPB6 (encoding the common subunit Rpb6p), even when it was on a centromeric vector (Fig. 2). Note that centromeric vectors have a low copy number, but are not necessarily single copy, and that genes may be expressed at higher levels than in a chromosomal context (2). Suppression was specific of the human allele (Hs8), with no effect on other conditional mutants (rpb8-G99D, rpb8-G99K, and rpb8-L122D) obtained by site-directed mutagenesis (see below). RPB6 also strongly suppressed rpa190-G728D, a Pol I with a temperature sensitivity mutation in the largest subunit (Fig. 3). This mutant, initially called rpa190-3 (41), corresponds to the invariant Gly of a highly conserved P.I.KP..LW.GKQ motif characteristic of domain e (38), a conserved region present on the three eukaryotic Pol's and on the archaeal enzyme but absent from bacteria. On the core Pol II structure, this motif corresponds to a short β18-β19-α16 fold where the invariant glycine marks the beginning of α16 (8). As outlined below, this motif is located across Rpb8p on the spatial structure (Fig. 4) and is part of a conserved domain that is specifically recognized by Rpb8p on the largest subunits of Pol's I, II, and III (13).
FIG. 3.
Suppression of rpa190-G728D by RPB6. (A) Strain NOY265 (41) was transformed by URA3+ plasmids bearing the yeast (2μm RPB6) or human (2μm Hs6) RPB6 genes or the centromeric URA3 plasmid bearing the yeast RPB6 gene (CEN RPB6). The control strain was NOY265 transformed with a void plasmid (pFL44L [vector]). Cells were streaked on YPD and incubated for 5 days at 37°C. (B) A local alignment between the largest subunits of yeast (Rpa190p, Rpb1p, Rpc160p), human Rpb1p (Rpb1p Hs), and archaeal (A′ Sa) RNA polymerases. The black triangle indicates the position of the rpa190-G728D mutation. ∗, identity; +, similarity.
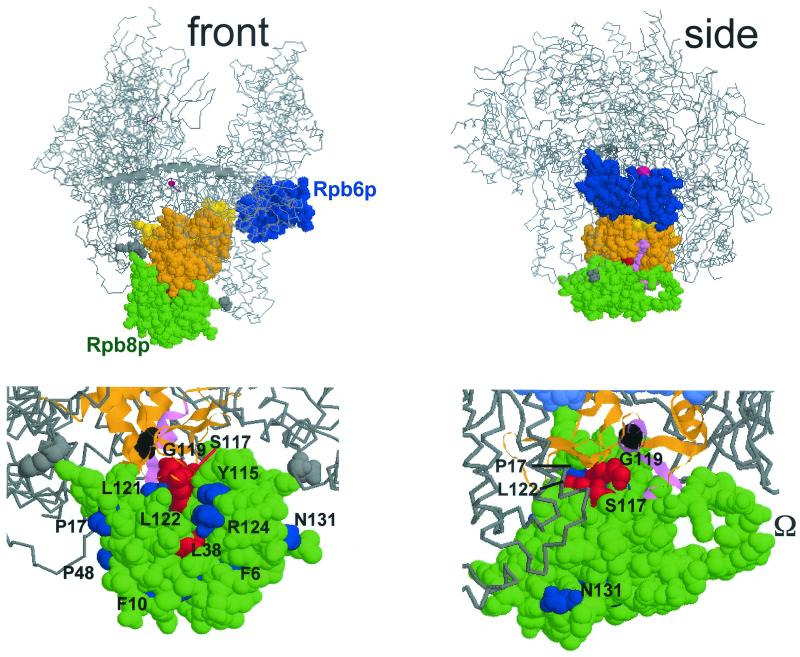
FIG. 4.
