Graphical abstract
Keywords: Breast cancer, Recurrence, HER2, H&E-stained histological images, Convolutional neural network
Abstract
HER2-positive breast cancer is a highly heterogeneous tumor, and about 30% of patients still suffer from recurrence and metastasis after trastuzumab targeted therapy. Predicting individual prognosis is of great significance for the further development of precise therapy. With the continuous development of computer technology, more and more attention has been paid to computer-aided diagnosis and prognosis prediction based on Hematoxylin and Eosin (H&E) pathological images, which are available for all breast cancer patients undergone surgical treatment. In this study, we first enrolled 127 HER2-positive breast cancer patients with known recurrence and metastasis status from Cancer Hospital of the Chinese Academy of Medical Sciences. We then proposed a novel multimodal deep learning method integrating whole slide H&E images (WSIs) and clinical information to accurately assess the risk of relapse and metastasis in patients with HER2-positive breast cancer. Specifically, we obtained the whole H&E staining images from the surgical specimens of breast cancer patients, and these images were adjusted to size 512 × 512 pixels. The deep convolutional neural network (CNN) was applied to these images to retrieve image features, which were combined with the clinical data. Based on the combined features. After that, a novel multimodal model was constructed for predicting the prognosis of each patient. The model achieved an area under curve (AUC) of 0.76 in the two-fold cross-validation (CV). To further evaluate the performance of our model, we downloaded the data of all 123 HER2-positive breast cancer patients with available H&E image and known recurrence and metastasis status in The Cancer Genome Atlas (TCGA), which was severed as an independent testing data. Despite the huge differences in race and experimental strategies, our model achieved an AUC of 0.72 in the TCGA samples. As a conclusion, H&E images, in conjunction with clinical information and advanced deep learning models, could be used to evaluate the risk of relapse and metastasis in patients with HER2-positive breast cancer.
1. Introduction
In 2020, breast cancer accounted for 12% of malignant tumors in all human population, overtaking lung cancer as the most common malignancy globally [1], [2]. Breast cancer is a kind of highly heterogeneous tumor, of which HER2-positive breast cancer accounts for about 25–30% incidences. HER2-positive breast cancer is highly aggressive, prone to brain metastasis, and poor in prognosis [3].
Recently, the emergence of trastuzumab, a kind of HER2-targeted drug, has greatly improved the survival and prognosis of patients with HER2-positive breast cancer [4], [5]. However, 20% of patients with HER2-positive breast cancer still develop recurrence and metastasis after adjuvant therapy including chemotherapy and trastuzumab [6], [7]. Dozens of new drugs, such as monoclonal antibodies (mAb), tyrosine kinase inhibitors (TKI) and antibody-drug conjugate (ADC), have been used for intensive treatment of patients with HER2-positive breast cancer, who are at a high risk of recurrence. How to identify patients with high risk of recurrence is a major difficulty. Currently, clinicians can only identify the risk of recurrence and make treatment plans based on clinicopathologic factors such as receptor expression, tumor size, lymph node metastasis, and age of onset. Fortunately, the Oncotype Dx and MammaPrint based on high-throughput sequencing to predict the risk of recurrence and metastasis have been verified by large-sample phase III clinical studies [2], [1], and have been written into the NCCN guidelines (www.nccn.org/patients), which are expected to accurately guide the treatment of HR-positive breast cancer.
They are only some tentative studies conducted to predict prognosis HER2-positive breast cancer based on genomics or radiomics. Cain et al. established a model to predict pathological complete response (pCR) in triple-negative/HER2-positive breast cancer patients through radiomics with an AUC value of 0.707 [8]. Prognostic prediction models based on genomics tend to have higher specificity and sensitivity, but they are often invasive, costly, and time-consuming, making it difficult to popularize [1]. Up to now, there is no mature product developed to predict the risk of recurrence and metastasis in patients with HER2-positive breast cancer. Therefore, it is urgent to establish a prediction model for this specific type of cancer to avoid overtreatment or undertreatment.
Hematoxylin and Eosin (H&E) staining is one of the commonly used staining methods in pathology. Studies have shown that the nuclear morphological characteristics of H&E histopathological images play an important role in the prognosis of various malignant tumors [9], [10]. With the continuous development of computer technology and the advent of Whole Slide imaging (WSI), computer-aided diagnosis and prognosis prediction based on H&E-stained histological and other images have received more and more attention. Because these pathological images not only contain the pathological characteristics of tumor morphology, growth, distribution and so on, but also have the advantages of radiomics, such as fast speed, non-invasive and low cost [11].
There are two computational methods for pathological images: traditional machine learning and deep learning [12], [13]. Machine learning algorithms can greatly reduce the time consuming of the diagnostic process, which are widely used in the field of prognostic prediction [14], [15], [16]. Convolutional neural network (CNN) is the most popular deep learning model for image processing at present [17], [18], [19]. It can be used not only for tumor detection and quantitative cell characteristics of pathological image analyses [20], [21], but also for the classification of small tissue images in pathological diagnosis [22], [23]. For example, Abdelzaher et al. used trained deep belief networks (DBN-NN) with similar structure weights to initialize the reverse propagation neural network to diagnose breast cancer [24]. Kather et al. established a deep residual learning model to predict Microsatellite instability (MSI) from H&E-stained histological images [25]. Zhi et al. used transfer learning with CNN to automatically diagnose breast cancer from histopathological images. They developed an ensemble model containing three customed CNN classifiers trained using transfer learning, and achieved higher accuracy than all other methods [26].In summary, CNN is a powerful algorithm that can directly process biomedical images, which overcomes the defects of subjective bias in the process of feature extraction from H&E-stained histological images.
In this study, we enrolled 127 HER2-positive breast cancer patients with known recurrence and metastasis information from Cancer Hospital of the Chinese Academy of Medical Sciences from 2010 to 2018. A predictive framework based on pathological images and clinical information was proposed to assess the risk of recurrence in HER2-postive breast cancer patients. Specifically, we first obtained whole slide images (WSIs) of H&E-stained sections from surgical specimens of the patients. These H&E WSIs were patched into images of size 512 × 512 pixels, which were then undergone a few image-preprocessing steps. Then, the image features were selected through the CNN algorithm, and combined with the original clinical data. Based on the combined features, a novel multimodal prognostic prediction model was constructed and validated by the 2-fold cross-validation (CV). Finally, we used all available HER2-positive breast cancer patients from The Cancer Genome Atlas (TCGA) as an independent test data set to evaluate the performance of this model.
