Summary
The maintenance of healthy and resilient gut microbiota is critical for the life quality and healthspan of the elderly. Fecal microbiota transplantation (FMT) has been increasingly used to restore healthy gut microbiota. We systemically studied the establishment and resilience of transplanted microbiota after autologous versus heterologous FMT in aged recipients. Gut microbiota of aged mice (20 months old) failed to restore their original diversity and composition over 8 weeks via spontaneous recovery after antibiotics treatment; in contrast, FMT using either autologous or heterologous (2 months old from a different vendor) donors facilitated the recovery successfully, established donor-like microbiota states, and affected host gene expression profile. Furthermore, the transplanted microbiota established by heterologous FMT is not resilient during chemical-induced colonic inflammation, in contrast to that of autologous FMT. Our findings highlighted the need to monitor the long-term stability of transplanted gut microbiota and to perform multiple FMT when necessary.
Subject areas: Microbiology, Microbiome
Graphical abstract
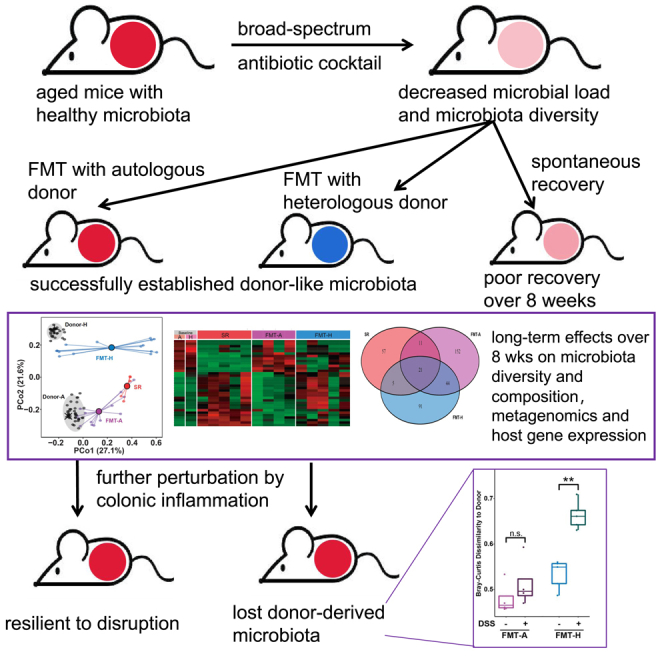
Highlights
-
•
Aged mice microbiota restores slowly after antibiotics treatment
-
•
Both autologous and heterologous FMT facilitate microbiota restoration in aged mice
-
•
FMT affects long-term homeostasis of gut metagenome and colon gene expression
-
•
Established microbiota after heterologous FMT is not resilient against colitis
Microbiology; Microbiome
Introduction
Gut microbiota is a complex and highly diverse ecosystem that maintains a symbiotic relationship with the host and regulates gastrointestinal (GI) homeostasis. The microbiota and the host establish homeostasis through lifelong interactions, resulting in such a highly compatible state that the composition of gut microbiota may serve as a biomarker of aging (Huang et al., 2020; Galkin et al., 2020). The maintenance of healthy and resilient gut microbiota is critical for the life quality and healthspan of the elderly. Dysbiosis of the gut microbiota is related to various types of GI diseases, including Clostridioides difficile infection (CDI), inflammatory bowel disease (IBD), and irritable bowel syndrome, many of which are more prevalent in the elderly (Asempa and Nicolau, 2017; Durazzo et al., 2017; Taleban et al., 2015).
Fecal microbiota transplantation (FMT), a procedure in which fecal material is transferred from a healthy donor to a patient with disrupted gut microbiota is now accepted as an effective treatment for CDI and increasingly tested for its potential to treat other GI or non-GI diseases (Zhang et al., 2018; Bakker and Nieuwdorp, 2017; Borody et al., 2013). Could FMT benefit the aged population with great clinical needs? One study showed that FMT is safe and effective in the treatment of CDI in elderly patients (Agrawal et al., 2016). However, another study of FMT for CDI patients found a higher relapse rate among elderly patients compared with younger counterparts during the 6-month period after FMT (Tseng et al., 2017). It remains elusive whether specific precautions need to be taken and how to rationally design the therapeutic strategy for the use of FMT in aged hosts.
The establishment of a diverse and balanced microbiota and its long-term stability is critical to the success of FMT treatment. Several studies have used mouse models to study the ecological changes of the microbiota following FMT. One recent study showed that autologous FMT improved the reconstitution of the post-antibiotic mucosal microbiome and gut transcriptome in young adult mice (8 weeks old) (Suez et al., 2018). Another study compared the short-term and long-term engraftment of transplanted microbiota in juvenile (3 weeks old) and adult (8 weeks old) mice, showing that donor microbiota engraftment was better in juvenile SPF mice than in adult SPF mice (Le Roy et al., 2019). These findings suggested that the establishment of the transplanted microbiota may be affected by the age of the recipients. Although several studies using mouse models have successfully performed heterologous FMT from young donor mice into aged recipient mice (Bárcena et al., 2019; Spychala et al., 2018; Stebegg et al., 2019), the long-term colonization of the transplanted microbiota is usually maintained by regularly repeating the FMT procedure (Bárcena et al., 2019) or by replenishing the cages of recipient mice with dirty bedding and fecal pellets from donor mice (Stebegg et al., 2019). To our knowledge, no previous publications have compared the long-term stability of transplanted microbiota after autologous versus heterologous FMT in the aged host.
The resilience of the newly established microbiota to further disturbance is another important criterion to evaluate the protective efficacy of FMT. For example, high rate of complications and opportunistic infections are seen in elderly patients with IBD (Lin et al., 2019; Bollegala et al., 2016), which may challenge the maintenance of transplanted microbiota. Especially in the case of heterologous FMT, when the highly compatible microbiota of the aged host is replaced by a new state with a different composition, it may be in a metastable state and thus the balance can be more easily tipped by environmental changes (Lahti et al., 2014). However, little has been studied about the resilience of transplanted microbiota to further disruption such as colitis, especially in aged hosts.
In this study, we used naturally aged mice (20 months old, considered equivalent to approximately 60 years old humans (Hagan, 2017; Dutta and Sengupta, 2016) as FMT recipients to study the establishment and resilience of transplanted gut microbiota. We treated aged mice with an antibiotic cocktail to disrupt the gut microbiota and monitored the recovery process either without FMT (spontaneous recovery, SR) or with autologous (FMT-A) or heterologous (FMT-H) FMT. Compared with the SR group which could not restore its microbiota diversity and composition by 8 weeks, both FMT-A and FMT-H facilitated the restoration of gut microbiota diversity as a donor-like microbiota established in the new host. The microbiota transplanted from different donors also had long-term effects (8 weeks after FMT) on the metagenomic gene pathways of the microbiota and the gene expression profiles of the host colon. Finally, we tested how the transplanted microbiota responded to dextran sulfate sodium (DSS) induced colitis, which caused a self-limiting inflammation in the gut. We found that in the FMT-A recipients, the transient perturbations in gut homeostasis did not affect the long-term stability of the transplanted microbiota. In contrast, the gut microbiota established after FMT-H failed to withstand the perturbations, suggesting lower resilience. Our findings suggested that the long-term stability of transplanted gut microbiota needs to be monitored, especially for aged recipients that received heterologous FMT.
Results
Spontaneous recovery of gut microbiota following antibiotic treatment in aged mice
Previous studies have shown that antibiotic treatment can disrupt the gut microbiota in young hosts, leading to a substantial decrease in microbial load (Reese et al., 2018a, 2018b) and microbiota diversity (Palleja et al., 2018; Suez et al., 2018; Ng et al., 2020). Afterward, there is a long-term restorative process, during which the gut microbial ecosystem is temporarily imbalanced and significantly distinct from the original state (Ng et al., 2020; Palleja et al., 2018; Suez et al., 2018; Dethlefsen and Relman, 2011). However, little is known about the recovery ability of gut microbiota in aged hosts.
