Abstract
Wastewater-based epidemiology (WBE) is utilized globally as a tool for quantifying the amount of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) within communities, yet the efficacy of community-level wastewater monitoring has yet to be directly compared to random Coronavirus Disease of 2019 (COVID-19) clinical testing; the best-supported method of virus surveillance within a single population. This study evaluated the relationship between SARS-CoV-2 RNA in raw wastewater and random COVID-19 clinical testing on a large university campus in the Southwestern United States during the Fall 2020 semester. Daily composites of wastewater (24-hour samples) were collected three times per week at two campus locations from 16 August 2020 to 1 January 2021 (n = 95) and analyzed by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) targeting the SARS-CoV-2 E gene. Campus populations were estimated using campus resident information and anonymized, unique user Wi-Fi connections. Resultant trends of SARS-CoV-2 RNA levels in wastewater were consistent with local and nationwide pandemic trends showing peaks in infections at the start of the Fall semester in mid-August 2020 and mid-to-late December 2020. A strong positive correlation (r = 0.71 (p < 0.01); n = 15) was identified between random COVID-19 clinical testing and WBE surveillance methods, suggesting that wastewater surveillance has a predictive power similar to that of random clinical testing. Additionally, a comparative cost analysis between wastewater and clinical methods conducted here show that WBE was more cost effective, providing data at 1.7% of the total cost of clinical testing ($6042 versus $338,000, respectively). We conclude that wastewater monitoring of SARS-CoV-2 performed in tandem with random clinical testing can strengthen campus health surveillance, and its economic advantages are maximized when performed routinely as a primary surveillance method, with random clinical testing reserved for an active outbreak situation.
Keywords: Random surveillance testing, College campus, Saliva testing, Wastewater-based epidemiology, Wi-fi data, Sewer, Wastewater collection system, Neighborhood-level monitoring, Maintenance hole
Graphical abstract
1. Introduction
The early stages of the COVID-19 pandemic introduced a myriad of testing and monitoring challenges associated with novel coronavirus infections, including development, production, distribution, cost, and reporting (Arvisais-Anhalt et al., 2021; Smyrlaki et al., 2020; Tromberg et al., 2020). Wastewater-based epidemiology (WBE), having successfully aided in monitoring pathogens such as influenza, hepatitis A, and norovirus (Choi et al., 2018; Lorenzo and Picó, 2019; Sims and Kasprzyk-Hordern, 2020), was utilized early in the pandemic to mitigate these barriers, providing near real-time, community-level assessments of SARS-CoV-2 in populations around the world (Ahmed et al., 2020a; Haramoto et al., 2020; Hart and Halden, 2020; Polo et al., 2020; Sherchan et al., 2020). At university campuses across the United States, wastewater-based epidemiology has been primarily used as an indicator for targeted clinical testing (Betancourt et al., 2021; Gibas et al., 2021; Reeves et al., 2021; Scott et al., 2021; Travis et al., 2021). This method has been celebrated for preventing outbreaks (Harris-Lovett et al., 2021), however, wastewater surveillance has only been limitedly evaluated in comparison to random surveillance COVID-19 testing.
As individual COVID-19 testing has become increasingly available, random testing has been hailed as an effective way to drastically reduce transmission (Müller et al., 2020; Padula, 2020; Piguillem and Shi, 2020). However, there are concerns surrounding the legitimacy of mandatory testing of individuals in the United States and elsewhere (ThunstrÖM et al., 2021), and economic constraints can create barriers to implementation (Lyng et al., 2021). University campuses may serve as close to ideal proving grounds for comparing the efficacy of WBE and universal random testing, due to their high degree of administrative autonomy and the short in-sewer travel durations of biomarkers between the time of excretion and sample collection (Gushgari et al., 2018; Gressman and Peck, 2020). University campuses also introduce the potential risk of increasing pathogen transmission in the school's community (Gressman and Peck, 2020; NPR, 2020) and in the students' local communities upon returning home (Mangrum and Niekamp, 2020), further emphasizing the importance of closely monitoring school campuses. However, during the Fall 2020 semester, only about one in four college campuses employed a random surveillance testing or regular testing program for its students (NPR, 2020).