2. Results
2.1. Description of entry data
In this study, 123 HER2-positive breast cancer patients with H&E-stained histological images were downloaded from the TCGA database. 26% of the patients were younger than 50 years old when breast cancer was diagnosed; 52% had positive lymph node. In addition, 127 HRE2-positive breast cancer patients from the Cancer Hospital of the Chinese Academy of Medical Sciences were enrolled. 49% of the patients were younger than 50 years old when breast cancer was diagnosed, 78% of the patients were stage I-II, 59% of the patients were ER positive, and 58% of the patients were PR positive. 49% of patients had positive lymph node, and 27% of patients had recurrence. All patients received radical surgery and obtained H&E-stained slides. After annotating the tumor area, each WSI was divided into 512x512 pixel patches. Finally, 199, 386 patches were used to train a CNN model. Clinical features of the patients enrolled in the study were shown in Table 1.
Table 1.
Summary of the general clinical information of breast patients.
| Clinicopathologic variable | Category | TCGA | CAMS data |
|---|---|---|---|
| Sample type | H&E | 123 | 127 |
| Age | <50 | 34 | 63 |
| ≥50 | 89 | 64 | |
| Tumor stage | I | 14 | 35 |
| II | 77 | 64 | |
| III | 32 | 28 | |
| PR | Positive | 73 | 74 |
| Negative | 50 | 53 | |
| ER | Positive | 90 | 74 |
| Negative | 33 | 53 | |
| Lymph nodes status | Positive (LMN+) | 65 | 60 |
| Negative (LMN-) | 58 | 67 | |
| Outcome | Non-recurrence | 118 | 95 |
| Recurrence | 5 | 32 |
2.2. A deep neural network framework to predict tumor recurrence
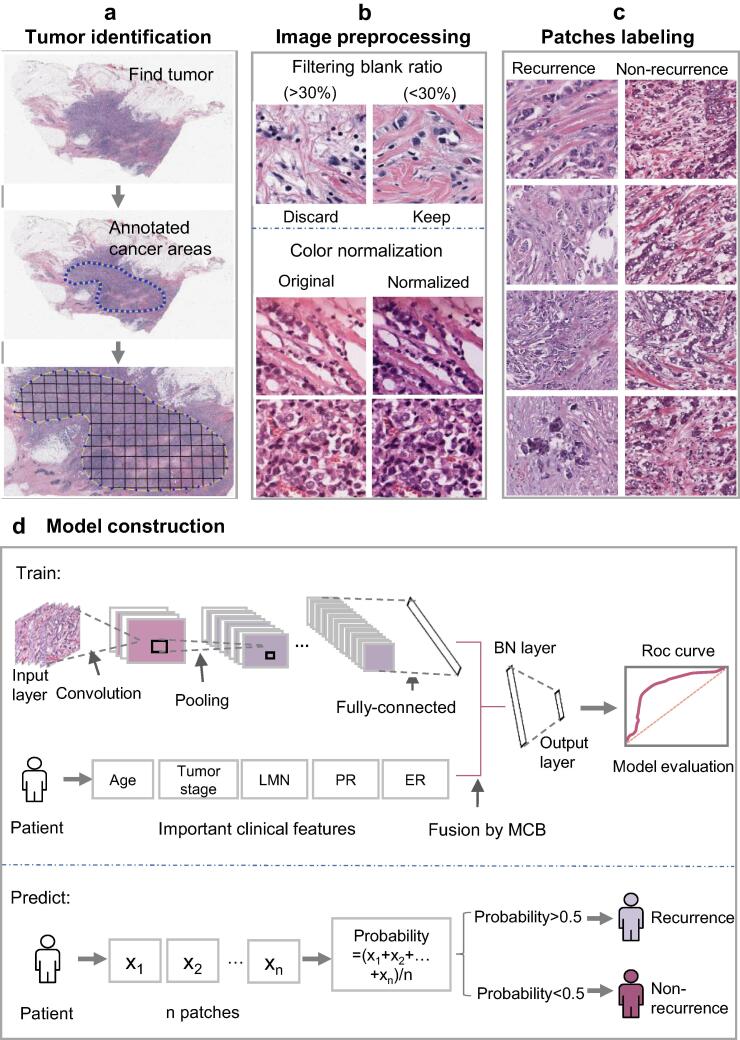
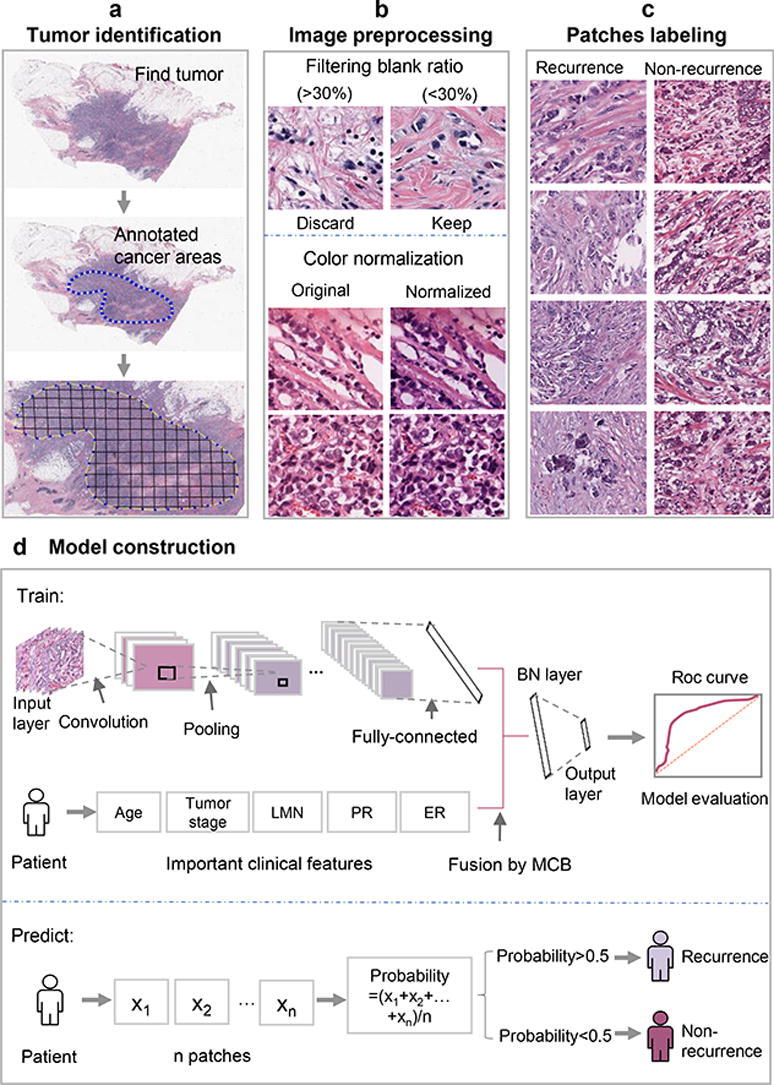
The complete process of predicting the risk of metastasis and recurrence in breast cancer patients was shown in Fig. 1. First, clinical data of breast cancer patients were downloaded from the TCGA database. After preprocessing the data, we used the random forest method to compare the importance of features on the clinical data set, and then used the logistic regression model for classification. Second, relevant case data and H&E-stained histological images of breast cancer patients who underwent surgery at the Cancer Hospital of the Chinese Academy of Medical Sciences from 2010 to 2018 were collected. We used CNN to build model on H&E-stained histological images. Third, the features selected from H&E images combined with the