Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA has been extensively detected in raw wastewater in studies exploring wastewater-based epidemiology (WBE) for early warning purposes. Nonetheless, only a few limited studies investigated the presence of SARS-CoV-2 in treated wastewaters to determine the potential health risks across the water cycle. The detection of SARS-CoV-2 has been done mostly by RT-qPCR and ddPCR, which only provides information on the presence of nucleic acids rather than information on potential infectivity. In this study, we set to develop and evaluate the use of viability RT-qPCR for the selective discrimination and surveillance of infectious SARS-CoV-2 in secondary-treated wastewater. Enzymatic (nuclease) and viability dye (Reagent D) pretreatments were applied to infer infectivity through RT-qPCR using porcine epidemic diarrhea virus (PEDV) as a CoV surrogate. Infectivity tests were first performed on PEDV purified RNA, then on infectious and heat-inactivated PEDV, and finally on heat inactivated PEDV spiked in concentrated secondary-treated wastewater. The two viability RT-qPCR methods were then applied to 27 secondary-treated wastewater samples positive for SARS-CoV-2 RNA at the outlet of five large urban wastewater treatment plants in Portugal. Reagent D pretreatment showed similar behavior to cell culture for heat-inactivated PEDV and both viability RT-qPCR methods performed comparably to VERO E6 cell culture for SARS-CoV-2 present in secondary-treated wastewater, eliminating completely the RT-qPCR signal. Our study demonstrated the lack of infectious SARS-CoV-2 viral particles on secondary-treated wastewater through the application of two pretreatment methods for the rapid inference of infectivity through RT-qPCR, showing their potential application in environmental screening. This study addressed a knowledge gap on the public health risks of SARS-CoV-2 across the water cycle.
Keywords: SARS-CoV-2 infectivity, Urban water cycle, Reagent D, Nuclease, Health risks, Viability RT-qPCR
Graphical abstract
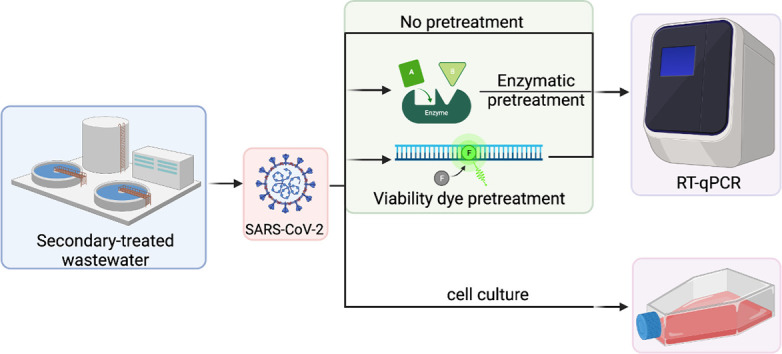
1. Introduction
Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2), responsible for the coronavirus disease 2019 (COVID-19), caused until December 9, 2021 more than 266,000,000 cases and almost 5,300,000 deaths worldwide (ECDC, 2021).
Although the most common routes of infection are aerosols and respiratory droplets, SARS-CoV-2 RNA has been commonly found in the feces of infected patients, regardless of the severity or absence of symptoms (Klompas et al., 2020; Wang et al., 2020). SARS-CoV-2 receptor is the angiotensin-converting enzyme 2 (ACE2), which although being detected in the upper respiratory tract samples, indicating nasopharynx as a site of replication (Qi et al., 2020; Zhao et al., 2020), has the highest expression in the brush border of intestinal enterocytes (Qi et al., 2020; The Human Protein Atlas, 2020). Viral RNA has been found, for instance, in rectal swabs even after the nasopharyngeal testing became negative, implying infection of the gastrointestinal tract (Holshue et al., 2020; Wang et al., 2020; Xiao et al., 2020a).
SARS-CoV-2 RNA has been detected worldwide in raw wastewater and in some cases in treated wastewater, which could imply potential environmental transmission via the water cycle (Gonzalez et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Westhaus et al., 2021; Monteiro et al., 2022). Historically, the gold standard for the isolation of infectious viral particles is cell culture, using distinct mammalian cell lines such as VERO, VERO E6 or BGM. However, the use of cell culture to determine the presence of SARS-CoV-2 in wastewater is hindered by several aspects: i) viable SARS-CoV-2 has rarely been isolated from the feces of infected patients despite the high levels of RNA detected, which suggests that the virus is already inactivated when excreted (Kim et al., 2020; Wölfel et al., 2020); ii) low throughput and significant costs of a cell culture system; iii) the need for availability of a biosafety level 3 (BSL-3) laboratory for the isolation of SARS-CoV-2 (CDC, 2021); iv) the need to concentrate large volume of wastewater for the detection of viruses, therefore co-concentrating contaminants that are difficult to remove prior to inoculation of samples in cell culture systems, thus impairing virus isolation. Due to these limitations, it is necessary to explore other approaches, namely based on molecular methods such as reverse transcription quantitative PCR (RT-qPCR). However, due to the nature of these techniques, they inform only on the presence of nucleic acids, providing no information on the infectivity. Such feature is not the most relevant when the main interest is to use wastewater-based epidemiology (WBE) for early warning purposes, but it is important to understand if the water cycle plays an important role in further disseminating SARS-CoV-2, namely to wastewater treatment plant workers and/or other individuals that might come into contact with contaminated water, such as bathers and other surface water users.
The virus envelope and capsid protect the viral genome from the external influence of nucleases exerted upon RNA, while the spike protein determines the ability of the virus to bind with high efficiency and stability to ACE2. In light of the current knowledge, the integrity of the envelope, and particularly the spike protein, is crucial for infectivity and for the virus' ability to establish infection in humans. In the last decade, methods based on nucleases (DNase or RNase) and on viability dyes have been tested as pretreatment to infer infectivity through qPCR in different matrices (Lamhoujeb et al., 2008; Nowak et al., 2011; Schielke et al., 2011; Monteiro and Santos, 2018; Puente et al., 2020; Leifels et al., 2021). The underlying principle is that the viral genome (e.g. RNA) in a given matrix may be degraded by nucleases. If the viral envelope or capsid is degraded (and the viral ligand to human receptors becomes impaired), then nucleic acids become exposed and susceptible to cleavage by endonucleases and, thus, amplification by PCR is greatly affected. If there are integer viruses, then the endonucleases will not come into contact with the nucleic acids that remain protected. Such pretreatments to infer infectivity using exposure and degradation of nucleic acids as proxy are extremely relevant in different contexts, including for environmental surveillance and for food and water safety assessments.
