Graphical abstract
Keywords: Antibiotics, Microbiomes, Natural products, Secondary metabolites, Nonribosomal peptides, Polyketides
Abstract
As we stand on the brink of the post-antibiotic era, we are in dire need of novel antimicrobial compounds. Microorganisms produce a wealth of so-called secondary metabolites and have been our most prolific source of antibiotics so far. However, rediscovery of known antibiotics from well-studied cultured microorganisms, and the fact that the majority of microorganisms in the environment are out of reach by means of conventional cultivation techniques, have led to the exploration of the biosynthetic potential in natural microbial communities by novel approaches. In this mini review we discuss how sequence-based analyses have exposed an unprecedented wealth of potential for secondary metabolite production in soil, marine, and host-associated microbiomes, with a focus on the biosynthesis of non-ribosomal peptides and polyketides. Furthermore, we discuss how the complexity of natural microbiomes and the lack of standardized methodology has complicated comparisons across biomes. Yet, as even the most commonly sampled microbiomes hold promise of providing novel classes of natural products, we lastly discuss the development of approaches applied in the translation of the immense biosynthetic diversity of natural microbiomes to the procurement of novel antibiotics.
1. Introduction
The increasing occurrence and spread of antibiotic resistant bacteria pose major threats to public health, and mitigation of the current development represents one of the most significant challenges to modern medicine. In 2019, approximately 2.6 million laboratory-confirmed urinary tract infections caused by Escherichia coli or Klebsiella pneumonia were reported to the Global Antimicrobial Resistance and Use Surveillance System (GLASS), of which 54.4% and 43.1%, respectively, were resistant to the first-line antibiotic co-trimoxazole. Notably, in the 500,000 reported bloodstream infections the occurrence of methicillin-resistant Staphylococcus aureus was 15% in high income countries and 33% in low and middle income countries [1]. While programs aiming at limiting the spread and reducing the emergence of antibiotic resistance are widely applied and under continuous development [2], it is of paramount importance that we can provide a continuous supply of new and effective antibiotics.
Decades ago, in the ‘Golden era of antibiotic discovery’, new compound classes were regularly discovered and nearly all antibiotics used today are derived from compounds described in the 1940s to 1960s [3], [4]. The principle approach in the bioprospecting for antibiotics during this period was based on agar overlay inhibition assays developed by Selman Waksman and colleagues [5]. With a primary focus on soil-derived actinobacteria, these initial systematic screening approaches, in combination with bioactivity guided fractionation, were instrumental for the identification of anti-microbial agents. In addition, they highlighted the fact that microorganisms from the soil microbial communities were proficient producers of antimicrobial compounds [6]. However, as re-discovery rates increased, bioprospecting hit the law of diminishing returns [7], and was in large part abandoned due to the consensus belief that the potential of cultured microbes had been exhausted. The search for novel antibiotics turned to chemical synthesis and class modification, where success rates were initially higher [8]. However, the chemical library approach has not been able to provide truly novel compounds in the long run and as the search continues, we have turned to nature yet again in the pursuit for structural novelty and truly novel classes of antibiotics.
While the screening of culturable bacteria and filamentous fungi has proven extremely useful in bioprospecting for novel antibiotics historically, cultivation-based discovery of novel secondary metabolites has two major limitations: first, the majority of microorganisms inhabiting natural niches are recalcitrant to cultivation as monocultures under standard manmade laboratory conditions, and although cultivation techniques have greatly improved in recent years, e.g. with the development of the iChip [9] and highly parallelized droplet cultivation [10], the majority of bacteria have ever only been observed through their DNA sequences [11], [12]. Second, even if microorganisms can be successfully cultured in the laboratory, they often only express a fraction of their biosynthesis potential under the conditions provided in standard screening programs [13], [14].
The last two decades have seen a remarkable development in high-throughput sequencing, allowing us to query the unculturable majority of microorganisms. By high-throughput amplicon sequencing approaches targeting the 16S rRNA gene, astounding amounts of information has become available describing microbiome composition and diversity in natural environments. Furthermore, sequencing targeting specific functional genes has shed light on the distribution and diversity of various functional groups of microorganisms in nature. As the sequencing output continues to increase, such targeted approaches are gradually replaced by broader, untargeted metagenomic sequencing of bulk environmental DNA (eDNA), and the subsequent generation of metagenome assembled genomes (MAGs) has allowed us to investigate the metabolism and ecology of unculturable microorganisms even further. Collectively, through the explosion in sequencing data and the in silico bioinformatic tools developed alongside, it has become evident that there is a large biosynthetic capacity in the unculturable majority of microorganisms, as well as a yet un-explored potential in the already cultured. However, the challenge of exploiting this genetic potential remains, and a more holistic and systematic understanding of the drivers of natural product diversity in different environments is needed. Biosynthetic domain-targeted amplicon sequencing and untargeted genome-resolved metagenomic studies have started to map this incredible diversity in environments ranging from arid soils to the ocean floors.
2. Evaluating the natural product biosynthesis potential of the unculturable majority
The collective microbiome of Earth encompasses a staggering 1030 bacterial cells [11], representing extensive microbial richness, which potentially represents as many as 1012 distinct species [15], [16]. These microorganisms drive the major element cycles on the planet, and are also master chemists capable of producing in excess of 109 distinct small (<1 kDa) bioactive molecules [17]. The natural roles of many bioactive small molecules are not elucidated [18], yet the untapped potential of these molecules as novel anti-microbial compounds is beyond a doubt enormous.
Assessing the potential of environmental microbial community members to produce bioactive compounds, and hence potential novel anti-microbial therapeutics, is by no means trivial. Cultivation recalcitrance and the enigmatic notion of “silent” or cryptic biosynthetic gene clusters (BGCs), complicates the holistic study of bioactive secondary metabolites produced by microbiomes in situ. Fortunately, recent advances in ‘-omics’ methodology have enabled reliable, high-throughput and cultivation-independent approaches to study the secondary metabolism of microorganisms in their environment. Some of the largest classes of bioactive secondary metabolites are the polyketides (PKs) and the nonribosomal peptides (NRPs). These compound classes have a wide range of biological activities and pharmacological properties; for example, the NRP actinomycin is a clinical anti-cancer drug and the PK erythromycin is a widely used antibiotic. The modular and iterative nature of the enzymatic biosynthesis of PKs and NRPs involves modules of highly conserved domains responsible for the incorporation of coenzyme A (CoA)- or amino acid-based building blocks in growing chains of compounds. The minimal module for a nonribosomal peptide synthetase (NRPS) consists of an adenylation domain (AD), selecting the respective amino acid, a condensation (C) domain, catalyzing the peptide bond formation, and finally a peptidyl carrier domain, which is carrying the growing chain. A polyketide synthase (PKS) has a very similar structure, in that a minimal module contains an acyltransferase (AT), a keto-synthase (KS) and an acyl carrier protein, however often additional domains such as a keto-reductase can be found. This high degree of conservation has facilitated the design of degenerate primers for high-throughput amplicon sequencing and subsequent clustering into operational biosynthetic units (OBUs), allowing targeted analyses of the biosynthesis potential for these two classes of compounds (Fig. 1) [19], [20]. The primers most widely used to target conserved domains in BGCs encoding PKSs, were originally designed from conserved regions in 20 known modular PKSs and were used to study the geldanamycin BGC in Streptomyces hygroscopicus NRRL 3602 [21]. Similarly, the most widely used NRPS primers were originally designed to be specific for actinomycetes based on six characterized NRPS gene clusters [22]. Additional primers have subsequently been made increasingly degenerate to target a broader range of PKS BGCs from taxonomically different microorganisms [23]. Initially, heterologous expression of such soil-derived PCR amplicons provided early evidence that the hidden potential of the unculturable majority could be tapped, and that it likely represented a trove of structural novelty [24]. Subsequently, within the past decade, targeted short- and long-read amplicon sequencing, as well as untargeted shotgun metagenomics have generated a multitude of sequence data describing the biosynthesis potential of some of nature’s prevalent microbiomes.
