Abstract
The c-fes locus encodes a 93-kDa non-receptor protein tyrosine kinase (Fes) that regulates the growth and differentiation of hematopoietic and vascular endothelial cells. Unique to Fes is a long N-terminal sequence with two regions of strong homology to coiled-coil oligomerization domains. We introduced leucine-to-proline substitutions into the coiled coils that were predicted to disrupt the coiled-coil structure. The resulting mutant proteins, together with wild-type Fes, were fused to green fluorescent protein and expressed in Rat-2 fibroblasts. We observed that a point mutation in the first coiled-coil domain (L145P) dramatically increased Fes tyrosine kinase and transforming activities in this cell type. In contrast, a similar point mutation in the second coiled-coil motif (L334P) was without effect. However, combining the L334P and L145P mutations reduced transforming and kinase activities by approximately 50% relative to the levels of activity produced with the L145P mutation alone. To study the effects of the coiled-coil mutations in a biologically relevant context, we expressed the mutant proteins in the granulocyte-macrophage colony-stimulating factor (GM-CSF)-dependent myeloid leukemia cell line TF-1. In this cellular context, the L145P mutation induced GM-CSF independence, cell attachment, and spreading. These effects correlated with a marked increase in L145P protein autophosphorylation relative to that of wild-type Fes. In contrast, the double coiled-coil mutant protein showed greatly reduced kinase and biological activities in TF-1 cells. These data are consistent with a role for the first coiled coil in the negative regulation of kinase activity and a requirement for the second coiled coil in either oligomerization or recruitment of signaling partners. Gel filtration experiments showed that the unique N-terminal region interconverts between monomeric and oligomeric forms. Single point mutations favored oligomerization, while the double point mutant protein eluted essentially as the monomer. These data provide new evidence for coiled-coil-mediated regulation of c-Fes tyrosine kinase activity and signaling, a mechanism unique among tyrosine kinases.
The human c-fes proto-oncogene encodes a 93-kDa non-receptor protein tyrosine kinase (Fes) with strong expression in myeloid hematopoietic cells. Fes has been implicated in signal transduction pathways for several hematopoietic cytokines, including interleukin-3 (IL-3), IL-4, IL-6, granulocyte-macrophage colony-stimulating factor (GM-CSF), and erythropoietin (reviewed in reference 37). Other evidence suggests that Fes may contribute to differentiation signaling from cytokine receptors. For example, expression of Fes in the human myeloid leukemia cell line K-562 results in growth suppression and terminal differentiation (4, 40). More recent work suggests that interaction with Bcr-Abl may be responsible for Fes activation in this chronic myelogenous leukemia (CML)-derived cell line, implicating Fes as a suppressor of CML progression as well (19). On the other hand, inhibition of Fes expression with antisense oligonucleotides blocks differentiation of HL-60 cells and triggers an apoptotic response instead (24). These results implicate Fes in both differentiation and survival signaling pathways in myeloid leukocytes.
The c-Fes protein consists of three distinct structural domains: a long unique N-terminal region, a central SH2 domain, and a C-terminal kinase domain (37). Fes lacks an SH3 domain, a negative regulatory tail tyrosine residue, and other structural features that contribute to the negative regulation of Src, Abl, and other non-receptor protein tyrosine kinases. However, like Src and Abl, Fes tyrosine kinase activity is tightly regulated in mammalian cells and induces little or no transforming activity upon ectopic expression in rodent fibroblasts (6, 8). The mechanism responsible for negative regulation of Fes tyrosine kinase activity in vivo is not clear.
One unique aspect of c-Fes structure is the presence of coiled-coil homology domains in the unique N-terminal region (4, 33). Coiled coils are composed of amphipathic α-helices that exhibit a heptad repeat pattern in which the first and fourth positions are occupied by hydrophobic amino acids (21). These residues align to form a hydrophobic interface between the supercoiled strands. Previous work from our laboratory has shown that the N-terminal region of Fes mediates its oligomerization in vitro, suggesting that interconversion of monomeric and oligomeric forms of the protein may regulate kinase activity in vivo. Consistent with this idea is our finding that overexpression of the isolated Fes N-terminal region can suppress the kinase and transforming activities of a full-length Fes mutant protein that is activated by membrane targeting. The suppressive effect may involve formation of an inactive heteromer comprised of full-length and truncated strands (4, 33).
In this study, we provide evidence that the Fes coiled-coil homology domains are required for both negative regulation and full activation of kinase function. We observed that a single point mutation of a key leucine residue in the first coiled-coil (CC1) domain results in strong kinase activation in vivo, leading to potent transforming activity in fibroblasts as well as survival and differentiation signaling in myeloid cells. Interestingly, when the activating mutation in CC1 is combined with a similar point mutation in the second coiled coil (CC2), tyrosine kinase and biological activities are significantly impaired, suggesting that CC2 may be required for mediating Fes oligomerization or downstream signaling in vivo. Gel filtration experiments show that the unique N-terminal region can adopt monomeric and oligomeric forms, suggesting that the coiled coils regulate kinase activity by controlling the oligomerization state of the protein. Use of coiled-coil domains is unique among non-receptor protein tyrosine kinases as a regulatory mechanism.
MATERIALS AND METHODS
Construction of Fes coiled-coil point mutant proteins.
Residues essential for coiled-coil structure were determined using the COILS algorithm (23) (see Fig. 1) and include Leu 145 in the CC1 domain and Leu 334 in the CC2 domain. These leucine residues were converted to proline using an Altered Sites mutagenesis kit according to the instructions of the manufacturer (Promega). Both individual-mutation (L145P, L334P) and double-mutation (2LP) proteins were created. The presence of each mutation was confirmed by automated DNA sequence analysis.
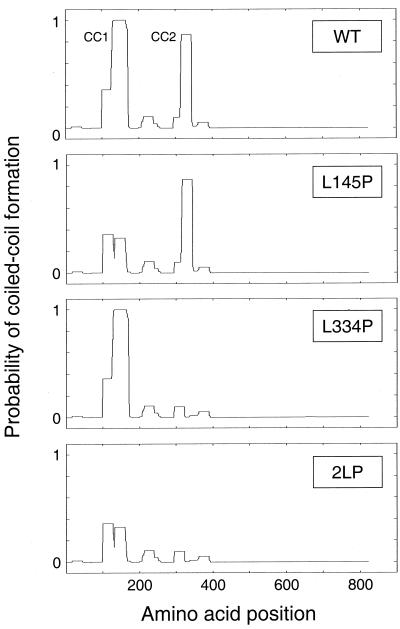
FIG. 1.
Identification of Fes N-terminal residues essential for coiled-coil formation. Wild-type and mutant forms of the 822-amino-acid Fes sequence were analyzed using the COILS algorithm. The probability of each residue contributing to coiled-coil formation is plotted as a function of its position within the overall sequence. The mutant proteins include one with a proline substitution for Leu 145 in the CC1 domain (L145P) protein, one with a proline substitution for Leu 334 in the CC2 domain (L334P protein), and a double mutant protein with both substitutions (2LP). For this analysis, a 28-residue sliding window was used along with the MTIDK matrix. For additional details, see http://www.ch.embnet.org/software/COILS_form.html. WT, wild type.