To study the disruption of gut microbiota by antibiotics and the ensuing spontaneous recovery in aged hosts, we treated aged (20 months old) female C57BL/6J mice with oral gavage of a broad-spectrum antibiotic cocktail (ampicillin, vancomycin, neomycin, and metronidazole) at a high dose (ABH group) or low dose (ABL group) or with water (Ctrl group). Fecal samples were collected before treatment and then regularly collected until 56 days after treatment (Figure 1A). We found that antibiotic treatment led to a substantial but transient decrease in bacterial load (the total copy number of the 16S rRNA gene) in feces (Figure 1B). In contrast, antibiotic treatment had a significant and long-term (8 weeks after treatment) impact on the number of observed ASVs (Figure 1C) and the Shannon diversity index (Figure 1D) of gut microbiota.
Figure 1.
Spontaneous recovery of gut microbiota following antibiotic treatment in aged mice
(A) Aged mice were treated with an antibiotic cocktail on Day 1. Ctrl, Control; ABH, antibiotics-high dose; ABL, antibiotics-low dose; 20mo: 20 months old mice. Circles mark the time points (Day) of fecal sample collection. N = 5 for each time point (except for the ABL group on Day 21, N = 3 due to storage problems with 2 samples).
(B) Total microbial load (16S copy number log/gram) in fecal samples assayed by qPCR.
(C and D) The number of observed ASVs (C) and the Shannon diversity index (D) did not recover to the baseline level. Error bars = SEM. Colored asterisks indicate statistically significant differences at p < 0.05 between the experimental groups and the control group. ∗∗p < 0.01, ∗∗∗p < 0.001.
(E–G) Changes in the relative abundance of microbial taxa during spontaneous recovery. O, order; F, family; G, genus.
(H and I) Principal Coordinate Analysis (PCoA) based on Bray-Curtis dissimilarity, showing the trajectory of microbiota composition for the ABH (H) and ABL (I) groups. Compositional profiles were used at the lowest taxonomy level. Baseline (gray dots, average shown as the black cross) indicates the microbiota composition averaged over aged mice in the control group from Day 1 to Day 56. For each time point, the central black dot indicates the average of five mice and the colored lines connect replicate samples.
(J) Bray-Curtis dissimilarity between the gut microbiota composition on Day 56 and the baseline composition. The FDR-adjusted p values are shown between various treatment groups (two-sided Wilcoxon rank-sum test). ∗∗p < 0.01.
The alpha diversity of gut microbiota in aged mice did not recover to the baseline level, suggesting the elimination of community members during the antibiotic treatment (Ng et al., 2020). These observations are consistent with previous studies in young mice (Suez et al., 2018) and in an independent experiment that we carried out in 2 months old mice (Figure S1).
We found that the composition of gut microbiota in aged mice was disrupted by antibiotic treatment and gradually recovered to a diverse community over 8 weeks (Figures 1E–1G). The microbiota composition of the control group (i.e., no antibiotic treatment) remained stable throughout the entire period (Figure 1E), whereas the gut microbiota in the antibiotic treatment groups (ABH and ABL) changed dramatically during the first 2 weeks. We observed a clear pattern of ecological succession in the absolute abundance profiles (Figure S2): some bacterial taxa transiently flourished and dominated the microbial community and then were replaced by other taxa. Consistent with previous studies on the post-antibiotic recovery of gut microbiota in young mice and humans (Lavelle et al., 2019; Chng et al., 2020; Gibbons, 2020), we observed some “recovery-associated bacterial taxa”, such as Bacteroides and Alistipes, which increased in abundance early during recovery. After Day 14, the relative abundance of most bacterial taxa stabilized, but some taxa (e.g., Muribaculaceae) took longer to return to their baseline level. Similar to the findings in young mice ((Suez et al., 2018) and Figure S1), we found that the gut microbiota in aged mice did not return to the baseline composition by 8 weeks after the antibiotic treatment (Figures 1H and 1I). On Day 56, the gut microbiota compositions of the treatment groups (ABH, ABL) were significantly different from the baseline composition (Figure 1J).
Establishment of transplanted gut microbiota in aged mice
Because heterologous FMT is usually the only FMT option for elderly patients in clinical situations, it is essential to investigate whether FMT donors with significantly different microbiota are equally successful in facilitating microbiota restoration in aged recipients. To verify whether donor-recipient compatibility affects the outcome, two parallel groups of aged mice were given a high-dose antibiotic cocktail treatment and then received FMT from the same batch of 20 months old donor mice (FMT-A), or a group of different aged (2 months old) donor mice from a different vendor (FMT-H). We confirmed that the two groups of donor mice had very distinct microbiota (Figure S3).
The change in the number of observed ASVs (Figure 2B) and Shannon index (Figure 2C) showed that FMT substantially accelerated the recovery of gut microbiota diversity in comparison with spontaneous recovery. The gut microbiota diversity of aged mice that received FMT was fully restored by Day 21 after antibiotic treatment (Figures 2B and 2C). This is consistent with a previous study in young hosts, which showed that autologous FMT improved the post-antibiotic reconstitution of the gut microbiome in both mice and human volunteers (Suez et al., 2018).
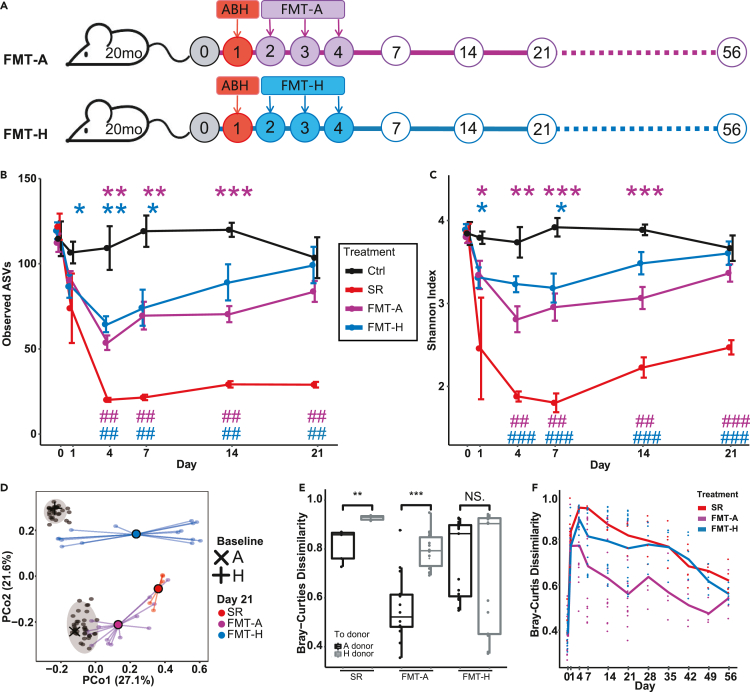
Figure 2.
Establishment of transplanted gut microbiota in aged mice
(A) Following high-dose antibiotics treatment, FMT was performed for aged mice between Day 2 and Day 4 using autologous or heterologous donors. Ctrl, Control. SR, spontaneous recovery. FMT-A, FMT using autologous donors. FMT-H, FMT using heterologous donors.
(B and C) FMT facilitates the restoration of gut microbiota diversity in aged mice. In contrast to the SR group, the number of observed ASVs (B) and Shannon diversity index (C) of the gut microbiota of the FMT groups returned to the baseline level in 3 weeks. Error bars = SEM.∗ indicates significant difference at p < 0.05 compared with the Ctrl group on the same day; # indicates significant difference at p < 0.05 compared with the SR group on the same day. ∗∗or ##, p < 0.01, ∗∗∗ or ###, p < 0.001.
(D) The post-FMT gut microbiota on Day 21 moved toward the baseline composition of the donors (A: autologous donor, H, heterologous donor). Compositional profiles were used at the lowest taxonomy level. The baseline microbiota composition is averaged over 30 mice on Day 0. The central black dot indicates the average of 5 mice (SR group) or 15 mice (FMT-A and FMT-H groups) and the colored lines connect replicate samples.
