Abstract
Well-orchestrated maternal–fetal cross talk occurs via secreted ligands, interacting receptors, and coupled intracellular pathways between the conceptus and endometrium and is essential for successful embryo implantation. However, previous studies mostly focus on either the conceptus or the endometrium in isolation. The lack of integrated analysis impedes our understanding of early maternal–fetal cross talk. Herein, focusing on ligand–receptor complexes and coupled pathways at the maternal–fetal interface in sheep, we provide the first comprehensive proteomic map of ligand–receptor pathway cascades essential for embryo implantation. We demonstrate that these cascades are associated with cell adhesion and invasion, redox homeostasis, and the immune response. Candidate interactions and their physiological roles were further validated by functional experiments. We reveal the physical interaction of albumin and claudin 4 and their roles in facilitating embryo attachment to endometrium. We also demonstrate a novel function of enhanced conceptus glycolysis in remodeling uterine receptivity by inducing endometrial histone lactylation, a newly identified histone modification. Results from in vitro and in vivo models supported the essential role of lactate in inducing endometrial H3K18 lactylation and in regulating redox homeostasis and apoptotic balance to ensure successful implantation. By reconstructing a map of potential ligand–receptor pathway cascades at the maternal–fetal interface, our study presents new concepts for understanding molecular and cellular mechanisms that fine-tune conceptus–endometrium cross talk during implantation. This provides more direct and accurate insights for developing potential clinical intervention strategies to improve pregnancy outcomes following both natural and assisted conception.
Keywords: proteomics, embryo–maternal cross talk, ligand–receptor pathway cascades, histone lactylation, implantation
Abbreviations: A1BG, alpha-1-B glycoprotein; ACN, acetonitrile; AGC, automatic gain control; AHSG, alpha 2-HS glycoprotein; AI, artificial insemination; ALPL, alkaline phosphatase, liver/bone/kidney; ART, assistant reproductive technology; BCAM, basal cell adhesion molecule; BSA, bovine serum albumin; BSG, basigin; C5, complement C5; C9, complement C9; C areas, caruncular areas; CAPN2, calpain 2; CHP1, calcineurin-like EF-hand protein 1; CID, collision-induced dissociation; CLDN4, claudin 4; COL6A1, collagen type VI alpha 1 chain; COL6A2, collagen type VI alpha 2 chain; CSTB, cathepsin B; DEPs, differentially expressed proteins; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; EMB, embigin; EMILIN1, elastin microfibril interfacer 1; ESI, electrospray ionization; FA, formic acid; FBS, fetal bovine serum; FC, fold change; FDR, false discovery rate; FITC, fluorescein isothiocyanate labeled; FKBP1A, FK506 binding protein 1A; FN1, fibronectin 1; GE, glandular epithelium; GO, Gene Ontology; GOT2, glutamic-oxaloacetic transaminase 2; GSEA, Gene Set Enrichment Analysis; HE, hematoxylin-eosin; HPLC, high-performance liquid chromatography; HSP90AB1, heat shock protein 90 alpha family class B member 1; HSPA8, heat shock protein family A (Hsp70) member 8; IAM, iodoacetamide; IC areas, intercaruncular areas; IDH2, isocitrate dehydrogenase (NADP(+)) 2; IFN-τ, interferon τ; ISG15, interferon-stimulated gene-15; ITGA9, integrin subunit alpha 9; IVF, in vitro fertilization; IVO, in vivo; LC, liquid chromatography; LC-ESI-MS/MS, liquid chromatography–electrospray ionization–tandem mass spectroscopy; LE, luminal epithelium; LTQ, linear ion trap; MCT1, monocarboxylate transporter 1; MCTs, monocarboxylate transporters; MS, mass spectrometry; myo, myometrium; NME1, nometastatic gene 23-H1; P/S, penicillin and streptomycin; PBS, phosphate-buffered saline; PC, PathwayConnector; PGF2α, prostaglandin F2α; PLG, plasminogen; PMSF, phenylmethanesulfonyl fluoride; PTK7, protein tyrosine kinase 7; ROS, reactive oxygen species; RPSA, 40S ribosomal protein SA; S, stroma; SCNT, somatic cell nuclear transfer; SLC2A1, solute carrier family 2 member 1; TCA, trichloroacetic acid; TGFBI, transforming growth factor beta induced; v, blood vessels; XICs, extracted ion currents
In mammals, successful implantation and healthy pregnancy depend on well-orchestrated cross talk between the developmentally competent conceptus and the receptive endometrium (1, 2, 3). Early pregnancy loss occurring during the peri-implantation period is an important and pervasive problem in both human and agricultural animals, especially low-ovulating species. In natural conception, it has been estimated that approximately 75% of failed pregnancies are caused by implantation failure (1, 4, 5). Failed implantation is also a major limiting factor in assisted reproduction (6). Therefore, a comprehensive understanding of the well-orchestrated conceptus–endometrium cross talk at the implantation stage is of importance for basic reproductive or developmental biology, clinical pregnancy intervention, and animal reproductive management.
Although current molecular and cellular mechanisms that govern conceptus–endometrium cross talk have been gleaned primarily from murine models (1, 3, 7), ruminant models have contributed key insights and updated our knowledge of conceptus–endometrium interactions. Compared with the short length of the implantation window in murine models, the prolonged period of apposition and attachment in cows and sheep makes them outstanding candidate models to study the conceptus–endometrium interactions (8, 9). The role of the interferon τ (IFNT), derived from the trophectoderm, in the maternal recognition of pregnancy, was initially shown in sheep (10). It has been well established that IFNT acts locally on the endometrium to prevent luteolysis by inhibiting the synthesis and release of prostaglandin F2α (PGF2α), thus allowing continued production of progesterone by the functional corpus luteum to establish and maintain pregnancy (11, 12, 13). Subsequent studies further reported that IFNT also fine-tunes a range of other physiological processes in endometrial remolding by stimulating or suppressing the expression of certain genes (14, 15, 16, 17). IFNT provides a classic illustration of how the establishment of uterine receptivity depends on a conceptus-originated biochemical signal. Other paracrine signals, such as cortisol, glycosylation-dependent cell adhesion molecule 1-like protein, secreted phosphoprotein 1, and prostaglandins, which contribute to successful implantation and the maintenance of pregnancy, were also deciphered using ruminant models (18, 19). In addition, using bovine embryos of different origins, it was shown that the endometrium responds differently to embryos with different developmental potential, and the limits of endometrial plasticity provided new insights into the contribution of embryo–maternal interactions to successful implantation (20, 21). More recently, paired conceptus–endometrium transcriptome analyses of individual pregnancies in bovine provided the first evidence that the conceptus and endometrium are fine-tuned and coordinated at the level of an individual pregnancy (22).
Fine-tuned and reciprocal cross talk is essential for embryo implantation and involves secreted ligands, their interacting receptors, and coupled pathways between the conceptus and endometrium. Until now, previous research studies have mostly focused on either the conceptus (23, 24, 25, 26) or the endometrium (20, 21, 27, 28, 29) in isolation, the lack of integrated analysis between the paired conceptus and endometrium has made it challenging to advance our understanding of the pathways and functions that govern conceptus–endometrium cross talk during implantation. Thus, despite the importance of ligand–receptor-based maternal–fetal cross talk, there are no reports of systematic studies trying to elucidate the repertoire of signaling routes essential for the cross talk. Recently, an emerging method for evaluation of ligand–receptor interactions based on high-throughput data provides a statistical tool to map the cell–cell communications of diverse physiological and pathological processes (30, 31, 32). However, ligand–receptor interaction maps, as well as coupled pathways at maternal–fetal interface by implantation stage, have never been characterized.
In the present study, via the screening method to identify potential ligand–receptor interactions, we used sheep as the model and present a comprehensive proteomic atlas of cross talk at maternal–fetal interface. This atlas reconstructed the first direct and accurate map of the ligand–receptor pathway cascades essential for implantation. In addition, by highlighting the enhanced glycolysis in the conceptus, we have firstly reported the novel role of lactate-induced histone lactylation, a newly identified important histone modification, in remodeling endometrial receptivity and achieving implantation.
Results
Identifying differentially abundant membrane and secreted proteins between the conceptus and endometrium
To profile the proteome in the conceptus and endometrium by implantation stage, we collected conceptuses, endometrial caruncular (C), and intercaruncular (IC) areas (Figs. 1A and S1A) from 36 pregnant sheep on day 17 of pregnancy, which is the time of filamentous conceptus attachment in the sheep uterus (33, 34) that is frequently selected to explore the mechanisms of conceptus–endometrium cross talk (35, 36, 37). In ruminants, uterine secreting glands are specifically localized in large endometrial areas (i.e., intercaruncular zones, IC), thus glandular IC areas are mainly responsible for the synthesis and secretion of histotroph, including cytokines, growth factors, and adhesion molecules, etc. By contrast, small aglandular caruncular areas of stromal origin (i.e., caruncles, C) are scattered over the endometrium surface. Aglandular C areas serve as the sites of superficial attachment and placentation (12, 38, 39). Therefore, given the structural and functional differences associated to the C and IC areas, these two distinct endometrial zones have to be analyzed separately (21, 22, 37, 40). We divided the 36 samples into equal three pools (12 samples/pool) as biological replicates and sequenced their proteomes, which showed high reproducibility (Fig. S1B). Finally, we identified 1468, 1494, and 1575 proteins in the conceptus, C area, IC area, respectively (Fig. 1A and Table S1). When hierarchical classification was applied, the endometrial C and IC areas clustered closely together, whereas the conceptus samples were categorized separately (Fig. S1C), which was also recapitulated by the results of principal component analysis (Fig. 1B). Comparative analysis of differentially abundant proteins (false discovery rate (FDR) < 0.05, fold change (FC) > 2) identified 196 and 232 proteins that were significantly more abundant in the conceptus compared with those in the C or IC areas; whereas 224 and 325 proteins were significantly more abundant in the C and IC areas compared with those in the conceptus, and a substantial proportion of differentially abundant proteins were specifically enriched in endometrial or conceptus tissues (Fig. S1, D and E).
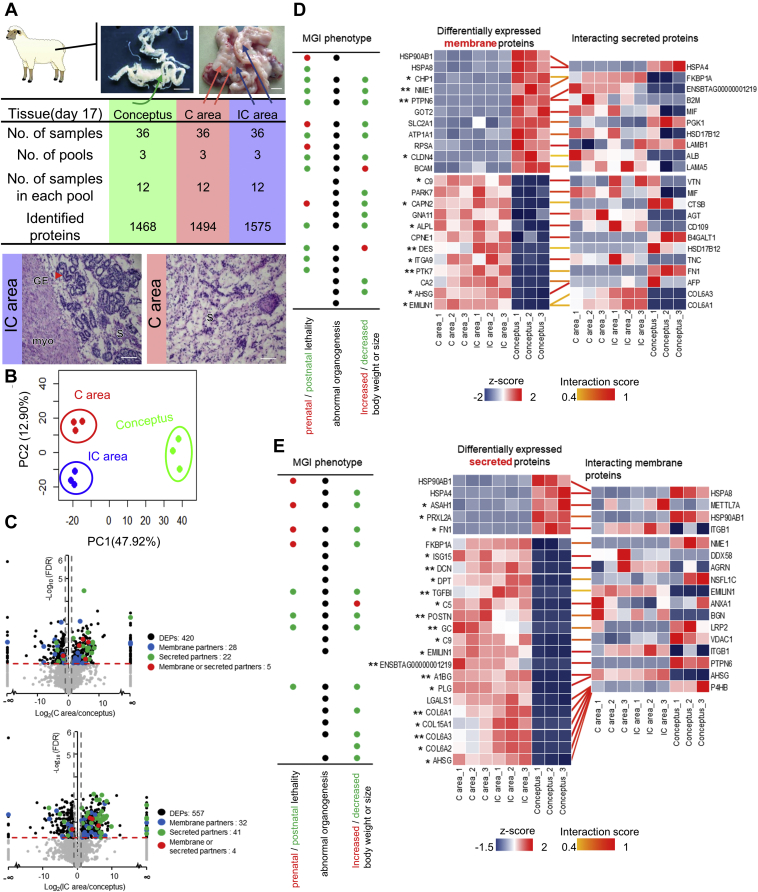
Figure 1.
