Abstract
The TIM22 protein import pathway of the yeast mitochondrion contains several components, including a family of five proteins (Tim8p, -9p, -10p, -12p, and -13p [Tim, for translocase of inner membrane]) that are located in the intermembrane space and are 25% identical. Tim9p and Tim10p have dual roles in mediating the import of inner membrane proteins. Like the Tim8p-Tim13p complex, the Tim9p-Tim10p complex functions as a putative chaperone to guide hydrophobic precursors across the intermembrane space. Like membrane-associated Tim12p, they are members of the Tim18p-Tim22p-Tim54p membrane complex that mediates precursor insertion into the membrane. To understand the role of this family in protein import, we have used a genetic approach to manipulate the complement of the small Tim proteins. A strain has been constructed that lacks the 70-kDa soluble Tim8p-Tim13p and Tim9p-Tim10p complexes in the intermembrane space. Instead, a functional version of Tim9p (Tim9S67Cp), identified as a second-site suppressor of a conditional tim10 mutant, maintains viability. Characterization of this strain revealed that Tim9S67Cp and Tim10p were tightly associated with the inner membrane, the soluble 70-kDa Tim8p-Tim13p and Tim9p-Tim10p complexes were not detectable, and the rate of protein import into isolated mitochondria proceeded at a slower rate. An arrested translocation intermediate bound to Tim9S67Cp was located in the intermembrane space, associated with the inner membrane. We suggest that the 70-kDa complexes facilitate import, similar to the outer membrane receptors of the TOM (hetero-oligomeric translocase of the outer membrane) complex, and the essential role of Tim9p and Tim10p may be to mediate protein insertion in the inner membrane with the TIM22 complex.
The mitochondrion has an elaborate set of translocons on the outer and inner membranes to mediate the import of proteins from the cytosol (19, 21, 25, 30). Most mitochondrial proteins are synthesized as cytosolic precursors containing a cleavable N-terminal presequence that directs their import into mitochondria via the general protein import pathway. The precursor is escorted through the cytosol by chaperones, and then the hetero-oligomeric translocase of the outer membrane (TOM) mediates translocation across the outer membrane. Several components function as receptors, while others form the translocation pore. After passage through the outer membrane, the Tim17p-Tim23p (Tim, for translocase of inner membrane) complex of the inner membrane, together with the ATP-dependent import motor Hsp70, Tim44p, and mGrpE, mediates translocation across the inner membrane. Finally, a number of soluble proteins in the matrix involved in the proteolytic maturation and folding of the imported proteins may be required to complete assembly (19, 21, 25, 30).
The mitochondrion has a separate import pathway for inner membrane proteins, with components residing in the intermembrane space and inner membrane (2, 16, 21). Proteins destined for the inner membrane are escorted by cytosolic chaperones and then pass through the TOM complex to the intermembrane space. The intermembrane space complexes Tim9p-Tim10p and Tim8p-Tim13p function as putative chaperones to transfer the hydrophobic precursors across the intermembrane space to an inner membrane machinery specialized for the insertion of membrane proteins (1, 13, 15, 32). The inner membrane complex consists of Tim12p, Tim18p, Tim22p, Tim54p, and a small fraction of Tim9p and Tim10p, which together form a 300-kDa complex (1, 11–13, 15, 17, 32). Components Tim9p, Tim10p, Tim12p, Tim22p, and Tim54p are essential for viability (1, 11–13, 15, 17, 32).
A typical protein imported by this pathway is the ADP/ATP carrier (AAC), which contains six membrane-spanning regions (22). AAC lacks a cleavable N-terminal targeting sequence, instead carrying targeting information in discrete regions throughout the polypeptide chain (5, 22). Yeast has about three dozen members of the mitochondrial metabolite carrier family (20). The TIM22 pathway probably imports all of these as well as many other integral proteins of the mitochondrial inner membrane, including the import components Tim22p and Tim23p (3, 5, 18).
The small Tim proteins (Tim8p, Tim9p, Tim10p, Tim12p, and Tim13p) are approximately 25% identical and 40 to 50% similar, yet Tim9p partners exclusively with Tim10p and Tim8p partners with Tim13p in soluble intermembrane space complexes (1, 3, 14, 15). The Tim9p-Tim10p complex is 10-fold more abundant than the Tim8p-Tim13p complex (1, 14, 15, 32). Approximately 5% of Tim9p and Tim10p is associated with the 300-kDa Tim18p-Tim22p-Tim54p complex at the inner membrane, whereas 95% is soluble in the 70-kDa intermembrane space complex (14, 15). Tim9p and Tim10p bind to translocation intermediates of the mitochondrial carrier family, Tim17p and Tim22p, whereas Tim8p and Tim13p bind to Tim23p (3, 5, 18), suggesting that the battery of small Tim proteins may have different substrate specificities.
While previous studies have focused on investigating direct interactions with translocation intermediates, we investigated the role of the small Tim proteins by using a genetic approach. We constructed a yeast strain that is deficient in both 70-kDa Tim9p-Tim10p and Tim8p-Tim13p complexes and is kept viable by a functional version of Tim9p, designated Tim9S67Cp, which was identified as a multicopy suppressor of a conditional tim10 mutant (15). Characterization of this strain revealed it was viable and that protein import into isolated mitochondria proceeded at a slower rate. Tim9p and Tim10p were bound to the inner membrane and seemingly were not present in a soluble 70-kDa complex. Both Tim9p and Tim10p could be cross-linked to an AAC translocation intermediate, indicating that they mediate protein import.