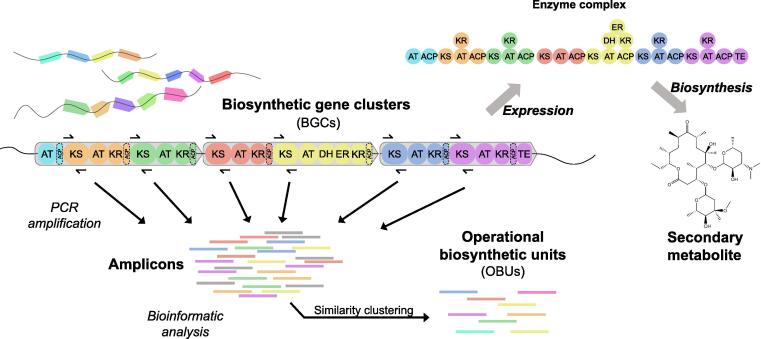
Fig. 1.
Working principle of biosynthetic gene clusters and their targeted analysis by amplicon sequencing. The genetic organization of an example PKS gene cluster is shown, along with the resulting enzyme complex and the final synthetized product. By targeted PCR amplification on environmental DNA, KS domains can be retrieved, sequenced and clustered into OBUs.
2.1. The biosynthesis potential of soil microbiomes
In addition to being readily accessible, soil is characterized by several features of significant importance for microbial secondary metabolite production. Often soil holds a significant amount of microbial biomass comprising approximately 109 bacteria, and 106 fungi per gram [25]. Especially, filamentous soil-dwelling actinomycetes exhibit a very high biosynthesis potential and account for the production of more than two thirds of known antibiotics [26]. Soil microbiomes have therefore rightly served as a starting point surveying the genetic potential for secondary metabolite production. More than 2000 soil samples have been queried using the PKS/NRPS amplicon approach described above [19], [27], [28], [29], [30], [31], [32], [33], and, perhaps not surprisingly, the majority of sequences, i.e. 80–99%, cannot be recovered in available databases. DNA extracted from around 0.25 g of soil is estimated to represent a richness of KS and AD domains in the range of 103-104 OBUs [19], [29], [32]. Thus, natural soil microbiomes hold an immense degree of biosynthetic diversity and consequently, structurally novel chemistry. While it is difficult to ascertain the spatial distribution of microorganisms in soil [34], the microscale heterogeneity is likely in part the reason why exploratory studies have reported a large inter-sample variation, even within the same collection site. Despite this variation, geographical distance, or dispersal limitation, and biome, or ‘habitat’, seem to be the most important drivers of biosynthetic diversity in soil [19], [29], [30], [32], [35]. Hence, widely interspersed terrestrial sampling sites should increase chances of unearthing the widest possible selection of metabolites in our pursuit of novel anti-microbial agents. However, in contrast to this line of thinking, the genetic capacity to synthesize specific biomedically relevant natural product families is widespread. For instance, the conserved domains originating from genes encoding the antibiotics erythromycin and teicoplanin, have been recovered from a single soil sample collected in the US, despite the fact that the compounds were originally isolated from microorganisms collected in the Philippines and India, respectively [29].
While the underlying physicochemical parameters in large parts shape the biomes and the small spatial scale heterogeneity, no single environmental driver of secondary metabolite richness or diversity has to date been identified using PKS/NRPS-targeted short-read amplicon sequencing. Weak correlations between the richness of NRPS and PKS sequences and the concentrations of potassium and calcium, and soil pH, have however been observed [19]. Other studies utilizing long-read sequencing methodology, have reported that biome may in some cases be a stronger driver than geographical distance. For instance, PKS fragments cloned from microbial community DNA extracted from soil and street sediment samples produced putative cosmopolitan KS and AT domain sequences, which consistently occurred in particular habitats separated by hundreds of kilometers [36]. In polar desert soil NRPS and PKS cluster richness, as determined using a third generation long-read sequencing approach, was negatively correlated with environmental parameters such as moisture, carbon, and nitrogen content. Hence, NRPS and PKS biochemistry may be an important physiological trait for survival in arid, nutrient-starved soils [37]. Whether the physicochemical drivers are habitat specific, remains to be determined.
While such targeted PCR-based approaches have inherent limitations, genome-resolved metagenomic approaches have corroborated the notion that the untapped secondary metabolite biosynthesis potential of soil microbiomes is immense. Crits-Christoph et al. reported that of 240 NRPS and PKS BGCs recovered from 376 MAGs from 120 grassland soil microbiomes, 220 did not share more than 50% of the genes of previously described clusters, and little sequence identity was observed at the amino acid level when compared to the ‘Minimum Information About a Biosynthetic Gene’ (MIBiG) [38] repository [39]. Furthermore, more than 900 additional BGCs were identified using the ‘Antibiotics and Secondary Metabolite Analysis Shell (AntiSMASH) [40], the majority of which encoded putative bioactive peptides, terpenes, and metabolites of unknown function. Similarly, genome-resolved metagenomics has revealed and compared thousands of diverse and novel BGCs from various grassland and forest soil microbiomes, underlining that habitat and soil depth are likely major drivers of overall BGC richness [41].
2.2. The biosynthesis potential of marine microbiomes
More than 70% of Earth’s surface is covered by oceans and marine environments represent the largest biosphere on the planet. Here, a plethora of niches distinct from any terrestrial environment exists. As a consequence, equally diverse microbiomes have evolved, and this diversity manifests itself in the compounds produced [42], [43] and comparisons of chemo-informatic data have shown that over 70% of molecular scaffolds in the Dictionary of Marine Natural Products (DMNP, 2007.6) are unique to marine organisms [44]. Nonetheless, marine microbiomes have not been scrutinized to the same extent as their terrestrial counterparts in the pursuit of novel secondary metabolites. As for soil microbiomes, most bioprospecting efforts in the marine environment have focused on lineages with culturable representatives, such as proteobacteria [43] and actinobacteria [45], resulting in the discovery of multiple bioactive compounds, of which many are antibiotics [46], [47], [48], [49], [50]. A few targeted metagenomic approaches have been used to assess the biosynthesis potential of the unculturable fraction of marine microbiomes, and especially marine sediments seem to hold a significant biosynthesis potential [20], [51], [52]. Coastal sediment microbiomes harbor twice as many KS and AD OBUs (97% sequence similarity cut-off) as compared to the coastal seawater above it, and in addition, 97.4–99.7% of OBUs from both seawater and sediments cannot be recovered from available databases [20]. Furthermore, seawater and sediment from the same location represent significantly different microbiomes in terms of their genetic capacity to synthesize PKs and NRPs indicating that habitat is a key driver of the diversity of the biosynthesis potential of marine microbiomes as well. Untargeted genome-resolved metagenomic analyses of a combined collection of ca. 35,000 MAGs, single amplified genomes (SAGs), and cultivated reference genomes from more than 1000 samples collected across the oceans, have in addition shown a significant differential biogeographic structuring of the oceanic biosynthetic potential [53]. In the oceans, tropical and epipelagic waters are enriched in terpenes as opposed to the less noticeable polar and deep waters, which conversely seem to harbor a more pronounced genetic potential for the biosynthesis of NRPSs and PKSs. Yet, as for soil microbiomes, the underlying drivers of the differentiation between habitats remain to be resolved.
