Abstract
Notch signaling dictates cell fate and critically influences cell proliferation, differentiation, and apoptosis in metazoans. Ligand binding initiates the signal through regulated intramembrane proteolysis of a transmembrane Notch receptor which releases the signal-transducing Notch intracellular domain (NICD). The HES/E(spl) gene family is a primary target of Notch and thus far the only known Notch effector. A newly isolated HERP family, a HES-related basic helix-loop-helix protein family, has been proposed as a potential target of Notch, based on its induction following NICD overexpression. However, NICD is physiologically maintained at an extremely low level that typically escapes detection, and therefore, nonregulated overexpression of NICD—as in transient transfection—has the potential of generating cellular responses of little physiological relevance. Indeed, a constitutively active NICD indiscriminately up-regulates expression of both HERP1 and HERP2 mRNAs. However, physiological Notch stimulation through ligand binding results in the selective induction of HERP2 but not HERP1 mRNA and causes only marginal up-regulation of HES1 mRNA. Importantly, HERP2 is an immediate target gene of Notch signaling since HERP2 mRNA expression is induced even in the absence of de novo protein synthesis. HERP2 mRNA induction is accompanied by specific expression of HERP2 protein in the nucleus. Furthermore, using RBP-Jk-deficient cells, we show that an RBP-Jk protein, a transcription factor that directly activates HES/E(spl) transcription, also is essential for HERP2 mRNA expression and that expression of exogenous RBP-Jk is sufficient to rescue HERP2 mRNA expression. These data establish that HERP2 is a novel primary target gene of Notch that, together with HES, may effect diverse biological activities of Notch.
The evolutionarily conserved Notch signaling pathway controls cell fate in metazoans through local cell-cell interactions (2, 14, 15). The transmembrane receptor Notch is activated by a cascade of regulated proteolytic cleavages (called Rip for regulated intramembrane proteolysis) (4) initiated by ligand binding, which liberates a cytosolic fragment, the Notch intracellular domain (NICD) (34, 41, 46). The NICD migrates into the nucleus, and nuclear NICD is physiologically maintained at an extremely low concentration that is below the threshold of current detection techniques (34, 41), indicating strict stoichiometric regulation of the signal. NICD associates with a number of proteins through its multiple protein-interacting domains (33).
Among them is a transcription factor, RBP-Jk [also called CSL/CBF1/Su(H)/Lag-1], that forms an NICD–RBP-Jk complex to up-regulate target gene expression (2, 14, 15, 33, 34). Thus, RBP-Jk when complexed with NICD acts as a transcriptional activator. However, RBP-Jk has a repression domain and can act as a transcriptional repressor from its DNA binding site by associating with a corepressor complex containing SMRT and a histone deacetylase, HDAC1, and this association is disrupted in the presence of NICD (22).
To date, the HES/E(spl) family of transcriptional repressors are the only known major target of the NICD–RBP-Jk complex (2, 14, 15, 20, 38), and they repress expression of downstream genes such as MASH1 (6, 12, 18) and neurogenin (1, 44). Thus, HES acts as a primary effector for Notch signaling and prevents cell differentiation. However, several studies show no or marginal induction of HES expression upon Notch stimulation by ligands such as Delta-like1 and Jagged1 (21, 27, 42). Furthermore, targeted disruption of Notch1 and RBP-Jk genes only partially affects the expression level or spatial distribution of HES in mouse embryos (12). These observations raise the possibility that yet-unidentified proteins may mediate the effects of Notch.
Several HES-related genes have been recently isolated, two of which we have named HERP1 and HERP2 (HES-related repressor protein) (also named Hesr/Hey/HRT/CHF/gridlock; Table 1 shows the relationship of different HERP family members). Their high sequence similarity with the HES/E(spl) family has raised the possibility that the HERP gene family might be new targets of Notch (Fig. 1; Table 2). In line with this, overexpression of NICD can stimulate expression of all studied HERP members in reporter gene assays following transient transfection (32, 35; our unpublished results). However, given the tight regulation that maintains native NICD at an extremely low concentration, the physiological relevance of such promiscuous HERP expression can only be determined by a study involving physiological Notch stimulation through ligand binding. Here, we show that ligand binding selectively induces endogenous HERP2 mRNA expression but not HERP1 mRNA, although overexpressed NICD indiscriminately induces expression of both. Surprisingly, HES1 mRNA was only marginally induced. Importantly, induction of HERP2 mRNA expression by ligand binding was observed even in the absence of de novo protein synthesis and was absolutely dependent on RBP-Jk. These findings provide the first evidence that HERP2 is a novel primary target of Notch that is directly up-regulated by a cellular signal transduction system (requiring RBP-Jk) activated by physiological Notch stimulation. The evidently stronger induction of HERP2 mRNA than of HES1 mRNA further supports the notion that HERP2 may play a more crucial role as a Notch effector.
TABLE 1.
HERP family nomenclature
| Full name | Abbreviationa
|
Reference | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| HES-related repressor protein | HERP1 | HERP2 | This paper | |
| Hairy/E(spl)-related | Hesr1 | 26 | ||
| Hairy/E(spl)-related with YRPW | Hey2 | Hey1 | HeyL | 29 |
| Hairy-related transcription factor | HRT2 | HRT1 | HRT3 | 36 |
| Cardiovascular helix-loop-helix factor | CHF1 | CHF2 | 8 | |
| gridlock | gridlock | 48 | ||
Each column shows proteins that are identical or homologous to other proteins in the column.
FIG. 1.
Conserved domains of HERP and HES with distinct features. Mouse HERP1, HERP2, and HES1 were aligned by Clustal W and presented using BOXSHADE. Identical amino acids are in black, and conserved residues are in gray. The basic helix-loop-helix dimerization domains and Orange repression domains (11) are indicated. An arrowhead indicates the invariant amino acid residues in the basic domain of HERP1 and HERP2 (glycine) and HES1 (proline). Asterisks indicate the conserved carboxyl-terminal tetrapeptide motifs, which are also divergent between the HERP (YRPW/YQPW) and HES (WRPW) families. The WRPW sequence of HES recruits the TLE/Groucho corepressor.
