Abstract
The coronavirus disease 2019 (COVID-19) pandemic continues worldwide with many variants arising, some of which are variants of concern (VOCs). A recent VOC, omicron (B.1.1.529), which obtains a large number of mutations in the receptor-binding domain (RBD) of the spike protein, has risen to intense scientific and public attention. Here, we studied the binding properties between the human receptor ACE2 (hACE2) and the VOC RBDs and resolved the crystal and cryoelectron microscopy structures of the omicron RBD-hACE2 complex as well as the crystal structure of the delta RBD-hACE2 complex. We found that, unlike alpha, beta, and gamma, omicron RBD binds to hACE2 at a similar affinity to that of the prototype RBD, which might be due to compensation of multiple mutations for both immune escape and transmissibility. The complex structures of omicron RBD-hACE2 and delta RBD-hACE2 reveal the structural basis of how RBD-specific mutations bind to hACE2.
Keywords: omicron, delta, variants of concern, VOCs, receptor-binding domain, RBD, hACE2, structure
Graphical abstract
Structural analysis of the complexes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RBD with the human ACE2 receptor for omicron and delta reveals variant-specific binding features.
Introduction
Receptor binding is a key step of virus invasion (Lu et al., 2015). Similar to severe acute respiratory syndrome coronavirus (SARS-CoV), SARS-CoV-2 uses its spike (S) protein to recognize the host receptor ACE2 (Wang et al., 2020). The C-terminal domain (CTD) of the S protein, also known as the receptor-binding domain (RBD), is responsible for ACE2 recognition and is an important determinant of host range (Wang et al., 2020; Wu et al., 2020). Additionally, the S protein is a major target for neutralizing antibodies (Liu et al., 2020; Piccoli et al., 2020). Mutations on the S protein, particularly the RBD, could lead to immune escape and undermine the efficacy of current therapies and vaccines (Garcia-Beltran et al., 2021).
SARS-CoV-2 keeps evolving into new variants due to sustained global transmission (Li et al., 2021a). Variants with evidence of increased transmissibility, severe disease, reduction in neutralization by antibodies of convalescents or vaccines, and a higher risk of eluding testing are classified as variants of concern (VOCs) (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html). Currently, five VOCs have been announced by the World Health Organization (WHO), namely alpha, beta, gamma, delta, and the recently identified omicron. Studies have reported that VOCs, especially the beta variant, are subject to immune escape when treated with most clinically available monoclonal antibodies (Abdool Karim and de Oliveira, 2021; Ferreira et al., 2021; Li et al., 2021b; Zhou et al., 2021). Compromised protective potency of current licensed vaccines targeting previous VOCs has also been reported, but the extent of decrease in efficacy is rather mild (Abdool Karim and de Oliveira, 2021; Cao et al., 2021a; Huang et al., 2021a). The delta VOC was shown to have much higher transmissibility (Liu and Rocklöv, 2021), which could be explained by its higher replication rates and S-mediated entry (Mlcochova et al., 2021).
In contrast to other VOCs that emerged when natural immunity was predominant, omicron has emerged under the circumstances when vaccination against SARS-CoV-2 is prevalent. Genome sequencing demonstrated that omicron carries an unusually high number of mutations, especially on the S protein (23 amino acid substitutions) and its RBD (15 amino acid substitutions). Putative analyses indicate that some of the mutations are associated with enhanced viral transmission, infectivity, and immune evasions (Genovese et al., 2021; Qin et al., 2021). Recent studies showed that omicron has extensive escape of known neutralizing antibodies and sera from convalescent patients or people vaccinated with the Pfizer vaccine BNT162b2 (Cao et al., 2021b; Cele et al., 2021; Wang et al., 2022). Unveiling the molecular mechanism of receptor recognition of VOCs is essential for understanding the impact of omicron on infection, transmission, and immune escape.
In this study, we examined several VOCs, including alpha, beta, gamma, delta, and omicron, and demonstrated that omicron variant RBD binds to human ACE2 (hACE2) with comparable affinity to that of the prototype. We then determined both the crystal and cryoelectron microscopy (cryo-EM) structures of omicron RBD complexed with hACE2 and identified the role of key residues for receptor recognition. Additionally, we solved the crystal structure of a delta RBD-hACE2 complex. Our results provide important molecular information about both omicron and delta VOCs and could provide guidance for development of novel prophylaxis/therapeutics targeting the dominant omicron variant.
Results
Receptor binding and efficient transduction of omicron engaged by hACE2
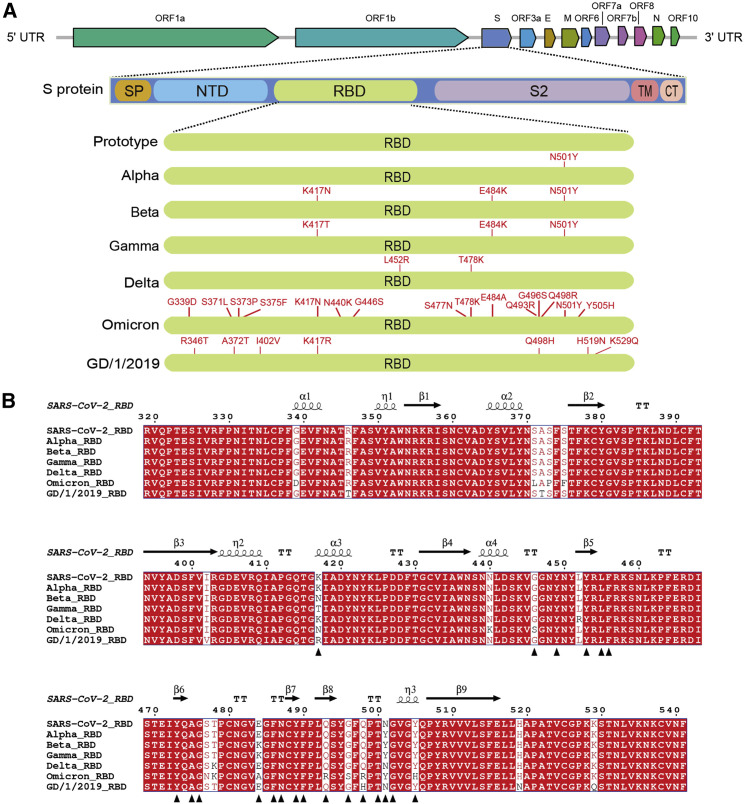
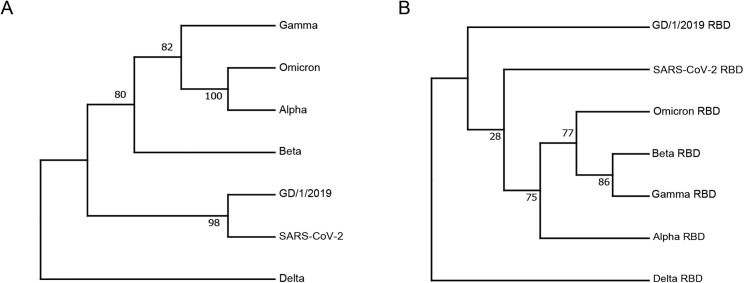
To demonstrate the substitution pattern of SARS-CoV-2 VOCs (alpha, beta, gamma, delta, and omicron), we mapped the substitutions of their RBDs and performed sequence alignment (Figures 1A and 1B). We also include the RBD of GD/1/2019, a pangolin-origin coronavirus closely related to SARS-CoV-2. The mapping demonstrated that omicron RBD carries an unusually high number of substitutions, among which substitution of K417, T478, E484, and N501 were previously observed in other VOCs, whereas 11 others were novel. Eight substitutions are located on the hACE2 binding residues of prototype RBD (Wang et al., 2020), some of which were previously observed on VOCs or bat-origin RaTG13 and pangolin-origin GD/1/2019 or GX/P2V/2017 (Liu et al., 2021a; Niu et al., 2021). In the phylogenetic analysis of genome, omicron is close to alpha (Figure S1 A). But for RBD region, omicron is closely related to beta and gamma (Figure S1B).
Figure 1.
Mutation mapping and sequence alignment of RBDs from prototype SARS-CoV-2, VOCs, and pangolin-origin GD/1/2019
(A) Architecture of SARS-CoV-2 genome and S protein and substitution mapping of RBDs from prototype SARS-CoV-2, VOCs, and pangolin-origin GD/1/2019.
(B) Sequence alignment of abovementioned RBDs. Interacting residues of prototype SARS-CoV-2 RBD to hACE2 are labeled with black triangles. The alignment is performed by T-COFFEE and visualized by ESPript 3.0.
See also Figure S1.
Figure S1.
Phylogenetic analysis of representative SARS-CoV-2, related VOCs, and GD/1/2019, Related to Figure 1
Phylogenetic trees are depicted for nucleotide sequences of the full genome (A) and RBD (B).
