Abstract
The current clinical diagnosis of COVID-19 requires person-to-person contact, needs variable time to produce results, and is expensive. It is even inaccessible to the general population in some developing countries due to insufficient healthcare facilities. Hence, a low-cost, quick, and easily accessible solution for COVID-19 diagnosis is vital. This paper presents a study that involves developing an algorithm for automated and noninvasive diagnosis of COVID-19 using cough sound samples and a deep neural network. The cough sounds provide essential information about the behavior of glottis under different respiratory pathological conditions. Hence, the characteristics of cough sounds can identify respiratory diseases like COVID-19. The proposed algorithm consists of three main steps (a) extraction of acoustic features from the cough sound samples, (b) formation of a feature vector, and (c) classification of the cough sound samples using a deep neural network. The output from the proposed system provides a COVID-19 likelihood diagnosis. In this work, we consider three acoustic feature vectors, namely (a) time-domain, (b) frequency-domain, and (c) mixed-domain (i.e., a combination of features in both time-domain and frequency-domain). The performance of the proposed algorithm is evaluated using cough sound samples collected from healthy and COVID-19 patients. The results show that the proposed algorithm automatically detects COVID-19 cough sound samples with an overall accuracy of 89.2%, 97.5%, and 93.8% using time-domain, frequency-domain, and mixed-domain feature vectors, respectively. The proposed algorithm, coupled with its high accuracy, demonstrates that it can be used for quick identification or early screening of COVID-19. We also compare our results with that of some state-of-the-art works.
Keywords: Cough sounds, COVID-19, Deep learning, Features, Signal processing, Voice pathology
1. Introduction
According to the global database maintained by John Hopkins University, more than 270 million COVID-19 (and its variants) cases and 5.3 million deaths have been reported till December 13, 2021 [1]. Social distancing, wearing masks, widespread testing, contact tracing, and massive vaccination are all recommended by the World Health Organization (WHO) to reduce the spreading of this virus [2]. To date, reverse transcription-polymerase chain reaction (RT-PCR) is considered the gold standard for testing coronavirus [3]. However, the RT-PCR test requires person-to-person contact to administer, needs variable time to produce results, and is still unaffordable to most global populations. Sometimes, it is unpleasant to the children. Not least, this test is not yet accessible to the people living in remote areas, where medical facilities are scarce [4]. Alarmingly, the physicians suspect that the general people refuse the COVID-19 test in fear of stigma [5].
Governments worldwide have initiated a free massive testing campaign to stop the spreading of this virus, and this campaign is costing them billions of dollars per day at the average rate of $23 per test [6]. Hence, easily accessible, quick, and affordable testing is essential to limit the spreading of the virus. The COVID-19 detection method, using human audio signals, can play an important role here.
Researchers and clinicians have suggested using the recordings of speech, breathing, and cough sounds to detect various diseases. The results published in the literature show that the speech samples can help clinicians to detect several diseases, including asthma [7], [8], [9], [10], Alzheimer's disease [11], [12], [13], Parkinson's disease [14], [15], [16], depression [17], [18], [19], schizophrenia [20], [21], [22], autism [23], [24], head or neck cancer [25], and emotional expressiveness of breast cancer patients [26]. A comprehensive survey on these works can be found in [27]. Among these diseases, respiratory diseases like asthma have some similarities to COVID-19. An extensive investigation on the detection of asthma using audio signal processing can be found in [7], [8], [9], [10]. These works show that asthma causes swollen and inflamed vocal folds, which do not vibrate appropriately during voice generation. Hence, the voice samples of asthma patients differ from that of healthy (i.e., control) subjects. For example, it is shown in [7] that asthmatic subjects show longer pauses between speech segments, produce fewer syllables per breath, and spend a more significant percentage of time in voiceless ventilator activity than their healthy counterpart.
Recently, researchers have been suggesting using cough sounds for the early detection of the COVID-19. However, there are still some challenges as the cough is also a symptom of 30 other diseases [28], [29]. Hence, it is very challenging to discriminate the cough sound of the COVID-19 patients from that of other patients. In [28], the authors considered three diseases: bronchitis, pertussis, and COVID-19. Investigation of 247 normal cough samples and 296 pathological samples was performed. The authors used a convolutional neural network (CNN) to implement a binary classifier and a multiclass classifier. The binary classifier discriminates pathological cough sounds from normal cough sounds, and the multiclass classifier categorizes the pathologies into one of the three pathology types. In a similar work [30], the authors considered bronchitis, bronchiolitis, and pertussis. They used a CNN to discriminate against these pathologies.
Various human audio samples, namely, sustained vowel ‘/a/’, counting (1-20), breathing, and cough samples, have been used in [31]. The authors considered nine acoustic voice features: spectral contrast, mel-frequency Cepstral Coefficients (MFCCs), spectral roll-off, spectral centroid, mean square energy, polynomial fit, zero-crossing rate, spectral bandwidth, and spectral flatness. They used a random forest (RF) algorithm to discriminate the COVID-19 samples from the control/healthy samples, and they have achieved an accuracy of 66.74%.
In [32], the authors used large samples (5320 samples) selected from the MIT open voice COVID-19 cough dataset [33]. They extracted the MFCC features from the cough sounds and classified them by using a CNN. The network consists of one Poisson biomarker layer and three pre-trained ResNet50s. The results showed that their proposed system achieved an accuracy of 97%.
Cough and breathing sounds have also been used in [34]. In that work, the authors used eleven acoustic features: RMS energy, spectral centroid, roll-off frequencies, zero-crossing rate, MFCC, Δ-MFCC, ∆^2-MFCC, tempo, duration onsets, and period. In addition, they used VGGish (a pre-trained CNN from Google) to classify the samples into COVID-positive/non-COVID, COVID-positive with cough/non-COVID with cough, and COVID-positive with cough/non-COVID asthma with cough. The achieved accuracy of that system was 80%, 82%, and 80%, respectively, for the classification tasks mentioned above.
In [35], the authors used Computational Paralinguistic Challenge (COMPARE) [36] features and extended Geneva Minimalistic Acoustic Parameter Set (eGeMAPS) [37] to discriminate the COVID-19 samples from the healthy samples. These features were extracted by using the OpenSMILE [38] tool kit. The voice samples were collected by using five sentences uttered by the patients. The authors classified the COVID-19 patients into three categories, namely, high, mild, and low. In that study, they used 260 samples, including 52 COVID-19 samples. The authors have used a support vector machine (SVM) and achieved an accuracy of 69%.
Three acoustic feature sets have been used in [39]. The first one was the COMPARE acoustic features, which were collected by using the OpenSmile software. The second one was a combination of acoustic feature sets extracted by freely available software, PRAAT [40] and LIBROSA [41]. The third one was an acoustic feature set consisting of 1024 embedded features extracted by a deep CNN. The samples used in the investigation comprised of three vowels (i.e., ‘/a/’, ‘/s/’, and ‘/z/’), cough sounds, six symptomatic questions, and counting from 50 to 80. The authors have used the SVM with radial basis function (RBF) and RF as the classifiers. Experimental results showed an average accuracy of 80% in discriminating the COVID-19 positive patients from the COVID-19 negative patients based on the features extracted from the cough sound and vowel ‘/a/’ recordings. They achieved even more accuracy (83%) by evaluating six symptomatic questions.
In [42], the authors used voice features, namely, cepstral peak prominence (CPP), harmonic-to-noise ratio (HNR), first and second harmonic (H1H2), fundamental frequency and its variations (F0SD), Jitter, Shimmer, and maximum phonation time (MPT) to discriminate the voice samples of the COVID-19 patients from that of the healthy subjects. The authors collected the sustained vowel sample ‘/a/’ from 70 healthy and 64 COVID-19 patients of Persian speakers. They revealed significantly higher F0SD, Jitter, shimmer, H1H2, and voice break numbers in the COVID-19 patients than the control/healthy group.
