Abstract
Eukaryotes have the ability to respond to changes in oxygen tension by alterations in gene expression. For example, OLE1 expression in Saccharomyces cerevisiae is upregulated under hypoxic conditions. Previous studies have suggested that the pathway regulating OLE1 expression by unsaturated fatty acids may involve Mga2p and Spt23p, two structurally and functionally related proteins. To define the possible roles of each of these genes on hypoxia-induced OLE1 expression, we examined OLE1 expression under normoxia, hypoxia, and cobalt treatment conditions in Δmga2 or Δspt23 deletion strains. The results of OLE1 promoter-lacZ reporter gene and Northern blot analyses showed that hypoxia- and cobalt-induced OLE1 expression was dramatically decreased in a Δmga2 strain but not in a Δspt23 strain. Further analyses using low-oxygen response element (LORE)-CYC1-lacZ fusion reporter assays and electrophoretic mobility shift assays (EMSAs) demonstrated that MGA2 significantly affects the LORE-dependent hypoxic induction pathway of gene expression. When MGA2 was supplied by a plasmid, the LORE-dependent hypoxia-inducible reporter expression was recovered, as was the hypoxia-inducible complex in EMSAs in the S. cerevisiae Δmga2 strain. Supershift analysis of EMSAs using crude extracts containing mycMga2p indicated that Mga2p is a component of the LORE-binding complex. Another LORE-dependent, hypoxia-inducible gene, ATF1, was similarly affected in the Δmga2 strain. These results indicate that MGA2 is required for the LORE-dependent hypoxic gene induction in S. cerevisiae.
Humans and other eukaryotes have developed sophisticated mechanisms to respond to decreased oxygen tension. These mechanisms are fundamentally important for developmental, physiological, and pathophysiological processes. Because of the importance of the response to hypoxic conditions, many mammalian cell types share a common mechanism of oxygen sensing and signal transduction (reviewed in references 3 and 22). One of the best-studied signal transduction pathways involved in the response to decreased oxygen tension contains the transcription factor, hypoxia-inducible factor 1 (HIF-1). Many hypoxia-inducible genes, such as erythropoietin and vascular endothelial growth factor, are upregulated by hypoxia via the activation of HIF-1 (reviewed in references 2 and 27). HIF-1 is a heterodimer consisting of an α subunit and a β subunit, two basic helix-loop-helix proteins in the PAS family of transcription factors. The β subunit is also known as the aryl hydrocarbon receptor nuclear translocator ARNT. ARNT mRNA and protein levels are not significantly affected by ambient oxygen tension. In contrast, though HIF-1α mRNA levels are not appreciably affected by oxygen tension, HIF-1α protein is rapidly degraded by the ubiquitin-proteosome pathway in normoxia (15, 26). However, HIF-1α protein accumulates in response to hypoxia, certain transition metals (e.g., cobalt and nickel), and iron chelators such as desferrioxamine. Once the heterodimer is formed under these conditions, it can interact with other DNA-binding proteins which function in part to provide tissue and developmental specificity (reviewed in references 3 and 7). HIF-1α has also been shown to associate with the coactivator protein p300 (also called CBP) (1), which interacts with the basal transcription machinery (19, 32).
The yeast Saccharomyces cerevisiae is a facultative anaerobic eukaryote, which differentially expresses a large number of genes in response to changes in oxygen availability (reviewed in references 3, 17, and 35). The oxygen-sensing and signal transduction pathways involved in the regulation of these genes have been the focus of several studies. Many yeast genes, such as ANB1, have been shown to be upregulated by complete anaerobiosis and are mediated in large part through the Rox1p protein, a DNA-binding protein which functions as a repressor (reviewed in reference 35). Recently, it has been shown that some genes in S. cerevisiae exhibit increased expression in response to hypoxia, cobalt, and iron chelation, mimicking hypoxia-regulated genes in higher organisms (18, 30). We have studied one such gene, OLE1, which encodes a Δ9 fatty acid desaturase gene in S. cerevisiae. A cis transactivation element, the low-oxygen response element (LORE), was identified and characterized in the promoter region of OLE1. The LORE sequence is also found in a family of yeast genes which may also be regulated via LORE (30). A similar but longer element has also been reported by Nakagawa et al. (23). The results of these studies strongly indicate that there is another hypoxia signal transduction pathway in yeast, in addition to the Rox1p-dependent repression mechanism.
MGA2 and SPT23 are two functionally and genetically related genes. MGA2 was identified as a multicopy suppressor of a transcription defect caused by a null mutation in the SNF2 gene in S. cerevisiae (33). SPT23 is also functionally related to SNF2 (4, 33). It has been shown that Snf2p is a key component of the SWI-SNF nucleosome remodeling complex, which plays an important role in activating the transcription of many genes (16, 28). Sequence analysis shows that MGA2 and SPT23 have considerable homology, with 43% of the amino acids overall being identical and 60% being similar (33). Deletion of either one of these genes has only a modest effect on cell growth. However, cells with a MGA2 SPT23 double mutation are not viable (33). Studies have also shown that both MGA2 and SPT23 can activate transcription when fused to a Gal4 DNA-binding domain (33). A subsequent search for genes which are functionally related to or controlled by MGA2 and SPT23 led to the identification of OLE1 as a gene which is positively influenced by MGA2 and SPT23 at the transcription level (34). It is possible that MGA2 and SPT23 control cell viability by stimulating OLE1 transcription (34).
In eukaryotes, regulated intracellular turnover of many proteins, such as the α subunit of HIF-1, is primarily mediated by the ubiquitin/proteasome pathway, which normally results in the complete proteolysis of a targeted protein (12). In a few cases, however, this pathway is involved in partial, rather than complete, proteolysis. Examples include the proteasome-dependent processing of the p105 precursor of the transcription factor NF-κB from mammalian cells and the processing of the precursors of certain yeast transcription factors (20). Recently, Hoppe et al. (14) identified a novel processing pathway in S. cerevisiae involving SPT23 and MGA2, which they have termed regulated ubiquitin/proteasome-dependent processing. Spt23p and Mga2p are initially made as dormant precursors that are firmly anchored in the endoplasmic reticulum or nuclear envelope membranes by their C-terminal tails. The shortage of unsaturated fatty acid leads to the RSP5-mediated ubiquitination of Spt23p and Mga2p, which leads to the release of the N-terminal transcription factor domain into the cytosol and finally to the enhanced expression of OLE1 mRNA. Addition of unsaturated fatty acids that contain more than one double bond almost completely blocks Spt23p precursor processing. Data suggest that this pathway controls the level of unsaturated fatty acids in S. cerevisiae by regulating OLE1 expression (14).
