Abstract
The telomerase enzyme exists as a large complex (∼1,000 kDa) in mammals and at minimum is composed of the telomerase RNA and the catalytic subunit telomerase reverse transcriptase (TERT). In Saccharomyces cerevisiae, telomerase appears to function as an interdependent dimer or multimer in vivo (J. Prescott and E. H. Blackburn, Genes Dev. 11:2790–2800, 1997). However, the requirements for multimerization are not known, and it remained unclear whether telomerase exists as a multimer in other organisms. We show here that human TERT (hTERT) forms a functional multimer in a rabbit reticulocyte lysate reconstitution assay and in human cell extracts. Two separate, catalytically inactive TERT proteins can complement each other in trans to reconstitute catalytic activity. This complementation requires the amino terminus of one hTERT and the reverse transcriptase and C-terminal domains of the second hTERT. The telomerase RNA must associate with only the latter hTERT for reconstitution of telomerase activity to occur. Multimerization of telomerase also facilitates the recognition and elongation of substrates in vitro and in vivo. These data suggest that the catalytic core of human telomerase may exist as a functionally cooperative dimer or multimer in vivo.
The catalytic subunit of telomerase, the telomerase reverse transcriptase (TERT), possesses the hallmark amino acid motifs of a reverse transcriptase (RT) (23, 29, 40, 47). However, unlike viral RTs, telomerase is a unique eukaryotic RT that carries an intrinsic RNA template essential for the de novo addition of telomere sequences (reviewed in reference 19). Proteins associated with telomerase activity include TEP1 (22, 48), hsp90/p23 (18, 25), dyskerin (42, 43), L22 (32), and hStau (32) in mammals; the Sm proteins (54) as well as Est1p and Est3p, (26, 56) in Saccharomyces cerevisiae; and p80, p95, and p43 in ciliates (1, 11, 21, 35). TEP1 is not essential for telomerase activity in vitro or in vivo (5, 37). A subset of these associated factors are known to serve distinct roles in telomerase assembly and telomere length maintenance (15, 16, 27, 34, 41, 43, 51, 54).
In S. cerevisiae, different telomerase RNAs can functionally cooperate to form an active telomerase complex in vivo. Prescott and Blackburn demonstrated the presence of at least two primer recognition-elongation sites within S. cerevisiae telomerase (50). In addition, they showed that a mutant telomerase RNA incapable of telomere elongation could nonetheless support elongation in a diploid strain containing one mutant and one wild-type telomerase RNA (50). These results provided the first evidence that telomerase could form an active multimer in vivo that might contain, at minimum, two active sites (50). A recombinant reconstitution assay for human telomerase showed that two separately inactive, nonoverlapping fragments of human telomerase RNA could reconstitute telomerase activity in vitro (59). While consistent with a model of telomerase RNA multimerization, these results are also consistent with reconstitution of a single active telomerase RNA from two inactive telomerase RNA fragments.
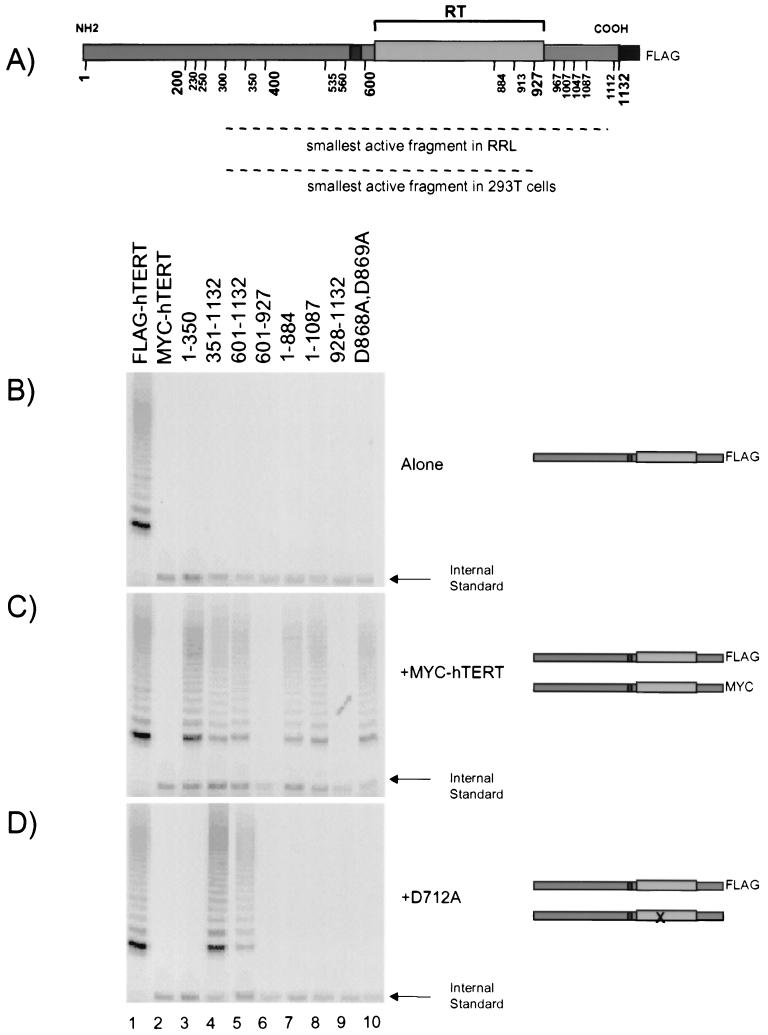
In vitro, the minimal requirements for telomerase activity appear to comprise the telomerase RNA and human TERT (hTERT) (3, 6, 9, 60). Previously, we found that the first 300 amino acid residues (aa) of hTERT were dispensable for telomerase activity in vitro and in vivo (5) (summarized in Fig. 1A). However, the telomerase activities associated with N-terminal truncations of hTERT were severely reduced in rabbit reticulocyte lysates (RRLs) relative to the activities achieved when the same hTERT truncation proteins were introduced into telomerase-positive 293T cells (5). Furthermore, deletion of the C-terminal 204 aa of hTERT did not affect telomerase activity in 293T cells, whereas all but the C-terminal 20 aa are absolutely required for telomerase activity in RRL (4, 5) (summarized in Fig. 1A). In this study, we set out to determine whether the observed discrepancies in activity between truncated hTERT proteins in RRL and 293T cells might be explained by the multimerization of specific hTERT fragments with endogenous hTERT.