(E) Bray-Curtis dissimilarity between the post-FMT gut microbiota on Day 21 and the baseline microbiota (i.e., autologous (A) donors or heterologous (H) donors).
(F) Bray-Curtis dissimilarity between the gut microbiota of treatment groups and the baseline microbiota of aged mice throughout the long-term recovery. For the FMT-A and FMT-H groups, N = 15 until Day 21 and N = 5 thereafter. N = 5 for the Ctrl and SR groups. The FDR-adjusted p values are shown between various treatment groups (two-sided Wilcoxon rank-sum test).
On Day 21, the microbiota composition of mice that received FMT-A or FMT-H was more similar to the baseline (average microbiota composition of donors on Day 0) of their donors, respectively (Figure 2D). Interestingly, although all recipients of FMT-A showing microbiota composition closer to the baseline of autologous donors compared to heterologous donors, there was larger variation in the gut microbiota of mice in the FMT-H group, only a subgroup have microbiota close to the baseline of heterologous donors (Figure 2E). Kinetic variations of microbiota composition recovery after antibiotic treatment have been observed in previous studies in conventional and humanized young mice (Ng et al., 2020). However, our results revealed that aged mice in the FMT-A group had more consistent compositional kinetics than those in the FMT-H group. The individual differences in gut microbial composition on Day 21 between mice in both the FMT-A and FMT-H groups were not fully attributable to cage effects (Figure S4).
We demonstrated that heterologous donors were as effective as autologous donors in restoring the diversity of the microbiota, with both groups showing a significant increase in diversity when compared with the SR group. Moreover, the composition of newly established microbiota was donor dependent. We followed five recipient mice in each FMT group until Day 56 (Figure S5). For each mouse, we calculated the Bray-Curtis dissimilarity between its gut microbiota at different time points and the original baseline gut microbiota (i.e., Day 0). We found that the gut microbiota of mice in the FMT-A group was closer to the baseline of aged mice than the other two groups throughout the entire experiment (Figures 2F and S5A).
Long-term effects of FMT on the gut metagenome and colon gene expression
Previous research has shown that functional restoration of bacteriomes and viromes revealed by metagenomic sequencing could be an indicator of successful FMT (Fujimoto et al., 2021). To evaluate the long-term effects of FMT in aged mice with different donors, we performed shotgun metagenomic sequencing of Day-56 fecal samples from each treatment group (SR, FMT-A, and FMT-H) together with the baseline of 20 months old mice (also used as autologous donors or AD) and heterologous donors (HD). We found that 35 out of 219 functional pathways were differentially abundant between the SR group and AD (Figure 3A). We then examined the levels of these pathways in the FMT-A and FMT-H groups after 56 days of recovery. We found that most of the 35 pathways that did not spontaneously recover to baseline levels were restored in the FMT-A group. In contrast, FMT-H was not as effective in facilitating recovery of microbiota function. When all 219 functional pathways were taken into consideration, endpoint samples of both the FMT-A and FMT-H groups were closer to the baseline samples compared with samples from the SR group on Day 56 (Figure S6).
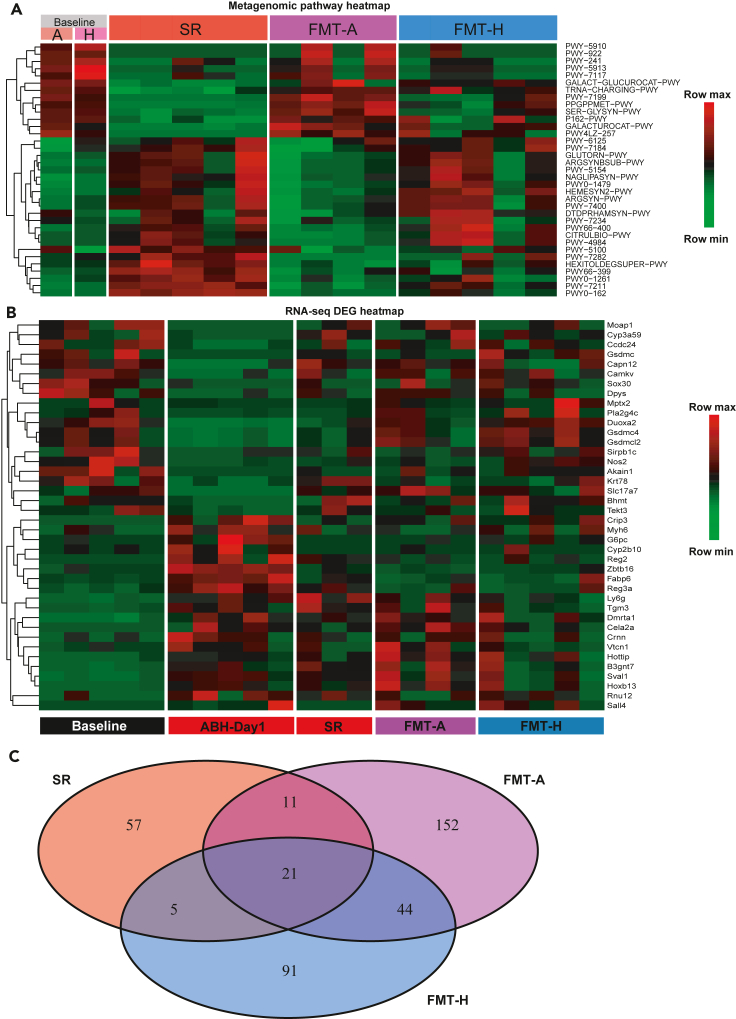
Figure 3.
Long-term effects of FMT on the gut metagenome and colon gene expression in aged mice
(A) Heatmap of metagenomic gene pathways that were differentially abundant (FDR-corrected p < 0.01) between the SR group on Day 56 and the baseline of aged mice. SR, spontaneous recovery. FMT-A, FMT using autologous donors. FMT-H, FMT using heterologous donors. SR/FMT-A/FMT-H samples were collected on Day 56. Baseline sample (autologous (A) or heterologous (H)) represents the average of 5 mice each, collected on Day 0. For the other groups, each column represents one mouse. The FMT-A group only has 4 mice because of the death of a mouse.
(B) Heatmap of differentially expressed genes (DEGs) between ABH-Day1 and Baseline, based on RNA-seq of colon samples. Fold change >2, p < 0.05, only the top 40 genes were presented. Baseline, aged mice on Day 0. ABH-Day1, samples collected on Day 1 after high-dose antibiotic treatment. SR/FMT-A/FMT-H samples were collected on Day 56. Each column represents one mouse. The FMT-A group only has 4 mice because of the death of a mouse. The SR group only has 3 mice because one mouse was euthanized because of tumor and RNA sample from another mouse had poor quality.
(C) Venn diagram showing the DEGs (fold change >2, P < 0.05) between the baseline and treatment groups. Most of the DEGs were present in only one group. Shared DEGs were rare.
The shift in the transcriptome of the host following antibiotics treatment can be dramatic (Suez et al., 2018; Lavelle et al., 2019; Morgun et al., 2015), and the response is determined by the combined effects of microbiota depletion and the remaining antibiotic-resistant microbes as well as the direct effects of antibiotics on host tissues (Morgun et al., 2015). It was shown that autologous FMT in young mice helped restore the gene expression profile across GI tracts to some extent (Suez et al., 2018). However, the effects of FMT donor on the transcriptional profiles of aged hosts remain unknown. We performed RNA sequencing (RNA-seq) of upper colon samples of aged mice before treatment (Baseline), Day 1 after antibiotics (ABH-Day1), and at the endpoint (Day 56) for the three treatment groups (SR, FMT-A, and FMT-H). We first examined the restoration of differentially expressed genes (DEGs) between ABH-Day1 and the Baseline on Day 56 in the SR, FMT-A, and FMT-H groups (Figure 3B). More genes that were affected by antibiotics treatment were reverted to the Baseline configuration in the FMT groups than in the SR group, suggesting that FMT facilitated the restoration of the host transcriptome, which is consistent with previous reports in young mice and human donors (Suez et al., 2018). We then examined the DEGs between each group and the Baseline and the overlap among them (Figures 3C and S7). Interestingly, a large fraction of DEGs were not shared between the different treatment groups (SR, FMT-A, and FMT-H), which demonstrated that the transcriptional landscapes on Day 56 were highly dependent on FMT and the choice of donor. We also found that the FMT from autologous donors did not outperform that from heterologous donors in the reversion of the transcriptional landscape (the number of total DEGs compared with the Baseline was 228 in FMT-A and 161 in FMT-H).