Previously, OLE1 expression has also been shown to be increased by oxygen deprivation (18, 23, 30). Oxygen is critical for Ole1p function. Given the critical need for unsaturated fatty acid production in S. cerevisiae to maintain membrane integrity, we hypothesized that MGA2 and/or SPT2 may also mediate OLE1 expression by hypoxia. Here, we demonstrate that MGA2, but not SPT23, is required for LORE-dependent hypoxic induction of gene expression in S. cerevisiae.
MATERIALS AND METHODS
Media, chemicals, and enzymes.
Yeast strains were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose) medium (Bio 101, Inc., Carlsbad, Calif.) or synthetic complete (SC) dropout medium, depending on the plasmid selectable markers. Luria broth (LB) was used to grow bacteria. Ampicillin (U.S. Biochemical Corp., Cleveland, Ohio) was used at a dose of 50 μg/ml unless indicated otherwise. o-Nitrophenyl-β-galactopyranoside (ONPG) was obtained from Sigma Chemical Co. (St. Louis, Mo.). Radiolabeled compounds were purchased from Perkin-Elmer (Boston, Mass.). Formamide was obtained from American Bioanalytical (Natick, Mass.). Ultrahyb Hybridization Solution Acrylamide was obtained from Ambion Inc. (Austin, Tex.). Bisacrylamide, N,N,N′,N′-tetramethylethylenediamine (TEMED), and protein molecular mass markers were from Bio-Rad (Richmond, Calif.). Ammonium sulfate, phenylmethylsulfonyl fluoride (PMSF), CoCl2, NiCl2, 1,10-phenanthroline, and Nonidet P-40 were obtained from Sigma Chemical Co. SeaKem ME agarose was obtained from FMC Bioproducts (Port Clyde, Maine). T4 polynucleotide kinase and deoxynucleoside triphosphates were purchased from Promega Corporation (Madison, Wis.). Shrimp alkaline phosphatase and Taq polymerase were purchased from Roche Molecular Biochemicals (Indianapolis, Ind.); other restriction enzymes were obtained from New England BioLabs (Beverly, Mass.). All enzymes were used according to the manufacturer's instructions.
Plasmid and plasmid construction.
The construction of several of the OLE1 promoter-lacZ fusion deletion series was described previously (5, 30). One of these plasmids is p62::934, in which the number following the two colons indicates the position of the nucleotide at the 5′ end of the OLE1 promoter fragment with respect to the start codon (A of ATG is +1). The reporter plasmid pAM6 contains a tandem repeat of the OLE1 LORE (sequences −347 to −328 relative to the ATG translational start codon with the A of the codon designated +1) in front of the CYC1 basal promoter-lacZ fusion in vector pTBA30. A centromeric plasmid with a LEU2 selection marker, pAM23, was constructed by subcloning a 5.1-kb HindIII fragment that contains MGA2 from pYK2 (gift of D. J. Garfinkel, Gene Regulation and Chromosome Biology Laboratory, National Cancer Institute, Frederick, Md.) into pRS315. A 2μm plasmid with a LEU2 selection marker, YEplac181-mycMGA2, contains a triple repeat of a myc epitope in front of MGA2. This was a gift from S. Jentsch, Department of Molecular Cell Biology, Max Planck Institute for Biochemistry, Martinsried, Germany (14).
Strains and growth conditions.
Table 1 contains the yeast strains used in these studies. The MGA2 and SPT23 deletion strains and their parental strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) were purchased from Research Genetics, Inc. (Huntsville, Ala.). Yeast cells containing lacZ fusion reporter plasmids were grown at 30°C on uracil dropout medium containing dextrose (or uracil and leucine double dropout medium) (29). Plasmid amplifications and bacterial transformations were performed using Escherichia coli strain DH5 (Invitrogen Corp., Carlsbad, Calif.). Yeast transformations were performed by the method of Elble (8). Preparative cultures were grown aerobically in a shaker at 200 rpm (Innova 4000 incubator shaker; New Brunswick Scientific., Edison, N.J.) at 30°C to mid-logarithmic phase. For experiments under hypoxic and cobalt-treated conditions, the procedure was as described previously (30). All experiments were performed with yeast in logarithmic growth phase in a shaker (200 rpm) at 30°C. Growth was monitored by measuring the yeast optical density at 600 nm (OD600) at the completion of each experiment.
TABLE 1.
S. cerevisiae strains and plasmids used in this study
| Strain or plasmid | Strain genotype or plasmid description | Source and/or reference |
|---|---|---|
| Strains | ||
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | RGa |
| Δspt23 | MATahis3Δ1 leu2Δ01 met15Δ0 ura3Δ0 ΔSPT23 | RG |
| Δmga2 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ΔMGA2 | RG |
| Plasmids | ||
| p62::934 | The OLE1 promoter-lacZ fusion | 5 |
| pTBA30 | Basal CYC1 promoter-lacZ fusion | A. K. Vershon |
| pAM6 | A tandem repeat LORE-basal CYC1 promoter-lacZ fusion | 30 |
| pANB1-lacZ | The ANB1 promoter-lacZ fusion | R. Zitomer |
| pAM23 | A centromeric plasmid containing a 5.1-kb fragment which contains MGA2 | This study |
| pRS315 | A parental centromeric plasmid of pAM23 | This lab |
| YEplac181-mycMGA2 | A 2μm plasmid containing triple myc-tagged MGA2 | 14 |
RG, Research Genetics, Inc. (31).
β-Galactosidase Assays.
Assays of cells containing plasmids derived from the OLE1 promoter-lacZ fusion p62 constructs were performed as described previously (25). Cell densities for these assays were determined by measurement at OD600. β-Galactosidase activities reported here are the results of at least two independent experiments. Each experimental assay was performed in quadruplicate.
DNA sequencing.