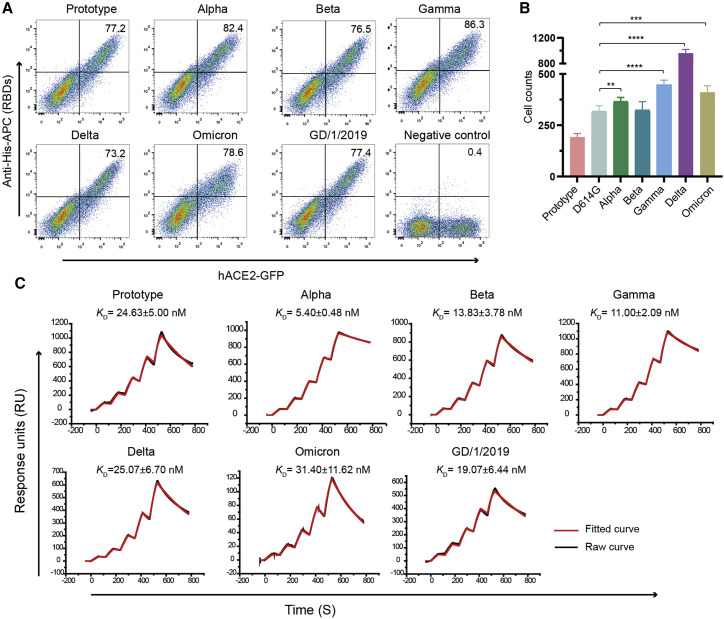
We then explored the binding capacity of omicron RBD to hACE2 in comparison to that of the prototype and other variants as well as to pangolin-origin GD/1/2019. We purified RBDs from the prototype SARS-CoV-2 (the first strain of SARS-CoV-2, which was isolated from a clinical patient on January 6, 2020; GISAID: EPI_ISL_402119; Tan et al., 2020) and the variants, as well as pangolin-origin GD/1/2019, and tested their binding to hACE2-expressing BHK-21 cells with flow cytometry assay (Figure 2 A). RBDs from VOCs and GD/1/2019 bound to hACE2-expressing cells to comparable levels with prototype RBD, with 73.3%–86.3% positive cells in staining experiments. We further measured the binding affinities of the RBDs to hACE2 with surface plasmon resonance (SPR) assay (Figure 2C; Table S1). The results showed that the prototype SARS-CoV-2 binds to hACE2 with a dissociation constant (K D) of 24.63 nM. RBDs from alpha, beta, and gamma demonstrated enhanced affinities to various extents, ranging from 1.78- to 4.56-fold increase. On the other hand, omicron, along with delta RBD, showed no significant change in binding affinities when compared with those of the prototype RBD (Figure 2C).
Figure 2.
Binding characteristics and transduction of SARS-CoV-2 VOCs
(A) BHK-21 cells stably expressing GFP and hACE2 were incubated with His-tagged RBDs from prototype SARS-CoV-2, VOCs, and pangolin-origin GD/1/2019. Allophycocyanin (APC)-conjugated anti-His antibodies were used to detect the His-tagged protein binding to the cells. Representative results from three experiments are shown. The frequency of RBD-binding positive cells in ACE2-GFP positive cells are labeled in the upright corner.
(B) Transduction efficiency of pseudotyped prototype SARS-CoV-2, D614G variant, and VOCs. The GFP-positive cells were quantified with CQ1 confocal image cytometer (Yokogawa), and representative results from three experiments are shown. Statistical significance was analyzed with a t test comparison between VOCs and D614G.
(C) SPR characterization of RBDs from prototype SARS-CoV-2, VOCs, and pangolin-origin GD/1/2019 interacting with hACE2. Dissociation constant (KD) indicates mean ± SD of three independent replicates. Actual and fitted curves are colored in black and red, respectively.
See also Figure S2.
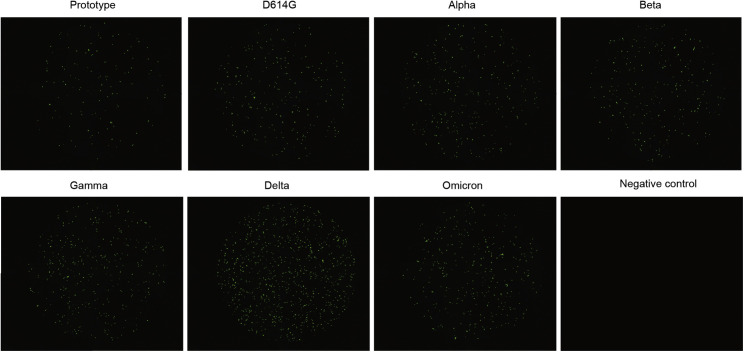
To test whether omicron S protein supports virus entry with different efficiency, we tested the potential influence of the SARS-CoV-2 variants on cell infection with an assay with pseudotyped virus. Vesicular stomatitis virus (VSV)-vectored pseudotyped viruses incorporated S proteins of the prototype, the aforementioned VOCs, and D614G, the first dominant SARS-CoV-2 variant (Hou et al., 2020). These pseudotyped viruses were then used to infect Vero cells, and the GFP signals were quantified by confocal image cytometer for the infection efficiency. The results showed that all variants infected Vero cells with significantly higher efficiency (Figure 2B; Figure S2 ). The delta strain was shown to infect cells with the highest efficiency. The omicron strain also demonstrated an increased trend in infection efficiency, although it was milder than that of the delta strain.
Figure S2.
Entry of the pseudovirus of prototype SARS-CoV-2 and its VOCs into Vero cells, Related to Figure 2
Green fluorescent Vero cells indicate pseudovirus-transducing cells. Untransfected Vero cells were used as negative controls.
Structures of hACE2 in complex with RBDs of omicron and delta variants
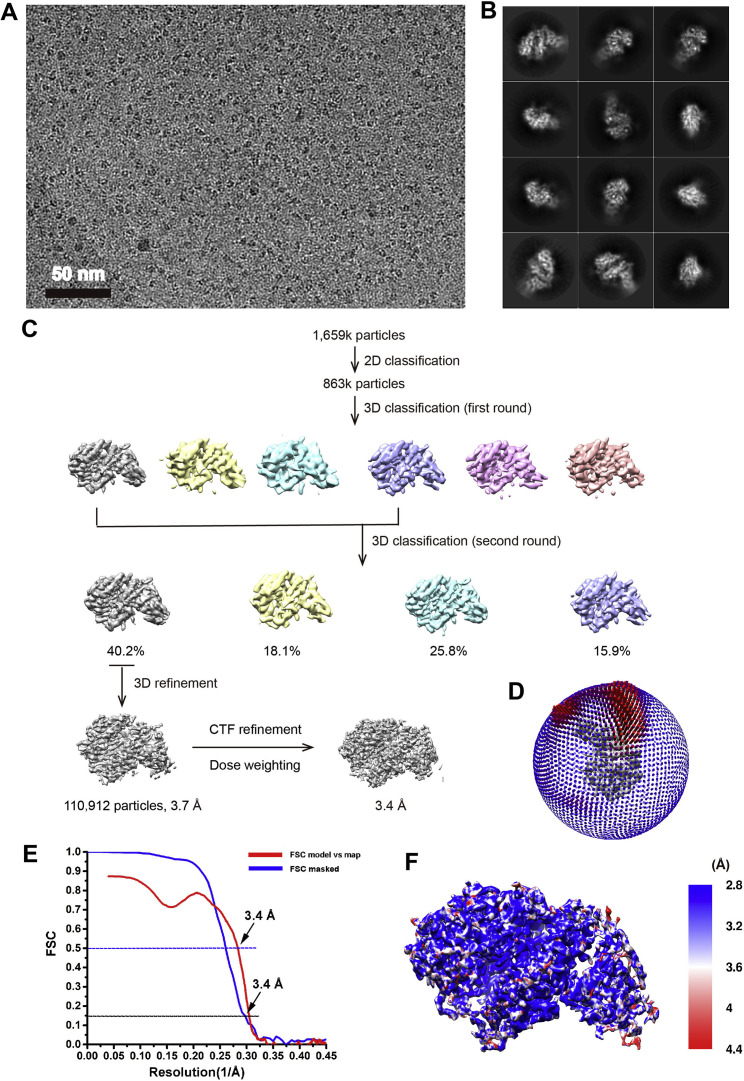
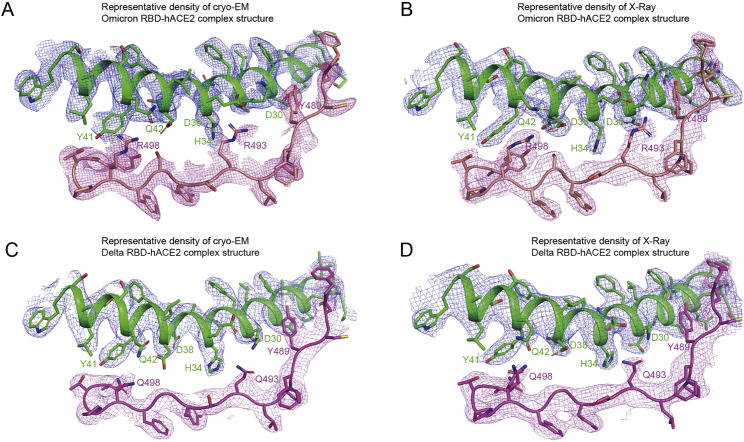
To further characterize the binding modes of hACE2 with omicron RBD or delta RBD, we used both the X-ray crystallography and cryo-EM to determine the complex structures. The crystal and cryo-EM complex structures of hACE2 with omicron RBD were solved at resolutions of 3.0 Å and 3.4 Å, respectively (Table 1 ; Figure S3 ; Tables S2 and S3). The crystal structure of hACE2-delta RBD complex was also determined at a resolution of 3.35 Å (Table 1; Table S2). The electron density map of the crystal structures clearly shows the binding details of omicron RBD-hACE2 and delta RBD-hACE2 interface (Figures 3D and 3E; Figure S4 ). Comparatively, local features of the cryo-EM map (electrostatic potential map) of the omicron RBD-hACE2 binding interface suggest that it is much more flexible in the cryo-EM structure than it is in the crystal structure, reflecting dynamic properties of the RBD-hACE2 interaction. Thus, we used the crystal structures for further analysis.