The goal of this study was to conduct SARS-CoV-2 monitoring in campus wastewater and compare those measurements to the results of COVID-19 random clinical testing in the student and staff population on a large U.S. university campus. We hypothesized that if the two independently collected sets of data presented as moderately or strongly correlated, wastewater virus monitoring would be further validated as an effective method for SARS-CoV-2 surveillance. Furthermore, the implications of the effectiveness and efficiency of wastewater SARS-CoV-2 monitoring could support new surveillance efforts, especially in communities without the tools or resources to conduct regular COVID-19 testing.
2. Materials and methods
2.1. Study location
The study was performed throughout the 2020 Fall academic semester on a large university campus in the southwestern U.S. with over 50,000 enrolled students and more than 10,000 employees. During the study, classes were available in a hybrid learning environment (online and in-person) and on-campus housing populations were ~10% lower than in pre-pandemic years. Two independent wastewater catchments cover approximately 90% of the total geographic campus area.
2.2. Wastewater sample collection
Flow-weighted, 24-hour composites of raw wastewater were collected using refrigerated autosamplers (Avalanche, Teledyne ISCO, Lincoln, NE) placed in permanent aboveground sampling locations across the participating campus. Each of the two locations were outfitted with a conduit through which a suction line of the autosampler traversed into an adjacent maintenance hole location, which also housed a laser flow meter. Samplers were timed to start collection early in the morning and operated for 24 h prior to sample collection on Tuesdays, Thursdays, and Saturdays of each week. Total collection volume within each sampler varied, however approximately 2 L of wastewater was transferred to high-density polyethylene bottles in the field, stored on ice after collection while transported to the laboratory, and processed on the same day to avoid degradation. Additional location details related to the pipe network are outlined in Table S1.
2.3. Wastewater sample processing and analysis
In the lab, each wastewater sample was mixed thoroughly and approximately 150 mL filtered through a 0.45 μm polyethersulfone vacuum filtration unit (Fisherbrand, Waltham, MA). The filtrate was then concentrated using two Millipore Sigma Amicon Ultra Centrifugal Filter Units (Burlington, MA) with a 10,000 molecular weight cut-off with ~15 mL volumes centrifuged consecutively five times at 2200g for 15 min. The resultant concentrate from each filter was aggregated and well-mixed, with 200 μL subsequently extracted for total RNA using a Qiagen RNeasy Mini Kit (Hilden, Germany) following the protocol for centrifugation of animal cells with molecular-grade β-mercaptoethanol (Burlington, MA). All resultant volumes (filtrate, filtrate concentrated, resultant concentrate) were recorded for quantification purposes. SARS-CoV-2 RNA was quantified using the Invitrogen SuperScript III Platinum One-Step qRT-PCR Kit (Carlsbad, CA) and Applied Biosystems QuantStudio 3 Real-Time PCR System (Foster City, CA) using Charité/Berlin (World Health Organization) designed primers and probe for E (envelope) SARS-CoV-2 RNA gene target; probes and primers were purchased from Integrated DNA Technologies (Coralville, IA) (Corman et al., 2020; Holland et al., 2020). The whole genome synthetic positive control was purchased from Twist Bioscience (San Francisco, CA). Standard curves ranged from 106 to 102 copies μL−1 with method efficiency of 92%. Triplicate standard curves were analyzed for each new batch of assay reagents and were used to quantify samples. Negative controls included whole and molecular process blanks with DNAase/RNAase-free water. Spike-and-recovery tests were performed using murine hepatitis virus as a surrogate (Ahmed et al., 2020b) (200,000 copies added into sewage before filtration) with an average recovery of 25 ± 17%. Additional qPCR method details are included in Tables S2 and S3.
2.4. Campus population estimates
Campus populations were estimated using online publicly available housing data updated monthly based on the total number of student housing contracts. Additionally, daily university affiliate Wi-Fi connections were also used to approximate commuting students and employees. Unique Wi-Fi connections from handheld devices or personal computers longer than 30 min and shorter than 16 h during the wastewater collection period of 7:00 a.m. to 6:59 a.m. were counted as commuting individuals present on campus that day. The on-campus resident populations were weighted by 30% to account for university residential buildings outside the catchment areas and those previously captured in campus Wi-Fi data. WBE researchers had access only to unique connection counts to protect the privacy of campus visitors and prevent any possibility of tracing unique users.