Screening of differentially abundant membrane or secreted proteins and their interacting partners between the conceptus and endometrium.A, data structure used in this study. Representative images of a pregnant uterus and conceptus (scale bar = 1 cm). Data of genome-wide protein abundance were obtained from the conceptus, as well as the C and IC areas from 36 pregnant uteri, which were equally divided into three pools, 12 samples in each pool. Lower panel, representative hematoxylin-eosin staining images of C and IC areas identifying the tissues from which proteomic data were used in this study. Scale bar = 50 μm. B, principal component analysis of protein expression patterns of three biological replicates of the conceptus, C area, and IC area. C, volcano plot of differentially abundant proteins between the conceptus and the C area (upper panel) or IC area (lower panel). The black dots represent the differentially abundant proteins with FDR <0.05 and FC >2. The red line represents FDR = 0.05, and the gray line represents FC = 2. The blue dots represent identified membrane partners. The green dots represent identified secretion partners. The red dots represent proteins that could be secreted or located in membrane. D and E, overview of the interactions of membrane (D) or secreted (E) proteins that were significantly changed in both C and IC areas compared with that in the conceptus, with their interacting secreted/membrane partners. The Z-score normalized protein abundance is represented in red (relatively high) or blue (relatively low). ∗ represents the fold change of the protein in the conceptus versus that in the C areas and in the conceptus versus that in the IC areas, both >10. ∗∗ represents the fold change >100. The interaction scores are represented in yellow (0.4) to red (1). Left panel, the representative Mouse Genome Informatics (MGI) phenotypic annotations of differentially abundant membrane proteins. FDR, false discovery rate; GE, glandular epithelium; myo, myometrium; S, stroma.
To explore the potential interactions between the conceptus and endometrium systematically, we screened out differentially abundant membrane and secreted proteins based on subcellular localization annotations from UniProt (Fig. 1C). Secreted proteins appeared to be more enriched in the C or IC areas than in the conceptus, which supported the previous notion that the endometrium is an active site of cytokine production and action (41). Next, we used 304 common differentially abundant proteins between the conceptus and endometrial tissues to construct a protein–protein network and found nine high-scoring proteins (interaction edges > 40) (Fig. S1F), including the secreted protein fibronectin (FN1) and the membrane protein 40S ribosomal protein SA (RPSA), both of which are well-known adhesion molecules that have been reported to mediate intercellular interaction in various cell types (42, 43, 44). To investigate the conceptus–endometrium cross talk deeply, we next focused on the following three aspects: differentially abundant membrane proteins, differentially abundant secreted proteins, and differentially enriched pathways.
Screening differentially abundant membrane proteins and their interacting secreted partners
Having identified differentially abundant membrane and secreted proteins of the maternal–fetal interface, we next attempted to screen potential interacting partners that might play roles in conceptus–endometrium cross talk during implantation. To this end, we first focused on the membrane proteins that changed commonly between the conceptus and endometrial tissues (FDR < 0.05, FC > 2). We screened out their interacting secreted partners based on the interaction score from Search Tool for Retrieval of Interacting Genes/Proteins (STRING), a well-established database of known and predicted protein–protein interactions (45), as well as the protein subcellular location from UniProt (Table S2). Many of the screened high-scoring interactions involve proteins that have been reported to participate in transmembrane transport (solute carrier family 2 member 1 (SLC2A1), calcineurin-like EF-hand protein 1 (CHP1), glutamic-oxaloacetic transaminase 2 (GOT2)), heat stress response (heat-shock protein 90 alpha family class B member 1 (HSP90AB1), heat shock protein family A member 8 (HSPA8)), and cell adhesion (claudin 4 (CLDN4), basal cell adhesion molecule (BCAM)) (Fig. 1D). Phenotype annotations based on Mouse Genome Informatics (MGI) database suggested that these membrane proteins may be associated with embryonic and fetal development and survival (Fig. 1D). In addition to differentially abundant membrane proteins common to endometrial tissues relative to the conceptus, we also examined differentially abundant membrane proteins (FDR < 0.05, FC > 2) specific to the C or IC areas and their interacting secreted partners (Fig. S2, A and B), which might provide further candidates to investigate the different roles of the C and IC areas in supporting conceptus implantation.
To further validate the role of the predicted membrane partners in supporting conceptus–endometrium cross talk, we next characterized the expression patterns of these candidates in the endometria of successful and failed pregnancies using our previously published proteomic data (40). We found that the levels of a majority of these candidates changed significantly (p < 0.05) in the endometrium that underwent pregnancy failure, implying the essential functions of these membrane partners in supporting a successful pregnancy (Fig. S2C). Interestingly, we noticed that protein tyrosine kinase 7 (PTK7), an evolutionarily conserved transmembrane receptor, was significantly more abundant in the endometrium that underwent pregnancy failure compared with that in the successful pregnancy (Fig. S2C). In addition, using transcriptomic data of human endometrium from the prereceptive to receptive phase, we found that the mRNA expression levels of PTK7, CLDN4, and STAT3 were also significantly change during establishment of human endometrial receptivity (Fig. S2D), suggesting that those candidates are essential for the normal physiological functions of the endometrium. Meanwhile, we also found that the levels of many of our screened membrane partners changed significantly (p < 0.05) in endometrial tissues of patients with endometriosis compared with those in the healthy endometrium (Fig. S2E), suggesting that these interacting membrane partners might also participate in uterine pathology.
Screening differentially abundant secreted proteins and their interacting membrane partners
Having screened potential interacting partners based on differentially abundant membrane proteins, we next focused on differentially abundant secreted proteins (FDR < 0.05, FC > 2) and their interacting membrane partners (Figs. 1E and S3, A and B). Many of the high-scoring interactions involved cell adhesion molecules (FN1, collagen type VI alpha 1 chain (COL6A1), collagen type VI alpha 2 chain (COL6A2)), glycoprotein family members (alpha 2-HS glycoprotein (AHSG), alpha-1-B glycoprotein (A1BG)), and complement components (complement C5 (C5), complement C9 (C9), plasminogen (PLG)). The MGI phenotype annotations suggested that these membrane partners might be essential for embryonic and fetal development and survival (Fig. 1E). Similarly, reanalysis of secreted partners using previously published proteomic or transcriptomic data also supported the physiological or pathological significance of our screened candidates (Fig. S3, C–E).
Construction of a repository of the potential ligand–receptor pathway cascade
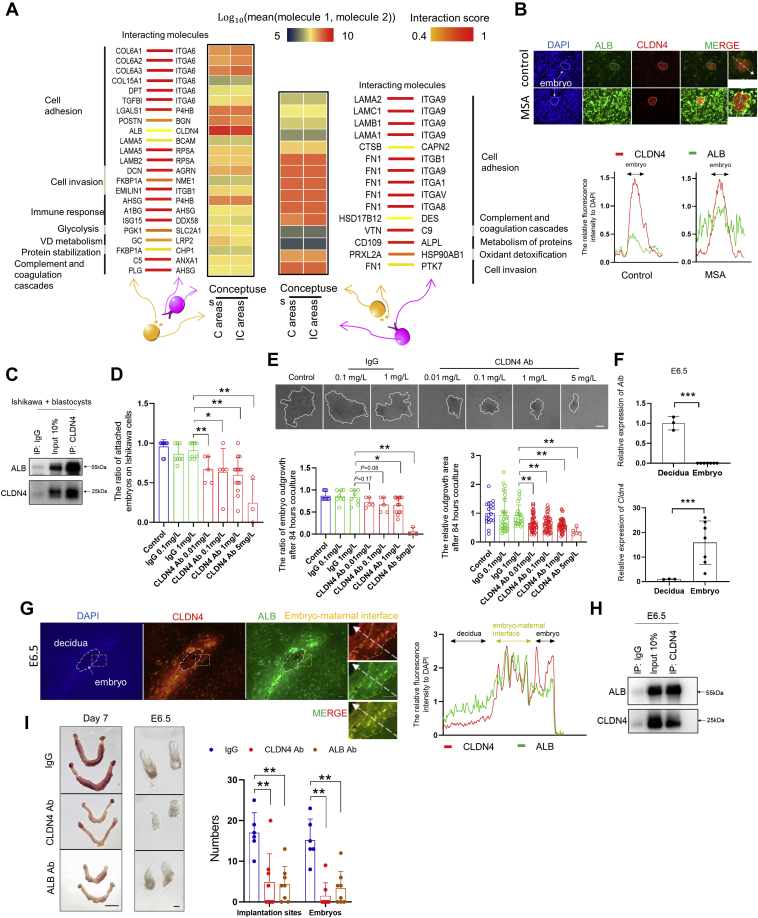
Having characterized conceptus–endometrium cross talk using interacting secreted and membrane partners, we next attempted to further screen out the associated pathways and biological processes that may couple to ligand–receptor complexes to support the cross talk at the maternal–fetal interface. Therefore, functional profiling was performed using proteins that were more abundant in endometrial or conceptus tissues (Fig. S4, A–D). We found that proteins that were abundant in endometrial tissues were functionally associated with glutathione metabolism, complement and coagulation cascades, and focal adhesion (Fig. S4A), as well as biological processes of cell adhesion, response to virus, and actin cross-link formation (Fig. S4B). By contrast, using proteins that were abundant in the conceptus, we identified that pathways or biological processes of energy metabolism, cellular oxidant detoxification, cell–cell adhesion, and protein stabilization were significantly enriched (Fig. S4, C and D). Based on the analyses of differentially abundant secreted proteins, membrane proteins, and enriched pathways between the conceptus and C or IC areas, we integrated them to reconstruct a repository of the potential ligand–receptor pathway cascade, which includes bidirectional cascades: Secreted proteins from the C or IC areas act on membrane partners on the conceptus and in turn promote the development of the conceptus. Secreted proteins of the conceptus act on membrane partners of the C or IC areas and in turn promote endometrial remodeling. Among these, some of candidate cascades have been identified, such as laminins-integrin-cell adhesion and collagens-integrin-cell adhesion (46); whereas many cascades have not been identified at embryo–maternal interface, such as ALB-CLDN4-cell adhesion and fibronectin 1 (FN1)-protein tyrosine kinase 7 (PTK7)-cell invasion (Fig. 2A). Particularly, the albumin is one of the most abundant proteins in uterine luminal fluid (47, 48) and was reported to support embryo development by carrying energy sources, osmoregulators, pH stabilizers, or scavenging ions and toxins (49), but its role in mediating conceptus–endometrium cross talk has never been determined. We therefore focused our analyses on ALB-CLDN4-cell adhesion cascade. Our results showed that 5 μg/ml albumin exposure not only significantly enhanced the adhesiveness of Ishikawa cells (a well-established human receptive endometrial cell line is widely used as an in vitro model for studying mechanism underlying uterine receptivity (50)) (Fig. S5A), but also significantly improved the attachment of blastocysts by using an in vitro implantation model based on the attachment and invasiveness of mouse blastocysts to Ishikawa cells (51, 52) (Fig. S5B). Notably, given there is no well-accepted in vitro model for ovine embryo–endometrium interactions, the use of this convenient method is technically feasible. The short-term (30 min) protein overload assay showed a significant increase in ALB binding interactions on the cell surface of blastocysts exposed to mouse serum albumin relative to control (Fig. S5C). More importantly, the increased binding interactions were also verified using the in vitro implantation model (Figs. 2B and S5D), and the Coimmunoprecipitation (Co-IP) of CLDN4 provided direct evidences of the physical interaction between ALB and CLDN4 (Fig. 2C), as predicted by the protein–protein docking model (Fig. S5E). Then we blocked the function of CLDN4 by supplementing CLDN4 mouse monoclonal antibody (CLDN4 Ab) to the coculture medium. We found the significant decreased ratio of embryo attachment in CLDN4 Ab 0.01, 0.1, 1, and 5 mg/l groups relative to IgG 1 mg/l group (Fig. 2D). Moreover, the blastocyst outgrowth ratios and areas also significantly decreased in CLDN4 Ab 1 mg/l and 5 mg/l groups (Fig. 2E). These indicated the important role of ALB-CLDN4-cell adhesion cascade at embryo implantation. Furthermore, in vivo results from E6.5 mouse embryo–maternal interface showed mutually exclusive expression patterns of Alb and Cldn4 during implantation (Fig. 2F), which was similar to those in day 17 sheep conceptus and C or IC areas (Fig. 1D). Notably, in situ immunostaining and Co-IP of E6.5 showed an obvious colocalization and direct interaction of ALB and CLDN4 at embryo–maternal interface (Figs. 2, G and H, and S5F). Then, to block the function of CLDN4 or ALB at implantation, we injected CLDN4 Ab, ALB mouse monoclonal antibody (ALB Ab), or mouse IgG into the uterus of day 4 pregnant mice. As we expected, the number of implantation sites and E6.5 embryos were significantly reduced in CLDN4 Ab and ALB Ab groups. What’s more interesting is that the morphological structure of some embryos developed to E6.5 was aberrant when CLDN4 or ALB blocked at implantation, which suggested the role of CLDN4 and ALB in embryo development (Figs. 2I and S5G). Collectively, we proved, in vitro and in vivo, the important role of ALB-CLDN4-cell adhesion cascade in embryo–maternal cross talk during implantation.