2.3. The biosynthesis potential of host-associated microbiomes
While the evolutionary and ecological forces selecting for an elaborate repertoire of bioactive metabolites are varied [39], the microbiomes associated with eukaryotic hosts are likely hotspots for biosynthetically talented bacteria. In terrestrial systems, the plant-associated microbiomes of the phyllo- and rhizospheres represent rich repositories for novel BGCs [54], [55]. However, even within identical plant cultivars, the biosynthetic gene composition is significantly different between individuals, and is additionally affected by plant health, growth stage, and geographical distance [56], [57]. This suggests that plants engage in host-microbe interactions with microorganisms producing distinct collections of diverse bioactive compounds. Similarly, in marine systems, microbiomes associated with invertebrates, e.g. ascidians and sponges, have been widely studied due to their production of exceptionally diverse secondary metabolites [58], [59], [60], [61], [62], [63]. An initial cultivation-independent approach seeking to evaluate the natural product biosynthesis potential of marine sponges showed that of 150 distinct KS OBUs, 127 formed an independent clade that was present in the majority of a diverse collection of marine sponges, suggesting that sponge microbiomes produce ubiquitous yet unexplored polyketides [64]. In contrast to this finding, the exceptionally prolific secondary metabolite-producing microbiomes of the lithistid sponge Theonella swinhoei produce largely non-overlapping metabolite profiles, resulting in distinct host chemotypes [58], [65]. Global genome-resolved metagenomic data has revealed that microbiomes associated with more complex eukaryotic host animals including arthropods, insects, and humans all harbor a broad assortment of BGCs encoding the most prevalent classes of secondary metabolites, i.e. PKs, NRPs, ribosomally synthesized and post-translationally modified peptides (RiPPs), and terpenoids [12], [66], yet in-depth comparisons of the richness and diversity of BGCs across host-associated microbiomes are currently lacking.
2.4. Comparisons across biomes and methods
Individual studies evaluating the natural product biosynthesis potential of microbiomes from divergent biomes are limited [12], [20], [67], and our extrapolation and presentation of the current state of the field in the subsections above compile data retrieved from multiple distinct environments using different methodologies, each with inherent limitations. Studies relying on amplicon sequencing targeting conserved domains within PKS and NRPS BGCs, which is the most widely applied high-throughput approach, suffer from the fact that the amplicons only cover parts of the domains of interest. Since PKSs and NRPSs consist of multiple domains, each amplicon in itself does not easily allow for the inference of a specific natural product, nor the taxonomy of the organism it originated from. Furthermore, there are differences in the choice of target domains, sequencing technology, and primer pairs across studies. An in silico PCR analysis using the most common primer pairs on: 1) an existing collection of MAGs from a soil microbiome [39], [41], and 2) all KS and AD domains from the AntiSMASH database [68], shows that some primers fail to amplify the majority, if not all target sequences in these two extensive datasets (Table 1). For KS domains, the highest number of observed hits are generated using the MDPQQRf/HGTGTr primer pair designed by Tae et al. (2006) [61]. However, the in silico generated amplicons vary in length, indicating a potential risk for unspecific amplification. The degKS2F/degKS2R primer pair initially designed by Schirmer et al. (2005) [23], [29], which has been used to generate the majority of KS sequencing reads from soil and marine systems [20], [29], [30], [32], is in contrast quite consistent in amplicon length and generates the second highest number of hits (Table 1). Similarly, the Schirmer et al. (2005) NRPS primers targeting AD domains, exhibit superior performance compared to other primer pairs in silico. However, due to the longer amplicons generated with this set of primers, it has been substituted with the A3F/A7R AD-targeting primers [22] in most high-throughput amplicon sequencing studies as these are based on short-read sequencing technologies [20], [29], [30], [32]. Interestingly, the more recently developed primer pairs designed to target divergent NRPS and PKS clusters from soil microbiomes across latitudes and climate zones [69], [70], perform poorly in silico, both against soil derived MAGs known to carry a broad repertoire of PKS and NRPS BGCs [39], [41], and the collective sequence data in the antiSMASH database. In extension of the difficulty of developing suitable primers, amplicon sequencing approaches are confined to the analysis of a select subset of compound classes, i.e. only NRPS and PKS clusters currently. Furthermore, comparisons with un-targeted metagenomic approaches, which in some cases can generate complete or near-complete BGCs [71], are hampered by the relatively limited information stored in the small sequences generated by amplicon sequencing. This issue can to some extent be resolved using bioinformatic tools such as dom2BGC [72], or the ‘Environmental Surveyor of Natural Product Diversity’ (eSNaPD), which compares the sequenced amplicons to a curated reference dataset of 450 unique gene clusters, resulting in around 10,000 signature domains [73]. However, only a small percentage of detected OBUs can be assigned to corresponding domains in the database and the results are highly dependent on the somewhat arbitrary set similarity cut-off. While amplicon sequencing currently remains the best tool for deep-sequencing of specific biosynthetic domains, un-targeted metagenomic approaches are gradually replacing this approach as the field progresses, hopefully fostering a more standardized approach across studies and microbiomes.
Table 1.
List of degenerate primers used in biosynthetic amplicon studies targeting various conserved domains in NRPS and PKS BGCs, and the number of in silico amplicons obtained with the listed primers. The publicly available program ‘in silico PCR’ (https://github.com/egonozer/in_silico_pcr) was used to generate amplicons form a set of 1334 metagenome-assembled genomes of a soil microbiome [41] and all KS and AD domains on the AntiSMASH database (stand: august 2019) allowing for one mismatch and one insertion per primer sequence.
| Name | Target domain | Theoretical amplicon length | Sequence | Reference | Hits in MAGs | Hits in AntiSMASH database | % detected of AntiSMASH domains | In silico amplicon length |
|---|---|---|---|---|---|---|---|---|
| degNRPS-1F | AD | 900–1100 | AARDSNGGNGSNGSNTAYBNCC | Schirmer 2005 [23] | 891 | 15,255 | 37 | 1010 ± 43 |
| degNRPS-4R | CKRWANCCNCKNANYTTNAYYTG | |||||||
| MTF2 | AD | 1000 | GCNGGYGGYGCNTAYGTNCC | Neilan et al. 1999 [99] | 342 | 7039 | 17 | 1004 ± 51 |
| MTR | CCNCGDATYTTNACYTG | |||||||
| A3F | AD | 700 | GCSTACSYSATSTACACSTCSGG | Ayuso sacido and Genilloud 2005 [22] | 178 | 8060 | 20 | 709 ± 25 |
| A7R | SASGTCVCCSGTSCGGTAS | |||||||
| F | AD | 480 | CGCGCGCATGTACTGGACNGGNGAYYT | Amos 2015 [69] | 0 | 0 | 0 | NA |
| R | GGAGTGGCCGCCCARNYBRAARAA | |||||||
| DnDmF | C | 480 | ATGCATCACATTRTNYYNGA | Woodhouse 2013 [60] | 16 | NA | NA | 481 ± 1.5 |
| DCCR | GTGTTNACRAARAANCCDAT | |||||||
| MDPQQRf | KS | 670 | RTRGAYCCNCAGCANCG | Tae 2006 [61] | 1517 | 7833 | 40 | 674 ± 179 |
| HGTGTr | VGTNCCNGTGCCRTG | |||||||
| degKS2F | KS | 700 | GCNATGGAYCCNCARCARMGNVT | Schirmer 2005 [23] | 93 | 4330 | 22 | 680 ± 10 |
| degKS2R | GTNCCNGTNCCRTGNSCYTCNAC | |||||||
| KSDPQQF | KS | 700 | MGNGARGCNNWNSMNATGGAYCCNCARCANMG | Piel 2002 [100] | 69 | 3513 | 18 | 705 ± 7 |
| KSHGTGR | GGRTCNCCNARNSWNGTNCCNGTNCCRTG | |||||||
| KSLF | KS | 700 | CCSCAGSAGCGCSTSYTSCTSGA | Courtois 2003 [101] | 32 | 2419 | 12 | 671 ± 11 |
| KSLR | GTSCCSGTSCCGTGSGYSTCSA | |||||||
| DKF | KS | 700 | GTGCCGGTNCCRTGNGYYTC | Moffitt 2003 [102] | 19 | 1125 | 6 | 683 ± 51 |
| DKR | GCGATGGAYCCNCARMG | |||||||
| MAK1 | KS | 320 | GACACSGCSTGYTCBTCGTCG | Savic and Vailjevic 2006 [103] | 0 | 656 | 3 | 317 ± 0 |
| MAK3 | CCGTTSGACGCRCCGTCCTGGTTSCA | |||||||
| P1 | KS | TSGAYCCSCAGCARCG | Zhao 2012 [56] | 78 | 25 | 0.1 | 591 ± 427 | |
| P2 | GTSGAYACNGCSTGYTC | |||||||
| K1F | KS - methyl-malonyl-CoA transferase | 1200–1400 | TSAAGTCSAACATCGGBCA | Ayuso-Sacido and Genilloud 2005 [22] | 9 | NA | NA | 1304 ± 47 |
| M6R | CGCAGGTTSCSGTACCAGTA | |||||||
| PKS_firmi_F | ACP | 340 | GCNGGNCAYWSNYTNGGNGARTAYA | Aleti 2017 [57] | 10 | NA | NA | 340 ± 4 |
| PKS_firmi_R | CATRWANCKNSWRTGRAANGCNCC | |||||||
| KSα-F | KS alpha (type II pks) | 613 | TSGCSTGCTTCGAYGCSATC | Metsä-Ketelä 1999 [104] | 15 | 28 | 0.1 | 613 ± 0.3 |
| KSα-R | TGGAANCCGCCGAABCCGCT | |||||||
| F | KS alpha (type II pks) | 350 | GGCAACGCCTACCACATGCANGGNYT | Amos 2015 [69] | 0 | 0 | 0 | NA |
| R | GGTCCGCGGGACGTARTCNARRTC |
3. Tapping into microbial biosynthetic diversity
Considering the diversity of microbial biosynthesis genes in nature, even the most commonly sampled microbiomes hold promise of providing novel classes of natural products. As classical bioprospecting has only been able to capture a miniscule amount of the true biosynthetic potential, alternative measures are needed to procure truly novel classes of antibiotics.