Vowels in ‘/ah/’, snoring consonants in ‘/z/’, cough sound, and counting samples from 50 to 80 have been used in [43]. The authors have used a recurrent neural network (RNN) based expert classifier in work. The authors have used three techniques: pre-training, bootstrapping, and regularization to avoid the over-fitting problem of RNN. They also used the leave-one-speaker-out validation technique to achieve a recall of 78%. In a similar work [44], the authors used the RNN algorithm with long short-term memory (LSTM) to detect the COVID-19 patients. In that investigation, the authors used several features, including spectral centroid, spectral roll-off, zero-crossing-rate, MFCCs, and ΔMFCCs from the cough sound, breathing sound, and voice samples of the COVID-19 patients. The authors used 60 healthy and 20 COVID-19 patients in the work. To improve accuracy, they removed the silence part from the samples using the PRAAT software. As a result, the authors achieved an accuracy of 98.2%, 97.0%, and 77.2% by using breathing, cough, and voice samples, respectively.
In [45], the authors have used the MFCC features of cough, breathing, and voice sounds to discriminate the COVID-19 patients from the non-COVID-19 patients. The authors concluded that the MFCCs of cough and breathing sounds for the COVID-19 patients and non-COVID-19 patients are similar. However, the MFCCs of voice sounds are very distinct between the COVID-19 and non-COVID-19 patients.
A cloud computing and artificial intelligence-based early detection of the COVID-19 patients have been presented in [46]. The authors used three-voice features, namely, HNR, Jitter, and Shimmer. In addition, they used the RBF algorithm as a classifier. The authors suggested that the HNR, Jitter, and Shimmer can be used to differentiate between healthy and asthma patients. They also indicated that the same parameters can be used to discriminate the healthy and COVID-19 patients.
Recurrence quantification measures in the MFCCs have been introduced in [47] to detect the COVID-19 patients using sustained vowel ‘/ah/' and cough sounds. The authors have used several classifiers, namely, decision trees, SVM, k-nearest neighbor, RF, and XGBoost. Among these classifiers, they achieved the best results with the XGBoost classifier. That model achieved accuracies of 97% (with an F1 Score of 91%) and 99% (with an F1 Score of 89%) for coughs and sustained vowels, respectively.
In [48], the authors used crowdsourced cough audio samples that were acquired on a smartphone from around the world. They collected three acoustic features: MFCCs, mel-frequency spectrum, and spectrogram from the cough sounds. The authors used an innovative ensemble classifier model (consisting of three networks) to discriminate the COVID-19 patients from the healthy subjects. The highest accuracy achieved was 77.1%.
This work is a preliminary investigation of Artificial Intelligence's (AI's) capability to detect COVID-19 by using acoustic features. The proposed algorithm has been developed based on the available data which is limited. Rigorous testing of the algorithm is required with more data before deploying the algorithm in practice for COVID-19 pre-screening. The main contributions of this paper are as follows:
-
•
To develop a novel algorithm based on signal processing and a deep neural network (DNN).
-
•
To compute the acoustic features and compare their uniqueness for the cough sound samples of control (i.e., healthy) and COVID-19 patients.
-
•
To form the feature vectors using three domains: time-domain, frequency-domain, and mixed-domain, to investigate the efficacy of these feature vectors.
-
•
To achieve a high classification accuracy (compared to other related works) without overwhelming computation burden on the system.
-
•
To use a dropout strategy in the proposed algorithm to make the training process faster and to overcome the overfitting problem.
-
•
To provide a detailed performance analysis of the proposed system in terms of the confusion matrix, Accuracy, Precision, Negative predictive value (NPV), and F1-Score.
The rest of the paper is organized as follows. The related background is presented in Section 2. The models, materials, and methods are explained in Section 3. Simulation results and discussions are presented in Section 4. Research applicability is explained in Section 5, and the paper is concluded with Section 6.
2. Background
The human voice generation system mainly consists of lungs, larynx, and articulators. Among them, the lungs are considered the power source of the voice generation system. Respiratory diseases prevent the lungs from working properly and hence affect the human voice generation system. Respiratory diseases can be classified into two main classes, namely, (a) obstructive and (b) restrictive [49]. Obstructive lung diseases make the pulmonary airways narrow and affect a patient's ability to expel air from the lungs completely. Hence, a significant amount of air remains in the lungs all the time. On the other hand, people suffering from restrictive lung diseases cannot fully expand their lungs to fill them with air. Hence, the lungs fail to fully expand. Some patients may suffer from a combination of both obstructive and restrictive respiratory diseases. Cough is the common symptom of obstructive, restrictive, and combined lung diseases. Hence, cough sounds are considered useful for detecting lung diseases caused by respiratory issues [50].
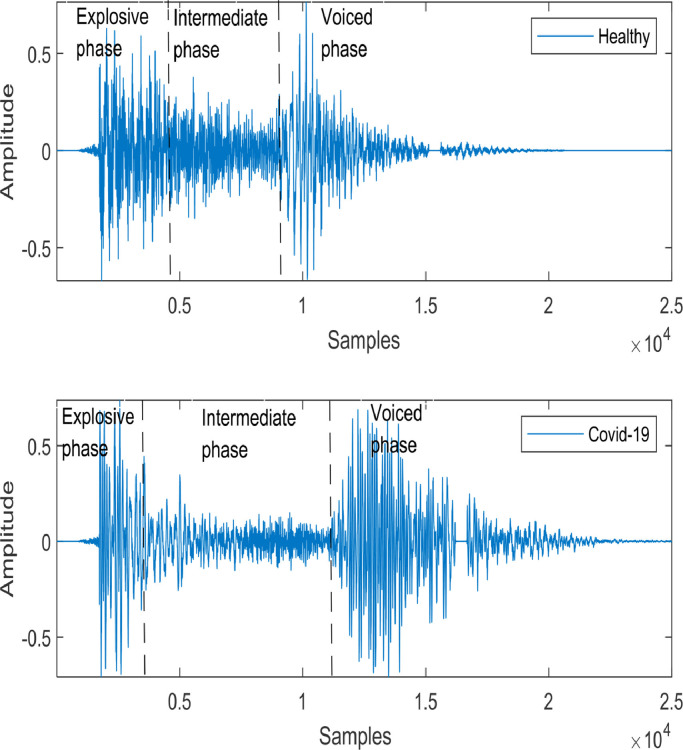
COVID-19 is also considered a respiratory disease. Like other respiratory diseases, COVID-19 can cause the lungs to fill with fluid and get inflamed. As a result, patients can suffer from breathing difficulty and need treatment at the hospital with severe onset. Untreated COVID-19 can progress and lead to acute respiratory distress syndrome (ARDS), a form of lung failure [51]. Although coughing is a common symptom of any respiratory illness, including COVID-19, recent studies suggest that the COVID-19 cough is characterized by dry, persistent, and hoarse at the earliest stage of coronavirus infected patients. Hence, the cough sound samples of COVID-19 patients differ from those of other patients suffering from some other respiratory diseases. Human cough samples contain three phases: explosive phase, intermediate phase, and voiced phase [52], as shown in Fig. 1 . These phases represent the glottal airflow variation in the vocal cord, and they differ depending on the pathological conditions of the patients.
Fig. 1.
A typical cough sound signal [52].
Two segmented cough sound samples are randomly selected from the Virufy database [53] to investigate the differences between the cough sound samples of a COVID-negative (i.e., healthy/control) subject and a COVID-positive patient. The cough sound samples of a healthy subject and a COVID-positive patient are shown in Fig. 2 . This figure demonstrates that the healthy sample is similar to the typical human cough sound signal presented in Fig. 1. However, the cough sound sample of the COVID-19 patient varies significantly from the typical human cough sound sample. For example, both the intermediate and voiced phases are longer for the COVID-positive patient than for the healthy subject.