In this study, we aimed to develop a specific viability RT-qPCR for the selective detection of infectious SARS-CoV-2 in secondary-treated wastewater and then apply this methodology to infer the infectivity of over 80 secondary-treated wastewater samples collected for a 32-week period in 2020, during the first two waves of the COVID-19 pandemic in Portugal. Porcine epidemic diarrhea virus (PEDV), a member of the Alphacoronavirus genus in the Coronaviridae family, was used as a model surrogate for SARS-CoV-2. Enzymatic reaction and a viability dye (monoazide dye; Reagent D) were used to infer infectivity through RT-qPCR. The optimized viability RT-qPCR was then applied to secondary-treated wastewater to evaluate the infectivity of detected SARS-CoV-2, thus helping to assess the potential risk exerted by the presence of this virus in treated wastewater and along the water cycle.
2. Materials and methods
2.1. Sampling sites and sample collection
Secondary-treated wastewater (n = 89) samples were collected over a 32-week period, between April 27thand December 2nd 2020, from five wastewater treatment plants (WWTP) located in the North of Portugal (Vila Nova Gaia (GA) and Serzedelo (SE)) and in Lisboa e Vale do Tejo (LVT; Alcântara (AL), Beirolas (BE), and Guia (GU)) region (Fig. S1).
Twenty-four-hour composite samples were collected using an automated sampler (ISCO, US). Samples were transported refrigerated to the laboratory, within 8 h of collection and processed immediately upon arrival to the laboratory, as described in Monteiro et al. (2022).
2.2. SARS-CoV-2 analysis of secondary-treated wastewater samples
Five litre of secondary-treated wastewater were concentrated using hollow-fiber filters inuvai R180 (inuvai, a division of Fresenius Medical Care, Germany), with a molecular weight cut-off of 18.8 kDa. Samples were eluted in 300 ml of 1× phosphate buffered saline (PBS) containing 0.01% sodium polyphosphate (NaPP; Sigma-Aldrich, US) and 0.01 Tween 80/0.001% antifoam and precipitated overnight with 20% polyethylene-glycol (PEG) 8000 (Sigma-Aldrich, US). Samples were then centrifuged at 10,000 xg for 30 min and resuspended in 5 ml 1× PBS, pH 7.4 (Blanco et al., 2019). Samples were kept at −80 (± 10) °C until further processing.
2.3. PEDV viral strain and infectivity assay
The PEDV strain CV777 (kindly provided by Dr. Gloria Sanchez, IATA-CSIC, Spain) was propagated in VERO cells (Puente et al., 2020). Briefly, VERO cells cultured in Dulbecco's Modified Eagles's Medium (DMEM, ThermoFisher Scientific, US) supplemented with 100 units/ml of penicillin (Lonza, Swiss), 100 units/ml of streptomycin (Lonza, Swiss), 0.25 mg/ml amphotericin B (Lonza, Swiss) and 10% heat-inactivated fetal bovine serum (FBS; Biological Industries, Israel), were assayed as complete confluent monolayers in 24-well plates (Corning, US). Briefly, ten-fold dilutions of PEDV were prepared in DMEM supplemented with 10 μg/μl trypsin (trypsin 1:250; SAFC, Sigma-Aldrich, US) and 100 μl per well were inoculated. Following 2 h post-infection, 100 μl of media (DMEM supplemented with 0.3% tryptose phosphate broth (TPB, Sigma, US), 100 units/ml of penicillin, 100 units/ml of streptomycin, 0.25 mg/ml amphotericin B, and 10 μg/μl trypsin) was added. Plates were incubated at 37 (± 1) °C in a 5% CO2 incubator and monitored for cytopathic effects (CPE) for 4 days. CPE are morphological changes in cells caused by a viral infection. After visual observation of cells for detection of CPE, the infectivity was calculated by determining the 50% tissue culture infectious dose (TCID50) using the Spearman-Karber method (Hierholzer and Killington, 1996).
2.4. Nuclease and viability dye pretreatment on purified PEDV RNA
Nucleic acid extraction was performed in DNA LoBind microcentrifuge tubes (Eppendorf, Germany) using the QIAamp DNA stool mini kit (Qiagen, Germany) according to the manufacturer's instructions. Viral RNA was eluted in a final volume of 100 μL.
Pierce Universal nuclease for cell lysis (Thermo Fischer Scientific, US) and viability dye (Reagent D, Biotecon, Germany) were tested primarily on purified PEDV RNA. Reagent D contains a light sensitive substance that upon exposure to visible light binds covalently to nucleic acids and prevents their amplification via PCR. This reagent is provided already reconstituted by the manufacturer and was used in accordance with the manufacturer's instructions. Briefly, to each sample, Reagent D was added in a proportion of 1:4 (v:v), the mixture incubated in the dark for 5 min at room temperature and the dye photoactivated for 15 min using a photoactivation system (PhAST Blue; GenIUL, Spain) (Fig. 1 ). For the enzymatic pretreatment, 50 units of Pierce universal nuclease for cell lysis, a genetically engineered endonuclease that degrades single-stranded, double-stranded, linear, and circular DNA and RNA and is effective for cell lysis over a wide range of temperatures and pH, was added to each sample and incubated for 15 min at room temperature (Fig. 1).
Fig. 1.
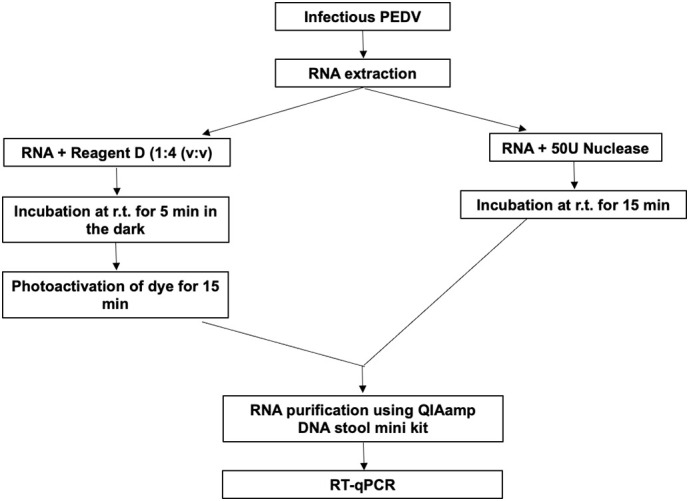
Schematics for the pretreatments (viability dye and nuclease) applied to purified PEDV RNA.