FIG. 1.
Physical and functional interactions of full-length and truncated hTERT proteins in vitro. (A) A schematic diagram of full-length hTERT, including a summary of the minimal fragments of hTERT that are sufficient for telomerase activity in RRL and 293T cells (5). The RT region of hTERT is depicted with a gray box, and the smaller, darker box depicts the telomerase-specific motif. (B to D) FLAG-tagged full-length hTERT (FLAG-hTERT), FLAG-tagged inactive hTERT truncated proteins (indicated in amino acids), and hTERT containing two point mutations (D868A and D869A) were each synthesized in RRL alone (B), with full-length hTERT containing a MYC epitope (C), or with an inactive hTERT substitution mutant (D712A) that contained no epitope tag (symbolized with an X in the RT domain) (D). Each translation reaction was carried out in the presence of excess telomerase RNA. The lysates were subjected to immunoprecipitation with an anti-FLAG antibody and assayed for telomerase activity by TRAP. The arrow at the bottom right of each panel shows the internal PCR standard for amplification.
MATERIALS AND METHODS
Plasmids and transfections.
All hTERT constructs were cloned into the vector pCR3.1 (Invitrogen, Carlsbad, Calif.) as described previously (5, 23). Full-length hTERT contained either a FLAG epitope or a MYC epitope at the C terminus (see text and figure legends for details). The truncated hTERT proteins contained a FLAG epitope at the C terminus, and the hTERT point mutant D712A did not contain an epitope tag. The hTERT constructs indicated in Fig. 4 were transfected into GM847 cells by using Lipofectamine (Life Technologies, Gaithersburg, Md.) as per the manufacturer's instructions.
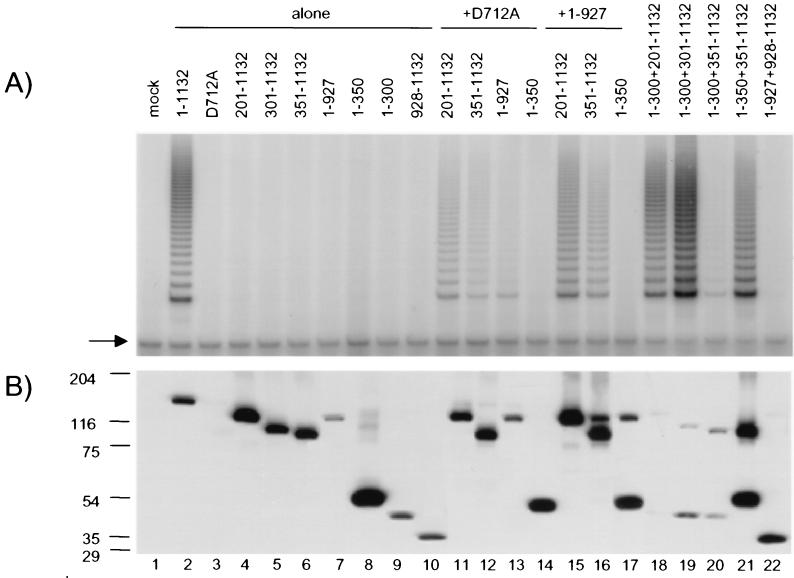
FIG. 4.
Functional hTERT multimerization in GM847 cells. (A) TRAP was performed (for 28 PCR cycles) on anti-FLAG immunoprecipitates from mock-transfected GM847 cells (lane 1), cells transfected with full-length, FLAG-tagged hTERT (lane 2), or cells transfected with different FLAG-tagged hTERT fragments either alone (lanes 3 to 10), with full-length hTERT containing the point mutation D712A (without a FLAG epitope) (lanes 11 to 14), or with a FLAG-tagged, truncated hTERT spanning aa 1 to 927 (lanes 15 to 17). Other nonoverlapping fragments of hTERT were also tested for their ability to reconstitute telomerase activity (lanes 18 to 22). The arrow at the left indicates the internal PCR standard for TRAP. (B) The lysates used in the experiment shown in panel A were analyzed by Western blotting with an anti-hTERT antibody. Arrows at the left indicate the molecular mass of protein standards. Note that the D712A mutant lacks a FLAG epitope and is therefore not immunoprecipitated (lane 3). Thus, the telomerase activity observed in lanes 11 to 13 is by virtue of an association of D712A with the appropriate FLAG-tagged hTERT fragment. The weak hTERT signals in lanes 18 and 22 are visible after a longer exposure (data not shown).
Synthesis and purification of human telomerase RNA.
Plasmid DNA containing the human telomerase RNA (hTER) gene (5, 23) was linearized by digestion with EcoRI to obtain a template for full-length hTER RNA synthesis. T7 transcription reactions were carried out with a MEGAscript in vitro transcription kit (Ambion Inc., Austin, Tex.). The transcription reaction products were extracted with phenol and then with chloroform-isoamyl alcohol (24:1), ethanol precipitated, and dissolved in water. The telomerase RNA was purified from a 4% (wt/vol) polyacrylamide gel (19:1 [wt/wt] acrylamide:bisacrylamide) containing 8 M urea by elution in water overnight at 4°C, filtered through a Supor membrane (0.8/0.2 μm pore size; Gelman Sciences, Ann Arbor, Mch.), precipitated in ethanol, and dissolved in water. All in vitro transcription and telomerase reconstitution experiments were carried out at the Amgen Institute/Ontario Cancer Institute.
In vitro reconstitution of telomerase activity.
All hTERT proteins were synthesized in vitro by using a rabbit reticulocyte T7-coupled transcription-translation system (Promega, Madison, Wis.) as per the manufacturer's instructions and as described previously (5, 6). Full-length hTERT cDNAs (or truncation versions thereof), at a concentration of 0.01 μg/μl, were added to the RRL in the presence of 0.01 μg of in vitro-transcribed, purified telomerase RNA/μl (except where indicated; see figure legends) and incubated at 30°C for 90 min.