Fig. 2.
Comparison of the cough sounds for a healthy subject and a COVID-19 patient collected from the Virufy database [53].
Moreover, the signal amplitude during the voiced phase is higher for the COVID-positive patient than for the healthy subject. The amplitudes in the explosive phase also differ between these two cough sound samples, as depicted in Fig. 2. The differences mentioned above indicate that the cough sound can be used as a valuable tool to discriminate the COVID-positive patient from the healthy subject. The power spectral densities (PSDs) of these two samples are plotted in Fig. 3 . It is observed in the figure that the healthy cough sound has prominent frequencies of continuously decreasing magnitudes. On the other hand, the COVID-positive cough sound samples do not contain very distinct frequencies.
Fig. 3.
Comparison of the power spectral densities (PSDs) of the cough sounds for a healthy subject and a COVID-19 patient.
3. Models, materials, and methods
The proposed system model is presented in Fig. 4 . It consists of four major steps: pre-processing, feature extraction, formation of feature vectors, and classification. The main functions of the pre-processing stage are audio segmentation and windowing. Afterward, the frames are formed. In the next step, the features are extracted from the framed samples. The extracted features are then grouped to form the feature vectors. Finally, the feature vectors are applied as the input to the classifier. The most crucial component of the proposed system is feature extraction (also called the data reduction procedure). It involves extracting features from the cough sound of interest. The main advantage of using features is that the analysis algorithm (i.e., classifier) needs to deal with small and transformed data compared to original voluminous cough sound data.
Fig. 4.
Block diagram of the proposed algorithm.
In practice, acoustic features are extracted, and a feature vector is formed, representing the original data. However, the selection of features and the formation of the appropriate feature vector is an open issue for ongoing research in pattern recognition. In this investigation, 33 acoustic features are considered to form three feature vectors. The acoustic features used in this work can be broadly classified into two major classes: time-domain and frequency-domain features. In this investigation, the cough sound samples are divided into small frames using a rectangular window, and the features are extracted from these frames. These features are explained in the following subsections.
3.1. Time-domain features
In this investigation, we consider the following time-domain features: (i) short-term energy, (ii) zero-crossing rate, and (iii) entropy of energy [54]. The short-term energy of the th frame is calculated by
| (1) |
where, is the th frame, with being the length of the frame. The energy expressed in (1) is normalized as
| (2) |
The normalized energy contents of the COVID-positive and healthy cough sounds are plotted in Fig. 5 (a). This figure shows that the energy contents of both samples are concentrated in a few frames, and they exhibit a high variation over successive frames. However, the energy content of the COVID-positive patient is much higher than that of the healthy subject. It indicates that the cough sound sample of the COVID-positive patient contains weak phonemes and a short period of silence between two coughs. Hence, the energy content also varies rapidly between two successive frames.
Fig. 5.
The time-domain features (a) Short time energy distribution, (b) Short time zero-crossing rate, and (c) Energy entropy.
The zero-crossing rate of a cough sound signal can be defined as the rate of sign changes of the movement over the frames. It is calculated by using the following equation
| (3) |
where, is the sign function defined by , when and , when . The zero-crossing rates of the COVID-positive patient and the healthy subject are plotted in Fig. 5(b), which shows that the healthy cough sound sample has a more zero-crossing rate than that of the COVID-positive patient. Since the zero-crossing rate measures the noisiness of a signal, it exhibits a higher zero-crossing rate for the unvoiced part of the cough sound sample and a lower zero-crossing rate for the voiced samples. As shown in Fig. 2, the voiced phase of the cough sound samples for the COVID-positive patient is longer than that of the healthy subject. Hence, the zero-crossing rate is lower for the COVID-positive patient than for the healthy subject, as depicted in Fig. 5(b). The short-term entropy of energy can be interpreted as a measure of the abrupt changes in the energy level of an audio signal. To compute it, we first divide each short-term frame into sub-frames of fixed duration. Then, for each sub-frame, , the energy is calculated by using (1) and divide it by the total energy, of the short-term frame. Then, the sub-frame energy values, , for j =1,2, …, K, is computed as a sequence of probabilities and is defined as
| (4) |
where, . At the final step, the entropy, , is calculated from the sequence by
| (5) |
The short-term entropy of energy for the COVID-positive patient and the healthy subject are plotted in Fig. 5(c). The short-term entropy of energy for the COVID-positive patient is greater than that of the healthy subject for most of the frames. Since the energy content of the COVID-positive patient varies more abruptly than that of the healthy subject, the energy entropy tends to be higher for the COVID-positive patient after frame 20, as shown in Fig. 5(c).
3.2. Frequency domain features
Frequency-domain acoustic features are extracted from the discrete Fourier transform (DFT) of a signal. The DFT of a frame of audio signal can be expressed as
| (6) |
where, is the size of the DFT, is the value of the DFT coefficients, and . The spectral centroid dictates a noise-robust estimate of the dominant frequency for the cough sound signal that varies over time. It is also called the center of gravity of the spectrum. The value of the spectral centroid, , for the th audio frame is calculated by
| (7) |
The spectral centroids of the COVID-19 positive and the healthy person are shown in Fig. 6 (a). It is shown in the figure that the spectral centroids of the cough sound for the healthy person are higher compared to those of the COVID-19 cough sound samples until approximately frame number 50. The highest value corresponds to the brightest sound practically. Usually, the existence of noise, silence, etc. signifies the lower values of the spectral centroid. This is noticeable for COVID-positive patient as opposed to the healthy person for the range mentioned above. From nearly 50-80 frames, the COVID-positive patient exhibits higher values of the spectral centroid. After frame number 80, both the samples show insignificant spectral components.
Fig. 6.
The frequency-domain features (a) Spectral centroid, (b) Spectral entropy, and (c) Spectral flux
Spectral entropy is a measure of irregularities in the frequency domain. The spectral entropy features are computed from the short-time Fourier transform (STFT) spectrum. Spectral entropy is widely used to detect the voiced regions of an acoustic signal. The flat distribution of silence or noise induces high entropy values. The spectral entropy is computed with the same method that follows to calculate the cough signal's energy entropy. First, the spectrum of the short-term frame is divided into sub-bands. The energy, , of the th sub-band, where , is normalized by the total spectral energy. The normalized energy is defined as , . Finally, the entropy of the normalized spectral energy, , is computed by
| (8) |
The spectral entropies of the COVID-positive and the healthy person are shown in Fig. 6(b). This figure shows that the spectral entropy of the healthy person is higher than that of the COVID-positive patient for most of the frames. The reason is that the voiced part of the signal contains less spectral entropy than the unvoiced one.
The spectral flux measures the spectral change between two successive frames. The spectral flux is computed as the squared difference between the normalized magnitudes of the spectra for the two subsequent short-term windows. It is defined by
| (9) |
where, . The spectral fluxes of the cough sound sample for the COVID-positive and the healthy person are plotted in Fig. 6(c). The magnitudes of the spectral flux are higher for the healthy person compared to the COVID-positive patient for most frames. The reason is the more frequent local spectral changes in the healthy cough sound samples than in the COVID-positive ones. This indicates more rapid spectral alternation among phonemes in the healthy cough sound sample than in the COVID-positive patient.
The spectral roll-off is the frequency below which a certain percentage (usually around 90%) of the magnitude distribution of the spectrum is concentrated. Therefore, if the th DFT coefficient corresponds to the spectral roll-off of the th frame, then it satisfies the following equation
| (10) |
where, is the adopted percentage (user parameter). The spectral roll-off frequency is usually normalized by dividing it with , so that it takes values between 0 and 1. The spectral roll-offs of the cough sound samples for the healthy person and the COVID-positive patient are shown in Fig. 7 (a). It can be easily observed that the cough sound samples of the healthy person show a higher spectral roll-off value than that of the COVID-positive patient for most of the frames. It means that the cough sound sample of the healthy person has a wider spectrum compared to that of the COVID-positive patient.