Construction of GFP-Fes fusion proteins.
To create green fluorescent protein (GFP)-Fes expression constructs, the enhanced GFP coding sequence was amplified from the vector pEGFP-C1 (Clontech) by PCR and subcloned into the retroviral expression vector pSRαMSVtkneo (27) to create plasmid pSRα-GFP. The cDNA clones encoding wild-type Fes, the coiled-coil domain point mutant proteins, and the other forms of Fes used in this study were subcloned downstream and in frame with GFP in this vector.
Production of recombinant retroviruses.
Recombinant retroviruses were produced by cotransfecting 293T cells with the pSRα-GFP–Fes constructs described above and either ecotropic or amphotropic packaging vectors as described elsewhere (2, 4, 18). A GFP retrovirus was prepared from the pSRα-GFP parent vector for use as a negative control. Retroviral supernatants were collected every 12 h for 3 days, and the pooled supernatants were aliquoted and stored at −80°C.
Retroviral infection of Rat-2 fibroblasts and focus-forming assay.
Rat-2 cells were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum. Cells were plated in six-well plates (2 × 105 cells per well) 1 day prior to infection. Retroviral stocks (4 ml) were thawed on ice, supplemented with Polybrene to 4 μg/ml, and added to the cells. The plates were centrifuged at 1,500 × g for 3 h at room temperature to enhance the efficiency of infection. The virus was replaced with fresh medium, and the infected cells were incubated for 48 h at 37°C prior to being plated in transformation assays.
For focus-forming assays, infected cells (2 × 104) were plated in 60-mm-diameter dishes in the presence of 800 μg of G418 per ml. The culture medium was changed every second or third day for 14 days. Transfomed foci were stained with Wright-Giemsa stain and counted from a scanned image using a Bio-Rad GS-710 calibrated imaging densitometer and Quantity One software.
Expression of Fes coiled-coil mutant proteins in TF-1 myeloid leukemia cells.
The human myeloid leukemia cell line TF-1 (17) was obtained from the American Type Culture Collection and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 50 μg of gentamicin per ml, and 1 ng of GM-CSF (BioSource International) per ml. Recombinant retroviruses for TF-1 cell infection were prepared using the 293T cell protocol described above, except that an amphotropic packaging plasmid was used. For TF-1 cell infection, 2 × 105 cells were plated in each well of a six-well plate and centrifuged at 500 × g for 15 min. The medium was aspirated and replaced with the retroviral supernatant plus Polybrene, and the infection protocol was continued as described above for the Rat-2 cells. Following infection, the cells were placed under G418 selection for 10 to 14 days. Expression of GFP or the GFP-Fes fusion proteins was confirmed by fluorescence microscopy, under which the drug-resistant cell population exhibited uniform green fluorescence. Expression of full-length GFP-Fes fusion proteins was also confirmed by immunoblotting (see below).
To monitor GM-CSF dependence, 2 × 105 cells were washed free of GM-CSF and plated in duplicate six-well plates. GM-CSF (1 ng/ml) was added to one plate, and viable cells were counted following trypan blue staining every 2 days until saturation density was reached.
For attachment experiments, 5 × 104 cells were plated in duplicate 24-well plates in a total volume of 1 ml in the presence or absence of GM-CSF. Cells were returned to the incubator for 5 to 7 days and fixed in situ with cold acetone-methanol (1:1) for 5 min prior to fluorescence microscopy to confirm c-Fes expression. Alternatively, cells were directly stained with Giemsa stain and visualized by light microscopy. The number of attached cells in three randomly chosen fields was counted, and the results were averaged. Data are presented as percentages of attached cells observed relative to the number of attached cells of the GFP–Fes L145P mutant protein, which gave the strongest response.
Analysis of Fes protein expression and autophosphorylation.
Aliquots of Rat-2 or TF-1 cells were pelleted, washed with phosphate-buffered saline, and lysed in 1 ml of radioimmunoprecipitation assay (RIPA), buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA, and 1% sodium deoxycholate). Fes proteins were immunoprecipitated from clarified cell lysates using either the M2 anti-FLAG monoclonal antibody resin (Sigma) or an anti-GFP antibody (Clontech) and protein G-Sepharose (Pharmacia). Immunoprecipitates were washed with RIPA buffer, resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and immunoblotted for Fes expression with the anti-FLAG antibody or for phosphotyrosine content with the antiphosphotyrosine monoclonal antibody PY99 (Santa Cruz).
Gel filtration analysis of the Fes N-terminal region.
The coding sequence of the Fes N-terminal region encompassing both coiled-coil motifs (Ala 91 to Gly 392; CC1+2) was amplified from wild-type and coiled-coil mutant Fes templates by PCR and subcloned into the baculovirus transfer vector pVL-GFP. The pVL-GFP vector was constructed by PCR amplification of the enhanced GFP coding sequence from pEGFP-C1 (Clontech) and subcloning downstream of the polyhedrin promoter. The resulting vectors were used to produce recombinant baculoviruses using Baculogold DNA (PharMingen) and the manufacturer's protocol. For gel filtration experiments, the GFP-Fes CC1+2 fusion proteins were expressed in Sf-9 insect cells and lysates were prepared by sonication in 1.0 ml of S-300 buffer (50 mM Tris-HCl [pH 7.4], 50 mM NaCl, 1 mM EDTA). Lysates were clarified by centrifugation at 100,000 × g for 1 h and applied to a Sephacryl S-300 column (1.6 by 77 cm) previously equilibrated with S-300 buffer. Fractions (0.25 ml) were collected directly in 96-well microtiter plates, and the elution profile of each GFP-CC1+2 fusion protein was determined by fluorescence detection using a SpectraMax Gemini XS fluorimetric microplate reader (Molecular Devices, Inc.). As an additional control, unfused GFP was run alone and found to be monomeric. Peak fractions were pooled and concentrated, and the presence of the GFP-CC1+2 fusion protein was verified by immunoblotting with anti-GFP antibodies (Clontech).
RESULTS
Identification of N-terminal residues critical for coiled-coil structure.
Coiled-coil domains are characterized by a heptad repeat of amino acids often denoted a-b-c-d-e-f-g, with hydrophobic residues at positions a and d (21). The hydrophobic residues pack against each other to form the supercoil interface, while polar residues generally occupy the other positions and interact with solvent. Analysis of the Fes sequence with the COILS algorithm (22) predicts two regions with a high probability of coiled-coil structure formation within the Fes N-terminal region (33) (Fig. 1). Using COILS, we identified Leu residues that occupy a central d position within each predicted coiled coil and appear to be critical for the integrity of the coiled-coil structure. As shown in Fig. 1, replacement of Leu 145 with Pro in the first coiled-coil motif reduced the probability of coiled-coil formation from nearly 100% to less than 40%. A similar Pro substitution for Leu 334 within the second coiled-coil region nearly abolished the predicted coiled-coil formation. These substitutions were introduced into the full-length c-Fes coding region either alone or in combination. The proteins with the resulting coiled-coil mutations (L145P, L334P, and 2LP) were fused to GFP in a retroviral vector for expression in fibroblasts and human myeloid progenitor cells. The structures of these Fes proteins and the other constructs used in this study are shown in Fig. 2.