Spatial organization of Rpb8p and Rpb6p on Pol II. This figure was prepared with the RASMOL software (ftp://ftp.dcs.ed.ac.uk/pub /rasmol), using yeast Pol II coordinates (PDB accession codes, 1I3Q and 1I50; see reference 8). (Top) Front and side views approximately corresponding to the synomymous views used by Cramer et al. (8, 9). Rpb8p (green), Rpb6p (blue) without its first 71 amino acids, forming a disorganized tail (9, 11), and part of Rpb1p (between positions 508 and 663) are space-filled, the rest of the core Pol II structure being given as a Cα backbone, with the β'F bridge domain (positions 809 to 845 of Rpb1p) shown as an α-helix (see references 8 and 9 for more details on the spatial structure). Orange, two-hybrid interacting domain recognized by Rpb8p (positions 516 to 639; 13); violet, positions 563 to 580, corresponding to the invariant P.I.KP..LW.GKQ motif (see Fig. 3); yellow, part of the pore 1 module (positions 508 to 663; see reference 8) that is not included in the two-hybrid interacting domain; grey, K738 and D974, two positions that closely contact Rpb8p on the structure but are not included in the two-hybrid fragment. The catalytic Mg2+ is represented by a red sphere (not drawn to scale). (Bottom) Scaled-up views of the front and side views shown above, with slightly different viewing angles. The two-hybrid interacting domain is shown as an α-helical and β-stranded ribbon structure, with the same color code as above. Position G574 (corresponding to the rpa190-G728D mutation of Pol I; see Fig. 3) is space-filled and shown in black. Some of the Rpb8p positions mutagenized in this study are individualized in blue (phenotypically silent mutations) or red (lethal or conditional growth defects). The dispensable Ω loop (partly disorganized on the Rpb8p structure) (8, 21) is also indicated.
Despite their strong physiological interaction, Rpb6p and Rpb8p do not directly contact each other on the high-resolution Pol II structure (8) (Fig. 4). However, dose-dependent suppression is mediated by a mass action effect of the overproduced suppressor gene and need not imply a direct protein contact. Moreover, the relevant interaction could occur at an early stage of enzyme assembly. A closer look at the spatial structure shows that Rpb8p and Rpb6p are essentially separated by a module of the largest subunit (positions 508 to 663 on Rpb1p), called pore 1 (9). Accordingly, the dose-dependent suppression effect of RPB6 on rpa190-G728D and on the temperature sensitivity phenotype associated with Hs8 could be mediated by an allosteric rearrangement involving the pore 1 module.
On the other hand, the present Pol II structure lacks the first 71 amino acids of Rpb6p that form a highly disordered and very acidic domain in the yeast subunit (9, 33) and in the NMR structure of the human subunit (11). The N end might conceivably protrude out of the Pol II structure to reach Rpb8p, notwithstanding the fact that the 72nd amino acid of Rpb6p (K72) is located away from Rpb8p on the Pol II structure. The human Rpb6p (Hs6), which essentially differs from the yeast Rpb6p by its shorter acidic N tail, complements an S. cerevisiae null mutant (26, 34). It has no suppressor effect on the temperature sensitivity phenotype of the human Rpb8p (Hs8) when borne on a centromeric plasmid (Fig. 2A, Hs8 + Hs6) and only weakly suppresses when expressed from a multicopy plasmid (data not shown). The yeast and human Rpb6p are phosphorylated (3), and this phosphorylation occurs at the N-terminal Ser2 position of the human subunit (19). However, a S2A replacement at this position effectively complements the yeast rpb6-Δ null mutant (Fig. 2C), indicating that phosphorylation is not strictly critical for growth, although it may have some subtle role in vivo.
Rpb8p interacts with conserved domain e on largest subunit of Pol's I, II, and III.
To search for more putative partners of Rpb8p, we used a systematic two-hybrid screening, based on a yeast genomic library containing random fusions to the Gal4p activation domain (14). In this approach, a pGBT9-RPB8 fusion of Rpb8p to the Gal4p DNA binding domain was used as bait, and it yielded 15 independent positive clones (see Materials and Methods and Fig. 5). Five of them encoded homologous segments of the largest subunit of Pol's II (Rpb1p, one clone) and III (Rpc160p, four clones), and an ad hoc construction bearing the homologous domain of the Pol I subunit Rpa190p also responds to pGBT9-RPB8 (13). Four other clones corresponded to overlapping fragments of the orphan gene YGR089w, which may thus conceivably encode a physiological partner of Rpb8p (see below). NUP82, encoding a yeast nucleoporin (18), was represented by two isolates. The remaining six clones were each represented by a different gene (GCR1, PET309, UBP14, and CUP1) and, given their known physiological role or extranuclear localization, almost certainly have no physiological interaction with Rpb8p.
FIG. 5.