Table 1.
Amino acid residues comparison of prototype RBD, omicron RBD, and delta RBD interacting with hACE2
| hACE2 | Prototype RBD | Omicron RBD | Delta RBD |
|---|---|---|---|
| S19 (7/17/3) | A475 (3, 1), G476 (4) | A475 (3, 1), G476 (3), N477 (11, 2) | A475 (2), G476 (1) |
| Q24 (24/22/13) | A475 (4), G476 (5), N487 (15, 1) | A475 (3), G476 (5), N477 (1), N487 (13, 1) | A475 (1), G476 (2), N487 (9, 1), Y489 (1) |
| T27 (15/13/13) | F456 (5), Y473 (1), A475 (2), Y489 (7) | F456 (5), Y473 (1), A475 (2), Y489 (5) | F456 (7), A475 (2), Y489 (4) |
| F28 (7/9/8) | Y489 (7) | Y489 (9) | Y489 (8) |
| D30 (10/2/10) | K417 (4, 1), L455 (2), F456 (4) | L455 (1), F456 (1) | K417 (3, 1), L455 (2), F456 (5) |
| K31 (19/15/14) | L455 (2), F456 (5), E484 (1), Y489 (6), F490 (2), Q493 (3) | L455 (1), F456 (4), Y489 (6), R493 (4), | L455 (1), F456 (4), Y489 (4), Q493 (5, 1) |
| H34 (20/29/13) | Y453 (5, 1), L455 (9), Q493 (6) | Y453 (10, 1), R493 (11), S494 (8) | Y453 (8, 2), L455 (5) |
| E35 (8/6/0) | Q493 (8) | R493 (6, 1) | - |
| E37 (7/2/7) | Y505 (7) | H505 (2) | Y505 (7, 1) |
| D38 (15/19/11) | Y449 (9, 1), G496 (5), Q498 (1) | Y449 (7, 1), S496 (6), R498 (5, 2), Y501 (1) | Y449 (9, 1), Q498 (2) |
| Y41 (23/25/28) | Q498 (8), T500 (7, 1), N501 (8, 1) | R498 (3), T500 (7, 1), Y501 (15) | Q498 (13), T500 (7, 1), N501 (8) |
| Q42 (16/9/15) | G446 (4, 1), Y449 (4, 1), Q498 (8, 3) | Y449 (2, 1), R498 (7) | G446 (4), Y449 (2, 1), Q498 (9, 2) |
| L45 (4/1/2) | Q498 (3), T500 (1) | T500 (1) | Q498 (2) |
| L79 (2/2/0) | F486 (2) | F486 (2) | - |
| M82 (9/7/7) | F486 (9) | F486 (7) | F486 (7) |
| Y83 (20/22/18) | F486 (11), N487 (8, 1), Y489 (1) | F486 (11), N487 (10, 1), Y489 (1, 1) |
F486 (8), N487 (9, 1), Y489 (1, 1) |
| N330 (8/8/4) | T500 (8) | T500 (8) | T500 (4) |
| K353 (50/57/57) | G496 (7, 1), N501 (11), G502 (4, 1), Y505 (28) |
Y495 (1), S496 (2), Y501 (21), G502 (6, 1), H505 (27) | G496 (7, 1), Q498 (4), N501 (16), G502 (5, 1), Y505 (25) |
| G354 (11/11/10) | G502 (7), Y505 (4) | G502 (7), H505 (4) | G502 (7), Y505 (3) |
| D355 (9/8/9) | T500 (8, 1), G502 (1) | T500 (7), G502 (1) | T500 (8, 1), G502 (1) |
| R357 (3/3/3) | T500 (3) | T500 (3) | T500 (3) |
| R393 (1/0/1) | Y505 (1) | - | Y505 (1) |
| Total | 288, 16 | 287, 14 | 247, 16 |
The numbers in parentheses for prototype RBD, omicron RBD, and delta RBD residues represent the number of Van der Waals’ contacts between the indicated residues with hACE2. The numbers with underline suggest numbers of potential H bonds between the pairs of residues. Van der Waals’ contacts were analyzed at a cutoff of 4.5 Å and H bonds at a cutoff of 3.5 Å."-" represents that these amino acids in the hACE2 do not interact with the corresponding RBD.
Figure S3.
Cryo-EM structrual analysis of hACE2-omicron RBD, Related to Figure 3
(A) A representative cryo-EM micrograph of hACE2-Omicron RBD. Scale bar, 50 nm. (B) 2D class average images of hACE2-Omicron RBD. (C) A brief workflow of cryo-EM image processing and reconstruction. (D) Euler angle distribution of the final reconstruction. (E) The FSC curve for the reconstruction. (F) Local resolution distribution for the density map of hACE2-Omicron RBD.
Figure 3.
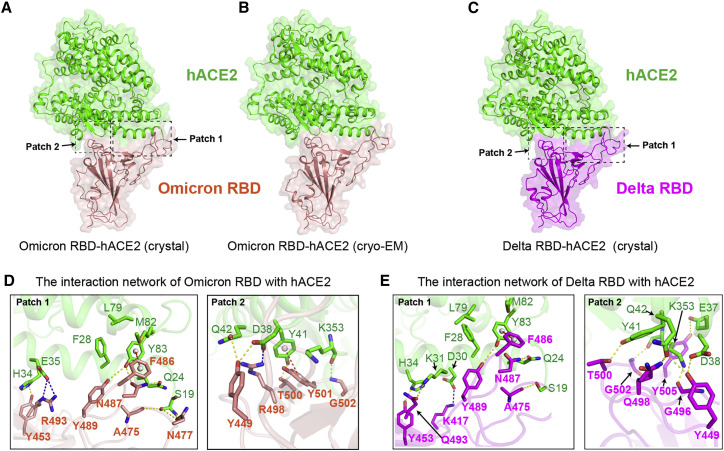
Overall architecture and interaction network of hACE2-omicron RBD and hACE2-delta RBD complexes
(A–C) Overall architecture of hACE2-omicron RBD crystal structure (A) and cryo-EM structure (B) and crystal structure of hACE2-delta RBD complex (C).
(D) Interaction patches in crystal structure of hACE2-omicron RBD. Side chains of interacting residues on hACE2 (green) and omicron RBD (pink) are shown as sticks and labeled appropriately. Yellow, red, and blue dashes present H bond, π-π stacking interaction, and salt bridges, respectively.
(E) Interaction patches in crystal structure of hACE2-delta RBD. Side chains of interacting residues on hACE2 (green) and omicron RBD (purple) are shown as sticks. Yellow, red, and blue dashes present H bond, π-π stacking interaction, and salt bridges, respectively.
See also Figures S3, S4, and S5.
Figure S4.
Representative densities and atomic models, Related to Figure 3
Representative densities and atomic models of Omicron RBD-hACE2 complex structures solved through cryo-EM (A) and X-ray (B) were shown. hACE2 was shown as green and Omicron RBD was colored by salmon. The representative densities and atomic models of Delta RBD-hACE2 complex structures determined by cryo-EM (PBD: 7V7V) (C) and X-ray (D) were shown. hACE2 was also shown as green and Delta RBD was labeled by magenta.
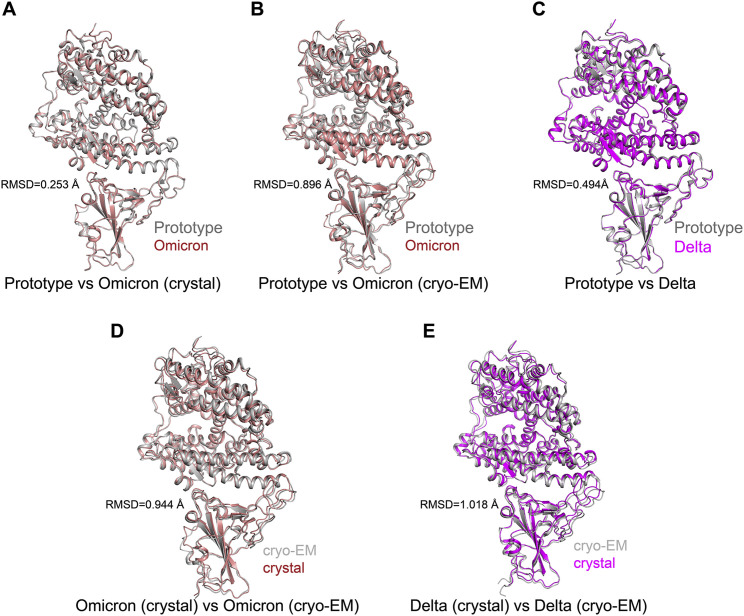
In comparison with those of the prototype RBD-hACE2 complex, omicron RBD-hACE2 and delta RBD-hACE2 crystal structures exhibit overall similar conformation, with root-mean-square deviation (RMSD) of 0.253 Å for 746 Cα atoms and 0.494 Å for 751 Cα atoms, respectively (Figures 3A–3C; Figures S5 A–S5C). When compared to the crystal structure with their own cryo-EM structure, the omicron RBD-hACE2 and delta RBD-hACE2 complexes have RMSDs of 0.944 Å and 1.018 Å, respectively.