Fig. 7.
The frequency-domain features (a) Spectral roll-off, (b) MFCC coefficient, (c) Chroma vector, and (d) Feature harmonics.
We also include the MFCCs to form the feature vector. The MFCCs have been widely used in respiratory disease detection algorithms for a long time [55], [56], [57]. The main advantage of the MFCCs over other acoustic features is that they can completely characterize the shape of the vocal tract configuration. Once the vocal tract is accurately characterized, we can estimate an accurate representation of the phonemes being produced by the vocal tract. The shape of the vocal tract manifests itself in the envelope of the short-time power spectrum, and the MFCCs accurately represent this envelope [58]. The following procedure is used to compute the MFCCs [59]. The voice sample, is first windowed with an analysis window and the STFT, is computed by
| (11) |
where, , with being the DFT length. The magnitude of is then weighted by a series of filter frequency responses whose center frequencies and bandwidth are roughly matched with the auditory critical band filters called mel scale filters. The next step is to compute the energy using the STFT, weighted by each mel scale filter frequency response. The energy for each speech frame at time, and for the th mel-scale filter is given by
| (12) |
where, is the frequency response of the th mel-scale filter, and and Ul are the lower and upper-frequency indices, respectively, over which each filter is nonzero, while is defined as
| (13) |
The cepstrum, associated with , is then computed for the speech frame at time, by
| (14) |
where is the number of filters. In this work, we consider 13 MFCCs. The plots for the arbitrarily chosen 7th coefficient of the MFCCs for both the healthy cough sound samples and COVID-positive cough sound samples are shown in Fig. 7(b). It is shown in the figure that the magnitude of the 7th MFCC coefficient is higher for the COVID-positive cough sound sample compared to that of the healthy cough sound for most of the frames.
The chroma vector used in this work is a 12-element representation of spectral energy. The chroma vector is computed by grouping the DFT coefficients of a short-term window into 12 bins. Each bin represents the 12 equal-tempered pitch classes of semitone spacing. Also, each bin produces the mean of the log-magnitudes of the respective DFT coefficients, defined by
| (15) |
where, is a subset of the frequencies that correspond to the DFT coefficients and is the cardinality of . In the context of a short-term feature extraction procedure, the chroma vector is usually computed on a short frame basis. This results in a matrix , with elements , where indices and represent pitch-class and frame-number, respectively. The chroma vector plots of the healthy and the COVID-positive cough sound samples are shown in Fig. 7(c). It is shown that the chroma vector of the healthy person shows one dominant coefficient, and the rest of the coefficients are of small magnitudes. On the other hand, the chroma vector of the COVID-positive cough sound sample is noisier and does not have any dominant coefficient. In addition, the chroma vector of the cough sound sample for the COVID-positive patient does not contain any nonzero coefficient.
The autocorrelation function for the th frame is computed by
| (16) |
Actually, is the correlation of the th frame with itself at time lag, . Then the autocorrelation function is normalized as
| (17) |
Afterward, the maximum value of , i.e., the harmonic ratio is calculated as
| (18) |
where , and are the minimum and maximum allowable values of the fundamental period. Here, is often defined by the user, whereas usually corresponds to the lag in time for which the first zero crossing of the occurs. The plots for the harmonic ratio of the healthy and the COVID-positive patients are shown in Fig. 7(d). It is depicted in the figure that the harmonic ratio of the cough sound sample for the healthy person is higher for most of the frames. However, the harmonic ratio shows nonzero values for all analysis frames of the COVID-positive cough sound samples. On the other hand, the harmonic ratio of the healthy person has zero values for some of the analysis frames.
In this work, the cough sound samples collected from the Virufy database [53] are used. The Virufy is a volunteer-run organization, which has built a global database to identify the COVID-19 patients using AI. The database contains both clinical data and crowdsourced data. The clinical data is accurate because it was collected and reprocessed at a hospital following a standard operating procedure (SOP). Qualified physicians monitored the whole process of data collection. The subjects were confirmed as healthy persons (i.e., COVID-19 negative) and COVID-19 patients (i.e., COVID-positive) by using the RT-PCR test, and the data was labeled accordingly. The database also contains the patients’ information, including age, gender, and medical history. Virufy provided 121 segmented cough samples from these 16 patients. The Virufy database contains both the original cough audio recordings and the segmented version of the cough sounds. The segmented cough sounds were created by identifying the periods of relative silence in the recordings and separating cough sound samples based on those silences. The segments with no coughing sound or having too much background noise were removed. The crowdsourced data, maintained by Virufy, is diverse and donated by patients from multiple countries. This database is significantly increasing in volume over time as more people are contributing their cough samples. In this work, only the clinically collected cough samples are used as they are more authentic than crowdsourced data and, also, the segmented cough samples are used.
A DNN discriminates the COVID-19 cough sound samples from the healthy cough sound samples, as shown in Fig. 4. The DNN model presented in [60] is used and modified to implement the proposed system. The DNN used in the network consists of three hidden layers. Each hidden layer consists of 20 nodes. The network has 500 input nodes for the matrix input. It has only one output node as the decision is binary. The output node employs the softmax activation function, whereas the hidden nodes consist of the sigmoid function. One of the limitations of the DNN is that they are vulnerable to overfitting. This problem worsens as the network includes more nodes. To solve the overfitting problem, we employ a dropout algorithm. This algorithm trains only some of the randomly selected nodes rather than all the entire network nodes. The dropout effectively prevents overfitting as it continuously alters the nodes and weights in the training process. In this work, a dropout ratio of 10% and 20% are used for the input and hidden layers, respectively.
4. Simulation results and discussion
For biomedical signals classification, findings are made in the context of medical prognosis [61]. Therefore, in COVID-19 cough sound sample detection, we need to provide a clinical or diagnostic interpretation of the rule-based classifications made with the acoustic features pattern. The following terminologies and performance parameters are used [55,62]:
True positive (TP) occurs when the predicted test is positive for COVID while the subject is also COVID-positive. True negative (TN) occurs when the predicted test is negative for COVID and the subject is COVID-negative as well.
Sensitivity or Recall is denoted by and it is defined by
| (19) |
Specificity is denoted by and it is given by
| (20) |
False-negative (FN) occurs when the test is negative for a subject who possesses the COVID. The probability of this error, known as the false-negative fraction (FNF), is given by
| (21) |
False-positive (FP) is defined as the case when the predicted result is COVID-positive, but the individual is COVID-negative. The probability of this type of error or a false alarm, known as the false-positive fraction (FPF), is given by
| (22) |
Accuracy is simply a ratio of the correctly predicted observations to the total number of observations. The accuracy is defined by
| (23) |
Precision or Positive Predictive Value (PPV) is the ratio of the correctly predicted positive observations to the total predicted positive observations. The precision is defined by
| (24) |
F1 Score is the weighted average of the Precision and Recall. Therefore, this score takes both false positives and false negatives into account. The F1 Score is defined by,
| (25) |
NPV (Negative Predictive Value) represents the percentage of the cases labelled as truly negative. The NPV is defined by
| (26) |
The samples are distributed into three parts: 70% are for training the DNN, the remaining 30% into validation, and testing with a ratio of 2:1. Five-fold validation is used. The data samples and patient information are listed in Table 1 . The proposed system's training, validation, and testing results with the three feature vectors are listed in Table 2 .
Table 1.
