Abstract
Acute pain that is associated with herpes zoster (HZ) can become long-lasting neuropathic pain, known as chronic post-herpetic neuralgia (PHN), especially in the elderly. HZ is caused by the reactivation of latent varicella-zoster virus (VZV), whereas PHN is not attributed to ongoing viral replication. Although VZV infection reportedly induces neuronal cell fusion in humans, the pathogenesis of PHN is not fully understood. A genome-wide association study (GWAS) revealed significant associations between PHN and the rs12596324 single-nucleotide polymorphism (SNP) of the heparan sulfate 3-O-sulfotransferase 4 (HS3ST4) gene in a previous study. To further examine whether this SNP is associated with both PHN and VZV reactivation, associations between rs12596324 and a history of HZ were statistically analyzed using GWAS data. HZ was significantly associated with the rs12596324 SNP of HS3ST4, indicating that HS3ST4 is related to viral replication. We investigated the influence of HS3ST4 expression on VZV infection in cultured cells. Fusogenic activity after VZV infection was enhanced in cells with HS3ST4 expression by microscopy. To quantitatively evaluate the fusogenic activity, we applied cytotoxicity assay and revealed that HS3ST4 expression enhanced cytotoxicity after VZV infection. Expression of the VZV glycoproteins gB, gH, and gL significantly increased cytotoxicity in cells with HS3ST4 expression by cytotoxicity assay, consistent with the fusogenic activity as visualized by fluorescence microscopy. HS3ST4 had little influence on viral genome replication, revealed by quantitative real-time polymerase chain reaction. These results suggest that HS3ST4 enhances cytotoxicity including fusogenic activity in the presence of VZV glycoproteins without enhancing viral genome replication.
Keywords: post-herpetic neuralgia, neuropathic pain, varicella-zoster virus, herpes zoster, single-nucleotide polymorphism, heparan sulfate 3-O-sulfotransferase 4, glycoproteins, fusogenic activity
Introduction
Post-herpetic neuralgia (PHN) is long-lasting neuropathic pain that is caused by the reactivation of latent varicella-zoster virus (VZV; i.e., herpes zoster (HZ)). Among HZ-afflicted patients, 27–73% develop PHN, which is age-dependent. 1 PHN causes severe pain that lasts for months or even more than 1 year. New ways need to be discovered to prevent PHN and further elucidate the pathogenesis of PHN.
In a previous study, PHN was significantly associated with the intronic rs12596324 single-nucleotide polymorphism (SNP) of the HS3ST4 gene. 2 PHN pain has been shown to not be attributable to ongoing viral replication. 3 More severe HZ leads to PHN, 3 implying that some damage that occurs in HZ affects the susceptibility to PHN. Indeed, VZV infection induces neuronal cell fusion in humans.4,5 Accordingly, virus-induced cytopathology in HZ likely contributes to the pathogenesis of PHN as well as HZ.
VZV is an enveloped double-stranded DNA virus that belongs to the varicellovirus genus in the alphaherpesvirus subfamily of the herpesviridae family. Typically, VZV begins initial replication in the mucosal epithelium of the upper respiratory tract. The virus then enters the blood circulation and localizes to the skin, leading to the formation of characteristic blisters. 1 Upon primary infection, the virus causes varicella (chickenpox). Subsequently, sensory neurons that project to the skin become infected, leading to latent infection with VZV that persists for an individual’s lifetime. Immunocompromised hosts and the elderly have a higher susceptibility to the reactivation of VZV, causing HZ.
The DNA genome of VZV encodes over 70 open reading frames (ORFs), and 11 ORFs encode glycoproteins. 1 Glycoproteins on the virion membrane, which are glycosylated through N- or O-linked sugars, enable attachment to cell surface proteins and the fusion and entry of VZV into cells. The VZV glycoproteins gB, gH, and gL are prerequisites for membrane fusion that is required for virion entry into cells. 6 After releasing nucleocapsid into the cytoplasm, the viral genome enters the nucleus, followed by viral replication that leads to latent or lytic infection. During latent infection, the viral genome resides in the nucleus as episomes.
Heparan sulfate (HS) is a linear polysaccharide that covalently attaches to core proteins, referred to as HS proteoglycans. These molecules exist in the extracellular matrix and at the cell surface. 7 HS on HS proteoglycans is modified by HS-modifying enzymes, resulting in the functional modification of HS proteoglycans. Heparan sulfate glucosamine 3-O-sulfotransferases (HS3STs) form a family of HS-modifying enzymes. In humans, seven HS3STs have been reported to date. Among these, HS3ST2, HS3ST3A, HS3ST3B, HS3ST4, and HS3ST6 can produce HS-binding motifs for the glycoprotein gD of HSV-1, which is closely related to VZV. 7
The present study was based on our previous findings that a SNP of the HS3ST4 gene is significantly associated with PHN. 2 We further clarified that this SNP is also associated with HZ. This indicates that HS3ST4 is related to the reactivation of VZV. We investigated whether HS3ST4 influences physiological events during VZV infection. We found that HS3ST4 significantly enhanced cytotoxicity including fusogenic activity that is induced by VZV glycoproteins, whereas HS3ST4 had little effect on viral replication. These results suggest that HS3ST4 accelerates cytopathology through virus-induced fusogenic activity, raising the possibility that the enhancement of cytopathology by HS3ST4 expression is related to the pathogenesis of HZ. The pathogenesis of HZ may lead to PHN, although further studies are needed.
Materials and methods
Human genetic association analysis of genome-wide genotyping data
Subjects with chronic pain and healthy subjects: The human GWAS was performed in our previous study. 2 We reanalyzed the genotyping data in the present study. Briefly, enrolled in the study were 194 adult patients who suffered from chronic pain, including PHN, and had a history of HZ and attended JR Tokyo General Hospital (Tokyo, Japan), Juntendo University Hospital (Tokyo, Japan), or Nihon University Itabashi Hospital (Tokyo, Japan) for the treatment of chronic pain. The patients were apparently Japanese. The detailed demographic data of the subjects and their statistics were provided in previous reports. 2
Enrolled in the study as controls were 282 adult healthy volunteers without any particular diseases or chronic pain who lived in or near the Kanto area in Japan. Detailed demographic data of the subjects and their statistics were provided in previous reports.8,9
The study protocol was approved by the Institutional Review Board of JR Tokyo General Hospital (Tokyo, Japan), Institutional Review Board of Juntendo University Hospital (Tokyo, Japan), Institutional Review Board of Nihon University Itabashi Hospital (Tokyo, Japan), and Institutional Review Board of Tokyo Metropolitan Institute of Medical Science (Tokyo, Japan). Written informed consent was obtained from all of the patients and healthy volunteers.