Figure S5.
Overall structure comparison of hACE2 with VOC RBDs and GD/1/2019 RBD, Related to Figure 3
(A-C) The crystal structure (A) and cryo-EM structure (B) of Omicron RBD-hACE2 complex and crystal structure of Delta RBD-hACE2 complex are compared with prototype RBD-hACE2 complex and labeled by salmon. Both crystal structure and cryo-EM structure of Omicron RBD-hACE2 complex were colored by salmon. Delta RBD-hACE2 complex was labeled by magenta. Prototype RBD-hACE2 complex was labeled by white. (D and E) The structural comparations between crystal structure and cryo-EM structure of Omicron RBD-hACE2 complex (D) and Delta RBD-hACE2 complex (E). The cryo-EM of both Omicron RBD-hACE2 complex and Delta RBD-hACE2 complex were colored by white and the corresponding crystal structures were labeled by salmon and magenta, respectively.
Structural analysis further shows the accessible surface areas (ASAs) for the omicron RBD-hACE2 crystal complex structure is larger than its ASA in cryo-EM, with 1,727.2 Å2 and 1,709.5 Å2, respectively. A similar result is also observed in the delta RBD-hACE2 complexes, with an ASA of 1,704.0 Å2 for the crystal structure and 1,656.6 Å2 for the cryo-EM structure, respectively.
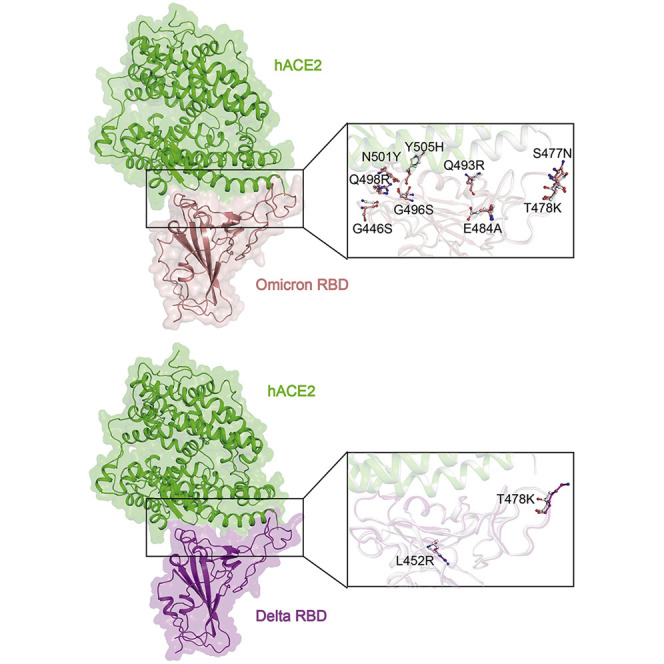
As previously reported (Wang et al., 2020), the binding interface of hACE2 for SARS-CoV-2 RBD is distributed over two patches. In patch 1 of omicron RBD-hACE2 complex, S19 of hACE2 forms H bonds with both A475 and N477 of RBD. Q24 of hACE2 interacts with N487 of RBD through an H bond. Y83 of hACE2 contacts Y489 and N487 of RBD by H bonds, and it also forms a π-π stacking interaction with F486 of RBD. H34 from hACE2 contacts Y453 of RBD through an H bond (Figure 3D). E35 of hACE2 forms a salt bridge with R493 from RBD (Figure 3D). Residue F486 from RBD packs against a small hydrophobic pocket in the interface formed by F28, L79, M82, and Y83 of hACE2 (Figure 3D). In patch 2 of omicron RBD-hACE2 complex, residue D38 from hACE2 forms an H bond with Y449 and a salt bridge with R498 of RBD (Figure 3D). Q42 of hACE2 also interacts with Y449 of RBD by an H bond. Y41 from hACE2 forms an H bond with T500 of RBD and also forms a π-π stacking interaction with Y501 of RBD (Figure 3D). K353 from hACE2 forms a hydrogen bond with G502 of RBD.
In patch 1 of delta RBD-hACE2 complex, S19, Q24, K31, H34, and Y83 from hACE2 contact residues of A475, N487, Q493, Y453, and Y489 from RBD through H bonds, respectively. Y83 of hACE2 forms an H bond with Y498 from RBD and a π-π stacking interaction with F486 of RBD. D30 from hACE2 binds to K417 of RBD with a salt bridge. F486 of RBD can also constitute to strong hydrophobic interactions with F28, L79, M82, and Y83 of hACE2 (Figure 3E). In patch 2, E37, D38, Y41, and K353 from hACE2 form H bonds with Y505, Y449, T500, and G496 on the RBD, respectively. Q42 of hACE2 contacts both Y449 and Q498 from RBD through H bonds (Figure 3E). It should be noted that both of the two substitutions, specifically the L452R and T478K, locate outside the RBD-hACE2 binding interface, which is consistent with the observation that similar binding affinities with ACE2 for both delta and prototype RBDs were obtained (Figure 3A; Table 1).
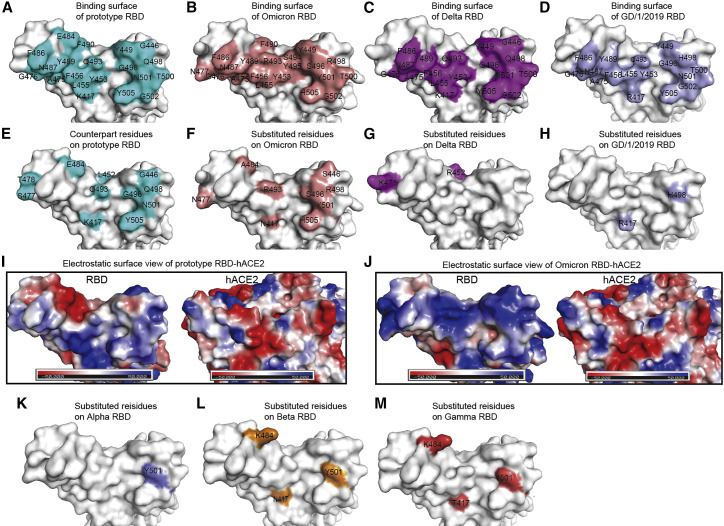
Next, we focused on the substitutions of RBD on the hACE2 binding interface. We labeled the residues involved in hACE2 binding on RBDs of prototype, omicron, delta, and GD/1/2019 (Figures 4 A–4D). In comparison with what was seen on prototype RBD, omicron RBD possesses eight substitutions on the hACE2 recognizing interface, namely K417N, G446S, E484A, Q493R, G496S, Q498R, N501Y, and Y505H. Another substitution, S477N, implicates N477 from hACE2 and expands area of the binding interface (Figures 4E and 4F). The two substitutions observed on delta RBD, namely L452R and T478K, do not participate in hACE2 interaction (Figure 4G). Two substitutions (R417 and H498) were observed on the interface of GD/1/2019 (Figure 4H), and their impact on hACE2 binding has already been analyzed and explained in a previous study (Niu et al., 2021). As a reference, we also labeled the substituted residues in the RBD from other VOCs (Figures 4K–4M), which demonstrated that the omicron variant possesses mutations on almost all the spots where substitutions were observed in previous VOCs.
Figure 4.
The RBMs and different residues on the RBDs
(A–D) The binding surfaces of hACE2 with prototype RBD (A), omicron RBD (B), delta RBD (C), and GD/1/2019 RBD (D) were labeled in cyan, salmon, magenta, and light blue, respectively.
(E and F) Nine different residues on hACE2 binding interface between omicron RBD and prototype RBD were labeled.
(I and J) Electrostatic surface view of prototype SARS-CoV-2 RBD and omicron RBD. The first panel represents the top view. The others are yielded by rotation of the former panel along a horizontal axis.
(K and L) The different residues on the RBMs between VOCs and prototype are labeled.
See also Figure S6.
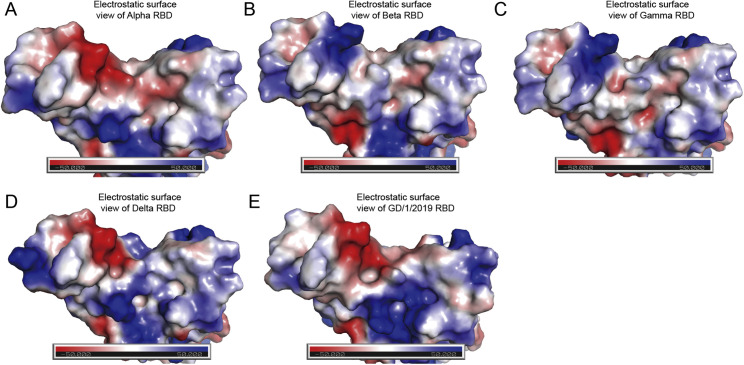
Notably, the substitutions in omicron RBD lead to drastic change of electrostatic charges (Figures 4I and 4J). Compared with other VOC RBDs and GD/1/2019 RBD, the binding surface of omicron RBD has the largest-scale positive charge region (Figure S4). Particularly, T478K, Q493R, and Q498R substitutions significantly increased positive changes, and, meanwhile, E484A decreases the negative charges. R493 and R498 of RBD might be attracted by the negative charges around E35 and D38 of hACE2, respectively. Similar results have also been observed in the previous study (Niu et al., 2021), and Q498 was mutated to H498, enhancing its binding affinity to human, mouse, or rat ACE2. On the other hand, K478 is located away from hACE2 binding surface and might only influence hACE2 binding allosterically.
