Abstract
The emergence of new SARS-CoV-2 variants poses a threat to the human population where it is difficult to assess the severity of a particular variant of the virus. Spike protein and specifically its receptor binding domain (RBD) which makes direct interaction with the ACE2 receptor of the human has shown prominent amino acid substitutions in most of the Variants of Concern. Here, by using all-atom molecular dynamics simulations we compare the interaction of Wild-type RBD/ACE2 receptor complex with that of the latest Omicron variant of the virus. We observed a very interesting diversification of the charge, dynamics and energetics of the protein complex formed upon mutations. These results would help us in understanding the molecular basis of binding of the Omicron variant with that of SARS-CoV-2 Wild-type.
Keywords: Covid19, Variants, SARS-CoV-2, Molecular dynamics
Graphical abstract
1. Introduction
The Covid19 pandemic started in the later part of 2019 claiming millions of lives and affecting several others physically and mentally [1,2]. Although several vaccines have been developed against the virus, we still have to remain observant due to emergence of novel variants of the virus. It is in the nature of RNA viruses, such as coronaviruses, to undergo frequent genomic mutations [3]. As a result of the mutation, the structure as well as the dynamics of the proteins, including the viral proteins get affected [4,5]. The WHO has briefly classified the variants of SARS-CoV-2 into Variants of Concern (VOC) and Variants of Interest (VOI). The VOCs are of particular interest due to higher infectivity and transmissibility [2]. After the Alpha, Beta, Gamma and Delta variants, recently, a new Omicron variant (B.1.1.529) has been identified [2]. The initial reports indicate a notable 32 mutations in the Spike glycoprotein of the virus [6]. The Spike glycoprotein is one of the largest structural proteins of the virus, whose stalk is embedded in the viral membrane while a large head interacts with the host Angiotensin Converting Enzyme 2 (ACE2) receptor [7]. The primary role of the Spike protein lies in the attachment of the virus to the host cells [8].
The Spike protein, constituted by three peptide chains, has a specific domain on the surface which is known as the Receptor Binding Domain or RBD [9]. This RBD is the interacting domain of the virus. Most of the reported mutations of VOCs have been found to occur in the RBD which vary from residue 333–527 [10]. For instance, in N501Y mutate in Alpha, Beta and Gamma variants; K417T in Alpha and Beta; and T478K in the Delta variant [2]. As per the initial reports, a total of 15 mutations have been identified on the RBD of the Omicron variant. Since the primary role of RBD lies in the binding with the ACE2 receptor, the identified mutations would also influence the nature of interaction in the protein-protein complex as observed in earlier studies [11]. Molecular studies have shown that mutations have resulted in variation in the residue wise interaction energies reported for SARS and SARS-CoV-2 as well as in other variants [12,13].
In our study we have considered the RBD/ACE2 complex taken from the crystal structure as the Wild-type system [14]. Due to the lack of available crystal structures of the Omicron variant and the urgency in understanding the molecular details of protein interactions, we have generated the Omicron variant model using homology modeling using Modeller [15]. We have omitted the glycans in our study, since earlier studies by Amaro et al. confirmed that the RBD of the Spike protein had significantly less glycans compared to the rest of the Spike protein and did not directly participate in the ACE2 interaction [16]. Subsequently both the systems were subjected to minimization, equilibration and production run using Gromacs MD Simulation package [17] and CHARMM36 force field [18] for 100 ns (ran in triplicates) to ascertain the accuracy of the structural-dynamics parameters (details below).
2. Materials and methods
The crystal structure of the Spike protein RBD associated with ACE2 was taken from Protein Data Bank (PDB ID: 6LZG) as the starting structure [14]. This structure was considered as the Wild-type system. The Omicron variant was modelled using Modeller 10.1 molecular modeling suite using the Wild-type RBD/ACE2 complex as the template [19]. All the residues that had mutated were modelled. Out of the 30 models that were generated, the model with the lowest DOPE score [20] was taken as the starting structure for the Omicron system.