2.5. Campus clinical testing data
COVID-19 clinical test results from the university were obtained from weekly public online releases. The participating university was conducting saliva secretion PCR processing at a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory. A statistically significant random selection of students and employees were required to complete a COVID-19 saliva test each week (1206 ± 166 tests per week, 11 ± 5% of total campus population estimates), and the number of total random tests and positive random tests for on-campus students and employees were used in this study.
2.6. Data analysis
Measured concentrations in each sewer catchment were transformed to viral load (VL) per day (SARS-CoV-2 genome copies per day) using the following equation:
| (1) |
where C (SARS-CoV-2 RNA copies L−1) is the measured concentration and Q is the total daily volumetric wastewater flow rate (L day−1). Population normalized viral load (SARS-CoV-2 genome copies day−1 per 1000 people) was calculated using the following equation:
| (2) |
where Pop is the total number of people contributing to the catchment defined by the methodology in Section 2.5. Statistical analysis was performed using Microsoft Excel (Redmond, WA) and MATLAB (MathWorks, Natick, MA) using population corrected SARS-CoV-2 wastewater data and percent positivity to control for the variable population on the university campus throughout the study period. Price estimates were taken directly from the internal SARS-CoV-2 analysis cost from the ASU laboratory and added to estimated labor costs for field collection. Clinical testing costs were estimated based on internal correspondence with the participating clinical laboratory.
3. Results and discussion
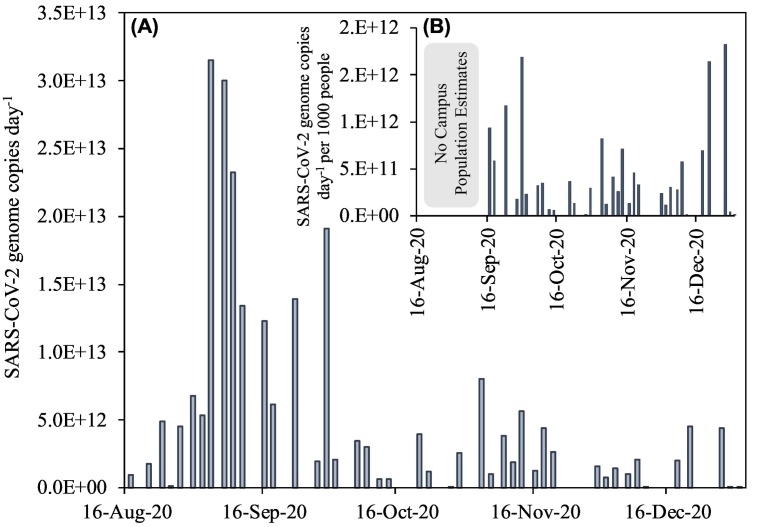
This study measured SARS-CoV-2 RNA in raw wastewater on a U.S. university campus with the goal of comparing wastewater-derived trends to the campus' COVID-19 random surveillance testing. Ninety-five samples were collected throughout the duration of the study with viral RNA detected in 68% of samples (Fig. SI). When detected, concentrations were on average 0.97 ± 1.14 million genome copies per day. Results showed a general trend of increasing quantities of SARS-CoV-2 viral loads in the wastewater early in the semester (mid-August through early September) (Fig. 1A), coinciding with the influx of students at the start of the academic year. The localized increases of coronavirus levels in campus wastewater coincided with new clinical cases within the city's zip code. This suggests that the incoming student population was driving the observed SARS-CoV-2 RNA signal increase in wastewater, likely through the influx of infected students at the start of the semester, and exacerbated by close living quarters and socialization at campus events. Similar trends of elevated SARS-CoV-2 measurements in wastewater were seen by other universities throughout the U.S. (Gibas et al., 2021; Travis et al., 2021), and counties with large student populations also had high virus presence in August/September despite lower state-wide values (New-York-Times, 2021). In this study, the remainder of the semester saw smaller increases in SARS-CoV-2 RNA viral loads in wastewater approximately 2 weeks after Halloween (31 October) and 1 week after Thanksgiving (26 November).
Fig. 1.