Statistical analysis: A GWAS was conducted for a subgroup of the 96 patient subjects with a history of HZ. A total of 282 control subjects were used in the analyses. To explore associations between the SNP and a history of HZ, Pearson’s χ 2 test or Fisher’s exact test was conducted to compare genotype data between patients with HZ and control subjects. Genotypic, dominant, and recessive genetic models were used for the analyses. The Cochran–Armitage trend test was performed to investigate linear trends using JMP 15.0.0 software (SAS Institute). The patients’ demographic and clinical data are expressed as the mean ± SD. All of the other statistical analyses of genotype data were performed using SPSS Statistics 24 software (IBM Japan). In all of the statistical tests, the criterion for significance was set at p < .05.
Plasmid construction
The human HS3ST4-expressing plasmid was provided by Dr Jian Liu (University of North Carolina, Chapel Hill). 10 Plasmids that encoded the VZV glycoproteins gB (ORF31), gH (ORF37), and gL (ORF60) with a pFN21A or pFC14A HaloTag CMV Flexi vector (Promega) backbone were constructed. Fragments of gB, gH, and gL were amplified from VZV clinical isolate 1710 strain genome DNA using KOD DNA polymerase (KOD FX Neo, Toyobo) with the following primer sets: gB (forward, cagagcgataacgcgatgtccccttgtggctatta; reverse, agcccgaattcgtttttacacccccgttacattct), gH (forward, actatagggctagcgatgtttgcgctagttttagc; reverse, gttggctcgagagctctcacggctccacagaga), and gL (forward, actatagggctagcgatggcatcacataaatggtt; reverse, gttggctcgagagctggcgcgggggcggacgggcc). The gB fragment was cloned into a pFN21A-HaloTag vector, which adds a HaloTag at the amino terminus, and gH and gL were cloned into the pFC14A-HaloTag vector, which adds a HaloTag at the carboxyl terminus using the In-Fusion HD Cloning Kit (Takara Bio).
Cells
The human skin MeWo melanoma cell line was cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM; Fujifilm Wako Pure Chemical Corporation) supplemented with 10% fetal bovine serum (FBS; Biological Industries) and a penicillin-streptomycin solution (Thermo Fisher Scientific). MeWo cells were seeded in 24-well culture dishes (Corning) 1 day before transfection. The plasmid that encoded HS3ST4-cDNA with a pcDNA3 backbone or the vector plasmid pcDNA3 (Thermo Fisher Scientific) was transfected in MeWo cells using FuGENE 6 Transfection Reagent (Promega). After incubation for 5 min at room temperature, the mixture was added to the cells and incubated in a 5% CO2 incubator at 37°C for 24 h, followed by medium exchange. Two days later, the medium was changed to DMEM that contained 0.3 mg/mL G418 (Thermo Fisher Scientific). Selection continued for 14 days. Before the end of selection, all of the control MeWo cells without plasmids were dead. MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) clones were then obtained by limiting dilution. We selected one clone for use in the experiments. We repeated the experiments using another clone and did not find significant differences between clones.
Virus
The VZV wildtype strain 1710, which was isolated in our laboratory from the blister contents of a 9-year-old girl with HZ, was used in this study. MeWo cells that were infected with VZV were collected and frozen using CELLBANKER1 (Takara Bio) until the experiments were conducted. For the infection experiments, the cells were infected with the frozen infected cells after thawing. The titers of infected cells were determined using a plaque assay of frozen infected cells after thawing on MeWo cells.
Western blot
To quantify protein expression, MeWo, MeWo-HS3ST4 (+), and MeWo-HS3ST4 (−) cells at passage eight (P8) or P17 in 10 cm dishes were washed and suspended in electrophoresis sample buffer (Santa Cruz Biotechnology), followed by sonication. The samples were loaded onto e-PAGEL(R) 5–20% precast gels (ATTO). The protein bands were transferred to a polyvinyl difluoride membrane (Trans-Blot Turbo Midi 0.2 μm PVDF Transfer Packs, Bio-Rad) using a Trans-Blot Turbo Transfer System (Bio-Rad), followed by blocking with Blocking One (Nakalai) and blotting with anti-HS3ST4 sheep antibodies (R&D) and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) 1E6D9 mouse antibodies (Proteintech) as primary antibodies and peroxidase donkey anti-sheep IgG (H+L) and peroxidase donkey anti-mouse IgG (H+L) (Jackson Immunoresearch) as secondary antibodies. The membrane was treated with horseradish peroxidase substrate (WBKLS0100 Immobilon Western Chemiluminescent HRP Substrate, Merck). Chemiluminescence was quantified using a Lumino Image Analyzer (ImageQuant LAS-4000mini, Cytiva).
Plaque assay
MeWo cells were seeded in 12-well culture dishes (Corning) 1 day before infection with VZV. Frozen infected cells were thawed immediately and transferred to DMEM, followed by centrifugation. The pellet was resuspended in fresh DMEM. The serially diluted infected cells were added to naive cells in the well, and then the cells were incubated in a 5% CO2 incubator at 37°C until fixation and staining. Six days after infection, the cells were fixed in crystal violet dyes with HCHO for 2 h at room temperature, followed by washing and drying and then observing and counting plaques.