3.1. Tying taxonomy to biosynthesis and focusing cultivation
In recent years, several new cultivation and isolation techniques have been developed, some implemented with the aim of discovering novel bioactive compounds. By applying the principle of in situ cultivation, the novel antibiotic teixobactin was identified through the screening of extracts from over 10,000 iChip-derived soil isolates [74]. The promising therapeutic candidate is the first antibiotic to bind lipid II, validating the potential for obtaining structural novelty from yet-to-be cultured microorganisms. Similarly, droplet cultivation techniques eliminate competition for space and nutrients by microencapsulation, allowing otherwise cultivation recalcitrant microorganisms to grow under controlled conditions [10], [75], [76]. While increasing the culturable fraction of environmental microorganisms, such approaches are untargeted and necessitate extensive downstream screening of isolates. Focusing on specific uncultured taxa exhibiting elaborate biosynthetic repertoires could increase the efficiency of such cultivation-based approaches substantially. Combining droplet cultivation with fluorescence in situ hybridization in living cells (liveFISH [77]) introduces the possibility for high-throughput and targeted isolation of single microbial cells belonging to promising taxonomic groups. A prerequisite is, however, that the interconnection between taxonomy and biosynthesis potential can be determined. The generation of complete, or near-complete, MAGs from metagenomes clearly allows for this interconnection and coupling taxonomic affiliations. Using the ribosomal S3 protein-encoding gene in combination with antiSMASH outputs for individual MAGs, have demonstrated that the biosynthesis potential varies greatly across phyla in soil microbiomes, and that actinobacteria, but also the less well-studied chloroflexi phylum, likely harbors an extensive secondary metabolite repertoire [41]. In addition, candidate phyla without cultured representatives, e.g. rokubacteria, eisenbacteria, and dormibacteraeota, may be promising candidates for targeted cultivation efforts. The size and repetitive nature of many BGCs, and the extensive diversity of low-abundant microorganisms, does however represent significant challenges for MAG-based approaches. In contrast, the application of linear correlation analyses on targeted amplicon sequencing data allows for the connection between taxonomy (16S operational taxonomic units; OTUs) and biosynthesis potential (OBUs) of low-abundant taxa [20]. Such analyses have demonstrated that most marine bacterial species have no, or very few associated OBUs, while taxa not previously associated with secondary metabolism may in fact be promising targets for cultivation efforts. Interestingly, in both soil and marine microbiomes, targeted approaches have demonstrated that the majority of the uncharacterized biosynthesis potential is associated with low-abundant taxa [20], [27]. Yet again other approaches have been able to link secondary metabolite biosynthesis potential to taxonomy in microbiomes. Coupling mass-spectrometry data to 16S OTUs have established the connection between staurosporine and the Salinispora genus in ocean sediments [51], and single-cell sequencing approaches have facilitated the assignment of individual PKS and NRPS genes to sponge-associated members of the Poribacteria, the Chloroflexi, and the candidate genus Entotheonella [58], [78]. Lastly, opposed to indirect correlation of 16S amplicon data with functional gene amplicon (or chemistry data), emulsion paired isolation and concatenated PCR (epicPCR) fuses phylogenetic and functional amplicons directly in vitro [79]. This technique, which makes use of the capture of single cells in emulsified droplets, has been used to taxonomically identify sulfate reducing bacteria and bacteria carrying antibiotic resistance genes [80], [81], and may in a similar manner be a promising tool in the determination of which taxa to bring into culture.
3.2. Culture-independent acquisition of novel secondary metabolites
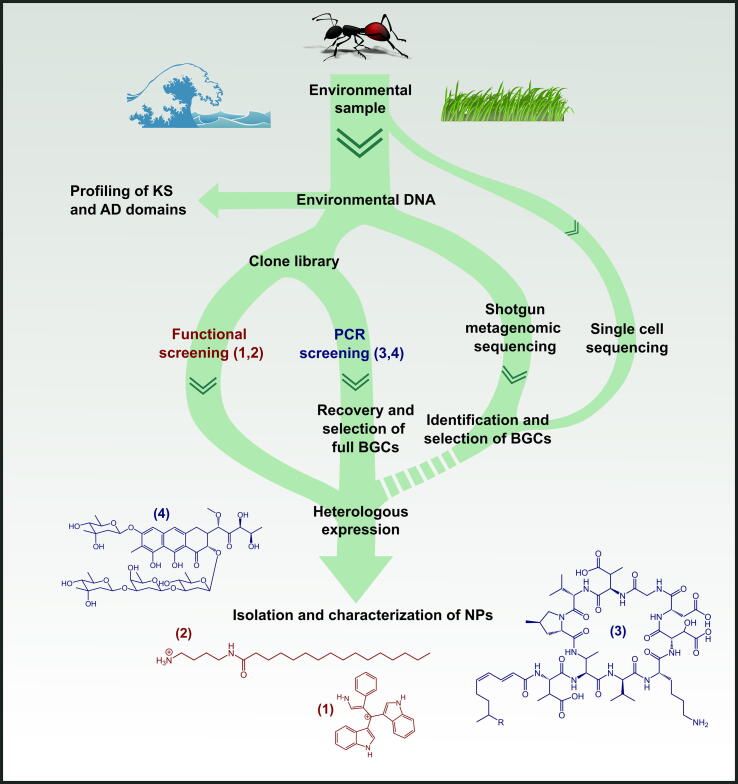
Culture-independent discovery of secondary metabolites can be facilitated by the use of metagenomic clone libraries in combination with heterologous expression [82]. In such approaches, eDNA is captured in e.g. a cosmid library and maintained in a host organisms [83]. The library can subsequently be screened either for functionality such as coloration, bioactivity or compound detection by high performance liquid chromatography - mass spectrometry (HPLC-MS) [84]. Two examples of novel antibiotics discovered by such approaches include the small molecule turbomycin (1) (Fig. 2), which was found by investigating melanin-like colored colonies of a large metagenomic soil eDNA clone library [85], and the compound palmitoylputrescine (2) (Fig. 2), which was found by bioactivity screening of a bromeliad tank water eDNA library [86].