Following pretreatment, a new RNA purification step using the QIAamp DNA stool mini kit (Qiagen, Germany) was conducted as previously described to remove potential interference of the enzyme and the viability dye in the following steps.
Each experiment was performed in triplicate and a purified PEDV RNA sample without pretreatment was included as a positive control.
2.5. Viability pretreatments to infer infectivity of heat-inactivated PEDV by RT-qPCR
Nuclease and viability dyes were additionally tested on heat-inactivated PEDV. PEDV suspensions were divided into two categories: i) non-treated infectious viral particles; and ii) heat-treated viral particles by incubation for 15 min at 72 °C (heat-inactivated). The heat-inactivated samples were subjected to the above-described pretreatments before RT-qPCR (Fig. 2 ).
Fig. 2.
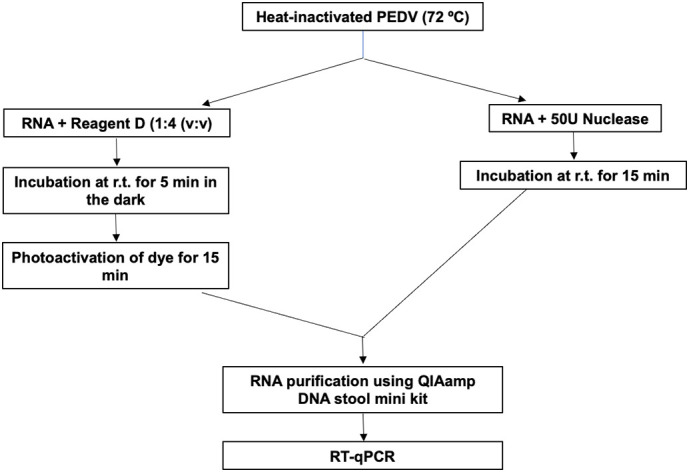
Schematics for the pretreatments (viability dye and nuclease) applied to heat-inactivated PEDV.
The experiments were conducted in triplicate and three controls were added: i) infectious virus with viability pretreatments; ii) infectious virus without pretreatment; and iii) heat-inactivated virus without pretreatment. All experiments were conducted in DNA LoBind microcentrifuge tubes. Following viability pretreatments, samples were extracted as described previously and quantified by RT-qPCR.
2.6. Artificial contamination of secondary-treated wastewater
Heat-inactivated PEDV suspensions (100 μL, final concentration ~ 104 TCID50/ml) were spiked into 5 ml of concentrated secondary-treated wastewater from two distinct WWTP: SE and GA. SE WWTP has the particularity of receiving a large input of industrial influent, namely from the tannery industry. It has been shown previously that having a high input of industrial wastewater impaired the detection of SARS-CoV-2 from raw wastewater (Gawlik et al., 2021) given that such wastewater generates a large amount of liquid waste constituted by pollutants such as organic and inorganic matter, total dissolved solids as well as a variety of synthetic compounds which can difficult the concentration and the final detection of the virus. Therefore, and taking into account such characteristics, SE WWTP was chosen to test the potential use of pretreatments to determine infectivity through RT-qPCR. The spiked secondary-treated wastewaters were subjected to the two viability pretreatments as described above. All experiments were conducted in DNA LoBind microcentrifuge tubes. Three controls were included: i) infectious viruses spiked into secondary-treated wastewater subjected to pretreatment; ii) infectious viruses spiked into secondary-treated wastewater without pretreatment; and iii) heat-inactivated viruses spiked into secondary-treated wastewater without pretreatment. Following pretreatment, samples were purified as described previously. Experiments were performed in triplicate.
2.7. SARS-CoV-2 infectivity in secondary-treated wastewater
SARS-CoV-2 RNA positive concentrated secondary-treated wastewater samples were tested for infectivity using VERO E6 cells, which are commonly used to isolate and propagate SARS-CoV-like viruses since they support viral replication to high titers. Cells were cultured in DMEM containing 10% FBS. Plates with freshly grown VERO E6 cells were inoculated with 1 ml volume from each secondary-treated wastewater sample following sterilization through a 0.22 μm polyvinylidene fluoride (PVDF) filter (Pall, UK) (Tartera et al., 1992). Samples were incubated for 1 h, the supernatant was removed, rinsed twice with phosphate-buffered saline (PBS) and 20 ml of fresh culture medium (DMEM supplemented with FBS, 50 units/ml penicillin, 50 units/ml streptomycin, and 2 mM l-glutamine (Sigma, US)) was added to the samples. Plates were incubated at 37 (± 1) °C for 5 days, inspected for CPE and the TCID50 was calculated according to the Spearman-Karber method. Negative controls (PBS) were included in each test batch.
Concentrates from secondary-treated wastewater were analyzed with and without viability pretreatment (previously tested on PEDV as previously described) to evaluate the usefulness of the viability RT-qPCR for SARS-CoV-2. To 200 μl of secondary-treated wastewater concentrated sample were added 600 μl of Reagent D (Biotecon, Germany) or 50 units of Pierce universal nuclease. Incubations and photoactivation of Reagent D were performed as described previously. Experiments were performed in triplicate.