The resilience of transplanted gut microbiota against colon inflammation
Whether the transplanted microbiota is resilient against perturbations to gut homeostasis reflects its long-term stability. We utilized a self-limiting colon inflammation model induced by administration of 3% dextran sulfate sodium (DSS) in the drinking water of the mice for 7 days, which is widely used to induce acute colitis in rodents (Ohkusa, 1985; Wirtz et al., 2017). Previous studies had shown that DSS-induced-colitis caused changes in the gut microbiota composition, which were spontaneously restored to the original state after the end of DSS treatment (Munyaka et al., 2016; Osaka et al., 2017; De Fazio et al., 2014).
In the sections above we showed that the microbiota diversity returned to baseline levels on Day 21 in the FMT-A and FMT-H groups (Figures 2B and 2C). In addition, microbial compositions largely stabilized after Day 21 (Figure S5). Hence, we chose Day 26 as the starting point for DSS treatment. From Day 26 to Day 31 after antibiotic treatment, aged recipients of FMT-A or FMT-H were given 3% DSS treatment for seven consecutive days and then switched back to normal drinking water for 42 days (Figure 4A). Aged recipient mice of FMT-A or FMT-H showed typical symptoms of acute colitis such as weight loss (Figure S8A), diarrhea and blood in feces (Figure S8B). These disease symptoms slowly resolved after the mice were switched back to normal drinking water, similar to observations in young and non-transplanted mice (Melgar et al., 2005; Tang et al., 2018; Cao et al., 2018).
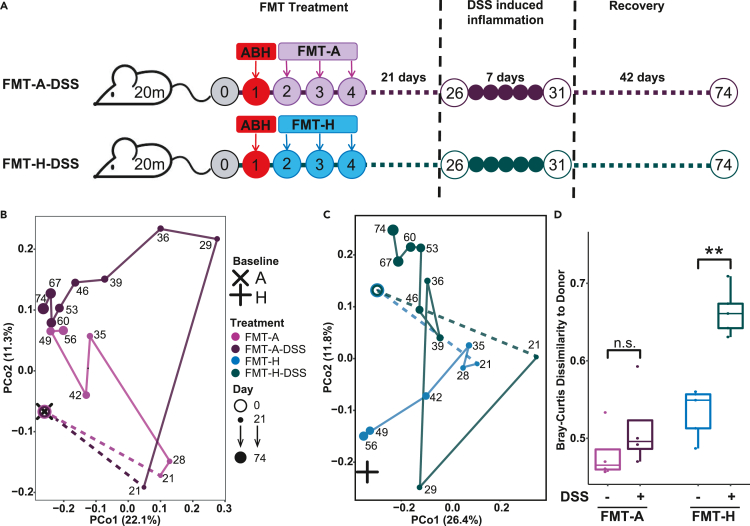
Figure 4.
The resilience of transplanted gut microbiota against colon inflammation in aged mice
(A) Three weeks after FMT treatment (Day 26), aged mice were treated with DSS for 7 days to induce acute colon inflammation. Fecal samples were collected every week until Day 74.
(B and C) PCoA analysis based on Bray-Curtis dissimilarity of microbiota composition at different time points. The comparison between the FMT-A group (FMT from autologous donor, without DSS perturbation) and FMT-A-DSS group (FMT from autologous donor, with DSS perturbation) are shown in (B). Similarly, the comparison between the FMT-H and FMT-H-DSS groups is shown in (C). Compositional profiles were used at the lowest taxonomy level. The baseline microbiota composition is averaged over 30 donor mice, respectively, collected on Day 0. For each group, the dot is the average of five mice (N = 5) at each time point.
(D) Bray-Curtis dissimilarity between the final microbiota composition of treatment groups and the baseline microbiota of the respective FMT donors (autologous baseline: FMT-A and FMT-A-DSS; heterologous baseline: FMT-H and FMT-H-DSS). Central line, median; box limits, first and third quartile; whiskers, 1.5 X interquartile range. The FDR-adjusted p values are shown between linked treatment groups (two-sided Wilcoxon rank-sum test). ∗∗ indicates significant difference at p < 0.01.
PCoA based on Bray-Curtis dissimilarity was used to compare the changes in microbiota composition in FMT recipients with or without DSS treatment (Figures 4B and 4C). For mice in the FMT-A-DSS group, the microbiota restoration process was only briefly disrupted by the induced inflammation; the microbiota composition after Day 53 was similar to that of FMT-A group (without DSS treatment) (Figure 4B). At the endpoint of the experiments, the microbiota composition of FMT-A mice and that of FMT-A-DSS mice showed no significant difference in the distance to that of their aged donors (Figure 4D).
In contrast, for mice in the FMT-H-DSS group, the microbiota composition after DSS treatment gradually shifted toward the original baseline of aged mice (Figure 4C). At the endpoint, compared with the FMT-H mice (without DSS treatment), the microbiota composition of FMT-H-DSS mice was significantly further from the baseline of its young donors (Figure 4D). These observations suggested that under the conditions of colon inflammation, the transplanted gut microbiota from heterologous donors may not be as stable as that from autologous donors.
Discussion
Heterologous FMT is a widely used gut microbiome intervention in clinical settings. Most research so far has focused on the efficacy and safety of FMT (Tabbaa et al., 2018; Ding et al., 2019; Xiao et al., 2020; Baruch et al., 2021; Davar et al., 2021). Several studies using animal models have reported that heterochronic FMT in aged hosts could result in the transfer of some physiological properties from the donor to the recipient, including lifespan and healthspan (Smith et al., 2017; Bárcena et al., 2019), the germinal center reaction (Stebegg et al., 2019), and neurogenesis and behavior (D'Amato et al., 2020; Kundu et al., 2019). However, less attention has been paid to the “pharmacokinetics” of FMT. Although FMT has become the most effective treatment for recurrent CDI, it was reported that about 20% of patients still experience recurrent CDI after the initial FMT therapy (Kelly and LaMont, 2008; Mamo et al., 2018). Previous studies investigating FMT treatment for IBD patients also suggested that multiple FMT procedures are needed for better clinical outcomes and that the timing of the subsequent FMT is critical in determining the treatment effects (He et al., 2017; Li et al., 2019). One study of ulcerative colitis patients reported that long-term post-FMT maintenance was significantly affected by the age difference between donors and patients (Okahara et al., 2020). Taken together, these results suggest that monitoring the long-term effects of FMT and identifying the determinants of relapse after FMT is critical in making rational clinical decisions.
In this study, we focused on the establishment and resilience of transplanted gut microbiota from autologous versus heterologous donors by following the post-FMT dynamics in aged mice for 8 weeks. One intriguing observation was that although a new state of microbiota composition could successfully establish in the aged hosts, the microbiota transplanted from heterologous donors could not resist the perturbation caused by colitis, in contrast to autologous donors. Our findings suggest that the choice of the donor may affect the resilience of gut microbiota after FMT.
Another interesting finding in our current study is that FMT from autologous or heterologous donors significantly reshapes the gene expression profiles of the recipient mice and the effect could extend to as long as 8 weeks after FMT. FMT caused dramatic and complex changes to the interaction within the microbiota and between the microbiota and the host. Transplanted microbiota stimulates physiological changes in the host, like immune responses and metabolism (Littmann et al., 2021; Chu and Mazmanian, 2013; Zheng et al., 2020), which may in turn affects the successful establishment of the transplanted microbiota and its resilience. For example, innate immune surveillance systems coevolve with the commensal microbiota-derived Toll-like receptors and Nucleotide oligomerization domain ligands, resulting in highly selective recognition and reactivity pattern in response to invasion by foreign species (Chu and Mazmanian, 2013; Zheng et al., 2020). Further animal and clinical studies are definitely needed to validate and understand the influences of hosts on the resilience of microbiota.