Fig. 2.
Flow chart for the culture independent discovery of natural products from environmental microbiomes. Starting with the environmental sample and the extracted microbial DNA, KS and AD domains can be profiled to gain insights into the genetic biosynthesis potential. A combination of different pathways can lead then to the isolation and characterization of novel natural products. Dashed arrows indicate methods not experimentally established yet. At the bottom four examples of bioactive natural products are shown and color coded according to the methodology of isolation.
Alternatively metagenomic clone libraries can be screened for the presence of conserved DNA sequences such as AD and KS domains by PCR [87]. The latter strategy recently led to the discovery of a new class of calcium-dependent antibiotics, the malacidins (3) (Fig. 2) [28]. These novel secondary metabolites were identified in AD amplicon sequenced soil samples based on homology and phylogenetic analysis of conserved AD domains of calcium-dependent NRP antibiotics. Assisted by the eSNaPD tool [73], a soil community especially rich in AD domains encoding for potentially novel calcium-dependent NRPs was selected. Via cloning of eDNA into a cosmid library, a novel BGC was identified, assembled, cloned and expressed in the heterologous host Streptomyces albus, eventually resulting in the discovery of the calcium-dependent malacidin antibiotics [28]. A similar workflow has been employed in the search for analogs of the anti-cancer compound mithramycin in soil [88]. Metagenomic library screening with degenerate primers targeting KSα genes resulted in the identification of a cluster with structural similarity, but sufficient sequence divergence to represent a novel analog. The BGC was cloned into S. albus for heterologous expression, and after bioassay-guided fractionation and pathway engineering, the novel compound metathramycin (4) (Fig. 2) was structurally elucidated and characterized as a potent anti-cancer compound. Hence, such examples demonstrate the possibility of procuring novel secondary metabolites from eDNA and, given the large untapped biosynthetic diversity, promise to be only the tip of the iceberg.
However, multiple technical challenges currently limit the effectiveness of large-scale culture independent drug discovery studies. Heterologous expression in a suitable host is not always straightforward as problems such as codon incompatibility [89], a lack of necessary regulatory elements [90] and toxicity of the encoded compound towards the host can arise [91]. Several of these challenges are being addressed both in heterologous expression and in induction efforts of silent BGCs from, and in, culturable bacteria. Development of methods such as transformation-associated recombination cloning of BGCs [92], promoter-swapping [93], and experiences from the induction by e.g. co-cultivation [94], [95] could aid in overcoming challenges associated with heterologous expression of metagenome-retrieved BGCs. Another challenge is the size limitation of DNA inserts in cosmid libraries, which means that BGCs larger than 40 kb will be distributed over two or more clones. This makes it more time consuming to screen, find and assemble BGCs [28]. Lastly, BGCs only constitute a small proportion of bacterial genomic DNA [82], [96], which necessitates sufficiently deep clone libraries in combination with extensive screening efforts to find them. Therefore, methods aiming at enriching the microbial community for biosynthetic talented bacteria before generating a cosmid or sequencing library may be a promising way to improve the discovery of novel BGCs and their corresponding secondary metabolites. This strategy was applied to extract an enriched fraction of the tunicate microbiome actively expressing bioactive compounds at the time of sampling. Through fluorescent labelling of the carrier proteins involved in NRP and PK biosynthesis and subsequent fluorescence activated cell sorting (FACS), it was possible to enrich the BGC content from 0.77 to 1.17 BGC per Mbp [97].
To what extent the need for clone libraries remains in the future is unknown, however promising full length BGCs identified by shotgun sequencing directly could potentially be extracted from eDNA using targeted PCR and subsequent cloning into a suitable host. This approach has been successful for silent BGCs of cultured representatives; for example, a new phenazine compound from Serratia fonticola has been successfully expressed in E. coli [98].
Lastly, single cell sequencing may prove a promising avenue for the identification of candidate secondary metabolites in the future. To date it has mainly been used to either identify the producer of a known compound, or the BGC responsible for its biosynthesis. One example of this approach is the identification of host-associated onnamide A producers [58], where members of a complex sponge-associated microbiome were isolated and the production of the compound could be tied to members of the Entotheonella genus.
4. Summary and outlook
Microorganisms have been our most generous source of natural products, and they have provided us with the majority of anti-microbial agents applied in our continuous battle against pathogenic bacteria. Regrettably, as antibiotic resistance is on the rise, the supply of novel antibiotics from established sources is running dry, and we must again turn to nature to mitigate this development.
The microbiomes of Earth harbor immense taxonomic diversity and the development within the meta-omics field has enabled the exposure of their genetic potential to produce novel secondary metabolites. Through the past decade, targeted and untargeted metagenomic analyses of soil, marine, and host-associated microbiomes have identified these as rich reservoirs of novel bioactive compounds, including PKs and NRPs. However, a lack of standardization in methodology has complicated comparisons across biomes and current approaches have not yet enabled us to identify key drivers of biosynthetic diversity. Nor are we currently able to predict, which microbiomes are the most promising to mine for truly novel anti-microbial agents.
To exploit the biosynthesis potential of natural microbiomes, both cultivation-dependent and cultivation-independent avenues need to be explored. Through the development of targeted cultivation methodology, we may be able to isolate representatives of the rare taxa, which seem to be proficient producers of uncharacterized bioactive compounds in nature. However, it is improbable that we will ever be able to cultivate the full phylogenetic range of microbes found in the environment. Instead, cultivation-independent function based and sequence based metagenomics have been successful in the identification and isolation of novel bioactive compounds. With improved sequencing technologies, more sensitive and precise bioinformatic pipelines, and a comprehensive understanding of the biosynthesis, we will likely be able to extract BGC sequences directly from environmental metagenomes and express them heterologously to find true chemical novelty. Additionally, single cell isolation and sequencing hold a great promise of identifying rich producers, for more focused efforts in cultivation or sequencing. Eventually, we will need to make use of the full potential of environmental microbiomes, using both cultivation and cultivation-independent approaches to procure the novel anti-microbial agents needed to avert the antibiotic resistance crisis.
Funding
This study was supported by The Independent Research Fund Denmark (grant DFF – 8048-00035B) and from the Danish National Research Foundation (grant DNRF137).
CRediT authorship contribution statement
Aileen Ute Geers: Conceptualization, Writing – original draft, Visualization. Yannick Buijs: Writing – original draft, Visualization. Mikael Lenz Strube: Writing – review & editing. Lone Gram: Writing – review & editing. Mikkel Bentzon-Tilia: Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Agnew E, Dolecek C, Hasan R, Lahra M, Merk H, Perovic O, et al. Global antimicrobial resistance and use surveillance system (GLASS) report; 2021.
- 2.CDC. Antibiotic Resistance Threats in the United States. 2019. https://doi.org/10.15620/cdc:82532.