2.8. Extraction and quantification of PEDV and SARS-CoV-2
Viral RNA from concentrated untreated secondary-treated wastewater (200 μl), Reagent D pretreatment concentrates (800 μl) and nuclease pretreatment concentrates (200 μl) was extracted using the QIAamp DNA stool mini kit, according to the manufacturer's instructions, with final elution in 100 μl. Molecular detection of PEDV and SARS-CoV-2 was performed in an Applied Biosystems 7300 Real-Time PCR (Applied Biosystems, US) using the AgPath-ID One-Step RT-PCR kit (Thermo Fischer Scientific, US), with primers and probes described by Zhou et al. (2017) and Corman et al. (2020) (Supplementary Table 1). For SARS-CoV-2, E_Sarbeco, RdRp, and N_Sarbeco were amplified as described by Monteiro et al. (2022). The 25 μl final volume reaction mixture consisted of 12.5 μl of 2× RT-PCR buffer, 1 μl of RT-PCR enzyme mix, 800 nM of each primer, 200 nM of probe, 5 μl of sample, with the final volume completed with nuclease-free water. PCR inhibition was evaluated by determining the concentration of PEDV and SARS-CoV-2 in the 10- and 100-fold sample dilutions. Cycle threshold differences (ΔCt) ≥ 3.50 between crude extracts and 10-fold dilution and between 10-fold dilution and 100-fold dilutions, were considered amplification inhibition free. Thermal cycling conditions were as follows: i) PEDV: reverse transcription for 10 min at 45 °C, initial denaturation for 10 min at 95 °C, followed by 45 cycles of 15 s at 95 °C and 1 min at 60 °C; ii) SARS-CoV-2: reverse transcription for 10 min at 45 °C, initial denaturation for 10 min at 95 °C, followed by 45 cycles of 15 s at 95 °C and 1 min at 58 °C. Reactions were considered positive only if the cycle threshold was below 40 cycles (Medema et al., 2020; Wu et al., 2020). Analysis of PEDV was performed qualitatively, therefore, positive and negative controls were added with each reaction. Quantification of E_Sarbeco and RdRp assays was performed through calibration curves using 10-fold dilutions of nCoV-ALL-Control plasmid (Eurofins Genomics, Germany), ranging from 1.94 to 1.94 × 106 and 1.00 to 1.00 × 106 GC per reaction respectively. Quantification of N_Sarbeco assay was performed using 2-fold and 10-fold dilutions (ranging between 2.00 and 2.00 × 104 GC per reaction) of the Amplirun SARS-CoV-2 RNA control (Vircell, Spain). Negative controls (extraction and RT-qPCR assay) were also performed using DNase/RNase free distilled water, following the same conditions as the samples.
2.9. Statistical analysis
Data analysis was conducted with GraphPad Prism (GraphPad Software, US). Each experiment was conducted in triplicate, and each sample was analyzed in duplicate. Normality test of the dataset was conducted using the Shapiro-Wilk test and the equality of variances was determined using the Levene's test. Kruskal-Wallis test (KW statistics) was conducted to compare differences between each test and pairwise comparison was performed with Dunn's test. Mann-Whitney test was used to compare between infectious and heat-inactivated PEDV, and between infectious and heat-inactivated PEDV following pretreatments (nuclease and Reagent D). Data was considered significant with values of p < 0.05.
3. Results
3.1. Viability of purified PEDV RNA
Pierce Universal nuclease for cell lysis and Reagent D were initially screened for their potential to discriminate between infectious and non-infectious viral particles in purified PEDV RNA extracts (Fig. 3 ). Treatment with Pierce Universal nuclease for cell lysis was able to completely remove the amplification signal (Ct) from purified PEDV RNA (mean removal ΔCt ≥ 21.3).
Fig. 3.
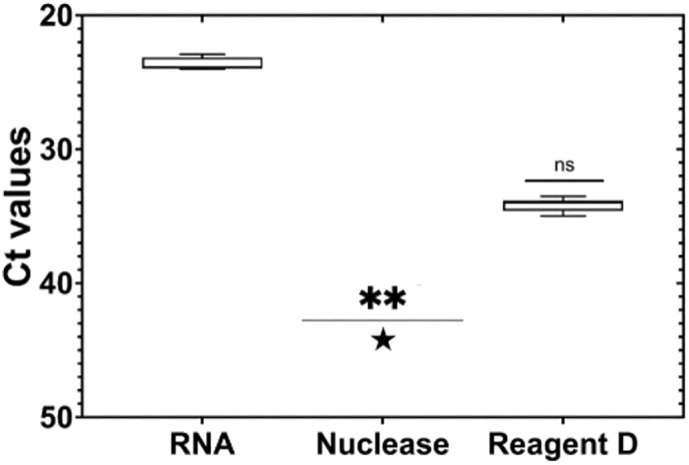
Number of cycles (Ct) as a function of different viability treatments applied to purified PEDV RNA. Asterisks represent statistically significant differences. ** p = 0.001; ns, no significant difference. Star () represent undetected results.
Applying Reagent D on purified PEDV RNA prior to RT-qPCR decreased on average the PCR signal by 10.5 Ct. Differences between control and the tested viability treatments were statistically significant (p = 0.001). Dilutions of 1:10 and 1:100 did not show inhibitory effects on RT-qPCR for both pretreatments.
3.2. Efficiency of viability pretreatments on heat-inactivated PEDV
Infectious and heat-inactivated (72 °C for 15 min) PEDV were treated with Pierce Universal nuclease for cell lysis and Reagent D before extraction and quantification by RT-qPCR (Fig. 4 ). Differences were obtained between infectious and heat-inactivated viral particles (p < 0.05), with mean ΔCt of 1.98.
Fig. 4.
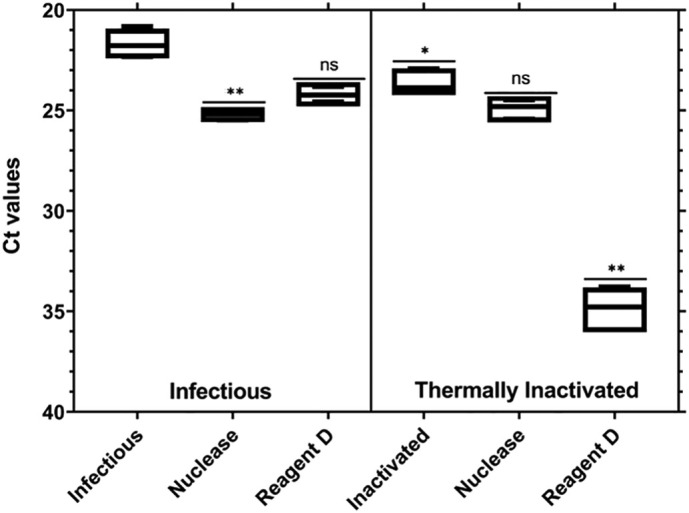
Number of cycles (Ct) as a function of different viability treatments applied to purified infectious and thermally inactivated PEDV. Asterisks represent statistically significant differences. *p < 0.05; **p = 0.001; ns, no significant difference.