To test whether distinct hTERT proteins could interact in vitro, each truncated hTERT protein was synthesized in a separate RRL reaction in the presence or absence of the telomerase RNA (see Fig. 2) or was synthesized together in the same RRL reaction in the presence of 0.01 μg of hTER/μl (see Fig. 1 and 3). Two microliters of each RRL reaction product containing one truncated hTERT protein was mixed on ice for 1 h and then assayed for telomerase activity. In the reactions in which only one truncated hTERT protein was present, 2 μl of CHAPS buffer {0.5% [wt/vol] 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 10 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 0.1 M NaCl, 5 mM β-mercaptoethanol, and 10% [vol/vol] glycerol} was added.
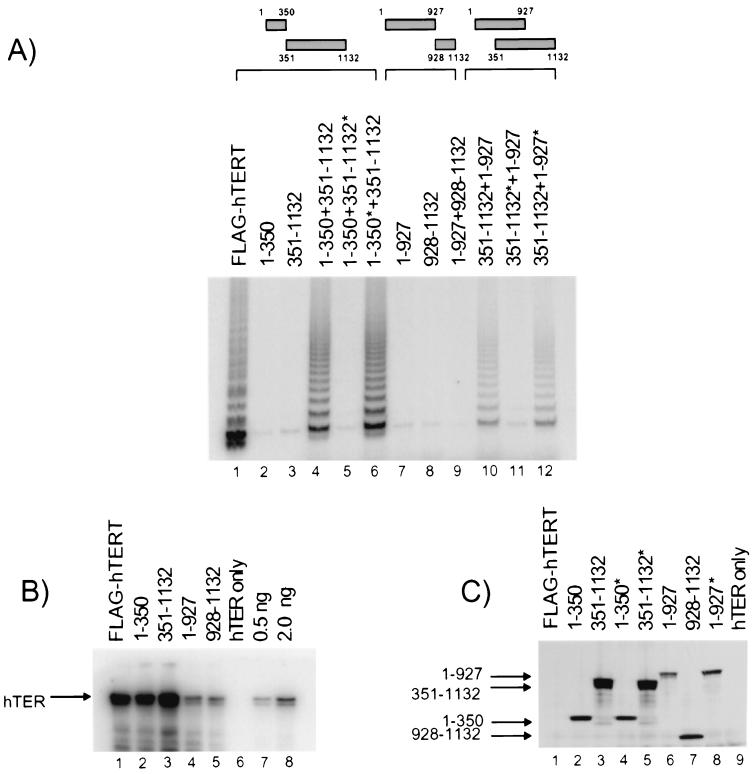
FIG. 2.
Functional complementation of distinct truncated hTERT proteins in vitro. Different fragments of hTERT containing a FLAG epitope were synthesized individually in the presence or absence of human telomerase RNA (hTER). hTERT truncations synthesized in the absence of hTER are indicated by an asterisk. (A) RRLs containing different truncated hTERT proteins (shown schematically in the top panel) were mixed on ice and assayed for telomerase activity by TRAP. (B) RRLs, as in panel A, were subjected to immunoprecipitation with anti-FLAG and analyzed for the presence of telomerase RNA by Northern blotting with an hTER-specific probe. As a control for nonspecific binding of telomerase RNA to anti-FLAG beads, RRL containing hTER only was also subjected to immunoprecipitation with anti-FLAG (lane 6). Lanes 7 and 8, respectively, contain 0.5 and 2.0 ng of in vitro-transcribed hTER as a standard. The results shown in panel B are representative; however, levels of hTER coimmunoprecipitated with hTERT differed between experiments, likely as a result of different amounts of precipitated hTERT (5). (C) hTERT protein levels were assessed by Western blotting with an anti-TERT antibody and an anti-FLAG antibody. In lane 1, we noted a previously published yet unexplained observation: in some instances, the levels of full-length hTERT in RRL are extremely low when synthesized together with hTER (5).
FIG. 3.
Functional interaction between distinct hTERT proteins. Different truncated hTERT proteins were cotranslated in RRL in the presence of human telomerase RNA (hTER). (A) A 2-μl volume of each RRL, containing different protein combinations, was assayed for telomerase activity by TRAP (A) and for protein expression by Western blotting with anti-hTERT and anti-FLAG antibodies (B). Full-length hTERT (lane 1) was included as a positive control. The numbers to the left of panel B indicate the molecular mass of protein standards (Bio-Rad, Hercules, Calif.).
Immunoprecipitations.
Twenty-five microliters of RRL was immunoprecipitated with 15 μl of M2 affinity resin (Sigma, St. Louis, Mo.) in 0.5% (wt/vol) CHAPS buffer. A 2-μl volume of beads was analyzed for telomerase activity by the telomere repeat amplification protocol (TRAP), and a 10-μl volume of beads was analyzed by Northern blot analysis.
Western analysis.
Fifteen microliters of sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading dye was added to 5 μl of RRL or 25 μg of GM847 cell extract. The samples were heated for 5 min at 100°C and resolved by Tris-glycine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (4 to 12% polyacrylamide). The gel contents were transferred onto a polyvinylidene difluoride membrane in a solution containing 48 mM Tris, 39 mM glycine, and 20% (vol/vol) methanol at 20V for 2 h. The membrane was then blocked with 5% (wt/vol) milk and probed with a 0.2-μg/ml solution of anti-hTERT peptide polyclonal antibody (23) or a 0.5-μg/ml solution of anti-FLAG polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.).
Northern analysis.
A 10-μl volume of the anti-FLAG beads from the RRL immunoprecipitation experiments was extracted once with phenol and once with chloroform-isoamyl alcohol (24:1) and then precipitated with 1/10 volume of 3 M sodium acetate and 2.5 volumes of ethanol. The RNA was subsequently electrophoresed, transferred to membrane, and probed for hTER as previously described (5, 17).
Telomerase assays.