MATERIALS AND METHODS
Plasmids and strains.
Standard genetic techniques were used for growth, manipulation, and transformation of yeast strains (8, 31). The Saccharomyces cerevisiae strains used in this study are listed in Table 1. CK13 and CK14 are strains that contain the temperature-sensitive tim10-1 allele (designated as tim10-1 in Fig. 1) and have been previously described (15). The strain CK18 contains allele tim9199A→T coding for protein Tim9S67Cp and previously was identified because it restored growth at the restrictive temperature of 37°C for temperature-sensitive tim10-1 and tim12-1 strains (15). The strain CK26 (designated as Tim9S67C) was constructed by transforming a centromeric plasmid harboring tim9199A→T into diploid strain CK22, in which one TIM9 allele is disrupted, followed by sporulation and auxotrophic selection.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Source or reference | Comments |
|---|---|---|---|
| GA74-1A | MATaade8 his3 leu2 trp1 ura3 | 10 | Parental strain |
| GA74-6A | MATα ade8 his3 leu2 trp1 ura3 | 10 | Parental strain |
| GA74-1A.d | MATa/α ade8/ade8 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 | 10 | Parental strain |
| CK13 | MATα ade8 his3 trp1 ura3 Δtim10::HIS3 tim10-1:LEU2 | 15 | Used for segregation analysis of second-site suppressor locus |
| CK14 | MATaade8 his3 leu2 ura3 Δtim10::HIS3 tim10-1:TRP1 | 15 | Original mutant used to generate second-site suppressors, designated tim10-1 |
| CK18 | MATaade8 his3 leu2 ura3 Δtim10::HIS3 tim10-1:TRP1 tim9199A→T | 15 | Extragenic suppressor strain, designated tim10-1 Ts+ |
| CK19 | MATa/α ade8/ade8 his3/his3 ura3/ura3 Δtim10::HIS3/Δtim10::HIS3 trp1/tim10-1:TRP1 leu2/tim10-1:LEU2 TIM9/tim9199A→T | 15 | CK18 × CK13 |
| CK22 | MATa/α ade8/ade8 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 TIM9/Δtim9::HIS3 | 15 | Used to generate CK26; one copy of TIM9 is deleted |
| CK26 | MATaade8 his3 trp1 ura3 leu2 Δtim9::TRP1 [ptim9199A→Ts:URA] | This study | Tim9p is replaced by Tim9S67Cp, designated Tim9S67C |
| CK73 | MATa/α ade8/ade8 his3/his3 ura3/ura3 Δtim10::HIS3/Δtim10::HIS3 leu2/tim10-1:LEU2 trp1/tim10-1 ts:TRP1 TIM8/Δtim8::URA3 TIM9/tim9199A→T | This study | CK19 in which one copy of TIM8 was disrupted with URA3 |
| CK79 | MATaade8 his3 trp1 ura3 Δtim10::HIS3 tim10-1ts:LEU2 Δtim8::URA3 tim9199A→T | This study | Spore from CK73, designated Δ70k |
| CK80 | MATaade8 his3 leu2 ura3 Δtim10::HIS3 tim10-1 ts:TRP1 Δtim8::URA3 tim9199A→T | This study | Spore from CK73 |
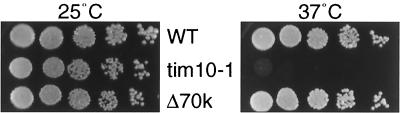
FIG. 1.
Strain Δ70k grows similar to the parental strain. The Δ70k and tim10-1 strains and the parental (wild-type [WT]) strain (Table 1) were grown to mid-log phase at 25°C in liquid YPD. Cultures were serially diluted by a factor of 3 and spotted onto YPD plates. Plates were incubated for 3 days at 25 or 37°C. WT, GA74-1A (10); tim10-1, temperature-sensitive tim10-1 allele integrated into LEU2 locus (15); Δ70k, strain in which 70-kDa Tim9p-Tim10p and Tim8p-Tim13p complexes are not detectable by immunoblot analysis.
The strain Δ70k was generated as follows. CK19 was generated by crossing CK18 and CK13. One TIM8 allele was disrupted with URA3 to generate strain CK73. Diploid CK73 was sporulated, and tetrads were separated on rich glucose medium at 25°C. Those that grew were screened for their ability to grow at 37°C on SC media lacking histidine, uracil, and tryptophan or leucine. In all cases, the segregation pattern of markers indicated that the strain grew well at 37°C when TIM8 was deleted and allele tim9199A→T cosegregated. Strains CK79 and CK80 were two spores in which initial characterization of growth rate and analysis of the abundance of Tim proteins by immunoblotting were identical. CK79 was used for further characterization.
For in vitro transcription and translation, the DNA fragment encoding Tim23p (9) was subcloned into pGEM3Z (Promega), and the AAC2 gene was subcloned into pSP65 (Promega).
Import of radiolabeled proteins into isolated mitochondria and cross-linking studies.