TABLE 2.
Summary of amino acid sequence similarity among mouse HES1 and human and mouse HERP1 and HERP2 proteins inferred from cDNA sequences
| Protein (no. of amino acids) | % Similarity with protein (no. of amino acids)
|
||
|---|---|---|---|
| Mouse HES1 (282) | Mouse HERP1 | Human HERP2 | |
| Human HERP1 (337) | 27 | 94 | 52 |
| Mouse HERP1 (339) | 30 | ||
| Human HERP2 (304) | 31 | ||
| Mouse HERP2 (299) | 33 | 53 | 94 |
MATERIALS AND METHODS
Cloning of HERP1 and HERP2 and sequence analysis.
The GenBank DNA sequence database was screened using the sequence corresponding to the basic helix-loop-helix region of mouse HES1. Three expressed sequence tag clones (GenBank AA116067, R60704, and W58864) were identified and named human HERP1, human HERP2, and mouse HERP2, respectively. Missing parts of cDNAs for all four clones (human and mouse HERP1 and HERP2) were isolated from cDNA libraries of human adult heart or mouse 17-day embryos (Clontech) by 5′ or 3′ rapid amplification of cDNA ends by the method described previously (3). The PCR products were verified by sequencing (Microchemical Core Facility, University of Southern California).
Plasmids, transfection, and Northern blot analyses.
Cells were transfected with the plasmids indicated in the figure legends with a 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid-buffered saline protocol as previously described (16). Parental pEF-BOS empty vector and the NICD (amino acids 1747 to 2531)-expressing vector were kindly provided by G. Weinmaster (37). Plasmids encoding wild-type RBP-Jk and R218H mutant were generously provided by T. Honjo (9). Total RNA was extracted with Trizol (Life Technologies). Aliquots (15 μg) of total RNA were size fractionated by electrophoresis, transferred onto Hybond-N+ membranes (Amersham), and then hybridized with radiolabeled probe. Radioactivities of each signal were measured by a PhosphorImager using ImageQuant software (STORM 840; Molecular Dynamics).
Probes used for the Northern blot were the following; nucleotides 576 to 1151 of mouse HERP1, nucleotides 465 to 1079 of mouse HERP2, and nucleotides 751 to 1085 of rat HES1. pSV2-CMV-HES1 was generously provided by R. Kageyama (40). The mouse glyceraldehyde-3-phosphate dehydrogenase probe was excised with HindIII and PstI and used as an internal control.
Coculture.
Jagged1-expressing L cells and Dll1-expressing QT6 cells were kindly provided by G. Weinmaster (30) and A. Israel (21), respectively. Jagged1-expressing L cells were cultured in growth medium consisting of Dulbecco Modified Eagle Medium supplemented with 10% fetal bovine serum. Dll1-expressing QT6 cells were grown in Ham F-10 medium supplemented with 1×tryptose phosphate, 5% fetal calf serum, and 1% chicken serum. An RBP-Jk−/− fibroblastic cell line (OT11) and a wild-type cell line (OT13) were kindly provided by T. Honjo (24) and were grown in Dulbecco Modified Eagle Medium supplemented with 10% fetal bovine serum and 100 U of mouse gamma interferon (Life Technologies) per ml. For coculture, C2C12, 10T1/2, OT11, or OT13 cells were plated in 100-mm-diameter tissue culture dishes in growth medium at a density such that they would be 70 to 80% confluent the next day. Then, either the Jagged1-or the Dll1-expressing cells were added at a density equal to the monolayer of these cells. Total RNA was extracted, and Northern blot analysis was performed as described above.
Immunohistochemistry.
An anti-HERP2 antibody was affinity purified from rabbit antisera directed against a HERP2-specific synthesized oligopeptide corresponding to the amino terminus of the mouse HERP2 sequence, DETIEVEKESADENG (Bethyl Laboratories). This antibody does not cross-react with either HERP1 or HES1 as confirmed by Western blot analysis (data not shown). For oligopeptide competition, the anti-HERP2 antibody was incubated with or without the specific oligopeptide (100 μg/ml) prior to use. Cells were cultured in two-well glass chamber slides and transiently transfected with the NICD expression vector. Three days later, cells were fixed, permeabilized, and incubated with goat anti-Notch1 antibody (2 μg/ml; C-20; Santa Cruz Biotech) plus the anti-HERP2 antibody (20 μg/ml), followed by fluorescein isothiocyanate-conjugated anti-goat immunoglobulin G (Vector Laboratories) and Cy3-conjugated anti-rabbit immunoglobulin G (Sigma). Signals were observed by confocal microscopy (LSM 510; Zeiss).
Reverse transcription-PCR (RT-PCR).
The first-strand cDNAs were made from 2 μg of total RNA from C2C12, 10T1/2, OT11, and OT13 cells using RETROscript (Ambion). Then, 1 μl of the reverse transcription reaction was used as a template for PCR amplification (Advantage2 polymerase mix; Clontech) in a volume of 50 μl containing 0.2 μM gene-specific primers. The gene-specific primers were the following: 5′-CCTTCCTAGGTGCTCTTGCG-3′ and 5′-TGCGGTCTGTCTGGTTGTGC-3′ for mNotch1, 5′-ACCCCTCCTGCTACCTGTCA-3′ and 5′-GATAGGGTCCCTTGGATGGC-3′ for mNotch2, and 5′-CAAATGGAGGTCGGTGCACCC-3′ and 5′-TGGGCTGCAGCTGACACTCAT-3′ for mNotch3. To exclude contaminating genomic DNA that would serve as a template, PCRs were performed both with and without reverse transcription. PCR products were run on a 1% agarose gel, and the images were captured under UV light. The PCR products were subjected to restriction digestion to verify their identities of distinct Notch receptors (data not shown).
Generation of recombinant adenoviruses.