Comparative analyses of hACE2 interaction with omicron RBD and prototype RBD
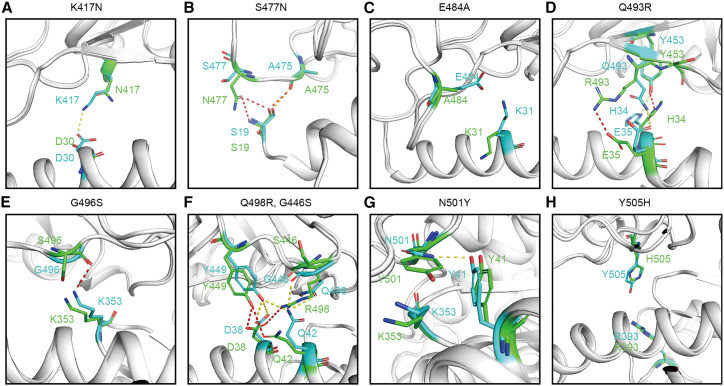
In order to better understand the differences of hACE2 binding with RBD variants and prototype, we investigated details of the substitutions on the RBD variant and compared their interaction patterns with those of prototype RBD (Table 1). For omicron RBD, the impact of K417N and N501Y substitutions has already been extensively analyzed by previous studies (Han et al., 2021), which showed that K417N destroyed the salt bridge with D30 of hACE2 and decreased hACE2 binding affinity, and the aromatic ring of the Y501 could make new favorable non-bonded interactions with hACE2, such as cation-π interaction with K353 of hACE2 (Figures 5A and 5G ). The substitution of S477 to N477 on omicron RBD confers two new H bonds, with S19 strengthening the hACE2 RBD interaction (Figure 5B). E484 in prototype RBD forms Van der Waals’ contacts with K31 from hACE2, and the substitution to A484 could exhibit mild impact on hACE2 binding (Figure 5C). Substitution of Q493 to R493 forms a salt bridge with E35 instead of the H bond (Figure 5D). Although S496 is located in the loop region and undergoes slight conformational change, the altered positioning deprived them of the H bonds with K353 (Figure 5E). Likewise, the position altering caused by G446S substitution deprives S446 of the H bond with Q42. The Q498R mutant, with its longer side chain, formed a new salt bridge with D38 (Figure 5F). Interestingly, Y505H has been observed on the bat-origin RaTG13 (Liu et al., 2021a), where the substitution led to fewer Van der Waals’ contacts. Mutated RaTG13 RBD carrying a H505Y substitution demonstrated strengthened interaction (Liu et al., 2021a). The H505 residue of omicron RBD also formed fewer Van der Waals’ interactions with hACE2 (Figure 5H; Table 1), indicating the substitution plays an unfavorable role during hACE2 binding.
Figure S6.
Electrostatic surface view, Related to Figure 4
Electrostatic surface view of SARS-CoV-2 VOCs Alpha, Beta, Gamma and Delta RBDs and GD/1/2019 RBD. The first panel represents the top view. The others are yielded by rotation of the former panel along a horizontal axis.
Figure 5.
Structural comparison of prototype RBD-hACE2 and omicron RBD-hACE2
(A–H) The nine substitutions on the binding surface of RBD are shown. The residues in the prototype RBD are shown as cyan sticks and the residues in the omicron RBD are shown as green sticks. The yellow dashes represent H bond and salt bridges between prototype RBD and hACE2. The red dashes represent H bond and salt bridges between omicron RBD and hACE2. H-bond interactions were analyzed at a cutoff of 3.5 Å.
Discussion
Although recombination mutations are common for coronaviruses (Su et al., 2016), the SARS-CoV-2 virus is rapidly evolving with the accumulation of multiple mutations, which will definitely worsen the situation of pandemic and threaten the public health. Both delta and omicron strains are highly contagious variants, raising the global concerns. In this study, we determined both X-ray crystallography and cryo-EM structures of omicron RBD-hACE2 complex as well as X-ray crystallography structure of delta RBD-hACE2 complex. We extensively analyzed both VOCs for hACE2-RBD binding interface and compared with that of the published delta-hACE2 cryo-EM structure (PDB ID: 7V7V).
In omicron RBD-hACE2 complex, some substituted residues decrease the binding affinity between RBD and hACE2, and others enhance the binding affinity. The K417N mutation in omicron RBD was also found in beta, gamma, and in some delta lineage RBDs. Delta (B.1.617.2) itself is not defined as containing the K417N mutation, but a sublineage (AY.2, or B.1.617.2.2) is defined as such (Baay et al., 2020; Han et al., 2021). In prototype RBD-hACE2 complex, K417 forms a salt bridge with D30 from ACE2 (Liu et al., 2021b; Wang et al., 2020). When it was mutated to N417 or T417, the absence of a salt bridge decreases the binding affinity with hACE2 (Han et al., 2021; Zhang et al., 2021b). The S477N mutation in omicron RBD has also been reported in B.1.620. S477N was shown to enhance the binding affinity of RBD with hACE2 (Singh et al., 2021). According to the omicron RBD-hACE2 complex structure, we found that N477, but not S477, forms an H bond with S19 from hACE2 to enhance the binding of RBD with hACE2. E484K mutation was reported in beta, gamma, zeta, eta, and theta variants, and E484Q mutation was reported in kappa variant (Han et al., 2021; Thye et al., 2021). As for omicron variant, E484 mutated to A484. E484 of RBD prototype forms weak contact with K31 in hACE2. When it was mutated to A484, the side chain is too short to contact with hACE2, resulting in decreased binding. Additionally, Q493K and Q498H mutations were observed in mouse-adapted strains (Huang et al., 2021b). Q498H also appears in two SARS-CoV-2-like viruses isolated from pangolins, GX/P2V/2017 and GD/1/2019 (Niu et al., 2021). Both Q493K and Q498H increase the binding affinity of SARS-CoV-2 RBD with hACE2 (Huang et al., 2021b; Niu et al., 2021). Q493Y and Q498Y were reported in the RBD of bat-origin virus RaTG13 (Liu et al., 2021a). Y493Q and Y498Q substitutions decrease the binding affinity of RaTG13 RBD with hACE2 (Liu et al., 2021a). In the omicron RBD-hACE2 complex, both Q493 and Q498 were substituted by a positive charged amino acid, arginine (R), and R493 and R498 form salt bridges with E35 and D38, respectively. N501Y is the key residue to enhance SARS-CoV-2 RBD binding to hACE2, which is also reported in alpha, beta, gamma, and theta variants (Han et al., 2021). N501 in SARS-CoV-2 RBD forms an H bond with Y41 from hACE2 and forms Van der Waals’ force with K353. Multiple complex structures of SARS-CoV-2 RBD-hACE2 possessing N501Y substitution show that Y501 forms a π-π stacking interaction with Y41 and forms H bonds with K353 of hACE2 (Han et al., 2021; Nabel et al., 2021; Zhu et al., 2021). Notably, Q498R and N501Y mutations of omicron RBD were previously observed during in vitro evolution, which exhibited ∼600-fold higher affinity with hACE2 (Zahradník et al., 2021). It was shown that Q498R is epistatic to N501Y and that R498 shifts the electrostatic feature to positive, thus attracting negative charges of R498 interacting residues. H505 is reported in the RaTG13 RBD-hACE2 complex (Liu et al., 2021a). Y505 forms van der Waals’ force with E37, K353, and G354, but H505 only contacts K353 and G354 and doesn’t contact E37 (Liu et al., 2021a). In our previous work, when we substituted H505 to Y505 in RaTG13 RBD, the binding affinity of RaTG13 RBD with hACE2 increased (Liu et al., 2021a). On the contrary, the Y505 in prototype RBD was substituted by H505 in omicron RBD. H505 forms fewer contacts with hACE2, which also indicates that Y505H substitution on omicron RBD could decrease the binding of omicron RBD with hACE2. In summary, K417N, G446S, E484A, G496S, and Y505H substitutions decrease the binding affinity of omicron RBD with hACE2, whereas S477N and N501Y compensatively increase the binding affinity of omicron RBD with hACE2.
Although the binding affinity of delta RBD with hACE2 is not significantly changed comparing with the prototype RBD, delta variant has higher transmissibility and a shorter incubation period, which indicates that there are some other factors affecting viral transmission. The mutations in other regions of S protein could also influence virus infection. For instance, the P681R mutation in the S protein of delta increased the level of the cleaved S2 subunit, and the level of the cleaved S2 subunit of the D614G/P681R mutation was significantly higher than that of D614G alone (Tian et al., 2021). For omicron variant, the multiple substitutions outside RBD in the S protein domain are needed to be evaluated in the future.