In summary, our present investigation sheds light on the establishment and long-term dynamics of the transplanted microbiota in aged hosts. The resilience of the transplanted gut microbiota needs to be monitored and multiple FMT may be necessary in the case of heterologous FMT.
Limitations of the study
In our current investigation, we used heterologous donors with both age and vendor differences from the autologous donors. The effect of FMT using age-matched donors from different vendors or diverse aged donors from the same vendor may be helpful to further address the sensitivity of FMT “pharmacokinetics” in aged recipients. Our findings highlight the need of testing the “donor-recipient compatibility” and how it affects the long-term stability and resilience of the microbiota after FMT by further studies.
The mechanism that underlies the differences of resilience between transplanted microbiota from heterologous and autologous donors is beyond the scope of our present investigation and remains to be further studied.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Ampicillin sodium | Sangon Biotech | Cat#A610028 CAS: 69-52-3 |
| Vancomycin hydrochloride | Sangon Biotech | Cat#A600983 CAS: 404-93-9 |
| Neomycin trisulfate salt hydrate | Sangon Biotech | Cat#A610366 CAS:1405-10-3 |
| Metronidazole | Sangon Biotech | Cat#A600633 CAS : 443-48-1 |
| Dextran sulfate sodium salt, colitis grade (36,000 - 50,000) | MP biomedicals | Cat#160110 CAS: 9011-18-1 |
| Critical commercial assays | ||
| QIAamp PowerFecal Pro DNA Kit | Qiagen | Cat#:51804 |
| E.Z.N.A. Gel Extraction Kit | Omega Bio-tek | Cat#:D2500 |
| NEBNext Ultra DNA Library Prep Kit for Illumina | New England Biolabs | Cat#E7370L |
| NEBNext Ultra RNA Library Prep Kit for Illumina | New England Biolabs | Cat#E7530L |
| TruSeq PE Cluster Kit v3-cBot-HS | Illumina | PE-401-3001 |
| Deposited data | ||
| Sequencing data | this paper | NCBI Sequencing Read Archive (BioProject accession: PRJNA716074) |
| SILVA_138_SSURef_Nr99 | SILVA database | https://www.arb-silva.de/documentation/release-138/ |
| Mouse reference genome C57BL_6NJ_v1 | NCBI | GenBank assembly accession: GCA_001632555.1 |
| Mouse reference genome GRCm38.p6 | NCBI | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.26/ |
| Genecode annotation : GRCm38.p6.genome, gencode.vM25.annotationgtf | Genecode | https://www.gencodegenes.org/mouse/release_M25.html |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6JNifdc | The Vital River Laboratories | Strain ID:219 |
| Mouse: C57BL/6J | The Model Organisms Center, Inc | Identified by strain name |
| Oligonucleotides | ||
| 16S rRNA gene primer 27F AGAGTTTGATCCTGGCTCAG | probeBase | www.microbial-ecology.net/probebase/ |
| 16S rRNA gene primer 1492R TACGGTTACCTTGTTACGACTT | probeBase | www.microbial-ecology.net/probebase/ |
| 16S rRNA gene primer 764F CAAACAGGATTAGATACCC | probeBase | www.microbial-ecology.net/probebase/ |
| 16S rRNA gene primer 907R CCGTCAATTCCTTTRAGTTT | probeBase | www.microbial-ecology.net/probebase/ |
| 16S rRNA gene V3-V4 region primer 338F ACTCCTACGGGAGGCAGCA | probeBase | www.microbial-ecology.net/probebase/ |
| 16S rRNA gene V3-V4 region primer 806R GGACTACCAGGGTATCTAAT | probeBase | www.microbial-ecology.net/probebase/ |
| Software and algorithms | ||
| original code | This paper | https://github.com/LD-Lab/FMT-project |
| QIIME2 (version 2019.7) | Bolyen et al. (2019) | https://docs.qiime2.org/2019.7/ |
| Cutadapt (via q2-cutadapt) | Martin (2011) | https://cutadapt.readthedocs.io/en/stable/ |
| DADA2 (via q2-dada2) | Callahan et al. (2016) | https://www.bioconductor.org/packages/release/bioc/html/dada2.html |
| fasttree2 (via q2-phylogeny) | Price et al. (2010) | http://www.microbesonline.org/fasttree/#Install |
| Vegan package in R | Dixon (2003) | https://CRAN.R-project.org/package=vegan |
| LEfSe | Segata et al. (2011) | http://huttenhower.sph.harvard.edu/lefse/ |
| Trimmomatic v.0.39 | Bolger et al. (2014) | http://www.usadellab.org/cms/index.php?page=trimmomatic |
| Bowtie2 | Langmead and Salzberg (2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Kraken2 v2.0.9-beta | Wood et al. (2019) | https://bear-apps.bham.ac.uk/applications/2019b/Kraken2/2.0.9-beta-gompi-2019b-Perl-5.30.0/ |
| HUMAnN2 | Franzosa et al. (2018) | https://huttenhower.sph.harvard.edu/humann2/ |
| STAR v2.7.4a | Dobin et al. (2013) | https://github.com/alexdobin/STAR |
| featureCounts (Subread-2.0.0) | Liao et al. (2014) | http://subread.sourceforge.net/ |
| naïve_bayes classifier (via q2-feature-classifier) | Bokulich et al. (2018) | https://github.com/qiime2/q2-feature-classifier |
| DESeq2 | Love et al. (2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| R ggplot2 package | Wickham (2016) | https://bioconductor.org/help/search/index.html?q=ggplot2+/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lei Dai (lei.dai@siat.ac.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
16 months old C57BL/6J mice were purchased from The Model Organisms Center, Inc. (Shanghai, CN) and naturally aged to 20 months old in specific pathogen-free vivarium at the Shenzhen Institutes of Advanced Technology (SIAT). 2 months old C57BL/6J mice were purchased from The Vital River Laboratories (Beijing, CN). All mice used in the present study were females. The mice were housed based on age before any treatments. After antibiotics and FMT treatment, mice receiving the same treatment were housed together. All animal experiments were approved by the Institutional Animal Care and Use Committee at SIAT.
Method details
Fecal and tissue collection from mice
Fecal pellets were collected and snap-frozen in liquid nitrogen and transferred to storage at -80C. Upon the termination of experiments, mice were sacrificed by isoflurane inhalation. The upper colon (between 1/4 to 1/2 of the total length from beneath the cecum to the rectal) was dissected and stored in liquid nitrogen to be used for RNA sequencing.
Antibiotic treatment
High-dose antibiotics cocktail (ABH) was prepared in sterile ddH2O at the following concentration: ampicillin (1 mg/mL), vancomycin (5 mg/mL), neomycin trisulfate (10 mg/mL), metronidazole (10 mg/mL). This antibiotics cocktail combines β-lactam (ampicillin), glycopeptide (vancomycin), aminoglycoside (neomycin) and nitroimidazole (metronidazole) antibiotics and was proved sufficient to decrease mouse gut microbial load within hours (Reese et al., 2018a, 2018b) and target against the full spectrum of bacteria including both Gram positive (ampicillin and vancomycin) and Gram negative (ampicillin and neomycin) strains. All antibiotics purchased form Sangon biotec, Shanghai. Low-dose antibiotics cocktail (ABL) was prepared by 10-fold dilution of ABH solution in ddH2O. Antibiotics cocktails were freshly prepared on the day of treatment and was administered by one-time oral gavage (250μL per mice) on Day 0 after the collection of fecal samples.