Empty adenovirus (Adeno-empty) and adenoviruses expressing NICD and RBP-Jk (Adeno-NICD and Adeno-RBP-Jk, respectively) were generated by using the AD EASY Vector System as described elsewhere (17). This cloning system is based on homologous recombination in Escherichia coli. Briefly, cDNAs of NICD and RBP-Jk were subcloned into the KpnI-XbaI site and the XbaI-EcoRV site of pShuttle-CMV vector, respectively. The hemagglutinin-(HA) tag sequence was introduced at the amino terminus of RBP-Jk. Parental empty pShuttle-CMV and pShuttle-CMV vectors bearing either NICD or RBP-Jk cDNA were first linearized with PmeI, and then, together with adenoviral backbone vector pAdEasy, introduced into E. coli BJ5183 by electroporation. Recombinant adenoviral plasmids were recovered from E. coli and introduced into 293A cells by Lipofectamine (Life Technologies). The recombinant virus was propagated in 293A cells. The viruses were purified and concentrated by CsCl banding. High titers of these viruses (1.0 × 1010 to 4.0 × 1010 PFU/ml) were routinely obtained.
Nucleotide sequence accession number.
The accession numbers for the sequences in GenBank are as follows: human HERP1, AF232238; human HERP2, AF232239; mouse HERP1, AF232240; and mouse HERP2, AF232241.
RESULTS AND DISCUSSION
Both HERP1 and HERP2 mRNA expression are induced by overexpression of constitutively active NICD.
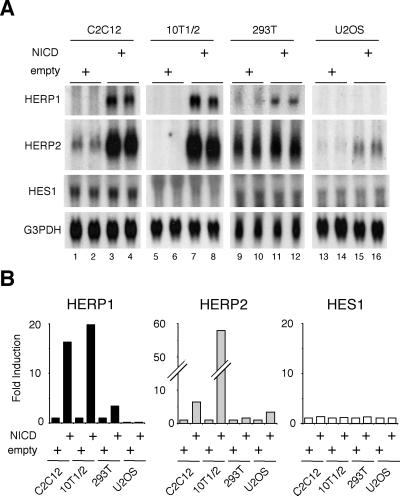
It has previously been shown that putative regulatory regions of HERP genes are responsive to overexpressed NICD in transient-transfection studies using reporter gene assays (32, 35; our unpublished results). We first studied whether endogenous HERP genes are similarly regulated by overexpressed NICD. Constitutively active NICD induced expression of both HERP1 and HERP2 mRNAs to different degrees depending on the cell type (Fig. 2A). Expression of both HERP mRNAs was up-regulated most strikingly in C2C12 cells (skeletal muscle) (Fig. 2A, lanes 1 to 4) and 10T1/2 cells (fibroblast) (lanes 5 to 8) but hardly at all in U2OS cells (osteosarcoma) (lanes 13 to 16), indicating cell-type-specific regulatory mechanisms. Interestingly, significant basal expression of HERP2 mRNA was observed in C2C12 (lanes 1 and 2) and 293T (lanes 9 and 10) cells but not in the others. Whether this reflects basal activities of Notch signals in these particular cells or the presence of a non-Notch pathway that maintains HERP2 basal expression remains to be clarified. In contrast, HES1 was only slightly up-regulated by overexpressed NICD in C2C12 cells but not up-regulated in any other cell types. The minor degree of HES1 up-regulation is consistent with previous studies (42). The Northern blot signals were quantitatively measured by a PhosphorImager, and the summary of the results is shown in Fig. 2B. Thus, both HERP1 and HERP2 impressively respond to NICD stimulation (up to 55-fold), whereas HES1 hardly does so at all in all the cell types tested, suggesting that HERP rather than HES1 may be a primary target of NICD in these cells.
FIG. 2.
The constitutively active form of Notch (NICD) indiscriminately up-regulates both HERP1 and HERP2. (A) C2C12, C3H10T1/2, 293T, and U2OS cells were transfected with 20 μg of pEF-BOS empty vector or NICD-expressing vector. Three days after the transfection, total RNA was extracted and Northern blot analysis was performed using the probes indicated in Materials and Methods. Data are from duplicate transfections. Note the marked induction of both HERP1 and HERP2 mRNA and marginal up-regulation of HES1. G3PDH, glyceraldehyde-3-phosphate dehydrogenase. (B) Radioactivities of each signal were measured.
Selective induction of HERP2 mRNA expression by ligand binding.
In marked contrast to a normally very low concentration of endogenous NICD, exogenously introduced NICD is expressed at easily detectable, supraphysiological levels (5) (see also Fig. 5A and E). Such unregulated promiscuous expression of NICD by transient transfection may cause artifactual biochemical and biological consequences derived from the multiple protein interaction domains present in NICD (33). We addressed this concern by studying whether natural stimulation of Notch receptors can induce HERP expression. Several Notch ligands and receptors have been identified for vertebrates (13, 43, 45, 46). Jagged1 activates both Notch1 and Notch2, whereas Delta-like 1 (Dll1) can activate only Notch1 efficiently (46). C2C12 cells naturally express Notch1, -2, and -3 receptors (31). It has previously been shown that coculture of ligand-expressing cells with C2C12 cells successfully stimulates Notch signaling and results in block of muscle differentiation by inhibiting myogenin and MLC2 expression (21, 30).
FIG. 5.
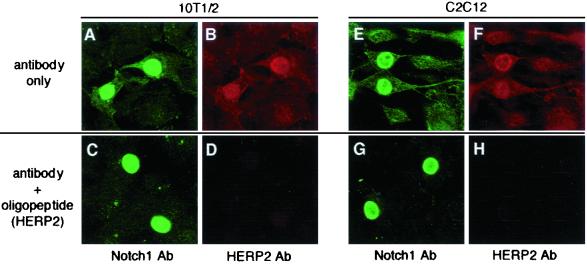
HERP2 protein accumulates in the nucleus by NICD stimulation. Cells were transiently transfected with an NICD expression vector, and a double-immunofluorescence assay was performed using specific anti-Notch1 antibody plus anti-HERP2 antibody. NICD shows as green; HERP2 shows as red. (A to D) 10T1/2 cells. (E to H) C2C12 cells. (C, D, G, and H) An anti-HERP2 antibody is blocked by the HERP2 oligopeptide competitor. Two irrelevant oligopeptides (specific for HERP1) did not block the HERP2 protein detection (data not shown). Ab, antibody.