Plasmid templates for sequencing were isolated using a QIAprep spin purification kit (Qiagen, Santa Clarita, Calif.). The fmol DNA sequencing system (Promega Corp.) was used for sequencing according to its technical manual. Reactions were run on 6% acrylamide sequencing gels, which were dried and exposed to X-OMAT AR film (Kodak, N.Y.) to visualize the sequence.
Yeast extract preparation.
Haploid yeast cells were cultured in 1-liter flasks containing 200 ml of YPD medium either under normoxic or hypoxic conditions, harvested at mid-log phase (OD600 of 0.8), and lysed by vortexing with glass beads according to published protocols (24). Following addition of ammonium sulfate to 40% and incubation on a rocker table at 4°C for 30 min, the precipitate was collected by centrifugation at 14,000 rpm in a microcentrifuge at 4°C for 10 min. The pellet was resuspended in storage buffer (20 mM HEPES [pH 8.0], 5 mM EDTA, 20% [vol/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride, 7 mM β-mercaptoethanol) and stored frozen at −80°C. The soluble protein concentration was determined using a Bradford dye binding assay (Bio-Rad).
EMSA.
Electrophoretic mobility shift assays (EMSAs) were performed essentially as described previously (30) utilizing synthetic paired oligonucleotides (LORE, 5′-GAACACTCAACAAACCTTAT-3′, or mutated LORE, 5′-GAACACTCAAaAAACCTTAT-3′ [lowercase indicates a mutation]) as a probe. Oligonucleotides were synthesized by Integrated DNA Inc. (Coralville, Iowa). and end labeled using polynucleotide kinase and purified using a Sephadex G-25 spin column (Roche Molecular Biochemicals) to remove unincorporated nucleotide. In each reaction mixture, 10 to 20 ng of probe was used. Binding reactions were in 40 μl of buffer H [25 mM HEPES (pH 7.5) at room temperature, 0.5 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM MgCl2, 1 mM CaCl2, 50 mM NaCl, 7% glycerol, 1% Nonidet P-40, 15 ng of poly(dA-dT) per μl] for 20 min at room temperature. Proteins were diluted into binding buffer on ice immediately before use. Reaction mixtures were loaded on 5% acrylamide gels (29:1, acrylamide-bis), electrophoresed in 0.5× Tris-borate-EDTA (TBE), and run for 3 h at 4°C at 15 V/cm. Gels were dried and exposed to X-OMAT AR film to visualize the shifted bands.
Antibody supershift analyses were performed with a monoclonal anti-c-myc antibody (clone 9E10; Sigma). The monoclonal anti-His tag antibody was obtained from Qiagen. The EMSA binding buffer was used to prepare the working dilution. Crude extracts, antibody, and probe were incubated at 25°C for 45 min before this reaction mixture was loaded on 5% acrylamide gels.
RNA isolation and Northern blot analysis.
Total yeast RNA was isolated as described previously (6). Equal amounts (15 μg) of total RNA were analyzed by Northern blotting according to standard procedures for separation of RNA using 1% formaldehyde gels (6). RNA from the gels was transferred to Nytran Plus membranes (Schleicher & Schuell Inc., Keene, N.H.) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight. Prehybridization, hybridization, and washing of membranes were performed as described previously (2). Northern blots were quantified using a PhosphorImager (Molecular Dynamics), and autoradiographs were also prepared on X-OMAT AR film (Kodak).
To make radiolabeled cDNA probes for other genes of interest (including ACT1 as a control), yeast genomic DNA prepared by the rapid isolation of yeast chromosomal DNA protocol (13) was subjected to PCR with appropriate pairs of primers for the particular genes of interest. The PCR products were first purified using a QIAquick spin PCR purification kit (Qiagen), separated by agarose gel electrophoresis in 1× Tris-acetic acid-EDTA (TAE), and then purified by a Qiagen gel extraction kit according to the manufacturer's recommendations.
For the detection of OLE1 mRNA, a radiolabeled DNA probe was made using a 0.5-kb EcoRI fragment from the OLE1 coding sequence. All DNA fragments were separated by agarose gel electrophoresis in 1× TAE and purified using a Qiagen gel extraction kit according to the manufacturer's recommendations. The purified DNA fragments were labeled to high specific activity with [32P]dCTP (DuPont NEN) by the random primer extension method using Ready to Go DNA labeling beads (Amersham Pharmacia Biotech, Piscataway, N.J.) reaction kit. Unincorporated nucleotides were removed from the sample using a Sephadex G-50 spin column (Roche Molecular Biochemicals). The specific activities of the labeled probes were determined by liquid scintillation counting.
RESULTS
MGA2 affects OLE1 expression under normoxic, hypoxic, and cobalt treatment conditions.
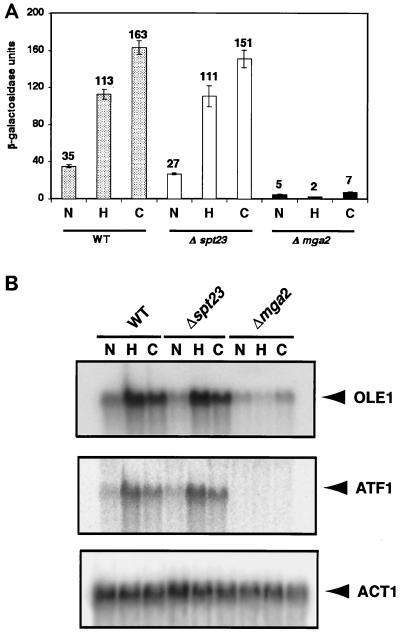
To investigate the roles of MGA2 and SPT23 in regulating OLE1 expression, we performed reporter gene assays with a series of plasmids containing OLE1 promoter elements fused to lacZ in Δmga2 and Δspt23 deletion mutants and their parental strain BY4741. As shown in Fig. 1A, in vivo β-galactosidase activity using reporter p62::934 was essentially the same in the Δspt23 deletion mutant as in its parent under normoxic, hypoxic, and cobalt treatment conditions. Expression of the p62::934 reporter gene construct in the Δmga2 deletion strain, however, was dramatically reduced under all conditions. Of note is that the hypoxic induction was completely abolished. These results suggested that MGA2, not SPT23, is the primary activator of OLE1 transcription under conditions of oxygen starvation and that Mga2p is involved in the basal expression of OLE1 under normoxic conditions. To confirm this, Northern blot analysis was performed. As shown in Fig. 1B, OLE1 mRNA levels in the Δspt23 deletion mutant were comparable to those in the wild type under normoxic, hypoxic, and cobalt treatment conditions. Consistent with the reporter gene activity results, the relative mRNA levels of hypoxic and cobalt-treated Δmga2 cells were also dramatically reduced. Although OLE1 mRNA levels in normoxic Δmga2 cells also appeared to be lower than that of the wild type, they were higher than would be predicted on the basis of reporter gene activity, suggesting that other mechanisms of regulation, such as posttranscriptional controls, may contribute to the overall levels of expression under those conditions. This observation is consistent with the results in this and previous studies (14, 34) which reported that the Δmga2 strain is viable and that, in the absence of MGA2, the activation of OLE1 expression by Spt23p sustains unsaturated fatty acid biosynthesis at levels necessary for normal growth. However, the reduced OLE1 mRNA levels in normoxic cells, taken together with the results from the reporter gene assays, imply that Mga2p is a major contributor to the basal expression of OLE1.