Microscopy imaging
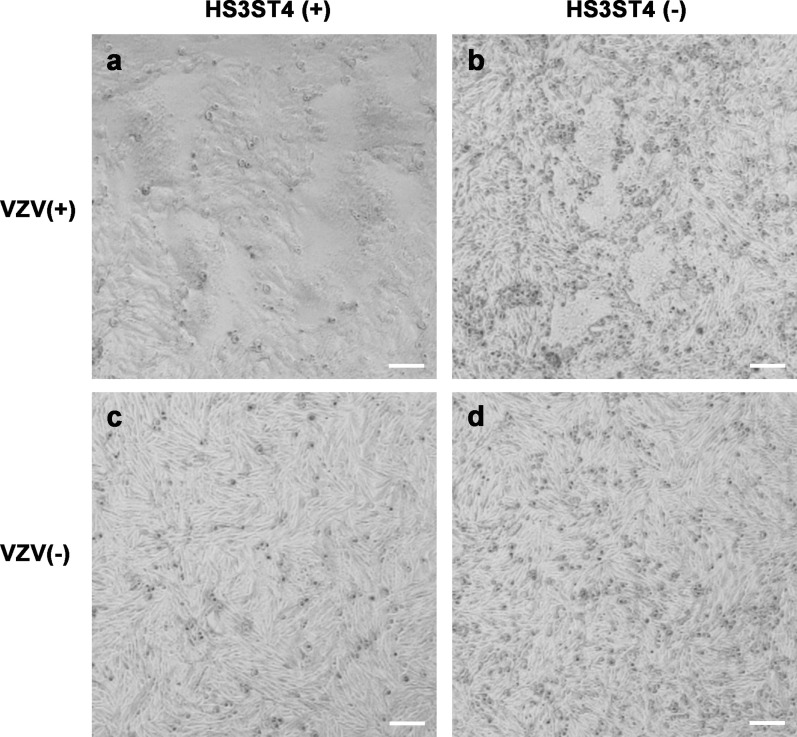
MeWo cells were seeded in 12-well culture dishes (Corning) 1 day before infection with VZV. Frozen infected cells were thawed immediately and transferred to DMEM, followed by centrifugation as described in the plaque assay section. The pellet was resuspended in fresh DMEM. The diluted infected cells were added to 4 × 105 naive cells in the well at a multiplicity of infection (MOI) of 0.005, and then the cells were incubated in a 5% CO2 incubator at 37°C. Live-cell images that were acquired with an inverted microscope (Axiovert 200, Carl Zeiss, Digital Sight DS-L1, Nikon Solutions) 2 days after infection in Figure 3.
Figure 3.
Virus-mediated cell fusion occurred robustly in cells with HS3ST4 infected with VZV. MeWo-HS3ST4 (+) (a and c) and MeWo-HS3ST4 (−) cells (b and d) were infected (a and b) or mock-infected with VZV (c and d) and observed under an inverted microscope 2 days after infection. Scale bar = 100 μm.
Cytotoxicity assay
As a cytotoxicity assay, a lactate dehydrogenase (LDH) release was quantified using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega) according to the manufacturer’s protocol. 11 MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells were seeded in 96-well culture dishes (Corning) 1 day before infection with VZV. The infected cells were added to the cells in 96-well culture dishes at an MOI of 0.05. After incubation for 2 h in a 5% CO2 incubator at 37°C, the medium was exchanged with 100 μL of fresh DMEM with FBS. Zero hours after infection (h.a.i.) was set at this point in the experiment. The dispensed supernatant (50 μL) and 100 μL of the exchanged fresh medium with the cells were frozen at −80°C at 0, 6, 24, 48, 72, and 96 h.a.i. The LDH release ratio of 50 μL of the supernatant with or without cells after the freeze-thaw cycle was examined.
Quantitative real-time polymerase chain reaction assay
MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells were seeded in 24-well culture dishes (Corning) 1 day before infection with VZV. The infected cells were added to cells in 24-well culture dishes at an MOI of 0.005. After incubation in a 5% CO2 incubator at 37°C for 4 h, the medium was exchanged.
Total DNA was extracted from the cells with and without infection in 24-well culture dishes (Corning) using the QIAamp DNA Mini Kit (Qiagen). VZV DNA and ribonuclease (RNase) P as a copy number reference were quantitatively examined using TaqMan Real-Time Polymerase Chain Reaction (PCR) Master Mix (Thermo Fisher) and the 7500 Fast Real-Time PCR System (Applied Biosystems). The probe (final concentration of 0.25 μM) and primers (final concentration of 0.9 μM) for the VZV genome were the following: ORF28 probe (TCC AGG TTT TAG TTG ATA CCA FAM-TAMRA), sense primer (CGA ACA CGT TCC CCA TCA A), and antisense primer (CCC GGC TTT GTT AGT TTT GG). 12 For RNase P, the TaqMan Copy Number Reference Assay, human, RNase P (VIC-TAMRA, Thermo Fisher) was used for detection as a copy number reference. The PCR conditions for VZV and RNase P were the following: one cycle of 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The copy numbers of the VZV genome were corrected relative to the copy numbers of RNase P, which represented the total number of nuclei. To determine cell proliferation rates, quantitative real-time PCR with RNase P detection was performed. Rates were similar among the cell lines.
Electroporation
MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells were treated with the Amaxa 4D Nucleofector Kit SF (Lonza) according to the manufacturer’s protocol and transferred to a 16-well cuvette. 2.5 × 106 cells per sample were electroporated with 0.4 μg of each of the plasmids that encoded the VZV glycoproteins gB, gH, and gL and 0.2 μg of the control plasmid pmaxGFP (Lonza) by the Amaxa 4D Nucleofector X unit (Lonza). After 10 min of incubation at room temperature, cells with fresh DMEM and FBS were transferred to 4 wells of a 96-well culture dish per sample. The medium in the 96-well culture dishes was exchanged with 100 μL of fresh medium 4–6 h after electroporation. As a control for electroporation efficiency, the control plasmid pmaxGFP was co-electroporated. Efficiency was monitored 24 hours after electroporation by observing fluorescence (Axiovert 200, Carl Zeiss, Digital Sight DS-L1, Nikon Solutions), which was sufficiently high in every well (∼95–98%). Glycoprotein expression was monitored by observing the fluorescence of glycoproteins that were tagged with HaloTag after the addition of HaloTag TMRDirect Ligand (Promega). Twenty-four hours after electroporation, 50 μL of the dispensed supernatant and 100 μL of the exchanged fresh medium with cells were frozen. The cytotoxicity of 50 μL of the supernatant with and without cells after the freeze-thaw cycle was examined using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega), similar to the cytotoxicity assay section.
Statistical analysis of assay data
Significant differences between two groups were evaluated using Student’s t-test for the time course of cytotoxicity assay and VZV genome copy number assay or Welch’s t-test for the cytotoxicity assay with VZV glycoproteins (JMP 15 software, SAS Institute). All of the experiments were repeated at least three times. The time course of the cytotoxicity assay was performed in triplicate. The cytotoxicity assay with VZV glycoproteins was performed in quadruplicate. The values in the supernatant of the cells were adopted after background correction according to the supernatant without cells followed by baseline correction. LDH release ratios were determined against cell numbers (that is, maximum LDH release) using the supernatant of the freeze-thaw cells. The VZV genome copy number assay was performed in duplicate. The VZV genome copy numbers were adopted after correcting for cell nucleus numbers using RNase P copy numbers.