All-atom Molecular Dynamics (MD) Simulation studies were carried out using Gromacs MD Simulation package [17]. CHARMM36 force field [18] with TIP3P waters and neutralizing Na + ions were used in the initial system setup [21]. After initial minimization and equilibration, production simulation was carried out for 100ns in triplicates to generate the trajectory for analysis. Berendson thermostat [25] and Parrinello-Rahman barostat [22] were used for running the simulations at 310K and 1 atm pressure in NPT conditions. The generated trajectory was saved at the interval of every 1000 ps. Particle-mesh Ewald method was used to treat the long-range electrostatic interactions [23]. Pymol [24] were used for visualization of the trajectories, generating the porcupine plots [25] and Adaptive Poisson Boltzmann surface Area calculations [26,27]. All the analyses were carried out using Gromacs tools [17].
G_mmpbsa module was used for the Molecular Mechanics/Poisson Boltzmann Surface Area (MM/PBSA) calculations [28,29]. Binding energies between the RBD/ACE2 proteins were calculated using the following theory:
where ΔGprotein, ΔGcomplex, and ΔGligand represent the total free energies of the complex, the ligand, and the protein also separately in the solvent. Free energy was calculated by the following equation:
where EMM stands for the average molecular mechanic's potential energy in the vacuum, Gsolvation denotes the free energy of solvation and TS stands for entropy and temperature, respectively. The binding energies of the complexes were calculated from 50 snapshots over the last 10 ns of the simulation trajectories.
3. Results and discussion
Although the RBD is the interacting domain of the Spike protein, the exact site of interaction lies between residues 438–506 known as the Receptor Binding Motif (RBM) [10]. Structurally, the RBD comprises a twisted beta sheet and a small alpha helix, while the RBM is made up of four loops and two small beta strands9 .
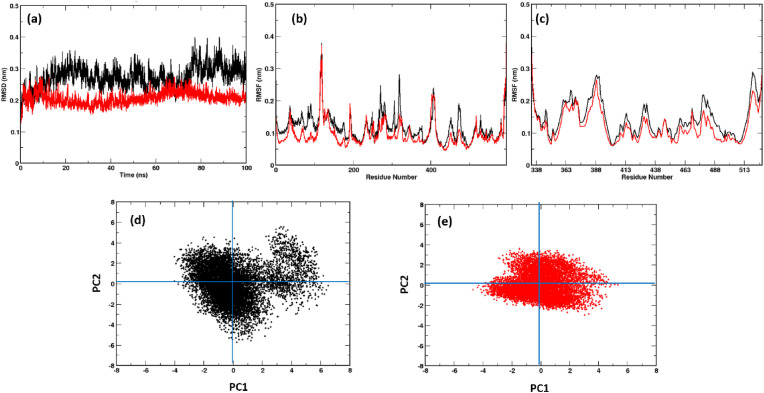
During the 100 ns simulation run time, we observed stability of both the systems (Fig. 1 a, Fig. S1). Surprisingly, the Omicron system was more stable than the Wild-type despite the mutations. Detailed RMS fluctuations of the Cα atoms of the residues shows some interesting observations. Compared with earlier studies with Alpha, Beta, Gamma and Delta [13], the RMSF values of the Omicron variant over the simulation time were lower than the Wild-type (Fig. 1b, c, Fig. S2). This is noteworthy since earlier mutational studies had shown the RMSF values to be either similar or higher than the Wild-type system. This phenomenon was observed for both the ACE2 (Fig. 1b) as well as the RBD (Fig. 1b) part of the Omicron protein complex clearly indicating the formation of a more stable protein complex. Not only the RBM in the RBD but the whole protein appears very relaxed with the ACE2 partner protein.
Fig. 1.
(a) RMSDs and RMSFs of (b) ACE2 and (c) RBD calculated over the simulation time and projection of the first and second principal components of the (d) Wild-type and (e) Omicron system displaying the difference in RBD/ACE2 complex flexibility. The Omicron system was found to be more stable than Wild-type. (Color Wild-type: black; Omicron: Red). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The peculiarity in the RMSF values, prompted us to investigate the dynamics of the RBD/ACE2 complex along the same lines. We performed Principal Component Analysis (PCA) of the Wild-type and Omicron RBD/ACE2 complex [30]. In Fig. 1d and e, a clear difference in the dynamics of the Omicron system was evident. In the Wild-type system the flexibility was found to be more along the first principal component. The trace values of 13.91 in Wild-type and 10.02 in Omicron also indicate compactness of the Omicron system.
