Abstract
Bacillus subtilis is a widely used bacterial model to decipher biofilm formation, genetic determinants and their regulation. For several years, studies were conducted on colonies or pellicles formed at the interface with air, but more recent works showed that non-domesticated strains were able to form thick and structured biofilms on submerged surfaces. Taking advantage of time-lapse confocal laser scanning microscopy, we monitored bacterial colonization on the surface and observed an unexpected biphasic submerged biofilm development. Cells adhering to the surface firstly form elongated chains before being suddenly fragmented and released as free motile cells in the medium. This switching coincided with an oxygen depletion in the well which preceded the formation of the pellicle at the liquid-air interface. Residual bacteria still associated with the solid surface at the bottom of the well started to express matrix genes under anaerobic metabolism to build the typical biofilm protruding structures.
Keywords: Bacillus subtilis, Biofilm, Pellicle, 4D-CLSM, Motility, Oxygen, Metabolism
1. Introduction
In their natural habitat, bacteria mostly live in biofilms, associated with surfaces and embedded in a complex mixture of exopolymers [1]. These structures provide to their inhabitants a protective environment in which they can resist harsh conditions such as desiccation, nutrients starvation or the action of toxic compounds [2]. Microbial biofilms can be considered useful since they are involved in natural biogeochemical cycles and increasingly used in biotechnologies for wastewater treatments or the production of green energies [3]. However, biofilms also facilitate pathogen persistence despite antimicrobial treatments and thus have a severe negative impact in human health being involved in up to 80% of chronic and recurrent infections [4].
These infectious concerns have driven the international research efforts for more than 30 years to unravel the mechanisms of biofilm formation and their control [[5], [6], [7]]. Most of the pioneer studies in this field have been carried out with axenic biofilms of pathogenic strains, such as Pseudomonas aeruginosa, grown in flow-cells and observed by in situ confocal imaging [8]. In the early 2000s, Bacillus subtilis emerged as a model of Gram-positive bacteria for the dissection of the genetic determinants of biofilm formation and their regulation [9,10]. Most of these studies used the strain NCIB3610, that has been shown to form spatially organized multicellular structures i.e. colony on agar, floating pellicle at the liquid-air interface, and submerged biofilm at the solid-liquid interface [9,[11], [12], [13]]. These experimental models have then been successfully used to dissect the complexity of B. subtilis multicellularity and their genetic circuits to identify the regulators controlling biofilm development, maturation and dispersion (e.g. Spo0A, SinR, SinI, AbrB, SlrR or SigB) [12,[14], [15], [16], [17], [18], [19]]. In response to external signals, individual motile cells switch to sessile chains by inactivating expression of the motility genes (hag, encoding the principal flagellar protein and lytABC and lytF, encoding the autolysins responsible of cell separation) and by activation of the matrix production genes [[20], [21], [22]]. This matrix of B. subtilis biofilms is essentially composed of the polysaccharides synthesized by the products of the 15-genes operon epsA-O (eps operon), the amyloid-like protein TasA, synthesized from the tapA-sipW-tasA operon, and the amphiphilic protein BslA [[23], [24], [25], [26], [27]].
Focusing on non-domesticated B. subtilis strains isolated from food or from medical environments, we showed that some strains, especially the NDmed strain isolated from an endoscope washer-disinfector, were able to form thick and structured biofilms on submerged surfaces [28]. Moreover, NDmed was able to protect Staphylococcus aureus from biocide action in a submerged mixed-species biofilm [29]. The gene ypqP (renamed spsM [30]) likely involved in the synthesis of polysaccharide, is inactivated in both the model strain NCIB3610 and the lab strain 168, and was identified as being responsible for these features of NDmed submerged biofilms [13].
These works point out the importance of the submerged biofilm as a model of growth in the study of B. subtilis social behavior. Moreover B. subtilis submerged biofilms are also considered to be representative of other B. subtilis natural habitats such as soil and plant roots surface [31,32]. Nevertheless, little is still known about the genetic pathways involved in the formation of B. subtilis submerged biofilms [28,33,34].
To better understand the molecular strategies that bacteria undergo to build biofilms, we used 4D confocal laser scanning imaging (4D-CLSM) to visualize, in situ, in time-lapse and at cell level, the biofilm structural dynamics. By this approach, we have discovered that the transition from sessile cells to a highly structured biofilm of B. subtilis on the submerged level involves an unexpected sudden and coordinated fragmentation of the sessile population to a motile one. The latter event has been shown to be closely connected with the pellicle formation at the liquid-air interface and with transition from aerobic to anaerobic metabolisms. This work points out sophisticated programs of cellular specialization and cell-cell communication within the microbial community.
2. Materials and methods
2.1. Bacterial strains and growth conditions
The strains used during this study are listed in Table 1. B. subtilis NDmed derivative strains were obtained by transformation with chromosomal DNA of various strains to introduce the corresponding suitable reporter fusion. Extraction of chromosomal DNA and transformation of B. subtilis were performed as described previously [34]; transformants were selected on Luria-Bertani (LB, Sigma, France) plates supplemented with appropriate antibiotics at the following concentrations: spectinomycin (spec), 100 μg/mL; chloramphenicol (cm), 5 μg/mL kanamycin (kan), 8 μg/ml. The B. subtilis strain GM2938 expressing mCherry was obtained by transforming for spectinomycin resistance strain BSB168, a trp + derivative of the reference strain 168 Marburg [37] with plasmid pIC630. This results in the integration by a double crossing over into the chromosomal amyE locus of the mCherry gene placed under the control of the Phyperspank promoter, an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter derived from the Escherichia coli lac operon. Plasmid pIC630 was constructed by placing the mCherry gene (codon-optimized for B. subtilis) under the control of Phyperspank through cloning into pDR111 of a HindIII–SphI restriction fragment obtained from a PCR on plasmid pDR201 with primers DC014 (5’-CCCAAGCTTACATAAGGAGGAACTACTATG-3’) and DC015 (5’-ACATGCATGCTTATTTGTATAATTC-3’) (both pDR111 and pDR201 are kind gifts from D. Rudner, Harvard Medical School). A similar IPTG-inducible fusion of the gfpmut2 gene under the control of the Phyperspank promoter was introduced into NDmed to give the B. subtilis strain NDmed-GFP expressing GFPmut2. The transcriptional fusions of the fnr or gapB promoter with gfpmut3 were constructed within the pBaSysBioII plasmid using ligation-independent cloning prior to integration into the chromosome of BSB168 in a non-mutagenic manner, resulting in strains BBA0184 and BBA9006, respectively [37,38]. Bacterial stock cultures were kept at −20°C in Tryptone Soy Broth (TSB, bioMerieux, France) containing 20% (vol/vol) glycerol. Prior to each experiment, frozen cells were sub-cultured twice in TSB at 30°C. The final overnight culture was used as an inoculum for the growth of biofilms.