TRAP was performed with the TRAPeze kit as per the manufacturer's (Intergen Inc., Purchase, N.Y.) instructions (30). Two different primers were used in the telomerase extension step: the TS primer (5′-AATCCGTCGAGCAGAGTT-3′) and a second primer identical to TS except that it lacked the 3′-terminal GTT nucleotides. Two microliters of the anti-FLAG beads or 4 μl of the RRL lysate was incubated with 0.1 μg of primer and assayed for telomerase activity by TRAP (25 PCR cycles). Subsequently, five microliters of each reaction product was electrophoresed on a nondenaturing 12% (wt/vol) polyacrylamide gel (29:1 [wt/wt] acrylamide-bisacrylamide) for 1 h at 800 V.
RESULTS
Physical association of distinct hTERT species in RRL.
We previously found a discrepancy between the minimal fragments of hTERT sufficient for telomerase activity in RRL and in telomerase-positive 293T cells (5) (Fig. 1A). To test the hypothesis that the hTERT truncations in 293T cells might interact with endogenous hTERT, we performed mixing experiments in RRL with a full-length active hTERT protein containing a MYC epitope and several inactive hTERT truncations that contained a FLAG epitope. If an interaction occurred, immunoprecipitation of the inactive FLAG-tagged hTERT would coprecipitate active MYC-hTERT. Indeed, immunoprecipitation of the inactive, truncated hTERT proteins (Fig. 1B) with anti-FLAG revealed that active MYC-hTERT could associate with certain inactive, truncated hTERT proteins (Fig. 1C). The truncated hTERT proteins that could associate with full-length hTERT included both N- and C-terminally truncated hTERT derivatives spanning aa 1 to 350, 351 to 1,132, 601 to 1,132, 1 to 884, and 1 to 1,087 (Fig. 1C). The interaction was specific, since hTERT proteins spanning aa 601 to 927 and aa 928 to 1,132, as well as a control anti-FLAG immunoprecipitation reaction mixture containing no FLAG-hTERT, did not coprecipitate MYC-hTERT (Fig. 1C, lanes 2, 6, and 9). While sufficient to detect telomerase elongation activity, the levels of associated MYC-hTERT were nonetheless below the level of detection by Western blotting of the anti-FLAG beads (data not shown). The inability to detect associated MYC-hTERT is consistent with our previous results suggesting that only a small percentage of hTERT is folded into an active conformation in RRL (references 5 and 6 and data not shown). We showed previously that hTERT proteins containing the single point mutation D712A or the double point mutation D868A D869A are completely inactive in vitro and in vivo (6, 23, 60). The catalytically inactive hTERT D868A D869A mutant was also able to associate with active MYC-hTERT (Fig. 1C, lane 10).
Functional complementation of distinct hTERT proteins in vitro.
We also found that inactive hTERT containing the point mutation D712A restored activity to other inactive FLAG-hTERT proteins—for example, the truncated hTERT proteins spanning aa 351 to 1,132 and aa 601 to 1,132 (Fig. 1D, lanes 4 and 5). This functional complementation was specific, since other hTERT fragments and the inactive hTERT D868A D869A mutant did not support activity when combined with the hTERT D712A mutant (Fig. 1D, lanes 3 and 6 through 10). Two other truncated hTERT proteins, spanning aa 401 to 1,132 and aa 536 to 1,132, were inactive when mixed with the hTERT D712A point mutant (data not shown). The inability of these two hTERT N-terminal truncations to functionally complement the hTERT D712A mutation is not explained by differences in telomerase RNA recognition, since all hTERT N-terminal truncations beyond aa 301 result in a comparable ability to bind the telomerase RNA in vitro (5). It is possible that a more subtle conformational alteration hinders the ability of these two truncated hTERT proteins to functionally interact with the hTERT D712A mutant.
To test whether two inactive hTERT truncations could also functionally interact, an N-terminally truncated hTERT protein that lacked the first 350 aa (351 to 1,132) was mixed with an hTERT fragment that lacked the C-terminal 205 aa (1 to 927). Each truncated protein alone was inactive, but when the two were mixed together, telomerase activity was restored (Fig. 2A, compare lanes 3, 7, and 10). In this mixture, two RT domains that share an overlapping region spanning aa 351 to 927 are present. The minimal overlap for a functional interaction between two truncated hTERT proteins was further narrowed to a region containing the first 927 aa of one hTERT protein (1 to 927) and the C-terminal 531 aa of the second hTERT protein (601 to 1,132) (Fig. 3A, lanes 6 and 7). The C-terminally deleted hTERT spanning aa 1 to 884 was not sufficient to restore telomerase activity to a truncated hTERT lacking the first 350 aa (Fig. 3A, lane 10). These results suggest that an important structural or catalytic determinant of the functional interaction may reside between aa 884 and 927 of hTERT.
We next tested whether the N terminus or C terminus of hTERT was sufficient to restore activity to hTERT mixtures containing a single RT domain (Fig. 2 and 3 and data not shown). An hTERT fragment spanning aa 928 to 1,132 did not restore telomerase activity to a truncated hTERT protein spanning aa 1 to 927 (Fig. 2A, lanes 7 to 9); however, the first 350 aa of hTERT restored telomerase activity to a truncated hTERT protein spanning aa 351 to 1,132 (Fig. 2A, lane 4). Similarly, aa 1 to 300 of hTERT restored telomerase activity to an hTERT protein spanning aa 301 to 1,132 (Fig. 3A, lanes 2 and 3). The weak but detectable activity in a mixture of truncated hTERT proteins spanning aa 1 to 300 and aa 351 to 1,132 suggests that aa 300 to 350 are not absolutely required for a functional interaction to occur (Fig. 3A, lanes 4 and 5). The absence of telomerase activity in hTERT mixtures that lacked a C-terminal or an N-terminal hTERT fragment (Fig. 3A, lanes 10 and 11) further indicates that C-terminal residues on one hTERT protein and N-terminal residues on another must be present for a functional interaction to occur. Furthermore, the lack of a functional interaction between truncated hTERT proteins spanning aa 301 to 927 and 351 to 1,132 (Fig. 3A, lane 11) suggests that aa 301 to 350 of hTERT are not sufficient to mediate a functional interaction.