FIG. 1.
Effects of SPT23 and MGA2 on OLE1 gene expression under normoxia, hypoxia, and cobalt treatment conditions. In both panels, N, H, and C represent normoxia, hypoxia, and cobalt treatment conditions, respectively, as described in Materials and Methods. (A) Histogram of β-galactosidase activity of the p62::934 reporter in different strain backgrounds. The Δspt23 and Δmga2 strains and the wild-type(WT) strain BY4741, all containing the plasmid p62::934, were exposed to normoxia, hypoxia, and cobalt treatment conditions as described in Materials and Methods. The absolute units of activity indicated are averages of at least three independent experiments that were performed in quadruplicate. (B) Northern blot assay of OLE1 gene expression in different strain backgrounds. OLE1 and ATF1 were tested for normoxia-, hypoxia-, and cobalt-induced gene expression by Northern blot analysis. Cells were grown to mid-logarithmic phase, and total RNAs were extracted as described in Materials and Methods. The RNA blot was first probed with one specific probe, and after the blot was stripped, it was reprobed with another probe. The ACT1 cDNA probe was prepared using PCR. The positions of OLE1, ATF1 and ACT1 mRNAs are shown to the right. Phosphorimages of the resulting blots are shown in the figure.
MGA2 affects LORE-dependent OLE1 expression under normoxic, hypoxic, and cobalt treatment conditions.
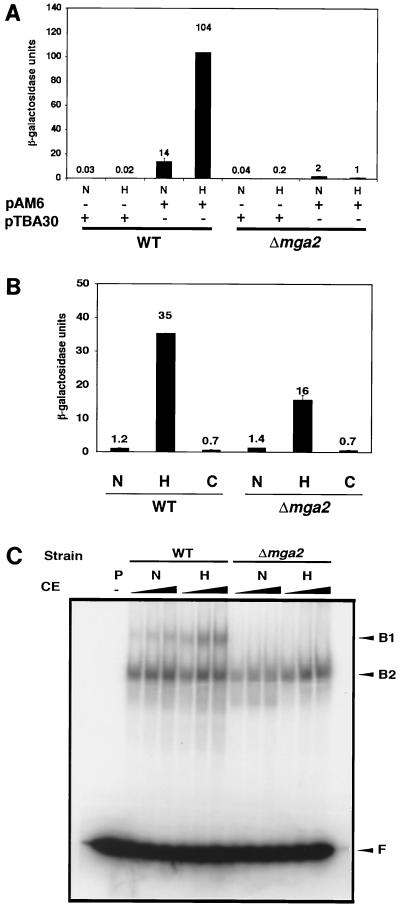
To test whether MGA2 affects the LORE-dependent hypoxic induction of gene expression, we transformed pAM6, a plasmid carrying two copies of the LORE in tandem fused to the basal CYC1 promoter-lacZ, into a Δmga2 strain. pTBA30, a plasmid which contains only the basal CYC1 promoter-lacZ, served as a control in both the wild-type and Δmga2 strains. As shown in Fig. 2A, the LORE-dependent reporter expression was markedly reduced under hypoxic conditions in the Δmga2 strain. The basal expression under normoxic conditions was also decreased from that of the wild type. The expression of the ROX1-dependent anoxia-inducible gene, ANB1, was tested as a control in a Δmga2 strain. An ANB1 promoter-lacZ fusion plasmid was transformed into a Δmga2 strain to measure its β-galactosidase activity under different conditions. As shown in Fig. 2B, there was an 11-fold hypoxic induction of reporter expression observed in the Δmga2 strain. However, the absolute value of hypoxia-induced β-galactosidase activity in the Δmga2 strain was about half of that in its parental wild-type strain; basal expression levels were virtually the same. As previously reported (30), incubation in CoCl2 did not affect ANB1 expression at all. These data suggest that MGA2 does not appear to dramatically affect ROX1-dependent hypoxic or anoxic gene expression; however, the fact that absolute β-galactosidase activity in the Δmga2 strain was decreased implies that MGA2 influences ANB1 gene expression, either directly or indirectly.
FIG. 2.
Effects of MGA2 on LORE-dependent hypoxic gene expression. In all panels, N, H, and C represent normoxia, hypoxia, and cobalt treatment conditions, respectively, as described in Materials and Methods. (A) Histogram of β-galactosidase activity of pAM6 and pTBA30 reporter gene assays under various conditions. The Δmga2 deletion strain and the wild-type (WT) strain, BY4741, both containing pAM6, were exposed to normoxic and hypoxic conditions as described in Materials and Methods. The absolute units of activity indicated are averages of two independent experiments that were performed in quadruplicate. (B) Histogram of β-galactosidase activity of reporter pANB1-lacZ under various conditions. The Δmga2 strain and the wild-type strain, BY4741, containing pANB1-lacZ were exposed to normoxia and hypoxia conditions as described in Materials and Methods. The absolute units of activity indicated are averages of two inde- pendent experiments that were performed in quadruplicate. (C) EMSA for crude extracts from the Δmga2 strain and BY4741 grown under different conditions. Increasing amounts of crude extract (CE) (5, 10, and 50 μg [the amount of CE is indicated by the height of the black triangle over the lanes]) from normoxia- and hypoxia-treated yeast cells were incubated for 20 min at 25°C with 32P-labeled paired LORE oligonucleotides and then subjected to electrophoresis at 4°C for ∼3 h. Free probe (F) and bound complexes (B1 and B2) were detected by autoradiography. Lane −, no CE was added. P, DNA probe.