Table 1.
Strains used in this study.
| Strain | Relevant genotype or isolation source | Reference or constructiona |
|---|---|---|
| B. subtilis NDmed | Undomesticated, isolated from endoscope washer-disinfectors | [35] |
| B. subtilis NDmed-GFP | NDmed amyE::Phyperspank-gfpmut2 (spec) | [29] |
| B. subtilis GM2938 | BSB168 amyE::Phyperspank-mCherry (spec) | This work |
| B. subtilis NDmed-mCherry | NDmed amyE::Phyperspank-mCherry (spec) | TF NDmed/DNA GM2938 |
| B. subtilis TMN547 | NCIB3610 amyE::Phag-gfp (cm) sacA::PtapA-mKate2 (kan) | [36] |
| B. subtilis NDmed 547 | NDmed amyE::Phag-gfp (cm) sacA::PtapA-mKate2 (kan) | TF NDmed/DNA TMN547 |
| B. subtilis BBA9006 | BSB168 PgapB-gfpmut3 (spec) | [37] |
| B. subtilis GM3378 | NDmed PgapB-gfpmut3 (spec) | TF NDmed/DNA BBA9006 |
| B. subtilis BBA0184 | BSB168 Pfnr-gfpmut3 (spec) | [38] |
| B. subtilis GM3361 | NDmed Pfnr-gfpmut3 (spec) | TF NDmed/DNA BBA0184 |
| B. subtilis 168 | trpC2 (Domesticated strain) | Bacillus genetics Stock Center |
| B. subtilis BSB168 | trp + derivative of 168 | [37] |
| B. subtilis NCIB3610 | Less domesticated strain | [9] |
| B. subtilis NDfood | Isolated from a dairy product | [28] |
| B. subtilis BSn5 | Isolated from a plant | [39] |
| B. subtilis BSP1 | Isolated from poultry | [40] |
| B. cereus 407 | [41] | |
| B. licheniformis LMG7559 | Isolated from flour | [42] |
| B. amyloliquefaciens 20P6 | Isolated from lettuce | This work |
TF NDmed/DNA stands for transformation of NDmed by chromosomal DNA of indicated strains.
2.2. Biofilm development in 96 well microplates
Submerged biofilms were grown on the surface of polystyrene 96-well microtiter plates with a μclear® base (Greiner Bio-one, France) enabling high-resolution fluorescence imaging [43]. 200 μL of an overnight culture in TSB (adjusted to an OD 600 nm of 0.02) were added in each well. The microtiter plate was then incubated at 30°C for 90 min to allow the bacteria to adhere to the bottom of the wells. Wells were then rinsed with TSB to eliminate non-adherent bacteria and refilled with 200 μL of sterile TSB. When appropriate, the medium was supplemented with 200 μM IPTG to induce the expression of the fluorescent reporters GFP or mCherry from the Phyperspank promoter. The vital stain FM4-64 (Invitrogen), added to the medium at a final concentration of 1 μg/mL, was used to label bacteria membranes when appropriate.
In order to acquire high resolution images of pellicle (Fig. 3B), floating biofilms grown in a 12-well microplate (Greiner bio-one, Germany) were detached by a tip from the sides of the well and placed on a slide to be observed under confocal microscopy.
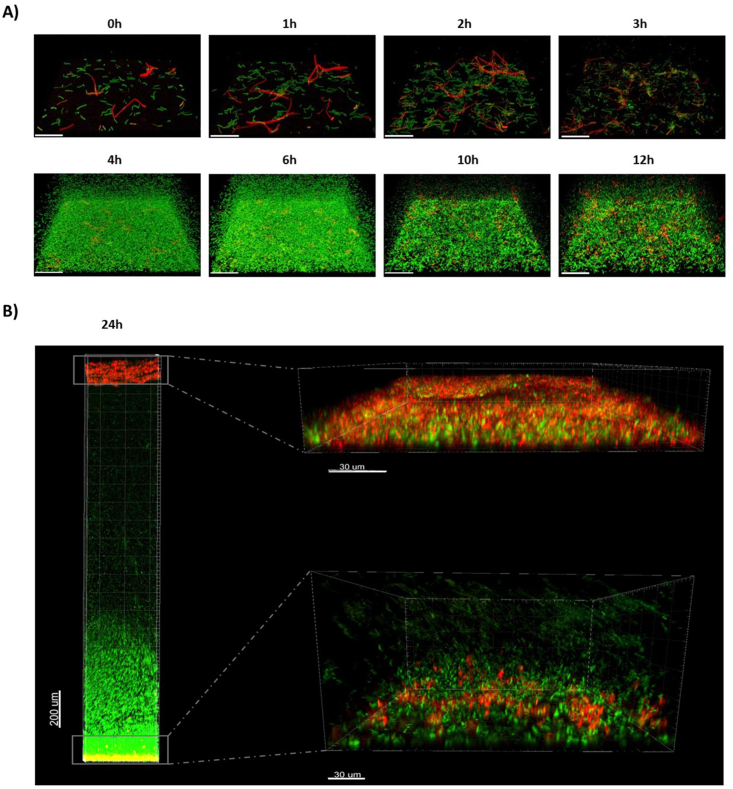
Fig. 3.