SARS-CoV-2 genome copies day−1 in wastewater on individual days across a large U.S. university campus (A), and population normalized viral load (SARS-CoV-2 genome copies L−1 per 1000 people) (B). Note viral loads are the summation of both the major and minor sewer catchment contributions on a given day; Wi-Fi population estimates were not available until mid-September 2020, so population normalized data for this time period are not shown.
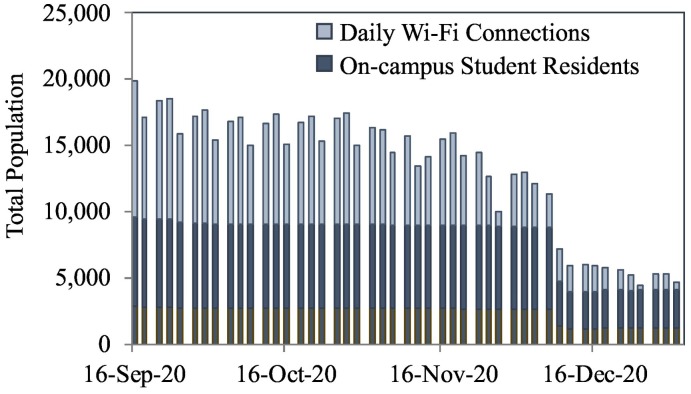
The result of normalizing wastewater derived SARS-CoV-2 viral load data with population had the effect of increasing virus data in the month of December month as compared to other months because of the lower campus population at that time (Fig. 1B). The campus residential population was generally stable from September through early December 2020, with population estimates averaging 9084 ± 182 (Fig. 2 ), whereas Wi-Fi data ranged from a high in September of 10,000 unique users, declining to 4000 by the end of the semester, with a low of 355 people on Christmas Day. These values revealed a higher population on campus during Mondays and Wednesdays and a significant drop on Fridays, as was expected with class and workweek schedules. The sewage contributions from the commuter populations are generally more challenging to estimate due to unpredictable in-person attendance. While previous studies have used other excreted biomarkers to estimate populations, metabolites can often be unstable or not validated in small populations (Thai et al., 2019). This challenge here was met successfully by combining the relatively stable campus resident population with unique, fully anonymized Wi-Fi connection data from personal devices such as cell phones or laptop computers.
Fig. 2.
Campus population estimates obtained by using de-identified unique Wi-Fi connections and total on-campus student resident data. Note that only 30% (outlined in brown) of the total residential population was used in the analysis because Wi-Fi captured a portion of those buildings. Each cluster of three bars represents the Monday, Wednesday, and Friday of the sampling week (representative day).
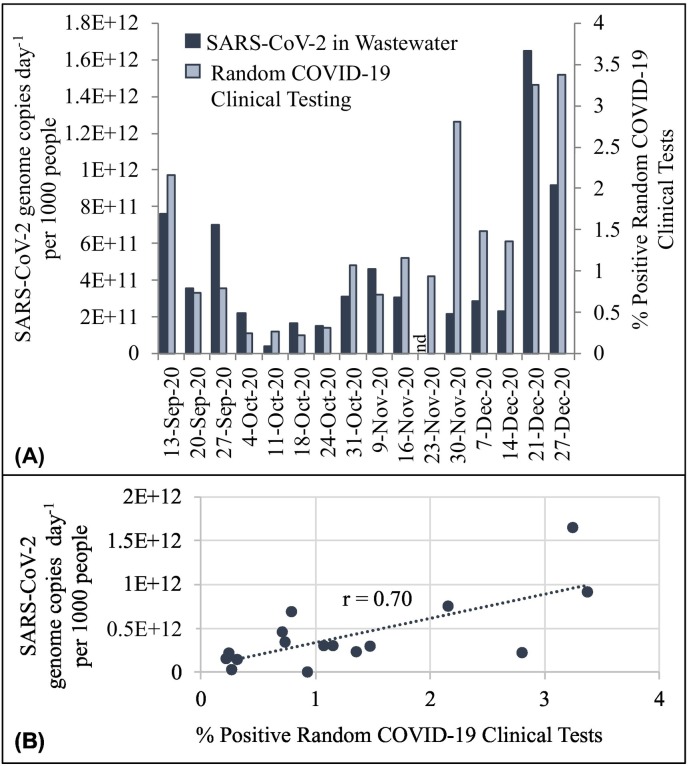
Weekly aggregated population normalized SARS-CoV-2 data were compared to the percentage of positive tests obtained in random COVID-19 clinical surveillance (Fig. 3 ). Random COVID-19 clinical test results were reported once a week by the university, so wastewater population normalized viral loads were aggregated over the three sampling days for a temporally comparable assessment. When trends in wastewater virus measurements were compared with random COVID-19 clinical test results, a strong positive correlation resulted, with a Pearson correlation coefficient of r = 0.71 (p < 0.01). It is important to note, that on the week of the Thanksgiving holiday, only a single sample was collected (Tuesday), which was non-detect for SARS-CoV-2 RNA. This data point was omitted from the correlation. There is a relatively high degree of correlation between these two entirely independent methods of monitoring, which suggests these two metrics could be used interchangeably or jointly to obtain more robust data, resources permitting.