EMSAs were performed using an end-labeled LORE sequence as a probe. Figure 2C demonstrates that the hypoxia-inducible complex (B1) formation using the crude extract from the Δmga2 strain was abolished and the basal B1 complex formed in normoxia was dramatically decreased as well. These results are consistent with the data from reporter assays in Fig. 2A in which the basal reporter expression was also reduced under normoxic conditions. Together, these data further support the previous conclusion that LORE is involved in OLE1 expression not only under hypoxic but also under normoxic conditions.
MGA2 is required for LORE-dependent hypoxia-induced gene expression.
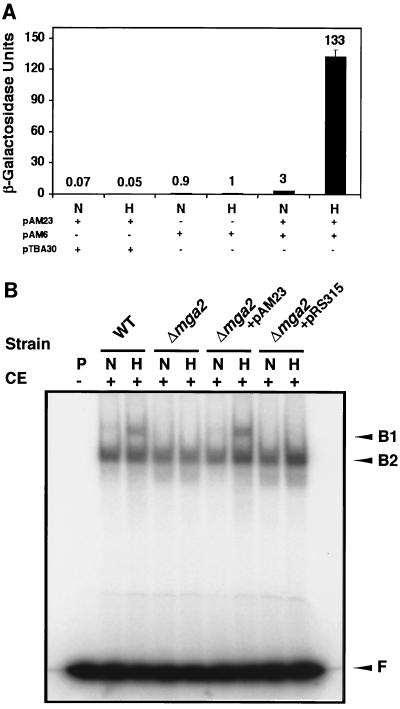
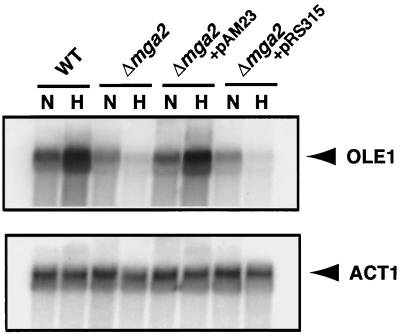
To confirm that MGA2 is required for LORE-dependent hypoxia-induced gene expression, we transformed pAM23, a plasmid carrying an intact MGA2 gene, into a Δmga2 strain already containing either pAM6, the LORE-CYC1 basal promoter-lacZ fusion, or the control plasmid, pTBA30, the CYC1 basal promoter-lacZ fusion. As shown in Fig. 3A, pAM6 reporter gene expression was significantly induced under hypoxic conditions only in the presence of pAM23. The absolute β-galactosidase units are very similar to that from the Δmga2 parental wild-type strain BY4741 (Fig. 2A). As expected, the Δmga2 strain carrying pAM23 and pTBA30 did not show any induction of reporter expression under the same hypoxic conditions. Additionally, the Δmga2 strain containing the LORE-CYC1 basal promoter-lacZ fusion reporter pAM6 and the backbone centromeric plasmid pRS315 did not exhibit increased reporter expression under hypoxic conditions (data not shown). These results clearly demonstrate that MGA2 is required for LORE-dependent hypoxic induction of gene expression in yeast. To further confirm this result, we performed an EMSA using LORE as a probe and crude extracts from the Δmga2 strain transformed with either pAM23 or pRS315 grown under normoxic and hypoxic conditions. As shown in Fig. 3B, there was a hypoxia-inducible complex B1 in the Δmga2 strain transformed with plasmid pAM23 but not in the Δmga2 strain transformed with pRS315. The intensity of the B1 band in the Δmga2 strain transformed with pAM23 was similar to that seen in the wild-type strain. The Northern blot analysis in Fig. 4 demonstrated that OLE1 mRNA levels were completely restored in the Δmga2 strain transformed with a centromeric plasmid (pAM23) containing MGA2, consistent with the results from the reporter and EMSA assays. Taken together, these data provide evidence that MGA2 is required for LORE-dependent hypoxic induction of OLE1.
FIG. 3.
MGA2 is required for LORE-dependent hypoxic gene expression. In both panels, N and H represent normoxic and hypoxic conditions, respectively, as described in Materials and Methods. (A) β-Galactosidase assays. Plasmid pAM23 carrying intact MGA2 was transformed into the Δmga2 deletion strain, which was transformed with either a LORE-CYC1 basal promoter-lacZ fusion (pAM6) or its backbone plasmid without LORE (pTBA30). Yeast cells were exposed to normoxic and hypoxic conditions as described in Materials and Methods. The absolute units of activity indicated are averages of two independent experiments that were performed in quadruplicate. (B) EMSA. Fifteen micrograms of crude extract (CE) from normoxia- and hypoxia-treated yeast cells was incubated for 20 min at 25°C with 32P-labeled paired LORE oligonucleotides and then subjected to electrophoresis at 4°C for ∼3 h. Free probe (F) and bound complexes (B1 and B2) were detected by autoradiography. Symbols: +, CE was added; −, no CE was added. Abbreviations: P, DNA probe; WT, wild type.
FIG. 4.
Northern blot analysis. OLE1 was tested for normoxic and hypoxic gene expression by Northern blot analysis in the wild-type (WT) and Δmga2 strains. The Δmga2 strain carrying plasmid pAM23 (containing intact MGA2) or the backbone vector pRS315 are indicated. Cells were grown to mid-logarithmic phase, and total RNA was extracted as described in Materials and Methods. The RNA blot was first probed with one specific probe, and then the blot was stripped and reprobed with another probe. The ACT1 cDNA probe was prepared using PCR. The positions of OLE1 and ACT1 mRNAs are indicated to the right. Phosphorimages of the resulting blots are shown in the figure.
Mga2p is a component of the LORE-binding complex.