Fecal microbiota transplantation
Fecal pellets were collected from autologous donor mice (20 months old, Shanghai) or heterologous donor mice (2 months old, Beijing). Fecal pellets were pooled, mixed, and homogenized in PBS at concentration of 1g feces/10mL PBS (D'Amato et al., 2020; Lai et al., 2018). Mixture was centrifuged at 500 rpm for 5 minutes at 4°C and the supernatant were collected and used for FMT. Each aged recipient mouse received 150ul of the supernatant by oral gavage once a day continuously for 3 days. For FMT with autologous donors (FMT-A), the same group of mice were used as donor and recipients by collecting feces samples before antibiotics treatment to prepare FMT supernatant.
DSS-induced colon inflammation
3% w/v dextran sulfate sodium (DSS) solution was prepared in autoclaved ddH2O and used to replace normal drinking water for 7 days. Mice had free access to water and the amount of water consumption was monitored to confirm that there was no difference among treatment groups. DSS solution was freshly made and changed for the mouse cages every 2-3 days. Weight of each mouse were measured at fixed time (9 am) every day. Disease activity index (DAI) were calculated as a combined score of weight loss (0 or positive=0, 0.1-5%=1, 5-10%=2, 10-15%=3, >15%=4), stool consistency (normal=0, partially loose=1, loose stool=2, mild diarrhea=3, diarrhea=4), and rectal bleeding (normal=0, dark stool with no fresh blood=1, fresh blood seen in <50% pallets =2, blood regularly seen= 3, active bleeding= 4).
Bacterial DNA extraction and qPCR
DNA was isolated from fecal samples using the QIAamp PowerFecal DNA Kit (Qiagen) according to manufacturer's instructions. Using DNA extracted from fecal samples of 2 months old mice, the 16S rRNA gene was amplified by primers 27F 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1492R 5′-TACGGTTACCTTGTTACGACTT-3′ with PrimeSTAR® Max DNA Polymerase and purified by gel extraction. The purified 16S rRNA gene is used as a standard in qPCR. The copy number of 16S rRNA gene in fecal samples was determined by qPCR using the TB Green Premix Ex Taq II (Tli RNaseH Plus) with primers 764F 5′-CAAACAGGATTAGATACCC-3′ and 907R 5′-CCGTCAATTCCTTTRAGTTT-3′.
16S rRNA amplicon and metagenomic sequencing
V3-V4 region of 16S rRNA gene was amplified using primers 338F 5′-ACTCCTACGGGAGGCAGCA-3′ and 806R 5′-GGACTACCAGGGTATCTAAT-3′ with 12bp barcode. PCR products were mixed in equidensity ratios. Then, mixture PCR products were purified with EZNA Gel Extraction Kit (Omega, USA). Sequencing libraries were generated using NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs, USA) and sequenced on an Illumina Hiseq2500 platform (250 bp paired-end reads), conducted by Guangdong Magigene Biotechnology Co.,Ltd. (Guangzhou, China). The average sequencing depth of each sample was around 70,000 reads. For metagenomic sequencing, DNA extracted from fecal samples (100 ng) was sheared with a Covaris E220X sonicator. Sequencing libraries were generated using NEB Next Ultra DNA Library Prep Kit for Illumina (New England Biolabs, USA) and sequenced on an Illumina Hiseq X-ten platform (150 bp paired-end reads), conducted by Guangdong Magigene Biotechnology Co.,Ltd. (Guangzhou, China).
RNA-seq of colon samples
Approximately 50 mg were used for RNA isolation. Frozen colon tissues were homogenized, and RNA was isolated using TRIzol Reagent (Invitrogen). A total amount of 1 μg RNA per sample was used as input material for the library preparations. Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina® (NEB, USA). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina). After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated. RNA extraction, library generation and sequencing were conducted by Novogene Co., Ltd. (Tianjin, China).
Analysis of 16S rRNA amplicon sequencing data
16S sequencing data were analyzed by QIIME2 (version 2019.7) (Bolyen et al., 2019). Primers of the raw sequence data were cut with Cutadapt (via q2-cutadapt) (Martin, 2011), followed by denoising, merging and removing chimera with DADA2 (via q2-dada2) (Callahan et al., 2016). All amplicon sequence variants (ASVs) from DADA2 were used to construct a phylogenic tree with fasttree2 (via q2-phylogeny) (Price et al., 2010). The ASVs were assigned to taxonomy with naïve_bayes classifier (via q2-feature-classifier) (Bokulich et al., 2018) against the SILVA database (SILVA_138_SSURef_Nr99). The ASV table was normalized by the sample with the fewest sequence reads, and rare ASVs (<0.1% in relative abundance) were filtered out. Alpha diversity was calculated by two measures, the Observed species based on the number of ASV and the Shannon diversity index per sample. Bray-Curtis dissimilarity and weighted unifrac distance was calculated by the Vegan package in R (https://CRAN.R-project.org/package=vegan) (Dixon, 2003). In stacking column plots, relative abundance was calculated to the lowest taxonomy level. If some reads from the same family were annotated at genus level, calculating these reads into family level. LEfSe (Segata et al., 2011) was used to identify microbial taxa with differential abundant across groups.
Analysis of metagenomics sequencing data
Raw reads were first trimmed using Trimmomatic v.0.39 (parameter setting: SLIDINGWINDOW:4:20, MINLEN:50) (Bolger et al., 2014), and then reads mapped to the host genome (mouse_C57BL_6NJ) or PhiX were filtered out using Bowtie2 (parameter setting: -very-sensitive-dovetail) (Langmead and Salzberg, 2012). Kraken2 v2.0.9-beta (with default parameters) was used to obtain the taxonomic classification of reads (Wood et al., 2019). HUMAnN2 (with default parameters) was used to profile functional gene pathways with MetaCyc annotations (Franzosa et al., 2018).
Analysis of RNA-seq data
Raw reads were trimmed using Trimmomatic v.0.39 (parameter setting: SLIDINGWINDOW:4:20, MINLEN:50) and mapped to GRCm38 genome using STAR v2.7.4a (default parameters) (Dobin et al., 2013). Gene-level read counts were generated with featureCounts (Subread-2.0.0) (Liao et al., 2014) and Genecode annotation (GRCm38.p6.genome, gencode.vM25.annotationgtf). Genes with a minimum of 5 reads in at least one sample were included for downstream analysis. Normalization of the gene counts and differential expression analysis was performed by DESeq2 (Love et al., 2014) to find differentially expressed genes. Statistical significance was assessed using a negative binomial Wald test, and then corrected for multiple hypothesis testing with the Benjamini–Hochberg method.
Quantification and statistical analysis
Statistical analysis was performed in R (version 3.6.1). Statistically significant findings were marked according to the following cutoffs: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. Data were plotted with R ggplot2 package (Wickham, 2016). To account for multiple comparisons at each day, we considered a false discovery rate (FDR)-adjusted P value. Sample size for each experiment and the statistical tests used are specified in figure legends.
Acknowledgments
We thank members of the LD lab for insightful discussions. L.D. is supported by the National Key R&D Program of China (2019YFA09006700) and National Natural Science Foundation of China (31971513). Y.W. is supported by National Natural Science Foundation of China (31900839) and China Postdoctoral Science Foundation (2020M672880). Y-Y Liu is supported by grants R01AI141529, R01HD093761, R01AG067744, UH3OD023268, U19AI095219, and U01HL089856 from National Institutes of Health, USA.
Author contributions
Y. Wang and L. Dai designed the research. Y. Wang, J. Tang, Q. Lv, X. Dong and H. Liu performed the experiments. J. Tang, Y. Wang, Y. Tan, X. Dong, N. Zhao, Y-Y Liu and L. Dai analyzed the data. J. Tang and Y. Wang made the figures. Y. Wang, Y-Y Liu and L. Dai wrote the manuscript. All authors critically revised and approved the final version.
Declaration of interests
L. Dai received financial support from Xbiome and China Mengniu Dairy.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103654.