Fig. 3.
Relationship between average weekly population normalized viral loads (SARS-CoV-2 genome copies per day per 1000 people) and the percent positive random COVID-19 clinical tests during each week of the Fall 2020 semester at a large U.S. university campus. Relationships are illustrated through (A) chronological trends and (B) linear correlations. Each presented wastewater value is six samples (major and minor catchments collected over three days). Note on the week of 23 November 2020, only one sample was collected due to the holiday and was below the detection limit (nd). The Pearson Correlation Coefficient was r = 0.71 throughout the entire duration of the study (n = 15 comparison, n = 76 wastewater samples).
To assess the feasibility of this based on cost, we compared the clinical and wastewater sample processing and analysis throughout the duration of the study. Random clinical testing for the university began the week of 10 September and continued throughout the conclusion of 2020 with approximately 19,300 people tested. With costs at $17.50 per test (including labor), this equates to ~$338,000 USD. The wastewater-based laboratory assay used in this study is $62.50 per sample (consumables and labor). An additional $17 in field-labor was added to account for sample collection time, for a total study cost of $6042.00 for samples processed during the equivalent time period. Consequently, the cost of wastewater testing was 1.7% that of its clinical testing counterpart and for the cost of clinical testing, wastewater testing could be increased by over 50×. The purpose of this comparison is not to rank one method as superior over the other, but rather to provide perspective for areas where implementing randomized clinical testing of individuals raises financial and ethical or cultural concerns (Jacobs et al., 2021). Wastewater could serve as a sentinel matrix to assess what is happening within a community at a relatively low cost before more targeted testing is implemented if a problem is identified.
There are important limitations that are worthy of noting in this study. Sampling within the sewershed brings challenges with a dynamic population; we were able to improve our estimates using Wi-Fi connectivity data, without which correlations with clinical testing were reduced (r = 0.47 vs. 0.71). However, challenges with residential population estimates occurred at the end of the semester, when the university transitioned from hybrid learning to online for the single remaining week after the Thanksgiving holiday. Anecdotally, students left the week of November 23 and did not return until January, even though housing contracts were active through the end of December 12. Additionally, ~10% of the geographic footprint of campus is not captured by those two wastewater sampling points, with ~5% of student housing residing in that area. Although small, if recorded cases occurred with those residents, it would not necessarily be reflected as a signal in wastewater, unless sewer contributions occurred elsewhere on-campus. More generally, wastewater captures all campus visitors, even those not included in the pool of individuals selected for random testing. Finally, viral shedding times vary over days to weeks (Yan et al., 2021), a challenge faced by other wastewater assessments when comparing to clinical cases (Bowes et al., 2021; Wu et al., 2020).
The type of monitoring performed in this study is considered neighborhood- or catchment-level monitoring, and is distinct from building-level monitoring that is also being implemented on university campuses (Brooks et al., 2021; Gibas et al., 2021). In this study, hundreds to thousands of people are contributing to the sewershed, the SARS-CoV-2 RNA signal is generally always measurable, and the challenge is discerning a potential outbreak from baseline measurements. Determining a trigger for action is often complex and specific to the catchment under investigation. This brings to the fore the importance of understanding changes in wastewater-derived measurements as the field of WBE moves into the future.