Pretreatment of infectious PEDV viral particles with nuclease significantly decreased the RT-qPCR signal by an average of 3.51 Ct (p < 0.05), whereas Reagent D reduced the signal, on average, by more than 2.50 Ct (p = 0.40). The difference in detection by RT-qPCR between heat-inactivated PEDV and nuclease-treated inactivated PEDV was 1.22 Ct. No differences between nuclease-treated infectious and heat-inactivated PEDV were detected by RT-qPCR (p = 0.31). Reagent D was able to significantly decrease the RT-qPCR signal of heat-inactivated PEDV (mean ΔCt = 11.2; p < 0.05). Statistically significant differences were determined between infectious and heat-inactivated PEDV pretreated with Reagent D (mean ΔCt = 10.65; p < 0.05). No inhibitory effect was detected for both pretreatments.
3.3. Performance of viability RT-qPCR in spiked secondary-treated wastewater
Secondary-treated wastewater was spiked with heat-inactivated PEDV (72 °C for 15 min) and subjected to pretreatment with nuclease and Reagent D prior to RT-qPCR to determine the influence of the matrix on the viability pretreatments. Viability pretreatments were tested in two very distinct secondary-treated wastewaters: GA and SE. GL WWTP receives mainly municipal wastewater with an average flow of 66,700 m3/day, and secondary-treated wastewater presented high turbidity. On the other hand, SE WWTP in addition to municipal wastewater, receives a large volume of industrial wastewater from the tannery industry which by itself represents an additional challenge, but the turbidity levels were lower.
Results from GA showed a RT-PCR signal reduction of heat-inactivated PEDV treated with nuclease when compared to the inactivated spiked control (4.59 Ct), but the difference was not statistically significant (p = 0.45) (Fig. 5 ). Conversely, treating spiked GA secondary-treated wastewater with Reagent D strongly decreased the RT-qPCR signal (ΔCt > 20.0; p < 0.05).
Fig. 5.
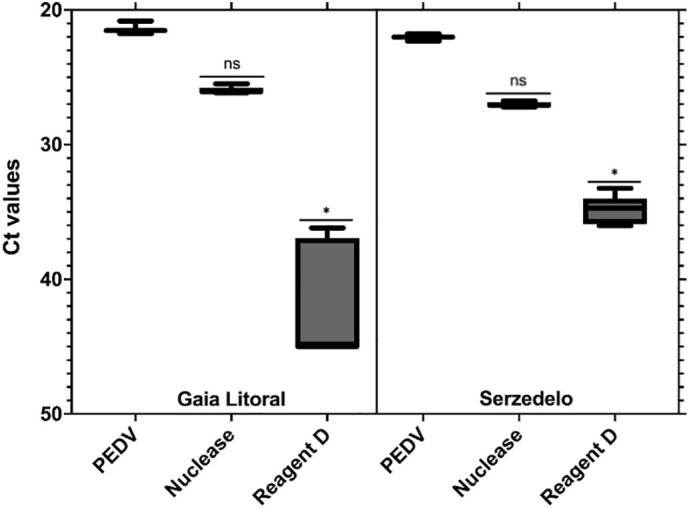
Number of cycles (Ct) as a function of different viability treatments applied to heat-inactivated PEDV spiked into two secondary-treated wastewater, GA and SE. Asterisks represent statistically significant differences. *ρ < 0.05; ns, no significant difference.
Similarly, data for PEDV spiked SE secondary-treated wastewater showed that Reagent D pretreatment performed at a higher level than nuclease (ΔCt = 12.8 and ΔCt = 5.00, respectively). Results for Reagent D differed significantly from the control (p < 0.05). Smaller variability was detected for the SE secondary-treated wastewater pretreated with Reagent D compared to GL, possibly due to the higher turbidity of the latter, which may have affected the performance of the dye, therefore increasing the variability of the results. Inhibition was not detected in the RT-qPCR assays, as indicated by molecular results of dilution testing.
3.4. Presence of SARS-CoV-2 RNA in secondary-treated wastewater using RT-qPCR
Throughout the 32-week study, a total of 89 secondary-treated wastewater samples were collected and tested for the presence of SARS-CoV-2 RNA, using the three assays described by Corman et al. (2020): E_Sarbeco, RdRp, and N_Sarbeco. SARS-CoV-2 RNA was present, as determined by RT-qPCR, in 30% (27/89) of the samples, with concentrations ranging from 1.71 × 102 in SE to 1.18 × 104 GC/L in BE (Fig. 6 ). From the 27 positive samples, 18 were positive for a single assay, 8 were positive for two assays (E_Sarbeco and RdRp) and a single sample was positive for all three assays.
Fig. 6.
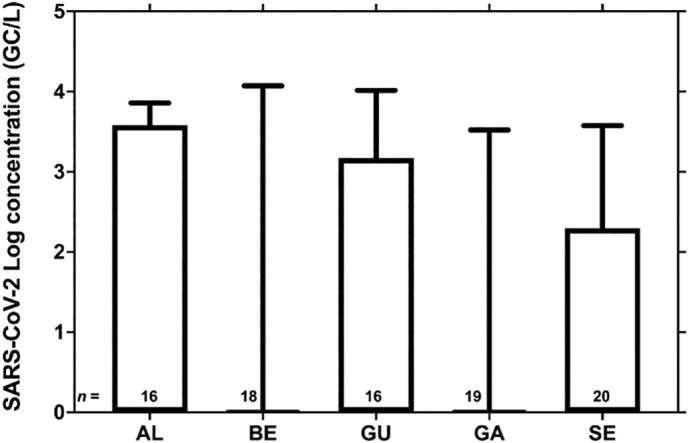
SARS-CoV-2 RNA concentration in the secondary-treated wastewater from LVT WWTP (AL- Alcântara; BE – Beirolas; GU – Guia) and the WWTP from the North region of Portugal (GA – Gaia Litoral; SE – Serzedelo), from April to December 2020. Boxes, 25th and 75th percentile; lines within the boxes, median; whiskers, lowest and highest SARS-CoV-2 RNA concentration; n, number of samples from each WWTP.
As the number of COVID-19 cases in the country increased by the end of our sampling period, an increase was also registered in the percentage of positive samples for this matrix (Fig. S2).