- 3.Davies J. Where have all the antibiotics gone? Can. J. Infect. Dis. Med. Microbiol., vol. 17, Hindawi Limited; 2006, p. 287–90. https://doi.org/10.1155/2006/707296. [DOI] [PMC free article] [PubMed]
- 4.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12(5):371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 5.Waksman S.A., Woodruff H.B. The soil as a source of microorganisms antagonistic to disease-producing bacteria. J Bacteriol. 1940;40(4):581–600. doi: 10.1128/jb.40.4.581-600.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waksman S.A., Schatz A., Reynolds D.M. Production of antibiotic substances by actinomycetes. Ann N Y Acad Sci. 2010;1213:112–124. doi: 10.1111/j.1749-6632.2010.05861.x. [DOI] [PubMed] [Google Scholar]
- 7.Livermore D.M., Blaser M., Carrs O., Cassell G., Fishman N., Guidos R., et al. Discovery research: the scientific challenge of finding new antibiotics. J Antimicrob Chemother. 2011;66(9):1941–1944. doi: 10.1093/jac/dkr262. [DOI] [PubMed] [Google Scholar]
- 8.Silver L.L. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24(1):71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berdy B., Spoering A.L., Ling L.L., Epstein S.S. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat Protoc. 2017;12(10):2232–2242. doi: 10.1038/nprot.2017.074. [DOI] [PubMed] [Google Scholar]
- 10.Mahler L, Niehs SP, Martin K, Weber T, Scherlach K, Hertweck C, et al. Highly parallelized droplet cultivation and prioritization of antibiotic producers from natural microbial communities. Elife 2021;10:e64774. https://doi.org/10.7554/eLife.64774. [DOI] [PMC free article] [PubMed]
- 11.Whitman W.B., Coleman D.C., Wiebe W.J. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95(12):6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nayfach S., Roux S., Seshadri R., Udwary D., Varghese N., Schulz F., et al. A genomic catalog of Earth’s microbiomes. Nat Biotechnol. 2021;39(4):499–509. doi: 10.1038/s41587-020-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao D., Okada B.K., Wu Y., Xu F., Seyedsayamdost M.R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr Opin Microbiol. 2018;45:156–163. doi: 10.1016/j.mib.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abrudan M.I., Smakman F., Grimbergen A.J., Westhoff S., Miller E.L., van Wezel G.P., et al. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci U S A. 2015;112(35):11054–11059. doi: 10.1073/pnas.1504076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennon J.T., Locey K.J., DeLong E.F., McFall-Ngai M.J. The underestimation of global microbial diversity. MBio. 2016;7(5) doi: 10.1128/mBio.01298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locey K.J., Lennon J.T. Scaling laws predict global microbial diversity. Proc Natl Acad Sci U S A. 2016;113(21):5970–5975. doi: 10.1073/pnas.1521291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies J. Small molecules: the lexicon of biodiversity. J Biotechnol. 2007;129(1):3–5. doi: 10.1016/j.jbiotec.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Traxler M.F., Kolter R. Natural products in soil microbe interactions and evolution. Nat Prod Rep. 2015;32(7):956–970. doi: 10.1039/C5NP00013K. [DOI] [PubMed] [Google Scholar]
- 19.Charlop-Powers Z., Owen J.G., Reddy B.V.B., Ternei M.A., Brady S.F. Chemical-biogeographic survey of secondary metabolism in soil. Proc Natl Acad Sci. 2014;111(10):3757–3762. doi: 10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bech PK, Lysdal KL, Gram L, Bentzon-Tilia M, Strube ML. Marine sediments hold an untapped potential for novel taxonomic and bioactive bacterial diversity. MSystems 2020;5. https://doi.org/10.1128/mSystems.00782-20. [DOI] [PMC free article] [PubMed]
- 21.Rascher A, Hu Z, Viswanathan N, Schirmer A, Reid R, Nierman WC, et al. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL 3602. FEMS Microbiol Lett 2003;218:223–30. https://doi.org/10.1016/S0378-1097(02)01148-5. [DOI] [PubMed]
- 22.Ayuso-Sacido A., Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol. 2005;49(1):10–24. doi: 10.1007/s00248-004-0249-6. [DOI] [PubMed] [Google Scholar]
- 23.Schirmer A., Gadkari R., Reeves C.D., Ibrahim F., DeLong E.F., Hutchinson C.R. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl Environ Microbiol. 2005;71(8):4840–4849. doi: 10.1128/AEM.71.8.4840-4849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seow K.T., Meurer G., Gerlitz M., Wendt-Pienkowski E., Hutchinson C.R., Davies J. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J Bacteriol. 1997;179(23):7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young I.M., Crawford J.W. Interactions and self-organization in the soil-microbe complex. Science. 2004;304(5677):1634–1637. doi: 10.1126/science:1097394. [DOI] [PubMed] [Google Scholar]
- 26.Barka E.A., Vatsa P., Sanchez L., Gaveau-Vaillant N., Jacquard C., Klenk H.-P., et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev. 2016;80(1):1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libis V., Antonovsky N., Zhang M., Shang Z., Montiel D., Maniko J., et al. Uncovering the biosynthetic potential of rare metagenomic DNA using co-occurrence network analysis of targeted sequences. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-11658-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hover B.M., Kim S.-H., Katz M., Charlop-Powers Z., Owen J.G., Ternei M.A., et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat Microbiol. 2018;3(4):415–422. doi: 10.1038/s41564-018-0110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charlop-Powers Z., Pregitzer C.C., Lemetre C., Ternei M.A., Maniko J., Hover B.M., et al. Urban park soil microbiomes are a rich reservoir of natural product biosynthetic diversity. Proc Natl Acad Sci. 2016;113(51):14811–14816. doi: 10.1073/pnas.1615581113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charlop-Powers Z, Owen JG, Reddy BVB, Ternei M, Guimaraes DO, De Frias UA, et al. Global biogeographic sampling of bacterial secondary metabolism. Elife 2015;4:e05048. https://doi.org/10.7554/eLife.05048. [DOI] [PMC free article] [PubMed]
- 31.Reddy B.V.B., Kallifidas D., Kim J.H., Charlop-Powers Z., Feng Z., Brady S.F. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl Environ Microbiol. 2012;78(10):3744–3752. doi: 10.1128/AEM.00102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemetre C., Maniko J., Charlop-Powers Z., Sparrow B., Lowe A.J., Brady S.F. Bacterial natural product biosynthetic domain composition in soil correlates with changes in latitude on a continent-wide scale. Proc Natl Acad Sci. 2017;114(44):11615–11620. doi: 10.1073/pnas.1710262114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen J.G., Reddy B.V.B., Ternei M.A., Charlop-Powers Z., Calle P.Y., Kim J.H., et al. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc Natl Acad Sci U S A. 2013;110(29):11797–11802. doi: 10.1073/pnas.1222159110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Donnell A.G., Young I.M., Rushton S.P., Shirley M.D., Crawford J.W. Visualization, modelling and prediction in soil microbiology. Nat Rev Microbiol. 2007;5(9):689–699. doi: 10.1038/nrmicro1714. [DOI] [PubMed] [Google Scholar]
- 35.Rego A., Sousa A.G.G., Santos J.P., Pascoal F., Canário J., Leão P.N., et al. Diversity of bacterial biosynthetic genes in maritime antarctica. Microorganisms. 2020;8(2):279. doi: 10.3390/microorganisms8020279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill P., Piel J., Aris-Brosou S., Krištůfek V., Boddy C.N., Dijkhuizen L. Habitat-specific type I polyketide synthases in soils and street sediments. J Ind Microbiol Biotechnol. 2014;41:75–85. doi: 10.1007/s10295-013-1362-7. [DOI] [PubMed] [Google Scholar]
- 37.Benaud N, Zhang E, van Dorst J, Brown M V, Kalaitzis JA, Neilan BA, et al. Harnessing long-read amplicon sequencing to uncover NRPS and Type I PKS gene sequence diversity in polar desert soils. FEMS Microbiol Ecol 2019;95. https://doi.org/10.1093/femsec/fiz031. [DOI] [PubMed]
- 38.Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, Van Der Hooft JJJ, et al. MIBiG 2.0: A repository for biosynthetic gene clusters of known function. Nucleic Acids Res 2020;48:D454–8. https://doi.org/10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed]