4. Conclusions
The correlations between SARS-CoV-2 RNA measured in wastewater and random clinical surveillance testing are promising in support of wastewater monitoring as an effective method for quantifying viral presence at the sewershed level on university campuses and other neighborhood-leveling sampling campaigns. As we move forward with wastewater monitoring within sewer systems, collection should be noted as a viable alternative to the more common city-level treatment plant sampling. The strong correlation of the two different approaches further suggests that in resource-poor situations it may be prudent to conduct WBE routinely and random clinical testing optionally only when WBE data hint at an uptick in infections that needs to be managed.
CRediT authorship contribution statement
Jillian Wright: Conceptualization, methodology, investigation, visualization, formal analysis, writing-original draft. Erin M. Driver: Conceptualization, methodology, investigation, supervision, validation, writing-review & editing. Devin A. Bowes: Methodology, investigation, resources, visualization, validation, writing-review & editing. Bridger Johnston: Methodology, investigation, writing-review & editing. Rolf U. Halden: Supervision, funding acquisition, writing-review & editing.
Declaration of competing interest
R.U.H. and E.M.D. are cofounders of AquaVitas, LLC, 9260 E. Raintree, Ste. 130, Scottsdale, AZ 85260, USA, a startup company providing commercial services in wastewater-based epidemiology. R.U.H., J.W., and D.A.B. are current or former members of OneWaterOneHealth, a non-profit project managed by the university's Foundation.
Acknowledgements
The authors would like to thank the local municipality and members of the study institute for their support and guidance related to this work. We would specifically like to thank personnel from the university's Chief Information Office, for their expertise and assistance with anonymizing and interpreting Wi-Fi data, and the Geographic Information System group for their knowledge of the campus building sewer connections. This work was supported in part by J.M. Kaplan Fund (Project 30009070). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Kaplan Fund.
Editor: Damià Barceló
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.152877.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739:139960. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvisais-Anhalt S., Lehmann C.U., Park J.Y., Araj E., Holcomb M., Jamieson A.R., et al. What the coronavirus disease 2019 (COVID-19) pandemic has reinforced: the need for accurate data. Clin. Infect. Dis. 2021;72:920–923. doi: 10.1093/cid/ciaa1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779:146408. doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes D.A., Driver E.M., Kraberger S., Fontenele R.S., Holland L.A., Wright J., et al. medRxiv; 2021. Unrestricted Online Sharing of High-frequency, High-resolution Data on SARS-CoV-2 in Wastewater to Inform the COVID-19 Public Health Response in Greater Tempe, Arizona. 2021.07.29.21261338. [Google Scholar]
- Brooks Y.M., Gryskwicz B., Sheehan S., Piers S., Mahale P., McNeil S., et al. Detection of SARS-CoV-2 in wastewater at residential college, Maine, USA, August–November 2020. Emerg. Infect. Dis. 2021;27:3111–3114. doi: 10.3201/eid2712.211199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., Apos Brien J.W., Grant S.C., et al. Wastewater-based epidemiology biomarkers: past, present and future. Trends Anal. Chem. 2018;105:453–469. [Google Scholar]
- Corman V., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Roppolo Brazell L., et al. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782:146749. doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressman P.T., Peck J.R. Simulating COVID-19 in a university environment. Math. Biosci. 2020;328:108436. doi: 10.1016/j.mbs.2020.108436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gushgari A.J., Driver E.M., Steele J.C., Halden R.U., et al. Tracking narcotics consumption at a Southwestern U.S. University campus by wastewater-based epidemiology. J. Hazard. Mater. 2018;359:437–444. doi: 10.1016/j.jhazmat.2018.07.073. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Lovett S., Nelson K.L., Beamer P., Bischel H.N., Bivins A., Bruder A., et al. Wastewater surveillance for sars-cov-2 on college campuses: initial efforts, lessons learned and research needs. Int. J. Environ. Res. Public Health. 2021;18:4455. doi: 10.3390/ijerph18094455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L.A., Kaelin E.A., Maqsood R., Estifanos B., Wu L.I., Varsani A., et al. An 81-nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (January to March 2020) J. Virol. 2020;94 doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs D., McDaniel T., Varsani A., Halden R.U., Forrest S., Lee H. Wastewater monitoring raises privacy and ethical considerations. IEEE Trans. Technol. Soc. 2021:1. [Google Scholar]