Figure 2.
Working model of secreted-membrane partner interactions and related essential biological processes that govern cross talk at the maternal–fetal interface.A, overview of selected secreted-membrane partner interactions-related biological processes. The interaction scores are represented in yellow (0.4) to red (1). The average expression level of interacting molecule 1 in cluster 1 and interacting molecule 2 in cluster 2 are indicated by colors (blue to red). B, the colocalization of ALB and CLDN4. Upper panel, representative fluorescent images of ALB (green), CLDN4 (red), and DAPI (blue) staining in mouse embryos cocultured 48 h with Ishikawa cells. Control and MSA group were incubated in FBS medium and MS medium 30 min before fixed, separately. Lower panel, intensity profiles of ALB and CLDN4 relative to DAPI obtained using ImageJ software, along the white dotted straight line crossing Ishikawa cells and blastomeres. White arrows represent the attached embryos. C, Western blot showing coimmunoprecipitated proteins in MSA group of Ishikawa cells cocultured with blastocysts immunoprecipitation experiments, which were repeated at least twice with similar results. D, the attachment of mouse blastocysts to Ishikawa cells was assayed after 48 h of coculture in FBS medium supplemented with different concentrations of CLDN4 Ab or IgG. E, upper panel, the representative image of blastocyst outgrowth on Ishikawa cells after 84 h coculture in FBS medium with different concentration of CLDN4 Ab or IgG. The white lines circled the blastocyst outgrowth. Lower panel, the statistics of the blastocyst outgrowth ratios and the relative outgrowth area after 84 h coculture. F, quantification of Alb and Cldn4 mRNAs relative to Actb in the E6.5 decidua and embryo. G, left panel, representative fluorescent immunohistochemistry images of DAPI (blue), ALB (green), and CLDN4 (red) staining in E6.5. The white dotted circles represent the embryo sites. The areas between two yellow dotted lines represent maternal–fetal interface. Right panel, intensity profiles of ALB and CLDN4 relative to DAPI obtained using ImageJ software, along the white dotted straight line crossing the decidua, embryo–maternal interface, and embryo. H, Western blot showing coimmunoprecipitated proteins in E6.5 immunoprecipitation experiments, which were repeated at least twice with similar results. I, embryo implantation and morphological structure in the presence of CLDN4 Ab or ALB Ab on day 7 of pregnancy. ∗ represents p < 0.05, ∗∗ represents p < 0.01. Scale bar = 200 μm (B, E, G, and middle panel of I), scale bar = 1 cm (left panel of I).
The key pathways and processes at the maternal–fetal interface
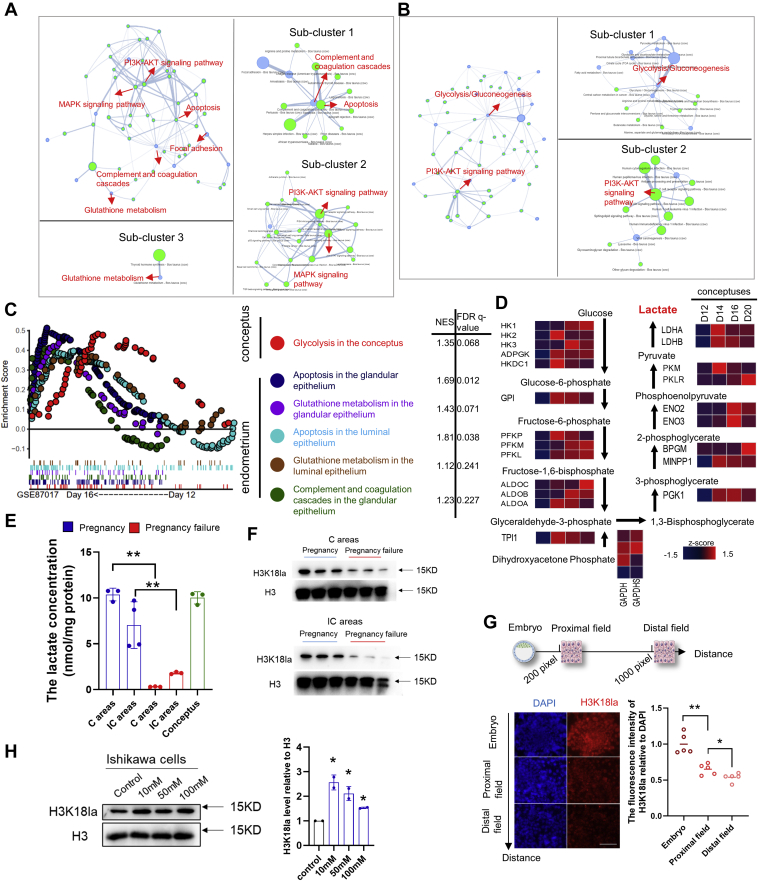
To further investigate the conceptus–endometrium cross talk deeply, we constructed and clustered a pathway network using PathwayConnector (PC) (53) to find key nodes functionally linked with an endometrial receptive status. Three subclusters and their key pathways were noteworthy: subcluster 1: complement and coagulation cascades and apoptosis; subcluster 2: phosphatidylinositol-3-kinase (PI3K)-protein kinase B (AKT) and mitogen-activated protein kinase (MAPK) signaling pathway; subcluster 3: glutathione metabolism (Fig. 3A). Similarly, we constructed the direct connections using the enriched pathways in the conceptus. Two subclusters and their key pathways were clustered: subcluster 1: glycolysis/gluconeogenesis; subcluster 2: the PI3K-AKT signaling pathway (Fig. 3B). Furthermore, the physiological significance of these enriched pathways for conceptus–endometrium cross talk during implantation was further supported via Gene Set Enrichment Analysis (GSEA) of the conceptus and endometrial transcriptomes on day 12 and 16 of pregnancy (48). Our results of reanalysis indicated that glycolysis/gluconeogenesis (normalized enrichment score (NES) = 1.35, FDR q-value = 0.068) is enriched in the conceptus on day 16 of pregnancy (Fig. 3C), while apoptosis, glutathione metabolism, and complement and coagulation cascades are enriched in the endometrium on day 16 of pregnancy (Fig. 3C).
Figure 3.
The key pathways and processes at the maternal–fetal interface.A, the pathway network enriched in the endometrium was constructed by PathwayConnector (PC). The key pathways are marked with red arrows, and the important subclusters are presented separately. The blue dots represent the input pathways, and the green dots represent the complementary pathways. B, the pathway network enriched in the conceptus was constructed using PC. C, differentially regulated pathways between day 16 and day 12 conceptuses, the glandular epithelium, and the luminal epithelium were identified by GSEA (NES > 1, FDR q-value < 0.25). D, dynamic expression of glycolytic enzymes in the conceptus from day 12 to day 20 of pregnancy (GSE87017). The Z-score normalized RPKM is represented in red (relatively high) and blue (relatively low). E, lactate concentrations of the sheep conceptus and endometrial tissues with successful and failed pregnancy by implantation stage. ∗∗ represents p < 0.01. F, immunoblots of H3K18la and H3 from the endometrium with successful and failed pregnancy by implantation stage. G, representative fluorescent images of H3K18la (red) and DAPI (blue) staining in mouse embryos cocultured 72 h with Ishikawa cells, which were chosen from Fig. S5I. The intensity profiles of H3K18la relative to DAPI of Ishikawa cells obtained using ImageJ software, which was stronger at proximal fields than that at distal fields. ∗ represents p < 0.05, ∗∗ represents p < 0.01. Scale bar = 200 μm. H, immunoblots of H3K18la and H3 in Ishikawa cells exposure to different concentrations of sodium L-lactate. Lower panel, the relative intensity of H3K18la. ∗ represents p < 0.05 relative to the control group.
The receptive response of the endometrium to the embryonic lactate signal
Based on the observation of enriched glycolysis/gluconeogenesis in the conceptus at the maternal–fetal interface, we next profiled gene expression dynamics of glycolysis-related enzymes in the conceptus from day 12 to 20 of pregnancy (Fig. 3D) using previously published transcriptomic data (48). The results showed a tendency toward enhanced glycolysis, which agreed with that reported in other species (54, 55, 56, 57). Given that glycolysis is less efficient in terms of ATP production compared with oxidative phosphorylation, it has been thought that there might be nonenergy providing functions of glycolysis in peri-implantation embryos (58, 59). This was reminiscent of the functions of lactate, the metabolic by-product of glycolysis. It has been reported that lactate production increases significantly at implantation in mice and humans (60, 61). In line with this, we detected relatively high levels of lactate in the conceptus and endometrial tissues in pregnant sheep (Fig. 3E), which were comparable to those in tumor cells (62). It should be also emphasized that in the endometrium undergoing pregnancy failure, lactate concentrations were much lower in both the C and IC areas compared with that in their counterparts in the successful pregnancy (Fig. 3E). This fact, together with the results that an important lactate-preferring transporter (monocarboxylate transporter, MCT1) was upregulated in the endometrium during implantation (Fig. S5H), suggested that lactate might play a critical role in manipulating the microenvironment for uterine implantation. Interestingly, we noticed that the chaperone glycoproteins, immunoglobulin family, which interact with monocarboxylate transporters (MCTs) to maintain their stability (63), showed tissue-biased expression patterns (Fig. S5H), indicating that MCT1 in the conceptus might interact preferentially with basigin (BSG), whereas MCT1 in the endometrium might interact preferentially with embigin (EMB).
Notably, a recent study showed that lactate-derived lactylation of histone residues is an important epigenetic modification that directly regulates gene transcription (64). Therefore, we hypothesized that increased lactate levels at the maternal–fetal interface might serve as the donor to stimulate histone lactylation and thus facilitate remodeling processes to prepare a receptive endometrium. To test this, we first determined the H3K18la level, a subtype of recently identified histone lactylation that can respond to lactate in mouse and human cells (64), in the sheep C and IC endometrial areas on day 17 of pregnancy. Western blotting detection showed high levels of H3K18la in the pregnant endometrial C and IC areas. By contrast, H3K18la levels were significantly lower in the endometrium undergoing pregnancy failure, implying the important role of H3K18la in establishing endometrial receptivity (Fig. 3F). Next, we used the in vitro implantation model to test if histone lactylation in the endometrium was associated with embryo-originated stimulations. H3K18la levels detected by immunofluorescence assays in Ishikawa cells showed a declining tendency from the proximal to the distal fields of the attached embryos (Figs. 3G and S5I), suggesting that embryos could stimulate H3K18la in the endometrium. In addition, exogenous supplementation of sodium L-lactate that mimicked physiological (65, 66) or higher concentrations directly induced a significant increase in H3K18la levels in human Ishikawa endometrial cells (Fig. 3H). These facts also suggested that lactate-induced H3K18la in the endometrium appears to be common across species.