Two-hybrid interactions of Rpb8p. (A) Effect of Rpb8 mutations on growth and two-hybrid interactions. Growth patterns were determined on YPD plates streaked and incubated for 3 to 5 days at 25, 30, and 37°C. cs, little or no growth at 25°C; ts, little or no growth at 37°C. Strengths of the two-hybrid interactions are as follows: ++, wild-type level of interaction; +, partly defective interaction; (−), weak interaction; −, no interaction. The plasmids bearing rpb8 mutant baits were all derived from pGBT9-RBP8 by site-directed mutagenesis. The prey plasmids were pACT2Δ-RPA190e, pACT2Δ-RPB1e, pACT2Δ-RPC160e, pACT2Δ-NUP82, and pACT2Δ-YGR089w (see Table 1). (B) β-Galactosidase plate assays of two-hybrid interactions. The assay is illustrated by the wild-type, human, and S. pombe RPB8 and rpb8-L122D alleles. The negative control corresponds to the void pGBT9 plasmid (13).
The Rpb1p fragment recognized by Rpb8p in this two-hybrid assay is comprised of amino acids 516 to 639. The N-terminal half of that domain contains part of the conserved domain d (shared by all Pol's) downstream of the invariant Mg2+-binding motif NADFGDG. Its C-terminal half corresponds to conserved domain e (38) and contains the eukaryotic consensus motif P.I.KP..LW.GKQ shared by all eukaryotic and archaeal Pol's (Fig. 2). This two-hybrid interaction is in excellent agreement with the high-resolution structure now available for Pol II (8), where the 516-to-639 fragment coincides almost exactly with a spatial module (positions 508 to 663) called pore 1. As seen in Fig. 4, this is the main Rpb1p region contacted by Rpb8p, and the latter subunit contacts no other subunit of the core enzyme. However, Rpb8p also contacts Rpb1p at Lys738 and Asp974, two positions that are not included in the two-hybrid interacting domain but could play an important role in the overall folding of Rpb1p.
The P.I.KP..LW.GKQ consensus motif noted above corresponds to a short β18-β19-α16 fold on the pore 1 module (Fig. 3 and 4) and is located across Rpb8p on the spatial structure. The invariant glycine of that motif marks the beginning of α16 and is mutated in the temperature-sensitive Pol I mutant rpa190-G728D (or rpa190-3) (41). The effect of this mutation is not alleviated by increasing the gene dosage in RPB8 but, as already mentioned, is strongly suppressed by RPB6 (Fig. 3). Taken together, these data strongly suggest that Rpb8p, Rpb6p, and the pore 1 module form a “ménage à trois” in terms of functional and structural interactions.
Site-directed mutagenesis of Rpb8p: effect on growth and on two-hybrid interactions.
Seventeen conserved positions scattered on the spatial structure and on the amino acid sequence of Rpb8p were submitted to site-directed mutagenesis, yielding 30 mutants that were tested for growth by plasmid shuffling in strain YGVS043 (34). As summarized in Fig. 1, 12 of these mutagenized positions are invariant (or highly conserved, L38) in all eukaryotes analyzed so far (F10, L38, P48, Y95, M97, G99, Y115, S117, G119, G120, L121, L122). The remaining five positions (F6, P17, D53, R124, N131) were chosen because they are also highly conserved in eukaryotes but not in the S. pombe sequence. Keeping in mind that the S. pombe subunit does not support growth in S. cerevisiae (35, 40), these positions were mutated to match the corresponding S. pombe amino acid.
Most of the rpb8 mutations were phenotypically indistinguishable from the wild-type allele, indicating a remarkable tolerance of Rpb8p to amino acid changes, even when strictly invariant positions are replaced in a nonconservative way (e.g., rpb8-P48V, rpb8-Y95E, rpb8-M97K, or rpb8-Y115E). While this mutagenesis was by no means exhaustive, the spatial distribution of lethal (rpb8-S117K, rpb8-S117D, rpb8-G119D, rpb8-G120K, rpb8-G120D) or conditionally lethal (rpb8-L38D, rpb8-G99D, rpb8-G99K, rpb8-L122D) mutations is clearly nonrandom, as shown in Fig. 4. With one exception (rpb8-L38D), these mutations are spatially close to each other and are located opposite the main Rpb1p interface, close to the P.I.KP..LW.GKQ consensus formed by β18, β19, and α16 on Rpb1p. On Rpb8p, they correspond to the β6 (G99) and β8 (S117) strands and to a highly conserved region immediately after β8, defined by the 119GGLLM123 signature shared by all Rpb8p. β6 forms a β-addition motif with the β18 of Rpb1p (8). These critical positions therefore appear to define the main interface between Rpb8p and Rpb1p. rpb8-L38D is the only critical mutation located away from Rpb1p. This mutation is likely to disrupt the β4-β5-sheet motif and could thus strongly affect the overall folding of Rpb8p.