Mitochondria were purified from lactate-grown yeast cells (7) and assayed for in vitro protein import as described previously (24). Proteins were synthesized in a rabbit reticulocyte lysate in the presence of [35S]methionine after in vitro transcription of the corresponding gene by SP6 or T7 polymerase. The reticulocyte lysate containing the radiolabeled precursor was incubated with isolated mitochondria at the indicated temperatures in import buffer (1-mg/ml bovine serum albumin, 0.6 M sorbitol, 150 mM KCl, 10 mM MgCl2, 2.5 mM EDTA, 2 mM ATP, 2 mM NADH, 20 mM K+-HEPES [pH 7.4]). Where indicated, the potential across the mitochondrial inner membrane was dissipated with 1 μM valinomycin. Nonimported radiolabeled proteins were removed by treatment with 100 μg of trypsin or 50 μg of proteinase K per ml for 15 to 30 min on ice; trypsin was inhibited with 200 μg of soybean trypsin inhibitor per ml, and proteinase K was inhibited with 1 mM phenylmethylsulfonyl fluoride (PMSF), respectively.
The translocation intermediates of AAC were cross-linked to adjacent proteins with 0.1 mM m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS) or 0.5 mM bis-maleimidohexane (BMH). The cross-linking protocol was performed as follows (15, 18), except as noted in Fig. 8. After import, protease was omitted, and mitochondria were washed, suspended at 1 mg/ml in import buffer, and incubated with the cross-linker on ice for 30 min followed by a quench with 100 mM Tris-HCl (pH 7.5) (for MBS) or 1 mM 2-mercaptoethanol (for MBS and BMH). For immunoprecipitation, solubilized mitochondria were incubated with the corresponding monospecific rabbit immunoglobulins G (IgGs) coupled to protein A-Sepharose (23).
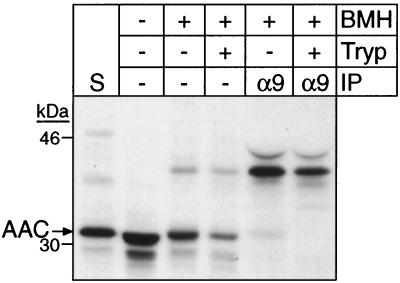
FIG. 8.
The cross-linked AAC import intermediate is protected by the outer membrane in strain Δ70k mitochondria. In vitro import and cross-linking with radiolabeled AAC into strain Δ70k mitochondria were performed as in Fig. 7A, followed by treatment with 100 μg of trypsin per ml on ice for 30 min. After the addition of 200 μg of soybean trypsin inhibitor (Tryp) per ml, immunoprecipitation (IP) with protein A-Sepharose beads containing immobilized rabbit IgGs monospecific for Tim9p (α9) was performed as in Fig. 7A. S, standard (as defined in the legend to Fig. 6).
Blue native gel electrophoresis.
Mitochondria (2.5 mg/ml) were solubilized in a mixture containing 20 mM K+-HEPES (pH 7.4), 50 mM NaCl, 10% glycerol, 2.5 mM MgCl2. 1 mM EDTA, 0.16% n-dodecylmaltoside (Boehringer Mannheim) for 30 min on ice. Insoluble material was removed by centrifugation at 100 000 × g for 10 min, and the solubilized proteins were analyzed by blue native gel electrophoresis on a 6 to 16% linear polyacrylamide gradient (4, 27, 28).
Coimmunoprecipitation assays.
Monospecific antibody (10 to 20 μl per mg of mitochondria) against Tim12p was bound to protein A-Sepharose (30 μl wet volume per mg of mitochondria; Amersham Pharmacia Biotech) for 1 h in 1.0 ml of wash buffer (20 mM HEPES-KOH [pH 7.4], 0.2 M sucrose, 50 mM NaCl, 1 mM PMSF). The beads were washed two times to remove unbound antiserum. Mitochondria were solubilized as described for blue native gel electrophoresis and incubated with the antibody-bound protein A-Sepharose beads by gentle rotation for 2 h at 4°C. After the beads had been washed twice with wash buffer, bound proteins were extracted at 65°C with sodium dodecyl sulfate (SDS)-containing sample buffer and analyzed by Tricine-SDS-polyacrylamide gel electrophoresis (PAGE).
Miscellaneous.
Submitochondrial localization of proteins was determined as described previously (6). Mitochondrial proteins were analyzed by SDS-PAGE with a 10 or 16% polyacrylamide gel and a Tricine-based running buffer (29). Proteins were detected by immunoblotting with nitrocellulose or polyvinylidene difluoride (PVDF) membranes and visualization of immune complexes with 125I-protein A. Protein concentration was assayed by the bicinchoninic acid method (Pierce) with bovine serum albumin as the standard.
RESULTS
A yeast strain with a minimal complement of small Tim proteins is viable.
Tim9p and Tim10p potentially have multiple roles in mediating the import of inner membrane proteins because they are partner proteins in a soluble 70-kDa complex in the intermembrane space and components of a 300-kDa inner membrane complex with Tim12p, Tim18p, Tim22p, and Tim54p. This raises the question of where the essential role of Tim9p and Tim10p is—in the intermembrane space or at the inner membrane? Furthermore, the intermembrane space contains the distinct 70-kDa Tim8p-Tim13p complex, which is not essential for viability. We have used a genetic approach to investigate whether the 70-kDa complexes are essential to mediate protein import or whether they facilitate import in a manner similar to the outer membrane receptors on the TOM complex.