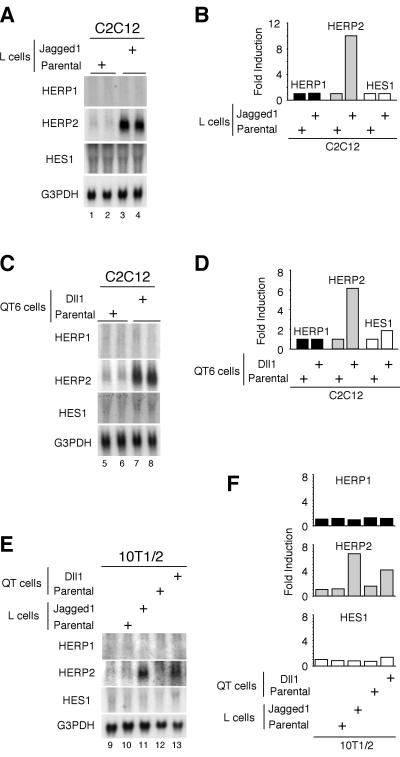
We used the same coculture approach to determine whether HERPs are induced by Notch stimulation with either of the two Notch ligands, Jagged1 and Dll1 (Fig. 3). Surprisingly, despite the strong induction of HERP1 mRNA by overexpressed NICD (Fig. 2), coculture of C2C12 cells with Jagged1-expressing cells did not induce HERP1 mRNA expression (Fig. 3A, lanes 1 to 4). In contrast, HERP2 mRNA expression was strongly induced by coculture. The absence of HERP1 induction, however, might reflect the presence of ligand-specific regulation for different members of the HERP family. Therefore, we next employed a different cell type that expresses another Notch ligand, Dll1, which showed essentially an identical result (Fig. 3C). A low level of HES1 was constitutively expressed in C2C12 cells, but either no (Fig. 3A) or only weak (Fig. 3C) induction was observed with Jagged1 or Dll1, respectively. The selective induction of HERP2 mRNA expression was not limited to C2C12 cells, as we obtained essentially the same results by coculturing 10T1/2 cells with Jagged1- or Dll1-expressing cells (Fig. 3E). Consistent with these findings, we found that 10T1/2 cells, like C2C12 cells, expressed at least three known mammalian Notch receptors (see Fig. 8). These results (summarized in Fig. 3B, D, and F) strongly suggest that coculture successfully stimulated Notch signaling and that only HERP2, not HERP1, is a physiological target of Notch signaling stimulated by Jagged1 and Dll1. Furthermore, the result clearly indicates the distinct nature of ligand-induced versus NICD overexpression-induced stimuli and justifies a cautionary note for interpreting the results of such experiments as that presented in Fig. 2A and in published work from other laboratories (32, 35). It remains to be determined whether HERP1 expression is regulated by other subtypes of Notch ligands such as Jagged2 and Dll2/3/4, and/or in other cell types, or by non-Notch signaling. These possibilities are now under investigation.
FIG. 3.
Selective up-regulation of HERP2 mRNA following stimulation by Notch ligands. (A to D) C2C12 cells were cocultured with either parental or Jagged1-expressing L cells (A and B) and parental or Dll1-expressing QT6 cells (C and D). Data are from duplicate cocultures. (E) 10T1/2 cells were cocultured with either parental or Jagged1-expressing L cells and parental or Dll1-expressing QT6 cells. Twenty-four hours later, total RNA was extracted and Northern blot analysis was performed. Note that HERP2 mRNA is selectively up-regulated in both C2C12 and 10T1/2 cells. (B, D, and F) The radioactivities of each signal were measured. G3PDH, glyceraldehyde-3-phosphate dehydrogenase.
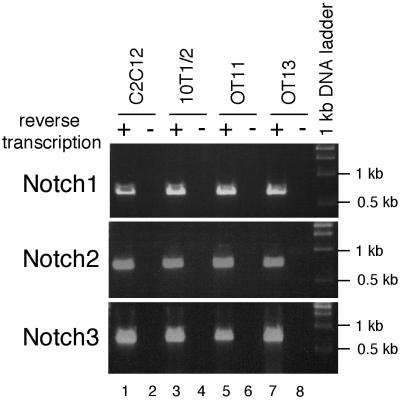
FIG. 8.
Notch receptors are expressed in RBP-Jk-deficient cells and the other cells tested. Two micrograms of total RNA from each indicated cell line was reverse transcribed and PCR amplified with gene-specific primers as described in Materials and Methods. Samples without reverse transcription were used as negative controls to show that PCR products were not derived from contaminating genomic DNA. Sizes of PCR products are 678 bp for Notch1, 728 bp for Notch2, and 789 bp for Notch3. Note that all cells tested expressed Notch1, -2 and -3 receptors.
HERP2 is an immediate target of Notch signaling.