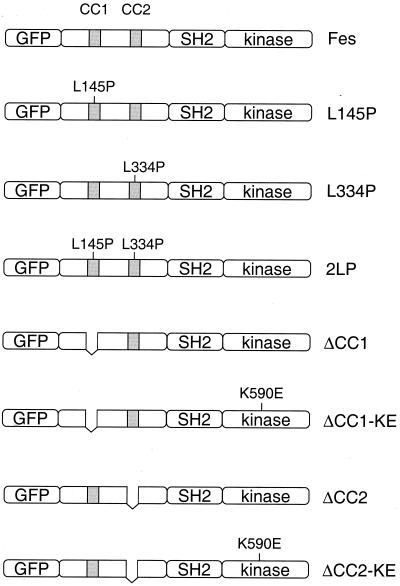
FIG. 2.
GFP-Fes constructs used in this study. The structure of the wild-type GFP-Fes fusion protein is shown at the top and includes a unique N-terminal region, an SH2 domain, and a C-terminal kinase domain. The locations of the two coiled-coil homology regions are indicated as the shaded boxes (CC1 and CC2). Mutant proteins include one with a proline substitution for Leu 145 in the first coiled-coil domain (L145P), one with a proline substitution for Leu 334 in the CC2 domain (L334P), and the corresponding double mutant protein with both substitutions (2LP). We also constructed GFP fusion proteins of the ΔCC1 and ΔCC2 deletion mutant proteins and the corresponding kinase-inactive forms of these proteins (ΔCC1-KE, ΔCC2-KE). All constructs have a C-terminal FLAG epitope tag (not shown).
Proline substitution for d-position leucine residues in the Fes coiled-coil domains have opposing actions on tyrosine kinase activity in Rat-2 fibroblasts.
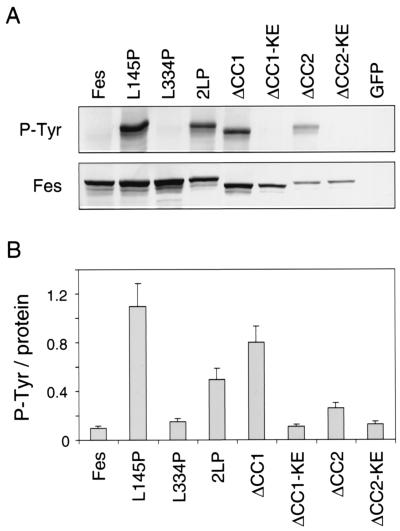
Previous studies have shown that wild-type c-Fes is nontransforming and that its tyrosine kinase activity is highly repressed following expression in Rat-2 fibroblasts, providing a convenient system to study the contribution of Fes structural features to the negative regulation of kinase activity (4, 8). Using recombinant retroviruses, the Fes coiled-coil domain point mutant proteins were expressed in Rat-2 fibroblasts as GFP fusion proteins. The GFP-Fes proteins were immunoprecipitated, and kinase activity was analyzed in terms of autophosphorylation by antiphosphotyrosine immunoblotting. As shown in Fig. 3, wild-type GFP-Fes tyrosine autophosphorylation was very low in this system, indicative of negative regulation as observed previously. However, proline substitution for Leu 145 within the CC1 domain enhanced autophosphorylation by more than 10-fold, indicating that the CC1 domain is required for negative regulation of kinase activity in vivo. Proline substitution for Leu 334 in the CC2 domain did not affect kinase activity relative to that of wild-type GFP-Fes. However, this same substitution reduced the kinase activity of the L145P mutant protein by more than 50%, suggesting that an intact CC2 domain is required for efficient autophosphorylation in vivo.
FIG. 3.
Autophosphorylation of GFP-Fes proteins in Rat-2 fibroblasts. Rat-2 fibroblasts stably expressing each of the GFP-Fes constructs shown in Fig. 2 were lysed, and GFP-Fes fusion proteins were precipitated with the M2 anti-FLAG monoclonal antibody. (A) Immunoprecipitates were washed with RIPA buffer, resolved by SDS-PAGE, and immunoblotted for Fes expression with the anti-FLAG antibody (Fes) or for phosphotyrosine content with the antiphosphotyrosine monoclonal antibody PY99 (P-Tyr). A representative blot is shown. (B) The experiment was repeated three times, and the relative levels of GFP-Fes protein and phosphotyrosine content were quantitated using an imaging densitometer. Ratios of tyrosine autophosphorylation to Fes protein were calculated and are presented as means ± standard deviations. KE, kinase-inactive form.
We next compared the activities of these point mutant proteins to those of coiled-coil deletion mutant proteins previously described by members of our laboratory (4). We observed that a GFP-Fes deletion mutant protein lacking the CC1 domain (GFP-ΔCC1) exhibited approximately 80% of the kinase activity of the GFP-L145P mutant protein (Fig. 3). Addition of a kinase-inactivating mutation abolished GFP-ΔCC1 protein autophosphorylation, indicating that CC1 deletion affects Fes kinase activity directly. Interestingly, deletion of the CC2 domain resulted in a slight increase in tyrosine phosphorylation, an effect that was also dependent upon the intact kinase domain. These results, as well as those obtained with the point mutant proteins, are consistent with a model in which CC1-CC2 interaction contributes to negative regulation by holding the kinase in an inactive conformation (see Discussion).
The Fes CC1 domain point mutant protein exhibits strong transforming activity in Rat-2 cells.
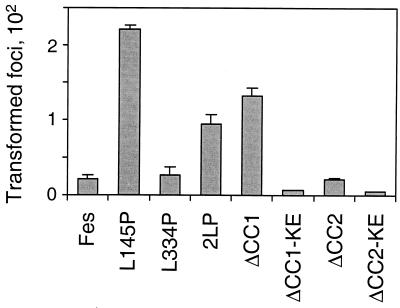
Results presented in the previous section show that disruption of the CC1 domain with a Leu-to-Pro substitution releases Fes kinase activity in vivo. To determine whether this mutant protein retains the capacity to generate a biological signal, we compared its transforming activity with those of wild-type Fes and the other coiled-coil domain mutant proteins. Rat-2 fibroblasts were infected with recombinant Fes retroviruses and placed under G418 selection for 2 weeks, at which time the foci were stained and counted. As shown in Fig. 4 and 5, the wild-type GFP-Fes fusion protein did not exhibit focus-forming activity, consistent with earlier studies of wild-type c-Fes (4, 8, 18). However, the L145P mutant protein exhibited very potent focus-forming activity, consistent with the effect of this mutation on Fes autophosphorylation. In contrast, proline substitution for Leu 334 in the CC2 domain did not release transforming activity. Combining the CC1 and CC2 mutations reduced focus-forming activity by more than 50% relative to that of the L145P single mutant protein. The focus-forming activities of the mutant proteins closely paralleled the relative levels of kinase activity (Fig. 3).