The total interaction energies of the generated protein complex formed in both Wild-type RBD/ACE2 and Omicron RBD/ACE2 variants was compared during the last 10ns of the simulation time (Table 1 ). A value of −2658 kJ/mol of the Omicron distinctly shows the strength of the formed protein complex. Wild-type complex shows a value of only −1022 kJ/mol, which is 2.5 times lower than the Omicron system. Major changes were observed in the electrostatic energies of the protein. This can be explained due to the change in the amino acid R-groups in the mutating residues as shown in Table 2 .
Table 1.
Total energy of binding of RBD with the human ACE2 receptor.
| Wild-type | Omicron | |
|---|---|---|
| van der Waal energy | −363.775 ± 20.446 kJ/mol | −335.186 ± 21.937 kJ/mol |
| Electrostatic energy | −1232.614 ± 64.543 kJ/mol | −3083.962 ± 89.676 kJ/mol |
| Polar solvation energy | 617.810 ± 161.796 kJ/mol | 803.804 ± 119.784 kJ/mol |
| SASA energy | −43.889 ± 3.469 kJ/mol | −42.889 ± 3.818 kJ/mol |
| Binding energy | −1022.467 ± 150.703 kJ/mol | −2658.233 ± 129.686 kJ/mol |
Table 2.
Residue-wise binding energy calculated for the mutated residues in RBD.
| Mutated Residue | Change in R-group | Wild-type (kJ/mol) | Omicron (kJ/mol) |
|---|---|---|---|
| ASP/GLY-339 | Negative to Neutral | 1.7602 | 157.4091 |
| LEU/SER-371 | Neutral to Positive | 2.6324 | 1.5017 |
| PRO/SER-373 | Neutral to Positive | 2.6935 | 2.7563 |
| PHE/SER-375 | Neutral to Positive | −0.2065 | −0.0565 |
| ASN/LYS-417 | Negative to Positive | −230.7012 | −4.1475 |
| LYS/ASN-440 | Positive to Negative | 2.7283 | −240.3853 |
| SER/GLY-446 | Positive to Neutral | −2.5231 | −1.1289 |
| ASN/SER-477 | Negative to Positive | −2.5193 | −0.3975 |
| LYS/THR-478 | Positive to Positive | 0.652 | −206.7846 |
| ALA/GLU-484 | Neutral to Negative | 233.7813 | −0.4936 |
| LYS/GLN-493 | Positive to Negative | −0.3732 | −222.7693 |
| SER/GLY-496 | Positive to Neutral | 1.2866 | 3.175 |
| ARG/GLN-498 | Positive to Negative | −0.2806 | −265.8465 |
| TYR/ASN-501 | Positive to Negative | −7.8806 | −14.9393 |
| HIS/TYR-505 | Positive to Positive | −16.9694 | −11.9467 |
The interface of RBD is lined by a number of charged residues such as ASP, GLU, LYS. Upon analyzing the mutations, we observe that several mutations resulted in change in the amino acid which could severely impact its binding potential. We calculated the per residue binding energies of Wild-type and Omicron variants using MM/PBSA. Since the RBD region alone shelters fifteen different mutations, we compared the binding energies of those mutated residues. There are three instances of mutations changing the residue from charged to neutral. While S496G and S446G do not show significant difference in the binding energies, the D339G mutation from a highly negative to a neutral residue makes it unfavorable for RBD to bind to the ACE2 (refer to Table S1). Similarly, not much difference could be seen for mutations that result in changing the residues from neutral to charged residues. Although, the change of A484E is favorable for the Omicron RBD to bind to ACE2. The most evident changes were seen when there is change of positive residues to negative such as K440N, K493Q and R498Q (Refer to Table S1). Here, more than 200 kJ/mol gain in per residue binding energy could be seen. Conversely, when mutation led to change in amino acid from negative to positive there was a loss in binding energy values. A significant difference in binding energy was observed when highly positive K478 mutates to polar threonine residue which adds around −206 kJ/mol towards the binding of the RBD in the Omicron system. Apart from the mutated residues, we observed changes in binding energies of residues R454 and K458 in the RBM, where the Omicron system had a higher energy of binding (Table S1).