Functional complementation requires the association of telomerase RNA with only one hTERT subunit.
We found that the reconstitution of telomerase activity via hTERT multimerization required the assembly of telomerase RNA with only one hTERT protein. For example, telomerase activity could be restored if the telomerase RNA was coassembled with a truncated hTERT protein spanning aa 351 to 1,132 and then mixed with the hTERT fragment spanning aa 1 to 927, in which no telomerase RNA was present (Fig. 2A, lane 12). However, activity was not restored if the telomerase RNA was instead assembled with the truncated hTERT protein spanning aa 1 to 927 (Fig. 2A, lane 11). This result could not be explained by a difference in telomerase RNA recognition, since the truncated hTERT spanning aa 1 to 927 retained the ability to bind the telomerase RNA in vitro, albeit at reduced levels relative to the aa 1 to 350 hTERT fragment (Fig. 2B) (5). As estimated by Western blotting of each hTERT truncation protein prior to mixing, the levels of different hTERT proteins were similar irrespective of whether telomerase RNA was present or absent during their synthesis (Fig. 2C).
We found that the ability to restore activity to the two hTERT proteins spanning aa 1 to 350 and aa 351 to 1,132 required the preassociation of the telomerase RNA only with the latter hTERT fragment (Fig. 2A, lanes 5 and 6). The lack of restoration of telomerase activity when the telomerase RNA was incubated with fragments spanning aa 1 to 350 or 1 to 927 also rules out the possibility that the telomerase RNA can shuttle between hTERT species in the RRL. However, it does not address the stoichiometry of telomerase RNA to each hTERT subunit, nor does it address whether one or more telomerase RNAs may form a bridge between the two hTERT subunits upon mixing.
Functional complementation of distinct hTERT proteins can occur in vivo.
To test whether multimerization of hTERT can occur in vivo, we examined the properties of exogenously introduced hTERT in an immortalized human cell line, GM847. GM847 cells express telomerase RNA hTER, but they do not express endogenous hTERT mRNA and therefore do not contain detectable telomerase activity (7) (Fig. 4A, lane 1). These cells use an alternate mechanism for telomere length maintenance (“ALT”) that is thought to involve recombination (7). GM847 cells were separately transfected with full-length FLAG-tagged hTERT (Fig. 4B, lane 2), the inactive hTERT mutant D712A (Fig. 4B, lane 3), and several truncated hTERT proteins alone (lanes 4 to 11) or in combination with the hTERT point mutant D712A (Fig. 4B, lanes 12 to 16) or the hTERT protein spanning aa 1 to 927 (Fig. 4B, lanes 17 to 20). The inability of several truncated hTERT proteins to support telomerase activity in GM847 cells paralleled results obtained in RRL (Fig. 4A, lanes 4 to 11, and data not shown), except that hTERT fragments spanning aa 201 to 1,132 and aa 301 to 1,132 were inactive in GM847 cells (compared with short elongation products in RRL) (5) (Fig. 4A, lanes 4 and 5). This difference might reflect the reduced levels of hTERT protein in transfected GM847 cells compared to RRL (data not shown). When the catalytically inactive hTERT mutant D712A was cotransfected into GM847 cells, a wild-type pattern of elongation activity was restored to several of the inactive hTERT proteins (Fig. 4A, lanes 11 to 13, and data not shown). The functional complementation was specific, since a truncated hTERT spanning aa 1 to 350 did not restore activity when cotransfected with the D712A hTERT mutant (Fig. 4A, lane 15). We also found that an hTERT protein spanning aa 1 to 927, which is inactive when mixed with the hTERT point mutant D712A in RRL (data not shown), was active in combination with the same hTERT point mutant in GM847 cells (Fig. 4A, lane 13). This difference between RRL and GM847 cells in the reconstitution potential of C-terminally truncated hTERT proteins may reflect differences in the conformation of hTERT fragments or other accessory factors in GM847 cells and RRL. In parallel with results obtained in RRL, we also observed an ability of hTERT fragments spanning aa 1 to 300 or 1 to 350, but not aa 928 to 1,132, to functionally complement other nonoverlapping fragments of hTERT (Fig. 4A, lanes 18 to 22). These data demonstrate that a similar functional interaction can occur between distinct hTERT proteins in vivo and are consistent with our previous hypothesis that certain truncated hTERT proteins are active only in telomerase-positive 293T cells because of their ability to interact with endogenous hTERT.
A role for hTERT multimerization in substrate utilization.
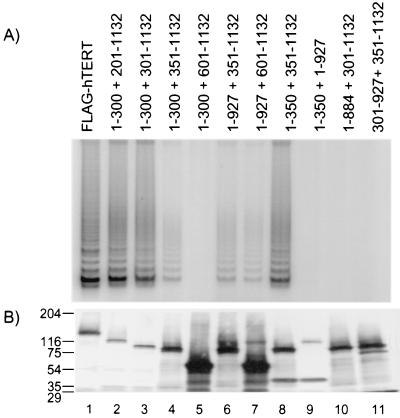
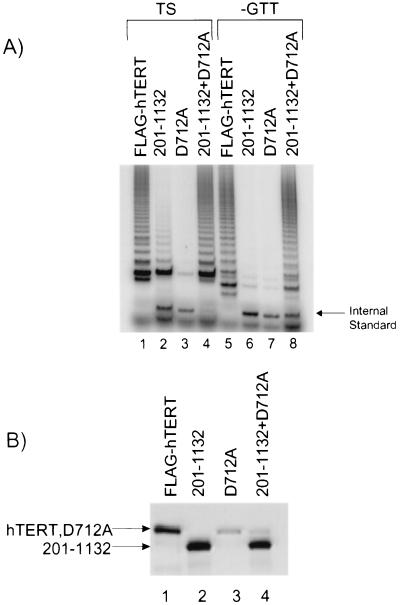
We observed previously that the telomerase activity of certain N-terminal deletion mutants of hTERT that were defective for substrate elongation in RRL could be restored to wild-type levels when introduced into telomerase-positive 293T cells (5). To test the hypothesis that hTERT multimerization may serve to promote substrate utilization and primer elongation, we mixed a truncated hTERT protein spanning aa 201 to 1,132 (which alone synthesizes predominantly short telomerase elongation products in RRL [5]) with the catalytically inactive hTERT mutant D712A (Fig. 5). When the hTERT D712A mutant was mixed with the hTERT fragment spanning aa 201 to 1,132, wild-type levels of telomerase activity were restored (Fig. 5A, compare lanes 2 and 4). In addition, the D712A mutant also restored the ability of the truncated hTERT to utilize a nontelomeric primer (Fig. 5A, compare lanes 6 and 8). These results are similar to the wild-type elongation activity that was observed upon introduction of the truncated hTERT protein spanning aa 201 to 1,132 into 293T cells (5) and is consistent with a role for hTERT multimerization in vivo in substrate recognition and elongation.