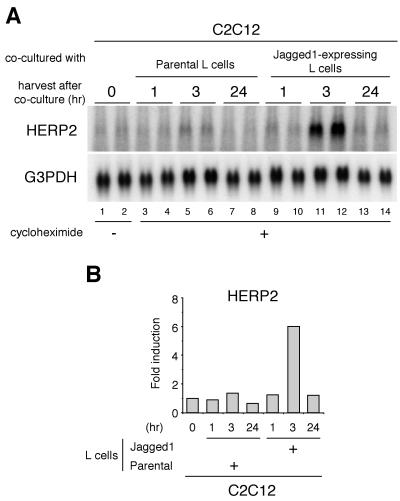
While these findings demonstrate that HERP2 is a main physiological target of Notch, they do not establish whether HERP2 mRNA expression is directly regulated by signal transduction by Notch or requires the expression of an additional gene(s). To address this, we studied HERP2 mRNA induction in the presence of cycloheximide (CHX) to block de novo protein synthesis. C2C12 cells were cocultured with Jagged1-expressing cells in the presence of CHX for various periods, and HERP2 mRNA expression was studied at each time point. A slight HERP2 mRNA induction was observed as early as 1 h after the coculture (Fig. 4A, compare lanes 3 and 4 and lanes 9 and 10), and maximum HERP2 expression was achieved at 3 h (Fig. 4A, lanes 11 and 12). At 24 h, expression of HERP2 mRNA was diminished, presumably due to depletion of general cellular proteins including those involved in Notch signaling in both the Jagged1-expressing L cells and C2C12 cells after the extended CHX treatment. These findings establish that HERP2 is a primary and direct target of ligand-stimulated Notch signaling and is not indirectly induced by the up-regulation of another gene and its protein product.
FIG. 4.
Ligand stimulation induces HERP2 mRNA without de novo protein synthesis. (A) C2C12 cells were cocultured with parental or Jagged1-expressing L cells in the presence of 10 μM CHX. Cells were harvested at indicated time points, and total RNA was extracted for Northern blot analysis. Data are from duplicate cocultures. G3PDH, glyceraldehyde-3-phosphate, dehydrogenase. (B) Radioactivities of each signal were measured.
HERP2 protein accumulates in the nucleus following NICD expression.
Since mRNA expression is not always coupled with expression of the corresponding protein (10), we evaluated whether HERP2 mRNA up-regulation is accompanied by expression of the cognate protein. For this purpose, we generated an antibody specific to HERP2. Cells transiently transfected with the NICD expression vector were subjected to double immunofluorescence with anti-Notch1 and anti-HERP2 antibodies (Fig. 5). HERP2 expression was clearly detected only in the nuclei of Notch1-positive cells (Fig. 5B and F). When the HERP2 antibody was competed with the specific oligopeptide against which the antibody was generated, the HERP2 signal became undetectable (Fig. 4D and H), although Notch1 signals were largely unaffected (Fig. 5C and G), indicating specific HERP2 protein expression. An irrelevant oligopeptide (from the HERP1 sequence) did not compete the HERP2 signal (data not shown). We have been unable to detect significant HERP2 staining following coculture with the ligand-expressing cells (data not shown), suggesting a low protein expression level. These data showed that the HERP2 protein is accumulated specifically in the cell nucleus, which is consistent with its expected role as a transcription factor.
Dominant negative RBP-Jk fails to verify a role for RBP-Jk in HERP2 mRNA expression.
The observed rise in HERP2 mRNA expression could be due to accelerated transcription and/or mRNA stabilization. Putative HERP promoters have consensus DNA sequences for RBP-Jk binding. Mutations of these sites resulted in the reduced gene expression in transiently transfected luciferase reporter gene assays, suggesting that RBP-Jk may be involved in the regulation of HERP gene transcription (32, 35; our unpublished results).
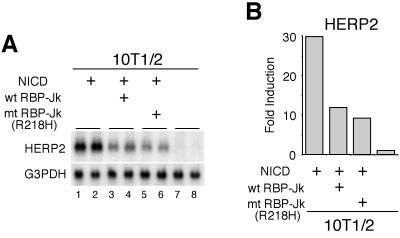
However, it remains to be determined whether endogenous HERP mRNA expression is regulated by RBP-Jk protein. To address this, we first employed a dominant negative RBP-Jk protein (RBP-Jk R218H) which does not bind DNA (9) and studied whether this RBP-Jk mutant can abolish HERP2 mRNA expression. As expected, HERP2 mRNA expression was reduced by expressing the R218H mutant (Fig. 6A, compare lanes 1 and 2 and lanes 5 and 6). Surprisingly, however, a similar degree of reduction was observed also following expression of wild-type exogenous RBP-Jk protein (wt RBP-Jk in Fig. 6A, lanes 3 and 4), presumably due to its uncontrolled expression, which squelches the signaling molecules and disrupts their proper stoichiometry (see also the discussion for Fig. 10 below). Consistent with this findings is that an overexpressed wild-type RBP-Jk protein functions as a transcriptional repressor on different promoters even under the condition where Notch signaling is stimulated (7, 23, 39, 47). Thus, a role for RBP-Jk in HERP mRNA expression remains inconclusive from studies using the dominant negative RBP-Jk.
FIG. 6.
HERP2 mRNA induction is equally reduced by overexpression of either wild-type or dominant negative RBP-Jk. (A) C3H10T1/2 cells were transfected with 4 μg of pEF-BOS NICD-expressing vector plus expressing vectors for 16 μg of either wild-type RBP-Jk or dominant negative R218H mutant as indicated. Three days after the transfection, total RNA was extracted and Northern blot analysis was performed. Data are from duplicate transfections. Note that the induction of HERP2 mRNA by NICD is equally suppressed by wild-type RBP-Jk and by the R218H mutant. G3PDH, glyceraldehyde-3-phosphate dehydrogenase. (B) The radioactivity of each signal was measured. wt, wild type; mt, mutant.
FIG. 10.
