Abstract
Human telomerase reverse transcriptase (hTERT), the essential catalytic subunit of telomerase, is associated with telomere homeostasis to prevent replicative senescence and cellular aging. However, hTERT reactivation also has been linked to the acquisition of several hallmarks of cancer, although the underlying mechanism beyond telomere extension remains elusive. This study demonstrated that hTERT overexpression promotes, whereas its inhibition by shRNA suppresses, epithelial-mesenchymal transition (EMT) in lung cancer cells (A549 and H1299). We found that hTERT modulates the expression of EMT markers E-cadherin, vimentin, and cytokeratin-18a through upregulation of the c-MET. Ectopic expression of hTERT induces expression of c-MET, while hTERT-shRNA treatment significantly decreases the c-MET level in A549 and H1299 through differential expression of p53 and c-Myc. Reporter assay suggests the regulation of c-MET expression by hTERT to be at the promoter level. An increase in c-MET level significantly promotes the expression of mesenchymal markers, including vimentin and N-cadherin, while a notable increase in epithelial markers E-cadherin and cytokeratin-18a is observed after the c-MET knockdown in A549.
Keywords: hTERT, c-MET, p53, c-Myc, EMT
hTERT, c-MET, p53, c-Myc and EMT.
1. Introduction
Cancer cells extended growth potential and bypassing cellular aging is critically dependent on sustained telomere homeostasis and integrity. Telomerase has a fundamental function in the maintenance of telomere length. It is an RNA-dependent DNA polymerase ribonucleoprotein complex that uses TERC as the template and adds TTAGGG repeats at the telomeric end. Typically somatic cells lack telomerase activity due to transcription suppression of hTERT. However, most cancers (∼90%) have a high expression of hTERT and active telomerase. Telomerase-mediated telomere maintenance is a canonical pathway to escape the Hayflick limit, a primary requirement of cellular immortalization [1, 2]. Recent studies suggest extracurricular activities of hTERT beyond its canonical function [3, 4]. Increased hTERT level is associated with cancer cell survivability, proliferation, and metastasis [5, 6]. However, the mechanistic basis of its role in cancer metastasis is not clearly understood. Thus, it becomes imperative to find out probable mediators of metastasis under the influence of hTERT expression.
c-MET is a disulfide-linked heterodimer activated through its natural ligand HGF and triggers various signaling pathways like PI3K, Gab1, STAT, and β-catenin [7, 8]. HGF/c-MET signaling plays an essential role in embryonic development, and disruption in HGF or c-MET gene leads to embryonic lethality and defect in organ generation [9, 10]. HGF/c-MET is also a potent angiogenic factor that incites endothelial growth and motility to promote angiogenesis [11]. HGF treatment induces EMT to favor the invasion and migration of cancer cells [12, 13]. Deregulation of the c-MET signaling due to mutation, gene amplification, or increased expression promotes cancer development. HGF/c-MET expression was found unproportioned in most types of tumors, including gliomas, sarcomas, carcinomas, and melanomas [14, 15, 16, 17].
This study aimed to explore the role of hTERT in the early metastasis stage, i.e., EMT and migration, and to determine the probable mediators of such response. Our experiments with cultured cells show that hTERT enhances the EMT and cell migration by upregulation of the c-MET expression.
2. Materials and methods
2.1. Cell culture and treatments
A549 (Human lung adenocarcinoma), H1299 (Human non-small cell lung carcinoma), and 293T (Human embryonic kidney cell) were purchased from NCCS, Pune. All the cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C under 5% CO2 in a humidified incubator. The 293T cell line was used to produce lentivirus carrying shRNA sequences to target hTERT, p53 c-Myc, and c-MET. Stably transfected cells were selected by puromycin. We used 2μg puromycin/ml for A549 and 1.0μg puromycin/ml for H1299 cells. For HGF treatment, A549 cells were seeded in 6 well plates and starved for 12 h, then treated with 20 ng/ml of HGF for 24 h A549 cells were also treated with 8 μM of the c-MET inhibitor (SU11274) for 24 h in complete media.
2.2. Reagents
HGF was procured from Peprotech and c-MET inhibitor (SU11274) from Santa Cruz. Primary antibodies were purchased from Abcam (hTERT), Cell Signalling Technology (Phospho-c-MET, vimentin, E-Cadherin, N-Cadherin), Santa Cruz (c-MET), Chemicon (Cytokeratin-18a), and Thermo Fisher Scientific (β-Actin). Secondary antibodies, HRP conjugated anti-mouse and anti-rabbit IgG, and HRP conjugated anti-mouse IgM were obtained from Santa Cruz. Secondary antibodies, FITC conjugated anti-mouse and anti-rabbit IgG were obtained from Santa Cruz.
2.3. Plasmids and transfection
pBABE-hTERT and control pBABE plasmids were purchased from Addgene. The pLVX-p53 expression vector was created by cloning of complete coding region of p53 in between EcoRI and BamHI of the pLVX-puro expression vector. The shRNA target sequence for hTERT, p53 c-Myc, and c-MET was synthesized from Sigma-Aldrich and cloned into the pLKO1-shRNA and pLVX-shRNA vectors. Lentiviral particles were synthesized by co-transfection of shRNA vectors (transfer vector), psPAX2, and pMD2.G (helper plasmid) in the 293T cells. The sequence of hTERT, p53, c-Myc, and c-MET shRNAs have been listed in the supplementary file (Supplementary Table 1). Expression vectors and shRNA construct were transfected using lipofectamine-3000 (Invitrogen) or metafectene (Biontex) as per the manufacturer's protocol.
2.4. Western blot analysis
Total cellular protein was extracted in cell lysis buffer [50 mM Tris-Cl (pH 7.4), 150mM sodium chloride (NaCl), 1mM EDTA, 1mM EGTA, 0.5% sodium deoxycholate, 1% NP40, 0.1% SDS and protease inhibitors]. Protein concentration was estimated by the Bradford method. 25–50 μg of protein was run on SDS-PAGE and transferred to PVDF membrane (Millipore) by wet transfer followed by blocking with 5% skimmed milk. Blots were incubated with primary antibody overnight at 4°C and with secondary antibody for 1 h at room temperature. Protein bands were visualized by chemiluminescence (Luminata Forte Millipore) and captured on X-ray film. β-Actin was used as an endogenous loading control.
2.5. Immunofluorescence
Cells were seeded on the tissue culture-treated coverslip and allowed to grow up to 30% confluence. Cells were washed with PBS and fixed in 4% paraformaldehyde for 15–30 min, followed by permeabilization of cells by Triton X-100 for 10 min and blocked with 5% BSA (in PBS + 0.05% Tween 20) for 1 h. Cells were incubated overnight with primary antibodies {rabbit anti-c-MET (1:100) and rabbit anti-cytokeratin (1:300)} at 4°C. After overnight incubation, cells were washed with PBST (PBS + 0.05% Tween 20) and incubated with FITC tagged anti-rabbit (1: 200) secondary antibody for 1 h at room temperature. Cells were observed under the confocal laser scanning microscope (Nikon TiE), and pictures were analyzed by Nikon NIS software.