From previous genetic studies, we isolated a temperature-sensitive tim10-1 strain, which lacked the soluble 70-kDa Tim9p-Tim10p complex (15). An extragenic suppressor of the tim10-1 mutant was identified that restored growth at 37°C; the suppressor contained a point mutation in the TIM9 locus, designated tim9199A→T, resulting in Ser-67 being replaced by Cys-67 (Tim9S67Cp) (15). In addition, allele tim9199A→T conferred growth to the temperature sensitive tim12-1 strain at 37°C, but could not support growth in a strain deleted for either TIM10 or TIM12 (15). Tim9S67Cp thus interacted genetically with both Tim10p and Tim12p. Because the common feature of Tim10p and Tim12p is their location at the inner membrane, these genetic interactions potentially suggest that the essential role for Tim10p is at the inner membrane.
The intermembrane space also contains the Tim8p-Tim13p complex, which unlike Tim9p-Tim10p is not essential for viability. However, deletion of TIM8, which results in loss of the Tim8p-Tim13p complex (14), resulted in synthetic lethality at 25°C with the tim10-1 mutant, indicating that the Tim8p-Tim13p complex is essential for viability under these conditions (14). Given that the intermembrane space contains two 70-kDa complexes, we investigated whether we could generate a viable yeast strain that lacked the 70-kDa intermembrane space complexes by deleting TIM8 from the temperature-sensitive tim10-1Ts+ strain, which does not contain the 70-kDa Tim9p-Tim10p complex. Deletion of TIM8 from the yeast tim10-1Ts+ strain was performed in a diploid followed by sporulation and tetrad analysis (see Materials and Methods for details). In all cases, allele segregation was as expected, and the strain, which was deleted for TIM8 and contained tim10-1 and tim9199A→T alleles, grew at a rate similar to the wild-type strain at 25 and 37°C (Fig. 1). This strain is designated Δ70k. Viability of this strain at 37°C was thus maintained by Tim9S67Cp.
The yeast strain Δ70k lacks the soluble 70-kDa intermembrane space complexes.
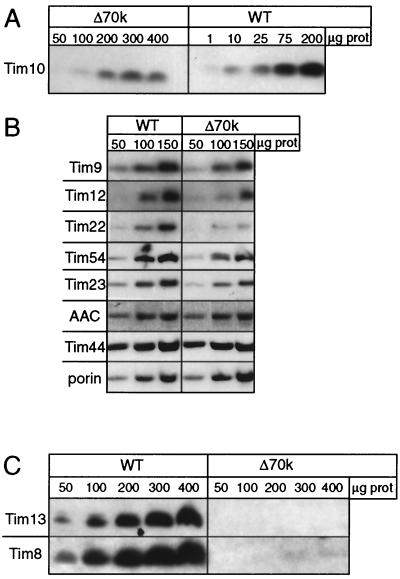
In strain Δ70k, the abundance of the Tim proteins was quantitated by immunoblotting to determine the relative abundance of the individual components. Mitochondria were purified from strain Δ70k and the parental strain grown at 37°C, and increasing amounts of mitochondrial protein were separated by SDS-PAGE as indicated in Fig. 2. Because 37°C is the nonpermissive temperature for the tim10-1 mutant, Tim10p should be nonfunctional. The amount of Tim10p was decreased by 94% (Fig. 2A); quantitation was performed by scanning laser densitometry (data not shown). In contrast, the levels of abundance of mitochondrial proteins porin, Tim23p, Tim44p, and AAC did not differ significantly (Fig. 2B). Previously, we estimated that mitochondria contain approximately 120 pmol of Tim10p and Tim9p per milligram of total mitochondrial protein (15). Given this decrease, the Δ70k mitochondria contain approximately 5 pmol of Tim10p/mg of mitochondria. Neither Tim8p nor Tim13p was detected by immunoblotting (Fig. 2C), because deletion of TIM8 leads to loss of the Tim8p-Tim13p complex (14). The abundance of Tim9p, Tim12p, and Tim54p was decreased by approximately 50%, and that of Tim22p was decreased by approximately 75%. The steady-state levels of Tim23p and AAC, substrates of the TIM22 import pathway, were not significantly affected in strain Δ70k mitochondria compared to wild-type mitochondria. This observation suggests that Tim23p and AAC seemingly are imported efficiently in vivo or, alternatively, may have an increased stability in strain Δ70k.
FIG. 2.
Abundance of the TIM22 import components is decreased in strain Δ70k. (A) The parental strain (wild type [WT]) and strain Δ70k were grown at 37°C in lactate medium. The mitochondria were isolated, and aliquots corresponding to total micrograms of mitochondrial protein (μg prot) were analyzed by SDS-PAGE and immunoblotting with rabbit antisera monospecific for Tim10. Blots were treated with 125I-protein A and subjected to autoradiography. (B) As in panel A, except that equal amounts (50, 100, and 150 μg) of mitochondrial proteins are loaded. Antibodies are listed on the left. AAC, ADP/ATP carrier. (C) As in panel A, except that equal amounts of mitochondrial proteins are loaded.