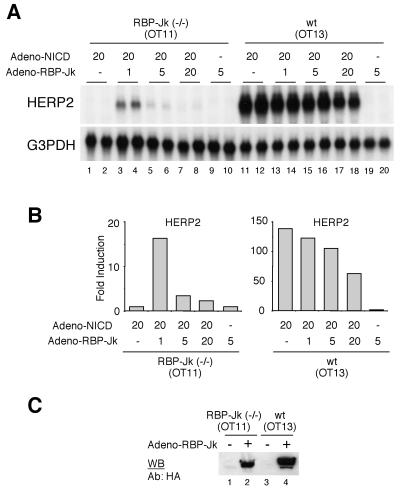
Rescue of HERP2 mRNA induction by exogenous RBP-Jk expression in RBP-Jk-deficient OT11 cells. (A) OT11 and OT13 cells were infected with recombinant adenovirus at the indicated MOI. Forty-eight hours later, total RNA was extracted and Northern blot analysis was performed. Data are from duplicate infections. Note that HERP2 mRNA is reinduced in OT11 cells only by coinfection with Adeno-NICD and Adeno-RBP-Jk at a low MOI. G3PDH, glyceraldehyde-3-phosphate, dehydrogenase. (B) The radioactivity of each signal was measured. (C) OT11 and OT13 cells were infected with Adeno-RBP-Jk at an MOI of 5. Nuclear proteins were extracted from infected and noninfected cells as described (19). Expression of RBP-Jk protein in OT11 cells as well as OT13 cells was confirmed by Western blot (WB) analysis using anti-HA antibody (Y-11; Santa Cruz Biotech). wt, wild type.
RBP-Jk-deficient cells do not express HERP2 mRNA in response to ligand binding.
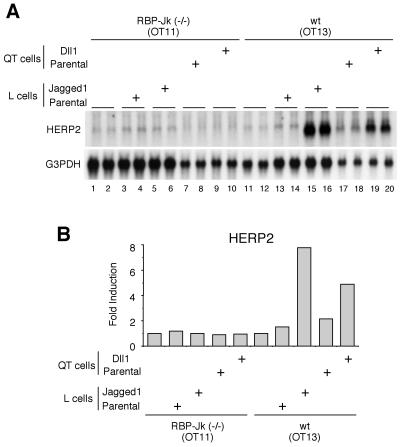
To address the role of RBP-Jk in HERP2 gene regulation, we next studied whether HERP2 mRNA expression can be induced in the absence of an RBP-Jk protein using RBP-Jk-deficient cells (OT11) derived from homozygous RBP-Jk null mice (24). Neither coculture with Jagged1-expressing cells nor coculture with Dll1-expressing cells induced HERP2 mRNA expression in these RBP-Jk-deficient cells (Fig. 7A, lanes 5, 6, 9, and 10). Interestingly, we did notice a low level of HERP2 mRNA expression without coculture in OT11 cells (lanes 1 and 2), suggesting the presence of an RBP-Jk-independent mechanism for basal HERP2 mRNA expression. In contrast, RBP-Jk-positive wild-type cells (OT13) showed a marked induction of HERP2 mRNA expression by coculture with either Jagged1- or Dll1-expressing cells (lanes 15, 16, 19, and 20). These data suggest that HERP2 mRNA expression is induced by ligand binding only in the presence of RBP-Jk protein.
FIG. 7.
RBP-Jk-deficient OT11 cells do not express HERP2 mRNA in response to Notch ligand stimulation. (A) RBP-Jk-deficient OT11 or wild-type OT13 cells were cocultured with either parental or Jagged1-expressing L cells and parental or Dll1-expressing QT6 cells as indicated. Twenty-four hours later, total RNA was extracted and Northern blot analysis was performed. Data are from duplicate cocultures. Note that HERP2 mRNA is up-regulated by ligand stimulation only in OT13 cells. G3PDH, glyceraldehyde-3-phosphate dehydrogenase. (B) The radioactivity of each signal was measured. wt, wild type.
Notch receptors are expressed in RBP-Jk-deficient OT11 cells.
The observed lack of HERP2 mRNA induction by ligand stimulation in OT11 cells could be due to additional phenotypic changes such as absence of functional Notch receptors. To address this point, we performed RT-PCR for the known mammalian Notch receptor mRNAs in OT11 and OT13 cells. We found that transcripts for at least three Notch receptors (Notch1, -2, and -3) were expressed in both OT11 and OT13 cells (Fig. 8, lanes 5 and 7). Although specific interactions between different Notch ligands and receptors have not been rigorously delineated, exogenously introduced Notch1 receptor can confer on cells responsiveness to Dll1 ligand (21), suggesting that Dll1 can bind Notch1 receptor. Therefore, our data suggest that the lack of HERP2 mRNA expression in OT11 cells is not caused by the absence of functional Notch receptors. The absence of PCR product without reverse transcription (lanes 6 and 8) excluded a contribution of contaminating genomic DNA as a PCR template. Additional RT-PCR studies using RNA from C2C12 and 10T1/2 cells demonstrated expression of three Notch receptors in these cells (lanes 1 to 4).
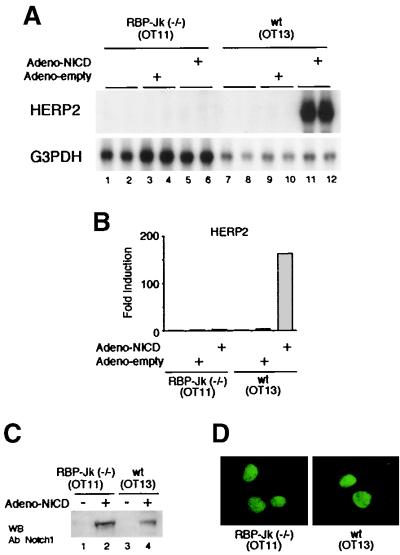
Constitutively active NICD does not induce HERP2 mRNA in RBP-Jk-deficient OT11 cells.
Although our data show that OT11 cells express Notch receptors, subsequent steps of Notch signaling such as regulated intramembrane proteolysis (Rip) of the Notch receptor upon ligand binding might be compromised in these cells, which could explain the observed absence of HERP2 mRNA induction (Fig. 7A). To bypass the process of Rip, we expressed constitutively active NICD using recombinant adenovirus, which infected most cells at the multiplicity of infection (MOI) used. Importantly, HERP2 mRNA expression was not induced by the NICD introduced by Adeno-NICD infection in RBP-Jk-deficient OT11 cells (Fig. 9A, lanes 5 and 6), whereas wild-type OT13 cells showed a dramatic HERP2 mRNA induction by the same stimulation (lanes 11 and 12). Infection with the Adeno-empty virus did not affect the expression level of HEPR2 mRNA in either OT11 or OT13 cells (Fig. 9A, lanes 3, 4, 9, and 10).