Glutathione (GSH)-based cell redox homeostasis is the target of lactate-induced histone lactylation
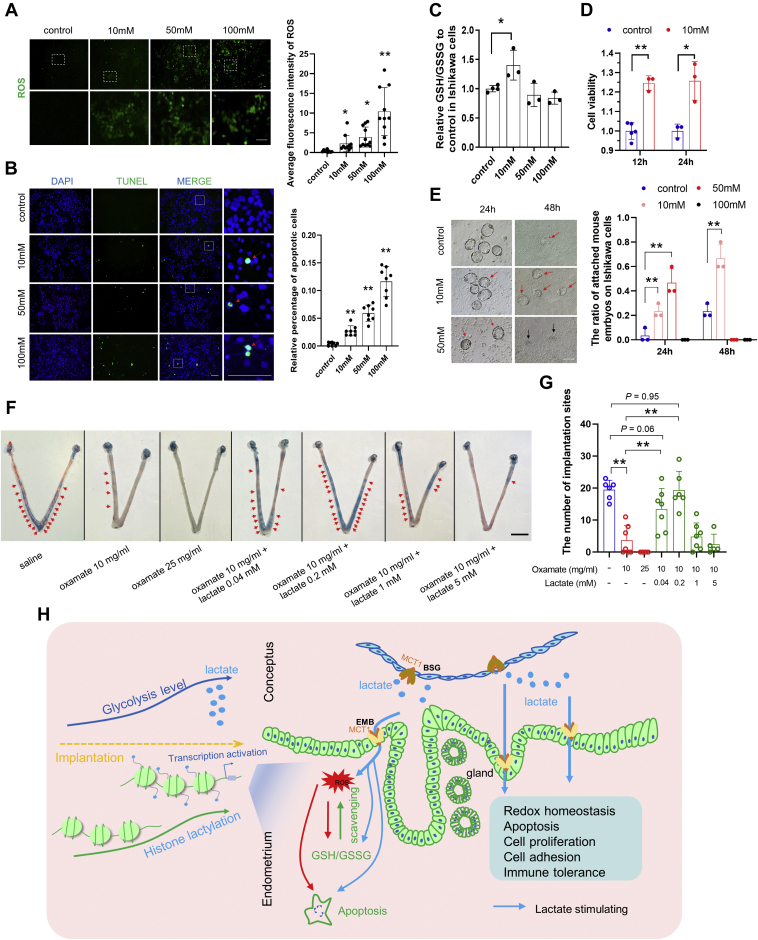
Next, we attempted to study the potential physiological role of lactate-induced lactylation in endometrial remolding during implantation. We focused on the potential H3K18la-associated endometrial genes and pathways via a Venn diagram consisting of the following data: (1) 1784 genes whose promoters were enriched with H3K18la (GSE115354), as the putative H3K18la-regulated targets; (2) 2077 genes whose expression changed significantly in the sheep luminal epithelium from day 12 to 16 of pregnancy (GSE87017), and (3) 2228 genes whose expression changed significantly in the sheep glandular epithelium from day 12 to 16 of pregnancy (GSE87017), as the potential candidates responsible for endometrial remodeling. This analysis identified 44 candidates that might be regulated by H3K18la in the endometrium during embryo implantation (Fig. S6A). Functional annotations showed that these genes are involved in the biological processes of cell redox homeostasis, apoptosis, cytoskeleton, proteasome, cell proliferation, cell migration, cell adhesion, and immune response (Fig. S6A). In addition, the significant responses of these processes to lactate were further validated by GSEA of the published transcriptome data (GSE115354): Apoptosis, cell adhesion molecules, negative regulation of immune response, the PI3K-AKT signaling pathway, and positive regulation of cell proliferation were significantly enriched in lactate-treated mouse cells (NES < −1, FDR q-value < 0.25) (Fig. S6B). Among these, glutathione (GSH)-mediated cell redox homeostasis, as well as related pathways, e.g., reactive oxygen species (ROS) positive response and apoptosis, attracted our attention. ROS at a controlled level has been reported to serve as the critical mediator of endometrial remodeling (67, 68). This was also supported by our observation that ROS positive response genes (69) were enriched significantly in the day 16 endometrium relative to that in the day 12 endometrium and in the endometrium on day 17 of pregnancy relative to that in pregnancy failure (NES > 1, FDR q-value < 0.25) (Fig. S6, C and D), while apoptosis, which is conducive to trophoblast invasion at the site of embryo attachment (70), was also upregulated in sheep endometrium during implantation (Fig. 3C). Thus, we next used Ishikawa cells, which can respond to lactate-induced lactylation, as the model to test if lactate could induce ROS production and apoptosis in the endometrium. As expected, sodium L-lactate supplementation in the culture medium increased ROS levels and apoptotic rate significantly in a dose-dependent manner (Fig. 4, A and B).
Figure 4.
The adaptive response of the endometrium to the embryonic lactate signal.A, representative fluorescent images of ROS production (green) in human endometrial cancer cells (Ishikawa line) exposed to different concentrations of sodium L-lactate. Right panel, normalized fluorescent intensity of ROS. ∗ represents p < 0.05, ∗∗ represents p < 0.01 relative to control group. B, representative fluorescent images of TUNEL (green) and DAPI (blue) staining in Ishikawa cells exposed to different concentrations of sodium L-lactate. Right panel, quantification of TUNEL-positive Ishikawa cells following different treatments. ∗∗ represents p < 0.01 relative to the control group. C, the GSH/GSSG ratio in different sodium L-lactate concentrations treated Ishikawa cells was measured. ∗ represents p < 0.05. D, proliferation assay of Ishikawa cells exposed to 10 mM sodium L-lactate for 12 h and 24 h. ∗∗ represents p < 0.01, ∗ represents p < 0.05. E, the attachment of mouse blastocysts to Ishikawa cells was assayed after 24 and 48 h of coculture in serum-free medium supplemented with different concentrations of sodium L-lactate. Representative images are shown and the red arrows represent the firmly attached embryos, and the black arrows represent the degraded embryos. ∗∗ represents p < 0.01. F and G, uterine morphology (F) and the number of implantation sites (G) on day 5 after uterine horns at day 4 were injected with different treatments. ∗∗ represents p < 0.01. Red arrows represent the implantation sites. Scale bar = 200 μm (A, B, and E). Scale bar = 1 cm (F). H, schematic diagram of the endometrial response to embryonic lactate signals during implantation. Glycolytic metabolism is upregulated in the conceptus, thus the conceptus-derived lactate would serve as an embryonic signal to promote histone lactylation in the endometrium, which in turn participates in the regulation of redox homeostasis, apoptosis, cell proliferation, cell adhesion, and immune tolerance. Blue arrows represent lactate stimulation, red arrows represent ROS stimulation, and the green arrow represents ROS scavenging by GSH/GSSG homeostasis. ROS, reactive oxygen species.
The GSH-based antioxidative system is functionally active at the maternal–fetal interface and is critical for the establishment of pregnancy (71, 72). In line with this, many genes involved in GSH metabolism and function were upregulated in the sheep endometrium during the implantation stage (Fig. S6E), which was consistent with the result of GSEA in Figure 3C. Correspondingly, the importance of the GSH-based antioxidative system in successful implantation was also supported by the analyses of GSEA and differentially abundant proteins related to glutathione metabolism of the endometrium in successful and failed pregnancies (Fig. S6, D and F). These results, together with clues suggesting that the GSH-based antioxidative system might be the target of lactate-induced H3K18la in the endometrium during implantation, led us to test whether lactate could stimulate GSH metabolism and function. Interestingly, we found that a relatively low level (10 mM), but not a high level (50 mM, 100 mM), of sodium L-lactate supplementation significantly promoted the ratio of GSH/GSSG (Fig. 4C), implying enhanced intracellular antioxidative activity (73, 74), while excessive ROS production induced by high-level lactate (Fig. 4A) might consume redundant GSH.
Finally, we attempted to determine if lactate production in the uterine microenvironment is beneficial for implantation. To this end, we first detected the proliferation of Ishikawa cells, because uterine cell proliferation is the prerequisite for establishing endometrial receptivity (7). We found that 10 mM sodium L-lactate stimulated significant and sustained proliferation of Ishikawa cells (Fig. 4D). More importantly, the in vitro mouse embryo attachment assay indicated that sodium L-lactate supplementation could improve embryo attachment in a time- and dose-dependent manner: 10 mM sodium L-lactate supplementation led to a significant increase in the attachment ratio, whereas excessive or prolonged lactate exposure resulted in unexpected embryonic degradation and endometrial cell death (Figs. 4E and S6G), implying that well-controlled lactate production may play an important role at implantation. These findings were further verified in vivo. Intrauterine injection of lactate dehydrogenase inhibitor (oxamate) on day 4 of pregnancy significantly reduced implantation sites on day 5 in a dose-dependent manner. Notably, the implantation defects could be rescued by exogenous addition of the relatively low level (0.04 mM and 0.2 mM), but not high level lactate (1 mM and 5 mM) (Fig. 4, F and G). Collectively, we suggested that a fine-tuned lactate production is essential and beneficial for embryo implantation.
Discussion
Despite the frequently reported high-throughput analyses to characterize the transcriptomic and proteomic features of the embryo/conceptus or endometrium at the implantation stage in mice (23, 75), humans (32, 76, 77), and domestic animals (22, 78, 79, 80), to the best of our knowledge, currently there has never been a report that focuses on the ligand–receptor pathway cascades at the maternal–fetal interface on the protein level. In addition, compared with transcriptomic data, proteomic interaction maps provide more direct and accurate clues for understanding this process. Using sheep as the model, we provided a comprehensive proteomic atlas of conceptus–endometrium cross talk at the implantation stage. Particularly, our study presents the potential ligand–receptor pathway cascades landscape at the maternal–fetal interface. Our reanalysis revealed an enhanced glycolysis in the ovine conceptus by implantation stage, which is line with results reported in mice, pigs, and cattle (54, 55, 56, 57). More recently, the bovine conceptus metabolomic profiling based on conditioned media analysis suggested that conceptus-derived glycolytic metabolite may diffuse to microenvironment at maternal–fetal interface by implantation stage (57). We further provided functional evidence of the potential role of lactate-induced lactylation in remodeling endometrial receptivity using in vivo and in vitro models.
Based on the well-established database of membrane and secreted proteins, we predicted a variety of ligand–receptor complexes at the protein level that might be essential for conceptus–endometrium cross talk. The predictability of our constructed interaction map was supported by a series of membrane or secreted partners that have been identified as key regulators of successful implantation or uterine pathology, such as calpain 2 (CAPN2) (81), integrin subunit alpha 9 (ITGA9) (46, 82), and nonmetastatic gene 23-H1 (NME1) (83, 84). For example, membrane protein CAPN2 is highly expressed in endometrial tissues, while its interacting secreted protein cathepsin B (CTSB) is enriched in the conceptus (Fig. 1D). CAPN2 is concentrated along the basal cell surface of rat uterine luminal epithelial cells at the time of implantation and plays a key role in focal adhesion disassembly and uterine receptivity (81), while CTSB has been reported to play a critical role in endometrial remodeling (85, 86). Our study also revealed the interactions between secreted and membrane adhesion molecules. We found that the expression of FN1 gradually increases in the conceptus from day 12 to 20 of pregnancy and is primarily enriched and secreted by the conceptus (Fig. S6H); whereas its high-scoring interacting membrane partner, integrin subunit beta 1 (ITGB1), a glycoprotein that is localized on the epithelial membrane (87, 88), is more abundant in endometrial tissues at the maternal–fetal interface. The integrin-mediated FN1 binding activity of trophoblast cells has been identified to strengthen trophoblast adhesion to the endometrial extracellular matrix (89, 90).
Our results also indicated candidate interactions associated with both implantation and uterine diseases: Membrane protein NME1, a wide-spectrum tumor metastasis suppressor, and its interacting secreted partner, FK506 binding protein 1A (FKBP1A), a member of the immunophilin protein family, are enriched in the conceptus and endometrial tissues respectively (Fig. 1E). Our reanalysis showed that the level of NME1 was significantly lower in endometrium of endometriosis (Fig. S2E), which is in line with the previous notion that NME1 plays an important role in regulating the invasiveness of trophoblast cells and participates in the pathogenesis of endometriosis (83, 84). Similarly, the level of FKBP1A is also significantly lower in endometrium of endometriosis (Fig. S3E), which agreed with the report that FKBP1A is upregulated in the endometrium of blocked or reduced the number of endometriotic vesicles (91).