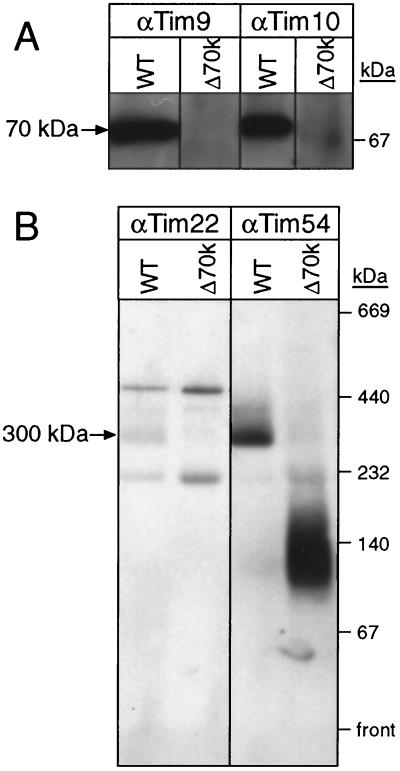
We next investigated whether Tim9p and Tim10p were present in a 70-kDa complex. Mitochondria were solubilized in 0.16% n-dodecylmaltoside and separated by blue-native gel electrophoresis followed by immunoblot analysis. While antibodies against Tim9p and Tim10p indicated both were in a 70-kDa complex in wild-type mitochondria, Tim9p and Tim10p were not present in the 70-kDa complex in strain Δ70k, even when the blots were overexposed several days (Fig. 3A). Tim22p and Tim54p were detected in the 300-kDa complex in wild-type mitochondria, but in strain Δ70k, Tim54p was present in a complex of 140 to 150 kDa as previously reported (Fig. 3B) (17, 18). Tim22p was not detectable by immunoblotting in the mutant strain.
FIG. 3.
Strain Δ70k lacks the soluble Tim9-Tim10 70-kDa complex. (A) Mitochondria from the parental (wild type [WT]) strain (10) and the Δ70k strain (Δ70k) were solubilized in 0.16% n-dodecylmaltoside and subjected to blue native gel electrophoresis (6 to 16% acrylamide) (27). Tim9 and Tim10 were detected by immunoblotting and incubation with 125I-protein A. (B) Blue native gel electrophoresis was performed as in panel A; the blot was incubated with monospecific sera against Tim22 and Tim54. In the immunoblot with anti-Tim22p antibody (α-Tim22p), bands located above and below the 300-kDa band resulted from proteins that cross-react with the antisera (17).
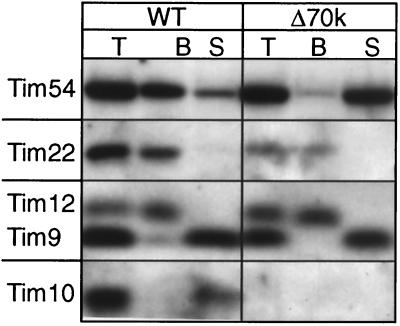
In wild-type mitochondria, approximately 5% each of Tim9p and Tim10p associates with the 300-kDa Tim12p-Tim22p-Tim54p complex. Are Tim9p and Tim10p partners with the inner membrane components in Δ70k mitochondria? Mitochondria were solubilized with 0.16% n-dodecylmaltoside and immunoprecipitated with antibodies against Tim12p (Fig. 4). Tim12p and Tim22p remained associated in Δ70k mitochondria as in wild-type mitochondria, but Tim54p did not coimmunoprecipitate. Furthermore, Tim9p and Tim10p did not coimmunoprecipitate with Tim12p, indicating that they are not associated stably with Tim12p and Tim22p in strain Δ70k. Therefore, the interaction of Tim9p and Tim10p with the 300-kDa membrane complex seemingly is not altered in strain Δ70k.
FIG. 4.
Tim22 and Tim12 remain associated in strain Δ70k. Mitochondria from the parental (wild type [WT]) strain and the Δ70k strain (Δ70k) were solubilized in 0.16% n-dodecylmaltoside. The lysate was subjected to immunoprecipitation with protein A-Sepharose beads containing immobilized rabbit IgGs monospecific for Tim12p. After centrifugation, 200 μg each of total (T), unbound (S), and bound (B) proteins was analyzed by SDS-PAGE and immunoblotting for Tim54p, Tim22p, Tim12p, Tim9p, and Tim10p.
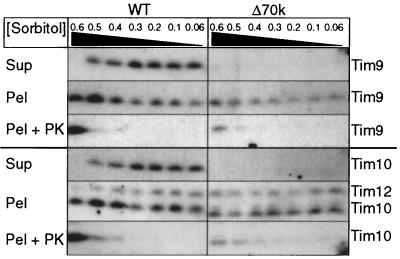
Because Tim9p and Tim10p were not present in a 70-kDa complex and were not associated with Tim12p and Tim22p, we selectively disrupted the mitochondrial outer membrane by osmotic shock to determine if Tim9p and Tim10p were associated with the inner membrane (Fig. 5), similar to Tim12p. In contrast to wild-type mitochondria, Tim9p and Tim10p only remained associated with the inner membrane in strain Δ70k. As expected, the mitochondrial outer membrane was completely disrupted because Tim9p and Tim10p were equally protease accessible. Based on these results, the Δ70k mitochondria seemingly do not have the soluble 70-kDa complexes in the intermembrane space; rather, Tim9p and Tim10p remain associated with the inner membrane.
FIG. 5.