Results
Herpes zoster was significantly associated with an HS3ST4 SNP
PHN was significantly associated with the rs12596324 SNP of the HS3ST4 gene. 2 This result raised the possibility that HS3ST4 contributes to both the pathology of PHN and the reactivation of VZV in HZ. To clarify the relationship between a history of HZ and the rs12596324 SNP, we statistically analyzed this relationship. The demographic data of the patients and healthy control subjects, stratified by rs12596324 SNP genotype, are presented in Table 1 and the dataset used are presented in Supplementary Dataset S1. As shown in Table 2, HZ was significantly associated with the rs12596324 SNP in both the genotypic model (GG, GT, and TT) and dominant genetic model (GG vs. GT + TT) for the minor T allele (genotypic model: p = .00108; dominant model: p = .000243). In the recessive model, HZ was not associated with rs12596324 (p > .05). These results suggest that the rs12596324 SNP of the HS3ST4 gene is associated with a history of HZ. To examine the linearity of trends toward HZ that depend on the major G allele, we performed the Cochran–Armitage trend test. This test with a genotypic model revealed a positive correlation between the number of HZ patients and copy number of the G allele of the SNP (p = .0030). Thus, the proportions of HZ patients linearly increased as the copy number of the G allele of the SNP increased. Overall, the rs12596324 SNP of the HS3ST4 gene was associated with HZ, indicating the possibility that HS3ST4 contributes to the reactivation of VZV in HZ.
Table 1.
Demographic data of patients and healthy control subjects, stratified by rs12596324 genotype.
| Patients and control subjects | rs12596324 SNP genotype | |||
|---|---|---|---|---|
| GG | GT | TT | GT + TT | |
| HZ | ||||
| Number of patients (males/females) | 50 (29/19) | 34 (14/19) | 12 (4/7) | 46 (18/26) |
| Age (years) | 71.6 ± 10.1 | 69.9 ± 12.0 | 72.6 ± 7.0 | 70.6 ± 11.0 |
| Healthy subjects | ||||
| Number of patients (males/females) | 88 (55/33) | 150 (81/69) | 44 (22/22) | 194 (103/81) |
| Age (years) | 35.5 ± 13.8 | 32.9 ± 11.9 | 34.8 ± 12.4 | 33.3 ± 12.1 |
The data are expressed as numbers and the mean ± SD (range).
Table 2.
Association between HZ and the rs12596324 SNP of HS3ST4 (p value).
| Gene and SNP | Model | HZ vs healthy subjects |
|---|---|---|
| HS3ST4 rs12596324 | GG, GT, TT | 0.00108* |
| GG vs GT + TT | 0.000243** | |
| GG + GT vs TT | 0.460 |
*p < .01, **p < .001.
Virus-mediated cell fusion occurred robustly in cells with HS3ST4 infected with VZV
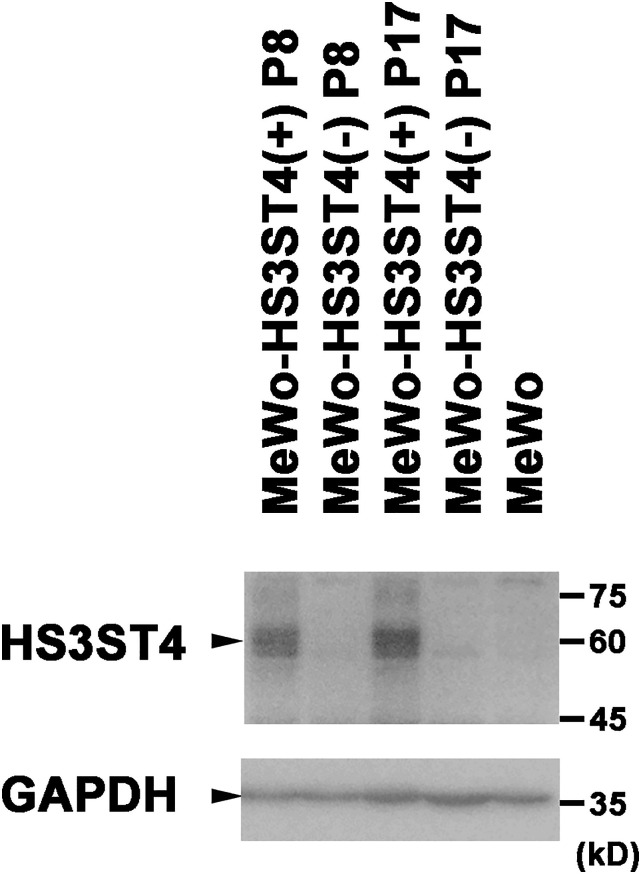
To clarify whether HS3ST4 contributes to VZV infection, we first produced cells that expressed HS3ST4. MeWo cells were introduced with a plasmid that encoded HS3ST4 cDNA (MeWo-HS3ST4 (+)) or a vector plasmid (MeWo-HS3ST4 (−)). To examine whether MeWo-HS3ST4 (+) cells stably express HS3ST4, the expression of HS3ST4 in MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells at passages eight and 17 were analyzed by Western blot (Figure 1). MeWo-HS3ST4 (+) cells showed a distinct band around 60 kD, detected by anti-human HS3ST4 antibodies, at passages eight and 17, whereas MeWo-HS3ST4 (−) cells and the original MeWo cells showed no band around 60 kD. These results suggested that MeWo-HS3ST4 (+) cells stably expressed HS3ST4 protein.
Figure 1.
HS3ST4 is expressed in MeWo-HS3ST4 (+) cells. HS3ST4 was analyzed in MeWo cells, MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells at passage eight (P8) or P17 by Western blot. Bands were detected around 60 kD by anti-HS3ST4 antibodies. GAPDH was detected as an internal control. Black triangles indicate the predicted positions of the antigens.