CLSM of NDmed547 [amyE::Phag-gfp sacA::PtapA-mKate2] reporting in green the expression of the hag gene (motility) and in red the expression of tapA (matrix synthesis). A) 4D-CLSM of the biphasic submerged biofilm formation process. See also movie S13. The scale bars represent 50 μm. B) CLSM visualization of the well colonization after 24h, both on the surface (with a zoom on submerged biofilm on the bottom right with a scale bar of 30 μm) and at the liquid air interface (with a zoom on a floating pellicle on the up right with a scale bar of 30 μm). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.3. 4D-CLSM
After the initial adhesion and washing steps, the 96 well microtiter plate was mounted on the motorized stage of a Leica SP8 AOBS inverter confocal laser scanning microscope (CLSM, LEICA Microsystems, Germany) at the MIMA2 platform (www6.jouy.inra.fr/mima2_eng/). Temperature was maintained at 30°C during all experiments. 4D (xyzt) acquisitions were performed with the following parameters: images of 246 × 246 μm were acquired at 600 Hz using a 63 × /1.2 N A. with a z-step of 1 μm and a thickness of 120 μm at intervals of 15 min. To detect GFP, an argon laser at 488 nm set at 10% of the maximal intensity was used, and the emitted fluorescence was collected in the range 495–550 nm using hybrid detectors (HyD LEICA Microsystems, Germany). To detect the red fluorescence of mKate2 or FM4-64, a 561 nm helium-neon laser set at 25% and 2% of the maximal intensity respectively was used, and fluorescence was collected in the range 590–720 nm and 605–705 nm respectively, using hybrid detectors.
To visualize simultaneously submerged biofilm and liquid-air pellicle dynamic (Fig. 1), the control software was set to take xyzt series of 1.5 × 1.5 mm images scanned at 600 Hz using a low-resolution long range 10 × /0.3 N A. air objective with a z-step of 5 μm and a thickness of 4.5 mm at intervals of 60 min. To detect mCherry emitted fluorescence, a 561 nm helium-neon laser set at 20% of the maximal intensity was used, and fluorescence was collected in the range 580–700 nm using hybrid detectors. High resolution imaging of 24h culture biofilms (Fig. 3B) was obtained with a HCX APO L U–V–I 40x/0.80 WATER objective lens in well filled only 100 μL of growth media to reduce the distance between both interfaces.
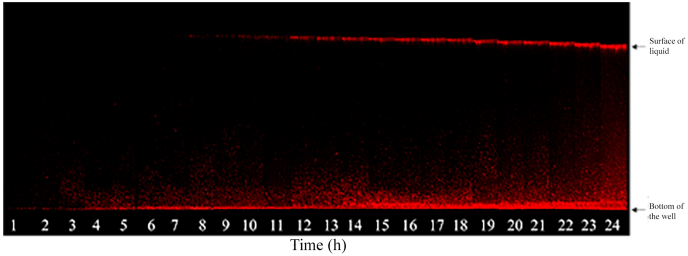
Fig. 1.
CLSM Section view of a microplate well colonized by B. subtilis NDmed-mCherry showing the relative dynamics of formation of submerged biofilm (bottom of the well) and the floating pellicle (surface of liquid); a representative experiment of three replicates is presented. Note that pellicle slightly falls over time due to liquid evaporation. The distance between the bottom of the well and the surface of the liquid is around 4.5 mm.
2.4. CLSM image analysis
Projections of the biofilm structural dynamic were constructed from xyzt images series using IMARIS 9.3 (Bitplane, Switzerland). Individual cell length and numbers were extracted from 4D-CLSM with the ImageJ (v1.53) particle analysis function. Space-time kymographs were constructed with the BiofilmQ visualisation toolbox [44]. Local density color code was calculated with BiofilmQ after Otsu segmentation and visualized with Paraview 5.9 [45].
2.5. Temporal transcriptome analysis
Biofilms were grown as previously explained. Briefly, 200 μL of an overnight culture of NDmed in TSB (adjusted to an OD 600 nm of 0.02) were added in each well. The microtiter plate was then incubated at 30°C for 90 min to allow the bacteria to adhere to the bottom of the wells. Wells were then rinsed with TSB to eliminate non-adherent bacteria and refilled with 200 μL of sterile TSB. For each time point (1h, 3h, 4h, 5h, 7h, 24h and 48h) 96-well plates were prepared. At each time point, the content of the wells was recovered and put into contact with the same volume of killing buffer [46].
Then, RNA was extracted following the method described by [46]. The RNA concentration was measured using Nanodrop and RNA quality was determined using an Agilent 2100 Bioanalyzer. Synthesis and Cy3-labelling of cDNA, and hybridization of the sample to the B. subtilis T3 2 × 400K tiling array (Agilent-044473) were performed following Agilent Technologies protocols, as previously described [47]. The microarray was scanned with Agilent Technologies Scanner, model G2505C. Grid: 044473_D_F_20121025. Protocol: GE1_107_Sep09. The tiling array data set is available from NCBI's Gene Expression Omnibus (GEO) database (accession number GSE190460).
2.6. Measurement of dissolved oxygen in the wells
A pO2 microelectrode (Lazard research laboratories, Inc) was used to measure over time oxygen concentration in the microtiter plate wells. Zero calibration was performed with a 2% sodium bisulfite solution and the results were expressed in ppm.
3. Results
-
1.
Coexistence of submerged biofilms and floating pellicles in microplates wells
In order to confirm that a B. subtilis non-domesticated strain was able to grow both as submerged biofilms and floating pellicles in microscopic grade microplate wells, we have combined 4D-CLSM and a long-range objective to monitor both interfacial communities simultaneously. For this, we used a B. subtilis NDmed strain derivative constitutively expressing mCherry red fluorescent protein to monitor its growth during 24 h starting from a single layer of adherent cells (Fig. 1, movies S1 and S2 in the supplemental material).