Tim9p and Tim10p remain associated with the inner membrane in strain Δ70k. Isolated mitochondria from the parental (wild type [WT]) strain and the Δ70k strain were incubated in 20 mM HEPES-KOH (pH 7.4) and the indicated sorbitol concentrations (0.6 to 0.06 M) at 4°C for 30 min, followed by addition of 1 mM phenylmethylsulfonyl chloride (PMSF) to lyse the outer membrane. After centrifugation, the supernatant (Sup) and pellet (Pel) were analyzed by SDS-PAGE and immunoblotting for Tim9p, Tim10p, and Tim12p (pellet fraction only). To confirm that the intermembrane space contents were released, proteinase K was added at 50 μg/ml (PEL + PK). In a separate control (unpublished data), the effectiveness of the protease was verified by addition of Triton X-100.
Tim9p and Tim10p mediate import of AAC in strain Δ70k mitochondria.
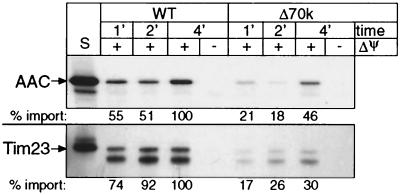
In mitochondria derived from the temperature-sensitive tim10-1mutant, the abundance and rate of in vitro protein import of the carrier proteins are decreased by 95% (13). Because Tim23p and AAC steady-state levels were not significantly reduced in strain Δ70k, the import of Tim23p and AAC may not be defective in vivo. For precursors AAC and Tim23p, we tested the rate of in vitro import into isolated mitochondria followed by protease treatment and carbonate extraction to confirm that AAC and Tim23p were inserted into the inner membrane (Fig. 6). Import of Tim23p and AAC was two- to threefold slower in Δ70k mitochondria than in wild-type mitochondria. Mitochondria from strain Δ70k thus are able to import substrates of the TIM22 import pathway.
FIG. 6.
Strain Δ70k mitochondria import inner membrane proteins AAC and Tim23p, but at a decreased rate. In separate experiments, radiolabeled AAC and Tim23p were synthesized in vitro and incubated for 1, 2, and 4 min at 25°C in the presence or absence of a membrane potential (ΔΨ) with wild-type (WT) or Δ70k mitochondria. Samples were treated with proteinase K to remove nonimported precursor, followed by addition of 1 mM PMSF. Samples were analyzed by SDS-PAGE and fluorography. S, standard (10% of the radioactive precursor added to each assay). The import rate was quantitated by densitometry, with 100% being set as the amount of precursor imported into wild-type mitochondria after 4 min.
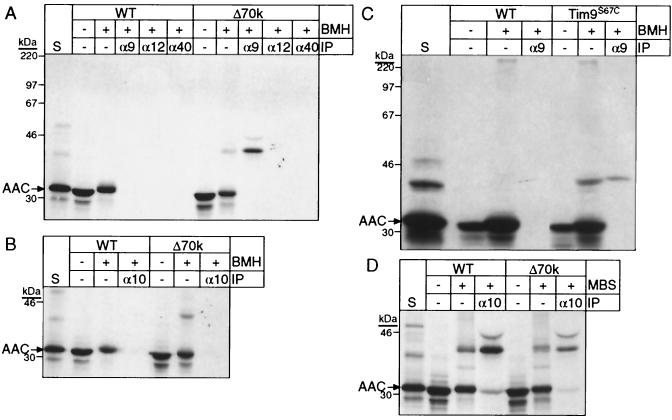
How do import intermediates negotiate the intermembrane space in mitochondria derived from strain Δ70k? Cross-linking experiments with an arrested AAC translocation precursor showed that Tim9p and Tim10p mediate translocation from the TOM complex to the TIM22 complex (5, 18). Similar studies were conducted with Δ70k mitochondria with AAC, by using a variety of cross-linkers followed by immunoprecipitation with antibodies monospecific for import components. The amino-reactive homobifunctional cross-linker dithiobis[succinimidyl propionate] (DSP), which previously resulted in abundant cross-linking between AAC and Tim9p and Tim10p (13, 15), failed to yield cross-links in Δ70k mitochondria (data not shown). However, using the sulfhydryl-reactive homobifunctional cross-linker BMH, AAC was cross-linked to Tim9p but not Tim10p, Tim12p, or Tom40p in Δ70k mitochondria (Fig. 7A and B); these cross-links also were not observed in wild-type mitochondria. Additional cross-links between AAC and Tom40p or Tom22p were not detected under a variety of conditions and with various cross-linkers (data not shown), indicating that the AAC translocation intermediate seemingly is not intimately associated with the TOM complex.
FIG. 7.
Tim9p and Tim10p, but not Tim12p and Tom40p, can be cross-linked to an AAC translocation intermediate in strain Δ70k. (A) Radiolabeled AAC was synthesized in vitro and imported into wild-type (WT) or Δ70k mitochondria. A fraction of the import reaction mixture was removed as an untreated control (−BMH), and the remainder was subjected to cross-linking with 0.5 mM BMH for 5 min on ice. After quenching, an aliquot was removed for direct analysis (+BMH). The remainder was denatured with SDS, and equal aliquots were incubated with protein A-Sepharose beads containing immobilized rabbit IgGs monospecific for Tim9p (α9), Tim12p (α12), and Tom40p (α40). After centrifugation, bound proteins were eluted with SDS-containing sample buffer and analyzed by SDS-PAGE and fluorography. The arrow marked “AAC” denotes the position of monomeric AAC. S, standard (as defined in the Fig. 6 legend). (B) As in panel A, except that antibodies against Tim10p were used for immunoprecipitation. (C) As in panel A, with isolated mitochondria from strain Tim9S67C, which expresses Tim9S67Cp, Tim8p, Tim10p, and Tim13p (Table 1), and wild-type mitochondria. Antibodies against Tim9p were used for immunoprecipitation (IP). (D) As in panel B, except that the cross-linker MBS (0.1 mM) was used.