More importantly, we also identified a variety of putative interactions that have not been shown previously to participate in cross talk at the maternal–fetal interface, including conceptus-secreted and endometrial membrane partners (CD109-ALPL, PRXL2A-HSP90AB1, and FN1-PTK7), as well as endometrium-secreted and conceptus-enriched partners (ALB-CLDN4, DCN-AGRN, and GC-LRP2).
Collectively, based on the well-established and potential receptor–ligand complexes, we constructed a map of conceptus–endometrium cross talk during implantation (Fig. 2A). On the one hand, conceptus-secreted proteins could interact with their endometrial membrane partners and remodel the endometrial receptivity by regulating the biological processes of cell adhesion, complement and coagulation cascades, metabolism, and oxidant detoxification. On the other hand, embryonic membrane proteins could respond to their endometrial secreted partners and orchestrate biological processes that are essential for conceptus survival and development, e.g., cell invasion, immune tolerance, and glycolysis. In addition, our in vitro and in vivo results of the physical interaction and physiological role of ALB-CLDN4-cell adhesion not only support the predictability of our constructed interaction map, but also suggest that our screened candidates may play a common role in mediating maternal–fetal cross talk among different species. Notably, though the in vitro implantation model used in our research provided convenient and direct evidence of successful or failed implantation, there remain more in-depth functional studies to unearth the role of our potential interactions at implantation.
Of interest, our constructed pathway network and functional experiments suggest a role of an enhanced glycolysis within the conceptus in stimulating endometrial remodeling. We proposed the cross talk model between the conceptus and endometrium (Fig. 4H): During conceptus implantation, glycolysis upregulation leads to enhanced lactate production. The appropriate concentration of lactate at the maternal–fetal interface may serve as an embryo-derived signal that can promote the histone lactylation modification in the endometrium, thus participating in the regulation of redox homeostasis, apoptosis, cell proliferation, cell adhesion, and immune tolerance in receptive endometrium. Particularly, embryonic lactate changed the ROS concentration, glutathione metabolism, and apoptosis in the endometrium, and the GSH/GSSG also scavenged ROS to prevent excessive ROS damage to the endometrium. It is also noteworthy that physiological functions of lactate in inducing histone lactylation and regulating downstream processes could be dose-dependent. High-level lactate over the physiological concentration could be toxic to both the embryo and the endometrium.
Lactate is the product of glycolysis, and sodium oxamate was extensively used as the lactate dehydrogenase inhibitor to block lactate production (64, 92, 93), despite its nonspecific inhibitory effect on pyruvate carboxylase (94). In our study, it is unlikely that possible inhibition of pyruvate carboxylase will contribute considerably to the reduced embryonic attachment and implantation following oxamate treatment. First, according to the public transcriptome data (48, 95), we showed that the expression level of pyruvate carboxylase is far less than lactate dehydrogenase both in sheep and mouse conceptuses by implantation stage (Fig. S6I). Second, only very little anaplerotic activity of pyruvate carboxylase was detected in nonstarved conditions (96, 97). Third, our gain-of-function experiments, in which we have rescued embryonic implantation by injecting lactate to oxamate-treated uterus, not only supported the important role of lactate in implantation, but also indicated that oxamate-induced pregnancy loss was mainly caused by inhibition of lactate production, rather than other nonspecific inhibitory effect of oxamate.
Taken together, our findings identified many putative molecular and cellular mechanisms that might be essential for achieving successful embryo implantation. A more comprehensive understanding of conceptus–endometrium cross talk at the protein level will provide important clues to develop clinical intervention strategies to improve pregnancy outcomes following both natural conception and assisted reproduction.
Experimental procedures
Experimental design
The proteomics of conceptuses, endometrial C areas, and IC areas in day 17 pregnant sheep were profiled. To avoid the ovine blastocysts of different quality implantation in natural pregnancy, causing aberrant cross talk between conceptuses and endometria, we carried the controlled and unified procedures of estrous synchronization, superovulation, artificial insemination (AI), good-quality blastocysts collected, selected, and transferred at day 6.5 of pregnancy, and sample collection at day 17. Considering the biological differences between individuals, we randomly divided samples to three biological replicates with 12 individuals each. In this way, individual differences could be minimized. Additionally, we conducted two technical replicates to avoid the technical error of proteomic detection. The schematic illustration of the experimental design is shown in Fig. S1A.
Ethics statement
The experiments were performed in accordance with the Guide for the Care and Use of Agricultural Animals in Agricultural Research and Teaching, and all procedures were approved by the Institutional Animal Care and Use Committee at the China Agricultural University (Beijing, China). Some publicly available data were analyzed to support our work. The endometrial proteomes of pregnant and pregnancy failed ewes during the peri-implantation period were obtained from Zhao et al. (40). Gene expression in endometrial samples from women with and without endometriosis was obtained from GSE135485. The ChIP-seq data of histone lactylation were from GSE115354 (64). The RNA-seq data of the endometrial luminal epithelium, endometrial glandular epithelium, and conceptus from ewes on day of pregnancy 12, 14, 16, and 20 were from GSE87017 (48). The RNA-seq data of the prereceptive to receptive human endometrium were from Hu et al. (77).
Animals and treatment
Chinese Small Tail Han ewes with normal estrous cycles were selected for the present study. The procedures of estrous synchronization, superovulation, artificial insemination (AI), and transfer of good-quality blastocysts were performed as described in our previous study (40).
ICR female mice aged 7 to 8 weeks and ICR male mice aged 10 to 12 weeks were fed ad libitum and housed under controlled lighting conditions (12 light:12 dark). They were maintained under specific pathogen-free conditions. All animal experiments were approved by and performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of China Agricultural University.
Sample collection
We collected good-quality embryos from 30 donors at day 6.5 of pregnancy. Then, two well-developed blastocysts were transferred into each synchronized recipient ewe (48 synchronized ewes). Sampling procedures were similar to the methods detailed in our previous study (40). Briefly, all recipients were slaughtered at day 17 of pregnancy, and then their uteri were collected and the conceptuses were flushed out using phosphate-buffered saline (PBS). Thirty-seven recipients had filamentous conceptuses. The endometrial caruncular (C) areas and intercaruncular (IC) areas were collected and processed as described (Attia et al. (21)). Opening the ipsilateral uterine horn longitudinally by scissors, the C areas were carefully cut out and collected, and then the IC areas were sampled. These samples were stored at liquid nitrogen until further analysis (Figs. 1A and S1A).
Protein extraction
We divided 36 samples into three equally pools, with 12 samples in each pool (Figs. 1A and S1A). Each pool was ground to powder in liquid nitrogen and stored overnight at −20 °C after adding a fivefold volume of chilled acetone containing 10% trichloroacetic acid (TCA) and 10 mmol/l dithiothreitol (DTT). The samples were then centrifuged at 4 °C, 16,000g for 20 min, and the supernatant was discarded. The precipitates were mixed with 1 ml of chilled acetone containing 10 mmol/l DTT, stored for 30 min at −20 °C, and centrifuged at 4 °C, 20,000g for 30 min. Centrifugation was repeated several times until the supernatant was colorless. The pellets were air-dried, dissolved in lysis buffer containing 1 mmol/l phenylmethanesulfonyl fluoride (PMSF), 2 mmol/l ethylenediaminetetraacetic acid (EDTA), and 10 mmol/l DTT, and sonicated at 200 W for 15 min before being centrifuged at 30,000g at room temperature for 30 min. The protein concentration in the supernatant was then detected by using the Bradford method.
Peptide digestion
An equal amount of protein (50 μg) was taken from each sample, and then equivalent protein samples were prepared by adding 8 mol/l urea solution. To reduce disulfide bonds, the samples were incubated with 10 mmol/l DTT at 56 °C for 1 h, and then cysteine bonding was blocked using 55 mmol/l iodoacetamide (IAM) in a dark room for 45 min. Thereafter, each sample was diluted eightfold with 50 mmol/l ammonium bicarbonate and digested with Trypsin Gold at a protein: trypsin ratio of 20:1 at 37 °C for 16 h. Following desalting using a Strata X C18 column (Phenomenex), the samples were vacuum dried. Peptides generated from digestion were directly loaded for liquid chromatography–electrospray ionization tandem mass spectroscopy (LC-ESI-MS/MS) analysis.
LC-ESI-MS/MS analysis with a linear ion trap-orbitrap (LTQ-orbitrap) collision induced dissociation (CID)
Each sample was resuspended in buffer A [2% acetonitrile (ACN), 0.1% formic acid (FA)] and centrifuged at 20,000g for 10 min. The final peptide concentration for each sample was approximately 0.5 μg/ml. The digested samples were fractionated using a Shimadzu LC-20AD nano-high-performance liquid chromatography (HPLC) system (Shimadzu). Each sample (10 μl) was loaded by the autosampler onto a 2 cm C18 trap column (200 μm inner diameter), and the peptides were eluted onto a resolving 10 cm analytical C18 column (75 μm inner diameter) prepared in-house. The samples were loaded at a flow rate of 15 μl/min for 4 min, and then a 91 min gradient from 2% to 35% buffer B (98% ACN, 0.1% FA) was run at a flow rate of 400 nl/min, followed by a 5 min linear gradient to 80% buffer B that was maintained for 8 min before finally returning to 2% buffer B within 2 min. The peptides were subjected to nano-electrospray ionization and then detected by MS/MS in an LTQ Orbitrap Velos (Thermo Fisher Scientific) coupled online to an HPLC system. Intact peptides were detected in the Orbitrap analyzer at a resolution of 60,000 m/z. Peptides were selected for MS/MS using the CID operating mode with a normalized collision energy setting of 35%, and ion fragments were detected in the LTQ. One MS scan followed by ten MS/MS scans was applied for the ten most abundant precursor ions above a threshold ion count of 5000 in the MS survey scan. Dynamic exclusion was used, with the following parameters: Repeat counts = 2; repeat duration = 30 s; and exclusion duration = 120 s. The applied electrospray voltage was 1.5 kV. Automatic gain control (AGC) was used to prevent overfilling of the ion trap; 1 × 104 ions were accumulated in the ion trap to generate CID spectra. For MS scans, the m/z scan range was 350 to 2000 Da.
Proteomic analysis
MaxQuant software (version 1.1.1.36) was used to analyze the mass spectra. And the detailed methods are in Supporting Information.
Cell culture
A human endometrial cancer cell line (Ishikawa, ATCC) and Ishikawa cells were grown at 37 °C in fetal bovine serum (FBS) medium (DMEM/F-12, HEPES (Gibco) supplemented with 10% (Hyclone) and 1% penicillin/streptomycin (Invitrogen)) in a humidified 5% CO2 incubator.
Fluorescent immunocytochemistry
Ishikawa cells were cocultured with blastocysts 72 h, then fixed with 4% paraformaldehyde for further detection of H3K18la. For albumin overload assay in Figure 2B and Fig. S5, C and D, control group was incubated 30 min with FBS-medium, while MSA group was incubated with MS-medium (DMEM/F-12, HEPES (Gibco) supplemented with 1% penicillin/streptomycin (Invitrogen) and 10% mouse serum) 30 min, then fixed with 4% paraformaldehyde for further detection of ALB and CLDN4. Immunostaining was performed according to standard protocols using the following primary antibodies: anti-H3K18la (1:1000, PTM-1406, PTM Bio Inc), anti-ALB (1:250, sc-271605, Santa Cruz Biotechnology), anti-CLDN4 (1:250, 16195-1-AP, Proteintech). And appropriate Alexa Fluor dye conjugated secondary antibodies (Invitrogen) were used. Nuclei were stained with DAPI (Life Technologies). The fluorescence signals were imaged using a confocal laser scanning microscope (Digital Eclipse C1; Nikon). Data analysis was performed by ImageJ.