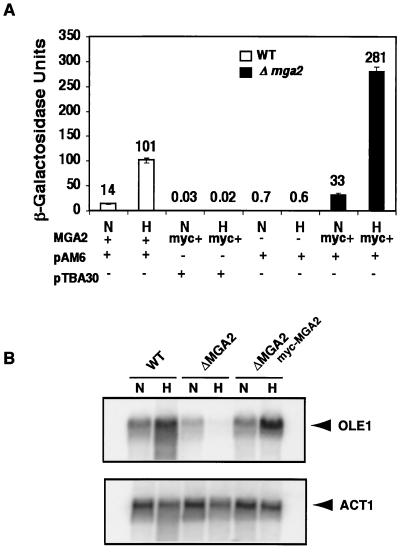
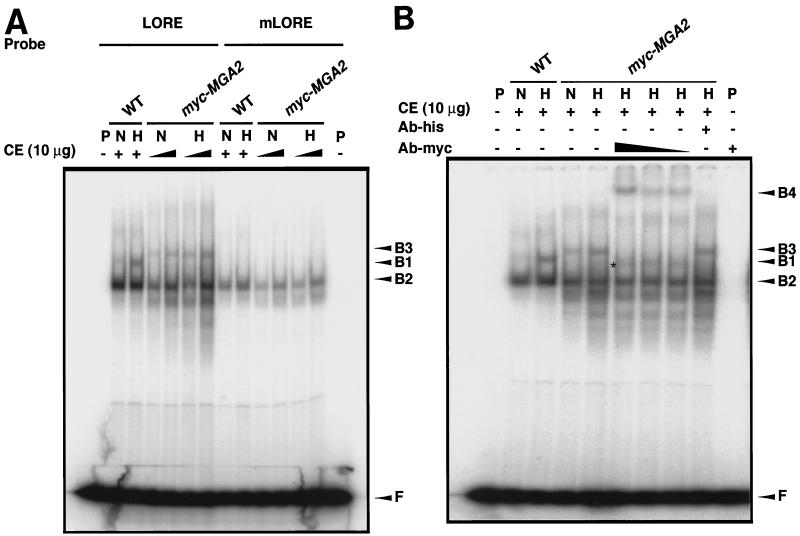
The results of the above studies clearly demonstrate that MGA2 is involved in LORE-dependent hypoxic induction of OLE1. Thus, we studied whether Mga2p is, in fact, a component of a LORE binding complex which is induced under hypoxic conditions. A 2μm-based myc-tagged MGA2 plasmid, YEplac181-mycMGA2 (14), was transformed into a Δmga2 strain. As expected, myc-MGA2 fully restored OLE1 expression as shown by in vivo reporter and Northern blot analyses in Fig. 5. As shown in Fig. 5A, the LORE-CYC1 basal promoter-lacZ fusion pAM6 reporter gene expression was significantly induced under hypoxic conditions. The absolute β-galactosidase units under both normoxic and hypoxic conditions were increased more than twofold compared to that of the wild-type strain. Consistent with the in vivo reporter analysis, OLE1 mRNA levels were also significantly increased under hypoxic conditions following transformation of the Δmga2 strain with the myc-MGA2 construct (Fig. 5B). Protein crude extracts were prepared for EMSA. As shown in Fig. 6A, the complex B3 from myc-MGA2 crude extracts and the hypoxia-inducible complex B1 from wild-type crude extracts disappeared when a mutated LORE sequence was used as a probe, suggesting that B1 and B3 are equivalent specific LORE-binding complexes. Mga2p participation in the hypoxia-induced LORE-binding complex might explain the slower migration of B3, as the mycMga2p has 30 additional amino acids at the N terminus of Mga2p. The B3 band was enhanced in hypoxic crude extract; however, due to the relatively high intensity of B3 under normoxic conditions, the degree of B3 induction appeared to be less than that in wild-type crude extracts. Again, this may be due to the high copy number of myc-MGA2 which was expressed from the 2μm plasmid.
FIG. 5.
myc-MGA2 confers functional LORE-dependent hypoxic gene expression. In both panels, N and H represent normoxic and hypoxic conditions, respectively, as described in Materials and Methods. (A) β-Galactosidase assays. Plasmid YEplac181-mycMGA2 carrying the myc-tagged MGA2 was transformed into the Δmga2 strain which was transformed with either a LORE-CYC1 basal promoter-lacZ fusion plasmid (pAM6) or its backbone plasmid without the LORE (pTBA30). The MGA2 row under the graph indicates the yeast strain used as follows: +, the wild-type strain BY4741; −, the Δmga2 strain; myc+, the Δmga2 strain transformed with YEplac181-mycMGA2. The pAM6 row indicates whether the yeast strain contains LORE-CYC1 basal promoter-lacZ fusion (pAM6). The pTBA30 row indicates whether the yeast strain contains backbone plasmid without LORE (pTBA30). The absolute units of activity indicated are averages of two independent experiments that were performed in quadruplicate. (B) Northern blot analysis. OLE1 was tested for normoxia- and hypoxia-induced gene expression by Northern blot analysis in the wild-type (WT) or Δmga2 strain. The Δmga2 strain transformed with plasmid YEplac181-mycMGA2 containing myc-tagged MGA2 is indicated (myc-MGA2). Cells were grown to mid-logarithmic phase, and total RNAs were extracted as described in Materials and Methods. The RNA blot was first probed with one specific probe, and then after the blot was stripped, it was reprobed with another probe. The ACT1 cDNA probe was prepared using PCR. The positions of OLE1 and ACT1 mRNAs are indicated to the right. Phosphorimages of the resulting blots are shown in the figure.
FIG. 6.
Mga2p is a component of the LORE-binding complex. In both panels, N and H represent normoxic and hypoxic conditions, respectively, as described in Materials and Methods. (A) EMSA. Ten micrograms of crude extracts (CE) from normoxia- and hypoxia-treated wild-type yeast (WT) or the Δmga2 strain containing plasmid YEplac181-mycMGA2 (myc-MGA2) were incubated for 20 min at 25°C with 32P-labeled paired LORE oligonucleotides or mutated LORE (C337A) (30) and then subjected to electrophoresis at 4°C for ∼3 h. Free probe (F) and bound complexes (B1, B2, and B3) were detected by autoradiography. Symbols: +, CE was added; −, no CE was added. P, DNA probe. (B) Supershift analysis. Ten micrograms of crude extracts (CE) from normoxia- and hypoxia-treated wild-type yeast (WT) or the Δmga2 strain containing plasmid YEplac181-mycMGA2 (myc-MGA2) were incubated for 45 min at 25°C with increasing amounts of anti-c-myc antibody (1:2,000, 1:1,600, and 1:1,000) and 32P-labeled paired LORE oligonucleotides and then subjected to electrophoresis at 4°C for ∼3 h. Free probe (F) and bound complexes (B1, B2, B3, and B4) were detected by autoradiography. Symbols: +, CE was added or 1:1,000 anti-c-myc antibody was added; −, no CE or no antibody was added. Abbreviations: P, DNA probe; Ab-his, a control antibody, anti-His tag, was used at a 1:40 dilution to the reaction mixture. The additional band observed between B1 and B2 after the addition of the antibody is indicated by an asterisk.