Because strain Δ70k contains Tim9S67Cp, the presence of an additional cysteine residue may account for the specific cross-link to AAC. To address this, yeast strain Tim9S67C was constructed in which Tim9S67Cp replaced Tim9p (Table 1); Tim8p, Tim10p, and Tim13p were present at wild-type levels (data not shown). The AAC translocation intermediate was cross-linked to Tim9S67Cp in this strain as in strain Δ70k. Cys-67 may be in close proximity to the AAC translocation intermediate.
Additional studies with an amino and sulfhydryl heterobifunctional cross-linker, MBS, indicated that Tim10p was bound to AAC (Fig. 7D), but not Tim12p or Tim22p (data not shown). The Tim23p translocation intermediate can be cross-linked to Tim8p and Tim13p, as well as, to a lesser extent, Tim9p and Tim10p, in wild-type mitochondria (3, 18). However, results from cross-linking studies with Δ70k mitochondria indicated that Tim23p was not cross-linked to Tim9p, Tim10p, or Tim22p (data not shown).
To confirm that the AAC translocation intermediate completely crossed the mitochondrial outer membrane and was not engaged in the TOM complex, we confirmed the location of the cross-linked AAC precursor by protease digestion (Fig. 8). Prior to cross-linking, trypsin was added to the import reaction to remove nonimported AAC. Cross-linked AAC was immunoprecipitated with antibodies against Tim9p, even after protease addition. The translocation intermediate remained associated with the inner membrane when the outer membrane was disrupted by osmotic shock (data not shown). Similar cross-linking experiments with wild-type mitochondria have shown that the AAC translocation intermediate is in the intermembrane space (13, 15; data not shown). Given that Tim9p is associated with the inner membrane, the AAC translocation intermediate is most likely present at the inner membrane and not engaged in the TOM complex.
DISCUSSION
Given the complexity of the TIM22 import pathway for inner membrane proteins, we have used a genetic approach to investigate whether the 70-kDa complexes are essential for mediating protein import or merely facilitate import, in a manner similar to the outer membrane receptors of the TOM complex. The TIM22 import pathway contains two soluble 70-kDa complexes, Tim9p-Tim10p and Tim8p-Tim13p, and the 300-kDa complex at the inner membrane, composed of Tim12p, Tim18p, Tim22p, and Tim54p with a fraction of Tim9p and Tim10p (1, 11–13, 15, 17, 32). We have constructed a strain, Δ70k, in which the soluble 70-kDa complexes are not detectable. Deletion of TIM8 results in loss of the Tim8p-Tim13p complex, and heat treatment at 37°C inactivates Tim10p. Instead, Tim9S67Cp seemingly maintains viability because this strain grows at a rate similar to the parental strain. Therefore, the reduced complement of small Tim proteins does not result in a decreased rate of growth. Although the rate of protein import for Tim23p and AAC into isolated Δ70k mitochondria is decreased by two- to threefold, protein import in vivo seemingly is not affected. Similar discrepancies between in vivo and in vitro protein import rates coupled with no observed differences in growth rate have been observed previously (33); alternatively, Tim23p and AAC may have increased stability in strain Δ70k. Taken together, the 70-kDa complexes seemingly facilitate import, perhaps in a manner similar to that of the outer membrane receptors of the TOM complex.
Tim9p and Tim10p in mitochondria from strain Δ70k remain associated with the inner membrane. While immunoprecipitation experiments failed to show that they were in a complex with Tim12p and Tim22p, they may be transiently associated with the TIM22 complex. In contrast, the abundance of Tim54p in strain Δ70k is not affected. Rather, Tim54p is in a smaller complex of approximately 140 kDa, as shown previously in a tim22 temperature-sensitive mutant and Δtim18 strain (17, 18).
Neupert and colleagues have shown that AAC can be arrested in the TOM complex by protease digestion (5), while Pfanner and colleagues reported that a protein generated by fusion between AAC and dihydrofolate reductase can be stably arrested with components of the TOM and TIM22 complexes (26). In our study, attempts to identify a translocation intermediate associated with the TOM complex or with Tim12p and Tim22p were not successful. Rather, the AAC translocation intermediate from strain Δ70k cross-linked with high specificity to Tim9p and Tim10p. The AAC translocation intermediate in strain Δ70k most likely is associated with the inner membrane, bound to Tim9p and Tim10p, because it was protected from exogenous protease and Tim9p and Tim10p were associated tightly with the inner membrane. Taken together, characterization of strain Δ70k strongly suggests that the 70-kDa Tim8p-Tim13p and Tim9p-Tim10p complexes are not essential to mediate protein import. Further genetic analysis will provide insights into how the small Tim proteins cooperate with the subunits of the TIM22 complex and TOM complex.
ACKNOWLEDGMENTS
C.M.K. is a Damon Runyon-Walter Winchell Scholar. This work was supported by the the Damon Runyon-Walter Winchell Cancer Research Foundation (DRS18), the American Heart Association (0030147N), Burroughs Wellcome Fund New Investigator Award in the Toxicological Sciences (1001120), Research Corporation (RI0459), and the National Institutes of Health (1R01GM61721–01).