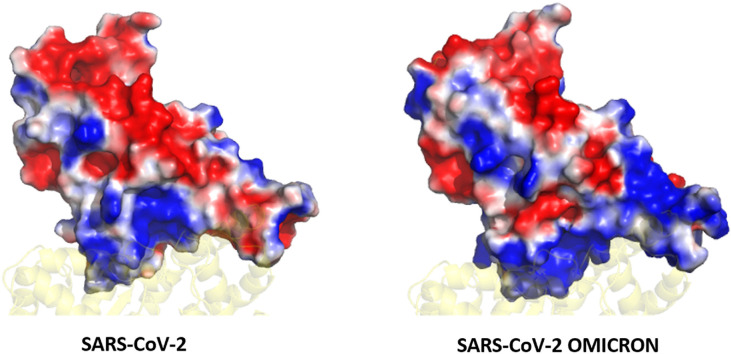
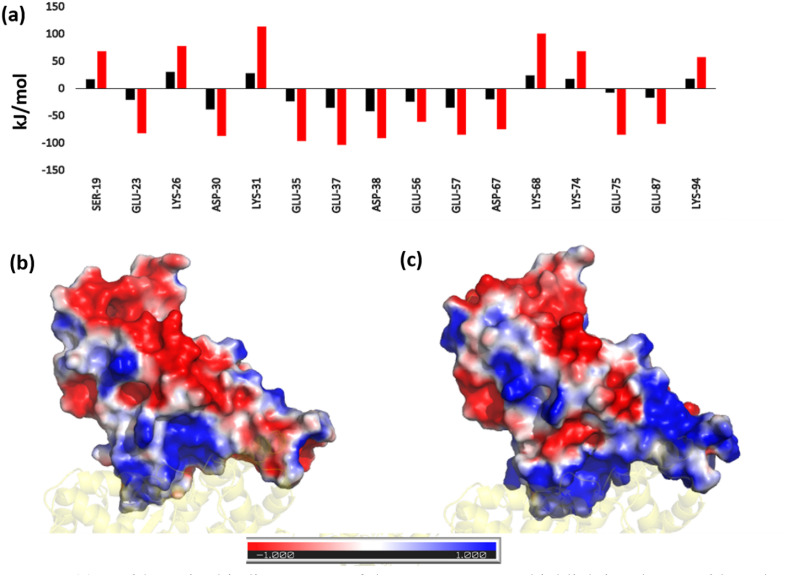
We also compared the difference in binding energies of the ACE2 residues (specifically the interface residues) and those that directly interact with the RBD. Those residues whose energies vary significantly are shown in Fig. 2 a. Here, we found reduced energies of residues S19, K26, K31, K68, K74 and K94. However, negatively charged residues E23, D30, E35, E37, D38, E56, E57, D67, E75, E87, all show an improved binding potential. The bias in the increase in binding energies of negatively charged residues instigated us to investigate the surface potential of the RBM in both the systems. Surface electrostatics of the RBM were calculated using the Adaptive Poisson Boltzmann Surface Area [[27], [31]]. Fig. 2b and c shows a clear difference in the interfacial surface potential of the RBD in both the complexes. The Wild-type had shown both positively and negatively charged regions at the binding surface. Conversely, the Omicron variant had a highly positive binding interface. This corresponds very well with the increase in binding energies of the negatively charged residues present at the ACE2 interface.
Fig. 2.