To determine whether Mga2p is a component of the LORE-binding complex, we performed a supershift assay using an anti-myc antibody. As shown in Fig. 6B, a clear supershifted band, B4, was observed when myc-MGA2 crude extracts were examined, and B3 disappeared. The anti-myc antibody had no effect on the band shift using crude extract from the wild-type strain (data not shown). Upon addition of antibody, an additional band between B1 and B2 was observed in Fig. 6B. Both this and the supershifted band could be competed out by adding cold LORE sequence (data not shown). The nature of this additional band is unknown. It could be an artifact that is related to the 2μm-based plasmid. When a cen-based myc-tagged MGA2 plasmid was transformed into a Δmga2 strain, a similar supershift assay was performed and a supershifted band equivalent to B4 in Fig. 6B was observed, but the additional band was eliminated (data not shown).
MGA2 is also involved in ATF1 expression.
Studies have shown that ATF1, encoding an alcohol acetyltransferase, is regulated by hypoxia and unsaturated fatty acid in a manner similar to OLE1 (9, 10). We previously demonstrated that a LORE sequence exists in the promoter region of ATF1 which is critical for hypoxic induction and hypothesized that the LORE-dependent hypoxia induction pathway plays an important role in the regulation of ATF1 expression (30). To determine whether MGA2 is also involved in ATF1 expression, we performed a Northern blot assay. As shown in Fig. 1B, similar to OLE1 expression, the ATF1 mRNA levels were significantly decreased in the Δmga2 strain. This result is consistent with the hypothesis that MGA2 plays a key role in the LORE-dependent hypoxia induction pathway in S. cerevisiae.
DISCUSSION
OLE1, which encodes the Δ-9 fatty acid desaturase, is critical for unsaturated fatty acid biosynthesis and thus, cell viability in S. cerevisiae. This enzyme introduces a double bond between carbons 9 and 10 of its substrates, palmitoyl (16:0) or stearoyl (18:0)–coenzyme A, with molecular O2 serving as an electron acceptor to form palmitoleic (16:1) or oleic (18:1) acid, respectively. Previous studies have demonstrated that OLE1 is upregulated under hypoxic conditions via a LORE-dependent pathway (23, 30). While the transactivation factors involved in the hypoxic induction of OLE1 were unknown, multicopy suppressor assays supported the hypothesis that MGA2 or SPT23 is required for the transcription of OLE1 (34).
The experiments presented here show that OLE1 expression is dramatically reduced in the Δmga2 strain. The hypoxia- and cobalt-induced OLE1 promoter-lacZ reporter expression is abolished in the Δmga2 strain but not in the Δspt23 strain (Fig. 1). Given that the basal expression of OLE1 in normoxic conditions is also decreased (Fig. 1), LORE may also be involved in the basal expression of OLE1. This hypothesis is supported by the decreased OLE1 reporter expression when mutations are introduced in the LORE sequence of the OLE1 promoter in both RZ53-6 (30) and BY4741(data not shown). Whether MGA2 may also be necessary for full OLE1 basal expression via elements outside the LORE remains to be explored.
Although MGA2 and SPT23 are two functionally and genetically related genes, they appear to have specific roles involving OLE1 expression. Recent studies reveal that Mga2p and Spt23p are membrane-bound transcription factors that can be activated by regulated ubiquitin/proteasome-dependent processing (14). Results from Hoppe et al. suggested that although structurally and functionally very similar to Spt23p, Mga2p appears to be regulated in a different manner (14). Specifically, unsaturated fatty acids had only a moderate influence on the processing reaction of Mga2p compared to that of Spt23p. In the case of cell viability, deletion of either of these genes had only modest effects on cell growth. However, cells with MGA2 SPT23 double mutations are nonviable (33). Multicopy suppressor analysis indicates that either MGA2 or SPT23 is required for transcription of OLE1 in normoxia (34). It is, therefore, quite surprising that the deletion of SPT23 has almost no effect on OLE1 expression under normoxic, hypoxic, and cobalt treatment conditions.
Our results here indicate that MGA2, not SPT23, is the dominant gene involved in OLE1 expression under the normoxic, hypoxic, and cobalt treatment conditions used in this study. Although it is possible that the drastically reduced OLE1 expression in Δmga2 Δspt23 strain is caused in large part by the deletion of the MGA2 locus, these results do not exclude the possibility that SPT23 is also involved in OLE1 expression. In fact, in other studies (34), Δmga2 ts-spt23 was found to exhibit fatty acyl compositions that are comparable to those of the wild type, indicating that Spt23p can independently activate basal OLE1 expression to levels sufficient for growth. Spt23p may be the major effector in response to certain nutritional or environmental signals, but not in response to others. Previous studies, for example, demonstrated that Spt23p proteolytic processing can be strongly regulated by unsaturated fatty acids (14), which are known to repress OLE1 expression (5, 11). The results of this study lend strong support to the idea that SPT23 and MGA2 have evolved distinct, but overlapping functions to maximize cellular responses to a range of stimuli that govern OLE1 expression.
Supershift analyses of EMSAs indicate that Mga2p is a component of the LORE-binding complex. It is not known whether Mga2p is a classical transcriptional activator. Mga2p could bind to LORE either directly or indirectly through an interaction with other proteins. Previous studies suggested that MGA2 encodes a protein which does not contain any recognized DNA-binding motifs (33). Therefore, if Mga2p binds to the LORE directly, it may possess a novel DNA-binding motif. Alternatively, Mga2p may bind to another as yet unidentified LORE-binding factor and form the LORE-binding complex which is normally observed.