2.6. Luciferase reporter assay
For the promoter-reporter assay, 1850 nucleotide-long region spanning from −1655 to +195 nucleotides, including transcription start site of the c-MET promoter was cloned in the pGL3 vector. A549 cells were seeded in 6 well plates in triplicates and co-transfected with the pGL3-c-MET promoter and different concentrations of shTERT plasmid (0.25 μg, 0.5 μg, and 1.0 μg). In another study, A549 and H1299, which stably express the hTERT were transfected with pGL3-c-MET promoter construct. After 36 h of pGL3-c-MET promoter transfection, total protein extracted in cell lysis buffer provided in the assay kit (Biovision). Luciferase activity was performed by luciferase reporter assay kit (Biovision) and measured by Thermo Varioscan plate reader. Luciferase values were normalized with total protein. All the experiments were performed in triplicates, and statistical analysis was done by graph-pad software. Bioinformatics analysis of c-MET promoter was performed for binding of different transcription factors using TRANSFAC.
2.7. Colony formation assay
Cells were seeded in 6 well plates (1000 cells per well) and allowed to grow. After 7–10 days, dishes were washed with PBS and fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. The number of colonies was counted in each well, and Clonogenic survivability was calculated.
2.8. Cell proliferation assay
Cells were knocked down for the expression of hTERT and seeded in 12 well plates in triplicate. In each well 2 × 104 cells were seeded and allowed to grow. At regular intervals, cells were suspended by trypsinization and counted by a haemocytometer.
2.9. Wound healing assay
Cells were seeded in 6 well plates and allowed to attach to the surface. Cells were starved for 12 h, and the cell monolayer was scratched with 200 μl tips. After scratch, wells were washed and replenished with medium supplemented with 1.0% FBS. Cell movement in the scratched zone was captured at 0, 12, 24, and 36 h after scratching.
2.10. Statistical analysis
The data presented in this study are the mean ± standard error (SE) of at least three independent experiments. Statistical significance of results was analyzed by unpaired t-test, two-way ANOVA followed by Tukey's multiple comparisons. P-value ≤ 0.05 was chosen to assess the significance of the results.
3. Results
3.1. hTERT promotes the epithelial to mesenchymal transition and cell migration in A549 and H1299
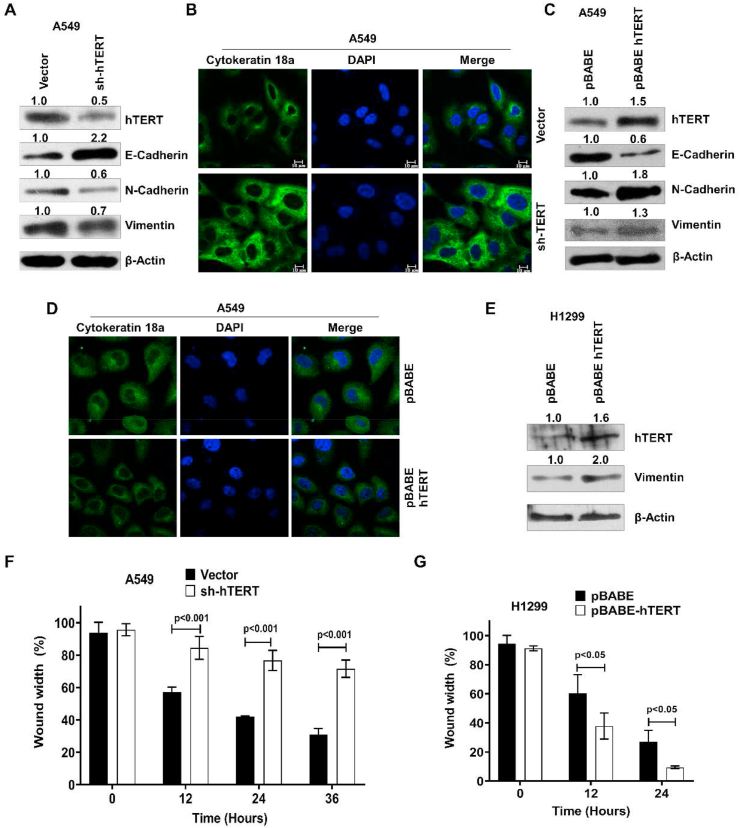
EMT plays a crucial role in cancer development by reverting epithelial cells to a mesenchymal state. Therefore, we assayed the expression of vimentin, N-cadherin, E-cadherin, and cytokeratin-18a as markers for the two cellular states after knocking down of hTERT in A549. Vimentin and N-cadherin are upregulated, while E-cadherin and cytokeratin-18a are downregulated during EMT. hTERT knockdown is accompanied by a significant reduction of vimentin and N-cadherin in A549; in contrast, the expression of epithelial protein E-cadherin was increased (Figure 1A). Cytokeratin-18a is a characteristic intermediate filament of epithelial cells, showed enhanced expression following hTERT knockdown in A549 (Figure 1B). Over-expression of hTERT causes up-regulation of mesenchymal proteins like N-cadherin and vimentin in A549 and H1299 (Figure 1C, E). However, the expression of E-cadherin and cytokeratin-18a were reduced in hTERT overexpressing A549 cells (Figure 1C, D). Further, the migratory potential of cells was studied by a wound-healing experiment in hTERT knockdown in A549 and H1299 cells. Wound healing experiment revealed that hTERT downregulation lowers cell movability in A549 cells, while hTERT overexpression enhanced cell migration in H1299 (Figure 1F, G). We also performed the cell proliferation and survivability assay and found that hTERT promotes cell proliferation. In the A549 cell line, about 25% inhibition was observed on hTERT shRNA treatment. Overexpression of hTERT promotes cell proliferation in the H1299 cell line (Supplementary Figure 1A, B). Knocking down hTERT with shRNA also reduced the cell survivability by about 50% in A549 cells (Supplementary Figure 1C).
Figure 1.
hTERT promotes the epithelial to mesenchymal transition and cell migration in A549 and H1299. A) Immunoblot analysis of E-cadherin, N-cadherin, and vimentin on hTERT knockdown B) Immunofluorescence assay showing knockdown of hTERT in A549 cells increased the expression level of cytokeratin-18a. C, D) hTERT overexpression in A549 leads to an increased expression of mesenchymal marker (N-cadherin and vimentin) and decreased epithelial markers (E-cadherin and cytokeratin-18a). E) hTERT expression plasmid was also transfected in H1299 cells, showing increased mesenchymal protein (vimentin) on hTERT overexpression. F, G) Migration assay in hTERT knockdown and overexpression in lung cancer cells (A549 and H1299), hTERT down-regulation reduced, while overexpression promoted the cell migration (Mean ± SE, n = 3). Full and non-adjusted images of immunoblots were shown in the supplementary material (Figure SM 1).