features of clinical data were used for training the recurrence and metastasis prediction model. In the process of training model, there are 4 parameters including optimazer, learning rate, momentum, loss function chosen as hyperparameters. According to the pytorch documentation (https://pytorch.org/docs/stable/optim.html; https://pytorch.org/docs/stable/generated/torch.nn.CrossEntropyLoss.html?highlight=crossentropyloss#torch.nn.CrossEntropyLoss), they were all set to empirical values. As a result, optimazer was set to be SGD; learning rate was set to be 0.01; momentum was set to be 0.9 and loss function was set to be nn.CrossEntropyLoss.
Fig. 1.
The protocol of whole process. (a) Find H&E-stained histological images of tumor and annotate cancer areas. (b) Color normalization was performed on areas where the filtering blank ratio was < 30%. (c) The patches were labeled according to the slide, recurrence (left) and non-recurrence (right). (d) The model was constructed after the fusion of image features and clinical features, and applied to predict on independent test set.
2.3. Many image features have different characteristics after image preprocessing
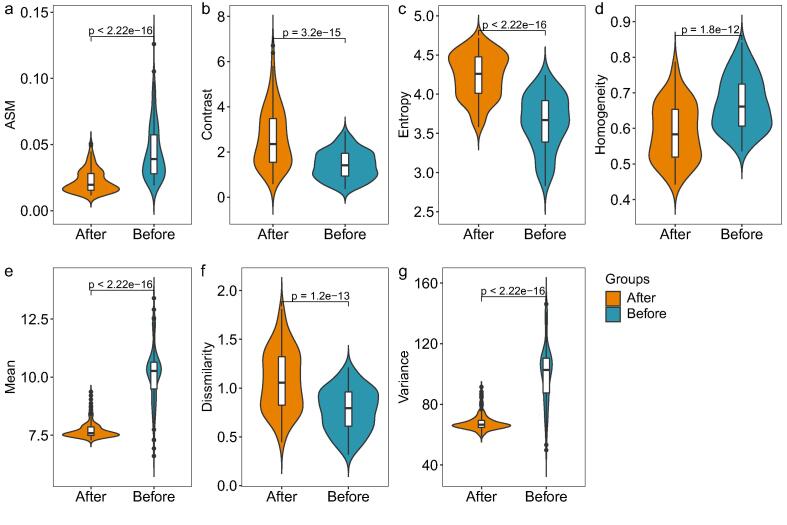
In the coloring process of H&E image, the color and intensity of histopathological image often change due to specimen preparation method, staining scheme (such as the temperature of the solution used), fixation characteristics, imaging equipment characteristics and other reasons. Therefore, before training the model, we normalized the image, and then used the rank-sum test to test the basic features of images before and after color normalization. The basic features involved included ASM (angular second moment-characteristics of gray level co-occurrence matrix), Contrast (The total amount of local gray changes in the image), Entropy (A measure of the amount of information an image has), Homogeneity (Inverse difference moment, a measurement of local gray level uniformity in image), Mean (Average value of gray scale), Dissimilarity (Local contrast of image), and Variance (Variance of image). The results were shown in Fig. 2. After color normalization, these basic features of the images changed significantly.
Fig. 2.
Violin diagram of image basic feature distribution before and after color normalization. (a) ASM. (b) Contrast. (c) Entropy. (d) Homogeneity. (e) Mean. (f) Dissimilarity. (g) Variance.