FIG. 5.
hTERT multimerization promotes elongation of telomerase substrates. (A) Anti-FLAG immunoprecipitation of RRL containing full-length hTERT (FLAG-hTERT), a truncated hTERT protein spanning aa 201 to 1,132 (201–1132), an untagged hTERT point mutant (D712A), or a mixture of the two mutants (201–1132+D712A) was analyzed for telomerase elongation activity by using a TS primer (TS) or a TS primer that lacked the 3′ GTT nucleotides (−GTT). The arrow at the right indicates the internal standard for the TRAP. (B) RRL samples were analyzed for hTERT protein levels by Western blotting with an anti-hTERT antibody. The arrows at the left indicate the positions of the hTERT mutant and the truncated hTERT protein.
DISCUSSION
hTERT multimerization occurs in trans.
Complementation is defined as the restoration of biological activity by the interaction of two or more different protein fragments (reviewed in reference 64). The phenomenon of intrasubunit (or cis) complementation has been observed with several proteins, including bovine pancreatic ribonuclease S and staphylococcal nuclease (2, 53). Evidence for cis complementation within RNA molecules has also been observed. For example, group I and group II self-splicing introns can be separated into two distinct catalytically inactive RNA chains that can then be combined to restore self-splicing activity (13, 28). The observation that nonoverlapping pieces of hTERT (aa, 1 to 350 and 350 to 1,132, aa 1 to 300 and 301 to 1,132, and aa 1 to 300 and 350 to 1,132) can restore activity is consistent with intrasubunit or cis complementation. Intrasubunit complementation of the telomerase RNA may also account for the ability of two separate, nonoverlapping fragments of hTER to form a functionally active complex in vitro (59).
In contrast to cis complementation, protein interactions in trans require the functional multimerization of at least two monomers. We have established that two mutants of hTERT that are inactive separately can reconstitute telomerase activity in a recombinant system or when introduced into a cell line that does not express endogenous hTERT. In these instances, the ability of a catalytically inactive hTERT to coprecipitate full-length active hTERT, combined with the observation that inactive, full-length hTERT can restore catalytic activity to other large, overlapping hTERT fragments, is most consistent with the ability of these hTERT subunits to functionally cooperate in trans.
Evolutionary conservation of RT multimerization.
The functional multimerization of RTs has been demonstrated in several systems. For example, human immunodeficiency virus (HIV) RT consists of a heterodimer of two polypeptides, p66 and proteolytic fragment p51, that together form a fully functional RT with one catalytic site (31, 52). The equine infectious anemia virus and Rous sarcoma virus RTs are also organized as asymmetric dimers containing one active polymerase site (55). An isolated fragment of the Moloney murine leukemia virus RT is monomeric in the absence of nucleic acid; however, upon addition of DNA, the enzyme forms an asymmetric dimer containing one active polymerase site (58). Since telomerase is most closely related to non-long terminal repeat retroposon RTs, which are thought to form a homodimer upon interaction with their target RNA and DNA (14, 63), one might anticipate functional and/or structural similarities between the telomerase RT multimer and this class of RTs (36, 46, 47).
Stoichiometry of the minimally active hTERT multimer.
The ability of the D712A hTERT mutant to support functional complementation of other truncated hTERT proteins, combined with the observation that only one hTERT subunit required preassembly with the telomerase RNA, suggests that one catalytic site within the hTERT multimer may be sufficient for catalytic activity in vitro and in vivo. One possibility consistent with our data is that the amino terminus of hTERT acts allosterically to modulate the conformation of one or more telomerase RNAs bound to a second hTERT protein containing the RT domain and C terminus. In HIV RT, for example, the p51 subunit elicits allosteric changes in the p66 active site that are essential for enzyme activation (45). However, our experimental design is set up to specifically test whether two inactive hTERT proteins can functionally restore a catalytically active multimer. Thus, our data do not rule out the possibility that endogenous hTERT dimers or multimers contain more than one active site, and our results do not contradict the two-active-site model first proposed by Prescott and Blackburn (50). The stoichiometry of the minimally active hTERT multimer is not precisely known. It remains possible that more than one telomerase RNA is present per hTERT dimer or that, in our experiments, multiple dimers form a higher-order complex in which more than one active site is present. Recently, Lingner and colleagues have noted a functional interaction between two distinct human telomerase RNAs (61). Furthermore, an N-terminal fragment of Euplotes crassus TERT is sufficient for a physical interaction with full-length E. crassus TERT (L. Wang and D. Shippen, personal communication). Arai et al. have also demonstrated that N-terminal and C-terminal fragments of hTERT can associate with one another in vitro (57; K. Masutomi and S. Murakami, personal communication). Taken together, these results are consistent with the notions that the hTERT multimer may form N-terminal and C-terminal protein-protein contacts that are independent of telomerase RNA and a that fully active telomerase multimer may contain two active sites. Niu and colleagues have proposed two models for the multimerization of S. cerevisiae telomerase: the first model suggests that there are two active sites with distinct roles in endonucleolytic cleavage and elongation, and the second model postulates that there exists a multimeric telomerase whose subunits are each able to perform both elongation and endonucleolytic cleavage (49). Additional experiments are required to address whether both catalytic sites perform the same telomere elongation function and whether they can catalyze telomere synthesis simultaneously.