3.2. c-MET expression is regulated by hTERT
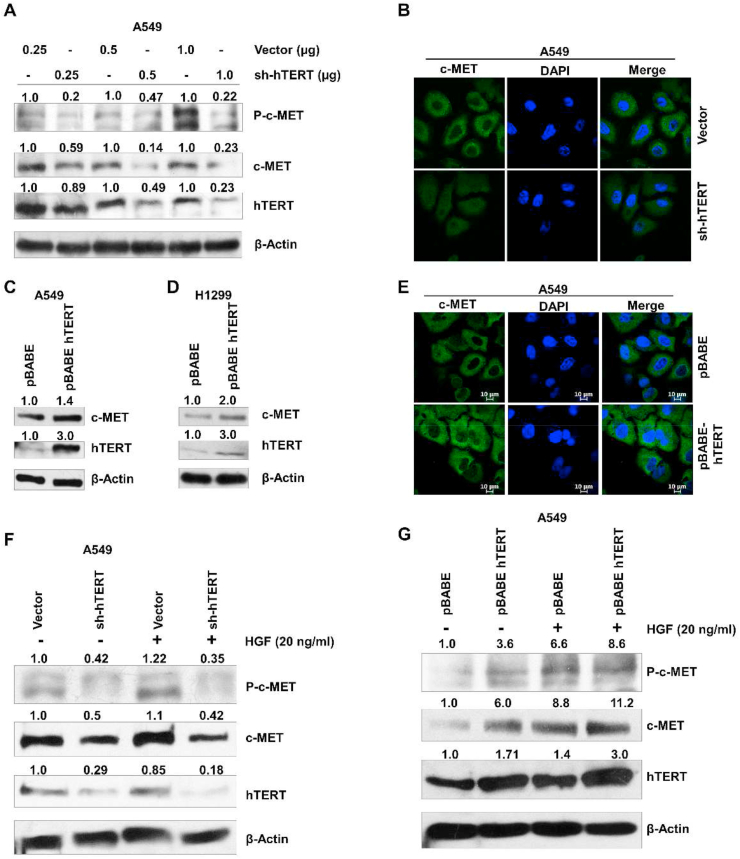
hTERT expression promotes cell proliferation and metastasis and inhibits cell death [18]. It has been reported that hTERT expression activates the Wnt/beta-catenin signaling and promotes the expression of growth controlling genes like EGFR, bFGF, and VEGF independent of telomerase activation [19, 20, 21]. The hepatocyte growth factor is also a growth-promoting factor secreted by the stromal cells, while its receptor is expressed on tumor cells [22]. Here, we have explored the possible crosstalk between HGF-c-MET signaling and hTERT expression in lung cancer cells. The expression level of total and phospho c-MET decreased gradually with the increase of hTERT-shRNA concentration (0.25–1.0 μg per ∼0.6 × 106 cells) (Figure 2A). Immunofluorescence of c-MET protein upon hTERT knockdown in A549 cells also shows decreased c-MET level (Figure 2B). Further, hTERT protein overexpressed in A549 and H1299 by transfecting the pBABE-hTERT expression vector. hTERT overexpression induced c-MET protein expression in A549 and H1299 cells (Figure 2C, D) as also confirmed by immunofluorescence assay (Figure 2E). HGF produced by stromal cells activates c-MET signaling and also promotes its expression through paracrine signaling [23, 24]. So, we treated the A549 cells with HGF ligand to explore the correlation between hTERT expression and HGF-c-MET signaling. HGF treatment in A459 cells induced phosphorylation and increased c-MET protein expression, hTERT downregulation with shRNA drastically reduced c-MET phosphorylation and expression (Figure 2F). However, HGF treatment in pBABE-hTERT transfected A549 cells synergistically promotes the expression and activation of c-MET protein (Figure 2G). We also used the inhibitor SU11274 to block the activation of the c-MET protein. SU11274 treatment in A549 and pBABE-hTERT transfected A549 cells drastically reduced the phosphorylation and expression of the c-MET protein (Supplementary Figure 2).
Figure 2.
c-MET expression is regulated by hTERT. A) Immunoblot analysis of phospho-c-MET, c-MET proteins after treatment with different doses of hTERT shRNA in A549. hTERT shRNA reduced the c-MET expression in a dose-dependent manner. B) Immunofluorescence of c-MET in hTERT knockdown A549 also shows the positive association between hTERT and c-MET expression. C, D) hTERT overexpression in A549 (left panel) and H1299 (right panel) shows the increased level of c-MET expression. E) Immunofluorescence of the c-MET protein in hTERT overexpressing A549 cells. F) hTERT knockdown A549 cells were starved for 12 h then stimulated by HGF (20 ng/ml) for 24 h. Expression of phospho and total c-MET were analyzed by western blotting. HGF treatment showed no significant promoting effect on the c-MET expression in cells knocked down for hTERT expression. G) Similar to the previous experiment, for pBABE-hTERT transfection, A549 cells were starved and treated with HGF. Western blots showed that hTERT overexpression has a synergistic stimulatory effect with HGF on the expression of c-MET. Full and non-adjusted images of immunoblots were shown in the supplementary material (Figure SM 2)
3.3. hTERT expression induced c-MET promoter activity
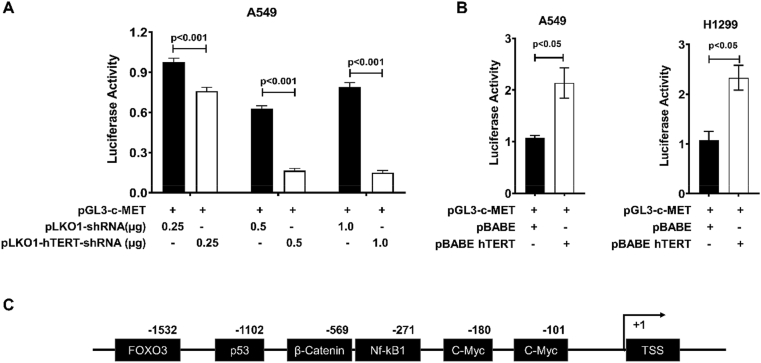
hTERT and c-MET overexpression were reported in lung cancer, and the expression of these two proteins are known to promote cancer progression [25, 26]. To demonstrate the effect of hTERT on c-MET promoter-based transcription, we co-transfected c-MET promoter-luciferase with hTERT shRNA plasmid (pLKO1-hTERT-shRNA) or hTERT expression plasmid (pBABE-hTERT) into A549 and H1299 lung cancer cells. Promoter luciferase assay showed that hTERT knockdown reduced the c-MET promoter activity in A549 cells in a shRNA concentration-dependent manner (Figure 3A). However, hTERT overexpression increased the promoter activity by about 2-fold in A549 and H1299 cells (Figure 3B). Further, the promoter region (1850 bp) of the c-MET gene was scanned employing the TRANSFAC database to find probable transcription factor binding sites. After analysis, we found total 1387 transcription binding elements (matrix score ≥8.5). Some of the transcription factor binding elements shown in Figure 3C and listed in Table 1 are known to interact or be regulated by hTERT [27, 28, 29].
Figure 3.
hTERT expression induced c-MET promoter activity. A) A549 cells were co-transfected with vector and hTERT shRNA plasmid with pGL3-c-MET promoter construct. c-MET promoter activity was assayed using a Luciferase reporter kit. c-MET promoter activity decreased with hTERT downregulation in a dose-dependent manner (Error bars represent mean ± SE, n = 3). B) The luciferase promoter-reporter assay in cells overexpressing hTERT, pBABE-hTERT and control plasmids were co-transfected with pGL3-c-MET promoter construct in A549 and H1299 cells. Results show an increase in c-MET promoter activity on hTERT overexpression in A549 and H1299 lung cancer cells (Mean ± SE, n = 3). Luciferase assay in A549 and H1299 was done independently, and the values of promoter activity were normalized for total cell protein and are shown relative to activity in control cells. C) Bioinformatics analysis of the c-MET promoter region showed many putative transcription factor-binding sites for FOXO3, p53, beta-catenin, NF-kB1, and c-Myc.
Table 1.