Supplemental information
Data and code availability
-
•
All sequencing data have been deposited at NCBI Sequencing Read Archive and are publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
All original code has been deposited at Github and is publicly available as of the date of publication. The link is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Agrawal M., Aroniadis O.C., Brandt L.J., Kelly C., Freeman S., Surawicz C., Broussard E., Stollman N., Giovanelli A., Smith B., et al. The long-term efficacy and safety of fecal microbiota transplant for recurrent, severe, and complicated Clostridium difficile infection in 146 elderly individuals. J. Clin. Gastroenterol. 2016;50:403–407. doi: 10.1097/MCG.0000000000000410. [DOI] [PubMed] [Google Scholar]
- Asempa T.E., Nicolau D.P. Clostridium difficile infection in the elderly: an update on management. Clin. Interv. Aging. 2017;12:1799–1809. doi: 10.2147/CIA.S149089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker G.J., Nieuwdorp M. Fecal microbiota transplantation: therapeutic potential for a multitude of diseases beyond Clostridium difficile. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.bad-0008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárcena C., Valdés-Mas R., Mayoral P., Garabaya C., Durand S., Rodríguez F., Fernández-García M.T., Salazar N., Nogacka A.M., Garatachea N., et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med. 2019;25:1234–1242. doi: 10.1038/s41591-019-0504-5. [DOI] [PubMed] [Google Scholar]
- Baruch E.N., Youngster I., Ben-Betzalel G., Ortenberg R., Lahat A., Katz L., Adler K., Dick-Necula D., Raskin S., Bloch N., et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., Huttley G.A., Gregory Caporaso J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollegala N., Jackson T.D., Nguyen G.C. Increased postoperative mortality and complications among elderly patients with inflammatory bowel diseases: an analysis of the national surgical quality improvement program cohort. Clin. Gastroenterol. Hepatol. 2016;14:1274–1281. doi: 10.1016/j.cgh.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borody T.J., Paramsothy S., Agrawal G. Fecal microbiota transplantation: indications, methods, evidence, and future directions. Curr. Gastroenterol. Rep. 2013;15:337. doi: 10.1007/s11894-013-0337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J., Mcmurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Wang K., Li Z., Tao F., Xu Y., Lan J., Chen G., Yang C. Bacillus amyloliquefaciens ameliorates dextran sulfate sodium-induced colitis by improving gut microbial dysbiosis in mice model. Front. Microbiol. 2018;9:3260. doi: 10.3389/fmicb.2018.03260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng K.R., Ghosh T.S., Tan Y.H., Nandi T., Lee I.R., NG A.H.Q., Li C., Ravikrishnan A., Lim K.M., Lye D., et al. Metagenome-wide association analysis identifies microbial determinants of post-antibiotic ecological recovery in the gut. Nat. Ecol. Evol. 2020;4:1256–1267. doi: 10.1038/s41559-020-1236-0. [DOI] [PubMed] [Google Scholar]
- Chu H., Mazmanian S.K. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 2013;14:668–675. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amato A., Di Cesare Mannelli L., Lucarini E., Man A.L., Le Gall G., Branca J.J.V., Ghelardini C., Amedei A., Bertelli E., Regoli M., et al. Faecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity- and neurotransmission-related proteins in young recipients. Microbiome. 2020;8:140. doi: 10.1186/s40168-020-00914-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davar D., Dzutsev A.K., Mcculloch J.A., Rodrigues R.R., Chauvin J.M., Morrison R.M., Deblasio R.N., Menna C., Ding Q., Pagliano O., et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio L., Cavazza E., Spisni E., Strillacci A., Centanni M., Candela M., Praticò C., Campieri M., Ricci C., Valerii M.C. Longitudinal analysis of inflammation and microbiota dynamics in a model of mild chronic dextran sulfate sodium-induced colitis in mice. World J. Gastroenterol. 2014;20:2051–2061. doi: 10.3748/wjg.v20.i8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., Relman D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U S A. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Li Q., Li P., Zhang T., Cui B., Ji G., Lu X., Zhang F. Long-term safety and efficacy of fecal microbiota transplant in active ulcerative colitis. Drug Saf. 2019;42:869–880. doi: 10.1007/s40264-019-00809-2. [DOI] [PubMed] [Google Scholar]
- Dixon P. Vegan, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazzo M., Campion D., Fagoonee S., Pellicano R. Gastrointestinal tract disorders in the elderly. Minerva Med. 2017;108:575–591. doi: 10.23736/S0026-4806.17.05417-9. [DOI] [PubMed] [Google Scholar]
- Dutta S., Sengupta P. Men and mice: relating their ages. Life Sci. 2016;152:244–248. doi: 10.1016/j.lfs.2015.10.025. [DOI] [PubMed] [Google Scholar]
- Franzosa E.A., Mciver L.J., Rahnavard G., Thompson L.R., Schirmer M., Weingart G., Lipson K.S., Knight R., Caporaso J.G., Segata N., et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods. 2018;15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K., Kimura Y., Allegretti J.R., Yamamoto M., Zhang Y.Z., Katayama K., Tremmel G., Kawaguchi Y., Shimohigoshi M., Hayashi T., et al. Functional restoration of bacteriomes and viromes by fecal microbiota transplantation. Gastroenterology. 2021;160:2089–2102.e12. doi: 10.1053/j.gastro.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin F., Mamoshina P., Aliper A., Putin E., Moskalev V., Gladyshev V.N., Zhavoronkov A. Human gut microbiome aging clock based on taxonomic profiling and deep learning. iScience. 2020;23:101199. doi: 10.1016/j.isci.2020.101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons S.M. Keystone taxa indispensable for microbiome recovery. Nat. Microbiol. 2020;5:1067–1068. doi: 10.1038/s41564-020-0783-0. [DOI] [PubMed] [Google Scholar]
- Hagan C. The Jackson Laboratory; 2017. When Are Mice Considered Old? [Online]https://www.jax.org/news-and-insights/jax-blog/2017/november/when-are-mice-considered-old# [Google Scholar]
- He Z., Li P., Zhu J., Cui B., Xu L., Xiang J., Zhang T., Long C., Huang G., Ji G., et al. Multiple fresh fecal microbiota transplants induces and maintains clinical remission in Crohn's disease complicated with inflammatory mass. Sci. Rep. 2017;7:4753. doi: 10.1038/s41598-017-04984-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Haiminen N., Carrieri A.P., Hu R., Jiang L., Parida L., Russell B., Allaband C., Zarrinpar A., Vázquez-Baeza Y., et al. Human skin, oral, and gut microbiomes predict chronological age. mSystems. 2020;5 doi: 10.1128/mSystems.00630-19. e00630–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C.P., LaMont J.T. Clostridium difficile--more difficult than ever. N. Engl. J. Med. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- Kundu P., Lee H.U., Garcia-Perez I., Tay E.X.Y., Kim H., Faylon L.E., Martin K.A., Purbojati R., Drautz-Moses D.I., Ghosh S., et al. Neurogenesis and prolongevity signaling in young germ-free mice transplanted with the gut microbiota of old mice. Sci. Transl. Med. 2019;11:eaau4760. doi: 10.1126/scitranslmed.aau4760. [DOI] [PubMed] [Google Scholar]