2.4. A few clinical characteristics are strongly associated with recurrence of breast cancer
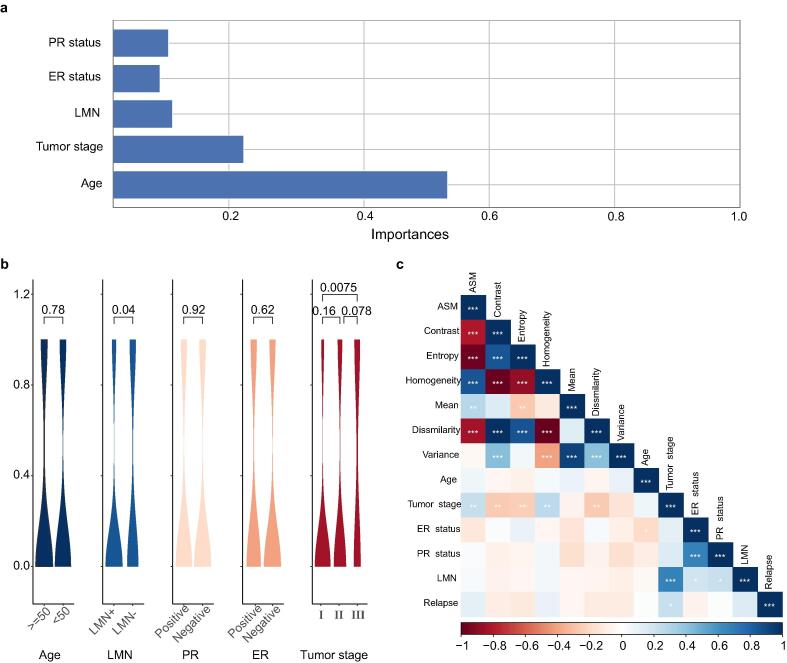
The random forest was used to verify the importance of characteristics of clinical data. Fig. 3(a) revealed the Mean Decrease Gini of all variables. As a result, tumor stage was the most important feature to model building. Correlation of 5 features with recurrence of breast cancer were examined by violin graphs and p-values were shown in Fig. 3(b). According to the characteristics of the training data set, the patients' tumor stage was strongly associated with the recurrence of breast cancer, especially in stage I and III. In addition, there were significant differences in LMN status. However, Age, ER and PR have almost no significant effect on the recurrence and metastasis of breast cancer. We also explored the correlation among clinical features and H&E image features as shown in Fig. 3(c). The results showed that some clinical features are related to H&E image features. For example, Tumor stage have strong correlation with LMN, and PR status have strong correlation with ER status. Finally, relapse have higher correlation with Tumor stage than with other clinical features and image features.
Fig. 3.
Features of clinical data. (a). Mean Decrease Gini corresponding of variables. (b). Correlation of several features with recurrence of breast cancer were examined by violin figure and p-values. (c). Correlation of image features and clinical features.
2.5. H&E staining image can be used to predict breast cancer recurrence with relatively good performance
2.5.1. The integration of image features and clinical features can improve the prediction accuracy
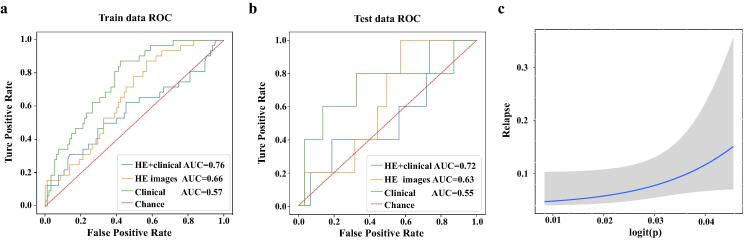
After downloading the H&E-stained histological images from the Cancer Hospital of the Chinese Academy of Medical Sciences, we took H&E images of size 512 × 512 as the input of CNN model. We divided the patches into two classes. Samples with tumor recurrence labels were marked as positive samples, while those without tumor recurrence labels were negative samples. Then the ResNet50 model was used to train the samples, and the 2-fold CV was used to split the samples and verify the results. The ROC curve and AUC were revealed in Fig. 4(a) and (b). The AUC of clinical data combined with H&E image information in training dataset and test dataset was 0.76 and 0.72, respectively, which was much higher than that predicted by clinical data alone. As an indication, the information in the H&E images helps improve the predictive power. A fitting curve for predicted breast cancer recurrence and true condition was shown in Fig. 4(c), it also proved that H&E image has a good performance in predicting the recurrence of breast cancer.
Fig. 4.
ROC and fitting curve. (a) ROC curve of 2-fold CV in training data set. (b) ROC curve in the test data set. (c) A fitting curve for predicting breast cancer recurrence and true condition.
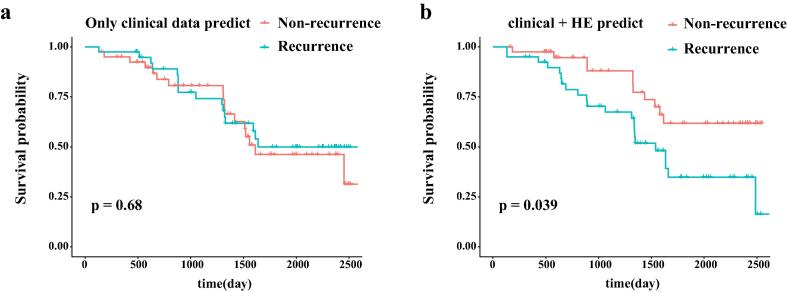
2.5.2. Survival analysis
In order to further verify the performance of the prediction method, we conducted survival analysis on the train data set (Fig. 5). As shown in Fig. 5(a), the survivals between recurrence and non-recurrence samples predicted by solely clinical data were not significantly different. However, there were significant differences after combining clinical data and H&E image information (Fig. 5(b), p = 0.039), which further indicated that H&E image can improve the prediction performance.
Fig. 5.
Survival analysis. (a). Survival analysis based on the predicted result of recurrence by only clinical data. (b). Survival analysis based on the predict result of recurrence by H&E-stained histological images combined with clinical data.
3. Discussion
As we all know, breast cancer is one of the most common malignancies in the world, especially in women, which poses a big threat to public health. HER2-positive breast cancer is highly invasive, prone to recurrence and metastasis, and has a poor prognosis. Identifying the risk of recurrence after surgical resection helps to develop a monitoring plan and provides personalized adjuvant treatment for them. Unfortunately, there is a lack of effective prediction model to identify the risk of recurrence. With the continuous development of computer technology, computer-aided diagnosis and prognostic prediction based on H&E staining images has attracted more and more attention because of its high speed, no extra cost, and no trauma. CNN has been used to distinguish benign and malignant breast tumors through imaging since the early 1990s. In many studies, the area under ROC curve can be as high as 0.9 [27], [28], [29], [30], [31]. In addition, image-based prognostic studies have been applied to colorectal cancer [32] and glial cancer [33] in the past, with an area under the ROC curve ranging from 0.5 to 0.8. All in all, the accuracy of image-based prognostic prediction model needs to be further improved.