Putative binding factors on the c-MET promoter.
| Transcription Factor | Sequence (core sequence bold) | site | Matrix similarity |
|---|---|---|---|
| c-Myc | gCCGCGcgcg | −101 | 0.943 |
| c-Myc | agacagaCACGTgctggggc | −180 | 0.992 |
| Nf-kB1 | atTTCCCt | −271 | 1 |
| β-catenin | gagagCAAAGc | −569 | 0.969 |
| TP53 | CATGAttgaacaagt | −1102 | 0.864 |
| FOXO3 | gcgTTGTTtattta | −1532 | 0.921 |
3.4. p53 and c-Myc play a critical role in hTERT mediated regulation of c-MET expression
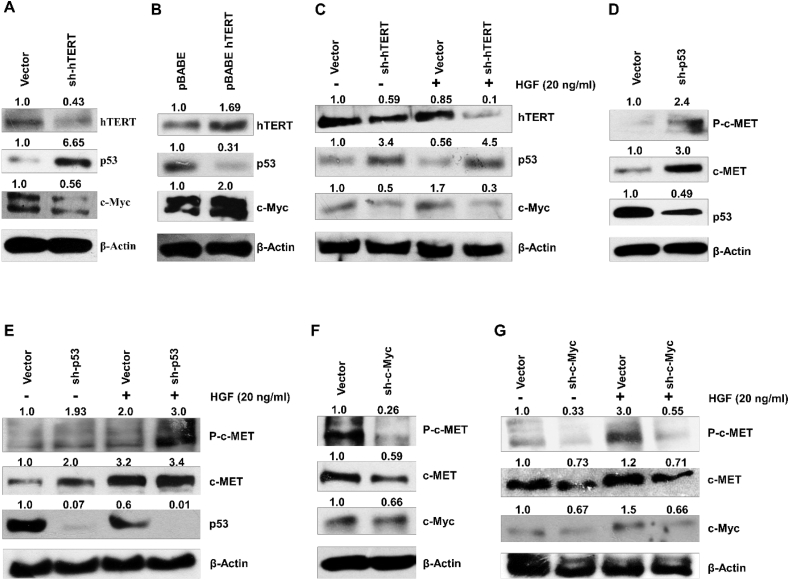
c-MET was closely associated with its promoter activity, and TRANSFAC based promoter analysis reveals the binding sites for some transcription factors like Nf-kb, c-Myc, p53, and FOXO3. Recent studies demonstrate the interaction of hTERT with NF-kB p65 subunit and c-Myc to regulate the expression of IL-6, TNF-α, and heparinase [27, 30]. hTERT also promotes MDM2 mediated ubiquitination and proteasomal degradation of FOXO3a to upregulate the ITGB1 in gastric cancer cells [31]. Similarly, hTERT expression antagonized the p53 induced apoptosis in colon carcinoma cells [32]. To explore whether p53 and c-Myc protein levels in lung cells are regulated by hTERT, we transfected the hTERT shRNA and pBABE-hTERT expression plasmid in A549 cells. shRNA-mediated hTERT knockdown leads to increased expression of tumor suppressor protein p53. In contrast, the c-Myc level was found downregulated in hTERT knockdown A549 cells (Figure 4A). Whereas ectopic overexpression of hTERT in A549 leads to the reduction of p53 expression, hTERT overexpression promotes the c-Myc expression in A549 cells (Figure 4B). There was no significant change in expression of c-Myc and p53 after HGF treatment in hTERT knockdown A549 cells (Figure 4C). Next, we investigated the role of p53 and c-Myc in the regulation of c-MET protein expression. We transfected the A549 cells with the p53 shRNA. Knocking down p53 leads to an increase in c-MET protein level in A549 cells (Figure 4D), and HGF treatment in p53 knockdown cells further increases the expression and phosphorylation of the c-MET protein (Figure 4E). In contrast, p53 overexpression in A549 and H1299 leads to a significant reduction in c-MET expression (Supplementary Figure 3A), irrespective of HGF treatment (Supplementary Figure 3B). In cell knocked down for both hTERT and p53, c-MET expression remained unaffected (Supplementary Figure 3C). The c-Myc is an oncoprotein which is known to activate the expression of several cell survival/proliferative genes [33]. shRNA-mediated downregulation of c-Myc caused concurrent reduction in c-MET expression in A549 (Figure 4F). HGF treatment in c-Myc knockdown A549 cells did not show any significant increase of phosphorylation and expression of c-MET (Figure 4G). Co-transfection of hTERT and c-Myc shRNA in A549 exhibited much lower level of c-MET expression than the individual hTERT and c-Myc shRNA (Supplementary Figure 3D). These results suggest that p53 and c-Myc play an important role in c-MET protein expression, which is induced by hTERT.
Figure 4.
p53 and c-Myc play a critical role in hTERT mediated regulation of c-MET expression. Immunoblot analysis is showing the p53 and c-Myc expression as influenced by (A) hTERT downregulation and (B) over-expression in A549. (C) p53 and c-Myc expression were observed in A549 cells after hTERT knockdown and treatment with HGF. (D) Immunoblot analysis of c-MET, phospho c-MET expression in p53 knockdown A549 cells. (E) Western blot analysis of c-MET and Phospho-c-MET in p53-shRNA transfected A549 cells followed by HGF treatment showed that p53 knockdown increased the c-MET level, and HGF treatment further elevated the expression and phosphorylation of c-MET. (F) Western blot analysis of Phospho-c-MET and c-MET in c-Myc knockdown A549. (G) Western blot analysis of c-MET and Phospho-c-MET in A549 cells knocked down for c-Myc and treated with HGF. c-Myc downregulation reduced the c-MET level, and further HGF treatment in c-Myc knockdown A549 cells does not significantly restore c-MET expression. Full and non-adjusted images of immunoblots were shown in the supplementary material (Figure SM 3)
3.5. c-MET induced EMT is positively associated with hTERT expression
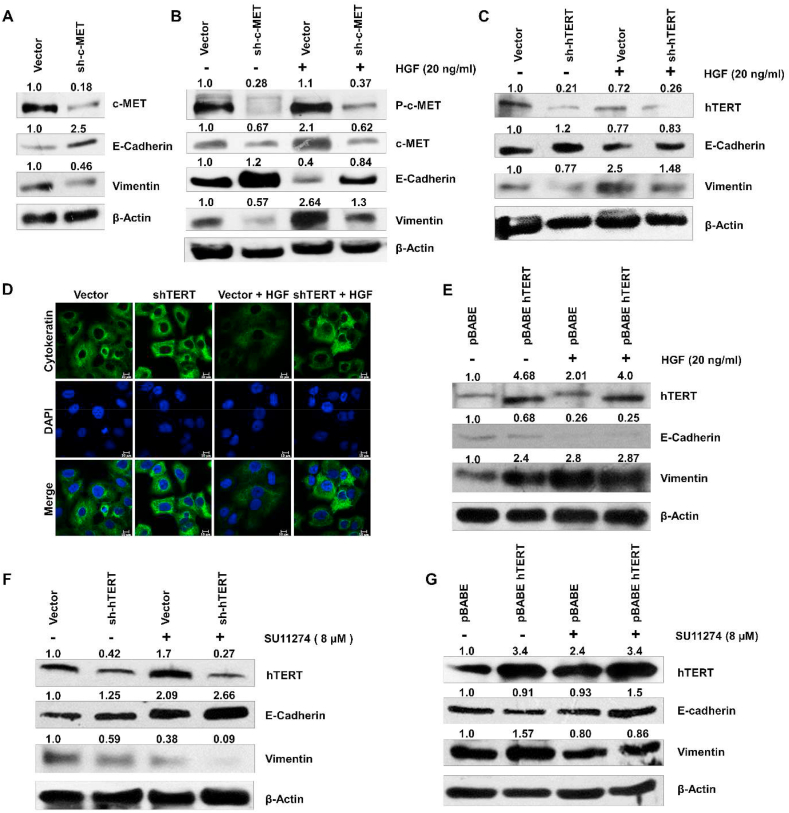
HGF/c-MET signaling triggers tumor growth and metastasis [34]. A549 cells knocked down for c-MET expression by shRNA showed decreased expression of mesenchymal protein vimentin and increased expression of the epithelial protein E-cadherin (Figure 5A). HGF treatment of A549 cells significantly reduced the expression level of E-cadherin and cytokeratin-18a, whereas it promotes the vimentin expression. However, knocking down c-MET expression with shRNA abolished the HGF induced EMT (Figure 5B & Supplementary Figure 4A). However, hTERT downregulation reversed the HGF mediated EMT by restoring expression of the epithelial markers E-cadherin and cytokeratin-18a (Figure 5C, D). In contrast, hTERT over-expression showed a synergistic effect with HGF and promoted the EMT by increasing the vimentin and suppressing E-cadherin (Figure 5E). To validate that HGF/c-MET mediated EMT is associated with hTERT expression, hTERT-shRNA or pBABE-hTERT transfected cells were treated with the SU11274 (c-MET inhibitor). The results showed that SU11274 treatment reduced the mesenchymal feature in A549 by reducing the vimentin