- Lahti L., Salojärvi J., Salonen A., Scheffer M., de Vos W.M. Tipping elements in the human intestinal ecosystem. Nat. Commun. 2014;5:4344. doi: 10.1038/ncomms5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z.L., Tseng C.H., HO H.J., Cheung C.K.Y., Lin J.Y., Chen Y.J., Cheng F.C., Hsu Y.C., Lin J.T., El-Omar E.M., et al. Fecal microbiota transplantation confers beneficial metabolic effects of diet and exercise on diet-induced obese mice. Sci. Rep. 2018;8:15625. doi: 10.1038/s41598-018-33893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle A., Hoffmann T.W., Pham H.P., Langella P., Guédon E., Sokol H. Baseline microbiota composition modulates antibiotic-mediated effects on the gut microbiota and host. Microbiome. 2019;7:111. doi: 10.1186/s40168-019-0725-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy T., Debédat J., Marquet F., Da-Cunha C., Ichou F., Guerre-Millo M., Kapel N., Aron-Wisnewsky J., Clément K. Comparative evaluation of microbiota engraftment following fecal microbiota transfer in mice models: age, kinetic and microbial status matter. Front. Microbiol. 2019;9:3289. doi: 10.3389/fmicb.2018.03289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Zhang T., Xiao Y., Tian L., Cui B., Ji G., Liu Y.Y., Zhang F. Timing for the second fecal microbiota transplantation to maintain the long-term benefit from the first treatment for Crohn's disease. Appl. Microbiol. Biotechnol. 2019;103:349–360. doi: 10.1007/s00253-018-9447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin E., Lin K., Katz S. Serious and opportunistic infections in elderly patients with inflammatory bowel disease. Gastroenterol. Hepatol. (N Y) 2019;15:593–605. [PMC free article] [PubMed] [Google Scholar]
- Littmann E.R., Lee J.J., denny J.E., Alam Z., Maslanka J.R., Zarin I., Matsuda R., Carter R.A., Susac B., Saffern M.S., et al. Host immunity modulates the efficacy of microbiota transplantation for treatment of Clostridioides difficile infection. Nat. Commun. 2021;12:755. doi: 10.1038/s41467-020-20793-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamo Y., Woodworth M.H., Wang T., Dhere T., Kraft C.S. Durability and long-term clinical outcomes of fecal microbiota transplant treatment in patients with recurrent Clostridium difficile infection. Clin. Infect Dis. 2018;66:1705–1711. doi: 10.1093/cid/cix1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Enet. J. 2011;17:10–12. [Google Scholar]
- Melgar S., Karlsson A., Michaëlsson E. Acute colitis induced by dextran sulfate sodium progresses to chronicity in C57BL/6 but not in BALB/c mice: correlation between symptoms and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G1328–G1338. doi: 10.1152/ajpgi.00467.2004. [DOI] [PubMed] [Google Scholar]
- Morgun A., Dzutsev A., Dong X., Greer R.L., Sexton D.J., Ravel J., Schuster M., Hsiao W., Matzinger P., Shulzhenko N. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut. 2015;64:1732–1743. doi: 10.1136/gutjnl-2014-308820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munyaka P.M., Rabbi M.F., Khafipour E., Ghia J.E. Acute dextran sulfate sodium (DSS)-induced colitis promotes gut microbial dysbiosis in mice. J. Basic Microbiol. 2016;56:986–998. doi: 10.1002/jobm.201500726. [DOI] [PubMed] [Google Scholar]
- Ng K.M., Aranda-Díaz A., Tropini C., Frankel M.R., van Treuren W., O'loughlin C.T., Merrill B.D., Yu F.B., Pruss K.M., Oliveira R.A., et al. Recovery of the gut microbiota after antibiotics depends on host diet, community context, and environmental reservoirs. Cell Host Microbe. 2020;28:628. doi: 10.1016/j.chom.2020.09.001. [DOI] [PubMed] [Google Scholar]
- Ohkusa T. [Production of experimental ulcerative colitis in hamsters by dextran sulfate sodium and changes in intestinal microflora] Nihon Shokakibyo Gakkai Zasshi. 1985;82:1327–1336. [PubMed] [Google Scholar]
- Okahara K., Ishikawa D., Nomura K., Ito S., Haga K., Takahashi M., Shibuya T., Osada T., Nagahara A. Matching between donors and ulcerative colitis patients is important for long-term maintenance after fecal microbiota transplantation. J. Clin. Med. 2020;9:1650. doi: 10.3390/jcm9061650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka T., Moriyama E., Arai S., Date Y., Yagi J., Kikuchi J., Tsuneda S. Meta-analysis of fecal microbiota and metabolites in experimental colitic mice during the inflammatory and healing phases. Nutrients. 2017;9:1392. doi: 10.3390/nu9121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleja A., Mikkelsen K.H., Forslund S.K., Kashani A., Allin K.H., Nielsen T., Hansen T.H., Liang S., Feng Q., Zhang C., et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018;3:1255–1265. doi: 10.1038/s41564-018-0257-9. [DOI] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese A.T., Cho E.H., Klitzman B., Nichols S.P., Wisniewski N.A., Villa M.M., Durand H.K., Jiang S., Midani F.S., Nimmagadda S.N., et al. Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut. Elife. 2018;7:e35987. doi: 10.7554/eLife.35987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese A.T., Pereira F.C., Schintlmeister A., Berry D., Wagner M., Hale L.P., Wu A., Jiang S., Durand H.K., Zhou X., et al. Microbial nitrogen limitation in the mammalian large intestine. Nat. Microbiol. 2018;3:1441–1450. doi: 10.1038/s41564-018-0267-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P., Willemsen D., Popkes M., Metge F., Gandiwa E., Reichard M., Valenzano D.R. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. Elife. 2017;6:e27014. doi: 10.7554/eLife.27014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spychala M.S., Venna V.R., Jandzinski M., Doran S.J., Durgan D.J., Ganesh B.P., Ajami N.J., Putluri N., Graf J., Bryan R.M., et al. Age-related changes in the gut microbiota influence systemic inflammation and stroke outcome. Ann. Neurol. 2018;84:23–36. doi: 10.1002/ana.25250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebegg M., Silva-Cayetano A., Innocentin S., Jenkins T.P., Cantacessi C., Gilbert C., Linterman M.A. Heterochronic faecal transplantation boosts gut germinal centres in aged mice. Nat. Commun. 2019;10:2443. doi: 10.1038/s41467-019-10430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suez J., Zmora N., Zilberman-Schapira G., Mor U., Dori-Bachash M., Bashiardes S., Zur M., Regev-Lehavi D., Ben-Zeev Brik R., Federici S., et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174:1406–1423.e16. doi: 10.1016/j.cell.2018.08.047. [DOI] [PubMed] [Google Scholar]
- Tabbaa O.M., Aboelsoud M.M., Mattar M.C. Long-term safety and efficacy of fecal microbiota transplantation in the treatment of Clostridium difficile infection in patients with and without inflammatory bowel disease: a tertiary care center's experience. Gastroenterol. Res. 2018;11:397–403. doi: 10.14740/gr1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleban S., Colombel J.F., Mohler M.J., Fain M.J. Inflammatory bowel disease and the elderly: a review. J. Crohns Colitis. 2015;9:507–515. doi: 10.1093/ecco-jcc/jjv059. [DOI] [PubMed] [Google Scholar]
- Tang C., Kakuta S., Shimizu K., Kadoki M., Kamiya T., Shimazu T., Kubo S., Saijo S., Ishigame H., Nakae S., et al. Suppression of IL-17F, but not of IL-17A, provides protection against colitis by inducing T(reg) cells through modification of the intestinal microbiota. Nat. Immunol. 2018;19:755–765. doi: 10.1038/s41590-018-0134-y. [DOI] [PubMed] [Google Scholar]
- Tseng A.S., Crowell M., Orenstein R., Patron R.L., Dibaise J. Older patient age is associated with similar safety but higher relapse after fecal microbiota transplantation for recurrent Clostridium difficile infection. Am. J. Gastroenterol. 2017;112:S51. [Google Scholar]
- Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wirtz S., Popp V., Kindermann M., Gerlach K., Weigmann B., Fichtner-Feigl S., Neurath M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017;12:1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Angulo M.T., Lao S., Weiss S.T., Liu Y.Y. An ecological framework to understand the efficacy of fecal microbiota transplantation. Nat. Commun. 2020;11:3329. doi: 10.1038/s41467-020-17180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Cui B., He X., Nie Y., Wu K., Fan D. Microbiota transplantation: concept, methodology and strategy for its modernization. Protein Cell. 2018;9:462–473. doi: 10.1007/s13238-018-0541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All sequencing data have been deposited at NCBI Sequencing Read Archive and are publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
All original code has been deposited at Github and is publicly available as of the date of publication. The link is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.