The emerging variants contain various important mutations in the RBD, enabling it to escape immune surveillance and thus decrease the protective potency of vaccines. Because there is only one mutation (N501Y) on the alpha variant, the efficacy of vaccine was slightly affected (Shen et al., 2021), and re-infection of alpha is found to be no greater than prototype SARS-CoV-2 virus (Graham et al., 2021). Beta variant has a great risk of re-infection because it escapes some neutralizing antibodies produced by dominant strains. The E484K substitution on RBD of beta stain is the key residue associated with a high rate of immune escape. Although there is only one amino acid (residue 417) difference in RBD between beta and gamma variants, gamma is less resistant to naturally acquired or vaccine-induced antibody responses. The transmission of delta increased a lot, but there is not much evidence for vaccine efficacy reduction. Compared with other variants, omicron possesses the largest-scale mutations in the receptor-binding motif (RBM) of RBD, in which E484 and Q493 have been observed to play important roles in immune escape. Notably, E484A, Q493K, and Q493R were repeatedly reported to emerge in immune-compromised patients or during monoclonal antibodies treatment, which is responsible for immune escape (Choi et al., 2020; Clark et al., 2021; Focosi et al., 2021; Guigon et al., 2021; Starr et al., 2021). Interestingly, in comparison with prototype SARS-CoV-2, the neutralization capacity of sera immunized by double mRNA1273-vaccine (non-boosted) and additionally BNT162b2-boosted vaccine were reduced by about 20- and 22.7-fold respectively for omicron (Wilhelm et al., 2021).
The in vitro receptor binding and pseudovirus entry assays indicate that SARS-CoV-2 has a broad range of hosts (Liu et al., 2021c; Wu et al., 2020). In nature, SARS-CoV-2 has been reported to infect lots of animals including cats, dogs, lions, tigers, minks, ferrets, snow leopards, pumas, gorillas, white-tailed deer, and hippopotamuses (Gao and Wang, 2021). SARS-CoV-2 from mink can also be transmitted back to human. Several mink-related SARS-CoV-2 variants were also detected. It indicates that SARS-CoV-2 can mutate to adapt to other hosts during cross-species transmission, which inevitably facilitates the evolution of the SARS-CoV-2. Residues 493, 498, and 501 in RBD have already been identified as key sites for the host range of SARS-CoV-2. For example, a single N501Y mutation of RBD allowed it to infect mice. Q493K and Q498H mutations were also detected in mice-adaptive strains of SARS-CoV-2. Studies showed Q498H mutation appeared after only one passage, and Q493K mutation was introduced to the S protein after five passages in BALB/C mice (Huang et al., 2021b). Therefore, residues 493, 498, and 501, as well as other mutations in the RBD of omicron, could probably change the host spectrum of SARS-CoV-2 and whether omicron variant would be able to cross the species barrier needs to be evaluated in the future.
Limitations of the study
Receptor binding is the key step for SARS-CoV-2 infection. In this study, we only focused on the interaction between the RBDs of two VOCs and hACE2. However, other mutations in the spike protein outside the RBD domain could also contribute to the transmissibility. In addition, other co-receptors and co-factors could also play a role in SARS-CoV-2 infection, which should be addressed in the future studies. Moreover, pseudovirus system only represents the function of S protein, and other proteins of SARS-CoV-2 could also influence the infectivity and pathogenicity. The authentic virus infection assay needs to be further studied.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| Escherichia coli (E. coli) strain DH5α | TIANGEN | Cat# CB101-02 |
| Escherichia coli (E. coli) strain BL21 (DE3) | Novagen | Cat# 69450 |
| MAX Efficiency DH10Bac Competent E. coli |
Invitrogen | Cat# 10361-012 |
| Chemicals, antibody and Recombinant proteins | ||
| PEI | Alfa | A04043896-1g |
| Anti-His/APC | Miltenyi Biotec | Cat# 130-119-820; RRID: AB_2751870 |
| SARS-CoV-2 prototype RBD protein with his-tag, spike residues 319-541 | This paper | accession number: EPI_ISL_402119 |
| SARS-CoV-2 Alpha RBD protein with his-tag, spike residues 319-541 | This paper | accession number: EPI_ISL_683466 |
| SARS-CoV-2 Beta RBD protein with his-tag, spike residues 319-541 | This paper | accession number: EPI_ISL_678615 |
| SARS-CoV-2 Gamma RBD protein with his-tag, spike residues 319-541 | This paper | accession number: EPI_ISL_833172 |
| SARS-CoV-2 Delta RBD protein with his-tag, spike residues 319-541 | This paper | accession number: EPI_ISL_6640916 |
| SARS-CoV-2 Omicron RBD protein with his-tag, spike residues 319-541 | This paper | accession number: EPI_ISL_410721 |
| GD/1/2019 RBD protein with his-tag, spike residues 319-541 | This paper | Accession number: EPI_ISL_410721 |
| hACE2 protein, residues 18-615 | This paper | accession number: NP_001358344 |
| Critical Commercial Assays | ||
| HisTrap HP 5 mL column | GE Healthcare | Cat# 17524802 |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare | Cat# 28989335 |
| Series S Sensor Chip CM5 | GE Healthcare | Cat# 29149603 |
| Membrane concentrator | Millipore | UFC901096 |
| Deposited Data | ||
| Omicron RBD/hACE2 complex (Cryo-EM) | This paper | Protein Data Bank: 7WBL |
| Omicron RBD/hACE2 complex (crystal) | This paper | Protein Data Bank:7WBP |
| Delta RBD/hACE2 complex | This paper | Protein Data Bank: 7WBQ |
| Experimental Models: Cell Lines | ||
| HEK293T cells | ATCC | ATCC CRL-3216 |
| BHK-21 cells | ATCC | ATCC CCL-10 |
| Vero | ATCC | ATCC CCL-81 |
| Recombinant DNA | ||
| pEGFP-N1 | MiaoLingPlasmid | Cat# P0133 |
| pEGFP-N1-hACE2 | This paper | accession number: BAJ21180 |
| pCAGGS | MiaoLingPlasmid | Cat# P0165 |
| Software and Algorithms | ||
| PyMOL software | Molecular Graphics System, Version 1.8 Schrö dinger | https://pymol.org/2/ |
| BIAcore® 8K Evaluation software | GE Healthcare | N/A |
| FlowJo V10 | FLOWJO | https://www.flowjo.com/solutions/flowjo/downloads |
| Motioncor2 | Zheng et al., 2017 | N/A |
| COOT | Emsley and Cowtan, 2004 | http://www.mrc-lmb.cam.ac.uk/personal/peemsley/coot/ |
| Phenix | Adams et al., 2010 | http://www.phenix-online.org/ |
| MolProbity | Duke Biochemistry | http://molprobity.biochem.duke.edu/index.php |
Resource availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peiyi Wang (wangpy@sustech.edu.cn).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cells
HEK293T cells (ATCC CRL-3216), BHK-21 cells (ATCC CCL-10), HuH7 cells and HeLa-hACE2 cells (prepared in this study) were cultured at 37°C in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Method details
Gene Cloning
The full-length sequence of hACE2 (GenBank: NP_001358344) was cloned in pEGFP-N1 vector for flow cytometry. The extracellular domain of hACE2 (residues 1-740, GenBank: NP_001358344) fused with the Fc domain of mouse IgG (mFc) were cloned into the pCAGGS vector for protein expression. The coding sequences of RBDs from original SARS-CoV-2 (residue 319-541) and VOCs (residue 319-541) and hACE2(residues 19-615, GenBank:NP_001358344) were cloned into the pCAGGS vector. The coding sequence S proteins from original SARS-CoV-2 and VOCs (residues 1-1233) were cloned into pCAGGS vectors.
Protein expression and purification
The hACE2 fused with mFc were expressed and purified from the culture supernatants of HEK293F cells using a Protein A affinity column (GE Healthcare) and further purified by gel filtration using a SuperdexTM 200 10/300 GL (GE Healthcare). Purified proteins were stored in a buffer containing 20 mM Tris-HCl and 150 mM NaCl (pH 8.0). Proteins for SPR assay were transferred to PBST (1.8 mM KH2PO4, 10 mM Na2HPO4 (pH 7.4), 137 mM NaCl, 2.7 mM KCl, and 0.005% (v/v) Tween 20) buffer.
The hACE2 and RBDs from original SARS-CoV-2 and VOCs cloned in pCAGGS were expressed in HEK293F cells. Cell culture supernatants were collected, filtered with a 0.22 μm filter, purified by His-Trap HP column (GE Healthcare), and SuperdexTM 200 Increase 10/300 GL column (GE Healthcare). Purified proteins were stored in protein buffer (20 mM Tris-HCl, pH 8.0 and 150 mM NaCl).
Flow Cytometry
The plasmids containing hACE2 fused with eGFP were transfected into BHK-21 cells. A mixture containing RBDs from prototype SARS-CoV-2 (1 μg/mL) or VOCs (1 μg/mL) were incubated with the BHK-21 cells at 4°C for 1 h. Subsequently, cells were washed with PBS thrice and stained with APC mouse anti-his secondary antibody for 1 h before being analyzed using BD FACS Canto FlowCytometer (BD Biosciences). The data of all samples were analyzed using FlowJo 7.6 (TreeStar Inc., Ashland, OR, USA)
SPR Analysis
The ACE2-mFC fusion proteins were transferred into PBST buffer (1.8 mM KH2PO4, 10 mM Na2HPO4 (pH 7.4), 137 mM NaCl, 2.7 mM KCl, and 0.05% (v/v) Tween 20) and immobilized on flow cell 2 of CM5 chip. Flow cell 1 was used as the negative control. Serially diluted RBDs from original SARS-CoV-2 and VOCs were then flowed over the chip in PBST buffer. Binding affinities were measured using a BIAcore 8K (GE Healthcare) at 25°C in the single-cycle mode. Binding kinetics were analyzed with Biacore™ Insight software (GE healthcare) using a 1:1 Langmuir binding model.