(a) Residue-wise binding energy of the ACE2 receptor highlighting those residues that show the most changes (Color coding similar to Fig. 1). Electrostatic potential Map of the RBD in (a) Wild-type and (c) Omicron Variant displaying the difference in charge distribution. The red and blue colors denote the negative and positive potentials respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
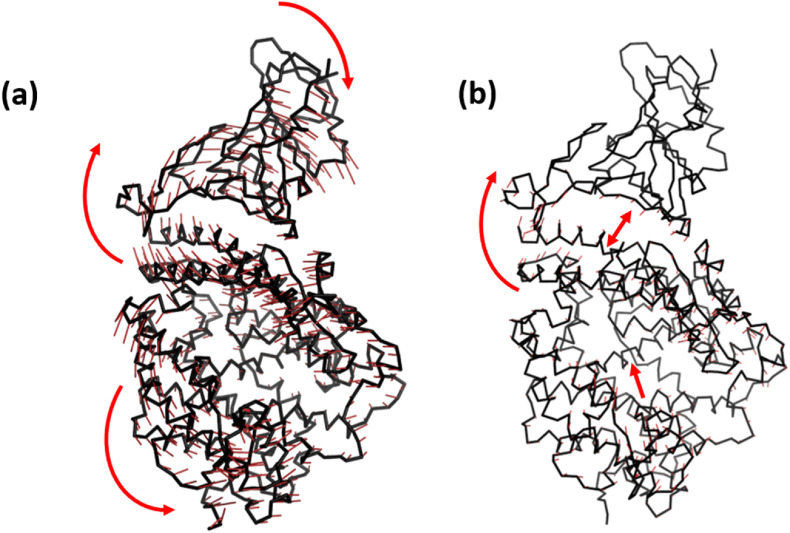
Superimposition of the final conformations of both the complexes show an RMSD value of 1.44 Å, implying not much change in the protein conformation. The mode of binding was also found to be similar after running the simulation for 100ns. However, we observed a slight displacement of the N-terminal helix of ACE2 towards RBD in the Omicron complex. Projections of the principal modes using porcupine plots show that both RBD and ACE2 are highly dynamic in the Wild-type (Fig. 3 ). Two different movements were observed along the first principal mode; the RBD and N-terminal ACE2 receptor of Wild-type move synchronously in clockwise direction. The second motion was of the C-terminal domain of ACE2 in the anticlockwise direction. The Omicron system, conversely shows remarkable reduction in the dynamics of Cα atoms (Fig. 3b). The magnitude as well as the direction of the principal modes were different. The inter and intramolecular movements were primarily observed in the Omicron complex. The magnitude of clockwise movement of RBD and N-terminal portion of ACE2 was also found to have drastically reduced. This indicates the stability of the Omicron RBD/ACE2 complex similar to Fig. 1d,e above.
Fig. 3.
Porcupine plots constructed by projecting the displacement of the backbone CA atoms along the first principal component in (a) Wild-type and (b) Omicron variant. The red arrows denote the direction of movements of the atoms. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
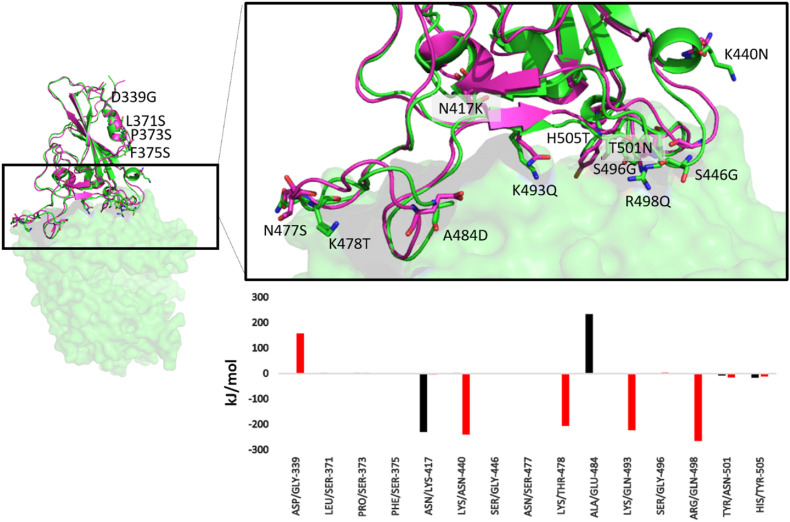
The relative orientation of the interfacial residues was also compared to gain more insights on the binding modes of both the proteins (Fig. 4 ). We found that four of the mutated residues do not directly interact with the ACE2. Moreover, the mutated residues namely L371S, P373S and F375S do not show significant change in their energetic contributions. The D339S mutation was found to be highly repulsive, however since the residue does not directly interact with the ACE2, it doesn't impact the protein complex formed. The most prominent changes were found for N417K, K440N, K478T, A484D, K493Q and R498Q. Except the N417K mutation which decreases the interaction energy, all the remaining residues massively increase the interaction energy of the RBD.