We observed an increase of red fluorescence from the bottom of the well up to a couple of hundred micrometers which corresponds to the development of the submerged biofilm. Increase in fluorescence intensity at the liquid-air interface occurred 7–8 h after the incubation starting point of the adhering cells, corresponding to the formation of the pellicle. It can be seen in Fig. 1 that the distance between the surface and the floating pellicle was initially 4.5 mm, but slightly decreased over time corresponding to liquid evaporation in the well and thereby a consequent drop of the liquid-air interface. Our observations demonstrate that a submerged biofilm and a floating pellicle can form consecutively in the same system. In addition, the fluorescent cells observed in the space between the attached submerged biofilm and the floating pellicles are swimming free cells (see movies S1 and S2), suggesting the existence of an interplay between both communities.
-
2.
A brutal and coordinated fragmentation of sessile elongating chains precede floating pellicle formation
Development of adherent B. subtilis NDmed GFP cells on the bottom of the microtiter plates wells was monitored by time-lapse confocal laser scanning microscopy (4D-CLSM) with images taken every 15 min during 14 h (Fig. 2 and movies S3 and S4 in the supplemental material). Fig. 2A represents tile images from the movie S3 at specific time points illustrating the two stages in the surface colonization. In a first stage, sessile cells proliferate as a dense network of long filaments covering the surface. Between 2 and 4 h after they started to proliferate, elongated chains with an average length of around 9 μm suddenly fragment and liberate a cloud of shorter free-swimming cells with an average length of 3 μm (Fig. 2B and C, movie S4). This transition occurred reproducibly in less than 30 min, and the construction of the typical B. subtilis protruding structures of sessile cells was visible only in a second kinetic, after 7–9 h (Fig. 2A and B, movie S3).
Fig. 2.
The biphasic process of submerged biofilm formation by B. subtilis NDmed. A) 4D-CLSM of B. subtilis NDmed GFP on submerged surfaces. Imaris Easy 3D reconstructions (top) and sections views as an XZ projection (bottom) at specific time points of a representative experiment of three independent experiments. The shadow on the right represents a vertical (YZ) projection of the submerged biofilm (scale bars represent 20 μm). B) Space-time kymograph generated with BiofilmQ from 4D-CLSM series showing the brutal apparition of free cell in all the well 3h after biofilm initiation and the late initiation of submerged biofilm after 7h. dz represents the distance to the surface in μm and Ich1 the GFP fluorescence intensity in relative arbitrary units. Representative of n = 3 independent biofilms. C) Individual cell length coordinately and brutally drop during chain fragmentation 2–3 h after biofilm initiation. Chains fragmentation is correlated with an increased number of detected individual objects in the medium. Mean cell length±SD calculated from n = 3 experiments.
Similar 4D-CLSM observations were acquired for other B. subtilis strains (movies S5-9 in the supplemental material), and strains of other related species (movies S10-12), using the vital FM4-64 fluorescent dye instead of GFP (see details of the strains in Table 1). The reference strains 168 and NCIB3610 displayed a similar behavior to that of the NDmed strain, as well as all other non-domesticated B. subtilis isolates tested (NDfood, BsN5 or BSP1). On the contrary, closely related but distinct Bacillus species strains such as Bacillus cereus 407, Bacillus licheniformis LMG7559 or Bacillus amyloliquefaciens 20P6 showed a continuous monophasic colonization of the surface without any coordinated liberation of free motile cells (movies S10-12 in the supplemental material).
-
3.
Temporal transcriptome analysis
To understand the mechanism behind sessile cells fragmentation into highly motile ones, a transcriptome analysis by tiling array was done for B. subtilis over a temporal scale. A global view of the results indicates that the genes encoding basic functions essential for cellular growth are expressed at a constant rate during the first hours (from 1h to 7h), with variation not exceeding at most a factor of 2 (see supplementary data S1). These ensure replication (DNA polymerase, primase, gyrase, topoisomerase, helicase, initiation/termination factors), transcription (RNA polymerase, sigma A factor, elongation/pause/termination factors), translation (ribosomal proteins, aminoacyl-tRNA synthetases, initiation elongation factors) or central carbon metabolism (enzymes of glycolysis and TCA cycle). Thus, this indicates that the process of fragmentation occurs while cells are growing at a constant rate. Results represented in Fig. S1 confirm the initial microscopic observations, where genes required for autolysis and motility start to be upregulated after 3 h to reach their maximum level after 4 h of incubation, the time in which elongated sessile chains fragment into motile short cells.
Nutrient or/and oxygen depletion were hypothesized to be possible signals triggering the early fragmentation of the sessile chains cells. To investigate the carbon source depletion as a triggering signal, we have monitored expression of gapB, a gene encoding a Glyceraldehyde-3-Phosphate-Dehydrogenase derepressed only under gluconeogenic conditions (supplementary data S1) [48]. The gapB gene expression was extremely downregulated during the first 7 h of incubation, and appeared strongly upregulated only in the late samples from 24 h to 48 h, indicating that glycolytic carbon source limitation occurred much later than the fragmentation process.
For oxygen sensing and respiration we have monitored the hemAT gene (encoding a soluble chemotaxis receptor oxygen sensor protein) [49], the cydABCD operon (encoding cytochrome bd oxidase induced under low oxygen tensions, represented as cydA in Fig. S1), and the qoxABCD operon (encoding cytochrome aa3 quinol oxidase, the major oxidase in aerated cultures, represented as qoxA gene in Fig. S1) [50]. For efficient aerobic growth, cells require either CydABCD or QoxABCD [50]. In the first hour only cydA appears to be upregulated (Fig. S1) and after 3 h none of the aerobic respiratory genes are expressed. Meanwhile, hemAT starts to be upregulated, in synchrony with the fragmentation of sessile cells to motile ones, to reach its maximum expression after 4 h. Aerobic respiration is regained and observed by the upregulation of qoxA. Shortly after, anaerobic respiration can be observed (Fig. S1) by upregulation of the self-regulated operon nark-fnr, encoding the Fnr transcriptional regulator for anaerobically induced genes, i.e. the narGHJI operon and the arfM gene encoding another anaerobic regulator. In addition, upregulation is also observed for the nasBCDEF operon (represented in Fig. S1 with nasD and nasF) encoding a nitrate- and a nitrite reductase [51].