These mutants were also examined for their effect on two-hybrid interactions between Rpb8p and the largest subunit of Pol's I (Rpa190p), II, (Rpb1p) and III (Rpc160p). As shown in Fig. 5, there is a good correlation between their growth pattern and their two-hybrid response. The nine point mutants with complete or conditional lethality were all strongly impaired in their interaction with one (rpb8-S117K, rpb8-G120D, rpb8-L122D), two (rpb8-S117D) or all three (rpb8-L38D, rpb8-G99D, rpb8-G99K, rpb8-G119D, rpb8-G120K) largest subunits. In contrast, the phenotypically silent mutations rpb8-R124A, rpb8-N131K, and rpb8-Ä21 (deleted Ω-loop) (40) fully retained the two-hybrid interaction pattern of the wild-type allele.
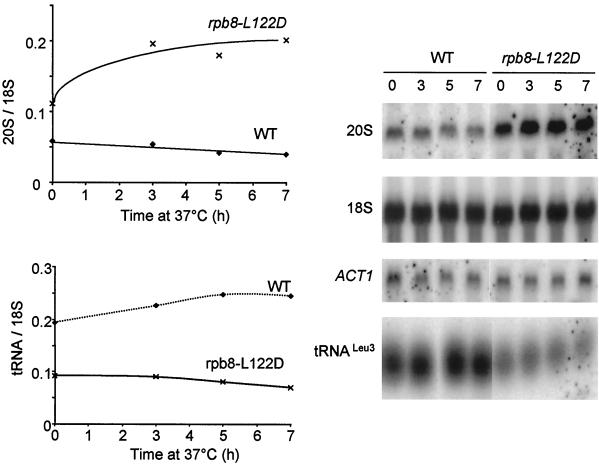
Rpb8-L122D, corresponding to the 119GGLLM123 invariant motif, was investigated in more detail, in view of its striking Pol III-specific defective response in the two-hybrid assay. This specificity is reflected in vivo, as shown by the low steady-state level in tRNA of rpb8-L122D cells grown at 30°C or shifted to 37°C (Fig. 6). Moreover, this mutant has a high steady-state level of 20S pre-rRNA, a property noted previously for bona fide Pol III mutants and probably reflecting a link between Pol III and rRNA maturation (4). A close link between growth and two-hybrid interaction data are also suggested by the properties of the human and S. pombe subunit in S. cerevisiae, since the former complements a rpb8-Δ null mutant in vivo (except at high temperature) and retains a wild-type interaction with all three subunits (Fig. 2), while the latter fails to complement at any temperature and fails to interact with Rpa190p and Rpc160p (40).
FIG. 6.
mRNA, tRNA, and rRNA levels in rpb8-L122D. Steady-state levels of the 20S pre-rRNA, 18S mature rRNA, tRNALeu3, and ACT1 mRNA analyzed by Northern blotting. RNAs were extracted from exponentially growing cells of YGVS043 (wild type) and YJFB025 (rpb8-L122D) shifted from 25 to 37°C for 7 h in YPD liquid medium. The oligonucleotide probes and Northern hybridization conditions were previously described (4). The time that each sample was taken (in hours) is shown at the top of the gel.
Ygr089p is a putative partner of Rpb8p.
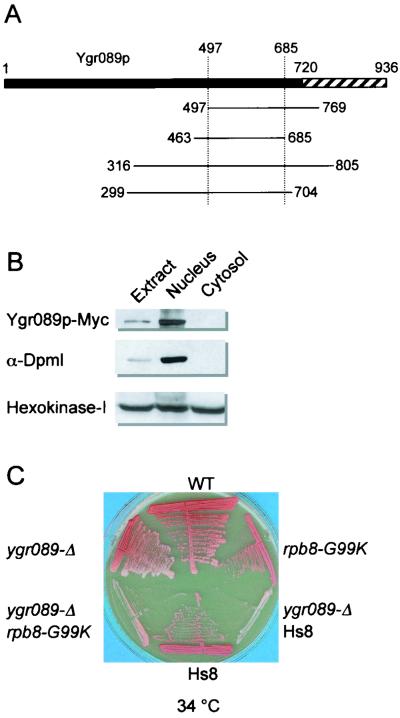
As noted above, our two-hybrid screen with pGBT9-RBP8 yielded two additional putative partners of Rpb8p. Two independent clones encode Nup82p, a poorly conserved but essential component of the yeast nucleopore (18). They overlap between amino acids 457 and 566. Four different clones encoding fragments of the orphan protein Ygr089p overlapping between positions 497 and 685 were also selected (Fig. 7A). The fact that overlapping fragments were selected in the screen implies that Rpb8p recognizes a specific protein domain on the target protein. However, this specificity is no proof that the corresponding interactions are physiologically relevant.