In this study, we proposed a predictive framework based on H&E images and clinical information to assess the risk of recurrence in HER2-positive breast cancer patients. We used H&E images and clinical information from 127 HER2 positive breast cancer patients from the Cancer Hospital of China Academy of Medical Sciences to construct a new multimodal prognostic prediction model based on combination characteristics, and evaluated the performance of the model using HER2-positive breast cancer patients from the TCGA database as an independent test set. H&E staining histopathological images were first used to predict the prognosis of breast cancer. The sensitivity and specificity were 67% and 83% respectively, and the area under the ROC curve was 0.72, which showed a strong performance in similar studies.
However, there are some limitations in this study. Firstly, clinical factors were relatively limited. Secondly, the race and genetic background between the two data sources were pretty different. Thirdly, heterogeneity of H&E images in the same patient was not considered and the quality of H&E images was different between the trainset and the test set. All the above limitations greatly limit the accuracy and reproducibility of the research results. Therefore, we will further optimize the following aspects in the follow-up research. Firstly, more clinical factors will be included in the modeling process, such as vascular tumor emboli, differentiation degree and family history of tumor. Secondly, multiple discontinuous H&E images were selected from the same patient for training.
4. Conclusion
In summary, we provided preliminary evidence in this study that deep learning based on H&E staining histopathological images and clinical information can predict breast cancer recurrence and metastasis, and provided a new direction for routine clinical application of deep learning. However, this study is still in the research stage. Only when the clinical effectiveness of the model is proven more rigorously can it be widely used to assist clinical diagnosis and treatment. In the future, we will integrate molecular biological information such as whole exon sequencing (WES), DNA methylation and RNA sequencing results with HE images, to conduct multi-omics analysis of the prognosis of breast cancer patients. In addition, we intend to conduct a prospective cohort study, in which patients predicted to relapse in this study will be divided into trastuzumab combined chemotherapy group and intensive targeted therapy combined chemotherapy group according to treatment methods. The purpose is to observe whether these patients have relapse, so as to verify the effectiveness of the prediction model and further promote the development of accurate treatment for HER2-positive breast cancer.
5. Materials and methods
5.1. TCGA data
TCGA is an open large-scale cancer genome database that contains many primary cancers and their pathological images, and its digitized form matches normal samples of multiple cancer types. It provides researchers with public data sets that can be searched, viewed, and downloaded to help improve diagnostic methods, treatment standards, and ultimately prevent cancer. We downloaded WSIs of H&E-stained sections of breast cancer from the TCGA database (https://portal.gdc.cancer.gov/repository/). All free breast cancer slide images were stored in SVS format. These H&E-stained pathological sections were scanned with a 40 × objective lens. The average slide size (height × width) was 80,386 ± 36,812 × 59,143 ± 25,060 (mean ± standard deviation). H&E-stained histological images can be opened and analyzed by the python package OpenSlide. 1 2 3 H & E-stained histological images downloaded from TCGA match the tags that contain information about metastasis and recurrence. We marked those H&E images with metastasis and recurrence risk as 1, and those without metastasis and recurrence risk as 0. We combined H&E-stained histological images and clinical data to study the prediction of “metastasis and recurrence of breast cancer”.
5.2. Clinical data
We collected clinical information of breast cancer patients from Cancer Hospital of Chinese Academy of Medical Sciences. A total of 127 patients were enrolled from 2010 to 2018. Patients were included with the following criteria: female patients undergone surgery; patients with corresponding case data and histological specimens; patients with primary unilateral breast cancer with neoplasm staging I-III; patients who received adjuvant trastuzumab; patients with invasive carcinoma; patients with HER2 3+ based on immunohistochemistry or HER2 gene amplification based on fluorescence in situ hybridization (FISH). Exclusion criteria included: male; patients with other malignant tumors; patients with primary bilateral breast cancer; patients without tissue sections clinical data or follow-up data; patients with severe cognitive impairment, communication disorders and mental illness. All patients underwent pathological examination, and the detailed deidentified clinical information and H&E images were transferred to the investigators.
5.3. Image preprocessing
5.3.1. Image patching and filtering
To predict breast cancer recurrence and metastasis, an expert pathologist first annotated the tumor area with a visual assessment. The solid blue dotted lines in the middle of Fig. 1a represent the boundaries of the tumor area. In order to ensure the consistency of the input image size, WSIs were divided into 512×512 pixel slices. Then those slides with a low amount of information (e.g. more than 30% of filtering blank ratio was covered by background) were discarded [34].
5.3.2. Color normalization
In the coloring process of H&E-stained histological images, the specimen preparation method, staining scheme, fixation characteristics, imaging equipment characteristics and other factors may make it difficult to standardize the images between different medical centers, or even between samples in different experimental periods in the same laboratory [35], [36]. Therefore, after obtaining the H&E image, we used a deep convolutional Gaussian mixture model (DCGMM) to conduct color normalization processing on images [37]. the (natural) log-likelihood function of the model is:
where is the total number of pixels in the input image (). In order to fulfill a valid probability definition, the mixing coefficient must satisfy together with . stands for a multivariate normal distribution with mean and covariance matrix . The objective is to maximize the likelihood function, all parameters of the DCGMM are jointly optimized by minimizing the negative log-likelihood with the gradient descent algorithm. After training the model, the responsibility vector for each pixel in the image can be calculated by applying DCGMM to any given test image.
5.3.3. The labeling of patched images
After a WSI was divided into 512x512 patches, the label of each patch was the same as that of the WSI, and labeled recurrence as 1, and 0 otherwise.