3.5. Evaluation of SARS-CoV-2 infectivity in secondary-treated wastewater by viability RT-qPCR
SARS-CoV-2 presence in secondary-treated wastewater was evaluated, in a first approach, using RT-qPCR alone. Nonetheless, RNA detection does not necessarily imply infectious potential and correspondent health risk, as discussed previously (Kim et al., 2020; Wölfel et al., 2020). Following confirmation of the presence of SARS-CoV-2 RNA in the tested effluents, we evaluated the infectivity potential in three ways: i) cell culture using Vero E6 cells; ii) enzymatic degradation of nucleic acids using Pierce Universal nuclease; and iii) viability dyes using Reagent D. Analysis of SARS-CoV-2 RNA positive secondary-treated wastewaters by cell culture provided negative results for infectivity. Secondary-treated wastewaters detected by viability RT-qPCR results are shown in Fig. 7 . This representation includes all the positive samples obtained at each site. Remarkably, Reagent D and nuclease pretreatment were able to completely remove the amplification signals obtained by RT-qPCR in all samples.
Fig. 7.
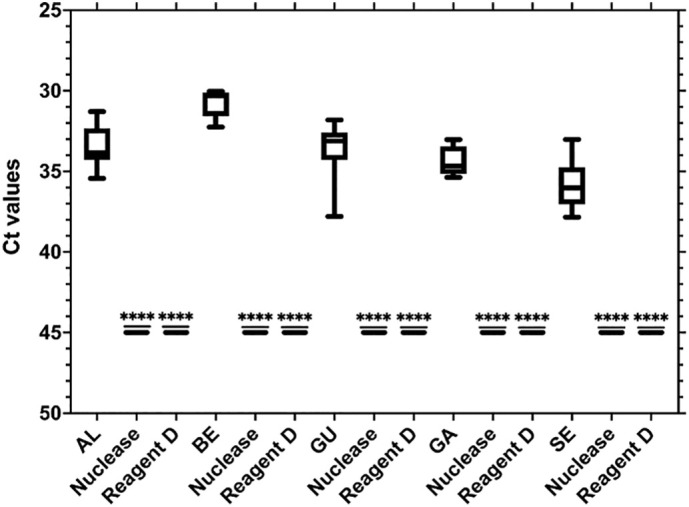
Number of cycles (Ct) as a function of different viability treatments applied to SARS-CoV-2 in secondary-treated wastewater. This representation includes the median Cts obtained for all positive samples at each location. Locations: AL – Alcântara (n = 7), BE – Beirolas (n = 3), GU – Guia (n = 7), GA – Gaia Litoral (n = 4), SE – Serzedelo (n = 6). Asterisks represent statistically significant differences. ****ρ < 0.0001.
Results for both viability RT-qPCR methods showed average Ct reductions of more than 9.00 with respect to RT-qPCR alone. The RT-qPCR pretreatments were able to completely remove the signal in all samples, with average decreases varying between 9.17 for SE and 14.22 for BE.
4. Discussion
The urgent situation the world has been facing for the last year and a half requires more in-depth research into enveloped viruses, including on the transmission and viral fate in the environment. Better analytical tools are thus necessary for monitoring potential routes of transmission. Although SARS-CoV-2 is preferentially transmitted via respiratory droplets (Qi et al., 2020; Zhao et al., 2020), excretion of the viruses in the feces have been confirmed in a high proportion of infected individuals (Holshue et al., 2020; Wang et al., 2020; Xiao et al., 2020a). Nonetheless, studies looked mainly at the presence of SARS-CoV-2 RNA in feces for two main reasons: (i) this is a problematic matrix due to the high concentration of microorganisms which may affect the performance of virus culture in cell lines; and (ii) SARS-CoV-2 isolation should be conducted at least in a BLS-3 laboratory using BSL-3 good practices (CDC, 2021) and most environmental laboratories do not have such facilities and protocols in place. At least two studies have investigated the infectivity of SARS-CoV-2 detected in the stools of infected patients with contradictory findings. While Xiao et al. (2020b) were able to detect infectious viral particles in the stools of an infected patient, using the Vero E6 cell line, Wölfel et al. (2020) were unable to isolate infectious viral particles, using the same cell line, in two separate laboratories, despite the high viral RNA load detected by RT-qPCR.
Due to the presence of SARS-CoV-2 in the feces of infected individuals, a WBE approach has been put in place in many locations of the world, with SARS-CoV-2 RNA being detected in raw wastewater. However, only a few studies have investigated the presence of SARS-CoV-2 RNA in treated wastewaters (Randazzo et al., 2020; Westhaus et al., 2021), with both studies confirming the presence of SARS-CoV-2 RNA in treated effluents. From these studies only Westhaus et al. (2021) looked at the potential presence of infectious viral particles in treated wastewater, using the CaCo-2 cell line. In agreement with the results from Wölfel et al. (2020), the authors were incapable of isolating infectious viral particles. In our study, SARS-CoV-2 RNA was detected in 30% of the secondary-treated wastewater samples in concentrations up to 104 GC/L, with the presence of RNA not implying immediate risks to public health. Following detection by RT-qPCR and to determine possible health risks across the water cycle, positive samples were tested in cell culture and using viability RT-qPCR techniques based on enzymatic and viability dyes. Our results suggest that SARS-CoV-2 detected in treated wastewater appears to be non-infectious. It is important to refer that until now, only a few studies described the usage of pretreatments to infer SARS-CoV-2 infectivity through RT-qPCR (Cuevas-Ferrando et al., 2021; Polo et al., 2021; Wurtzer et al., 2021). Polo et al. (2021) was capable of fully eliminating the RT-qPCR signal by using a combination of PMAxx with a surfactant in clam and sediment samples, a similar result to that obtained in our study. Wurtzer et al. (2021) was capable to differentiate between total viral genome and protected RNA by using a viability dye. On the other hand, in the study by Cuevas-Ferrando et al. (2021) PMAxx, although showing significant reduction in the signal from 8 replicates of purified SARS-CoV-2 RNA using the E gene (one of the targets used in our study), treatment of the viral RNA with a platinum compound (PtCl4), produced increased results. The authors have also shown that, regardless of the concentration of the platinum compound used, a complete removal of the RT-qPCR signal was achieved in samples with an initial low viral concentration (Ct values ≥30), which agrees with our results as all secondary-treated wastewater samples were detected in Ct values above 30. A study by Cuevas-Ferrando et al. (2021) on the use of viability dyes and platinum compounds to determine infectivity of PEDV by RT-qPCR has shown that PMAxx followed more closely the inactivation rates of PEDV at different temperatures determined by cell culture and that the combination of PMAxx with a surfactant (Triton X-100) sharply improved the results from the viability RT-qPCR. The authors have found similar results to those of our study, with PMAxx performing at a higher level than the other tested viability RT-qPCR.