FIG. 7.
Characteristics of the YGR089w gene product. (A) Schematic representation of the four inserts recognized by pGBT9-RPB8 in a two-hybrid test. The hatched area denotes a hydrophilic C-terminal domain. The coordinates of the Ygr089p fragments encoded by the clones selected in the Rpb8p two-hybrid screen are indicated. (B) Cellular localization of the YGR089w gene product. Cell fractionation was done as described in Materials and Methods. Extract, crude cell extract; nucleus, P13 fraction; cytosol, S100 fraction. Antibodies raised against Dolichol phosphate mannose synthase and hexokinase I were used as markers of the endoplasmic reticulum-nucleus and cytosolic fractions, respectively. Twenty micrograms of total proteins was loaded in each case. (C) Growth pattern of the ygr089w-Δ::URA3 null mutants. Cells of the isogenic strains YGVS043 (WT), YGVS045 (replacement of the yeast Rpb8p by Hs8), YPR2 (ygr089w-Δ), YJB021 (rpb8-G99K), YPR6 (ygr089w-Δ and Hs8), and YPR8 (ygr089w-Δ rpb8-G99K) were streaked on YPD and incubated for 3 days at 34°C.
YGR089w encodes a protein with a mass of 106 kDa and with no obvious structural features except for its hydrophilic C end. No related gene product can be recognized from presently available genomic sequences, even for the ascomycetous yeast S. pombe. A Myc(13X)-tagged version of Ygr089p copurifies with the endoplasmic reticulum-nucleus fraction and was excluded from the cytosol (Fig. 7B), but attempts to localize the tagged protein more precisely by immunofluorescence have been inconclusive. The ygr089w-Δ::URA3 null mutant is phenotypically silent in terms of growth, respiration, and competence for meiosis (not shown). However, the fact that it has a moderate but distinct adverse effect on growth in an rpb8-G99K context, or when yeast Rpb8p is replaced by the human subunit (Fig. 7C), supports the idea that the Rpb8p-Ygr089p interaction may be physiologically relevant.
DISCUSSION
In this study, we sought physiological partners of Rpb8p by genetic screens based on extragenic suppression and on two-hybrid interactions. The putative partners thus identified include the largest subunit of each nuclear Pol (Rpa190p, Rpb1p, and Rpc160p), the common subunit Rpb6p, and more tentatively, the Nup82p nucleoporin and the orphan gene product Ygr089p. The interaction with the largest subunits is in excellent agreement with the high-resolution crystal structure of yeast Pol II recently published by Cramer et al. (8). It involves a spatial cluster of amino acids containing the invariant 119GGLLM123 motif, shared by Rpb8p in all eukaryotes investigated so far. Mutants with a defect in this interaction have a severe growth defect or are lethal. Moreover, an rpb8-L122D mutant at the 119GGLLM123 motif specifically fails to interact with Rpc160p and has a preferential Pol III defect in vivo.
The target two-hybrid domain in the large subunits (domain e) (38) contains an invariant P.I.KP..LW.GKQ motif, located across the 119GGLLM123 Rpb8p signature, and essentially corresponds to the pore 1 module on the Pol II crystal structure (8). Our data strongly suggest that similar contacts holds Rpb8p to Rpa190p and Rpc160p on Pol I and Pol III. This interaction must be evolutionarily robust, since the yeast and human subunits, differing by 107 out of 150 amino acids, are largely indistinguishable in their ability to interact with all three RNA Pol's, as documented by heterospecific complementation and two-hybrid data and by the intact subunit composition of the purified chimeric Pol I. On the other hand, minor differences between the target domain on Rpa190p, Rpb1p, and Rpc160p may account for the Pol III-specific response noted above for rpb8-L122D and for Pol-specific effects also observed when replacing Rpb8p by the S. pombe subunit (40).