Then after 7 h of incubation, biofilm matrix genes (i.e. epsA, tasA, dhbA, yvcA, pgsB, and bslA) are upregulated, the time where the biofilms (submerged and pellicle) are in the process of formation and stabilization. Expression follows of late biofilm genes (i.e. yxaB, veg, ymcA, and ypqP) for the complex architectural biofilm formation after 24 and 48 h, as well as genes related to sporulation.
-
4.
In situ cell visualization for the fate switching with fluorescent reporters
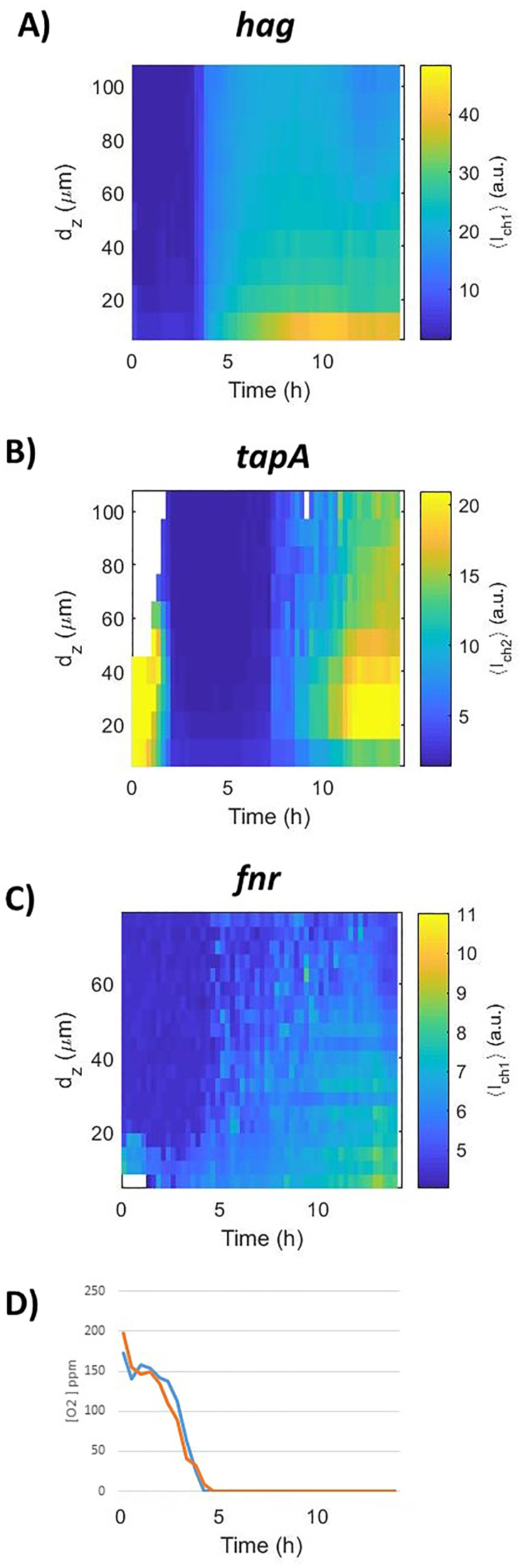
Using the NDmed547 strain harboring two transcriptional reporter constructions, we could monitor both motility and matrix production (Phag-gfp reporting the expression of flagella genes in green and PtapA-mKate2 reporting the expression of matrix genes in red). The submerged biofilm formation was monitored over 14 h (image every 15 min) for the spatio-temporal patterns of these two subpopulations of cell fate (Fig. 3A and movie S13 in the supplementary material). Promoter activities are illustrated in Fig. 4A and B, as a kymograph representing the fluorescence intensity as a function of time and altitude above. Moreover, the GM3361 strain allowed to report in the NDmed context expression of fnr, encoding a regulator of the global response to oxygen depletion (Pfnr-gfpmut3) (Fig. 4C).
Fig. 4.
Space-time kymographs for reporters (A)hag,(B)tapA, (C)fnr transcription during submerged biofilm formation of B. subtilis NDmed. Representative of n = 3 independent biofilms for each reporter. Kymographs were constructed with BiofilmQ visualization toolbox from 4D-CLSM image sequences with fluorescent transcriptional fusions (NDmed547 [amyE::Phag-gfp sacA::PtapA-mKate2] and GM3361 [Pfnr-gfpmut3]). dz represents the distance to the surface in μm and Ich1 the fluorescent reporter intensity in relative arbitrary units. The graph in panel (D) represents the oxygen concentration measured in two wells with a microelectrode showing a sharp decrease of oxygen concentration that drops from around 185 ppm at t = 0 below the probe detection limit after less than 5 h.
Initially, a subpopulation of sessile chains of cells expressing tapA (red) coexists with a subpopulation of motile cells expressing hag (green). During the first 3 h of submerged biofilm formation, sessile chains have elongated and propagated, discern visually by the high fluorescent intensity recorded in Fig. 4B, while the fraction of cells expressing hag was less abundant (Fig. 3, Fig. 4A). After 3 h, red cell chains (expressing tapA) suddenly and coordinately fragment into green individual motile cells (expressing hag), corresponding to the sudden change of the fluorescent intensity color observed in Fig. 4A–a more intense one. In parallel, a clear signal of fluorescence was detectable as early as 3–4 h with GM3361 [Pfnr-gfpmut3]), indicating that oxygen limitation occurred in early stages of the biofilm development (Fig. 4C). This was confirmed by a direct and continuous measurement of oxygen concentration using a microelectrode (Fig. 4D). Oxygen concentration in the well strictly decreased below the detection limit as early as 4 h after incubation of adherent cells. This oxygen limitation was correlated to trigger the population of red elongated chains, expressing the tapA operon, to fragment and acquire motility (Fig. 4). The tapA expression was highly regained after ∼7h (Fig. S1 and Fig. 4B) to structure the typical surface-associated protruding submerged biofilm and initiate the floating pellicle of B. subtilis, which leads to two biofilms in a same well of static liquid culture (Fig. 3B).