- Lorenzo M., Picó Y. Wastewater-based epidemiology: current status and future prospects. Curr. Opin. Environ. Sci. Health. 2019;9:77–84. [Google Scholar]
- Lyng G.D., Sheils N.E., Kennedy C.J., Griffin D.O., Berke E.M. Identifying optimal COVID-19 testing strategies for schools and businesses: balancing testing frequency, individual test technology, and cost. PloS one. 2021;16 doi: 10.1371/journal.pone.0248783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangrum D., Niekamp P. JUE insight: college student travel contributed to local COVID-19 spread. J. Urban Econ. 2020:103311. doi: 10.1016/j.jue.2020.103311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Derlet P.M., Mudry C., Aeppli G. Testing of asymptomatic individuals for fast feedback-control of COVID-19 pandemic. Phys. Biol. 2020;17:065007. doi: 10.1088/1478-3975/aba6d0. [DOI] [PubMed] [Google Scholar]
- New-York-Times . 2021. Tracking the Coronavirus at U.S. Colleges and Universities. 2021. [Google Scholar]
- NPR . National Public Radio, Inc. (NPR); 2020. Many Colleges Aren't Aggressively Testing Students For Coronavirus. [Google Scholar]
- Padula W.V. Why only test symptomatic patients? Consider random screening for COVID-19. Appl. Health Econ. Health Policy. 2020;18:333–334. doi: 10.1007/s40258-020-00579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguillem F., Shi L. Einaudi Institute for Economics and Finance (EIEF); 2020. Optimal COVID-19 Quarantine and Testing Policies. (EIEF Working Paper Series 2004). [Google Scholar]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., et al. Making waves: wastewater-based epidemiology for COVID-19 – approaches and challenges for surveillance and prediction. Water Res. (Oxford) 2020;186(116404):1–7. doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves K., Liebig J., Feula A., Saldi T., Lasda E., Johnson W., et al. High-resolution within-sewer SARS-CoV-2 surveillance facilitates informed intervention. Water Res. (Oxford) 2021;204:117613. doi: 10.1016/j.watres.2021.117613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.C., Aubee A., Babahaji L., Vigil K., Tims S., Aw T.G. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 2021;200:111374. doi: 10.1016/j.envres.2021.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., et al. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrlaki I., Ekman M., Lentini A., Rufino de Sousa N., Papanicolaou N., Vondracek M., et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat. Commun. 2020;11:4812. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai P.K., O'Brien J.W., Banks A.P.W., Jiang G., Gao J., Choi P.M., et al. Evaluating the in-sewer stability of three potential population biomarkers for application in wastewater-based epidemiology. Sci. Total Environ. 2019;671:248–253. doi: 10.1016/j.scitotenv.2019.03.231. [DOI] [PubMed] [Google Scholar]
- ThunstrÖM L., Ashworth M., Shogren J.F., Newbold S., Finnoff D. Testing for COVID-19: willful ignorance or selfless behavior? Behav. Public Policy. 2021;5:135–152. [Google Scholar]
- Travis S.A., Best A.A., Bochniak K.S., Dunteman N.D., Fellinger J., Folkert P.D., et al. Providing a safe, in-person, residential college experience during the COVID-19 pandemic. Front. Public Health. 2021;9:672344. doi: 10.3389/fpubh.2021.672344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromberg B.J., Schwetz T.A., Pérez-Stable E.J., Hodes R.J., Woychik R.P., Bright R.A., et al. Rapid scaling up of Covid-19 diagnostic testing in the United States — the NIH RADx initiative. N. Engl. J. Med. 2020;383:1071–1077. doi: 10.1056/NEJMsr2022263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., et al. SARS-CoV-2 titers in wastewater are higher thanexpected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. e00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Zhang X., Chen C., Jiang D., Liu X., Zhou Y., et al. Characteristics of viral shedding time in SARS-CoV-2 infections: a systematic review and meta-analysis. Front. Public Health. 2021;9:652842. doi: 10.3389/fpubh.2021.652842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material