FIG. 4.
Focus-forming activity of GFP-Fes proteins in Rat-2 fibroblasts. Rat-2 fibroblasts were infected with recombinant GFP-Fes retroviruses as described in Materials and Methods. For focus-forming assays, infected cells were plated in 60-mm-diameter dishes and incubated in the presence of G418 for 14 days. Scanned images of the stained cultures are shown, which appear as confluent monolayers with transformed foci (where present) growing on top.
FIG. 5.
Quantitative comparison of GFP-Fes coiled-coil mutant protein transforming activities. Each of the GFP-Fes proteins illustrated in Fig. 2 was tested for focus-forming activity as described in Materials and Methods and the legend to Fig. 4. Foci were visualized by Wright-Giemsa staining and counted using a Bio-Rad model GS-710 scanning densitometer and colony-counting software. Foci from three independent cultures were counted; the bar graph shows average values ± standard deviations. KE, kinase-inactive form.
Comparable transformation results were obtained with GFP fusion proteins of the Fes CC1 and CC2 deletion mutant proteins. GFP-Fes ΔCC1 protein exhibited strong focus-forming activity, although the number of foci observed was about 50% of that observed with the L145P mutant protein (Fig. 5). Transforming activity was not observed with the kinase-defective GFP-ΔCC1 mutant protein, indicating that transformation is directly dependent upon an active c-Fes kinase domain and is not the result of endogenous kinase activation. Interestingly, deletion of the CC2 domain caused a slight increase in GFP-Fes transforming activity, consistent with the autophosphorylation result.
The Fes CC1 domain point mutant protein generates signals for survival and attachment in myeloid leukemia cells.
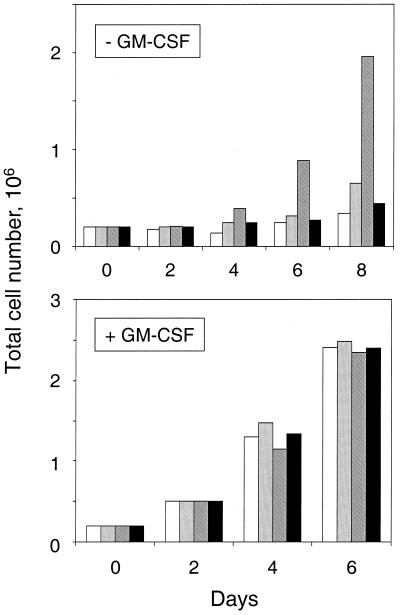
Previous studies have suggested that Fes may transduce signals for differentiation and survival in response to cytokines or other upstream signals (37). To determine whether the Fes CC1 domain point mutant protein can generate signals in a physiological context, we expressed this mutant protein in the myeloid leukemia cell line TF-1. This cell line requires GM-CSF for growth and differentiates into macrophage-like cells in response to phorbol esters (17). Wild-type GFP-Fes as well as each of the coiled-coil domain mutant proteins were introduced into TF-1 cells using recombinant retroviruses along with GFP alone as a negative control. Infected cell populations were selected in the presence of G418 and GM-CSF, and uniform expression of the transduced proteins in each of the infected cell populations was verified by fluorescence microscopy (data not shown). To determine whether expression of the Fes coiled-coil point mutant proteins affected the requirement of TF-1 cells for cytokine, levels of growth of the infected cell populations were compared in the presence and absence of GM-CSF. As shown in Fig. 6, no proliferation was observed in cells expressing wild-type GFP-Fes or GFP alone. However, cells expressing the Fes protein with the CC1 domain point mutation (L145P) grew rapidly in the absence of cytokine, providing direct evidence that Fes kinase activity is sufficient to generate a signal for survival and proliferation in myeloid cells. Interestingly, the protein with point mutations in both CC domains was unable to generate this response, suggesting that similar mechanisms regulate Fes activity in myeloid cells and upon ectopic expression in fibroblasts. All of the cell populations grew rapidly in the presence of GM-CSF. The protein with the CC2 domain point mutation (L334P) was unable to support cytokine-independent proliferation of TF-1 cells, consistent with the fibroblast result (data not shown).
FIG. 6.
Cytokine-independent survival and proliferation of TF-1 myeloid leukemia cells expressing the GFP-Fes CC1 domain point mutant protein. TF-1 cells were infected with recombinant retroviruses carrying GFP as a negative control (open bars), GFP-Fes (gray bars), GFP-Fes L145P protein (hatched bars), or GFP-Fes 2LP (solid bars). Following infection, the cells were placed under G418 selection in the presence of GM-CSF for 10 to 14 days, at which point each of the drug-resistant cell populations exhibited uniform green fluorescence (data not shown). To monitor GM-CSF dependence, cells were washed free of cytokine and replated in the absence (top panel) or presence (bottom panel) of GM-CSF and viable cells were counted following trypan blue staining every 2 days. Two independent experiments yielded comparable results.
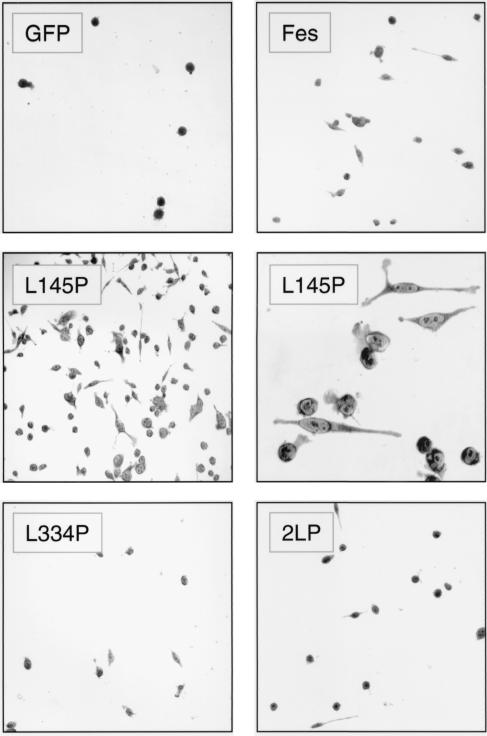
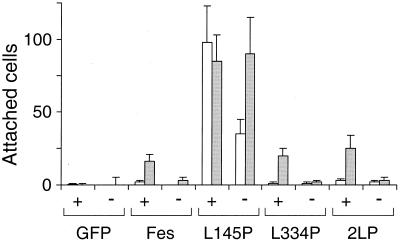
In addition to cytokine-independent survival, we observed that a significant subpopulation of TF-1 cells expressing the CC1 point mutant protein attached to the culture plate and spread to form macrophage-like cells (Fig. 7). These cells also expressed cell-surface CD13 and CD33, which are myeloid differentiation markers induced by Fes in other cell lines (4, 34; data not shown). This effect was unique to the GFP-Fes L145P protein, as cells expressing GFP alone, wild-type GFP-Fes, the CC2 point mutant protein, or the CC1+2 mutant protein did not attach or spread. We also investigated the effect of GM-CSF on the adherence of each of the TF-1 cell populations (Fig. 8). Adherent cells appeared more rapidly in the presence of GM-CSF, reaching a maximum at 5 days. In addition, a low attachment response was observed with GM-CSF-treated cells expressing wild-type Fes or the coiled-coil domain mutant proteins but not with GFP alone. These results suggest that Fes may transduce signals for morphological aspects of differentiation downstream of the GM-CSF receptor.