4. Discussion
We observed previously that the non-domesticated B. subtilis NDmed strain was able to form robust submerged biofilms in the bottom of microtiter-plates under static conditions [28]. In the present work, using 4D-CLSM, we visualized the formation dynamics of these submerged biofilms, which surprisingly appeared to be a discontinuous process. After a first stage of development on the surface, and concomitantly with oxygen limitation, sessile chains suddenly fragment, liberating a massive number of free motile cells. These planktonic cells partially migrate towards the liquid-air interface to initiate a floating pellicle. It is only in a second kinetics that the characteristic surface-associated protruding structures of B. subtilis NDmed rise, along with strong expression of the tapA-sipW-tasA matrix operon. To our knowledge, such biphasic biofilm formation has never been explored and when pellicles and submerged biofilms have been studied, it was frequently under different conditions.
In other bacterial species like Pseudomonas aeruginosa, detachment from the surface and dispersion has been described as a possible end to the biofilm lifestyle cycle. The active cells scattering from biofilm to new habitats seem to be driven by limitations of resources or the emergence of stressful conditions in the cell microenvironment. Depletion in nutrient availability or the accumulation of waste metabolic products (such as acids issuing from fermentation in oxygen-depleted zones) have been demonstrated to induce biofilm dispersal [52]. In the work presented here, we demonstrate that early oxygen limitation in the well is concomitant with massive liberation of planktonic cells from early submerged biofilm after approximately 4 h of development. In B. subtilis, oxygen depletion has been shown previously to induce matrix production by increasing transcription of the tapA operon in NCIB3610 colonies [53]. In our submerged system, expression in NDmed of tapA was also correlated with that of fnr, a gene induced under deep oxygen depletion. These observations suggest that, after a first step of proliferation of sessile cells as filaments covering the surface, the limitation of oxygen triggered their fragmentation into motile cells able to migrate to the air-medium interface to form the pellicle. Indeed, a temporal transcriptome analysis of biofilm showed that genes involved in oxygen sensing, autolysis and motility were highly expressed after 4 h of development, which corresponded to the sudden transition between filaments and motile cells observed with 4D-CLSM. This is in accordance with a previous report identifying oxygen as a putative trigger for active movement towards the air–liquid interface of B. subtilis NCIB3610 cells, since a ΔhemAT mutant was outcompeted by the wild type during pellicle formation in static co-cultures [54]. Similar behaviors were also described for the Gram-negative lake sediment bacterium Shewanella oneidensis MR-1, as biofilms formed by this species showed a rapid detachment from the surface upon a sudden downshift in oxygen concentration [55].
Interestingly, all other B. subtilis strains tested, including thick biofilm-forming isolates such as NDfood, BsN5 and BsP1, or weaker submerged biofilm-forming strains 168 and NCIB3610, showed a similar biphasic submerged biofilm dynamics (Movies S5-9). In contrast, other related species, such as B. cereus, B. amyloliquefaciens or B. licheniformis exhibited a continuous colonization of the surface (Movies S10-12). Some of these Bacilli are also able to form chains of cells, but which did not fragment during the time the biofilm formation was monitored. B. subtilis chain formation was first visualized in pellicles [11], however without observation of coordinated return to planktonic state in these conditions. The relation between submerged biofilm and liquid-air pellicle shown here could suggest an interplay, with co-metabolism between the populations of both interfaces: the population in contact with air could liberate metabolites used by the surface-associated population to grow in anaerobic condition. This is consistent with the described ability of B. subtilis to grow without oxygen, respiring nitrate or nitrite instead of oxygen as electrons acceptor [56]. Nevertheless, this potential relation between populations requires further investigation.
Understanding the triggers and effectors of this coordinated multicellular behavior could also contribute to identifying new actors involved in biofilm disruption. This has been largely studied in recent years because of its potential as an alternative treatment to promote biofilm cell detachment [57,58]. Some natural molecules produced by mature biofilms have been found to induce biofilm disassembly, such as nitric oxide in Pseudomonas spp. biofilms [59] or acidic amino acids in S. aureus biofilms [60]. The combination of such biofilm disruptors with antibiotics treatment to improve drug effectiveness could be a promising approach to tackle biofilm chronic infections.
The experimental approach proposed in this work allowed visualization, at the cell level, of the dynamic interactions between subpopulations in a B. subtilis community. The multidimensional description of spatio-temporal patterns of gene expression has been recently facilitated by the availability to biologists of advanced quantitative microscopic analysis tools [44]. The implementation of this system to more complex samples, such as multispecies biofilms, could provide an experimental approach for the study of spatial interactions between species, co-metabolism or cell-to-cell signaling.
CRediT authorship contribution statement
Pilar Sanchez-Vizuete: Conceptualization, experiments, Writing – original draft. Yasmine Dergham: Conceptualization, experiments, Data curation, Writing – original draft, Writing – review & editing. Arnaud Bridier: Conceptualization, experiments, Writing – review & editing. Julien Deschamps: Assistance with 4D-CLSM microscopy, Formal analysis. Etienne Dervyn: Transcriptomic experiments. Kassem Hamze: Supervision, reviewing. Stéphane Aymerich: Conceptualization, reviewing. Dominique Le Coq: Conceptualization, Methodology, Supervision, Writing – review & editing. Romain Briandet: Conceptualization, Methodology, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by INRAE. P. Sanchez-Vizuete was the recipient of a PhD grant from the Région Ile-de-France (DIM ASTREA). Y. Dergham is the recipient of fundings from the Union of Southern Suburbs Municipalities of Beirut, INRAE, Campus France PHC CEDRE 42280 PF and Fondation AgroParisTech. L. Tournier and V. Fromion (INRAE) are acknowledged for fruitful discussions, R. Losick for the gift of the strain TMN547, and M. Jules for the gift of strains BBA0184 and BBA9006.