We performed a coiled-coil structure analysis (21) to examine whether there is a potential coiled-coil protein interaction motif in Mga2p. The results predicted that amino acids 93 to 120 near the C terminus exhibit a high possibility of possessing a coiled-coil motif. We believe that this potential coiled-coil motif may play a role in protein-protein interactions if MGA2 binds to LORE through another protein. It may also be involved in homodimer (oligomer) formation.
Understanding how hypoxia affects the processing and mechanism of LORE-dependent hypoxic gene induction via MGA2 will likely provide significant insights into the cellular response to hypoxic stress in both lower and higher eukaryotic species.
ACKNOWLEDGMENTS
We thank H. Franklin Bunn and Fred Winston for their invaluable support through all phases of this project.
This work was supported in part by NIH grants DK45098 to M.A.G. and GM45768 to C.E.M.
REFERENCES
- 1.Arany Z, Huang L E, Eckner R, Bhattacharya S, Jiang C, Goldberg M A, Bunn H F, Livingston D M. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci USA. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown T, Mackey K. Analysis of RNA by Northern and slot blot hybridization. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1997. pp. 4.9.1–4.9.14. [Google Scholar]
- 3.Bunn H F, Poyton R O. Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 4.Burkett T J, Garfinkel D J. Molecular characterization of the SPT23 gene: a dosage-dependent suppressor of Ty-induced promoter mutations from Saccharomyces cerevisiae. Yeast. 1994;10:81–92. doi: 10.1002/yea.320100108. [DOI] [PubMed] [Google Scholar]
- 5.Choi J Y, Stukey J, Hwang S Y, Martin C E. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J Biol Chem. 1996;271:3581–3589. doi: 10.1074/jbc.271.7.3581. [DOI] [PubMed] [Google Scholar]
- 6.Collart M A, Oliviero S. Preparation of yeast RNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1997. pp. 13.12.1–13.12.5. [DOI] [PubMed] [Google Scholar]
- 7.Ebert B L, Bunn H F. Regulation of the erythropoietin gene. Blood. 1999;94:1864–1877. [PubMed] [Google Scholar]
- 8.Elble R. A simple and efficient procedure for transformation of yeast. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 9.Fujiwara D, Yoshimoto H, Sone H, Harashima S, Tamai Y. Transcriptional co-regulation of Saccharomyces cerevisiae alcohol acetyltransferase gene, ATF1 and delta-9 fatty acid desaturase gene, OLE1 by unsaturated fatty acids. Yeast. 1998;14:711–721. doi: 10.1002/(SICI)1097-0061(19980615)14:8<711::AID-YEA263>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara D, Kobayashi O, Yoshimoto H, Harashima S, Tamai Y. Molecular mechanism of the multiple regulation of the Saccharomyces ATF1 gene encoding alcohol acetyltransferase. Yeast. 1999;15:1183–1197. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1183::AID-YEA444>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez C I, Martin C E. Fatty acid-responsive control of mRNA stability. Unsaturated fatty acid-induced degradation of the Saccharomyces OLE1 transcript. J Biol Chem. 1996;271:25801–25809. doi: 10.1074/jbc.271.42.25801. [DOI] [PubMed] [Google Scholar]
- 12.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman C S. Rapid isolation of yeast chromosomal DNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1997. pp. 13.11.2–13.11.4. [Google Scholar]
- 14.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich H D, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 15.Huang L E, Gu J, Schau M, Bunn H F. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornberg R D, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 17.Kwast K E, Burke P V, Poyton R O. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J Exp Biol. 1998;201:1177–1195. doi: 10.1242/jeb.201.8.1177. [DOI] [PubMed] [Google Scholar]
- 18.Kwast K E, Burke P V, Staahl B T, Poyton R O. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc Natl Acad Sci USA. 1999;96:5446–5451. doi: 10.1073/pnas.96.10.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 20.Lin L, Ghosh S. A glycine-rich region in NF-κB p105 functions as a processing signal for the generation of the p50 subunit. Mol Cell Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell P H, Pugh C W, Ratcliffe P J. Inducible operation of the erythropoietin 3′ enhancer in multiple cell lines: evidence for a widespread oxygen-sensing mechanism. Proc Natl Acad Sci USA. 1993;90:2423–2427. doi: 10.1073/pnas.90.6.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa Y, Sugioka S, Kaneko Y, Harashima S. O2R, a novel regulatory element mediating Rox1p-independent O2 and unsaturated fatty acid repression of OLE1 in Saccharomyces cerevisiae. J Bacteriol. 2001;183:745–751. doi: 10.1128/JB.183.2.745-751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeifer K, Arcangioli B, Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987;49:9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- 25.Reynolds A, Lundblad V, Dorris D, Keaveney M. Yeast vectors and assays for expression of cloned genes. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1997. pp. 13.6.2–13.6.3. [DOI] [PubMed] [Google Scholar]
- 26.Salceda S, Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 27.Semenza G L. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 28.Travers A. An engine for nucleosome remodeling. Cell. 1999;96:311–314. doi: 10.1016/s0092-8674(00)80543-7. [DOI] [PubMed] [Google Scholar]
- 29.Treco D A, Winston F. Growth and manipulation of yeast. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1997. pp. 13.2.1–13.2.12. [Google Scholar]
- 30.Vasconcelles M J, Jiang Y, McDaid K, Gilooly L, Wretzel S, Porter D L, Martin C E, Goldberg M A. Identification and characterization of a low oxygen response element (LORE) involved in the hypoxic induction of a family of S. cerevisiae genes: implications for the conservation of oxygen sensing in eukaryotes. J Biol Chem. 2001;276:14374–14384. doi: 10.1074/jbc.M009546200. [DOI] [PubMed] [Google Scholar]
- 31.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, Chu A M, Connelly C, Davis K, Dietrich F, Dow S W, El Bakkoury M, Foury F, Friend S H, Gentalen E, Giaever G, Hegemann J H, Jones T, Laub M, Liao H, Davis R W. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 32.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S, Burkett T J, Yamashita I, Garfinkel D J. Genetic redundancy between SPT23 and MGA2: regulators of Ty-induced mutations and Ty1 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4718–4729. doi: 10.1128/mcb.17.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S, Skalsky Y, Garfinkel D J. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics. 1999;151:473–483. doi: 10.1093/genetics/151.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zitomer R S, Lowry C V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]