REFERENCES
- 1.Adam A, Endres M, Sirrenberg C, Lottspeich F, Neupert W, Brunner M. Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J. 1999;18:313–319. doi: 10.1093/emboj/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer M F, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 2000;10:25–31. doi: 10.1016/s0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- 3.Davis A J, Sepuri N B, Holder J, Johnson A E, Jensen R E. Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J Cell Biol. 2000;150:1271–1282. doi: 10.1083/jcb.150.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker P J, Muller H, Rassow J, Pfanner N. Characterization of the preprotein translocase of the outer mitochondrial membrane by blue native electrophoresis. Biol Chem. 1996;377:535–538. [PubMed] [Google Scholar]
- 5.Endres M, Neupert W, Brunner M. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J. 1999;18:3214–3221. doi: 10.1093/emboj/18.12.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glick B S, Brandt A, Cunningham K, Muller S, Hallberg R L, Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- 7.Glick B S, Pon L. Isolation of highly purified mitochondria from S. cerevisiae. Methods Enzymol. 1995;260:213–233. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 8.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 9.Haucke V, Schatz G. Reconstitution of the protein insertion machinery of the mitochondrial inner membrane. EMBO J. 1997;16:4560–4567. doi: 10.1093/emboj/16.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarosch E, Tuller G, Daum G, Waldherr M, Voskova A, Schweyen R J. Mrs5p, an essential protein of the mitochondrial intermembrane space, affects protein import into yeast mitochondria. J Biol Chem. 1996;271:17219–17225. doi: 10.1074/jbc.271.29.17219. [DOI] [PubMed] [Google Scholar]
- 11.Kerscher O, Holder J, Srinivasan M, Leung R S, Jensen R E. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol. 1997;139:1663–1675. doi: 10.1083/jcb.139.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerscher O, Sepuri N B, Jensen R E. Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol Biol Cell. 2000;11:103–116. doi: 10.1091/mbc.11.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koehler C M, Jarosch E, Tokatlidis K, Schmid K, Schweyen R J, Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- 14.Koehler C M, Leuenberger D, Merchant S, Renold A, Junne T, Schatz G. Human deafness dystonia syndrome is a mitochondrial disease. Proc Natl Acad Sci USA. 1999;96:2141–2146. doi: 10.1073/pnas.96.5.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koehler C M, Merchant S, Oppliger W, Schmid K, Jarosch E, Dolfini L, Junne T, Schatz G, Tokatlidis K. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 1998;17:6477–6486. doi: 10.1093/emboj/17.22.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koehler C M, Merchant S, Schatz G. How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem Sci. 1999;24:428–432. doi: 10.1016/s0968-0004(99)01462-0. [DOI] [PubMed] [Google Scholar]
- 17.Koehler C M, Murphy M P, Bally N, Leuenberger D, Oppliger W, Dolfini L, Junne T, Schatz G, Or E. Tim18p, a new subunit of the TIM22 complex that mediates insertion of imported proteins into the yeast mitochondrial inner membrane. Mol Cell Biol. 2000;20:1187–1193. doi: 10.1128/mcb.20.4.1187-1193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leuenberger D, Bally N A, Schatz G, Koehler C M. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 1999;17:4816–4822. doi: 10.1093/emboj/18.17.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 20.Palmieri F, Bisaccia F, Capobianco L, Dolce V, Fiermonte G, Iacobazzi V, Indiveri C, Palmieri L. Mitochondrial metabolite transporters. Biochim Biophys Acta. 1996;1275:127–132. doi: 10.1016/0005-2728(96)00062-x. [DOI] [PubMed] [Google Scholar]
- 21.Pfanner N. Mitochondrial import: crossing the aqueous intermembrane space. Curr Biol. 1998;8:R262–R265. doi: 10.1016/s0960-9822(98)70168-x. [DOI] [PubMed] [Google Scholar]
- 22.Pfanner N, Neupert W. Distinct steps in the import of ADP/ATP carrier into mitochondria. J Biol Chem. 1987;262:7528–7536. [PubMed] [Google Scholar]
- 23.Rospert S, Muller S, Schatz G, Glick B S. Fusion proteins containing the cytochrome b2 presequence are sorted to the mitochondrial intermembrane space independently of Hsp60. J Biol Chem. 1994;269:17279–17288. [PubMed] [Google Scholar]
- 24.Rospert S, Schatz G. Protein translocation into mitochondria. In: Celis J E, editor. Cell biology: a laboratory handbook. 2nd ed. Vol. 2. San Diego, Calif: Academic Press; 1998. pp. 277–285. [Google Scholar]
- 25.Ryan K R, Jensen R E. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 26.Ryan M T, Muller H, Pfanner N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J Biol Chem. 1999;274:20619–20627. doi: 10.1074/jbc.274.29.20619. [DOI] [PubMed] [Google Scholar]
- 27.Schägger H, Cramer W A, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 28.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 29.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 30.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 31.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sirrenberg C, Endres M, Folsch H, Stuart R A, Neupert W, Brunner M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- 33.Yaffe M P, Ohta S, Schatz G. A yeast mutant temperature-sensitive for mitochondrial assembly is deficient in a mitochondrial protease activity that cleaves imported precursor polypeptides. EMBO J. 1985;4:2069–2074. doi: 10.1002/j.1460-2075.1985.tb03893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]