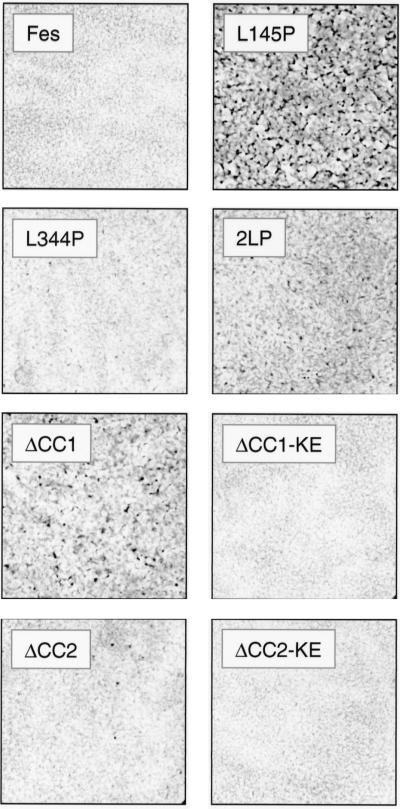
FIG. 7.
A point mutation in the GFP-Fes CC1 domain induces attachment and spreading of TF-1 myeloid leukemia cells. TF-1 cells were infected with a recombinant GFP retrovirus or the indicated GFP-Fes retroviruses and selected as described in the legend to Fig. 6. Following selection, the cells were replated in 24-well plates in the presence of GM-CSF and returned to the incubator for 5 days. Cells were stained in situ with Giemsa stain and visualized by light microscopy. Phase-contrast images of TF-1 cells expressing GFP, GFP-Fes, GFP-Fes L334P, and GFP-2LP were recorded under low-power magnification (×100). Images of cells expressing the GFP-Fes CC1 domain point mutant (L145P) protein were recorded under low-power (middle left panel) and high-power (×400; middle right panel) magnification.
FIG. 8.
Quantitative comparison of the effects of GFP-Fes CC domain mutant proteins on TF-1 cell morphology in the presence and absence of GM-CSF. TF-1 cells were infected with a recombinant GFP retrovirus or the indicated GFP-Fes retroviruses and selected as described in the legend to Fig. 6. For attachment experiments, 5 × 104 cells were plated in duplicate 24-well plates in the presence (+) or absence (−) of GM-CSF. Cells were returned to the incubator for 5 days (open bars) or 7 days (shaded bars), stained with Giemsa stain, and visualized by light microscopy. The number of attached cells was counted in three randomly chosen fields, and results are presented as average values ± standard deviations. This entire experiment was performed twice and produced the same pattern of responses in each case.
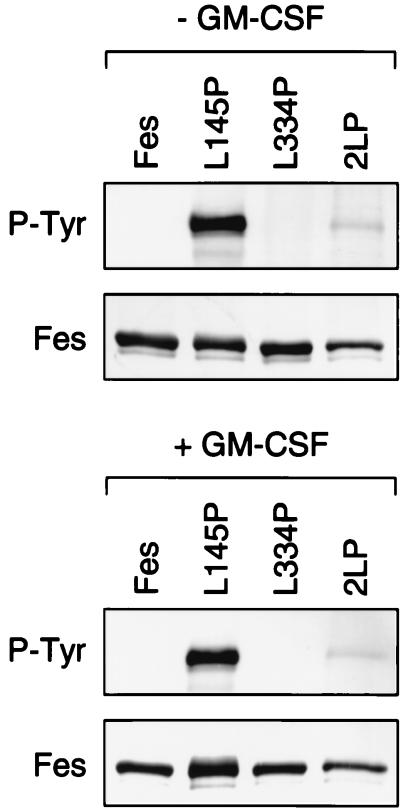
We also examined the tyrosine kinase activity of wild-type Fes and the coiled-coil domain mutants in the context of TF-1 cell expression. Fes proteins were immunoprecipitated and kinase activity was evaluated in terms of autophosphorylation on antiphosphotyrosine immunoblots. As shown in Fig. 9, autophosphorylation of both wild-type Fes and the CC2 domain point mutant protein was undetectable. In contrast, the CC1 point mutant protein exhibited a very high level of autophosphorylation in both the presence and the absence of GM-CSF. Addition of the L334P mutation in the CC2 domain substantially reduced the kinase activity of GFP-Fes L145P protein. These results correlate with both the survival and the differentiation responses in TF-1 cells and with the transforming and kinase activities of these mutant proteins in the fibroblast transformation assay, although the suppressive effect of the L334P mutation is more pronounced in TF-1 cells (see Discussion).
FIG. 9.
Autophosphorylation of GFP-Fes CC domain mutant proteins in TF-1 myeloid cells. TF-1 cells stably expressing each of the GFP-Fes fusion proteins indicated were washed free of GM-CSF and incubated in the presence or absence of cytokine for 48 h. Cells were collected by centrifugation and lysed, and GFP-Fes fusion proteins were precipitated with the M2 anti-FLAG monoclonal antibody. Immunoprecipitates were washed with RIPA buffer, resolved by SDS-PAGE, and immunoblotted for Fes expression with the anti-FLAG antibody (Fes) or for phosphotyrosine content with the antiphosphotyrosine monoclonal antibody PY99 (P-Tyr). This experiment was repeated twice and produced comparable results in each case; a representative blot is shown.
Analysis of the wild-type and mutant Fes coiled-coil regions by gel filtration.