Production and quantification of VSV pseudotyped viruses
The original SARS-CoV-2, and VOC pseudoviruses were constructed with a GFP encoding replication-deficient vesicular stomatitis virus (VSV) vector backbone (VSV-ΔG-GFP) and the coding sequence of corresponding spike proteins, as previously described (Muik et al., 2021). Briefly, HEK293T cells were transfected by 30 ng of spike protein expression plasmids. The VSV-ΔG-GFP pseudovirus was added 24 h post-transfection. The inoculum was removed after incubation for 1 h at 37°C. The culture medium was then changed into DMEM supplemented with 10% FBS and 10 lg/mL of anti-VSV-G antibody (I1-Hybridoma ATCC CRL2700TM) after washing cells with PBS. The pseudoviruses were harvested 20 h post-inoculation, passed through a 0.45-lm filter (Millipore, Cat#SLHP033RB) before aliquoted, and stored at −80°C.
All pseudoviruses were treated with 0.5 U/μL BaseMuncher Endonuclease (Abcam, ab270049) for 1.5 h at 37°C to remove unpackaged RNA before quantification. Viral RNA was extracted (Bioer Technology, Cat# BYQ6.6.101711-213) and quantitated by quantitative RT–PCR (qPCR) using 7500 fast real-time PCR system (Applied Biosystems) with the primers and probe for detecting the P protein coding sequence of VSV.
Pseudovirus infection assay
The pseudovirus particles of original SARS-CoV-2 and Omicron variant were normalized to the same amount for quantitation by qRT–PCR. Then, 100 μL of each pseudovirus was added to each well of 96-well plate containing Vero cells. Uninfected Vero cells were used as control. Plates were imaged 15 h post-transfection. The numbers of fluorescent cells were determined on a CQ1 confocal image cytometer (Yokogawa). Each group contains 5 replicates. Statistical significance was determined by a two-sided unpaired Student’s t test.
Crystallization
The sitting-drop method was used to obtain the Omicron RBD-hACE2 and Delta RBD-hACE2 complex crystals. In detail, purified complex proteins were concentrated to 5 and 10 mg/mL. Then, 0.8 μL protein was mixed with 0.8 μL reservoir solution. The resulting solution was sealed and equilibrated against 100 μL of reservoir solution at 18°C and 4°C. High resolution Omicron RBD-hACE2 complex crystal was grown in 0.15 M ammonium sulfate, 0.1 M Na HEPES, 20% w/v polyethylene glycol 4000, and Delta RBD-hACE2 was grown complex in 0.1 M sodium cacodylate, 25% w/v polyethylene glycol 4000.
Data collection and structure determination
The diffraction data were collected at Shanghai Synchrotron Radiation Facility (SSRF) BL02U1 (wavelength, 0.97919 Å). For data collection, the crystals were cryo-protected by briefly soaking in reservoir solution supplemented with 20% (v/v) glycerol before flash-cooling in liquid nitrogen. The dataset was processed with HKL2000 software (Otwinowski and Minor, 1997). The structure of 2 complexes were determined by the molecular replacement method using Phaser (Adams et al., 2010) with previously reported complex structure SARS-CoV-2-RBD complex with hACE2 (PDB: 6LZG). The atomic models were completed with Coot (Emsley and Cowtan, 2004) and refined with phenix.refine in Phenix (Adams et al., 2010), and the stereochemical qualities of the final models were assessed with MolProbity (Williams et al., 2018). Data collection, processing, and refinement statistics were summarized in Table S2. All structural figures were generated using Pymol software (https://pymol.org/2/).
Cryo-EM sample preparation and data collection
For the hACE2-Omicorn RBD complex, an aliquot of 3.5 μL solution (0.25 mg/mL) was applied to glow-discharged home-made graphene grids (Quantifoil Au R1.2/1.3, 300 mesh) blotted for 1.5 s with a humidity of 90% at 4°C before being plunged into liquid ethane using a Vitrobot Mark IV (Thermo Fisher). The frozen grids were loaded onto a Titan Krios transmission electron microscope (Thermo Fisher) and operated at 300 kV. The microscope is equipped with a BioQuantum energy filter and k3 direct electron detector (Gatan). A slit width of 10 eV was used for data collection. Automatic data collection was performed by using EPU software (Thermo Fisher). Movies were recorded in super-resolution counting mode at pixel size of 0.335 Å. The exposure was performed with a dose rate of 15 e-/pixel/s and an accumulative dose of ∼60 e-/Å2 for each movie, of which was fractionated into 40 sub-frames. The final defocus ranges of these two datasets were approximately −1.3∼-2.3 μm.
Image processing
The drift correction of all stacks was performed with MotionCor2 (Zheng et al., 2017) to generate 2 × binned micrographs. Initial contrast transfer function (CTF) values for each micrograph were calculated with CTFFIND4.1 (Rohou and Grigorieff, 2015). Micrographs with an estimated resolution limit worse than 5 Å were discarded in the initial screening. A set of ∼150,000 particles were auto-picked by Laplacian-of-Gaussian from RELION-3.1 (Zivanov et al., 2018) and then subjected to 2D classification to generate templates for auto-picking against the entire dataset. The subsequent data processing and reconstruction were performed using RELION-3.1.
For the Omicron RBD-hACE2 dataset, 1,659,778 particles were picked from 6,596 micrographs. The picked particles were then extracted and subjected to three rounds of reference-free 2D classification. A clean dataset with 863,953 particles from good 2D classes were selected to generate the initial model in RELION. Subsequently, the model was used as reference in RELION 3D classification. After the second round of 3D classification without applying symmetry, the predominant class containing a subset of 110,912 best particles shows the clear features of secondary structural elements and the high accuracy of particle alignment. These particles were subjected to 3D refinement, which yielded a reconstruction at 3.7 Å resolution. To further improve the resolution, dose-weighted images were generated by MotionCor2 with the first 2 frames and last 14 frames discarded for each stack, resulting in a reduced dose of ∼35 e-/Å2. In addition, CTF refinement was performed to correct the local CTF values of each particle. Combining these parameters, a new round of 3D refinement was performed to obtain the density map at 3.4 Å resolution determined by the Fourier shell correlation (FSC) 0.143 cut-off value (Figure S3). Local resolution estimation was performed with ResMap (Kucukelbir et al., 2014).
Model building and refinement
The crystal structure of the hACE2-SARS-CoV-2 RBD complex (PDB: 6LZG) was docked into the cryo-EM density maps using CHIMERA (Pettersen et al., 2004). The model was manually corrected for local fit in COOT (Emsley et al., 2010) and the sequence register was updated based on alignment. The model was refined against the corresponding map in real space using PHENIX (Adams et al., 2010), in which the secondary structural restraints and Ramachandran restrains were applied. The stereochemical quality of each model was assessed using MolProbity (Chen et al., 2010). Statistics for model refinement and validation are shown in Table S3.
Quantification and statistical analysis
Flow cytometry analysis
All experiments were performed three times; one representative of each experiment is shown in Figure 2.
Binding affinity analysis
KD values of SPR experiments were obtained with BIAcore 8K Evaluation Software (GE Healthcare), using a 1:1 binding model. The values indicate the mean ± SD of three independent experiments.
Acknowledgments
We are grateful to Drs G. Wong and G. Shang for revising the manuscript. We are grateful to the PMI-IMCAS Public Technology Service Center for its support on FACS assay. We thank Tong Zhao from Institute of Microbiology, Chinese Academy of Sciences, for her technical support in FACS assay. We acknowledge the staff of beamline BL02U1 and BL10U2 at the Shanghai Synchrotron Radiation Facility for assistance during data collection. We thank all staff at the Cryo-EM Center of Southern University of Science and Technology for their assistances. We thank Y.Q. Mi, H. Feng, and X.X. Bai at the Cryo-EM Center, Shanxi Academy of Advanced Research and Innovation, for their technical support on the cryo-EM. We thank Shuimu BioSciences for cryo-EM facility access and technical support during image acquisition. This work was supported by the National Key R&D Program of China (2021YFC2301401, 2020YFA0907102, 2020YFA0509202, 2021YFC0863300), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29010202, XDB37030204), the Queensland-Chinese Academy of Sciences Collaborative Science Fund 2020 (153211KYSB20200001), the intramural special grant for SARS-CoV-2 research from the Chinese Academy of Sciences, and the Science and Technology Innovation Committee of Shenzhen Municipality (JSGG20200207155251653). S. Liu is funded by President’s Excellent Postdoctoral Program of SUSTech.