A moderate increase in the gene dosage of RPB6 suffices to suppress the temperature sensitivity defect associated with the human Rpb8p-like subunit (Hs8). Moreover, this also suppresses rpa190-G728D, a Pol I mutant located at the invariant Glycyl of the Rpb8p target motif P.I.KP..LW.GKQ. An in vivo connection between Rpb6p and Rpb8p is thus clearly established by these data. Since RPB6 acts in a dose-dependent way and not as a result of a mutational change, the suppressible mutants are likely to affect the assembly or stability of Pol's rather than their activity. Rpb6p and Rpb8p do not contact each other on the spatial model of Pol II, where they are actually separated by a large spatial domain corresponding to the pore 1 module (8). Thus, Rpb8p, Rpb6p, and the pore 1 module may form a functionally integrated domain on the Pol II structure, with structural changes at the Rpb8p side (rpa190-G728D or the replacement of Rpb8p by the human Hs8 subunit) ultimately altering the folding of Rpb6p and perhaps facilitating its dissociation form the core enzyme. Alternatively, we also note that the present crystal structure lacks the entire N-terminal half of Rpb6p. The latter is highly disorganized both in the Pol II crystal data (8, 9) and in the NMR structure of the human subunit (11) and essentiallly contains acidic amino acids that could form a flexible helix stretching far out of Pol II and thus possibly contacting Rpb8p. This domain is essential for growth in S. cerevisiae (29) and is as uniquely eukaryotic as Rpb8p itself (23). It is phosphorylated at the N-terminal Ser of the human subunit (19), but the S2A mutant fully retains its ability to support growth in S. cerevisiae.
How do these data relate to the essential role played by Rpb8p in eukaryotic transcription, keeping in mind that this subunit is shared by all three Pol's and is uniquely eukaryotic? One clue may be given by the fact that Rpb8p is targeted to the pore 1 module, initially recognized as eukaryotic specific on the basis of sequence alignments (domain e; 38) and having different foldings in the bacterial and yeast enzyme (8). Cramer et al. (8, 9) have speculated that pore 1 could facilitate the entry of nucleoside triphosphates and allow the 3′ end of the nascent transcript to move backward in halted elongation complexes. Interestingly, the P.I.KP..LW.GKQ motif located opposite Rpb8p in the pore 1 fold (and mutated in the Pol I rpa190-G728D mutant) is also present on archaeal Pol's, although the latter has no recognizable Rpb8p subunit (10, 23). This motif is thus unlikely to be a mere docking site for Rpb8p. Moreover, the 119GGLLM123 motif of Rpb8p appears to form a flexible or partly disorded small loop across the P.I.KP..LW.GKQ motif. It would be worth examining in more detail how the corresponding rpa190-728D (P.I.KP..LW.GKQ) and rpb8-L122D (GGLLM) mutants affect transcriptional activity in vitro.
On the other hand, Rpb8p may not be directly required for the activity of eukaryotic Pol's but could rather determine the stability of their heteromultimeric structure or their correct nuclear assembly. As noted above, the suppression of rpa190-728D by an increased RPB6 gene dosage points indeed to such a stability or assembly defect. In a more speculative vein, the interaction noted between Rpb8p and the Nup82p nucleoporin (18) could also reflect a role of Rpb8p in the nuclear transport or assembly of Pol's I, II, and III. Finally, we note that most of Rpb8p is exposed to the solvent in the three-dimensional model of Pol II, thus forming an external surface available for nonessential interactions with other nuclear components, such as the one tentatively suggested here for the orphan yeast gene product Ygr089p.
ACKNOWLEDGMENTS
We thank O. Gadal for very useful suggestions, M. Riva and C. Carles for a sample of purified Pol I and for Pol I antibodies, A. Voutsina for S. pombe plasmids, M. Nomura for the rpa190-3 mutant, and P. Cramer, D. A Bushnell, and R. Kornberg for communicating their Pol II coordinates prior to publication. We heartily thank sharp-eyed and patient referees for improving our manuscript.
J.-F.B. had a Fellowship from the Fondation de la Recherche Médicale, and F.N. held a European Marie Curie Fellowship. This work was partly funded by a Training and Mobility Program of the European Union (grant FMRX-CT96-0064).