Fig. 4.
Superimposed structure of Wild-type (magenta) and Omicron (green) showing the mutated residues in stick representation and colored by CPK. The residue-wise binding energy is shown as a bar graph (Color scheme similar to Fig. 1). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Earlier studies have emphasized the difference in dynamics of SARS-CoV and SARS-CoV-2 on the interfacial interactions between RBD and ACE2 [15]. To elucidate the difference in hydrogen bonding and hydrophobic interactions among both the systems we compared the interfacial interactions by using Ligplot+ [32] (Fig. S3). Here we observed ten hydrogen bonded interactions in Wild-type while only eight in the Omicron variant (Figs. S3a and b). Apart from a common H-bond between D483 of RBD and Y83 of ACE2, most of the H-bonds formed were remarkably different. The hydrophobic interactions were also observed to be relatively more in the Wild-type RBD/ACE2 complex. However, the number of residues participating in hydrophobic interactions in both RBD and ACE2 were higher in Omicron as can be seen in Figs. S3c and d.
To summarize, we have made a detailed molecular analysis of the binding of the RBD domain of Wild-type SARS-CoV-2 and Omicron Variant with the human ACE2 receptor. Enhanced binding with the human receptor is one of the crucial factors in transmissibility of the virus. Apart from atomistic details related to the binding mode, we have meticulously analyzed the residue wise interaction energies of the mutated residues. The complementary changes were also observed in the human ACE2 receptor. Detailed surface electrostatics imply the change in the nature of the binding interface to a highly positive patch in RBD of Omicron. This resulting change is favorable for the binding of ACE2 which is lined by several negatively charged ASP and GLU residues. The overall relaxed dynamics of the protein complex further supports the stability of the Omicron when compared to Wild-type. The study not only provides a first-hand rationale for the high rate of transmission of the variant but would also prove crucial for the drug development studies as well as in the designing of antibodies.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
SLR thanks NIT Warangal for research seed grant (P1131) and National Energy Research Scientific Computing Center of the Ernest Orlando Lawrence Berkeley National Laboratory, a DOE Office of Science User Facility supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231 and the Extreme Science and Engineering Discovery Environment (XSEDE). We extend our gratitude to the Covid19 HPC Consortium for providing resources and helping researchers work for a noble cause.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2022.01.006.
Appendix A. Supplementary data
The following is the Supplementary data to this article:Supporting Information Available. Includes description of Materials and Methods and Table S1, Figs. S1–S3. This information is available free of charge via the internet.
References
- 1.Cecilia C., Wang Hsin-yi, Ebrahimi Omid V. Adjustment to a “new normal:” coping flexibility and mental health issues during the COVID-19 pandemic. Front. Psychiatr. 2021;12:353. doi: 10.3389/fpsyt.2021.626197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. 2021. [Google Scholar]
- 3.Peck Kayla M., Lauring Adam S. Complexities of viral mutation rates. J. Virol. 2021;92 doi: 10.1128/JVI.01031-17. e01031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponzoni L., Bahar I. Structural dynamics is a determinant of the functional significance of missense variants. Proc. Natl. Acad. Sci. Unit. States Am. 2018;115:4164–4169. doi: 10.1073/pnas.1715896115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu S., Tian C., Liu P., Guo D., Zheng W., Huang X., Zhang Y., Liu L. Effects of SARS-CoV-2 mutations on protein structures and intraviral protein-protein interactions. J. Med. Virol. 2021;93:2132–2140. doi: 10.1002/jmv.26597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulliam J., R C., et al. Preprint at medRxiv. 2021. [DOI]