Author contributions
J.Q., G.F.G., and P.W. initiated and designed the project. Specifically, J.Q. supervised and coordinated the experiments for crystals preparations, the diffraction data collection, and structure determinations. G.F.G. supervised the structural analysis, SPR and FACS assays, and pseudoviruses transductions. P.W. supervised and coordinated the samples preparations for the cryo-EM structural analysis, data collection, and cryo-EM structure determinations. Pengcheng Han, L.L., and D.Z. purified the proteins and grew the crystals with the help of B.H. and M.T. Pengcheng Han and L.L. performed the SPR analysis. D.Z. conducted the flow cytometry assay. Q.W., Pengcheng Han, and J.Q. collected the crystal structural data and solved the structures. S.L., Pu Han, and P.W. collected the cryo-EM structural data and solved the structure with the help of X.L., Q.P., L.F., and Y.G. D.L., R.Z., and X.Z. performed pseudotyped virus-related assays. Z.X., C.S., K.L., J.Q., G.F.G., and P.W. wrote and revised the manuscript.
Declaration of interests
The authors declare that they have no competing interests.
Published: January 6, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.01.001.
Supplemental information
Data and Code Availability
The atomic coordinates for the Cryo-EM structure of the omicron RBD-hACE2 complex (PDB code: 7WBL), the crystal structures of the omicron RBD-hACE2 complex (PDB code: 7WBP) and Delta RBD-hACE2 complex (PDB code: 7WBQ) have been deposited in the Protein Data Bank (www.rcsb.org).
This study did not generate custom computer code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants—clinical, public health, and vaccine implications. N. Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baay M., Lina B., Fontanet A., Marchant A., Saville M., Sabot P., Duclos P., Vandeputte J., Neels P. SARS-CoV-2: Virology, epidemiology, immunology and vaccine development. Biologicals. 2020;66:35–40. doi: 10.1016/j.biologicals.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Yisimayi A., Bai Y., Huang W., Li X., Zhang Z., Yuan T., An R., Wang J., Xiao T., et al. Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines. Cell Res. 2021;31:732–741. doi: 10.1038/s41422-021-00514-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.R., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R. B. 1.1. 529 escapes the majority of SARS-CoV-2 neutralizing antibodies of diverse epitopes. bioRxiv. 2021 doi: 10.1101/2021.12.07.470392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D., et al. NGS-SA. COMMIT-KZN Team SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv. 2021 doi: 10.1101/2021.12.08.21267417. 2021.12.08.21267417. [DOI] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.A., Clark L.E., Pan J., Coscia A., McKay L.G.A., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J., et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021;184:2605–2617.e18. doi: 10.1016/j.cell.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira I., Datir R., Papa G., Kemp S., Meng B., Rakshit P., Singh S., Pandey R., Ponnusamy K., Radhakrishnan V.S. SARS-CoV-2 B. 1.617 emergence and sensitivity to vaccine-elicited antibodies. bioRxiv. 2021 doi: 10.1101/2021.05.08.443253. [DOI] [Google Scholar]
- Focosi D., Novazzi F., Genoni A., Dentali F., Gasperina D.D., Baj A., Maggi F. Emergence of SARS-CoV-2 spike protein escape mutation Q493R after treatment for COVID-19. Emerg. Infect. Dis. 2021;27:2728–2731. doi: 10.3201/eid2710.211538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G.F., Wang L. COVID-19 expands its territories from humans to animals. China CDC Wkly. 2021;3:855–858. doi: 10.46234/ccdcw2021.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9, e2379. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese L., Zaccaria M., Farzan M., Johnson W., Momeni B. Investigating the mutational landscape of the SARS-CoV-2 Omicron variant via ab initio quantum mechanical modeling. bioRxiv. 2021 doi: 10.1101/2021.12.01.470748. [DOI] [Google Scholar]
- Graham M.S., Sudre C.H., May A., Antonelli M., Murray B., Varsavsky T., Kläser K., Canas L.S., Molteni E., Modat M., et al. COVID-19 Genomics UK (COG-UK) Consortium Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health. 2021;6:e335–e345. doi: 10.1016/S2468-2667(21)00055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigon A., Faure E., Lemaire C., Chopin M.-C., Tinez C., Assaf A., Lazrek M., Hober D., Bocket L., Engelmann I., Alidjinou E.K. Emergence of Q493R mutation in SARS-CoV-2 spike protein during bamlanivimab/etesevimab treatment and resistance to viral clearance. J. Infect. 2021 doi: 10.1016/j.jinf.2021.08.033. S0163-4453(21)00435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., Su C., Zhang Y., Bai C., Zheng A., Qiao C., Wang Q., Niu S., Chen Q., Zhang Y., et al. Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants. Nat. Commun. 2021;12:6103. doi: 10.1038/s41467-021-26401-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Dai L., Wang H., Hu Z., Yang X., Tan W., Gao G.F. Serum sample neutralisation of BBIBP-CorV and ZF2001 vaccines to SARS-CoV-2 501Y.V2. Lancet Microbe. 2021;2:e285. doi: 10.1016/S2666-5247(21)00082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Zhang Y., Hui X., Zhao Y., Gong W., Wang T., Zhang S., Yang Y., Deng F., Zhang Q., et al. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021;67:103381. doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lai S., Gao G.F., Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371.e9. doi: 10.1016/j.cell.2021.02.042. e2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Rocklöv J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J. Travel Med. 2021;28:taab124. doi: 10.1093/jtm/taab124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu K., Pan X., Li L., Yu F., Zheng A., Du P., Han P., Meng Y., Zhang Y., Wu L., et al. Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell. 2021;184:3438–3451.e10. doi: 10.1016/j.cell.2021.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Tan S., Niu S., Wang J., Wu L., Sun H., Zhang Y., Pan X., Qu X., Du P., et al. Cross-species recognition of SARS-CoV-2 to bat ACE2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020216118. e2020216118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hu G., Wang Y., Ren W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2025373118. e2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., Datir R., Collier D.A., Albecka A., Singh S., et al. Indian SARS-CoV-2 Genomics Consortium (INSACOG) Genotype to Phenotype Japan (G2P-Japan) Consortium. CITIID-NIHR BioResource COVID-19 Collaboration SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.-K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel K.G., Clark S.A., Shankar S., Pan J., Clark L.E., Yang P., Coscia A., McKay L.G.A., Varnum H.H., Brusic V., et al. Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science. 2021:l6251. doi: 10.1126/science.abl6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S., Wang J., Bai B., Wu L., Zheng A., Chen Q., Du P., Han P., Zhang Y., Jia Y., et al. Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin. EMBO J. 2021;40:e107786. doi: 10.15252/embj.2021107786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Cui M., Sun S., Zhou J., Du Z., Cui Y., Fan H. Genome characterization and potential risk assessment of the novel SARS-CoV-2 variant Omicron (B. 1.1. 529) Zoonoses. 2021 doi: 10.15212/ZOONOSES-2021-0024. [DOI] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O., et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29:529–539.e3, e523. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Steinkellner G., Köchl K., Gruber K., Gruber C.C. Serine 477 plays a crucial role in the interaction of the SARS-CoV-2 spike protein with the human receptor ACE2. Sci. Rep. 2021;11:4320. doi: 10.1038/s41598-021-83761-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Zhao X., Ma X., Wang W., Niu P., Xu W., Gao G.F., Wu G. A novel coronavirus genome identified in a cluster of pneumonia cases - Wuhan, China 2019-2020. China CDC Wkly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- Thye A.Y., Law J.W., Pusparajah P., Letchumanan V., Chan K.G., Lee L.H. Emerging SARS-CoV-2 variants of concern (vocs): an impending global crisis. Biomedicines. 2021;9:1303. doi: 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Sun Y., Zhou J., Ye Q. The global epidemic of the SARS-CoV-2 Delta variant, key spike mutations and immune escape. Front. Immunol. 2021;12:751778. doi: 10.3389/fimmu.2021.751778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Li Q., Liang Z., Li T., Liu S., Cui Q., Nie J., Wu Q., Qu X., Huang W. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg. Microbes Infect. 2022;11:1–5. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., Metzler M., Kohmer N., Hoehl S., Helfritz F.A., et al. Reduced neutralization of SARS-CoV-2 omicron variant by vaccine sera and monoclonal antibodies. medRxiv. 2021 doi: 10.1101/2021.12.07.21267432. [DOI] [Google Scholar]
- Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Chen Q., Liu K., Wang J., Han P., Zhang Y., Hu Y., Meng Y., Pan X., Qiao C., et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradník J., Marciano S., Shemesh M., Zoler E., Harari D., Chiaravalli J., Meyer B., Rudich Y., Li C., Marton I., et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021;6:1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Liu K., Li Y., Lu Q., Wang Q., Zhang Y., Wang L., Liao H., Zheng A., et al. The molecular basis for SARS-CoV-2 binding to dog ACE2. Nat. Commun. 2021;12:4195. doi: 10.1038/s41467-021-24326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6, e2346. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Mannar D., Srivastava S.S., Berezuk A.M., Demers J.P., Saville J.W., Leopold K., Li W., Dimitrov D.S., Tuttle K.S., et al. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 2021;19:e3001237. doi: 10.1371/journal.pbio.3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates for the Cryo-EM structure of the omicron RBD-hACE2 complex (PDB code: 7WBL), the crystal structures of the omicron RBD-hACE2 complex (PDB code: 7WBP) and Delta RBD-hACE2 complex (PDB code: 7WBQ) have been deposited in the Protein Data Bank (www.rcsb.org).
This study did not generate custom computer code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.