REFERENCES
- 1.Archambault J, Schappert K T, Friesen J D. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol Cell Biol. 1990;10:6123–6131. doi: 10.1128/mcb.10.12.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonneaud N, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 3.Bréant B, Buhler J M, Sentenac A, Fromageot P. On the phosphorylation of yeast RNA polymerases A and B. Eur J Biochem. 1983;130:247–251. doi: 10.1111/j.1432-1033.1983.tb07143.x. [DOI] [PubMed] [Google Scholar]
- 4.Briand J F, Navarro F, Gadal O, Thuriaux P. Cross-talk between tRNA and rRNA synthesis. Mol Cell Biol. 2001;21:189–195. doi: 10.1128/MCB.21.1.189-195.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buhler J M, Iborra F, Sentenac A, Fromageot P. Structural studies on yeast RNA polymerases. J Biol Chem. 1976;251:1712–1717. [PubMed] [Google Scholar]
- 7.Carles C, Treich I, Bouet F, Riva M, Sentenac A. Two additional common subunits, ABC10α and ABC10β, are shared by yeast RNA polymerases. J Biol Chem. 1991;266:24092–24096. [PubMed] [Google Scholar]
- 8.Cramer P, Bushnell D A, Kornberg R. Structural basis of transcription: RNA polymerase II at 2.8 ångstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 9.Cramer P, Bushnell D A, Fu J, Gnatt A L, Maier-Davis B, Thompson N E, Burgess R R, Edwards A M, David P R, Kornberg R D. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 10.Darcy T J, Hausner W, Awery D E, Edwards A M, Thomm M, Reeve J N. Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro. J Bacteriol. 1999;181:4424–4429. doi: 10.1128/jb.181.14.4424-4429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Rio-Portilla F, Gaskell A, Gilbert D, Ladias J A, Wagner G. Solution structure of the hRPABC14.4 subunit of human RNA polymerases. Nat Struct Biol. 1999;6:1039–1042. doi: 10.1038/14923. [DOI] [PubMed] [Google Scholar]
- 12.Dezélée S, Wyers F, Sentenac A, Fromageot P. Two forms of RNA polymerase B in yeast. Eur J Biochem. 1976;65:543–552. doi: 10.1111/j.1432-1033.1976.tb10372.x. [DOI] [PubMed] [Google Scholar]
- 13.Flores A, Briand J F, Boschiero C, Gadal O, Andrau J C, Rubbi L, Van Mullem V, Goussot M, Marck C, Carles C, Thuriaux P, Sentenac A, Werner M. A protein-protein interaction map of yeast RNA polymerase III. Proc Natl Acad Sci USA. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromont-Racine M, Rain J C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 15.Gadal O, Chédin S, Quémeneur E, Carles C, Sentenac A, Thuriaux P. A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machinery. Mol Cell Biol. 1997;17:1787–1795. doi: 10.1128/mcb.17.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadal O, Shpakovski G V, Thuriaux P. Mutants in ABC10b, a conserved subunit shared by all three yeast RNA polymerases, specifically affect RNA polymerase I assembly. J Biol Chem. 1999;274:8421–8427. doi: 10.1074/jbc.274.13.8421. [DOI] [PubMed] [Google Scholar]
- 17.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 18.Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt E C. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J Cell Biol. 1995;130:1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayukawa K, Makino Y, Yogosawa S, Tamura T. A serine residue in the N-terminal acidic region of rat RPB6, one of the common subunits of RNA polymerases, is exclusively phosphorylated by casein kinase II in vitro. Gene. 1999;234:139–147. doi: 10.1016/s0378-1119(99)00164-x. [DOI] [PubMed] [Google Scholar]
- 20.Kimura M, Ishiguro A, Ishihama A. RNA polymerase II subunits 2,3 and 11 form a core subassembly with DNA binding activity. J Biol Chem. 1997;272:25851–25855. doi: 10.1074/jbc.272.41.25851. [DOI] [PubMed] [Google Scholar]
- 21.Krapp S, Kelly G, Reischl J, Weinzierl R O, Matthews S. Eukaryotic RNA polymerase subunit RPB8 is a new relative of the OB family. Nat Struct Biol. 1998;5:110–114. doi: 10.1038/nsb0298-110. [DOI] [PubMed] [Google Scholar]