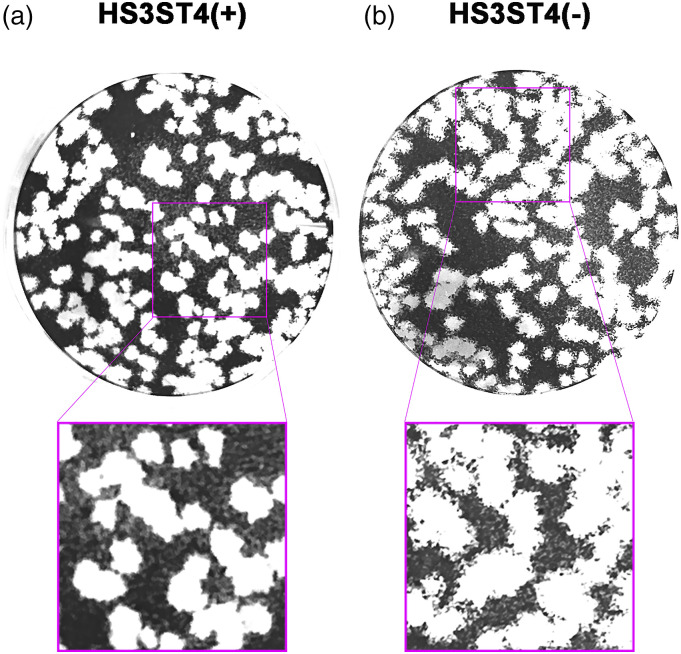
To determine whether HS3ST4 contributes to VZV infection, we performed a plaque assay and observed plaque phenotypes of cells with and without HS3ST4 infected with VZV. MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells that were infected with VZV in 12-well plates were fixed 6 days after infection. As shown in Figure 2, plaques on MeWo-HS3ST4 (+) cells had distinct borders (Figure 2(a)), whereas plaques on MeWo-HS3ST4 (−) cells had unclear borders (Figure 2(b)). These results suggested that infected MeWo-HS3ST4 (+) cells peeled off with engulfed surrounding cells.
Figure 2.
Plaque phenotype of MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells infected with VZV. MeWo-HS3ST4 (+) (a) and MeWo-HS3ST4 (−) cells (b) that were infected with VZV in 12-well plates were fixed 6 days after the infection. The images below are twice-enlarged images of pink rectangles in the upper images.
To determine why plaques of infected MeWo-HS3ST4 (+) cells had distinct borders, we observed cells with and without HS3ST4 after infection under an inverted microscope 2 days after infection (Figure 3). MeWo-HS3ST4 (+) cells that were infected with VZV had larger cell fusions (Figure 3(a)) compared with MeWo-HS3ST4 (−) cells that were infected with VZV (Figure 3(b)). Mock-infected cells had no cell fusion (Figures 3(c) and (d)). These results suggested that HS3ST4 accelerated cell fusion after VZV infection.
Higher cytotoxicity in cells with HS3ST4 after infection
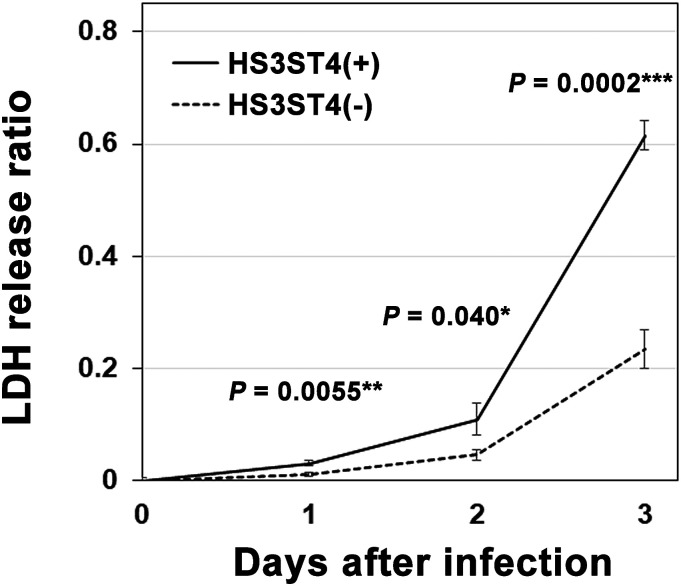
To quantitatively determine the time course of cell fusion after infection, cells with and without HS3ST4 were infected or mock-infected with VZV, and supernatants were collected on each day after infection and analyzed using the cytotoxicity assay (Supplementary Dataset S2). As shown in Figure 4, infected MeWo-HS3ST4 (+) cells had a higher LDH release ratio, that is, cytotoxicity, 1–3 days after infection compared with infected MeWo-HS3ST4 (−) cells (p = .0055 for day 1, p = .040 for day 2, p = .0002 for day 3). On day 4, the cells were almost overwhelmed by cell fusion and cytopathic effect. Therefore, we did not analyze the cytotoxicity for day 4. These results suggested that the cytotoxicity including fusogenic activity was higher in MeWo-HS3ST4 (+) cells than in MeWo-HS3ST4 (−) cells from 1 to 3 days after infection under the present experimental conditions.
Figure 4.
Higher cytotoxicity in cells with HS3ST4 after infection with VZV. MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells were infected or mock-infected with VZV, and supernatants were collected on each day after infection and analyzed by the cytotoxicity assay. *p < .05, **p < .01, ***p < .001.