However, it should be noted that the application of pretreatments to infer infectivity through RT-qPCR may be impaired in situations where disinfection with UV light occurs. The impact of free chlorine and UV254 in Phi6, an enveloped bacteriophage, has shown that UV254 inactivates Phi6 primarily by reacting with the genome (Ye et al., 2016). To be able to work, viability dyes must first enter the cell, and therefore it is necessary that damage to the envelope occur. Many publications have already shown, for non-enveloped viruses, that viability dyes are not effective at removing the signal of UV-inactivated viruses (Karim et al., 2015; Leifels et al., 2015).
A recent publication showed that SARS-CoV-2 RNA continued to be detected even when infectious SARS-CoV-2 was below the detection limit of the cell culture assay (Bivins et al., 2021). Times for 90% reduction (T 90) of viable SARS-CoV-2 in frozen untreated wastewater at room temperature varied between 1.5 and 2.1 days. The authors showed that, at high titers, SARS-CoV-2 could be detected for the entire 7-day duration (105 TCID50 ml−1), and at low titers (103 TCID50 ml−1) detection fell below the limit of detection after only 72 h, with both virus titers being highly improbable to be found in real world scenarios. Nonetheless, in the study by Bivins et al. (2021), the authors might have extended the survivability of the virus due to several experimental design choices made: i) the study was conducted in frozen/thawed wastewater that may have altered the microbiota usually contributing to the inactivation of viruses in water due to proteolytic activity (Gerba et al., 1978; Kim and Unno, 1996; John and Rose, 2005; Gundy et al., 2009; Ye et al., 2016); ii) the study was performed in a single wastewater from a single WWTP in a laboratory setting, therefore excluding the contribution of factors that are known to promote varying inactivation rates including the pH, mixing conditions, and suspended solids (Ye et al., 2016; Aquino de Carvalho et al., 2017). A recent systematic review and meta-analysis of the persistence of coronavirus and surrogates in water determined a 99% reduction of approximately 2 days in wastewater at room temperature (Silverman and Boehm, 2020). Likewise, a meta-analysis concluded that the persistence of different enveloped viruses varied widely for comparable conditions being highly dependent upon virus type, matrix composition and temperature (Aquino de Carvalho et al., 2017). Moreover, the authors concluded that differences in persistence in water are also dependent on the virus strain.
Considering data on the persistence of SARS-CoV-2 and other enveloped viruses in raw wastewater, the residence times in sewage systems (in the range of hours), and in the WWTP (varying between 24 and 48 h depending on the WWTP treatment line), SARS-CoV-2 detected in secondary-treated wastewater should already be mostly non-infectious, a premise supported by the results from our study, either by using cell culture or viability RT-qPCR.
5. Conclusion
To our knowledge, this is the first study applying viability RT-qPCR to infer SARS-CoV-2 infectivity in secondary-treated wastewater. Our study highlights the potential of viability RT-qPCR as a suitable, scalable and easy approach to infer infectivity of SARS-CoV-2 in the water cycle, with potential use in environmental applications used for risk analyses and prevention/control contingency plans as well.
CRediT authorship contribution statement
Sílvia Monteiro: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review and editing, Visualization. Daniela Rente: Investigation. Mónica V. Cunha: Conceptualization, Funding acquisition, Methodology, Formal analysis, Writing – review and editing. Tiago A. Marques: Formal analysis, Writing – review and editing. Eugénia Cardoso: Review and editing, Sampling. João Vilaça: Review and editing, Sampling. Nuno Brôco: Project administration, Funding acquisition, Review and editing; Marta Carvalho: Project administration, Funding acquisition, Review and editing; Ricardo Santos: Conceptualization, Methodology, Resources, Formal analysis, Writing – review and editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank all the workers from Águas de Portugal Group who contributed to wastewater sampling. We also thank the project's Advisory Board (EPAL, Águas do Douro e Paiva, National Environment Agency (APA), National Health Authority (DGS) and Portuguese Water and Waste Services Regulation Authority (ERSAR).
Strategic funding from Fundação para a Ciência e a Tecnologia (FCT), Portugal, to cE3c and BioISI Research Units (UIDB/00329/2020 and UIDB/04046/2020) and to CEAUL (UIDB/00006/2020) are also gratefully acknowledged.
Funding
This work was funded by COMPETE (Programa Operacional Competitividade e Internacionalização & Programa Operacional Regional de Lisboa), Portugal 2020 and FEDER funds, in the scope of project “COVIDETECT: Deteção, quantificação e modelação de SARS-CoV-2 em Águas residuais como ferramenta de alerta precoce para a disseminação do vírus na comunidade (Ref. 048467, Aviso N.° 15/SI/2020).
Editor: Thomas Kevin V
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.152914.