Analyses of differentially abundant proteins
To facilitate data analysis, all proteins were mapped to the Ensembl Bos Taurus gene ID. p values from student’s t test were corrected for multiple hypothesis tests using the FDR procedure (98). For each comparison, gene expression levels were considered significantly different when FDR <0.05 and fold change (FC) >2. The protein quantification values of the conceptus, C area, IC area, and differentially abundant proteins of each comparison (conceptus versus C area and conceptus versus IC area) are shown in Table S1.
Annotations of differentially abundant membrane and secreted proteins
The annotations of membrane and secreted proteins were processed as described (Vento-Tormo et al. (32)). Briefly, differentially abundant proteins were mapped to UniProt (https://www.uniprot.org/), then KW-0964 (secreted) was used to screen out the secreted partners. KW-1003 (cell membrane) was used to screen out the plasma membrane proteins. Peripheral proteins from the plasma membrane were annotated using the UniProt Keyword SL-9903, and the remaining proteins were annotated as membrane proteins, which act as extracellular signal receptors. Interestingly, some proteins were annotated as both secreted proteins and membrane proteins, such as heat shock protein 90 alpha family class B member 1 (HSP90AB1) and elastin microfibril interfacer 1 (EMILIN1). The differentially abundant secreted proteins or membrane proteins are shown in Table S2. Phenotype annotations of differentially abundant membrane proteins or secreted proteins were analyzed based on the MGI database (Mouse Genome Informatics, http://www.informatics.jax.org/phenotypes.shtml).
Construction of membrane-secreted partner interactions
To screen out valuable ligand–receptor complexes that may be essential for maternal–fetal cross talk, we used the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING version 11.0; https://string-db.org/), a well-established database of known and predicted protein–protein interactions that includes direct (physical) and indirect (functional) associations (45) to build the membrane-secreted partner interactions using edge information from three separate forms of evidence: databases, experiments, and text mining. Firstly, we inputted a secreted protein (or a membrane protein) to acquire its interacting partners and interaction scores. Then we mapped its interacting partners to UniProt to screen out the membrane partners (or secreted partners). Finally, we chose the membrane partner (or secreted partner) with the highest interaction score. In this way, we constructed the interactions of differentially abundant membrane proteins (or secreted proteins) with their secreted partners (or membrane partners). All the interactions are shown in Table S3.
Statistical analysis
The p value of Student’s t test was calculated using GraphPad Prism 7.0 software (GraphPad Inc) or R for individual analysis. It was considered significant when the p value <0.05. Error bars represent the means ± SD (standard deviation). Details of individual tests are outlined within each figure legend.
The full details can be found in Supporting Information, including experimental procedures and reagents: proteomic analysis; Western blotting; the detection of lactate concentration; the detection of GSH/GSSG; measurement of ROS; cell viability assay; the detection of cell apoptosis; cell adhesion assay; preparation of mouse embryos; in vitro embryo implantation model and mouse embryo attachment assay; intrauterine injection; gene ontology (GO) and KEGG pathway analysis and pathway network construction; hierarchical clustering analysis, principal components analysis, GSEA, and protein–protein network construction; constructed the protein–protein docking model of ALB-CLDN4; real-time quantitative PCR analysis; fluorescent immunohistochemistry
Data availability
All relevant data are within the paper and its Supporting Information files.
Supporting information
This article contains supporting information (98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the members of our laboratory for their helpful comments on the manuscript. This work was supported by the grants from National Key R&D Program (2017YFD0501905 and 2017YFD0501901), National Natural Science Foundation of China (No. 3167246 and 31972573), National Support Program for Youth Top-notch Talents and the Earmarked Fund for the Innovative Teams of Beijing Swine Industrialization Research Program, National transgenic major program (2009ZX08006-008B and 2008ZX08006-002, 2011ZX08006-002, 2013ZX08006-002).
Author contributions
Q. Y., L. A., and J. T. conceptualization; Q. Y., J. L., L. A., and W. Z. formal analysis; J. T. and L. A. funding acquisition; Q. Y., J. L., Y. W., W. Z., W. W., J. C., J. Y., Y. Y., S. Z., M. C., Q. L., L. M., Y. T., and Y. H. investigation; K. M. and H. Z. resources; Q. Y. writing—original draft; Q. Y., J. L., L. A., and J. T. writing—review and editing.
Edited by Qi-Qun Tang
Supporting information
References
- 1.Wang H., Dey S.K. Roadmap to embryo implantation: Clues from mouse models. Nat. Rev. Genet. 2006;7:185–199. doi: 10.1038/nrg1808. [DOI] [PubMed] [Google Scholar]
- 2.Bazer F.W., Spencer T.E., Johnson G.A., Burghardt R.C. Uterine receptivity to implantation of blastocysts in mammals. Front. Biosci. (Schol. Ed.) 2011;3:745–767. doi: 10.2741/s184. [DOI] [PubMed] [Google Scholar]
- 3.Cha J., Sun X., Dey S.K. Mechanisms of implantation: Strategies for successful pregnancy. Nat. Med. 2012;18:1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zinaman M.J., Clegg E.D., Brown C.C., O'Connor J., Selevan S.G. Estimates of human fertility and pregnancy loss. Fertil. Steril. 1996;65:503–509. [PubMed] [Google Scholar]
- 5.Norwitz E.R., Schust D.J., Fisher S.J. Implantation and the survival of early pregnancy. N. Engl. J. Med. 2001;345:1400–1408. doi: 10.1056/NEJMra000763. [DOI] [PubMed] [Google Scholar]
- 6.Edwards R.G. Human implantation: The last barrier in assisted reproduction technologies? Reprod. Biomed. Online. 2006;13:887–904. doi: 10.1016/s1472-6483(10)61039-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S., Lin H., Kong S., Wang S., Wang H., Wang H., Armant D.R. Physiological and molecular determinants of embryo implantation. Mol. Aspects Med. 2013;34:939–980. doi: 10.1016/j.mam.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morel O., Laporte-Broux B., Tarrade A., Chavatte-Palmer P. The use of ruminant models in biomedical perinatal research. Theriogenology. 2012;78:1763–1773. doi: 10.1016/j.theriogenology.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Lee K.Y., DeMayo F.J. Animal models of implantation. Reproduction. 2004;128:679–695. doi: 10.1530/rep.1.00340. [DOI] [PubMed] [Google Scholar]
- 10.Martal J.L., Chêne N.M., Huynh L.P., L’Haridon R.M., Reinaud P.B., Guillomot M.W., Charlier M.A., Charpigny S.Y. IFN-tau: A novel subtype I IFN1. Structural characteristics, non-ubiquitous expression, structure-function relationships, a pregnancy hormonal embryonic signal and cross-species therapeutic potentialities. Biochimie. 1998;80:755–777. doi: 10.1016/s0300-9084(99)80029-7. [DOI] [PubMed] [Google Scholar]
- 11.Bazer F.W. Mediators of maternal recognition of pregnancy in mammals. Exp. Biol. Med. 1992;199:373–384. doi: 10.3181/00379727-199-43371a. [DOI] [PubMed] [Google Scholar]
- 12.Spencer T.E., Johnson G.A., Bazer F.W., Burghardt R.C. Implantation mechanisms: Insights from the sheep. Reproduction. 2004;128:657–668. doi: 10.1530/rep.1.00398. [DOI] [PubMed] [Google Scholar]
- 13.Spencer T.E., Johnson G.A., Bazer F.W., Burghardt R.C., Palmarini M. Pregnancy recognition and conceptus implantation in domestic ruminants: Roles of progesterone, interferons and endogenous retroviruses. Reprod. Fertil. Dev. 2007;19:65. doi: 10.1071/rd06102. [DOI] [PubMed] [Google Scholar]
- 14.Choi Y., Johnson G.A., Burghardt R.C., Berghman L.R., Joyce M.M., Taylor K.M., David Stewart M., Bazer F.W., Spencer T.E. Interferon regulatory factor-two restricts expression of interferon-stimulated genes to the endometrial stroma and glandular epithelium of the ovine uterus. Biol. Reprod. 2001;65:1038–1049. doi: 10.1095/biolreprod65.4.1038. [DOI] [PubMed] [Google Scholar]
- 15.Rosenfeld C.S., Han C.-S., Alexenko A.P., Spencer T.E., Roberts R.M. Expression of interferon receptor subunits, IFNAR1 and IFNAR2, in the ovine uterus. Biol. Reprod. 2002;67:847–853. doi: 10.1095/biolreprod.102.004267. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., Antoniou E., Liu Z., Hearne L.B., Roberts R.M. A microarray analysis for genes regulated by interferon-τ in ovine luminal epithelial cells. Reproduction. 2007;134:123–135. doi: 10.1530/REP-07-0387. [DOI] [PubMed] [Google Scholar]
- 17.Oliveira J.F., Henkes L.E., Ashley R.L., Purcell S.H., Smirnova N.P., Veeramachaneni D.N.R., Anthony R.V., Hansen T.R. Expression of interferon (IFN)-stimulated genes in extrauterine tissues during early pregnancy in sheep is the consequence of endocrine IFN-τ release from the uterine vein. Endocrinology. 2008;149:1252–1259. doi: 10.1210/en.2007-0863. [DOI] [PubMed] [Google Scholar]
- 18.Dorniak P., Welsh T.H., Bazer F.W., Spencer T.E. Endometrial HSD11B1 and cortisol regeneration in the ovine uterus: Effects of pregnancy, interferon tau, and prostaglandins. Biol. Reprod. 2012;86:124. doi: 10.1095/biolreprod.111.097063. [DOI] [PubMed] [Google Scholar]
- 19.Spencer T.E., Bartol F.F., Bazer F.W., Johnson G.A., Joyce M.M. Identification and characterization of glycosylation-dependent cell adhesion molecule 1-like protein expression in the ovine uterus. Biol. Reprod. 1999;60:241–250. doi: 10.1095/biolreprod60.2.241. [DOI] [PubMed] [Google Scholar]