Expression of VZV glycoproteins increased the cytotoxicity in cells with HS3ST4
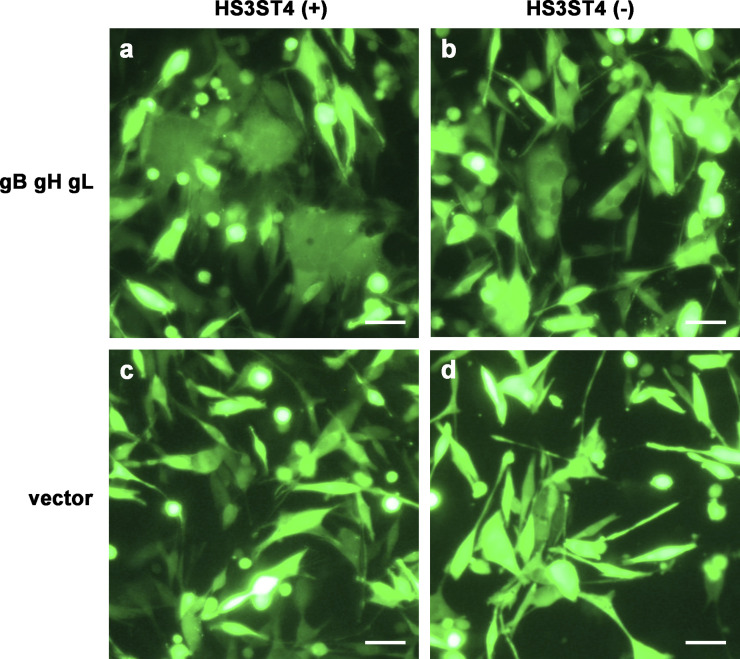
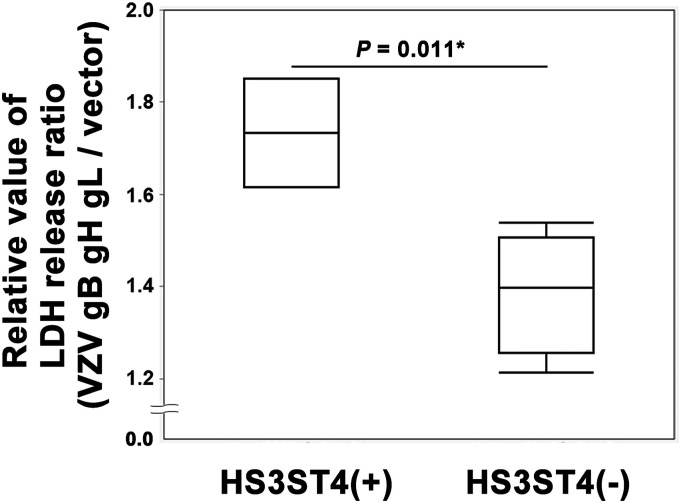
Glycoproteins gB, gH, and gL of VZV were reported to contribute to cell fusion. 6 To determine whether these glycoproteins play essential roles in cell fusion after infection in MeWo-HS3ST4 (+) cells, glycoprotein-encoding or vector plasmids were expressed in cells with and without HS3ST4. Twenty-four hours after electroporation, the cells were observed under a fluorescent microscope (Figure 5). MeWo-HS3ST4 (+) cells with gB, gH, and gL showed large fused cells (Figure 5(a)), whereas MeWo-HS3ST4 (−) cells with gB, gH, and gL showed smaller fused cells (Figure 5(b)). MeWo-HS3ST4 (+) nor MeWo-HS3ST4 (−) cells with vector plasmid showed no fused cells (Figure 5(c) and (d)). These results implied that the expression of HS3ST4 with the glycoproteins accelerated cell fusion. In parallel with fluorescent microscopy, the supernatants were collected and quantitatively analyzed using the cytotoxicity assay (Supplementary Dataset S3). As shown in Figure 6, glycoprotein expression significantly increased a relative value of the LDH release ratio (LDH release ratio in cells that expressed glycoprotein-encoding plasmid per LDH release ratio in cells that expressed the vector plasmid) in MeWo-HS3ST4 (+) cells compared with MeWo-HS3ST4 (−) cells (p = .011). These results suggested that these glycoproteins induced more severe cytotoxicity, including fusogenic activity, in MeWo-HS3ST4 (+) cells compared with MeWo-HS3ST4 (−) cells.
Figure 5.
Expression of HS3ST4 along with VZV glycoproteins accelerated cell fusion. Twenty-four hours after electroporation, the cells were observed under a fluorescent microscope. (a–d) Fluorescent images of MeWo-HS3ST4 (+) cells with gB, gH, and gL (a), MeWo-HS3ST4 (−) cells with gB, gH, and gL (b), MeWo-HS3ST4 (+) cells with the vector plasmid (c), and MeWo-HS3ST4 (−) cells with the vector plasmid (d). Scale bar = 100 μm.
Figure 6.
Expression of VZV glycoproteins increased cytotoxicity in MeWo-HS3ST4 (+) cells. The glycoproteins gB, gH, and gL of VZV and vehicle were expressed in MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells by electroporation. Twenty-four hours after electroporation, supernatants were collected and analyzed using the cytotoxicity assay. *p < .05.
VZV genome replication had similar efficiency in the cells with HS3ST4
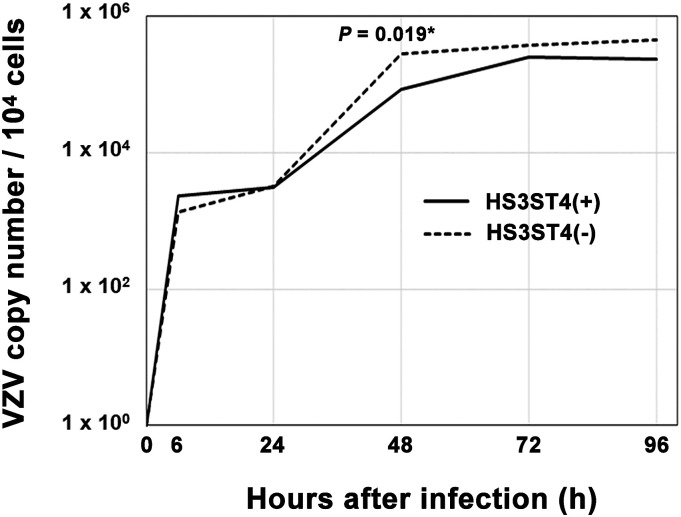
To investigate the way in which HS3ST4 contributes to viral genome replication, we analyzed copy numbers of the viral genome chronologically. Cells with and without HS3ST4 infected with VZV were collected at the indicated time points. Copy numbers of the VZV genome were compared with 0 hour, based on quantitative real-time PCR (Figure 7, Supplementary Dataset S4). From 0 to 96 h after infection, no significant difference in copy numbers of the VZV genome was found between MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells (p > .05), with the exception of 48 h after infection (p = .019). Although the difference was statistically significant at 48 h, it was only around three times lower in MeWo-HS3ST4 (+) cells compared with MeWo-HS3ST4 (−) cells. These results suggested that the expression of HS3ST4 had little effect on viral genome replication.
Figure 7.
VZV genome replication had similar efficiency in cells, regardless of HS3ST4 expression. MeWo-HS3ST4 (+) and MeWo-HS3ST4 (−) cells that were infected with VZV were collected at the indicated time points. Copy numbers of the VZV genome were analyzed by quantitative real-time PCR. *p < .05.