5.4. Definitions
Tumor recurrence included locoregional recurrence and distant tissue or organ metastasis. Locoregional recurrence was defined as the recurrence of ipsilateral breast, chest wall, or regional lymph nodes. Tumor stage was determined according to the 8th edition of American Joint Committee on Cancer (AJCC) staging system [38]. All the patients with positive lymph nodes (LMN+) were confirmed by pathology. According to the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) Panel, ER and PR were considered positive if there are at least 1% positive tumor nuclei in the sample [39]. HER2 status was defined according to 2018 American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines [40]. Immunohistochemical scores of 3+ were considered positive, and scores of 2+ were considered positive if HER2 gene amplification was evaluated by fluorescence in situ hybridization (FISH).
5.5. Clinical feature ranking based on random forest
Variable selection is important for interpretation and prediction, especially for high-dimensional data sets. Random forest is a combinatorial classifier model composed of decision tree classifier sets, which belongs to an integrated learning model [41]. It uses multiple decision trees to classify and predict samples and each decision tree is an unpruned decision tree constructed by the Classification And Regression Tree (CART) algorithm [42]. Random forest can also be used to estimate the importance of variables in a model and the importance of variables is measured by Mean Decrease Gini caused by variable . At the classification node , the calculation formula for Mean Decrease Gini is as follows:
where represents the total number of classes of the target variable, and represents the conditional probability that the target variable is of class at node . According to the formula, Gini index is calculated. Finally, the larger the value of is, the more important the is.
5.6. Image feature representation based on ResNet50
Convolution neural network (CNN) is a state-of-the-art algorithm in image recognition and classification because of its stable learning performance [43]. The structure of CNN includes input layer, hidden layer, and output layer. Its workflow was shown in Fig. 1(d): when an image is fed into the input layer, the data is entered the hidden layer and the low-level features is extracted. After that, the middle-level feature is extracted in the following convolutional layer and pooling layer, In the convolutional layer, some small trainable convolution kernel can be learned to extract convolution feature on the previous layer :
where, and represent the input and output at the layer , respectively; was a nonlinear function; Rectified Linear Unit (ReLU) is used in CNN as activation function; and represents the bias. Consequently, the extracted features in the convolutional layer will be transmitted to the pooling layer for feature dimensionality reduction and information filtering, and the high-level features will be extracted through the following hidden layers in turn. Finally, they are feed into the output layer.
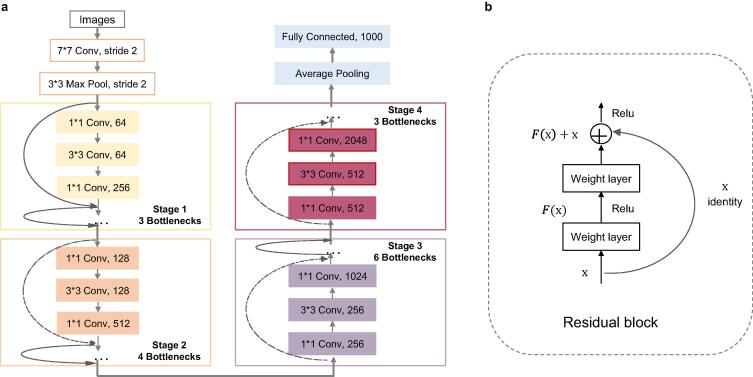
ResNet is a large-scale CNN constructed by residual blocks, which was proposed by the winner of ILSVRC image classification and object recognition algorithm in 2015 (Microsoft Research, the ARTIFICIAL intelligence team of Microsoft) [44]. The hidden layer of ResNet50 contains 16 residual blocks (Fig. 6(b)) in total [45]. The stack of the residual blocks mitigated the gradient-vanishing problem commonly seen in deep neural networks, and has been used by many subsequent algorithms [46]. We used ResNet50 to select features of breast cancer H&E images, and its workflow was shown in Fig. 6(a). The architecture of ResNet50 was divided into 4 stages. Every ResNet architecture performed the initial convolution and max-pooling using 7 × 7 and 3 × 3 kernel sizes respectively. Afterward, stage 1 of the network started and it has 3 bottlenecks containing 3 layers with 1 × 1, 3 × 3 and 1 × 1 convolutions. The 1 × 1 convolution layers were responsible for reducing and then restoring the dimensions; the 3 × 3 layer was a bottleneck with smaller input/output dimensions; and the kernel sizes used to perform convolution operations in all 3 layers were 64, 64 and 128, respectively. Curved arrows refer to the connection; the dashed arrow indicates that the convolution operation in the residual blocks was performed with stride 2; so, the size of the input will be reduced by half in height and width, but the channel width will be doubled. When the image advanced from one stage to another, the channel width will be doubled and the input size will be reduced by half. Finally, the network has an average pool layer, followed by a fully connected layer with 1000 neurons.
Fig. 6.
Network architecture of ResNet50. (a) ResNet50 structure diagram. (b) The basic structure of ResNet50 residual network.
Because there are not many positive samples of recurrence and metastasis, data augmentation is used to enrich the data images. We applied a fresh set of random operations with RandomCrop package of Python to crop the image data randomly. Instead of using the exact same items at every epoch, we showed a variant that has been changed in a different way. Here, for the training, we were randomly cropping the image and re-sizing it to shape (224, 224) in each epoch in order to enrich the data images.
5.7. Features fusion
In order to improve the performance of the model for predicting recurrence risk, it is necessary to fuse the features with two different modes (H&E image features and clinical features). The most common methods are concatenation, element wise product, and element wise sum. These simple operations are not as effective as outer products, which can establish a complex relationship between the two modes. However, the complexity of outer product calculation is too high. N-dimensional vector, the outer product is calculated to get a vector of n2. Therefore, Multimodal Compact Bilinear (MCB) is proposed. MCB maps the results of outer product into low dimensional space, and does not need to explicitly calculate outer product.
MCB algorithm was used to fuse the penultimate layer of ResNet50 with clinical features, and the fusion results were fed into the BN layer before linear layer. (Fig. 1(d)).
5.8. Sample classification
As shown in the predict part of Fig. 1(d), each patient’s slide was divided into many different patches, and each patch may get a predicted value by the trained model. The average of these values was used as the recurrence probability of the slide. After that, the slide was classified: if the recurrence probability was greater than 0.5, then this slide was determined to recurrence, otherwise it was determined to non-recurrence.