and increasing the E-cadherin (Supplementary Figure 4B). The combined treatment with SU11274 and hTERT shRNA in A549 additively downregulates the mesenchymal protein vimentin and promotes E-cadherin and cytokeratin-18a expression (Figure 5F and Supplementary Figure 4C). In contrast, treatment of SU11274 in pBABE-hTERT transfected A549 cells inhibits the hTERT-induced EMT by increasing the E-cadherin and cytokeratin-18a and inhibiting the vimentin expression (Figure 5G and Supplementary Figure 4D). A549 cells were transfected with p53-shRNA showed enhanced vimentin expression and decreased expression of E-cadherin and cytokeratin-18a in response to treatment with HGF (Supplementary Figures 5A & B). However, p53 over-expression promotes the epithelial feature (an increase of E-cadherin and cytokeratin-18a, and decreased vimentin expression) in HGF stimulated A549 cells compared to control cells treated with HGF ligand (Supplementary Figure 5C & D). Knocking down c-Myc expression promoted expression of the epithelial markers E-cadherin and cytokeratin-18a and suppressed vimentin. HGF treatment in the vector control A549 promoted the expression of the mesenchymal features (higher vimentin level and lower E-cadherin and cytokeratin-18a compared to the non-HGF treated A549). In contrast, HGF treatment in c-Myc-shRNA transfected A549 cells did not show a significant stimulatory effect on EMT markers (Supplementary Figure 5E & F). These results reflect that c-MET induced EMT is positively associated with hTERT expression in A549 cells.
Figure 5.
c-MET induced EMT is positively associated with hTERT expression. (A) Western blot showing increased E-cadherin expression and decreased vimentin expression following c-MET knockdown A549 cells. (B) c-MET knocked down A549 cells were starved for 12 h and treated with HGF (20 ng/ml) for 24 h. Western blots show inverse relationship between expression of c-MET and epithelial marker E-cadherin and a direct relationship with mesenchymal marker vimentin. C) hTERT knockdown A549 cells were treated with HGF and analyzed for expression of E-cadherin and vimentin. shRNA targeting hTERT. Western blots show that knocking down hTERT expression promoted the expression of epithelial marker E-cadherin and decreased the mesenchymal protein vimentin in cells. D) Immunofluorescence staining of cytokeratin-18a was done in hTERT knockdown A549 cells treated with HGF. HGF treatment reduced the cytokeratin level, while hTERT-shRNA treatment restored the cytokeratin expression. E) Stably hTERT over-expressing A549 cells were starved and then stimulated by HGF (20 ng/ml), leading to increased expression of vimentin and suppression of E-cadherin synergistically. F) hTERT downregulated A549 cells were treated with the 8 μM SU11274 (c-MET inhibitor), and expression of E-cadherin and vimentin were detected by western blotting. G) Immunoblot analysis of epithelial marker E-cadherin and vimentin following SU11274 (8 μM) treatment in hTERT over-expressing A549 cells showed lower expression of this mesenchymal marker. Full and non-adjusted images of immunoblots were shown in the supplementary material (Figure SM 4)
4. Discussion
Telomerase activation is crucial for genomic integrity and cellular immortalization. Its activation is prominent in dividing cells such as stem and cancer cells [35]. Telomerase consists of RNA (TERC) and protein components (hTERT); TERC works as a template for hTERT; together, they constitute RNA-dependent DNA polymerase activity. Unlike in the somatic cells, cancer cells show high telomerase activity due to increased transcriptional activity of hTERT [36]. Recent studies suggest the telomere-independent function of the telomerase protein component (hTERT) [3, 37]. hTERT has been shown to play a crucial role in the regulation of EMT and invasion process [18, 38, 39]; however, the precise pathway or mechanism apart from its canonical role remains elusive. Some studies have shown that hTERT is associated with the regulation of gene expression involved in cell survivability, angiogenesis, differentiation, cell cycle, and proliferation [40]. hTERT interaction with chromatin remodeling protein BRG1 and β-catenin activates the Wnt/β-catenin target genes, giving a probable explanation for hTERT mediated gene regulation [19]. hTERT also forms a complex with the p65 subunit of Nf-kB and activates the NF-kB target genes like IL-6, IL-8, and TNF-α inflammatory factors. A recent study also showed that hTERT interacts with MDM2 (E3 ubiquitin ligase) and increases the ubiquitination of FOXO3a, which subsequently induces ITGB1 expression [31].
hTERT promotes the epithelial to mesenchymal transition through activation of wnt/β-catenin signaling, increasing the snail, vimentin, and other EMT proteins [18]. However, hTERT/ZEB1 interaction directly suppresses the epithelial protein E-cadherin in colorectal cancer [41]. hTERT expression also promotes the remodeling of extracellular matrices (ECM) by activating MMPs and heparinase [29, 42]. Our study provides a new molecular pathway of hTERT induced epithelial to mesenchymal transition and migration. Here, we have demonstrated that hTERT promotes the EMT by activating the c-MET signaling. hTERT mediated activation of c-MET promotes the expression of vimentin (mesenchymal protein) while inhibiting E-cadherin and cytokeratin-18a (epithelial markers).