Discussion
In addition to significant associations that were found between the rs12596324 SNP of HS3ST4 and PHN in our previous study, 2 this SNP was also significantly associated with a history of HZ (Table 2). This indicates that the HS3ST4 SNP is associated with the pathogenesis of VZV reactivation in HZ as well as PHN. Based on these associations, we investigated the contribution of HS3ST4 to virus-induced cytopathology and viral replication using human MeWo cells. We found that HS3ST4 significantly augmented fusogenic activity that was accelerated by VZV glycoproteins, although the expression of HS3ST4 had little effect on viral genome replication.
Introns occasionally function to regulate transcription. 13 Although it remains unclear whether the G allele on intronic rs12596324 SNP of the HS3ST4 gene is positively correlated with increased HS3ST4 expression in humans, our main idea is that the HS3ST4 expression is possibly associated to pathogenesis of VZV. We demonstrated that expression of HS3ST4 augmented fusogenic activity induced by VZV infection. From these data, it can be easily speculated that in individuals who exhibit more robust HS3ST4 expression, reactivated VZV may more easily trigger disease pathogenesis. However, further studies are needed to prove the function of this SNP including the correlation among the genotypes of the SNP, the expression levels of HS3ST4, and the intensity of cell fusion in vivo.
VZV-infected MeWo cells fuse with the human neuronal somata. 4 Similarly, VZV infection was reported to provoke neural cell fusion in vivo. 5 Furthermore, we observed significant fusogenic activity even in cultured human neural cells after VZV infection (data not shown). Thus, neuronal cell fusion after VZV infection can be observed in humans. VZV latently resides in the human trigeminal ganglia (TG) and dorsal root ganglia (DRG) after primary infection. 14 In the TG, HS3ST4 is a major isoform that is expressed in sensory neurons and adjacent satellite cells. 15 HS3ST4 is expressed above median expression levels in the TG and DRG. 16 Consequently, HS3ST4 expression could accelerate virus-induced fusogenic activity in the TG and DRG, although we did not investigate whether HS3ST4 expression accelerates fusogenic activity in neural cells. Further studies are needed to investigate this possibility.
HS3ST4 has minimal effects on VZV replication and markedly enhances fusogenic activity in the presence of VZV glycoproteins. Hyperfusogenic measles virus and other paramyxovirus mutants induced strong cytopathology through cell fusion in infected cells, despite the suppression of viral replication. 11 Accordingly, it is reasonable that HS3ST4 accelerates cytopathology through virus-induced fusogenic activity despite almost no changes in the viral replication rate.
In Figure 6, MeWo-HS3ST4 (−) cells had a relative value > 1, indicating that the glycoproteins induced cytotoxicity even in MeWo-HS3ST4 (−) cells, consistent with Figure 5(b) compared with Figure 5(d). MeWo-HS3ST4 (+) cells had more robust cytotoxicity after expressing these glycoproteins, consistent with Figure 5(a) compared with Figure 5(c). These results suggested that these glycoproteins induced cytotoxicity, including fusogenic activity, in MeWo-HS3ST4 (+) cells. This implies that the glycoproteins gB, gH, and gL are sufficient to accelerate cell fusion through the HS3ST4-related pathway. HS that is modified by HS3ST4 has been shown to assist herpes simplex virus (HSV)-1 entry in Chinese hamster ovarian (CHO-K1) cells, which are not naturally susceptible to HSV-1.15,17 Since HSV-1 is closely related to VZV, HS3ST4 likely plays similar essential roles in VZV entry. In the present study, we assayed cell fusion using human cells that are naturally susceptible to VZV. The assumed receptors for VZV glycoproteins on the cell surface would be modified by HS3ST4 solely in cells with HS3ST4. This modification of HS in receptors on the cell surface would promote cell-cell attachment, followed by cell fusion.
Genes that encode the glycoproteins gB, gH, and gL are classified into late genes for VZV. 18 In the human TG, these glycoproteins are expressed only at low levels, if at all, during latent infection. 19 After robust reactivation of the virus, these glycoproteins are sufficiently expressed on the cell surface to enhance fusogenic activity, possibly in cells with higher HS3ST4 expression.
Supplemental Material
Supplemental Material, sj-xlsx-1-mpx-10.1177_17448069211052171 for Heparan sulfate 3-O-sulfotransferase 4 is genetically associated with herpes zoster and enhances varicella-zoster virus–mediated fusogenic activity by Seii Ohka,Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo and Kazutaka Ikeda in Molecular Pain
Supplemental Material, sj-xlsx-2-mpx-10.1177_17448069211052171 for Heparan sulfate 3-O-sulfotransferase 4 is genetically associated with herpes zoster and enhances varicella-zoster virus–mediated fusogenic activity by Seii Ohka,Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo and Kazutaka Ikeda in Molecular Pain
Supplemental Material, sj-xlsx-3-mpx-10.1177_17448069211052171 for Heparan sulfate 3-O-sulfotransferase 4 is genetically associated with herpes zoster and enhances varicella-zoster virus–mediated fusogenic activity by Seii Ohka,Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo and Kazutaka Ikeda in Molecular Pain
Supplemental Material, sj-xlsx-4-mpx-10.1177_17448069211052171 for Heparan sulfate 3-O-sulfotransferase 4 is genetically associated with herpes zoster and enhances varicella-zoster virus–mediated fusogenic activity by Seii Ohka,Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo and Kazutaka Ikeda in Molecular Pain
Acknowledgments
We thank Mr Michael Arends for editing the manuscript. We are grateful to the volunteers for their participation in the study and anesthesiologists and surgeons for collecting the clinical data.
Author contributions: Conceptualization: Seii Ohka, Kazutaka Ikeda.Data curation: Seii Ohka, Daisuke Nishizawa, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Kazutaka Ikeda. Formal analysis: Seii Ohka. Funding acquisition: Seii Ohka, Daisuke Nishizawa, Masayuki Saijo, Kazutaka Ikeda. Investigation: Seii Ohka, Kazutaka Ikeda. Methodology: Seii Ohka, Souichi Yamada, Daisuke Nishizawa, Shuetsu Fukushi, Masayuki Saijo, Kazutaka Ikeda. Project administration: Seii Ohka, Kazutaka Ikeda. Resources: Seii Ohka, Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo, Kazutaka Ikeda. Supervision: Shuetsu Fukushi, Masayuki Saijo, Kazutaka Ikeda. Validation: Seii Ohka. Visualization: Seii Ohka. Writing-original draft preparation: Seii Ohka. Writing-review & editing: Seii Ohka, Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo, Kazutaka Ikeda.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI [JP16H06276 (AdAMS) KI, 20K07774 SO, 17K09052 SO, 20K09259 DN, 17K08970 DN, 17H04324 KI, 18K07894 MS]; and Japan Agency for Medical Research and Development (AMED) [JP19ek0610011 KI].