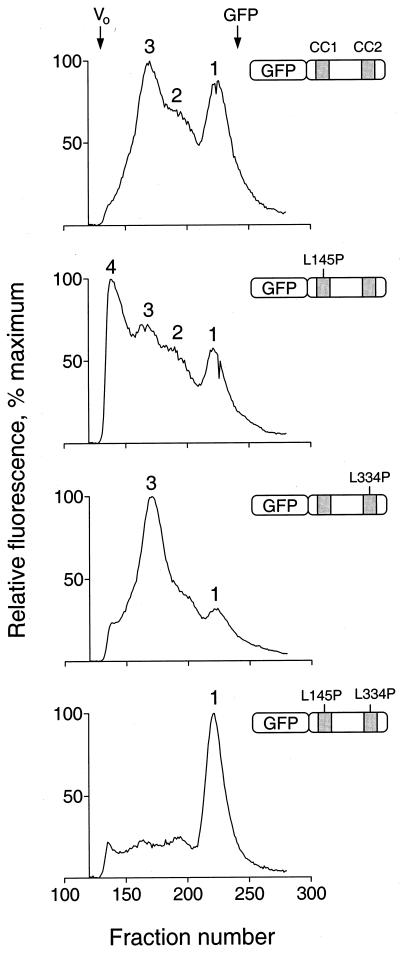
Data presented here and in our previous studies indicate that the Fes N-terminal coiled-coil domains may control Fes kinase activity by regulating the oligomerization state of the protein (4, 33). To investigate the effects of the coiled-coil domain point mutations on N-terminal oligomerization, we performed a series of gel filtration experiments. For these experiments, wild-type and mutant Fes N-terminal sequences encompassing both coiled-coil motifs (Ala 91 to Gly 392) were expressed as GFP fusion proteins in Sf-9 insect cells. Each GFP-CC1+2 fusion protein was then resolved on a Sephacryl S-300 gel filtration column, and 288 fractions were collected for each sample in 96-well plates. The positions of the GFP-CC1+2 fusion proteins in the elution profiles were determined as the relative fluorescence intensity of each well. As shown in Fig. 10, the wild-type GFP-CC1+2 fusion protein eluted as a mixture of monomeric and oligomeric forms. The peak intensities of the monomer (peak 1) and major oligomer (peak 3) were approximately equal. The monomer (peak 1) eluted with an apparent molecular mass lower than expected (40 kDa observed versus 52 kDa calculated), which may reflect a tight conformation of the protein. However, SDS-PAGE shows that this peak is full-length GFP-CC1+2 (see below). The major oligomer (peak 3) eluted with a mass of approximately 300 kDa, consistent with at least a pentamer; a similar result was observed previously for full-length Fes as well as the complete N-terminal region (33). At least one additional intermediate form was apparent (peak 2), suggesting that Fes can form multiple oligomeric structures. This result provides the first evidence that the Fes coiled-coil region can exist as a monomer and suggests that it can interconvert between monomeric and oligomeric states. Note that GFP itself elutes as a single symmetrical peak, consistent with a monomer, indicating that it does not contribute to oligomer formation (data not shown). The position of GFP in the chromatogram is indicated at the top of Fig. 10.
FIG. 10.
Analysis of N-terminal-Fes–GFP fusion proteins by gel filtration. Recombinant fusion proteins of GFP and Fes N-terminal amino acids encoding the CC1 and CC2 domains as well as the intervening sequence were expressed in Sf-9 cells. Cells were lysed by sonication, and lysates were clarified by ultracentrifugation and applied to a Sephacryl S-300 column. Fractions (0.25 ml) were collected in 96-well plates and analyzed for fluorescence using a microplate reader. Four fusion proteins were examined, and the resulting fluorescence elution profiles are shown together with an illustration of each protein. From top to bottom these include wild-type GFP-Fes CC1+2, the CC1 point mutant (L145P) protein, the CC2 point mutant (L334P) protein, and the double mutant protein. The void volume was determined independently using blue dextran, which eluted as a single peak at the position indicated as Vo (fraction 134). All fractions that appeared before the blue dextran peak were devoid of fluorescence and are omitted for clarity. Unfused GFP was also run as a control; it eluted as a single symmetrical peak, the position of which is indicated at the top (fraction 242). The following standards, with their molecular masses and elution positions noted, were also run: thyroglobulin, 634 kDa, fraction 149; ferritin, 439 kDa, fraction 165; catalase, 200 kDa, fraction 185; aldolase, 178 kDa, fraction 189; albumin, 62 kDa, fraction 202; ovalbumin, 48 kDa, fraction 207; and chymotrypsinogen A, 21 kDa, fraction 246. The entire experiment was repeated twice and produced the same pattern of fluorescent peaks for each protein. Note that the absolute levels of fluorescence varied less than twofold among the four fusion proteins.
The GFP-CC1+2 fusion protein with the CC1 point mutation (L145P) showed a dramatic shift from the monomeric to multiple oligomeric forms, including the peak 2 and peak 3 forms observed with wild-type Fes CC1+2 as well as an additional oligomer of higher molecular weight (peak 4). This result shows that the L145P mutation promotes oligomerization of the N-terminal region and is consistent with the hypothesis that this mutation activates Fes in vivo by disrupting intramolecular CC1-CC2 interaction and allowing oligomerization through CC2 (see Discussion).
Mutation of the CC2 domain (L334P) within GFP-CC1+2 also favored oligomerization, almost exclusively to the peak 3 form. This result also supports the model of intramolecular CC1-CC2 interaction as a mechanism for the suppression of kinase activity in vivo. In this case, oligomerization may be driven by the wild-type CC1 domain. However, the full-length L334P mutant protein lacks activity both in fibroblasts and in TF-1 cells, suggesting that a wild-type CC2 domain is essential for kinase activity and/or interaction with signaling partners in vivo (see Discussion).
We also investigated the oligomerization status of a GFP-CC1+2 fusion protein with point mutations in both coiled-coil domains. The double mutant protein eluted almost exclusively as the monomer (peak 1). This finding suggests that the full-length Fes double mutant protein may be impaired in its capacity to oligomerize, which may account in part for its reduced biological and kinase activities both in fibroblasts and in TF-1 myeloid cells.
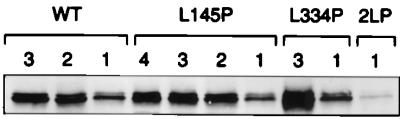
To verify that each of the fluorescent peaks observed in Fig. 10 corresponds to a GFP-CC1+2 fusion protein, peak fractions were pooled, concentrated, and analyzed by immunoblotting with anti-GFP antibodies. As shown in Fig. 11, all of the peaks contained GFP-immunoreactive bands of identical apparent molecular weights consistent with the expected size of the GFP-CC1+2 fusion protein. This result shows that the different peaks observed upon gel filtration reflect different oligomeric states of the Fes N-terminal protein.
FIG. 11.
Immunoblot analysis of GFP-Fes CC1+2 fusion proteins following gel filtration. Peak fractions from Sephacryl S-300 chromatography of the GFP-Fes CC1+2 fusion proteins shown in Fig. 10 were pooled, concentrated, and resolved by SDS-PAGE. The proteins were transferred to polyvinylidene difluoride membranes and probed with an antibody to GFP. Results are shown for wild-type GFP-Fes CC1+2 (WT), the CC1 mutation (L145P) protein, the CC2 mutation (L334P) protein, and the double mutant protein (2LP). The numbers above each lane refer to the major peaks shown in Fig. 10.
DISCUSSION
Cooperation of the CC1 and CC2 domains in the regulation of c-Fes tyrosine kinase activity.