Data Samples
| Sample | Corona test | Age | Gender | Medical history | Reported symptoms | Cough file name |
|---|---|---|---|---|---|---|
| 1 | Negative | 53 | Male | None | None | neg-0421-083-cough-m-53.mp7 |
| 2 | Positive | 50 | Male | Congestive heart failure | Shortness of breath | pos-0421-084-cough-m-50.mp3 |
| 3 | Negative | 43 | Male | None | Sore throat | neg-0421-085-cough-m-43.mp3 |
| 4 | Positive | 65 | Male | Asthma/chronic lung disease | Shortness of breath, new or worsening cough | pos-0421-086-cough-m-65.mp3 |
| 5 | Positive | 40 | Female | None | Sore throat, loss of taste, loss of smell | pos-0421-087-cough-f-40.mp3 |
| 6 | Negative | 66 | Female | Diabetes with complication | None | neg-0421-088-cough-f-66.mp3 |
| 7 | Negative | 20 | Female | None | None | neg-0421-089-cough-f-20.mp3 |
| 8 | Negative | 17 | Female | None | Shortness of breath, sore throat, body aches | neg-0421-090-cough-f-17.mp3 |
| 9 | Negative | 47 | Male | None | New or worsening cough | neg-0421-091-cough-m-47.mp3 |
| 10 | Positive | 53 | Male | None | Fever, chills, or sweating, shortness of breath, new or worsening cough, sore throat, loss of taste, loss of smell | pos-0421-092-cough-m-53.mp3 |
| 11 | Positive | 24 | Female | None | None | pos-0421-093-cough-f-24.mp3 |
| 12 | Positive | 51 | Male | Diabetes with complication | Fever, chills, or sweating, new or worsening cough, sore throat | pos-0421-094-cough-m-51.mp3 |
| 13 | Negative | 53 | Male | None | None | neg-0422-095-cough-m-53.mp3 |
| 14 | Positive | 31 | Male | None | Shortness of breath, new or worsening cough | pos-0422-096-cough-m-31.mp3 |
| 15 | Negative | 37 | Male | None | None | neg-0422-097-cough-m-37.mp3 |
| 16 | Negative | 24 | Female | None | New or worsening cough | neg-0422-098-cough-f-24.mp3 |
Table 2.
Training and Testing Accuracy of the Feature Vectors
| Feature Vector | Training Accuracy (%) | Validation Accuracy (%) | Testing Accuracy (%) |
|---|---|---|---|
| Time-domain feature vector | 100 | 93.27 | 89.20 |
| Frequency-domain feature vector | 100 | 98.50 | 97.50 |
| Mixed feature vector | 100 | 96.37 | 93.80 |
First, the time-domain feature vector is used that has three acoustic features, namely, zero-crossing rate, energy, and energy entropy. Then, the DNN (with five-fold cross-validation) is trained, and the system is tested with the time-domain feature vector. The results are shown in Table 2, with an average training accuracy of 100%, validation accuracy of 93.27%, and testing accuracy of 89.20%. The classification matrix [61] of the time-domain feature vector is provided in Table 3 . Based on the data presented in Table 3, it can be concluded that the DNN can correctly detect the COVID-positive cough sound samples with a sensitivity of 86.67% by using the time-domain features. On the other hand, it can accurately identify healthy cough sound samples with a specificity of 91.67%.
Table 3.
The Classification Matrix of the Time-Domain Feature Vector
| Actual | Prediction (%) | |
|---|---|---|
| Healthy | COVID-19 | |
| Healthy | 91.67% ( | 8.33% ( |
| COVID-19 | 13.33% ( | 86.67% ( |
Simulations are repeated by using the frequency-domain feature vector. As mentioned before, the features considered are spectral centroid, spectral entropy, spectral flux, spectral roll-offs, MFCCs, and chroma vector. The training, validation, and testing results are also listed in Table 2. The data shows that the DNN achieves training accuracy of 100%, validation accuracy of 98.50%, and testing accuracy of 97.50% by using the frequency-domain feature vector. It can be concluded that the testing accuracy of the frequency-domain feature vector is higher than that of the time-domain feature vector. The classification matrix of the frequency-domain feature vector is presented in Table 4 , which shows that the frequency-domain feature vector boosts the DNN's ability to detect the COVID-positive cough sound samples with a sensitivity of 95%. Moreover, the DNN can accurately identify the healthy samples with a specificity of 100%. Both parameters are higher than those of the time-domain feature vector.
Table 4.
The Classification Matrix of the Frequency-Domain Feature Vector
| Actual | Prediction (%) | |
|---|---|---|
| Healthy | COVID-19 | |
| Healthy | 100.00% ( | 0.00% ( |
| COVID-19 | 5.00% ( | 95.00% ( |
Lastly, time-domain and frequency-domain features are combined to form a mixed-feature vector. The training, validation, and testing accuracies for the mixed feature vector are listed in Table 2. The achieved training, testing, and validation accuracies are 100%, 96.37%, and 93.80%, respectively. The classification matrix of the mixed-feature vector is presented in Table 5 . The DNN can detect COVID-positive cough sound samples with a sensitivity of 93.34%. On the other hand, it can accurately identify the healthy cough sound samples with a specificity of 94.17%.
Table 5.
The Classification Matrix of the Mixed Feature Vector
| Actual | Prediction (%) | |
|---|---|---|
| Healthy | COVID-19 | |
| Healthy | 94.17% ( | 5.82% ( |
| COVID-19 | 6.67% ( | 93.34% ( |
The performances of the proposed system in terms of Accuracy, Precision, F1 Score, and NPV for the time-domain feature vector, frequency-domain feature vector, and mixed domain feature vector are listed in Table 6 . This table shows that the proposed system achieves the highest accuracy of 97.5% using the frequency-domain feature vector. On the other hand, the lowest accuracy of 89.2% is achieved using the time-domain feature vector. The other performance scores, including Precision, F1 Score, and NPV, are the highest for the frequency-domain feature vector.
Table 6.
The Performance Comparison
| Measures | Time-domain feature vector | Frequency- domain feature vector | Mixed feature vector |
|---|---|---|---|
| Accuracy | 0.892 | 0.975 | 0.938 |
| Precision/ PPV | 0.912 | 1.000 | 0.941 |
| F1 Score | 0.889 | 0.974 | 0.937 |
| NPV | 0.873 | 0.952 | 0.934 |
Cough is regarded as a natural defense mechanism of some respiratory disorders, including COVID-19. The human audible hearing range impaired existing subjective clinical approaches of cough sound analysis [63]. Exploration of noninvasive diagnostic approaches well above the audible frequency range (i.e., 48000 Hz) used for sample data can overcome this limitation as demonstrated in this study. The non-stationary characteristics of cough sound samples impose additional challenges for signal processing-based approaches. Also, cough patterns show variability in human subjects under the same pathological state. The cough features that are closely tied to the intensity levels as in the time domain can have dissimilarity for the identical pathology. The cough sound is characterized by the fundamental frequency and significant harmonics when pathology is involved. The restriction of airways causes turbulence in the cough sound that constitutes the harmonics [52]. More realistically, a method that captures both time and frequency changes over the cough sound samples should associate the investigated respiratory disorder, i.e., COVID-19, with greater accuracy. The best diagnostic performance with the frequency-domain feature vector as in Table 6 justifies that the cough sound features distributed in the frequency domain should possess greater significance.
Finally, the performance of the proposed system is compared with other related works available in the literature, as listed in Table 7 . The comparison table shows that the proposed system achieves a higher accuracy of 97.5% with the frequency-domain feature vector using the cough sound samples compared to [44]. The system achieves even higher accuracy with the time-domain and mixed-feature vector than the works published in [31, [34], [35], 64].
Table 7.