Supplemental Material: Supplemental material for this article is available online.
ORCID iDs
Seii Ohka https://orcid.org/0000-0003-2394-6440
Hideko Arita https://orcid.org/0000-0002-4151-5962
Kazutaka Ikeda https://orcid.org/0000-0001-8342-0278
References
- 1.Oliver SL, Yang E, Arvin AM. Varicella-zoster virus glycoproteins: Entry, replication, and pathogenesis. Curr Clin Microbiol Rep 2016; 3: 204–215. doi: 10.1007/s40588-016-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishizawa D, Iseki M, Arita H, et al. Genome-wide association study identifies candidate loci associated with chronic pain and postherpetic neuralgia. Mol Pain 2021; 17: 1744806921999924. doi: 10.1177/1744806921999924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett GJ, Watson CP. Herpes zoster and postherpetic neuralgia: past, present and future. Pain Res Manag 2009; 14: 275–282. doi: 10.1155/2009/380384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grigoryan S, Yee MB, Glick Y, et al. Direct transfer of viral and cellular proteins from varicella-zoster virus-infected non-neuronal cells to human axons. PLoS One 2015; 10: e0126081. doi: 10.1371/journal.pone.0126081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reichelt M, Zerboni L, Arvin AM. Mechanisms of varicella-zoster virus neuropathogenesis in human dorsal root ganglia. J Virol 2008; 82: 3971–3983. doi: 10.1128/JVI.02592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vleck SE, Oliver SL, Brady JJ, et al. Structure-function analysis of varicella-zoster virus glycoprotein H identifies domain-specific roles for fusion and skin tropism. Proc Natl Acad Sci U S A 2011; 108: 18412–18417. doi: 10.1073/pnas.1111333108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denys A, Allain F. The emerging roles of heparan sulfate 3-O-sulfotransferases in cancer. Front Oncol 2019; 9: 507. doi: 10.3389/fonc.2019.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishizawa D, Fukuda K, Kasai S, et al. Genome-wide association study identifies a potent locus associated with human opioid sensitivity. Mol Psychiatry 2014; 19: 55–62. doi: 10.1038/mp.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishizawa D, Fukuda K, Kasai S, et al. Association between KCNJ6 (GIRK2) gene polymorphism rs2835859 and post-operative analgesia, pain sensitivity, and nicotine dependence. J Pharmacol Sci 2014; 126: 253–263. doi: 10.1254/jphs.14189fp. [DOI] [PubMed] [Google Scholar]
- 10.Shworak NW, Liu J, Petros LM, et al. Multiple isoforms of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. Isolation, characterization, and expression of human cDNAs and identification of distinct genomic loci. J Biol Chem 1999; 274: 5170–5184. doi: 10.1074/jbc.274.8.5170. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe S, Ohno S, Shirogane Y, et al. Measles virus mutants possessing the fusion protein with enhanced fusion activity spread effectively in neuronal cells, but not in other cells, without causing strong cytopathology. J Virol 2015; 89: 2710–2717. doi: 10.1128/JVI.03346-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, Seitz S, Pointon T, et al. Varicella zoster virus infection of highly pure terminally differentiated human neurons. J Neurovirol 2013; 19: 75–81. doi: 10.1007/s13365-012-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chorev M, Carmel L. The function of introns. Front Genet 2012; 3: 55. doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy PG, Grinfeld E, Gow JW. Latent varicella-zoster virus is located predominantly in neurons in human trigeminal ganglia. Proc Natl Acad Sci U S A 1998; 95: 4658–4662. doi: 10.1073/pnas.95.8.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence R, Yabe T, Hajmohammadi S, et al. The principal neuronal gD-type 3-O-sulfotransferases and their products in central and peripheral nervous system tissues. Matrix Biol 2007; 26: 442–455. doi: 10.1016/j.matbio.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.BaseSpace Correlation Engine. Illumina Inc. [Google Scholar]
- 17.Tiwari V, O'Donnell CD, Oh MJ, et al. A role for 3-O-sulfotransferase isoform-4 in assisting HSV-1 entry and spread. Biochem Biophys Res Commun 2005; 338: 930–937. doi: 10.1016/j.bbrc.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 18.Cohen JI. Genomic Structure and Organization of Varicella-Zoster Virus. Karger, 1999. [DOI] [PubMed] [Google Scholar]
- 19.Depledge DP, Sadaoka T, Ouwendijk WJD. Molecular aspects of Varicella-Zoster virus latency. Viruses 2018; 10: 349. doi: 10.3390/v10070349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-xlsx-1-mpx-10.1177_17448069211052171 for Heparan sulfate 3-O-sulfotransferase 4 is genetically associated with herpes zoster and enhances varicella-zoster virus–mediated fusogenic activity by Seii Ohka,Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo and Kazutaka Ikeda in Molecular Pain
Supplemental Material, sj-xlsx-2-mpx-10.1177_17448069211052171 for Heparan sulfate 3-O-sulfotransferase 4 is genetically associated with herpes zoster and enhances varicella-zoster virus–mediated fusogenic activity by Seii Ohka,Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo and Kazutaka Ikeda in Molecular Pain
Supplemental Material, sj-xlsx-3-mpx-10.1177_17448069211052171 for Heparan sulfate 3-O-sulfotransferase 4 is genetically associated with herpes zoster and enhances varicella-zoster virus–mediated fusogenic activity by Seii Ohka,Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo and Kazutaka Ikeda in Molecular Pain
Supplemental Material, sj-xlsx-4-mpx-10.1177_17448069211052171 for Heparan sulfate 3-O-sulfotransferase 4 is genetically associated with herpes zoster and enhances varicella-zoster virus–mediated fusogenic activity by Seii Ohka,Souichi Yamada, Daisuke Nishizawa, Yoshiko Fukui, Hideko Arita, Kazuo Hanaoka, Masako Iseki, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida, Shuetsu Fukushi, Masayuki Saijo and Kazutaka Ikeda in Molecular Pain