Effect of hTERT on c-MET expression has been observed at promoter level through luciferase reporter assay. Our finding suggests the promotional effect of hTERT on c-MET expression. Further, c-MET promoter (1850 bp) analysis reveals the binding sites for transcription factors known to interact or regulate through hTERT like p53 and c-Myc. p53 is a tumor suppressor protein, targets the genes associated with growth, survivability, angiogenesis, and metastasis [43, 44, 45, 46]. Activation of p53 occurs in response to DNA damage, which further stimulates the transcription of cyclin-dependent kinase (Cdk) inhibitory protein p21 and cell cycle arrest [47]. hTERT expression antagonizes the p53 induced apoptosis independently of the telomerase activity [32]. Consistent with these reports, our findings also suggest the role of hTERT in p53 protein expression. shRNA-mediated silencing of hTERT increases the expression of p53 protein in lung cancer cells (A549 and H1299), while hTERT overexpression reduced the p53 level in cells.
c-Myc is a proto-oncogene belonging to the Myc family, which regulates cell cycle, proliferation, EMT, migration, apoptosis, and metabolism. c-Myc is involved in regulating a broad spectrum of genes; 10–15% of all promoter regions contain c-Myc binding sites [48]. This protein is overexpressed/activated in most cancers and is considered as a molecular hallmark of carcinogenesis [49]. c-Myc is known to induce the transcription activation of the catalytic subunit (hTERT) of telomerase in transformed or proliferating cells [50]. Bo Tang et al. had shown that hTERT interacts with c-Myc and regulates the expression of heparinase in gastric cancer [27]. In our study, we have shown the positive cooperation of hTERT on c-Myc expression. We found decreased c-Myc expression on shRNA-mediated hTERT knockdown; however, hTERT overexpression promotes the c-Myc expression in A549 cells.
Wild type p53 and c-Myc exerts opposing influences on c-MET protein expression in lung cancer cells. The c-MET promoter contains binding sites for p53 (−1102) and c-Myc (−101, −180) upstream to the transcription start site. Wild-type p53 is reported to promote the c-MET transcriptional repression through physical interaction with SP1 (c-MET transcription activation factor) to inhibit its binding to the c-MET promoter DNA [51]. However, p53 mutant exerts a dominant-negative effect over wild-type p53 [52]; mutant p53 acquires oncogenic potential and promotes the expression of c-MET [53]. Similarly, we found higher expression of c-MET in cancer cells that lack p53 or express mutant p53 over wild-type p53 expressing cells (Supplementary Figure 6A and B). Further, shRNA-mediated downregulation of p53 leads to an increased cellular level of c-MET protein in A549. In contrast, ectopic expression of p53 drastically reduced the c-MET expression. On the other hand, c-Myc protein expression is positively correlated with c-MET expression in large-cell medulloblastoma [54]. In our finding, shRNA-mediated downregulation of c-Myc reduced the c-MET level in lung cancer cells. Thus, it seems the availability of c-MET in cells depends on the fine-tuning between p53 and c-Myc expression. In cancer cells, high-level hTERT favors the c-Myc expression to promote the c-MET transcription activation.
c-MET, receptor tyrosine kinase regulates the invasion and migration during embryonic and cancer development [55, 56]. This protein is upregulated in most tumors and is correlated to increased invasion and migration with poor survival [57, 58]. Increased c-MET signaling stimulates ERK, PI3K, and FAK to facilitate cell motility and invasion [59, 60]. Furthermore, c-MET expression and signaling dysregulation promote angiogenesis through PI3K/Akt/mTOR pathway [61]. In our study, we have shown that c-MET downregulation reduces cell proliferation and survivability. Further study shows that c-MET silencing increases epithelial proteins (E-cadherin and cytokeratin-18a) and represses the mesenchymal protein (vimentin). More importantly, c-MET knockdown inhibits HGF induced EMT. Similarly, c-MET targeting through SU11274 also reduces EMT in a concentration-dependent manner.
5. Conclusion
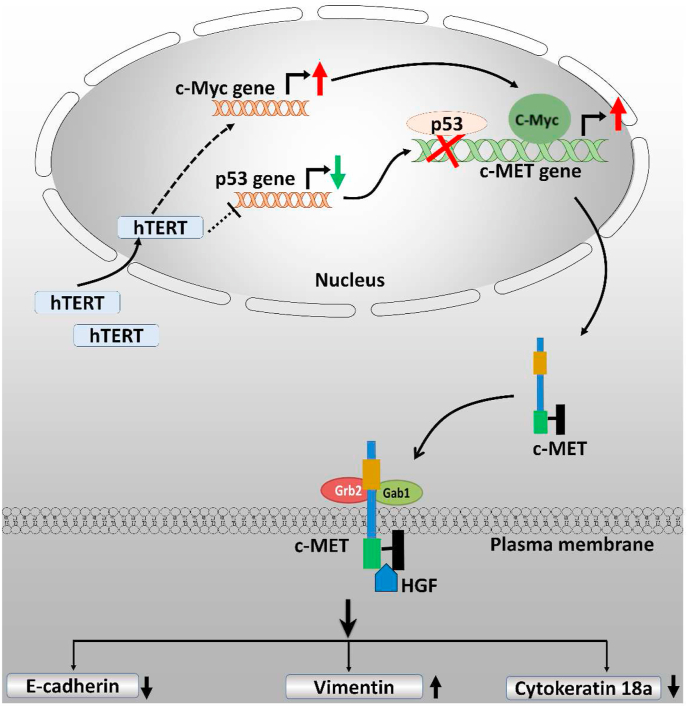
Here, we have reported an essential axis of molecular interactions determining EMT induced by hTERT. We found that hTERT promotes c-MET expression at the transcription level in a p53 and c-Myc dependent manner (Figure 6). c-MET is an essential regulator of cancer malignancy; Its expression helps the tumor cells to acquire epithelial to mesenchymal transition.
Figure 6.
A model is showing the role of hTERT in epithelial to mesenchymal transition by enhancing the c-MET upregulation.
Declarations
Author contribution statement
Ram Raj Prasad: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Deepak Kumar Mishra: Analyzed and interpreted the data.
Manoj Kumar: Performed the experiments.
Pramod Kumar Yadava: Conceived and designed the experiments.