The Performance Comparison with Existing Works
| Research Work | Samples | Phonemes | Features | Classifier | Accuracy |
|---|---|---|---|---|---|
| N. Sharma [31] | Healthy and COVID-positive: 941 | Cough, Breathing, Vowel, and Counting (1-20) | Spectral contrast, MFCC, Spectral roll-off, Spectral centroid, Mean squareenergy, Polynomial fit, zero-crossing rate, Spectralbandwidth, and Spectral flatness | RF | 66.74% |
| C. Brown et al. [34] | COVID-positive: 141, Non-COVID: 298, COVID-positive with Cough:54, Non-COVID with Cough:32, Non-COVID asthma: 20 | Cough, and Breathing | RMS energy, Spectral centroid, Roll-off frequencies, Zero-crossing, MFCC, Δ-MFCC, -MFCC, Duration, Tempo Onsets, Period | CNN | 80% |
| J. Han [35] | COVID-Positive: 52, Healthy: 208 | Voice | COMPARE, and eGeMAPS | SVM | 69% |
| A.Hassan [44] | COVID-Positive: 20, Healthy: 60 | Breathing, Cough, and Voice | Spectral centroid, Roll-off frequencies, Zero-crossing, MFCC, and Δ-MFCC | RNN | 98.2% (Breathing), 97% (Cough), 88.2% (Voice) |
| V. Espotovic [64] | COVID-Positive: 84, COVID-Negative: 1019 | Voice, Cough, and Breathing | Wavelet | Ensemble Boosted | 88.52% |
| Proposed System (time-domain) | COVID-Positive: 50, Healthy: 50 | Cough | zero-crossing rate, energy, and energy entropy | DNN | 89.2% |
| Proposed System (Frequency-domain) | COVID-Positve:50, Healthy: 50 | Cough | Spectral centroid, spectral entropy, spectral flux, spectral roll-offs, MFCC, and chroma vector | DNN | 97.5% |
| Proposed System (Mixed- feature) | COVID-Positve:50, Healthy: 50 | Cough | zero-crossing rate, energy, energy entropy, spectral centroid, spectral entropy, spectral flux, spectral roll-offs, MFCC, and chroma vector | DNN | 93.8% |
5. Research applicability
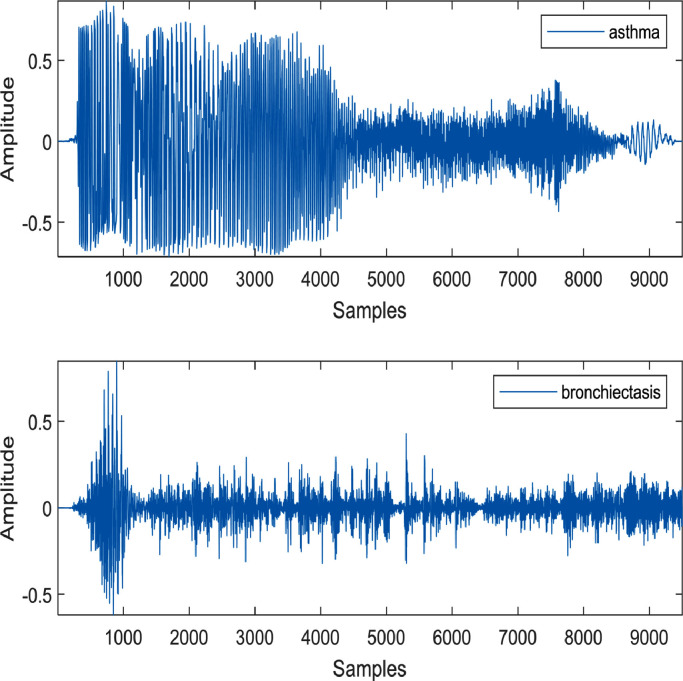
Since the publicly available databases are restricted to COVID-positive and COVID-negative (i.e., healthy/control) cases, this study focuses on discriminating COVID-19 cough sound from the healthy cough sound. However, the proposed algorithm can have a possibility to differentiate pathological cough sounds into distinct pulmonary/respiratory diseases, including COVID-19, asthma, bronchiectasis, etc. The pathophysiology and acoustic property of cough sounds can provide significant information in the frequency domain to characterize them for multi-classification purpose. Asthma causes the airways of the patient to be inflamed and narrower. On the other hand, bronchiectasis damages the airways and widens them abnormally. Few randomly selected cough sound samples of some respiratory disorders are investigated in [65]. The samples available in [66] are not sufficient to apply the proposed deep learning-based algorithm. One sample of asthma and bronchiectasis cough sound each is shown in Fig. 8 to demonstrate their uniqueness in the time domain. Bronchiectasis cough sound has longer cough sequences compared to asthma cough sound.
Fig. 8.
The cough sound samples of asthma and bronchiectasis.
Additionally, the bronchiectasis cough sound demonstrates more flow spikes than the asthmatic cough sound [52]. These flow spikes indicate more severe inflammation in bronchiectasis patients than in an asthmatic patient. Comparing Fig. 2 and Fig. 8, it can be concluded that the explosive, intermediate, and voiced phases are very distinct in the COVID-19 cough sample; however, these phases are hardly visible in asthma and bronchiectasis cough sounds.
As demonstrated in this study, some of the frequency-domain features of COVID-19, asthma, and bronchiectasis cough samples are plotted in Fig. 9 to show their uniqueness. The spectral entropy of the bronchiectasis sample is much higher for most of the frames compared to COVID-19 and asthma cough samples. The other features including spectral flux, MFCC, and feature harmonics are also non-identical for the mentioned three respiratory disorders. The distinct differences for the frequency domain features indicate that the proposed algorithm can also be applied to differentiate COVID-19 from asthma and bronchiectasis cough samples, provided a good number of datasets are available for each class.
Fig. 9.
The frequency domain features of (a) Spectral entropy, (b) Spectral flux, (c) MFCC coefficient (6th), and (d) Feature harmonics for COVID-19, asthma, and bronchiectasis cough samples.
6. Conclusion
In this paper, a DNN-based study for the early detection of COVID-19 patients has been presented using cough sound samples. This study proposed a system that extracts the acoustic features from the cough sound samples and forms three feature vectors. A rigorous, in-depth investigation has been provided in this work to show that the cough sound samples can be a valuable tool to discriminate the COVID-19 cough sound from other healthy cough sound samples for preliminary noninvasive assessment. In this work, it has been shown that some acoustic features are unique in the cough sound samples of the COVID-19 patients and hence can be used by a classifier like DNN to discriminate them from the healthy cough sound samples successfully. However, there has always been an argument about selecting the appropriate acoustic features for the classifications. The major challenges are (a) to decide whether to use a single feature (like MFCC, spectrogram, etc.) or feature vector, (b) to select the appropriate combination of acoustic features to form the feature vector, and (c) to choose the appropriate domain (i.e., time-domain, frequency-domain, or both). Three feature vectors have been investigated in this work to address this issue. It is shown and justified that the frequency-domain feature vector has provided the highest accuracy compared to the time-domain or mixed-domain feature vector.
The performance of the proposed system has been compared with those of other existing state-of-the-art methods that are presented in the literature for the diagnosis of COVID-19 patients from audio samples. The proposed noninvasive pre-diagnosis technique can enhance the screening of COVID-positive cases, including asymptomatic and pre-symptomatic patients. Also, early diagnosis can help them to stay in touch with healthcare providers for a better prognosis to avoid the critical consequences of COVID-19.