FIG. 9.
Forced expression of constitutively active NICD does not induce HERP2 mRNA expression in RBP-Jk-deficient OT11 cells. (A) OT11 and OT13 cells were infected with Adeno-empty or Adeno-NICD at an MOI of 20. Forty-eight hours later, total RNA was extracted and Northern blot analysis was performed. Data are from duplicate infections. Note the impressive induction of HERP2 mRNA by Adeno-NICD only in OT13 cells, not in OT11 cells. G3PDH, glyceraldehyde-3-phosphate dehydrogenase. (B) The radioactivity of each signal was measured. (C) OT11 and OT13 cells were infected with Adeno-NICD at an MOI of 20. Nuclear proteins were extracted from infected and noninfected cells as described (19). A high-level expression of NICD protein in OT11 cells as well as OT13 cells was confirmed by Western blot (WB) analysis using anti-Notch1 antibody (C-20; Santa Cruz Biotech). (D) OT11 and OT13 cells were infected with Adeno-NICD at an MOI of 5. Forty-eight hours later, an immunofluorescence assay was performed using the anti-Notch1 antibody as described in Materials and Methods. Note that only nuclei were equally stained in OT11 and OT13 cells. wt, wild type.
In this study, NICD was expressed in OT11 cells at least at a level comparably high with that of OT13 cells, as determined by Western blot analysis (Fig. 9C, lanes 2 and 4), and therefore, poor NICD expression was not a reason for the lack of HERP2 mRNA expression in OT11 cells. It has previously been suggested for Drosophila melanogaster that Suppressor of Hairless (RBP-Jk homologue) plays a role in nuclear entry of NICD (25). This raises the possibility that the inability of HERP2 mRNA to be expressed in OT11 cells may be due to impaired nuclear entry of NICD because of a lack of RBP-Jk. However, we observed similar degrees of NICD protein in the nucleus of OT11 and OT13 cells by immunofluorescence (Fig. 9D). Thus, impaired nuclear translocation is not a reason for the failure of NICD to induce HERP2 mRNA expression in OT11 cells, either.
Expression of RBP-Jk is sufficient to rescue HERP2 mRNA induction in RBP-Jk-deficient OT11 cells.
The absence of HERP2 mRNA induction in OT11 cells (Fig. 9A and B) could be due to phenotypic changes that are not directly associated with a lack of RBP-Jk. Therefore, we attempted to rescue the inability of OT11 cells to induce HERP2 mRNA expression by simply providing exogenous RBP-Jk protein. OT11 cells were infected at different MOIs with recombinant adenovirus carrying RBP-Jk cDNA. Neither NICD nor RBP-Jk alone induced HERP2 mRNA expression in OT11 cells (Fig. 10A, lanes 1, 2, 9, and 10). Strikingly, when both NICD and RBP-Jk were coexpressed, HERP2 mRNA expression was clearly induced (lanes 3 and 4). Thus, the expression of RBP-Jk protein was sufficient to rescue HERP2 mRNA induction in OT11 cells. When RBP-Jk protein was excessively expressed by infecting the cells at a higher MOI, we consistently observed a reduced HERP2 mRNA induction in both OT11 cells (lanes 5 to 8) and OT13 cells (lanes 13 to 18), indicating that HERP2 expression is tightly controlled by a proper concentration of an RBP-Jk protein. Although the exact reasons for the reduced HERP2 mRNA expression at a higher MOI remain to be clarified, it might be due to sequestration by excess RBP-Jk of positive cofactors such as SKIP, which binds RBP-Jk and activates NICD function (49), and/or to facilitation of the association of RBP-Jk with the SMRT/HDAC1-containing corepressor complex (22), which may interfere with the function of NICD–RBP-Jk in a dominant negative fashion. Expression of the HA–RBP-Jk protein was confirmed by Western blot analysis (Fig. 10C).
To date, only HES/E(spl) has been implicated as a primary target gene of Notch signaling (2, 14, 15). However, its role in the Notch pathway in mammalian cells is not entirely understood. Our demonstration that HERP2 expression is directly induced by ligand stimulation provides for a new, alternative Notch pathway. The striking induction of HERP2 expression would enable HERP2 to greatly amplify Notch signals and raises the intriguing possibility that HERP2 might be a primary effector for certain biological activities of Notch, such as the inhibition of muscle differentiation. In agreement with this idea is the genetic evidence which shows that HERP2 expression is perturbed in certain (but not all) tissues including presomitic mesoderm and nascent somites of Dll1 and Notch1 null mutant mice (26, 28).
The differential expression of HES and HERP in certain tissues (8, 26, 29, 36, 40) supports the idea that they may work separately in distinct cell types. Alternatively, the observed simultaneous expression of both HES and HERP in a single cell line (i.e., C2C12 muscle cells) raises the question whether they might independently regulate different sets of downstream genes or whether there might be cross talk between them for the regulation of common target genes. In this regard, we have recently shown that HERP and HES associate with each other in solution and form a stable heterodimer upon binding to specific DNA sequences with a markedly elevated DNA binding activity that results in a synergistic repression of target gene expression (19). These findings further reinforce the idea that HERP2 cooperates with HES as a new effector of Notch signaling. The Notch pathway involves multiple ligands and receptors. The identification of HERP2 as a new target of Notch adds another degree of regulation and opens a new avenue to our understanding of this exquisitely regulated signaling pathway.
ACKNOWLEDGMENTS
We are grateful to Gerry Weinmaster, Alain Israel, Tasuku Honjo, Ryoichiro Kageyama, and Bert Vogelstein for critical reagents. We thank T. Saluna for technical assistance and members of IGM for useful discussions. T.I. thanks Nobuko I. for her understanding and encouragement. Y.H. thanks M. D. Schneider, R. J. Schwartz, and A. I. Schafer for support.