- 20.Bauersachs S., Ulbrich S.E., Zakhartchenko V., Minten M., Reichenbach M., Reichenbach H.-D., Blum H., Spencer T.E., Wolf E. The endometrium responds differently to cloned versus fertilized embryos. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5681–5686. doi: 10.1073/pnas.0811841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mansouri-Attia N., Sandra O., Aubert J., Degrelle S., Everts R.E., Giraud-Delville C., Heyman Y., Galio L., Hue I., Yang X., Tian X.C., Lewin H.A., Renard J.-P. Endometrium as an early sensor of in vitro embryo manipulation technologies. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5687–5692. doi: 10.1073/pnas.0812722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biase F.H., Hue I., Dickinson S.E., Jaffrezic F., Laloe D., Lewin H.A., Sandra O. Fine-tuned adaptation of embryo–endometrium pairs at implantation revealed by transcriptome analyses in Bos taurus. PLoS Biol. 2019;17:1–20. doi: 10.1371/journal.pbio.3000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie J., An L., Miao K., Hou Z., Yu Y., Tan K., Sui L., He S., Liu Q., Lei X., Wu Z., Tian J. Comparative analysis of dynamic proteomic profiles between in vivo and in vitro produced mouse embryos during postimplantation period. J. Proteome Res. 2013;12:3843–3856. doi: 10.1021/pr301044b. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z., Hagen D.E., Elsik C.G., Ji T., Morris C.J., Moon L.E., Rivera R.M. Characterization of global loss of imprinting in fetal overgrowth syndrome induced by assisted reproduction. Proc. Natl. Acad. Sci. U. S. A. 2015;112:4618–4623. doi: 10.1073/pnas.1422088112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan K., An L., Miao K., Ren L., Hou Z., Tao L., Zhang Z., Wang X., Xia W., Liu J., Wang Z., Xi G., Gao S., Sui L., Zhu D.-S., et al. Impaired imprinted X chromosome inactivation is responsible for the skewed sex ratio following in vitro fertilization. Proc. Natl. Acad. Sci. U. S. A. 2016;113:3197–3202. doi: 10.1073/pnas.1523538113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malo Estepa I., Tinning H., Rosas Vasconcelos E.J., Fernandez-Fuertes B., Sánchez J.M., Burns G.W., Spencer T.E., Lonergan P., Forde N. Protein synthesis by day 16 bovine conceptuses during the time of maternal recognition of pregnancy. Int. J. Mol. Sci. 2020;21:2870. doi: 10.3390/ijms21082870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Wang C., Hou Z., Miao K., Zhao H., Wang R., Guo M., Wu Z., Tian J., An L. Comparative analysis of proteomic profiles between endometrial caruncular and intercaruncular areas in ewes during the peri-implantation period. J. Anim. Sci. Biotechnol. 2013;4:39. doi: 10.1186/2049-1891-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moraes J.G.N., Behura S.K., Geary T.W., Spencer T.E. Analysis of the uterine lumen in fertility-classified heifers: I. Glucose, prostaglandins, and lipids. Biol. Reprod. 2020;102:456–474. doi: 10.1093/biolre/ioz191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moraes J.G.N., Behura S.K., Bishop J.V., Hansen T.R., Geary T.W., Spencer T.E. Analysis of the uterine lumen in fertility-classified heifers: II. Proteins and metabolites†. Biol. Reprod. 2020;102:571–587. doi: 10.1093/biolre/ioz197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramilowski J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., Forrest A.R.R. A draft network of ligand–receptor-mediated multicellular signalling in human. Nat. Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar M.P., Du J., Lagoudas G., Jiao Y., Sawyer A., Drummond D.C., Lauffenburger D.A., Raue A. Analysis of single-cell RNA-seq identifies cell-cell communication associated with tumor characteristics. Cell Rep. 2018;25:1458–1468.e4. doi: 10.1016/j.celrep.2018.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.-E., Stephenson E., Polański K., Goncalves A., Gardner L., Holmqvist S., Henriksson J., Zou A., Sharkey A.M., et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spencer T., Burghardt R., Johnson G., Bazer F. Conceptus signals for establishment and maintenance of pregnancy. Anim. Reprod. Sci. 2004;82–83:537–550. doi: 10.1016/j.anireprosci.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Wan P.-C., Bao Z.-J., Wu Y., Yang L., Hao Z.-D., Yang Y.-L., Shi G.-Q., Liu Y., Zeng S.-M. αv β3 integrin may participate in conceptus attachment by regulating morphologic changes in the endometrium during peri-implantation in ovine. Reprod. Domest. Anim. 2011;46:840–847. doi: 10.1111/j.1439-0531.2011.01752.x. [DOI] [PubMed] [Google Scholar]
- 35.Song G., Satterfield M.C., Kim J., Bazer F.W., Spencer T.E. Gastrin-releasing peptide (GRP) in the ovine uterus: Regulation by interferon tau and progesterone. Biol. Reprod. 2008;79:376–386. doi: 10.1095/biolreprod.108.068403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bazer F.W., Spencer T.E., Ott T.L. Interferon tau: A novel pregnancy recognition signal. Am. J. Reprod. Immunol. 1997;37:412–420. doi: 10.1111/j.1600-0897.1997.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang Q., Fu W., Wang Y., Miao K., Zhao H., Wang R., Guo M., Wang Z., Tian J., An L. The proteome of IVF-induced aberrant embryo-maternal crosstalk by implantation stage in ewes. J. Anim. Sci. Biotechnol. 2020;11:7. doi: 10.1186/s40104-019-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wimsatt W.A. New histological observations on the placenta of the sheep. Am. J. Anat. 1950;87:391–457. doi: 10.1002/aja.1000870304. [DOI] [PubMed] [Google Scholar]
- 39.Bazer F.W. Uterine protein secretions: Relationship to development of the conceptus. J. Anim. Sci. 1975;41:1376–1382. doi: 10.2527/jas1975.4151376x. [DOI] [PubMed] [Google Scholar]
- 40.Zhao H., Sui L., Miao K., An L., Wang D., Hou Z., Wang R., Guo M., Wang Z., Xu J., Wu Z., Tian J. Comparative analysis between endometrial proteomes of pregnant and non-pregnant ewes during the peri-implantation period. J. Anim. Sci. Biotechnol. 2015;6:1–14. doi: 10.1186/s40104-015-0017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabibzadeh S. Human endometrium: An active site of cytokine production and action. Endocr. Rev. 1991;12:272–290. doi: 10.1210/edrv-12-3-272. [DOI] [PubMed] [Google Scholar]
- 42.Li F., Redick S.D., Erickson H.P., Moy V.T. Force measurements of the α5β1 integrin–fibronectin interaction. Biophys. J. 2003;84:1252–1262. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groulx J.F., Gagné D., Benoit Y.D., Martel D., Basora N., Beaulieu J.F. Collagen VI is a basement membrane component that regulates epithelial cell-fibronectin interactions. Matrix Biol. 2011;30:195–206. doi: 10.1016/j.matbio.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Lefebvre T., Rybarczyk P., Bretaudeau C., Vanlaeys A., Cousin R., Brassart-Pasco S., Chatelain D., Dhennin-Duthille I., Ouadid-Ahidouch H., Brassart B., Gautier M. TRPM7/RPSA complex regulates pancreatic cancer cell migration. Front. Cell Dev. Biol. 2020;8:549. doi: 10.3389/fcell.2020.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., von Mering C. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh H., Aplin J.D. Adhesion molecules in endometrial epithelium: Tissue integrity and embryo implantation. J. Anat. 2009;215:3–13. doi: 10.1111/j.1469-7580.2008.01034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leese H.J. The formation and function of oviduct fluid. Reproduction. 1988;82:843–856. doi: 10.1530/jrf.0.0820843. [DOI] [PubMed] [Google Scholar]
- 48.Brooks K., Burns G.W., Moraes J.G.N., Spencer T.E. Analysis of the uterine epithelial and conceptus transcriptome and luminal fluid proteome during the peri-implantation period of pregnancy in sheep. Biol. Reprod. 2016;95:88. doi: 10.1095/biolreprod.116.141945. [DOI] [PubMed] [Google Scholar]
- 49.Dobrinsky J.R., Johnson L.A., Rath D. Development of a culture medium (BECM-3) for porcine embryos: Effects of bovine serum albumin and fetal bovine serum on embryo development. Biol. Reprod. 1996;55:1069–1074. doi: 10.1095/biolreprod55.5.1069. [DOI] [PubMed] [Google Scholar]
- 50.Hannan N.J., Paiva P., Dimitriadis E., Salamonsen L.A. Models for study of human embryo implantation: Choice of cell lines? Biol. Reprod. 2010;82:235–245. doi: 10.1095/biolreprod.109.077800. [DOI] [PubMed] [Google Scholar]
- 51.Ruane P.T., Berneau S.C., Koeck R., Watts J., Kimber S.J., Brison D.R., Westwood M., Aplin J.D. Apposition to endometrial epithelial cells activates mouse blastocysts for implantation. Mol. Hum. Reprod. 2017;23:617–627. doi: 10.1093/molehr/gax043. [DOI] [PubMed] [Google Scholar]
- 52.Berneau S.C., Ruane P.T., Brison D.R., Kimber S.J., Westwood M., Aplin J.D. Investigating the role of CD44 and hyaluronate in embryo-epithelial interaction using an in vitro model. Mol. Hum. Reprod. 2019;25:265–273. doi: 10.1093/molehr/gaz011. [DOI] [PubMed] [Google Scholar]
- 53.Minadakis G., Zachariou M., Oulas A., Spyrou G.M. PathwayConnector: Finding complementary pathways to enhance functional analysis. Bioinformatics. 2019;35:889–891. doi: 10.1093/bioinformatics/bty693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merkle S., Pretsch W. A glucosephosphate isomerase (GPI) null mutation in Mus musculus: Evidence that anaerobic glycolysis is the predominant energy delivering pathway in early post-implantation embryos. Comp. Biochem. Physiol. Part B Comp. Biochem. 1992;101:309–314. doi: 10.1016/0305-0491(92)90004-b. [DOI] [PubMed] [Google Scholar]
- 55.Kelly A., West J.D. Genetic evidence that glycolysis is necessary for gastrulation in the mouse. Dev. Dyn. 1996;207:300–308. doi: 10.1002/(SICI)1097-0177(199611)207:3<300::AID-AJA7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 56.Kramer A.C., Steinhauser C.B., Gao H., Seo H., McLendon B.A., Burghardt R.C., Wu G., Bazer F.W., Johnson G.A. Steroids regulate SLC2A1 and SLC2A3 to deliver glucose into trophectoderm for metabolism via glycolysis. Endocrinology. 2020;161:1–19. doi: 10.1210/endocr/bqaa098. [DOI] [PubMed] [Google Scholar]
- 57.Simintiras C.A., Sánchez J.M., McDonald M., O’Callaghan E., Aburima A.A., Lonergan P. Conceptus metabolomic profiling reveals stage-specific phenotypes leading up to pregnancy recognition in cattle†. Biol. Reprod. 2021;104:1022–1033. doi: 10.1093/biolre/ioab021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith D.G., Sturmey R.G. Parallels between embryo and cancer cell metabolism. Biochem. Soc. Trans. 2013;41:664–669. doi: 10.1042/BST20120352. [DOI] [PubMed] [Google Scholar]
- 59.Gardner D.K. Lactate production by the mammalian blastocyst: Manipulating the microenvironment for uterine implantation and invasion? Bioessays. 2015;37:364–371. doi: 10.1002/bies.201400155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu W., Liang Y.X., Luo J.M., Gu X.W., Chen Z.C., Fu T., Zhu Y.Y., Lin S., Diao H.L., Jia B., Yang Z.M. Nucleolar stress regulation of endometrial receptivity in mouse models and human cell lines. Cell Death Dis. 2019;10:831. doi: 10.1038/s41419-019-2071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu X.W., Yang Y., Li T., Chen Z.C., Fu T., Pan J.M., Ou J.P., Yang Z.M. ATP mediates the interaction between human blastocyst and endometrium. Cell Prolif. 2019;53 doi: 10.1111/cpr.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belt J.A., Thomas J.A., Buchsbaum R.N., Racker E. Inhibition of lactate transport and glycolysis in Ehrlich ascites tumor cells by bioflavonoids. Biochemistry. 1979;18:3506–3511. doi: 10.1021/bi00583a011. [DOI] [PubMed] [Google Scholar]
- 63.Payen V.L., Mina E., Van Hée V.F., Porporato P.E., Sonveaux P. Monocarboxylate transporters in cancer. Mol. Metab. 2020;33:48–66. doi: 10.1016/j.molmet.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., Liu W., Kim S., Lee S., Perez-Neut M., Ding J., Czyz D., Hu R., Ye Z., He M., et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harris S.E., Gopichandran N., Picton H.M., Leese H.J., Orsi N.M. Nutrient concentrations in murine follicular fluid and the female reproductive tract. Theriogenology. 2005;64:992–1006. doi: 10.1016/j.theriogenology.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 66.Madaan A., Nadeau-Vallée M., Rivera J.C., Obari D., Hou X., Sierra E.M., Girard S., Olson D.M., Chemtob S. Lactate produced during labor modulates uterine inflammation via GPR81 (HCA1) Am. J. Obstet. Gynecol. 2017;216:60.e1–60.e17. doi: 10.1016/j.ajog.2016.09.072. [DOI] [PubMed] [Google Scholar]
- 67.Leitao B., Jones M.C., Fusi L., Higham J., Lee Y., Takano M., Goto T., Christian M., Lam E.W.-F., Brosens J.J. Silencing of the JNK pathway maintains progesterone receptor activity in decidualizing human endometrial stromal cells exposed to oxidative stress signals. FASEB J. 2010;24:1541–1551. doi: 10.1096/fj.09-149153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu J., Hansen J.M., Hao L., Taylor R.N., Sidell N. Retinoic acid stimulation of VEGF secretion from human endometrial stromal cells is mediated by production of reactive oxygen species. J. Physiol. 2011;589:863–875. doi: 10.1113/jphysiol.2010.200808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 70.Armant D.R. Life and death responses to trophinin-mediated adhesion during blastocyst implantation. Cell Cycle. 2011;10:574–575. [PubMed] [Google Scholar]