For future work, more focus will be given to investigate the progression level of the COVID-19 patients by using the cough sound analysis. Furthermore, since some other respiratory diseases produce similar cough sounds, it is imperative to compare the cough sound features of the COVID-19 patients with those of the other respiratory diseases. Currently, we are actively seeking data to investigate the mentioned issues.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Worldometer Corona Virus Cases. 2021 https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1? available at. accessed on November 23. [Google Scholar]
- 2.Coronavirus disease (COVID-19) technical guidance: Maintaining Essential Health Services and System. 2021 https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/maintaining-essential-health-services-and-systems available at. accessed on November 23. [Google Scholar]
- 3.Udugama B., et al. Diagnosing COVID-19: The Disease and Tools for Detection. American Chemical Society (ACS) Nano. April 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. no. [DOI] [PubMed] [Google Scholar]
- 4.Word Bank and WHO Half of the world lacks access to essential health services, 100 million still pushed into extreme poverty due because of health expenses. 2021 https://www.who.int/news/item/13-12-2017-world-bank-and-who-half-the-world-lacks-access-to-essential-health-services-100-million-still-pushed-into-extreme-poverty-because-of-health-expenses available at. accessed on November 23. [Google Scholar]
- 5.More than the virus, fear of stigma is stopping people from getting tested: Doctors. The New Indian Express. 2021 https://www.newindianexpress.com/states/karnataka/2020/aug/06/more-than-virus-fear-of-stigma-is-stopping-people-from-getting-tested-doctors-2179656.html available at. accessed on November 22. [Google Scholar]
- 6.Kliff S. Most Coronavirus Tests Cost About $100. Why Did One Cost $2,315? The New York Times. 2021 https://www.nytimes.com/2020/06/16/upshot/coronavirus-test-cost-varies-widely.html available at. accessed on November 23. [Google Scholar]
- 7.Lee L., et al. Speech segment durations produced healthy and asthmatic subject. Journal of Speech Hear Disorder. May 1988;53(2):186–193. doi: 10.1044/jshd.5302.186. [DOI] [PubMed] [Google Scholar]
- 8.Yadav S., et al. Analysis of acoustic features for speech sound-based classification of asthmatic and healthy subjects. Proceedings of the IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP), May 4-8, Barcelona. 2020:6789–6793. doi: 10.1109/ICASSP40776.2020.9054062. [DOI] [Google Scholar]
- 9.Kutor J., et al. Speech Signal Analysis as an alternative to spirometry in asthma diagnosis: investing the linear and polynomial correlation coefficients. International Journal of Speech Technology. September 2019;22(3):611–620. doi: 10.4172/2475-7586.1000136. [DOI] [Google Scholar]
- 10.Nathan V., et al. Assessment of chronic pulmonary disease patients using biomarkers from natural speech recorded by mobile devices. Proceedings of the IEEE 16th International Conference on Wearable and Implantable Body Sensor Networks (BSN), May 19-20. 2019:1–4. doi: 10.1109/BSN.2019.8771043. [DOI] [Google Scholar]
- 11.Kertesz A., Appell J., Fisman M. The dissolution of language in Alzheimer's disease. Canadian Journal of Neurological Science. November 1986;13:415–418. doi: 10.1017/s031716710003701x. [DOI] [PubMed] [Google Scholar]
- 12.Faber-Langendoen K., et al. Aphasia in senile dementia of the Alzheimer type. Annals of Neurology. April 1988;23(4):365–370. doi: 10.1002/ana.410230409. vol.no. [DOI] [PubMed] [Google Scholar]
- 13.Shirvan R.A., Tahami E. Proceedings of the 18th Iranian Conference on Biomedical Engineering. 2011. Voice Analysis for Detecting Parkinson's Disease using Genetic Algorithm and KNN; pp. 278–283. December 4-16Tehran, Iran. [DOI] [Google Scholar]
- 14.Rosen K.M., et al. Parametric quantitative acoustic analysis of conversation produced by speakers with dysarthria and healthy speakers. Journal of Speech, Language, and Hearing Research. April 2006;49(2):395–411. doi: 10.1044/1092-4388(2006/031. vol.no. [DOI] [PubMed] [Google Scholar]
- 15.Hare B., Peter M.C., Snyder J. Variability in fundamental frequency during speech in prodromal and incipient Parkinson's disease: A longitudinal case study. Brain and Cognition. November 2004;56(1):24–29. doi: 10.1016/j.bandc.2004.05.002. vol.no. [DOI] [PubMed] [Google Scholar]
- 16.LeWitt P.A. In: Parkinson's Disease and Movement Disorders. Current Clinical Practice. Adler C.H., Ahlskog J.E., editors. Humana Press; Totowa, NJ: 2000. Parkinson's Disease: Etiologic Considerations; pp. 91–100. [DOI] [Google Scholar]
- 17.Nilsonne A. Measuring the rate of change of voice fundamental frequency in fluent speech during mental depression. Journal of Acoustic Society of America. February 1988;83(2):716–728. doi: 10.1121/1.396114. [DOI] [PubMed] [Google Scholar]
- 18.France D.J., et al. Acoustical properties of speech as indicators of depression and suicidal risk. IEEE Transaction on Biomedical Engineering. July 2000;47(7):829–837. doi: 10.1109/10.846676. [DOI] [PubMed] [Google Scholar]
- 19.Mundt J., et al. Voice acoustic measures of depression severity and treatment response collected via interactive voice response (IVR) technology. Journal of Neurolinguistics. January 2007;20(1):50–64. doi: 10.1016/j.jneuroling.2006.04.001. no. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinberger D.R. Implications of normal brain development for the pathogenesis of schizophrenia. Archives of General Psychiatry. July 1987;44(7):660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 21.Elvevåg B. An automated method to analyze language use in patients with schizophrenia and their first-degree relatives. Journal of Neurolinguistics. May 2010;23(3):270–284. doi: 10.1016/j.jneuroling.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhangi J., et al. Clinical investigation of speech signal features among patients with schizophrenia. Shanghai Archives of Psychiatry. April 2016;28(2):95–102. doi: 10.11919/j.issn.1002-0829.216025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryson S.E. Brief Report: Epidemiology of autism. Journal of Autism and Developmental Disorder. April 1996;26:165–167. doi: 10.1007/BF02172005. [DOI] [PubMed] [Google Scholar]
- 24.Shriberg L.D., et al. Speech and prosody characteristics of adolescents and adults with high-functioning autism and Asperger syndrome. Journal of Speech, Language, and Hearing. Oct. 2001;44(5):1097–1115. doi: 10.1044/1092-4388(2001/087). [DOI] [PubMed] [Google Scholar]
- 25.Maier A., et al. Automatic Speech Recognition Systems for the Evaluation of Voice and Speech Disorders in Head and Neck Cancer. EURASIP Journal on Audio, Speech, and Music Processing. December 2009;1:1–7. doi: 10.1155/2010/926951. vol. [DOI] [Google Scholar]
- 26.Graves K. Emotional expression and emotional recognition in breast cancer survivor. Journal of Psychology and Health. January 2010;20:579–595. doi: 10.1080/0887044042000334742. [DOI] [Google Scholar]
- 27.Islam R., Tarique M., Raheem E. A survey on signal processing based pathological voice detection systems. IEEE Access. April 2020;8:66749–66776. doi: 10.1109/ACCESS.2020.2985280. vol. [DOI] [Google Scholar]
- 28.Imran A., et al. AI4COVID: AI-enabled preliminary diagnosis for COVID-19 from cough samples via an app. Informatics in Medicine Unlocked. June 2020;20:1–13. doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniel More, “Causes and Risk Factors of Cough Health Conditions Linked to Acute, Sub-Acute, or Chronic Coughs” available at https://www.verywellhealth.com/causes-of-cough-83024.
- 30.Bales C., et al. Proceedings of the IEEE International Conference on E-Health and Bioengineering (EHB) 2020. Can Machine Learning Be Used to Recognize and Diagnose Coughs? October 29-30, Lasi, Romania. [DOI] [Google Scholar]
- 31.Sharma N., et al. Proceedings of INTERSPEECH. 2020. Coswara- A Database of Breathing, Cough, and Voice Sounds for COVID-19 Diagnosis. October 25-29Shanghai. [DOI] [Google Scholar]
- 32.Laguarta J., Hueto F., Subirana B. COVID-19 Artificial Intelligence Diagnosis using Only Cough Recording. IEEE Open Journal of Engineering in Medicine and Biology. December 2020;1:275–281. doi: 10.1109/OJEMB.2020.3026928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subirana B., et al. Hi Sigma, do I have the Coronavirus?: Call for a new artificial intelligence approach to support healthcare professionals dealing with the COVID-19 pandemic. 2020 arXiv:2004.06510. [Google Scholar]
- 34.C. Brown et al., “Exploring Automatic Diagnosis of COVID-19 from Crowdsourced Respiratory Sound,” Proceedings of the ACM Knowledge Discovery and Data Mining (Health Day), August 23-27, Virtual, pp. 3474-3484, DOI: 10.1145/3394486.3412865.