Previous studies have shown that c-Fes and the closely related tyrosine kinase Fer exhibit several motifs with strong coiled-coil homology in their unique N-terminal regions (5, 16, 33). In c-Fes, the N-terminal region has been shown to mediate oligomerization of the kinase, leading to autophosphorylation by a trans mechanism (33, 35). These findings suggest that Fes oligomerization may be required for autophosphorylation of Tyr 713 within the activation loop, a prerequisite for kinase activation both in vitro and vivo (13, 35). In this report, we provide evidence demonstrating opposing actions of the coiled-coil domains in the regulation of Fes tyrosine kinase activity in vivo. Proline substitution for Leu 145 in the more N-terminal coiled-coil domain released Fes tyrosine kinase activity in Rat-2 fibroblasts, leading to transformation. This result suggests that the CC1 domain primarily serves a negative regulatory function, a conclusion consistent with our earlier work demonstrating that CC1 deletion can enhance the transforming and tyrosine kinase activities of a membrane-targeted form of c-Fes (4). The activating effect of the L145P mutation was reversed when combined with a proline substitution for Leu 334 in the more C-terminal region of coiled-coil homology. This result indicates that the CC2 domain is required for full kinase activation. Given its close proximity to the SH2 and kinase domains, the CC2 domain may act to align the kinase domains in an orientation favoring efficient trans phosphorylation within the active oligomer. Interestingly, deletion of the CC2 domain within GFP-Fes resulted in a small increase in kinase activity in fibroblasts, suggesting that the CC1 domain may partially complement the loss of CC2 (Fig. 3).
How does the CC1 domain act to repress Fes tyrosine kinase activity? One possibility is that it interacts intramolecularly with the CC2 domain, thereby repressing CC2-mediated formation of the active oligomer. The results of gel filtration experiments (Fig. 10 and 11) support this possibility. A GFP fusion protein with the N-terminal region encompassing both coiled coils was found to exist as a mixture of monomeric and oligomeric forms. The major wild-type GFP-CC1+2 oligomer elutes with an apparent molecular mass of about 300 kDa, which suggests that it forms at least a pentamer. This finding is consistent with the results of previous gel filtration experiments using full-length Fes as well as the isolated N-terminal region (33). Mutation of either CC1 or CC2 favored the elution of oligomers at the expense of the monomer, supporting the existence of CC1-CC2 interactions in the monomer. However, only the L145P mutant protein is active in cells, suggesting that oligomers formed by the L334P mutant protein are not compatible with kinase activation or signaling function. Interestingly, when point mutations were introduced into both coiled-coils, the GFP-CC1+2 fusion protein eluted as a monomer. This result suggests that the reduced kinase activity of the full-length double mutant protein may be due to a reduced oligomerization capacity in vivo. Whether or not CC1-CC2 interaction occurs within the context of full-length inactive Fes will require structural analysis.
The L145P mutation in the CC1 domain represents the first gain-of-function mutation described for Fes that is independent of an artificial membrane-targeting signal. However, N-terminal fusion of these mutant proteins to GFP is very important for stabilizing their expression in Rat-2 fibroblasts. Although a Fes L145P mutant protein without GFP could be transiently expressed in 293T cells and exhibited very high levels of tyrosine kinase activity relative to that of the wild-type kinase (data not shown), attempts to produce lines of Rat-2 cells that stably express this mutant were unsuccessful. The mechanism by which fusion to GFP allows for stabilization is unknown. One possibility is that this modification protects the active kinase from ubiquitination or other pathways for degradation. In this regard, active forms of Src have recently been shown to undergo ubiquitin-mediated degradation (10, 12).
Fes activation is sufficient for survival and differentiation signaling in myeloid cells.
Identification of an activated mutant enabled us to investigate the signaling function of Fes in a physiologically relevant cell type. The cell line used for this study was TF-1, a myeloid leukemia cell line that retains a cytokine requirement for growth and survival (17). Expression of the L145P mutant protein of Fes resulted in cytokine-independent proliferation of TF-1 cells, providing evidence that Fes may activate downstream signaling pathways that regulate cellular survival. These results are consistent with earlier work using v-Fps, a transforming retroviral homolog of c-Fes, and other tyrosine kinase oncogenes (25, 30). In addition, constitutive activation of Fes as a result of the L145P mutation caused attachment and spreading of TF-1 cells, consistent with previous work implicating Fes in the terminal differentiation of myeloid leukemia cells (4, 34, 40). Both the survival and differentiation effects of the L145P mutation were almost completely reversed in the double mutant protein (2LP), suggesting that an intact CC2 domain is required for efficient kinase activation or downstream signaling in myeloid cell types. Interestingly, the CC2 mutation did not have as dramatic an impact on the transforming function of the GFP-Fes L145P protein following ectopic expression in Rat-2 cells. One implication of this difference is that CC2-mediated interaction of Fes with a specific signaling partner may be required for the generation of physiological signals in myeloid cells. It also possible that cell-specific trans-acting factors may impose more strict control on Fes kinase activity in physiological sites of expression.
Previous studies have shown that overexpression of wild-type Fes is sufficient to induce terminal differentiation in the myeloid leukemia cell line K-562 (4, 40). These cells are derived from the blast crisis phase of CML and express the oncogenic tyrosine kinase Bcr-Abl (20). Recently, we observed that Bcr-Abl interacts directly with and phosphorylates Fes, suggesting that Bcr-Abl may serve as an upstream activator of Fes and that the differentiation response may be restricted to K-562 cells (19). However, data presented here with TF-1 cells show that Fes-induced differentiation is independent of the presence of Bcr-Abl. Introduction of the activated CC1 mutant of Fes induced a dramatic effect on TF-1 cell morphology, promoting cell attachment and spreading (Fig. 7). We have recently observed a similar effect in this cell line with another activated form of Fes in which SH2 domain specificity has been changed to that of Src (34). Interestingly, activation of Fes by either mechanism (SH2 domain substitution or CC1 mutation) correlates with a dramatic subcellular redistribution of Fes, from diffuse cytoplasmic sites to strong focal sites both in TF-1 cells and in fibroblasts (34; H. Cheng and T. Smithgall, unpublished data). Other work has shown that overexpression of wild-type Fes induces phosphorylation of p130 Cas and other proteins associated with cell adhesion and cell-cell contact in macrophages (1, 15). These results suggest that Fes may regulate morphological aspects of differentiation by targeting proteins involved in adhesion-related processes.
Constitutive activation of the Fes tyrosine kinase may promote TF-1 cell survival and morphological differentiation through a number of downstream signal transduction pathways. One possibility is Stat3, which is a direct substrate for Fes and associates with Fes in response to GM-CSF treatment (29, 31). Strong support for a Fes-Stat3 connection comes from studies of mice engineered to express kinase-defective Fes in place of the wild-type locus through gene-targeting techniques (36). GM-CSF was unable to induce activation of Stat3 in macrophages from these animals, implying a nonredundant role for Fes in Stat3 activation downstream of the GM-CSF receptor. In contrast, Stat3 activation is enhanced in cells from fes homozygous null animals, suggesting that the presence of the Fes protein is essential for effective control of Stat3 signaling in vivo (9). Stat3 has been linked to the induction of Bcl-XL and suppression of apoptosis in other systems and is required for terminal differentiation in some myeloid leukemia cell lines (3, 26, 28). Fes and related transforming oncogenes have also been shown to activate PI3K, which is linked to the suppression of apoptosis via the Akt kinase (7, 14). A final possibility involves activation of the small GTPases Ras, Rac, and Cdc42. Activation of all three of these small GTPases is required for transformation of Rat-2 fibroblasts by activated forms of Fes (18). Rac and Cdc42 directly regulate the actin cytoskeleton and may contribute to the observed effects of Fes on TF-1 cell attachment and spreading reported here (11, 38). In addition, constitutive activation of the Ras–mitogen-activated protein kinase pathway is sufficient to induce differentiation of myeloid leukemia cells and other cell types (32, 39). Future experiments will address the signaling pathways responsible for Fes-induced survival and differentiation responses in TF-1 cells.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (CA 58667) and the American Cancer Society (RPG-96-052-04-TBE).