- 39.Crits-Christoph A., Diamond S., Butterfield C.N., Thomas B.C., Banfield J.F. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature. 2018;558(7710):440–444. doi: 10.1038/s41586-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 40.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, et al. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 2019;47:W81–7. https://doi.org/10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed]
- 41.Sharrar A.M., Crits-Christoph A., Méheust R., Diamond S., Starr E.P., Banfield J.F., et al. Bacterial secondary metabolite biosynthetic potential in soil varies with phylum, depth, and vegetation type. MBio. 2020;11(3) doi: 10.1128/mBio.00416-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romano G., Costantini M., Sansone C., Lauritano C., Ruocco N., Ianora A. Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar Environ Res. 2017;128:58–69. doi: 10.1016/j.marenvres.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Buijs Y., Bech P.K., Vazquez-Albacete D., Bentzon-Tilia M., Sonnenschein E.C., Gram L., et al. Marine Proteobacteria as a source of natural products: advances in molecular tools and strategies. Nat Prod Rep. 2019;36(9):1333–1350. doi: 10.1039/C9NP00020H. [DOI] [PubMed] [Google Scholar]
- 44.Kong D.-X., Jiang Y.-Y., Zhang H.-Y. Marine natural products as sources of novel scaffolds: achievement and concern. Drug Discov Today. 2010;15(21-22):884–886. doi: 10.1016/j.drudis.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Gontang E.A., Gaudêncio S.P., Fenical W., Jensen P.R. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl Environ Microbiol. 2010;76(8):2487–2499. doi: 10.1128/AEM.02852-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaysser L., Bernhardt P., Nam S.-J., Loesgen S., Ruby J.G., Skewes-Cox P., et al. Merochlorins A-D, cyclic meroterpenoid antibiotics biosynthesized in divergent pathways with vanadium-dependent chloroperoxidases. J Am Chem Soc. 2012;134(29):11988–11991. doi: 10.1021/ja305665f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haste N.M., Hughes C.C., Tran D.N., Fenical W., Jensen P.R., Nizet V., et al. Pharmacological properties of the marine natural product marinopyrrole A against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55(7):3305–3312. doi: 10.1128/AAC.01211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bentzon-Tilia M., Gram L. Bioprospecting. Springer; 2017. Biotechnological applications of the Roseobacter clade; pp. 137–166. 10.1007/978-3-319-47935-4_7. [Google Scholar]
- 49.Porsby C.H., Webber M.A., Nielsen K.F., Piddock L.J.V., Gram L. Resistance and tolerance to tropodithietic acid, an antimicrobial in aquaculture, is hard to select. Antimicrob Agents Chemother. 2011;55(4):1332–1337. doi: 10.1128/AAC.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dittmann K.K., Porsby C.H., Goncalves P., Mateiu R.V., Sonnenschein E.C., Bentzon‐Tilia M., et al. Tropodithietic acid induces oxidative stress response, cell envelope biogenesis and iron uptake in Vibrio vulnificus. Environ Microbiol Rep. 2019;11(4):581–588. doi: 10.1111/1758-2229.12771. [DOI] [PubMed] [Google Scholar]
- 51.Tuttle R.N., Demko A.M., Patin N.V., Kapono C.A., Donia M.S., Dorrestein P., et al. Detection of natural products and their producers in ocean sediments. Appl Environ Microbiol. 2019;85(8) doi: 10.1128/AEM.02830-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Y., Zhang L., Zhou Z., Yan X. Diversity of gene clusters for polyketide and nonribosomal peptide biosynthesis revealed by metagenomic analysis of the yellow sea sediment. Front Microbiol. 2018;9:295. doi: 10.3389/fmicb.2018.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paoli L, Ruscheweyh H-J, Forneris CC, Kautsar S, Clayssen Q, Salazar G, et al. Uncharted biosynthetic potential of the ocean microbiome. BioRxiv 2021:2021.03.24.436479.
- 54.Müller C.A., Oberauner-Wappis L., Peyman A., Amos G.C.A., Wellington E.M.H., Berg G., et al. Mining for nonribosomal peptide synthetase and polyketide synthase genes revealed a high level of diversity in the Sphagnum bog metagenome. Appl Environ Microbiol. 2015;81(15):5064–5072. doi: 10.1128/AEM.00631-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blair PM, Land ML, Piatek MJ, Jacobson DA, Lu T-YS, Doktycz MJ, et al. Exploration of the biosynthetic potential of the Populus microbiome. MSystems 2018;3. https://doi.org/10.1128/mSystems.00045-18. [DOI] [PMC free article] [PubMed]
- 56.Zhao B., Gao Z., Shao Y., Yan J., Hu Y., Yu J., et al. Diversity analysis of type I ketosynthase in rhizosphere soil of cucumber. J Basic Microbiol. 2012;52(2):224–231. doi: 10.1002/jobm.201000455. [DOI] [PubMed] [Google Scholar]
- 57.Aleti G., Nikolić B., Brader G., Pandey R.V., Antonielli L., Pfeiffer S., et al. Secondary metabolite genes encoded by potato rhizosphere microbiomes in the Andean highlands are diverse and vary with sampling site and vegetation stage. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-02314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson M.C., Mori T., Rückert C., Uria A.R., Helf M.J., Takada K., et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506(7486):58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 59.Rust M., Helfrich E.J.N., Freeman M.F., Nanudorn P., Field C.M., Rückert C., et al. A multiproducer microbiome generates chemical diversity in the marine sponge Mycale hentscheli. Proc Natl Acad Sci U S A. 2020;117(17):9508–9518. doi: 10.1073/pnas.1919245117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woodhouse J.N., Fan L.u., Brown M.V., Thomas T., Neilan B.A. Deep sequencing of non-ribosomal peptide synthetases and polyketide synthases from the microbiomes of Australian marine sponges. ISME J. 2013;7(9):1842–1851. doi: 10.1038/ismej.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim T.K., Fuerst J.A. Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: Culture-dependent and culture-independent approaches. Environ Microbiol. 2006;8(8):1460–1470. doi: 10.1111/j.1462-2920.2006.01040.x. [DOI] [PubMed] [Google Scholar]
- 62.Fusetani N., Matsunaga S. Bioactive sponge peptides. Chem Rev. 1993;93(5):1793–1806. doi: 10.1021/cr00021a007. [DOI] [Google Scholar]
- 63.Trindade-Silva A.E., Rua C.P.J., Andrade B.G.N., Vicente A.C.P., Silva G.G.Z., Berlinck R.G.S., et al. Polyketide synthase gene diversity within the microbiome of the sponge Arenosclera brasiliensis, endemic to the southern Atlantic Ocean. Appl Environ Microbiol. 2013;79(5):1598–1605. doi: 10.1128/AEM.03354-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fieseler L., Hentschel U., Grozdanov L., Schirmer A., Wen G., Platzer M., et al. Widespread occurrence and genomic context of unusually small polyketide synthase genes in microbial consortia associated with marine sponges. Appl Environ Microbiol. 2007;73(7):2144–2155. doi: 10.1128/AEM.02260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wegerski C.J., Hammond J., Tenney K., Matainaho T., Crews P. A serendipitous discovery of isomotuporin-containing sponge populations of Theonella swinhoei. J Nat Prod. 2007;70(1):89–94. doi: 10.1021/np060464w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 2014;158:1402–14. https://doi.org/10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed]
- 67.Medema M.H., Cimermancic P., Sali A., Takano E., Fischbach M.A., Ouzounis C.A. A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput Biol. 2014;10(12):e1004016. doi: 10.1371/journal.pcbi.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blin K, Pascal Andreu V, De Los Santos ELC, Del Carratore F, Lee SY, Medema MH, et al. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res 2019;47:D625–30. https://doi.org/10.1093/nar/gky1060. [DOI] [PMC free article] [PubMed]