Distinct regions for physical or functional hTERT multimerization.
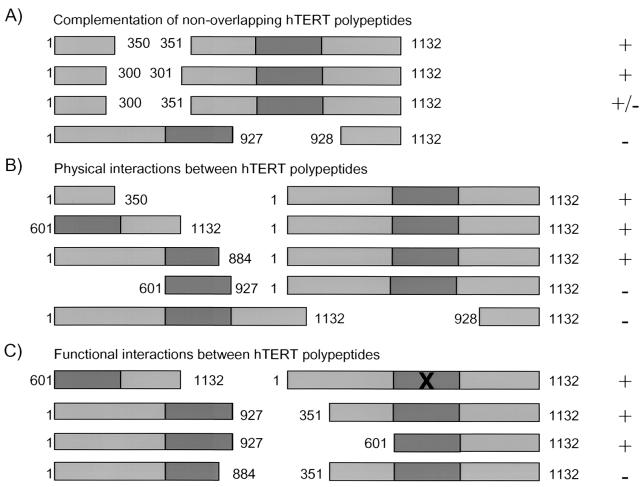
We found that the N terminus of hTERT was necessary and sufficient for an association with a second, full-length hTERT protein (Fig. 6B). However, functional multimerization of hTERT required a second hTERT protein containing an intact RT domain and C terminus (Fig. 6C). The RT domain of hTERT alone was not sufficient for a physical or functional interaction with a second hTERT protein (Fig. 1C and D, lanes 6). Since hTERT multimerization promotes the efficient elongation of different telomere substrates in vitro and in vivo, residues 1 to 350 of hTERT may facilitate primer recognition and elongation by altering the conformation of the hTERT-telomerase RNA complex. Consistent with this hypothesis, mutations within the N terminus of Est2p (S. cerevisiae TERT) can affect substrate utilization in vitro (62). Our results do not address the precise orientation of the hTERT proteins within the complex or whether the functional multimerization of hTERT occurs directly or indirectly via other factors in either the RRL or cells. For example, the foldasome protein p23, which interacts with aa 1 to 195 of hTERT in a two-hybrid assay, might participate in the assembly of an hTERT multimer in RRL and human cells (18, 25).
FIG. 6.
A schematic model for the interaction between distinct hTERT proteins. hTERT is represented by a light-gray box, the RT domain is indicated with a dark-gray box, and the X denotes the D712A point mutation. +, telomerase positive; +/−, weak but detectable telomerase activity; −, no detectable telomerase activity. (A) A summary of results obtained with nonoverlapping fragments of hTERT. (B) A summary of the minimal hTERT fragments required to coprecipitate full-length, MYC-tagged hTERT in vitro. (C) A summary of a subset of the functional interactions between distinct hTERT polypeptides.
Implications for hTERT multimerization in vivo.
Functional multimerization of hTERT may reflect an intrinsic biological role for telomerase catalysis or telomere length maintenance. In HIV RT, the association of p66 and p51 results in a protein interface that is necessary for RNase H cleavage activity (52). By analogy, hTERT multimerization might permit the formation of a unique tertiary interface that allows endonucleolytic cleavage of certain telomerase substrates (10, 39, 49). Alternatively, hTERT multimerization might affect the formation or activity of the anchor site, a G-rich DNA binding site within telomerase that is distinct from the catalytic site (10, 20, 24, 33, 36, 44). Two primer-binding sites within the hTERT multimer might also allow for increased processivity of telomerase during repeated cycles of template translocation (50, 61). The homodimerization of DNA polymerase delta is thought to facilitate the coordination of leading- and lagging-strand DNA synthesis (8). Since telomere DNA synthesis occurs in late S phase or early mitosis in S. cerevisiae (12, 38), multimerization of hTERT might serve to facilitate the coupling of DNA replication to telomere DNA elongation. The precise role of telomerase multimerization in catalysis and telomere length maintenance awaits further investigation.
ACKNOWLEDGMENTS
We thank members of the Harrington lab, B. Blencowe, H. Kha, K. Riabowol, V. Skalski, and M. Tyers for helpful discussion and comments on the manuscript; members of the Riabowol lab and J. Cruickshank for technical assistance; and J. Lingner, D. Shippen, L. Wang, K. Arai, K. Masutomi, and S. Murakami for communication of unpublished results.
T.L.B. is a research fellow of the National Cancer Institute of Canada and is funded by the Terry Fox Run. This work was funded in part by a grant to L.H. from the CIHR.