Funding statement
This work was supported by Jawaharlal Nehru University that provided access to its Central Instruments Facility and essential consumable grant out of grants received from UGC (176), DST (2-DST-PURSE 2015- 16), and ICMR.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick L. The cell biology of aging. Clin. Geriatr. Med. 1985;1:15–27. [PubMed] [Google Scholar]
- 3.Stewart S.A., Hahn W.C., O’Connor B.F., Banner E.N., Lundberg A.S., Modha P., Mizuno H., Brooks M.W., Fleming M., Zimonjic D.B., Popescu N.C., Weinberg R.A. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez P., Blasco M.A. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 5.Guo W., Lu J., Dai M., Wu T., Yu Z., Wang J., Chen W., Shi D., Yu W., Xiao Y., Yi C., Tang Z., Xu T., Xiao X., Yuan Y., Liu Q., Du G., Deng W. Transcriptional coactivator CBP upregulates hTERT expression and tumor growth and predicts poor prognosis in human lung cancers. Oncotarget. 2014;5:9349–9361. doi: 10.18632/oncotarget.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi J., Southworth L.K., Sarin K.Y., Venteicher A.S., Ma W., Chang W., Cheung P., Jun S., Artandi M.K., Shah N., Kim S.K., Artandi S.E. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4:124–138. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gherardi E., Gray J., Stoker M., Perryman M., Furlong R., Gough J., Bandyopadhyay A., Hartmann G., Butler P.J.G. Purification of scatter factor, a fibroblast-derived basic protein that modulates epithelial interactions and movement. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5844–5848. doi: 10.1073/pnas.86.15.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiss A., Wang N.J., Xie J.P., Thorgeirsson S.S. Analysis of transforming growth factor (TGF)-alpha/epidermal growth factor receptor, hepatocyte growth Factor/c-met,TGF-beta receptor type II, and p53 expression in human hepatocellular carcinomas. Clin. Cancer Res. 1997;3:1059–1066. [PubMed] [Google Scholar]
- 9.Uehara Y., Minowa O., Mori C., Shiota K., Kuno J., Noda T., Kitamura N. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature. 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt C., Bladt F., Goedecke S., Brinkmann V., Zschiesche W., Sharpe M., Gherardi E., Birchmeler C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 11.Bussolino F., Di Renzo M.F., Ziche M., Bocchietto E., Olivero M., Naldini L., Gaudino G., Tamagnone L., Coffer A., Comoglio P.M. Hepatocyte growth factor is a potent angiogenic factor which stimulates endothelial cell motility and growth. J. Cell Biol. 1992;119:629–641. doi: 10.1083/jcb.119.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vande Woude G.F., Jeffers M., Cortner J., Alvord G., Tsarfaty I., Resau J. Wiley-Blackwell; 2007. Met-HGF/SF: Tumorigenesis, Invasion and Metastasis; pp. 119–132. [DOI] [PubMed] [Google Scholar]
- 13.Farrell J., Kelly C., Rauch J., Kida K., García-Muñoz A., Monsefi N., Turriziani B., Doherty C., Mehta J.P., Matallanas D., Simpson J.C., Kolch W., von Kriegsheim A. HGF induces epithelial-to-mesenchymal transition by modulating the Mammalian Hippo/MST2 and ISG15 pathways. J. Proteome Res. 2014;13:2874–2886. doi: 10.1021/pr5000285. [DOI] [PubMed] [Google Scholar]
- 14.Giordano S., Di Renzo M.F., Narsimhan R.P., Tamagnone L., Gerbaudo E.V., Chiadó-Piat L., Comoglio P.M. Evidence for autocrine activation of a tyrosine kinase in a human gastric carcinoma cell line. J. Cell. Biochem. 1988;38:229–236. doi: 10.1002/jcb.240380402. [DOI] [PubMed] [Google Scholar]
- 15.Koochekpour S., Jeffers M., Rulong S., Taylor G., Klineberg E., Hudson E.A., Resau J.H., Vande Woude G.F. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57:5391–5398. [PubMed] [Google Scholar]
- 16.Maulik G., Kijima T., Ma P.C., Ghosh S.K., Lin J., Shapiro G.I., Schaefer E., Tibaldi E., Johnson B.E., Salgia R. Modulation of the c-Met/hepatocyte growth factor pathway in small cell lung cancer. Clin. Cancer Res. 2002;8:620–627. [PubMed] [Google Scholar]
- 17.Kuhnen C., Muehlberger T., Honsel M., Tolnay E., Steinau H.U., Miller K.-M. Impact of c-Met expression on angiogenesis in soft tissue sarcomas: correlation to microvessel-density. J. Cancer Res. Clin. Oncol. 2003;129:415–422. doi: 10.1007/s00432-003-0456-4. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z., Li Q., Li K., Chen L., Li W., Hou M., Liu T., Yang J., Lindvall C., Björkholm M., Jia J., Xu D. Telomerase reverse transcriptase promotes epithelial–mesenchymal transition and stem cell-like traits in cancer cells. Oncogene. 2013;32:4203–4213. doi: 10.1038/onc.2012.441. [DOI] [PubMed] [Google Scholar]
- 19.Park J.-I., Venteicher A.S., Hong J.Y., Choi J., Jun S., Shkreli M., Chang W., Meng Z., Cheung P., Ji H., McLaughlin M., Veenstra T.D., Nusse R., McCrea P.D., Artandi S.E. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L., Zheng D., Wang M., Cong Y.-S. Telomerase reverse transcriptase activates the expression of vascular endothelial growth factor independent of telomerase activity. Biochem. Biophys. Res. Commun. 2009;386:739–743. doi: 10.1016/j.bbrc.2009.06.116. [DOI] [PubMed] [Google Scholar]
- 21.Smith L.L., Coller H.A., Roberts J.M. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 22.Gao C.F., Vande Woude G.F. HGF/SF-Met signaling in tumor progression. Cell Res. 2005;15:49–51. doi: 10.1038/sj.cr.7290264. [DOI] [PubMed] [Google Scholar]
- 23.Ahn S.Y., Kim J., Kim M.A., Choi J., Kim W.H. Increased HGF expression induces resistance to c-MET tyrosine kinase inhibitors in gastric cancer. Anticancer Res. 2017;37(3):1127–1138. doi: 10.21873/anticanres.11426. [DOI] [PubMed] [Google Scholar]
- 24.Knowles L.M., Stabile L.P., Egloff A.M., Rothstein M.E., Thomas S.M., Gubish C.T., Lerner E.C., Seethala R.R., Suzuki S., Quesnelle K.M., Morgan S., Ferris R.L., Grandis J.R., Siegfried J.M. HGF and c-Met participate in paracrine tumorigenic pathways in head and neck squamous cell cancer. Clin. Cancer Res. 2009;15:3740–3750. doi: 10.1158/1078-0432.CCR-08-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu C.-Q., Cutz J.-C., Liu N., Lau D., Shepherd F.A., Squire J.A., Tsao M.-S. Amplification of telomerase (hTERT) gene is a poor prognostic marker in non-small-cell lung cancer. Br. J. Cancer. 2006;94:1452–1459. doi: 10.1038/sj.bjc.6603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sierra J.R., Tsao M.-S. c-MET as a potential therapeutic target and biomarker in cancer. Ther. Adv. Med. Oncol. 2011;3:S21–S35. doi: 10.1177/1758834011422557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang B., Xie R., Qin Y., Xiao Y.-F., Yong X., Zheng L., Dong H., Yang S.-M. Human telomerase reverse transcriptase (hTERT) promotes gastric cancer invasion through cooperating with c-Myc to upregulate heparanase expression. Oncotarget. 2016;7:11364–11379. doi: 10.18632/oncotarget.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin X., Beck S., Sohn Y.-W., Kim J.-K., Kim S.-H., Yin J., Pian X., Kim S.-C., Choi Y.-J., Kim H. Human telomerase catalytic subunit (hTERT) suppresses p53-mediated anti-apoptotic response via induction of basic fibroblast growth factor. Exp. Mol. Med. 2010;42:574. doi: 10.3858/emm.2010.42.8.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding D., Xi P., Zhou J., Wang M., Cong Y.-S. Human telomerase reverse transcriptase regulates MMP expression independently of telomerase activity via NF-κB-dependent transcription. Faseb. J. 2013;27:4375–4383. doi: 10.1096/fj.13-230904. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh A., Saginc G., Leow S.C., Khattar E., Shin E.M., Yan T.D., Wong M., Zhang Z., Li G., Sung W.-K., Zhou J., Chng W.J., Li S., Liu E., Tergaonkar V. Telomerase directly regulates NF-κB-dependent transcription. Nat. Cell Biol. 2012;14:1270–1281. doi: 10.1038/ncb2621. [DOI] [PubMed] [Google Scholar]