5.9. Evaluation criteria
5.9.1. 2-fold CV
In the modeling process, we used 2-fold CV to verify the accuracy of the training model. The workflow of building model for clinical data is shown in Table 2.
Table 2.
The workflow of building model.
| Algorithm: Build model | |
|---|---|
| Input: Training data set, test data set | |
| Output: Trained model and performance of the model | |
| 1 | for feature type in clinical data, H&E images, clinical + H&E images do |
| 2 | for 2-fold cross validation process do |
| 3 | if feature type = clinical data do |
| 4 | sort importance of features in Random forest; |
| 5 | train model on train data set; |
| 6 | compute AUC from ROC curve for one subset of cross validation; |
| 7 | draw ROC curve based on the results of 2-fold CV; |
| 8 | train the model with whole train data set; |
| 9 | test the performance of the model on test data set; |
| 10 | final; |
5.9.2. Independent validation
We trained the model with 127 samples from Cancer Hospital of Chinese Academy of Medical Sciences. Then TCGA data sets were used as independent test set to verify the prediction performance of the model.
5.10. Statistical analysis
We used statistical methods to analyze the clinical characteristics including age, LMN, PR, ER and tumor stage of 123 cases from TCGA. Specifically, age was divided into ≥50 years old and <50 years old. LMN was divided into positive lymph nodes (LMN+)and negative lymph nodes (LMN−), PR and ER were divided into positive and negative respectively, while tumor stage was divided into three groups of I, II, and III. A t-test was used to compare the agreement between the observers, and bilateral statistics was performed, p-value < 0.05 was considered statistically significant. All statistical analyses were performed using R software.
Funding
This work was supported by the CSCO Pilot Oncology Research Fund (Y-2019AZMS-0377), the Capital's Funds for Health Improvement and Research (2018-2-4023), the Clinical Translation and Medical Research Fund of Chinese Academy of Medical Sciences (12019XK320071), and the Beijing Hope Run Special Fund of Cancer Foundation of China (LC2019A13).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Yuebin Liang, Email: liangyb@geneis.cn.
Peng Yuan, Email: yuanpengyp01@163.com.
Data availability
The data and codes used in this study are publicly available from Github (https://github.com/bensteven2/HE_breast_recurrence).
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Liu H., Qiu C., Wang B.o., Bing P., Tian G., Zhang X., et al. Evaluating DNA methylation, gene expression, somatic mutation, and their combinations in inferring tumor tissue-of-origin. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.619330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung K. National Center for Biotechnology Information; 2012. Cy5.5-8-Amino-octanoic acid-Ser-Cys-Pro-Pro-Trp-Gln-Glu-Trp-His-Asn-Phe-Met-Pro-Phe-NH2 - Molecular Imaging and Contrast Agent Database (MICAD) - NCBI Bookshelf. [PubMed] [Google Scholar]
- 4.Perez E.A., Romond E.H., Suman V.J., Jeong J.-H., Sledge G., Geyer C.E., et al. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2–positive breast cancer: planned joint analysis of overall survival From NSABP B-31 and NCCTG N9831. J Clin Oncol. 2014;32(33):3744–3752. doi: 10.1200/JCO.2014.55.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slamon D.J., Eiermann W., Robert N.J., Giermek J., Martin M., Jasiowka M., et al. Abstract S5-04: Ten year follow-up of BCIRG-006 comparing doxorubicin plus cyclophosphamide followed by docetaxel (AC→T) with doxorubicin plus cyclophosphamide followed by docetaxel and trastuzumab (AC→TH) with docetaxel, carboplatin and trastuzumab (T. Cancer Res. 2016;76 S5-04-S05-04. [Google Scholar]
- 6.Piccart M., Procter M., Fumagalli D., de Azambuja E., Clark E., Ewer M.S., et al. Adjuvant Pertuzumab and Trastuzumab in Early HER2-Positive Breast Cancer in the APHINITY Trial: 6 Years' Follow-Up. J Clin Oncol. 2021;39(13):1448–1457. doi: 10.1200/JCO.20.01204. JCO.20.01204. [DOI] [PubMed] [Google Scholar]
- 7.Cameron D., Piccart-Gebhart M.J., Gelber R.D., Procter M., Goldhirsch A., de Azambuja E., et al. 11 years' follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive early breast cancer: final analysis of the HERceptin Adjuvant (HERA) trial. Lancet. 2017;389(10075):1195–1205. doi: 10.1016/S0140-6736(16)32616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cain E.H., Saha A., Harowicz M.R., Marks J.R., Marcom P.K., Mazurowski M.A. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Cancer Res Treat. 2019;173(2):455–463. doi: 10.1007/s10549-018-4990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo X., Zang X., Yang L., Huang J., Liang F., Rodriguez-Canales J., et al. Comprehensive computational pathological image analysis predicts lung cancer prognosis. J Thoracic Oncol Off Publ Int Assoc Study Lung Cancer. 2017;12(3):501–509. doi: 10.1016/j.jtho.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng Y., Ji L., Yuan X., Jiang Z., Zeng N., Zhan Nuclear shape, architecture and orientation features from H&E images are able to predict recurrence in node-negative gastric adenocarcinoma. J Transl Med. 2019 doi: 10.1186/s12967-019-1839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X., Xi B., Zhang Y., Zhu L., Sui X., Tian G., et al. A machine learning-based diagnosis of thyroid cancer using thyroid nodules ultrasound images. Curr Bioinform. 2020;15(4):349–358. [Google Scholar]
- 12.Hinton G.E., Osindero S., Teh Y.-W. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18(7):1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- 13.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang S., Weigelt B., Wen H.-C., Pareja F., Raghavendra A., Martelotto L.G., et al. IDH2 mutations define a unique subtype of breast cancer with altered nuclear polarity. Cancer Res. 2016;76(24):7118–7129. doi: 10.1158/0008-5472.CAN-16-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B., Lang J., Wang B., Liu X., Lu Q., He J., et al. TOOme: a novel computational framework to infer cancer tissue-of-origin by integrating both gene mutation and expression. Front Bioeng Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 18.Poplin R., Varadarajan A.V., Blumer K., Liu Y., McConnell M.V., Corrado G.S., et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nature Biomed Eng. 2018;2(3):158–164. doi: 10.1038/s41551-018-0195-0. [DOI] [PubMed] [Google Scholar]
- 19.Kumar N., Verma R., Arora A., Kumar A., Gann P.H. Convolutional neural networks for prostate cancer recurrence prediction. SPIE Medical Imaging. International Society for Optics and Photonics; 2017. [Google Scholar]
- 20.Ehteshami Bejnordi B., Veta M., Johannes van Diest P., van Ginneken B., Karssemeijer N., Litjens G., et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318(22):2199. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litjens G., Sánchez C.I., Timofeeva N., Hermsen M., Nagtegaal I., Kovacs I., et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016;6(1) doi: 10.1038/srep26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araújo T., Aresta G., Castro E., Rouco J., Aguiar P., Eloy C., et al. Classification of breast cancer histology images using Convolutional Neural Networks. PLoS ONE. 2017;12(6) doi: 10.1371/journal.pone.0177544. e0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y., Jia Z., Wang L.-B., Ai Y., Zhang F., Lai M., et al. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinf. 2017;18(1) doi: 10.1186/s12859-017-1685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdel-Zaher A.M., Eldeib A.M. Breast cancer classification using deep belief networks. Expert Syst Appl. 2016;46:139–144. [Google Scholar]
- 25.Kather J.N., Pearson A.T., Halama N., Jäger D., Krause J., Loosen S.H., et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25(7):1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhi W, Yueng H, Chen Z, Zandavi SM and Chung YY. The 24th International Conference On Neural Information Processing; 2017.