Footnotes
‘Given his role as Editor, Romain Briandet had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to Ákos T. Kovács’.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2021.100065.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Karygianni L., Ren Z., Koo H., Thurnheer T. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 2020;28:668–681. doi: 10.1016/j.tim.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 3.Mukherjee M., Cao B. Engineering controllable biofilms for biotechnological applications. Microb Biotechnol. 2021;14:74–78. doi: 10.1111/1751-7915.13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mah T.-F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012;7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 5.Nickel J.C., Ruseska I., Wright J.B., Costerton J.W. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother. 1985;27:619–624. doi: 10.1128/AAC.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 7.da Silva R.A.G., Afonina I., Kline K.A. Eradicating biofilm infections: an update on current and prospective approaches. Curr Opin Microbiol. 2021;63:117–125. doi: 10.1016/j.mib.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Pamp S.J., Sternberg C., Tolker-Nielsen T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytometry A. 2009;75:90–103. doi: 10.1002/cyto.a.20685. [DOI] [PubMed] [Google Scholar]
- 9.Branda S.S., González-Pastor J.E., Ben-Yehuda S., Losick R., Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piggot P.J., Hilbert D.W. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi K. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J Bacteriol. 2007;189:4920–4931. doi: 10.1128/JB.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlamakis H., Chai Y., Beauregard P., Losick R., Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Vizuete P., Le Coq D., Bridier A., Herry J.-M., Aymerich S., Briandet R. Identification of ypqP as a New Bacillus subtilis biofilm determinant that mediates the protection of Staphylococcus aureus against antimicrobial agents in mixed-species communities. Appl Environ Microbiol. 2015;81:109–118. doi: 10.1128/AEM.02473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kearns D.B., Chu F., Branda S.S., Kolter R., Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 15.Cairns L.S., Hobley L., Stanley-Wall N.R. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Mol Microbiol. 2014;93:587–598. doi: 10.1111/mmi.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartolini M., Cogliati S., Vileta D., Bauman C., Rateni L., Leñini C., et al. Regulation of biofilm aging and dispersal in Bacillus subtilis by the alternative sigma factor SigB. J Bacteriol. 2019;201 doi: 10.1128/JB.00473-18. e00473-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milton M.E., Draughn G.L., Bobay B.G., Stowe S.D., Olson A.L., Feldmann E.A., et al. The solution structures and interaction of SinR and SinI: elucidating the mechanism of action of the master regulator switch for biofilm formation in Bacillus subtilis. J Mol Biol. 2020;432:343–357. doi: 10.1016/j.jmb.2019.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishikawa M., Kobayashi K. Calcium prevents biofilm dispersion in Bacillus subtilis. J Bacteriol. 2021;203 doi: 10.1128/JB.00114-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnaouteli S., Bamford N.C., Stanley-Wall N.R., Kovács Á.T. Bacillus subtilis biofilm formation and social interactions. Nat Rev Microbiol. 2021 doi: 10.1038/s41579-021-00540-9. [DOI] [PubMed] [Google Scholar]
- 20.Chai Y., Kolter R., Losick R. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol. 2010;78:218–229. doi: 10.1111/j.1365-2958.2010.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López D., Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. 2010;34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- 22.Diethmaier C., Pietack N., Gunka K., Wrede C., Lehnik-Habrink M., Herzberg C., et al. A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation. J Bacteriol. 2011;193:5997–6007. doi: 10.1128/JB.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero D., Aguilar C., Losick R., Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostrowski A., Mehert A., Prescott A., Kiley T.B., Stanley-Wall N.R. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J Bacteriol. 2011;193:4821–4831. doi: 10.1128/JB.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi K., Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 26.Roux D., Cywes-Bentley C., Zhang Y.-F., Pons S., Konkol M., Kearns D.B., et al. Identification of poly-N-acetylglucosamine as a major polysaccharide component of the Bacillus subtilis biofilm matrix. J Biol Chem. 2015;290:19261–19272. doi: 10.1074/jbc.M115.648709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Mammeri N., Hierrezuelo J., Tolchard J., Cámara-Almirón J., Caro-Astorga J., Álvarez-Mena A., et al. Molecular architecture of bacterial amyloids in Bacillus biofilms. FASEB J. 2019;33:12146–12163. doi: 10.1096/fj.201900831R. [DOI] [PubMed] [Google Scholar]
- 28.Bridier A., Le Coq D., Dubois-Brissonnet F., Thomas V., Aymerich S., Briandet R. The spatial architecture of Bacillus subtilis biofilms deciphered using a surface-associated model and in situ imaging. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bridier A., Sanchez-Vizuete M.D.P., Le Coq D., Aymerich S., Meylheuc T., Maillard J.-Y., et al. Biofilms of a Bacillus subtilis hospital isolate protect Staphylococcus aureus from biocide action. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe K., Kawano Y., Iwamoto K., Arai K., Maruyama Y., Eichenberger P., et al. Developmentally-regulated excision of the SPβ prophage reconstitutes a gene required for spore envelope maturation in Bacillus subtilis. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y., Yan F., Chai Y., Liu H., Kolter R., Losick R., et al. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol. 2013;15:848–864. doi: 10.1111/j.1462-2920.2012.02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandin C., Le Coq D., Canette A., Aymerich S., Briandet R. Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microb Biotechnol. 2017;10:719–734. doi: 10.1111/1751-7915.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terra R., Stanley-Wall N.R., Cao G., Lazazzera B.A. Identification of Bacillus subtilis SipW as a bifunctional signal peptidase that controls surface-adhered biofilm formation. J Bacteriol. 