- 31.Hu C., Ni Z., Li B., Yong X., Yang X., Zhang J., Zhang D., Qin Y., Jie M., Dong H., Li S., He F., Yang S. hTERT promotes the invasion of gastric cancer cells by enhancing FOXO3a ubiquitination and subsequent ITGB1 upregulation. Gut. 2015:1–12. doi: 10.1136/gutjnl-2015-309322. [DOI] [PubMed] [Google Scholar]
- 32.Rahman R., Latonen L., Wiman K.G. hTERT antagonizes p53-induced apoptosis independently of telomerase activity. Oncogene. 2005;24:1320–1327. doi: 10.1038/sj.onc.1208232. [DOI] [PubMed] [Google Scholar]
- 33.Dang C.V. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeon H.-M., Lee J. MET: roles in epithelial-mesenchymal transition and cancer stemness. Ann. Transl. Med. 2017;5 doi: 10.21037/atm.2016.12.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janknecht R. On the road to immortality: hTERT upregulation in cancer cells. FEBS Lett. 2004;564:9–13. doi: 10.1016/S0014-5793(04)00356-4. [DOI] [PubMed] [Google Scholar]
- 36.Komiya T., Kawase I., Nitta T., Yasumitsu T., Kikui M., Fukuoka M., Nakagawa K., Hirashima T. Prognostic significance of hTERT expression in non-small cell lung cancer. Int. J. Oncol. 2000;16:1173–1177. doi: 10.3892/ijo.16.6.1173. [DOI] [PubMed] [Google Scholar]
- 37.Bednarek A., Budunova I., Slaga T.J., Aldaz C.M. Increased telomerase activity in mouse skin premalignant progression. Cancer Res. 1995;55:4566–4569. [PubMed] [Google Scholar]
- 38.He B., Xiao Y.-F., Tang B., Wu Y.-Y., Hu C.-J., Xie R., Yang X., Yu S.-T., Dong H., Zhao X.-Y., Li J.-L., Yang S.-M. hTERT mediates gastric cancer metastasis partially through the indirect targeting of ITGB1 by microRNA-29a. Sci. Rep. 2016;6:21955. doi: 10.1038/srep21955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L., Lü M.-H., Zhang D., Hao N.-B., Fan Y.-H., Wu Y.-Y., Wang S.-M., Xie R., Fang D.-C., Zhang H., Hu C.-J., Yang S.-M. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2013.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Low K.C., Tergaonkar V. Telomerase: central regulator of all of the hallmarks of cancer. Trends Biochem. Sci. 2013;38:426–434. doi: 10.1016/j.tibs.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Qin Y., Tang B., Hu C.-J., Xiao Y.-F., Xie R., Yong X., Wu Y.Y., Dong H., Yang S.-M. An hTERT/ZEB1 complex directly regulates E-cadherin to promote epithelial-to-mesenchymal transition (EMT) in colorectal cancer. Oncotarget. 2016;7:351–361. doi: 10.18632/oncotarget.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang B., Xie R., Qin Y., Xiao Y.-F., Yong X., Zheng L., Dong H., Yang S.-M., Tang B., Xie R., Qin Y., Xiao Y.-F., Yong X., Zheng L., Dong H., Yang S.-M. Human telomerase reverse transcriptase (hTERT) promotes gastric cancer invasion through cooperating with c-Myc to upregulate heparanase expression. Oncotarget. 2016;7:11364–11379. doi: 10.18632/oncotarget.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker S.J., Markowitz S., Fearon E.R., Willson J.K., Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Sci. Res. Libr. Pg. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 44.Yu J., Zhang L. No PUMA, no death: implications for p53-dependent apoptosis. Cancer Cell. 2003;4:248–249. doi: 10.1016/s1535-6108(03)00249-6. [DOI] [PubMed] [Google Scholar]
- 45.Teodoro J.G., Evans S.K., Green M.R. Inhibition of tumor angiogenesis by p53: a new role for the guardian of the genome. J. Mol. Med. 2007;85:1175–1186. doi: 10.1007/s00109-007-0221-2. [DOI] [PubMed] [Google Scholar]
- 46.Powell E., Piwnica-Worms D., Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov. 2014;4:405–414. doi: 10.1158/2159-8290.CD-13-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lakin N.D., Jackson S.P. Regulation of p53 in response to DNA damage. Oncogene. 1999;18(53):7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 48.Lüscher B., Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494:145–160. doi: 10.1016/j.gene.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 49.Gabay M., Li Y., Felsher D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014;4 doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu K.-J., Grandori C., Amacker M., Simon-Vermot N., Polack A., Lingner J., Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat. Genet. 1999;212(21):220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 51.Hwang C.-I., Matoso A., Corney D.C., Flesken-Nikitin A., Körner S., Wang W., Boccaccio C., Thorgeirsson S.S., Comoglio P.M., Hermeking H., Nikitin A.Y. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14240–14245. doi: 10.1073/pnas.1017536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S.V., Hainaut P., Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 53.Grugan K.D., Vega M.E., Wong G.S., Diehl J.A., Bass A.J., Wong K.K., Nakagawa H., Rustgi A.K. A common p53 mutation (R175H) activates c-Met receptor tyrosine kinase to enhance tumor cell invasion. Cancer Biol. Ther. 2013;14:853–859. doi: 10.4161/cbt.25406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Guessous F., Johnson E.B., Eberhart C.G., Li X.-N., Shu Q., Fan S., Lal B., Laterra J., Schiff D., Abounader R. Functional and molecular interactions between the HGF/c-Met pathway and c-Myc in large-cell medulloblastoma. Lab. Invest. 2008;88:98–111. doi: 10.1038/labinvest.3700702. [DOI] [PubMed] [Google Scholar]
- 55.Birchmeier C., Gherardi E. Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase. Trends Cell Biol. 1998;8:404–410. doi: 10.1016/s0962-8924(98)01359-2. [DOI] [PubMed] [Google Scholar]
- 56.Han Y., Luo Y., Zhao J., Li M., Jiang Y. Overexpression of c-Met increases the tumor invasion of human prostate LNCaP cancer cells in vitro and in vivo. Oncol. Lett. 2014;8:1618–1624. doi: 10.3892/ol.2014.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ichimura E., Maeshima A., Nakajima T., Nakamura T. Expression of c-met/HGF receptor in human non-small cell lung carcinomas in vitro and in vivo and its prognostic significance. Jpn. J. Cancer Res. 1996;87:1063–1069. doi: 10.1111/j.1349-7006.1996.tb03111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Organ S.L., Tsao M.-S. An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol. 2011;3:S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin B., Liu Z., Wang Y., Wang X., Liu W., Yu P., Duan X., Liu C., Chen Y., Zhang Y., Pan X., Yao H., Liao Z., Tao Z. RON and c-Met facilitate metastasis through the ERK signaling pathway in prostate cancer cells. Oncol. Rep. 2017;37:3209–3218. doi: 10.3892/or.2017.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Usatyuk P.V., Fu P., Mohan V., Epshtein Y., Jacobson J.R., Gomez-Cambronero J., Wary K.K., Bindokas V., Dudek S.M., Salgia R., Garcia J.G.N., Natarajan V. Role of c-met/phosphatidylinositol 3-kinase (PI3k)/Akt signaling in hepatocyte growth factor (HGF)-mediated lamellipodia formation, reactive oxygen species (ROS) generation, and motility of lung endothelial cells. J. Biol. Chem. 2014;289:13476–13491. doi: 10.1074/jbc.M113.527556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong S.-W., Jung K.H., Lee H.-S., Son M.K., Yan H.H., Kang N.S., Lee J., Hong S.-S. SB365, Pulsatilla saponin D, targets c-Met and exerts antiangiogenic and antitumor activities. Carcinogenesis. 2013;34:2156–2169. doi: 10.1093/carcin/bgt159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.