This work was supported by a research fellowship from the American Heart Association, Western States Affiliate (to T.I.), and in part by grants from the National Institutes of Health (to L.K.).
REFERENCES
- 1.Apelqvist A, Li H, Sommer L, Beatus P, Anderson D J, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Brown M S, Ye J, Rawson R B, Goldstein J L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 5.Capobianco A J, Zagouras P, Blaumueller C M, Artavanis-Tsakonas S, Bishop J M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, Thiagalingam A, Chopra H, Borges M W, Feder J N, Nelkin B D, Baylin S B, Ball D W. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc Natl Acad Sci USA. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Fischer W H, Gill G N. Regulation of the ERBB-2 promoter by RBPJkappa and NOTCH. J Biol Chem. 1997;272:14110–14114. doi: 10.1074/jbc.272.22.14110. [DOI] [PubMed] [Google Scholar]
- 8.Chin M T, Maemura K, Fukumoto S, Jain M K, Layne M D, Watanabe M, Hsieh C M, Lee M E. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J Biol Chem. 2000;275:6381–6387. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- 9.Chung C N, Hamaguchi Y, Honjo T, Kawaichi M. Site-directed mutagenesis study on DNA binding regions of the mouse homologue of Suppressor of Hairless, RBP-J kappa. Nucleic Acids Res. 1994;22:2938–2944. doi: 10.1093/nar/22.15.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daubas P, Tajbakhsh S, Hadchouel J, Primig M, Buckingham M. Myf5 is a novel early axonal marker in the mouse brain and is subjected to post-transcriptional regulation in neurons. Development. 2000;127:319–331. doi: 10.1242/dev.127.2.319. [DOI] [PubMed] [Google Scholar]
- 11.Dawson S R, Turner D L, Weintraub H, Parkhurst S M. Specificity for the Hairy/Enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Pompa J L, Wakeham A, Correia K M, Samper E, Brown S, Aguilera R J, Nakano T, Honjo T, Mak T W, Rossant J, Conlon R A. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 13.Dunwoodie S L, Henrique D, Harrison S M, Beddington R S. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124:3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- 14.Egan S E, St.-Pierre B, Leow C C. Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr Top Microbiol Immunol. 1998;228:273–324. doi: 10.1007/978-3-642-80481-6_11. [DOI] [PubMed] [Google Scholar]
- 15.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 16.Hamamori Y, Wu H Y, Sartorelli V, Kedes L. The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, Twist. Mol Cell Biol. 1997;17:6563–6573. doi: 10.1128/mcb.17.11.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishibashi M, Ang S L, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 19.Iso T, Sartorelli V, Poizat C, Iezzi S, Wu H-Y, Chung G, Kedes L, Hamamori Y. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol Cell Biol. 2001;21:6080–6089. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 21.Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong C F, Brou C, Seidah N G, Israel A. Delta-1 activation of Notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao H Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato H, Sakai T, Tamura K, Minoguchi S, Shirayoshi Y, Hamada Y, Tsujimoto Y, Honjo T. Functional conservation of mouse Notch receptor family members. FEBS Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- 24.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 25.Kidd S, Lieber T, Young M W. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kokubo H, Lun Y, Johnson R L. Identification and expression of a novel family of bHLH cDNAs related to Drosophila hairy and enhancer of split. Biochem Biophys Res Commun. 1999;260:459–465. doi: 10.1006/bbrc.1999.0880. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 28.Leimeister C, Dale K, Fischer A, Klamt B, de Angelis M H, Radtke F, McGrew M J, Pourquie O, Gessler M. Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev Biol. 2000;227:91–103. doi: 10.1006/dbio.2000.9884. [DOI] [PubMed] [Google Scholar]
- 29.Leimeister C, Externbrink A, Klamt B, Gessler M. Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech Dev. 1999;85:173–177. doi: 10.1016/s0925-4773(99)00080-5. [DOI] [PubMed] [Google Scholar]
- 30.Lindsell C E, Shawber C J, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 31.Luo B, Aster J C, Hasserjian R P, Kuo F, Sklar J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol. 1997;17:6057–6067. doi: 10.1128/mcb.17.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maier M M, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun. 2000;275:652–660. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- 33.Miele L, Osborne B. Arbiter of differentiation and death: Notch signaling meets apoptosis. J Cell Physiol. 1999;181:393–409. doi: 10.1002/(SICI)1097-4652(199912)181:3<393::AID-JCP3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 34.Mumm J, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa O, McFadden D G, Nakagawa M, Yanagisawa H, Hu T, Srivastava D, Olson E N. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci USA. 2000;97:13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa O, Nakagawa M, Richardson J A, Olson E N, Srivastava D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- 37.Nofziger D, Miyamoto A, Lyons K M, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 38.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oswald F, Liptay S, Adler G, Schmid R M. NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol Cell Biol. 1998;18:2077–2088. doi: 10.1128/mcb.18.4.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 41.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 42.Shawber C, Nofziger D, Hsieh J J, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 43.Shutter J R, Scully S, Fan W, Richards W G, Kitajewski J, Deblandre G A, Kintner C R, Stark K L. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14:1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 44.Takke C, Dornseifer P, von Weizsacker E, Campos-Ortega J A. her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development. 1999;126:1811–1821. doi: 10.1242/dev.126.9.1811. [DOI] [PubMed] [Google Scholar]
- 45.Weinmaster G. The ins and outs of notch signaling. Mol Cell Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- 46.Weinmaster G. Notch signaling: direct or what? Curr Opin Genet Dev. 1998;8:436–442. doi: 10.1016/s0959-437x(98)80115-9. [DOI] [PubMed] [Google Scholar]
- 47.Wettstein D A, Turner D L, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- 48.Zhong T P, Rosenberg M, Mohideen M A, Weinstein B, Fishman M C. gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]
- 49.Zhou S, Fujimuro M, Hsieh J J-D, Chen L, Miyamoto A, Weinmaster G, Hayward S D. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol Cell Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]