- 7.Raghuvamsi P.V., Tulsian N.K., Samsudin F., Qian X., Purushotorman K., Yue G., Kozma M.M., Hwa W.Y., Lescar J., Bond P.J., MacAry P.A., Anand G.S. SARS-CoV-2 S protein:ACE2 interaction reveals novel allosteric targets. Elife. 2021;10 doi: 10.7554/eLife.63646. PMID: 33554856; PMCID: PMC7932696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y., Yang C., Xf Xu, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature (London) 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 10.Xiaofeng Z., Jiumeng S., Ziqing Y., et al. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2021;94 doi: 10.1128/JVI.00831-20. e00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey W.T., Carabelli A.M., Jackson B., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spinello A., Saltalamacchia A., Magistrato A. Is the rigidity of SARS-CoV-2 spike receptor-binding Motif the hallmark for its enhanced infectivity? Insights from all-atom simulations. J. Phys. Chem. Lett. 2020;11:4785–4790. doi: 10.1021/acs.jpclett.0c01148. https://dx.doi.org/10.1021/acs.jpclett.0c01148. [DOI] [PubMed] [Google Scholar]
- 13.Mandal N., Padhi A.K., Rath S.L. vol. 10. bioRxiv; 2021. p. 465272. (Molecular Insights into the Differential Dynamics of SARS-CoV-2 Variants of Concern (VOC)). 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;14:894–904. doi: 10.1016/j.cell.2020.03.045. e9, Epub 2020 Apr 9. PMID: 32275855; PMCID: PMC7144619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformat. 2016;20 doi: 10.1002/cpbi.3. 5.6.1-5.6.37, PMID: 27322406; PMCID: PMC5031415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., A M., Fogarty A.M., Harbison C.A., Barros E.P., Taylor B.C., McLellan J.S., Fadda E., Amaro R.E. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham M., Murtola T., Schulz R., et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 18.Brooks B.R., Brooks C.L., III, MacKerell A.D., Jr., Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A.R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R.W., Post C.B., Pu J.Z., Schaefer M., Tidor B., Venable R.M., Woodcock H.L., Wu X., Yang W., York D.M., Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformat. 2016;20 doi: 10.1002/cpbi.3. 5.6.1-5.6.37, PMID: 27322406; PMCID: PMC5031415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen M., Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. PMID: 17075131; PMCID: PMC2242414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F.T.K., Mattos C., Michnick S., Ngo T., Nguyen D.T., Prodhom B., Reiher W.E., Roux B., Schlenkrich M., Smith J.C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 22.Lemak A.S., Balabaev N.K. On the berendsen thermostat. Mol. Simulat. 1994;13:177–187. doi: 10.1080/08927029408021981. [DOI] [Google Scholar]
- 23.Martonák R., Laio A., Parrinello M. Predicting crystal structures: the parrinello-rahman method revisited. Phys. Rev. Lett. 2003;90 doi: 10.1103/PhysRevLett.90.075503. [DOI] [PubMed] [Google Scholar]
- 24.Essmann U., Perera L., Berkowitz M., Darden T., Lee H., Pedersen L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 25.Lilkova E., et al. Schrödinger & LLC; 2015. The PyMOL Molecular Graphics System, Version 2.0. [Google Scholar]
- 26.Mourik T.V., Snoek L., Knapen T., Norris D.G., Porcupine A visual pipeline tool for neuroimaging analysis. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fogolari F., Brigo A., Molinari H. The Poisson–Boltzmann equation for biomolecular electrostatics: a tool for structural biology. J. Mol. Recogn. 2002;15(6):379–385. doi: 10.1002/jmr.577. [DOI] [PubMed] [Google Scholar]
- 28.Matteo A., Michael J.B., Stefan K., Philip C.B. Statistical analysis on the performance of molecular mechanics Poisson–Boltzmann surface Area versus absolute binding free energy calculations: bromodomains as a case study. J. Chem. Inf. Model. 2017;57:2203–2221. doi: 10.1021/acs.jcim.7b00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumari R., Kumar R. Open source drug discovery Consortium, lynn A. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 30.Pearson K., F.R S. LIII. On lines and planes of closest fit to systems of points in space. The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science. 2010;1901:559–572. doi: 10.1080/14786440109462720. [DOI] [Google Scholar]
- 31.Engel D.J.E., Star K., Monson K., Brandi J., Felberg L.E., Brookes D.H., Wilson R., Chen L., Liles J., Chun K., Li M., Gohara P., Dolinsky D.W., Konecny T., Koes D.R., Nielsen J.E., Head-Gordon T., Geng W., Krasny R., Wei G.-W., Holst M.J., McCammon J.A., Baker N.A. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [PubMed id: 21919503] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.