REFERENCES
- 1.Aigner S, Lingner J, Goodrich K J, Grosshans C A, Shevchenko A, Mann M, Cech T R. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 2000;19:6230–6239. doi: 10.1093/emboj/19.22.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anfinsen C B. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 3.Bachand F, Autexier C. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J Biol Chem. 1999;274:38027–38031. doi: 10.1074/jbc.274.53.38027. [DOI] [PubMed] [Google Scholar]
- 4.Bachand F, Autexier C. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol Cell Biol. 2001;21:1888–1897. doi: 10.1128/MCB.21.5.1888-1897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie T L, Zhou W, Robinson M O, Harrington L. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol Biol Cell. 2000;11:3329–3340. doi: 10.1091/mbc.11.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 7.Bryan T M, Marusic L, Bacchetti S, Namba M, Reddel R R. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum Mol Genet. 1997;6:921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]
- 8.Burgers P M. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma. 1998;107:218–227. doi: 10.1007/s004120050300. [DOI] [PubMed] [Google Scholar]
- 9.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins K, Greider C W. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 11.Collins K, Kobayashi R, Greider C W. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 12.Diede S J, Gottschling D E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 13.Doudna J A, Cech T R. Self-assembly of a group I intron active site from its component tertiary structural domains. RNA. 1995;1:36–45. [PMC free article] [PubMed] [Google Scholar]
- 14.Eickbush T H. Telomerase and retrotransposons: which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 15.Evans S K, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 16.Evans S K, Lundblad V. Positive and negative regulation of telomerase access to the telomere. J Cell Sci. 2000;113:3357–3364. doi: 10.1242/jcs.113.19.3357. [DOI] [PubMed] [Google Scholar]
- 17.Feng J, Funk W D, Wang S S, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 18.Forsythe H L, Jarvis J L, Turner J W, Elmore L W, Holt S E. Stable association of hsp90 and p23, but not hsp70, with active human telomerase. J Biol Chem. 2001;276:15571–15574. doi: 10.1074/jbc.C100055200. [DOI] [PubMed] [Google Scholar]
- 19.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 20.Hammond P W, Lively T N, Cech T R. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington L, Hull C, Crittenden J, Greider C. Gel shift and UV cross-linking analysis of Tetrahymena telomerase. J Biol Chem. 1995;270:8893–8901. doi: 10.1074/jbc.270.15.8893. [DOI] [PubMed] [Google Scholar]
- 22.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 23.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S, Mar V, Bass M B, Robinson M O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington L A, Greider C W. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- 25.Holt S E, Aisner D L, Baur J, Tesmer V M, Dy M, Ouellette M, Trager J B, Morin G B, Toft D O, Shay J W, Wright W E, White M A. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes T R, Evans S K, Weilbaecher R G, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- 27.Hughes T R, Morris D K, Salinger A, Walcott N, Nugent C I, Lundblad V. The role of the EST genes in yeast telomere replication. Ciba Found Symp. 1997;211:41–47. doi: 10.1002/9780470515433.ch4. [DOI] [PubMed] [Google Scholar]
- 28.Jarrell K A, Dietrich R C, Perlman P S. Group II intron domain 5 facilitates a trans-splicing reaction. Mol Cell Biol. 1988;8:2361–2366. doi: 10.1128/mcb.8.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilian A, Bowtell D D, Abud H E, Hime G R, Venter D J, Keese P K, Duncan E L, Reddel R R, Jefferson R A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 30.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 31.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 32.Le S, Sternglanz R, Greider C W. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol Biol Cell. 2000;11:999–1010. doi: 10.1091/mbc.11.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M S, Blackburn E H. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lingner J, Cech T R. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Snow B E, Hande M P, Baeriocher G, Kickhoefer V A, Yeung D, Wakeham A, Itie A, Siderovski D P, Lansdorp P M, Robinson M O, Harrington L. Telomerase-associated protein TEP1 is not essential for telomerase activity or telomere length maintenance in vivo. Mol Cell Biol. 2000;20:8178–8184. doi: 10.1128/mcb.20.21.8178-8184.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcand S, Brevet V, Mann C, Gilson E. Cell cycle restriction of telomere elongation. Curr Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- 39.Melek M, Greene E C, Shippen D E. Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, Bacchetti S, Haber D A, Weinberg R A. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 41.Miller C M, Collins K. The Tetrahymena p80/p95 complex is required for proper telomere length maintenance and micronuclear genome stability. Mol Cell. 2000;6:827–837. doi: 10.1016/s1097-2765(05)00078-x. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell J R, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell J R, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 44.Morin G B. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 45.Morris M C, Berducou C, Mery J, Heitz F, Divita G. The thumb domain of the P51-subunit is essential for activation of HIV reverse transcriptase. Biochemistry. 1999;38:15097–15103. doi: 10.1021/bi9914558. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura T M, Cech T R. Reversing time: origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 48.Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 49.Niu H, Xia J, Lue N F. Characterization of the interaction between the nuclease and reverse transcriptase activity of the yeast telomerase complex. Mol Cell Biol. 2000;20:6806–6815. doi: 10.1128/mcb.20.18.6806-6815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prescott J, Blackburn E H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi H, Zakian V A. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 52.Restle T, Muller B, Goody R S. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J Biol Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 53.Richards F M, Vithayahthil P J. The preparation of subtilisin-modified ribonuclease and the separation of the peptide and protein components. J Biol Chem. 1959;234:1459–1465. [PubMed] [Google Scholar]
- 54.Seto A G, Zaug A J, Sobel S G, Wolin S L, Cech T R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. . (Erratum, 402:898, 1999.) [DOI] [PubMed] [Google Scholar]
- 55.Souquet M, Restle T, Krebs R, Le Grice S F, Goody R S, Wohrl B M. Analysis of the polymerization kinetics of homodimeric EIAV p51/51 reverse transcriptase implies the formation of a polymerase active site identical to heterodimeric EIAV p66/51 reverse transcriptase. Biochemistry. 1998;37:12144–12152. doi: 10.1021/bi9731596. [DOI] [PubMed] [Google Scholar]
- 56.Steiner B R, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stewart J L, Baird K M, Farr C J. All's well that ends well. Trends Cell Biol. 2001;11:279–280. doi: 10.1016/s0962-8924(01)02029-3. [DOI] [PubMed] [Google Scholar]
- 58.Sun D, Jessen S, Liu C, Liu X, Najmudin S, Georgiadis M M. Cloning, expression, and purification of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase: crystallization of nucleic acid complexes. Protein Sci. 1998;7:1575–1582. doi: 10.1002/pro.5560070711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tesmer V M, Ford L P, Holt S E, Frank B C, Yi X, Aisner D L, Ouellette M, Shay J W, Wright W E. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol Cell Biol. 1999;19:6207–6216. doi: 10.1128/mcb.19.9.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, Taylor R D, Carlos R, Andrews W H, Wright W E, Shay J W, Harley C B, Morin G B. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 61.Wenz C, Enenkel B, Amacker M, Kelleher C, Damm K, Lingner J. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20:3526–3534. doi: 10.1093/emboj/20.13.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia J, Peng Y, Mian I S, Lue N F. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol Cell Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang J, Eickbush T H. RNA-induced changes in the activity of the endonuclease encoded by the R2 retrotransposable element. Mol Cell Biol. 1998;18:3455–3465. doi: 10.1128/mcb.18.6.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zabin I, Villarejo M R. Protein complementation. Annu Rev Biochem. 1975;44:295–313. doi: 10.1146/annurev.bi.44.070175.001455. [DOI] [PubMed] [Google Scholar]