2012;194:2781–2790. doi: 10.1128/JB.06780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dergham Y., Sanchez-Vizuete P., Le Coq D., Deschamps J., Bridier A., Hamze K., et al. Comparison of the genetic features involved in Bacillus subtilis biofilm formation using multi-culturing approaches. Microorganisms. 2021;9:633. doi: 10.3390/microorganisms9030633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin D.J.H., Denyer S.P., McDonnell G., Maillard J.-Y. Resistance and cross-resistance to oxidising agents of bacterial isolates from endoscope washer disinfectors. J Hosp Infect. 2008;69:377–383. doi: 10.1016/j.jhin.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Norman T.M., Lord N.D., Paulsson J., Losick R. Memory and modularity in cell-fate decision making. Nature. 2013;503:481–486. doi: 10.1038/nature12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rühl M., Le Coq D., Aymerich S., Sauer U. 13C-flux analysis reveals NADPH-balancing transhydrogenation cycles in stationary phase of nitrogen-starving Bacillus subtilis. Journal of Biological Chemistry. 2012;287:27959–27970. doi: 10.1074/jbc.M112.366492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Botella E., Fogg M., Jules M., Piersma S., Doherty G., Hansen A., et al. pBaSysBioII: an integrative plasmid generating gfp transcriptional fusions for high-throughput analysis of gene expression in Bacillus subtilis. Microbiology (Reading) 2010;156:1600–1608. doi: 10.1099/mic.0.035758-0. [DOI] [PubMed] [Google Scholar]
- 39.Deng Y., Zhu Y., Wang P., Zhu L., Zheng J., Li R., et al. Complete genome sequence of Bacillus subtilis BSn5, an endophytic bacterium of Amorphophallus konjac with antimicrobial activity for the plant pathogen Erwinia carotovora subsp. carotovora. J Bacteriol. 2011;193:2070–2071. doi: 10.1128/JB.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schyns G., Serra C.R., Lapointe T., Pereira-Leal J.B., Potot S., Fickers P., et al. Genome of a gut strain of Bacillus subtilis. Genome Announc. 2013;1 doi: 10.1128/genomeA.00184-12. e00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houry A., Briandet R., Aymerich S., Gohar M. Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology (Reading) 2010;156:1009–1018. doi: 10.1099/mic.0.034827-0. [DOI] [PubMed] [Google Scholar]
- 42.De Clerck E., De Vos P. Genotypic diversity among Bacillus licheniformis strains from various sources. FEMS Microbiol Lett. 2004;231:91–98. doi: 10.1016/S0378-1097(03)00935-2. [DOI] [PubMed] [Google Scholar]
- 43.Bridier A., Dubois-Brissonnet F., Boubetra A., Thomas V., Briandet R. The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. J Microbiol Methods. 2010;82:64–70. doi: 10.1016/j.mimet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Hartmann R., Jeckel H., Jelli E., Singh P.K., Vaidya S., Bayer M., et al. Quantitative image analysis of microbial communities with BiofilmQ. Nat Microbiol. 2021;6:151–156. doi: 10.1038/s41564-020-00817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayachit U. 2015. “The ParaView guide: a parallel visualization application,” in. Clifton Park, USA: Kitware. [Google Scholar]
- 46.Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 47.Rath H., Sappa P.K., Hoffmann T., Salazar M.G., Reder A., Steil L., et al. Impact of high salinity and the compatible solute glycine betaine on gene expression of Bacillus subtilis. Environ Microbiol. 2020;22:3266–3286. doi: 10.1111/1462-2920.15087. org/10.1111/1462-2920.15087. [DOI] [PubMed] [Google Scholar]
- 48.Fillinger S., Boschi-Muller S., Azza S., Dervyn E., Branlant G., Aymerich S. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J Biol Chem. 2000;275:14031–14037. doi: 10.1074/jbc.275.19.14031. [DOI] [PubMed] [Google Scholar]
- 49.Hou S., Larsen R.W., Boudko D., Riley C.W., Karatan E., Zimmer M., et al. Myoglobin-like aerotaxis transducers in archaea and bacteria. Nature. 2000;403:540–544. doi: 10.1038/35000570. [DOI] [PubMed] [Google Scholar]
- 50.Sachla A.J., Luo Y., Helmann J.D. Manganese impairs the QoxABCD terminal oxidase leading to respiration-associated toxicity. Mol Microbiol. 2021 doi: 10.1111/mmi.14767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakano M.M., Zuber P. Anaerobic growth of a “strict aerobe” (Bacillus subtilis) Annu Rev Microbiol. 1998;52:165–190. doi: 10.1146/annurev.micro.52.1.165. [DOI] [PubMed] [Google Scholar]
- 52.Rumbaugh K.P., Sauer K. Biofilm dispersion. Nat Rev Microbiol. 2020;18:571–586. doi: 10.1038/s41579-020-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolodkin-Gal I., Elsholz A.K.W., Muth C., Girguis P.R., Kolter R., Losick R. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev. 2013;27:887–899. doi: 10.1101/gad.215244.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hölscher T., Bartels B., Lin Y.-C., Gallegos-Monterrosa R., Price-Whelan A., Kolter R., et al. Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. J Mol Biol. 2015;427:3695–3708. doi: 10.1016/j.jmb.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thormann K.M., Saville R.M., Shukla S., Spormann A.M. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol. 2005;187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakano M.M., Hulett F.M. Adaptation of Bacillus subtilis to oxygen limitation. FEMS Microbiol Lett. 1997;157:1–7. doi: 10.1111/j.1574-6968.1997.tb12744.x. [DOI] [PubMed] [Google Scholar]
- 57.Kostakioti M., Hadjifrangiskou M., Hultgren S.J. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med. 2013;3:a010306. doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cascioferro S., Carbone D., Parrino B., Pecoraro C., Giovannetti E., Cirrincione G., et al. Therapeutic strategies to counteract antibiotic resistance in MRSA biofilm-associated infections. ChemMedChem. 2021;16:65–80. doi: 10.1002/cmdc.202000677. [DOI] [PubMed] [Google Scholar]
- 59.Barraud N., Kelso M.J., Rice S.A., Kjelleberg S. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr Pharm Des. 2015;21:31–42. doi: 10.2174/1381612820666140905112822. [DOI] [PubMed] [Google Scholar]
- 60.Warraich A.A., Mohammed A.R., Perrie Y., Hussain M., Gibson H., Rahman A. Evaluation of anti-biofilm activity of acidic amino acids and synergy with ciprofloxacin on Staphylococcus aureus biofilms. Sci Rep. 2020;10:9021. doi: 10.1038/s41598-020-66082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.