- 69.Amos G.C.A., Borsetto C., Laskaris P., Krsek M., Berry A.E., Newsham K.K., et al. Designing and implementing an assay for the detection of rare and divergent NRPS and PKS clones in European. Antarctic and Cuban soils. PLoS One. 2015;10(9):e0138327. doi: 10.1371/journal.pone.0138327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borsetto C., Amos G.C.A., da Rocha U.N., Mitchell A.L., Finn R.D., Laidi R.F., et al. Microbial community drivers of PK/NRP gene diversity in selected global soils. Microbiome. 2019;7(1) doi: 10.1186/s40168-019-0692-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waschulin V., Borsetto C., James R., Newsham K.K., Donadio S., Corre C., et al. Biosynthetic potential of uncultured Antarctic soil bacteria revealed through long-read metagenomic sequencing. ISME J. 2021:1–11. doi: 10.1038/s41396-021-01052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tracanna V, Ossowicki A, Petrus MLC, Overduin S, Terlouw BR, Lund G, et al. Dissecting disease-suppressive rhizosphere microbiomes by functional amplicon sequencing and 10× metagenomics. MSystems 2021;6. https://doi.org/10.1128/mSystems.01116-20.. [DOI] [PMC free article] [PubMed]
- 73.Reddy B., Milshteyn A., Charlop-Powers Z., Brady S. eSNaPD: a versatile, web-based bioinformatics platform for surveying and mining natural product biosynthetic diversity from metagenomes. Chem Biol. 2014;21(8):1023–1033. doi: 10.1016/j.chembiol.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P., et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517(7535):455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaminski T.S., Scheler O., Garstecki P. Droplet microfluidics for microbiology: techniques, applications and challenges. Lab Chip. 2016;16(12):2168–2187. doi: 10.1039/C6LC00367B. [DOI] [PubMed] [Google Scholar]
- 76.Mahler L., Tovar M., Weber T., Brandes S., Rudolph M.M., Ehgartner J., et al. Enhanced and homogeneous oxygen availability during incubation of microfluidic droplets. RSC Adv. 2015;5(123):101871–101878. doi: 10.1039/C5RA20118G. [DOI] [Google Scholar]
- 77.Batani G., Bayer K., Böge J., Hentschel U., Thomas T. Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria. Sci Rep. 2019;9:18618. doi: 10.1038/s41598-019-55049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siegl A., Kamke J., Hochmuth T., Piel J., Richter M., Liang C., et al. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011;5(1):61–70. doi: 10.1038/ismej.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spencer S.J., Tamminen M.V., Preheim S.P., Guo M.T., Briggs A.W., Brito I.L., et al. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 2016;10(2):427–436. doi: 10.1038/ismej.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hultman J, Tamminen M, Pärnänen K, Cairns J, Karkman A, Virta M. Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol Ecol 2018;94:fiy038. https://doi.org/10.1093/femsec/fiy038. [DOI] [PMC free article] [PubMed]
- 81.Qin H., Wang S., Feng K., He Z., Virta M.P.J., Hou W., et al. Unraveling the diversity of sedimentary sulfate-reducing prokaryotes (SRP) across Tibetan saline lakes using epicPCR. Microbiome. 2019;7(1) doi: 10.1186/s40168-019-0688-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katz M., Hover B.M., Brady S.F. Culture-independent discovery of natural products from soil metagenomes. J Ind Microbiol Biotechnol. 2016;43:129–141. doi: 10.1007/s10295-015-1706-6. [DOI] [PubMed] [Google Scholar]
- 83.Brady S.F. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat Protoc. 2007;2(5):1297–1305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- 84.Brady S.F., Chao C.J., Handelsman J.o., Clardy J. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org Lett. 2001;3(13):1981–1984. doi: 10.1021/ol015949k. [DOI] [PubMed] [Google Scholar]
- 85.Gillespie D.E., Brady S.F., Bettermann A.D., Cianciotto N.P., Liles M.R., Rondon M.R., et al. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl Environ Microbiol. 2002;68(9):4301–4306. doi: 10.1128/AEM.68.9.4301-4306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brady S.F., Clardy J. Palmitoylputrescine, an antibiotic isolated from the heterologous expression of DNA extracted from bromeliad tank water. J Nat Prod. 2004;67(8):1283–1286. doi: 10.1021/np049976610.1021/np0499766.s001. [DOI] [PubMed] [Google Scholar]
- 87.Feng Z., Kallifidas D., Brady S.F. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc Natl Acad Sci U S A. 2011;108(31):12629–12634. doi: 10.1073/pnas.1103921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stevenson L.J., Bracegirdle J., Liu L., Sharrock A.V., Ackerley D.F., Keyzers R.A., et al. Metathramycin, a new bioactive aureolic acid discovered by heterologous expression of a metagenome derived biosynthetic pathway. RSC Chem Biol. 2021;2(2):556–567. doi: 10.1039/D0CB00228C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng Q., Gao G., Lü J., Long Q., Chen X., Zhang F., et al. Engineered Streptomyces lividans strains for optimal identification and expression of cryptic biosynthetic gene clusters. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.03042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang J.J., Tang X., Zhang M., Nguyen D., Moore B.S., Davies J.E., et al. Broad-host-range expression reveals native and host regulatory elements that influence heterologous antibiotic production in Gram-negative bacteria. MBio. 2017;8(5) doi: 10.1128/mBio.01291-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Linares-Otoya L, Linares-Otoya V, Armas-Mantilla L, Blanco-Olano C, Crüsemann M, Ganoza-Yupanqui ML, et al. Identification and heterologous expression of the kocurin biosynthetic gene cluster. Microbiology 2017;163:1409–14. https://doi.org/10.1099/mic.0.000538. [DOI] [PubMed]
- 92.Yamanaka K., Reynolds K.A., Kersten R.D., Ryan K.S., Gonzalez D.J., Nizet V., et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A. 2014;111(5):1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laureti L., Song L., Huang S., Corre C., Leblond P., Challis G.L., et al. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci U S A. 2011;108(15):6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adnani N., Chevrette M.G., Adibhatla S.N., Zhang F., Yu Q., Braun D.R., et al. Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, keyicin. ACS Chem Biol. 2017;12(12):3093–3102. doi: 10.1021/acschembio.7b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Buijs Y, Zhang S Da, Jørgensen KM, Isbrandt T, Larsen TO, Gram L. Enhancement of antibiotic production by co-cultivation of two antibiotic producing marine Vibrionaceae strains. FEMS Microbiol Ecol 2021;97:fiab041. https://doi.org/10.1093/femsec/fiab041. [DOI] [PubMed]
- 96.Cimermancic P., Medema M., Claesen J., Kurita K., Wieland Brown L., Mavrommatis K., et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158(2):412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim W.E., Charov K., Džunková M., Becraft E.D., Brown J., Schulz F., et al. Synthase-selective exploration of a tunicate microbiome by activity-guided single-cell genomics. ACS Chem Biol. 2021;16(5):813–819. doi: 10.1021/acschembio.1c00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Greunke C., Duell E.R., D’Agostino P.M., Glöckle A., Lamm K., Gulder T.A.M. Direct Pathway Cloning (DiPaC) to unlock natural product biosynthetic potential. Metab Eng. 2018;47:334–345. doi: 10.1016/j.ymben.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 99.Neilan B.A., Dittmann E., Rouhiainen L., Bass R.A., Schaub V., Sivonen K., et al. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J Bacteriol. 1999;181(13):4089–4097. doi: 10.1128/JB.181.13.4089-4097.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci U S A. 2002;99(22):14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Courtois S., Cappellano C.M., Ball M., Francou F.-X., Normand P., Helynck Gérard, et al. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl Environ Microbiol. 2003;69(1):49–55. doi: 10.1128/AEM.69.1.49-55.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moffitt M.C., Neilan B.A. Evolutionary affiliations within the superfamily of ketosynthases reflect complex pathway associations. J Mol Evol. 2003;56(4):446–457. doi: 10.1007/s00239-002-2415-0. [DOI] [PubMed] [Google Scholar]
- 103.Savic M., Vasiljevic B. Targeting polyketide synthase gene pool within actinomycetes: new degenerate primers. J Ind Microbiol Biotechnol. 2006;33(6):423–430. doi: 10.1007/s10295-006-0080-9. [DOI] [PubMed] [Google Scholar]
- 104.Metsä-Ketelä M, Salo V, Halo L, Hautala A, Hakala J, Mäntsälä P, et al. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol Lett 2006;180:1–6. https://doi.org/10.1111/j.1574-6968.1999.tb08770.x. [DOI] [PubMed]