- 22.Lalo D, Carles C, Sentenac A, Thuriaux P. Interactions between three common subunits of yeast RNA polymerases I and III. Proc Natl Acad Sci USA. 1993;90:5524–5528. doi: 10.1073/pnas.90.12.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in Archaea: similarity to that in Eucarya. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lillie S H, Brown S S. Smy1p, a kinesin-related protein that does not require microtubules. J Cell Biol. 1998;140:873–883. doi: 10.1083/jcb.140.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann C, Buhler J M, Treich I, Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerase A and C. Cell. 1987;48:627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- 26.McKune K, Moore P A, Hull M W, Woychik N A. Six human RNA polymerase subunits functionally substitute for their yeast counterpart. Mol Cell Biol. 1995;15:6895–6900. doi: 10.1128/mcb.15.12.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milkereit P, Tschochner H. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during trancription. EMBO J. 1998;17:3692–3703. doi: 10.1093/emboj/17.13.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minakhin L, Bhagat S, Brunning A, Campbell E A, Darst S A, Ebright R H, Severinov K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc Natl Acad Sci USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nouraini S, Archambault J, Friesen J D. Rpo26p, a subunit common to yeast RNA polymerases, is essential for the assembly of RNA polymerase I and II and is essential for the stability of the largest subunits of these enzymes. Mol Cell Biol. 1996;16:5985–5996. doi: 10.1128/mcb.16.11.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with rrn3. EMBO J. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riva M, Buhler J M, Sentenac A, Fromageot P, Hawthorne D C. Natural variation in yeast RNA polymerase A. J Biol Chem. 1982;257:4570–4577. [PubMed] [Google Scholar]
- 32.Rubbi L, Labarre-Mariotte S, Chédin S, Thuriaux P. Functional characterization of ABC10α, an essential polypeptide shared by all three forms of eukaryotic DNA-dependent RNA polymerases. J Biol Chem. 1999;274:31485–31492. doi: 10.1074/jbc.274.44.31485. [DOI] [PubMed] [Google Scholar]
- 33.Shpakovski G V. The fission yeast Schizosacharomyces pombe rpb6 gene encodes the common phosphorylated subunit of RNA polymerase and complements a mutation in the corresponding gene of Saccharomyces cerevisiae. Gene. 1994;147:63–69. doi: 10.1016/0378-1119(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 34.Shpakovski G V, Acker J, Wintzerith M, Lacroix J F, Thuriaux P, Vigneron M. Four subunits shared by the three classes of RNA polymerases are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shpakovski G V, Gadal O, Labarre-Mariotte S, Lebedenko E N, Miklos I, Sakurai H, Proshkin S A, Van Mullem V, Ishihama A, Thuriaux P. Functional conservation of RNA polymerase II in fission and budding yeasts. J Mol Biol. 2000;295:1119–1127. doi: 10.1006/jmbi.1999.3399. [DOI] [PubMed] [Google Scholar]
- 36.Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol Gen Genet. 1993;239:169–176. doi: 10.1007/BF00281615. [DOI] [PubMed] [Google Scholar]
- 37.Thuillier V, Stettler S, Sentenac A, Thuriaux P, Werner M. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995;14:351–359. doi: 10.1002/j.1460-2075.1995.tb07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thuriaux P, Sentenac A. Yeast nuclear RNA polymerases. In: Jones E W, Pringle J R, Broach J R, editors. The molecular biology of yeast. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 1–45. [Google Scholar]
- 39.Valenzuela P, Bell G I, Weinberg F, Rutter W J. Yeast DNA-dependent RNA polymerases I, II and III. The existence of subunits common to the three enzymes. Biochem Biophys Res Commun. 1976;71:1319–1325. doi: 10.1016/0006-291x(76)90799-3. [DOI] [PubMed] [Google Scholar]
- 40.Voutsina A, Riva M, Carles C, Alexandraki D. Sequence divergence of the RNA polymerase shared subunit ABC14.5 (Rpb8) selectively affects RNA polymerase III assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:1047–1055. doi: 10.1093/nar/27.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittekind M, Dodd L, Vu J M, Kolb J M, Buhler J M, Sentenac A, Nomura M. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:3997–4008. doi: 10.1128/mcb.8.10.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woychik N A, Liao S M, Kolodziej P A, Young R A. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 1990;4:313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- 43.Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. . Present address: Department of Experimental Biology, University of Jaén, Paraje las Lagunillas, E-23071, Jaén, Spain. [DOI] [PubMed] [Google Scholar]