- 35.Han J., et al. An early-stage on Intelligent Analysis of Speech under COVID19: Severity, Sleep Quality, Fatigue, and Anxiety. Proceedings of the INTERSPEECH, Shanghai, China, October 25-29. 2020 [Google Scholar]
- 36.B. Schuller et al., “The INTERSPEECH 2014 Computational Paralinguistic Challenge: Cognitive and Physical Load,” Proceedings of the INTERSPEECH 2014, September 14-18, Singapore.
- 37.Eyben F., et al. The Geneva Minimalistic Acoustic Parameter Set (GeMAPS) for Voice Research and Affective Computing. IEEE Transaction on Affective Computing. July 2015;7(2):10–202. doi: 10.1109/TAFFC.2015.2457417. no. [DOI] [Google Scholar]
- 38.openSMILE 3.0. 2021 https://www.audeering.com/research/opensmile available at. accessed on November 24. [Google Scholar]
- 39.C. Shimon et al., “Artificial Intelligence enabled preliminary diagnosis for COVID-19 from voice cues and questionnaires,” Journal of Acoustic Society of America, vol. 149, no.2, pp.120-1124, DOI: 10.1121/10.0003434. [DOI] [PMC free article] [PubMed]
- 40.PRAAT: doing phonetics by computer. 2021 https://www.fon.hum.uva.nl/praat/accessed available at. on November 24. [Google Scholar]
- 41.Librosa: audio and music processing in Python. 2021 https://librosa.org/ available at. accessed on November 24. [Google Scholar]
- 42.Asiaee M., et al. Voice Quality Evaluation in Patients with COVID-19: An Acoustic Analysis. Journal of Voice, Article in Press. October 2020:1–7. doi: 10.1016/j.jvoice.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinkas G., et al. SARS-COV-2 Detection from Voice. IEEE Open Journal of Engineering in Medicine and Biology. 2020;1:268–274. doi: 10.1109/OJEMB.2020.3026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hassan A., Shahin I., Bader M. COVID-19 Detection System Using Recurrent Neural Networks. Proceedings of International Conference on Communication, Computing, Cybersecurity, and Informatics, November 3-5. 2020 doi: 10.1109/CCCI49893.2020.9256562. Sharjah. [DOI] [Google Scholar]
- 45.Alsabek M.B., Shahin I., Hassan A. Studying the Similarity of COVID-19 Sound based on Correlation Analysis of MFCC. Proceedings of International Conference on Communication, Computing, Cybersecurity, and Informatics, November 3-5. 2020 doi: 10.1109/CCCI49893.2020.9256700. [DOI] [Google Scholar]
- 46.A. O. Papdina, A. M. Salah, and K. Jalel, “Voice Analysis Framework for Asthma-COVID-19 Early Diagnosis and Prediction: AI-based Mobile Cloud Computing Application,” Proceedings of the IEEE Conference of Russian Young Researchers in Electrical and Electronic Engineering (ElConRus), 26-29 January, St Petersburg, Moscow, DOI: 10.1109/ElConRus51938.2021.9396367.
- 47.P. Mouawad, T. Dubnov, and S. Dubnov, “Robust Detection of COVID-19 in Cough Sounds Using Recurrence Dynamics and Viable Markov Model,” SN Computer Science, vol. 2, no. 34, pp. 1-13, DOI: 10.1007/s42979-020-00422-6. [DOI] [PMC free article] [PubMed]
- 48.G. Chaudhari et al., “Virufy: Global Applicability of Crowdsourced and Clinical datasets for AI Detection of COVID-19 from Cough,” arXiv: 2011.13320.
- 49.The Difference between Obstructive and Restrictive Lung Diseases is. November 24, 2021 https://centersforrespiratoryhealth.com/blog/the-difference-between-obstructive-and-restrictive-lung-disease/ available at. accessed on. [Google Scholar]
- 50.Morris F. Spirometry in the evaluation of pulmonary function. Western Journal of Medicine. Aug 1976;125(2):110–118. [PMC free article] [PubMed] [Google Scholar]
- 51.Galiatsatos P. COVID-19 Lung Damage. 2021 https://www.hopkinsmedicine.org/health/conditions-and-diseases/coronavirus/what-coronavirus-does-to-the-lungs available at. accessed on November 24. [Google Scholar]
- 52.Korpas J., Sadlonova J., Vrabec M. Analysis of the Cough Sound: an Overview. Pulmonary Pharmacology. 1996;9:261–268. doi: 10.1006/pulp.1996.0034. 1996. [DOI] [PubMed] [Google Scholar]
- 53.Virufy available at https://github.com/virufy/virufy-data.
- 54.Giannakopouls T. 1st Edition. Academic Press; 2014. Introduction to Audio Analysis; pp. 59–98. [Google Scholar]
- 55.Sreeram A.S.K, Ravishankar U., Sripada N.R., Mamidgi B. Investigating the potential of MFCC features in classifying respiratory diseases. Proceeding of the 7th International Conference on Internet of Things: Systems, Management, and Security (IOTSMS), December 14-16. 2020 doi: 10.1109/IOTSMS52051.2020.9340166. France. [DOI] [Google Scholar]
- 56.G. Chambres et al., “Automatic detection of patient with respiratory diseases using lung sound analysis,” Proceedings of the International Conference on Content-Based Multimedia Indexing, September 4-6, Rochelle, DOI: 10.1109/CBMI.2018.8516489.
- 57.Aykanat1 M., et al. Classification of lung sounds using convolutional neural network. EUROSIP Journal on Image and Video Processing. 2017;65:1–9. doi: 10.1186/s13640-017-0213-2. [DOI] [Google Scholar]
- 58.Quatieri T.E. Discrete-Time Speech Signal Processing: Principles and Practices. Prentice-Hall; Upper Saddle River: 2001. Production and Classification of Speech Sounds; pp. 72–76. [Google Scholar]
- 59.Rabiner L.R., Schafer R.W. Theory and Applications of Digital Speech Processing. International Edition, Pearson. 2011:477–479. [Google Scholar]
- 60.P. Kim, “MATLAB Deep Learning: With Machine Learning, Neural Networks and Artificial Intelligence,” Academic Press, pp. 121-14.
- 61.R. M. Rangayyan, “Biomedical Signal Analysis,” Second Edition, John Wiley and Sons, 111 River Street, NJ, pp. 598-606.
- 62.Y. Jiaa and P. Du, “Performance measures in evaluating machine learning-based bioinformatics predictors for classifications,” Quantitative Biology, vol. 4, no. 4, pp. 320-330, DOI 10.1007/s40484-016-0081-2.
- 63.Kosasih K., Abeyratne U.R., Swarnkar V. High Frequency Analysis of Cough Sounds in Pediatric Patients with Respiratory Diseases. Proceedings of the 34th Annual International Conference of the IEEE EMBS, San Diego, California USA. August 28-September 01, 2012:5654–5657. doi: 10.1109/EMBC.2012.6347277. [DOI] [PubMed] [Google Scholar]
- 64.Despotovic V., et al. Detection of COVID-19 from voice, cough and breathing patterns: Dataset and preliminary results. Computers in Biology and Medicine. November 2021;138 doi: 10.1016/j.compbiomed.2021.104940. vol.pp. 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith J.A., et al. The description of cough sounds by healthcare professionals. Cough. January 2006;2(1):1–9. doi: 10.1186/1745-9974-2-1. no. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.COVID-19-train-audio available at COVID-19-train-audio/not-covid19-coughs/PMID-16436200 at masterhernanmd/COVID-19-train-audioGitHub.