Appendix A. Supplementary data
Supplementary material
References
- Aquino de Carvalho N., Stachler E.N., Cimabue N., Bibby K. Evaluation of Phi6 persistence and suitability as an enveloped virus surrogate. Environ. Sci. Technol. 2017;51(15):8692–8700. doi: 10.1021/acs.est.7b01296. [DOI] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2021;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco A., Abid I., Al-Otaibi N., Pérez-Rodriguez F.J., Fuentes C., Guix S., Pintó R.M., Bosch A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ. Virol. 2019;11(2):184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2021. Biosafety for Specimen Handling.https://www.cdc.gov/coronavirus/2019-ncov/lab/lab-biosafety-guidelines.html (last accessed April, 2021) [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D., Haagmans B.L., van der Veer B., van der Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ferrando E., Randazzo W., Péres-Cataluña A., Falcó I., Navarro D., Martin-Latil S., Díaz-Reolid A., Girón-Gúzman I., Allende A., Sánchez G. Platinum chloride-based viability RT-qPCR for SARS-CoV-2 detection in complex samples. Sci. Rep. 2021;11:18120. doi: 10.1038/s41598-021-97700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC COVID-19 situation update worldwide, as of week 48, updated 9 December 2021. 2021. https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases (last accessed December, 2021)
- Gawlik B., Tavazzi S., Mariani G., Skejo H., Spona M., Higgins T., Medema G., Wintgens T. Publications Office of the European Union; Luxembourg: 2021. SARS-CoV-2 Surveillance Employing Sewage – Towards a Sentinel System, EUR 30684 EN. JRC125065. [DOI] [Google Scholar]
- Gerba C.P., Stagg C.H., Abadie M.G. Characterization of sewage-associated viruses in natural waters. Water Res. 1978;12:805–812. doi: 10.1016/0043-1354(78)90031-3. [DOI] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1:10–14. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Hierholzer J., Killington R. In: Virology Methods Manual. Mahy B., Kangro H., editors. Elsevier; Amsterdam: 1996. Virus isolation and quantification; pp. 25–46. [DOI] [Google Scholar]
- Holshue M.L., deBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K., Washington State 2019-nCoV case investigation team First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John D.E., Rose J.B. Review of factors affecting microbial survival in groundwater. Environ. Sci. Technol. 2005;39(19):7345–7356. doi: 10.1021/es047995w. [DOI] [PubMed] [Google Scholar]
- Karim M.R., Fout G.S., Johnson C.H., White K.M., Parshionikar S.U. Propidium monoazide reverse transcriptase PCR and RT-qPCR for detecting infectious enterovirus and norovirus. J. Virol. Methods. 2015;219:51–61. doi: 10.1016/j.jviromet.2015.02.020. [DOI] [PubMed] [Google Scholar]
- Kim J.-M., Kim H.M., Lee E.J., Jo H., Yoon Y., Lee N.-J., Son J., Lee Y.-J., Kim M.S., Lee Y.-P., Chae S.-J., Park K., Cho S.-R., Park S., Kim S., Wang E., Woo S., Lim A., Park S.-J., Jang J., Chung Y.-S., Chin B.S., Lee J.-S., Lim D., Han M.-G., Yoo C. Detection and isolation of SARS-CoV-2 in serum, urine, and stool specimens of COVID-19 patients from the Republic of Korea. Osong Public Health Res. Perspect. 2020;11(3):112–117. doi: 10.24171/j.phrp.2020.11.3.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-D., Unno H. The roles of microbes in the removal and inactivation of viruses in a biological wastewater treatment system. Water Sci. Technol. 1996;33:243–250. doi: 10.1016/0273-1223(96)00426-X. [DOI] [Google Scholar]
- Klompas M., Baker M., Rhee C. Airborne transmission of SARS-CoV-2 theoretical considerations and available evidence. JAMA. 2020;324(5):441–442. doi: 10.1001/jama.2020.12458. [DOI] [PubMed] [Google Scholar]
- Lamhoujeb S., Fliss I., Ngazoa S.E., Jean J. Evaluation of the persistence of infectious human noroviruses on food surfaces by using real-time nucleic acid sequence-based amplification. App. Environ. Microbiol. 2008;74:3349–3355. doi: 10.1128/AEM.02878-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifels M., Jurzik L., Wilhelm M., Hamza I.A. Use of ethidium monoazide and propidium monoazide to determine viral infectivity upon inactivation by heat, UV-exposure and chlorine. Int. J. Hyg. Environ. Health. 2015;218(8):686–693. doi: 10.1016/j.ijheh.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Leifels M., Cheng D., Sozzi E., Shoults D.C., Wuertz S., Mongkolsuk S., Sirikanchana K. Capsid integrity quantitative PCR to determine virus infectivity in environmental and food applications – a systematic review. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2020.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Monteiro S., Santos R. Enzymatic and viability RT-qPCR assays for evaluation of enterovirus, hepatitis a virus and norovirus inactivation: implications for public health risk assessment. J. Appl. Microbiol. 2018;124(4):965–976. doi: 10.1111/jam.13568. [DOI] [PubMed] [Google Scholar]
- Monteiro S., Rente D., Cunha M.V., Gomes M.C., Marques T.A., Lourenço A.B., Cardoso E., Álvaro P., Silva M., Coelho N., Vilaça J., Meireles F., Brôco N., Carvalho M., Santos R. A wastewater-based epidemiology tool for COVID-19 surveillance in Portugal. Environ. Sci. Technol. 2022;804 doi: 10.1016/j.scitotenv.2021.150264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak P., Topping J.R., Fotheringham V., Gallimore C.I., Gray J.J., Iturriza-Gómara M., Knight A.I. Measurement of the virolysis of human GII.4 norovirus in response to disinfectants and sanitisers. J. Virol. Methods. 2011;174:7–11. doi: 10.1016/j.jviromet.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Polo D., Lois M., Fernández-Núñez M.T., Romalde J.L. Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente H., Randazzo W., Falcó I., Carvajal A., Sánchez G. Rapid selective detection of potentially infectious porcine epidemic diarrhea coronavirus exposed to heat treatments using viability RT-qPCR. Front. Microbiol. 2020;11:1911. doi: 10.3389/fmicb.2020.01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronavirus. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044pmid:32199615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schielke A., Filter M., Appel B., Johne R. Thermal stability of hepatitis E virus assessed by a molecular biology approach. Virol. J. 2011;8:487. doi: 10.1186/1743-422X-8-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman A.I., Boehm A.B. Systematic review and meta-analysis of the persistence and disinfection of human coronavirus and their viral surrogates in water and wastewater. Environ. Sci. Technol. 2020;7:544–553. doi: 10.1021/acs.estlett.0c00313. [DOI] [PubMed] [Google Scholar]
- Tartera C., Araujo R., Michel T., Jofre J. Culture and decontamination methods affecting enumeration of phages infecting Bacteroides fragilis in sewage. Appl. Environ. Microbiol. 1992;58(8):2670–2673. doi: 10.1128/aem.58.8.2670-2673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Protein Atlas ACE2 protein expression summary. 2020. https://www.proteinatlas.org/ENSG00000130234-ACE2
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rother C., Hoelscher M., Bleicker, Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-19. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. MedRxiv. 2020 doi: 10.1101/2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Ferrier-Rembert A., Frenois-Veyrat G., Mouchel J.M., Boni M., Maday Y., OBEPINE consortium. Marechal V., Moulin L. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: implications for wastewater-based epidemiology and risk assessment. Water Res. 2021;198 doi: 10.1016/j.watres.2021.117183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.055. S0016-5085(20)30282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patients with severe COVID-19. Emerg. Infect. Dis. 2020;26(8):1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202(5):756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zhang T., Song D., Huang T., Peng Q., Chen Y., Li A., Zhang F., Wu Q., Ye Y., Tang Y. Comparison and evaluation of conventional RT-PCR SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Mol. Cell. Probes. 2017;33:36–41. doi: 10.1016/j.mcp.2017.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material