- 27.Wang J., Yang X., Cai H., Tan W., Jin C., Li L. Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci Rep. 2016;6:27327. doi: 10.1038/srep27327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chougrad H., Zouaki H., Alheyane O. Deep Convolutional Neural Networks for breast cancer screening. Comput Methods Programs Biomed. 2018;157:19–30. doi: 10.1016/j.cmpb.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Seokmin Ha, Ho K., Kang Ja Y., Jeong, Moon H., Park Wonsik, Kim A deep learning framework for supporting the classification of breast lesions in ultrasound images. Phys Med Biol. 2017 doi: 10.1088/1361-6560/aa82ec. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y., Han L., Dou H., Luo H., Yuan Z., Liu Q., et al. Two-stage CNNs for computerized BI-RADS categorization in breast ultrasound images. Biomed Eng Online. 2019;18(1) doi: 10.1186/s12938-019-0626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herent P., Schmauch B., Jehanno P., Dehaene O., Saillard C., Balleyguier C., et al. Detection and characterization of MRI breast lesions using deep learning. Diagn Interv Imaging. 2019 doi: 10.1016/j.diii.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Pushpanjali G., Sum F., Chiang P., Kumar S., Suvendu K., Mohapatra Prediction of colon cancer stages and survival period with machine learning approach. Cancers. 2019;11 doi: 10.3390/cancers11122007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L., Zhang S., Chen H., Luo L. Brain tumor segmentation and survival prediction using multimodal MRI scans with deep learning. Front Neurosci. 2019;13:810. doi: 10.3389/fnins.2019.00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z., Wang L., Li C., Cai Y., Liang Y., Mo X., et al. DeepLRHE: a deep convolutional neural network framework to evaluate the risk of lung cancer recurrence and metastasis from histopathology images. Front Genet. 2020;11 doi: 10.3389/fgene.2020.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehteshami Bejnordi B., Litjens G., Timofeeva N., Otte-Holler I., Homeyer A., Karssemeijer N., et al. Stain Specific Standardization of Whole-Slide Histopathological Images. IEEE Trans Med Imaging. 2016;35(2):404–415. doi: 10.1109/TMI.2015.2476509. [DOI] [PubMed] [Google Scholar]
- 36.Bautista P., Hashimoto N., Yagi Y. Color standardization in whole slide imaging using a color calibration slide. J Pathol Inf. 2014;5(1):4. doi: 10.4103/2153-3539.126153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghazvinian Zanjani F., Zinger S., de With P.H.N., Bejnordi E.B., van der Laak J.A.W.M. 1st Conference on Medical Imaging with Deep Learning (MIDL) 2018. Histopathology stain color normalization using deep generative models; pp. 1–11. [Google Scholar]
- 38.Giuliano A.E., Connolly J.L., Edge S.B., Mittendorf E.A., Rugo H.S., Solin L.J., et al. Breast Cancer—Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA: A Cancer J Clinicians. 2017;67:290–303. doi: 10.3322/caac.21393. [DOI] [PubMed] [Google Scholar]
- 39.Hammond American Society of clinical oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (vol 134, pg 907, 2010) Arch Pathol Lab Med. 2010;134:1101. doi: 10.5858/134.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antonio C.W., Hammond M.E.H., Kimberly H.A., Brittany E.H., Pamela B.M., John M.S.B., et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice guideline focused Update. J Clin Oncol. 2018;36:2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 41.Prusa JD, Khoshgoftaar TM, Napolitano A. Using Feature Selection in Combination with Ensemble Learning Techniques to Improve Tweet Sentiment Classification Performance. IEEE International Conference on Tools with Artificial Intelligence. 2015:186–193. [Google Scholar]
- 42.Denison D.G.T., Mallick B.K., Smith A. A Bayesian CART algorithm. Biometrika. 1998;85:363–377. [Google Scholar]
- 43.Egmont-Petersen M., de Ridder D., Handels H. Image processing with neural networks—a review. Pattern Recogn. 2002;35(10):2279–2301. [Google Scholar]
- 44.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vision. 2015;115(3):211–252. [Google Scholar]
- 45.He K, Zhang X, Ren S, SunDeep J. Residual Learning for Image Recognition. IEEE. 2016:770–778. [Google Scholar]
- 46.Szegedy C, Ioffe S, Vanhoucke V, Alemi, AA. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. 2016, p. 4278-4284.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and codes used in this study are publicly available from Github (https://github.com/bensteven2/HE_breast_recurrence).