- 71.Al-Gubory K.H., Garrel C. Antioxidative signalling pathways regulate the level of reactive oxygen species at the endometrial–extraembryonic membranes interface during early pregnancy. Int. J. Biochem. Cell Biol. 2012;44:1511–1518. doi: 10.1016/j.biocel.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 72.Al-Gubory K.H., Faure P., Garrel C. Different enzymatic antioxidative pathways operate within the sheep caruncular and intercaruncular endometrium throughout the estrous cycle and early pregnancy. Theriogenology. 2017;99:111–118. doi: 10.1016/j.theriogenology.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 73.Canestrari F., Buoncristiani U., Galli F., Giorgini A., Albertini M.C., Carobi C., Pascucci M., Bossù M. Redox state, antioxidative activity and lipid peroxidation in erythrocytes and plasma of chronic ambulatory peritoneal dialysis patients. Clin. Chim. Acta. 1995;234:127–136. doi: 10.1016/0009-8981(94)05990-a. [DOI] [PubMed] [Google Scholar]
- 74.Kim Y.-J. Antimelanogenic and antioxidant properties of gallic acid. Biol. Pharm. Bull. 2007;30:1052–1055. doi: 10.1248/bpb.30.1052. [DOI] [PubMed] [Google Scholar]
- 75.Sui L., An L., Tan K., Wang Z., Wang S., Miao K., Ren L., Tao L., He S., Yu Y., Nie J., Liu Q., Xing L., Wu Z., Hou Z., et al. Dynamic proteomic profiles of in vivo- and in vitro-produced mouse postimplantation extraembryonic tissues and placentas. Biol. Reprod. 2014;91:1–16. doi: 10.1095/biolreprod.114.124248. [DOI] [PubMed] [Google Scholar]
- 76.Carson D.D., Lagow E., Thathiah A., Al-Shami R., Farach-Carson M.C., Vernon M., Yuan L., Fritz M.A., Lessey B. Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Mol. Hum. Reprod. 2002;8:871–879. doi: 10.1093/molehr/8.9.871. [DOI] [PubMed] [Google Scholar]
- 77.Hu S., Yao G., Wang Y., Xu H., Ji X., He Y., Zhu Q., Chen Z., Sun Y. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-Seq. J. Clin. Endocrinol. Metab. 2014;99:E2744–E2753. doi: 10.1210/jc.2014-2155. [DOI] [PubMed] [Google Scholar]
- 78.Romero J.J., Liebig B.E., Broeckling C.D., Prenni J.E., Hansen T.R. Pregnancy-induced changes in metabolome and proteome in ovine uterine flushings. Biol. Reprod. 2017;97:273–287. doi: 10.1093/biolre/iox078. [DOI] [PubMed] [Google Scholar]
- 79.Mamo S., Mehta J.P., Forde N., McGettigan P., Lonergan P. Conceptus-endometrium crosstalk during maternal recognition of pregnancy in cattle. Biol. Reprod. 2012;87:1–9. doi: 10.1095/biolreprod.112.099945. [DOI] [PubMed] [Google Scholar]
- 80.Chae J.-I., Kim J., Lee S.G., Jeon Y.-J., Kim D.-W., Soh Y., Seo K.S., Lee H.K., Choi N.-J., Ryu J., Kang S., Cho S.-K., Lee D.-S., Chung H.M., Koo A.D.-B. Proteomic analysis of pregnancy-related proteins from pig uterus endometrium during pregnancy. Proteome Sci. 2011;9:41. doi: 10.1186/1477-5956-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaneko Y., Murphy C.R., Day M.L. Calpain 2 activity increases at the time of implantation in rat uterine luminal epithelial cells and administration of calpain inhibitor significantly reduces implantation sites. Histochem. Cell Biol. 2014;141:423–430. doi: 10.1007/s00418-013-1165-y. [DOI] [PubMed] [Google Scholar]
- 82.Lessey B.A. Adhesion molecules and implantation. J. Reprod. Immunol. 2002;55:101–112. doi: 10.1016/s0165-0378(01)00139-5. [DOI] [PubMed] [Google Scholar]
- 83.Xie K.M., Hou X.F., Li M.Q., Li D.J. NME1 at the human maternal-fetal interface downregulates titin expression and invasiveness of trophoblast cells via MAPK pathway in early pregnancy. Reproduction. 2010;139:799–808. doi: 10.1530/REP-09-0490. [DOI] [PubMed] [Google Scholar]
- 84.Li M.Q., Shao J., Meng Y.H., Mei J., Wang Y., Li H., Zhang L., Chang K.K., Wang X.Q., Zhu X.Y., Li D.J. NME1 suppression promotes growth, adhesion and implantation of endometrial stromal cells via Akt and MAPK/Erk1/2 signal pathways in the endometriotic milieu. Hum. Reprod. 2013;28:2822–2831. doi: 10.1093/humrep/det248. [DOI] [PubMed] [Google Scholar]
- 85.Quinn C.E., Simmons D.G., Kennedy T.G. Expression of cystatin C in the rat endometrium during the peri-implantation period. Biochem. Biophys. Res. Commun. 2006;349:236–244. doi: 10.1016/j.bbrc.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 86.Song G., Bailey D.W., Dunlap K.A., Burghardt R.C., Spencer T.E., Bazer F.W., Johnson G.A. Cathepsin B, cathepsin L, and cystatin C in the porcine uterus and placenta: Potential roles in endometrial/placental remodeling and in fluid-phase transport of proteins secreted by uterine epithelia across placental areolae. Biol. Reprod. 2010;82:854–864. doi: 10.1095/biolreprod.109.080929. [DOI] [PubMed] [Google Scholar]
- 87.Simon C. Embryonic regulation of integrins 3, 4, and 1 in human endometrial epithelial cells in vitro. J. Clin. Endocrinol. Metab. 1997;82:2607–2616. doi: 10.1210/jcem.82.8.4153. [DOI] [PubMed] [Google Scholar]
- 88.Wadehra M., Forbes A., Pushkarna N., Goodglick L., Gordon L.K., Williams C.J., Braun J. Epithelial membrane protein-2 regulates surface expression of αvβ3 integrin in the endometrium. Dev. Biol. 2005;287:336–345. doi: 10.1016/j.ydbio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 89.Goossens K., Van Soom A., Van Zeveren A., Favoreel H., Peelman L.J. Quantification of fibronectin 1 (FN1) splice variants, including two novel ones, and analysis of integrins as candidate FN1 receptors in bovine preimplantation embryos. BMC Dev. Biol. 2009;9:1–16. doi: 10.1186/1471-213X-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.George E.L., Georges-Labouesse E.N., Patel-King R.S., Rayburn H., Hynes R.O. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 91.Seguinot-Tarafa I., Luna N., Suarez E., Appleyard C.B., Flores I. Inhibition of histone methyltransferase EZH2 suppresses endometriotic vesicle development in a rat model of endometriosis. Reprod. Sci. 2020;27:1812–1820. doi: 10.1007/s43032-020-00257-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu J., Chai P., Xie M., Ge S., Ruan J., Fan X., Jia R. Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021;22:85. doi: 10.1186/s13059-021-02308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Muramatsu H., Sumitomo M., Morinaga S., Kajikawa K., Kobayashi I., Nishikawa G., Kato Y., Watanabe M., Zennami K., Kanao K., Nakamura K., Suzuki S., Yoshikawa K. Targeting lactate dehydrogenase-a promotes docetaxel-induced cytotoxicity predominantly in castration-resistant prostate cancer cells. Oncol. Rep. 2019;42:224–230. doi: 10.3892/or.2019.7171. [DOI] [PubMed] [Google Scholar]
- 94.Marlier J.F., Cleland W.W., Zeczycki T.N. Oxamate is an alternative substrate for pyruvate carboxylase from Rhizobium etli. Biochemistry. 2013;52:2888–2894. doi: 10.1021/bi400075t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y., Xiang Y., Yin Q., Du Z., Peng X., Wang Q., Fidalgo M., Xia W., Li Y., Zhao Z.A., Zhang W., Ma J., Xu F., Wang J., Li L., et al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 2018;50:96–105. doi: 10.1038/s41588-017-0003-x. [DOI] [PubMed] [Google Scholar]
- 96.Merritt M.E., Harrison C., Sherry A.D., Malloy C.R., Burgess S.C. Flux through hepatic pyruvate carboxylase and phosphoenolpyruvate carboxykinase detected by hyperpolarized 13C magnetic resonance. Proc. Natl. Acad. Sci. U. S. A. 2011;108:19084–19089. doi: 10.1073/pnas.1111247108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jin E.S., Moreno K.X., Wang J.X., Fidelino L., Merritt M.E., Sherry A.D., Malloy C.R. Metabolism of hyperpolarized [1-13C]pyruvate through alternate pathways in rat liver. NMR Biomed. 2016;29:466–474. doi: 10.1002/nbm.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 99.Koch J.M., Ramadoss J., Magness R.R. Proteomic profile of uterine luminal fluid from early pregnant ewes. J. Proteome Res. 2010;9:3878–3885. doi: 10.1021/pr100096b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 101.Graumann J., Hubner N.C., Kim J.B., Ko K., Moser M., Kumar C., Cox J., Schöler H., Mann M. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics. 2008;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- 102.Feng J., Naiman D.Q., Cooper B. Probability-based pattern recognition and statistical framework for randomization: Modeling tandem mass spectrum/peptide sequence false match frequencies. Bioinformatics. 2007;23:2210–2217. doi: 10.1093/bioinformatics/btm267. [DOI] [PubMed] [Google Scholar]
- 103.Li G.-Z., Vissers J.P.C., Silva J.C., Golick D., Gorenstein M.V., Geromanos S.J. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 104.Waanders L.F., Chwalek K., Monetti M., Kumar C., Lammert E., Mann M. Quantitative proteomic analysis of single pancreatic islets. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Behringer R., Gertsenstein M., Vintersen Nagy K., Nagy A. Cold Spring Harbor Laboratory Press; New York, United States: 2014. Manipulating the Mouse Embryo: A Laboratory Manual, Fourth Edition. [Google Scholar]
- 106.Kang Y.J., Forbes K., Carver J., Aplin J.D. The role of the osteopontin-integrin αvβ3 interaction at implantation: Functional analysis using three different in vitro models. Hum. Reprod. 2014;29:739–749. doi: 10.1093/humrep/det433. [DOI] [PubMed] [Google Scholar]
- 107.Kong C., Sun L., Zhang M., Ding L., Zhang Q., Cheng X., Diao Z., Yan Q., Zhang H., Fang T., Zhen X., Hu Y., Sun H., Yan G. miR-133b reverses the hydrosalpinx-induced impairment of embryo attachment through down-regulation of SGK1. J. Clin. Endocrinol. Metab. 2016;101:1478–1489. doi: 10.1210/jc.2015-1588. [DOI] [PubMed] [Google Scholar]
- 108.Gu X.-W., Chen Z.-C., Yang Z.-S., Yang Y., Yan Y.-P., Liu Y.-F., Pan J.-M., Su R.-W., Yang Z.-M. Blastocyst-induced ATP release from luminal epithelial cells initiates decidualization through the P2Y2 receptor in mice. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aba3396. [DOI] [PubMed] [Google Scholar]
- 109.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 111.Wickham H. Springer-Verlag; New York, NY: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 112.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M.J., Patterson N., Mesirov J.P., Golub T.R., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 114.Chin C., Chen S., Wu H. cyto-Hubba: A cytoscape plug-in for hub object analysis in network biology. Genome Informatics. 2009;5:2–3. [Google Scholar]
- 115.Desta I.T., Porter K.A., Xia B., Kozakov D., Vajda S. Performance and its limits in rigid body protein-protein docking. Structure. 2020;28:1071–1081.e3. doi: 10.1016/j.str.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vajda S., Yueh C., Beglov D., Bohnuud T., Mottarella S.E., Xia B., Hall D.R., Kozakov D. New additions to the ClusPro server motivated by CAPRI. Proteins. 2017;85:435–444. doi: 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kozakov D., Beglov D., Bohnuud T., Mottarella S.E., Xia B., Hall D.R., Vajda S. How good is automated protein docking? Proteins. 2013;81:2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.