REFERENCES
- 1.Areces L B, Sbarba P D, Jucker M, Stanley E R, Feldman R A. Functional specificity of cytoplasmic and transmembrane tyrosine kinases: identification of 130- and 75-kilodalton substrates of c-fps/fes tyrosine kinase in macrophages. Mol Cell Biol. 1994;14:4606–4615. doi: 10.1128/mcb.14.7.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs S D, Sharkey M, Stevenson M, Smithgall T E. SH3-mediated Hck tyrosine kinase activation and fibroblast transformation by the Nef protein of HIV-1. J Biol Chem. 1997;272:17899–17902. doi: 10.1074/jbc.272.29.17899. [DOI] [PubMed] [Google Scholar]
- 3.Catlett-Falcone R, Landowski T H, Oshiro M M, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernández-Luna J L, Nuñez G, Dalton W S, Jove R. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 4.Cheng H Y, Rogers J A, Dunham N A, Smithgall T E. Regulation of c-Fes tyrosine kinase and biological activities by N-terminal coiled-coil oligomerization domains. Mol Cell Biol. 1999;19:8335–8343. doi: 10.1128/mcb.19.12.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig A W, Zirngibl R, Greer P. Disruption of coiled-coil domains in Fer protein-tyrosine kinase abolishes trimerization but not kinase activation. J Biol Chem. 1999;274:19934–19942. doi: 10.1074/jbc.274.28.19934. [DOI] [PubMed] [Google Scholar]
- 6.Feldman R A, Lowy D R, Vass W C, Velu T J. A highly efficient retroviral vector allows detection of the transforming activity of the human c-fps/fes proto-oncogene. J Virol. 1989;63:5469–5474. doi: 10.1128/jvi.63.12.5469-5474.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukui Y, Saltiel A R, Hanafusa H. Phosphatidylinositol-3 kinase is activated in v-Src, v-Yes, and v-Fps transformed chicken embryo fibroblasts. Oncogene. 1991;6:407–411. [PubMed] [Google Scholar]
- 8.Greer P A, Meckling-Hansen K, Pawson T. The human c-fps/fes gene product expressed ectopically in rat fibroblasts is nontransforming and has restrained protein-tyrosine kinase activity. Mol Cell Biol. 1988;8:578–587. doi: 10.1128/mcb.8.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hackenmiller R, Kim J, Feldman R A, Simon M C. Abnormal Stat activation, hematopoietic homeostasis, and innate immunity in c-fes−/− mice. Immunity. 2000;13:397–407. doi: 10.1016/s1074-7613(00)00039-x. [DOI] [PubMed] [Google Scholar]
- 10.Hakak Y, Martin G S. Ubiquitin-dependent degradation of active Src. Curr Biol. 1999;9:1039–1042. doi: 10.1016/s0960-9822(99)80453-9. [DOI] [PubMed] [Google Scholar]
- 11.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 12.Harris K F, Shoji I, Cooper E M, Kumar S, Oda H, Howley P M. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc Natl Acad Sci USA. 1999;96:13738–13743. doi: 10.1073/pnas.96.24.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hjermstad S J, Peters K L, Briggs S D, Glazer R I, Smithgall T E. Regulation of the human c-fes protein-tyrosine kinase (p93c-fes) by its src homology 2 domain and major autophosphorylation site (tyr 713) Oncogene. 1993;8:2283–2292. [PubMed] [Google Scholar]
- 14.Izuhara K, Feldman R A, Greer P, Harada N. Interleukin-4 induces association of the c-fes proto-oncogene product with phosphatidylinositol-3 kinase. Blood. 1996;88:3910–3918. [PubMed] [Google Scholar]
- 15.Jucker M, McKenna K, Da Silva A J, Rudd C E, Feldman R A. The Fes protein-tyrosine kinase phosphorylates a subset of macrophage proteins that are involved in cell adhesion and cell-cell signaling. J Biol Chem. 1997;272:2104–2109. doi: 10.1074/jbc.272.4.2104. [DOI] [PubMed] [Google Scholar]
- 16.Kim L, Wong T W. The cytoplasmic tyrosine kinase FER is associated with the catenin-like substrate pp120 and is activated by growth factors. Mol Cell Biol. 1995;15:4553–4561. doi: 10.1128/mcb.15.8.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitamura T, Tange T, Terasawa T, Chiba S, Kuwaki T, Miyagawa K, Piao Y-F, Miyazono K, Urabe A, Takaku F. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3 or erythropoietin. J Cell Physiol. 1989;140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Smithgall T E. Fibroblast transformation by Fps/Fes tyrosine kinases requires Ras, Rac and Cdc42 and induces extracellular signal-regulated and c-Jun N-terminal kinase activation. J Biol Chem. 1998;273:13828–13834. doi: 10.1074/jbc.273.22.13828. [DOI] [PubMed] [Google Scholar]
- 19.Lionberger J M, Smithgall T E. The c-Fes protein-tyrosine kinase suppresses cytokine-independent outgrowth of myeloid leukemia cells induced by Bcr-Abl. Cancer Res. 2000;60:1097–1103. [PubMed] [Google Scholar]
- 20.Lozzio B B, Lozzio C B, Bamberger E G, Feliu A S. A multipotential leukemia cell line (K-562) of human origin. Proc Soc Exp Biol Med. 1981;166:546–550. doi: 10.3181/00379727-166-41106. [DOI] [PubMed] [Google Scholar]
- 21.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 22.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 23.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 24.Manfredini R, Grande A, Tagliafico E, Barbieri D, Zucchini P, Citro G, Zupi G, Franceschi C, Torelli U, Ferrari S. Inhibition of c-fes expression by an antisense oligomer causes apoptosis of HL60 cells induced to granulocytic differentiation. J Exp Med. 1993;178:381–389. doi: 10.1084/jem.178.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meckling-Gill K A, Yee S P, Schrader J W, Pawson T. A retrovirus encoding the v-fps protein-tyrosine kinase induces factor-independent growth and tumorigenicity in FDC-P1 cells. Biochim Biophys Acta. 1992;1137:65–72. doi: 10.1016/0167-4889(92)90101-g. [DOI] [PubMed] [Google Scholar]
- 26.Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller A J, Young J C, Pendergast A M, Pondel M, Landau R N, Littman D R, Witte O N. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive human leukemias. Mol Cell Biol. 1991;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson K, Rogers J A, Bowman T L, Jove R, Smithgall T E. Activation of STAT3 by the c-Fes protein-tyrosine kinase. J Biol Chem. 1998;273:7072–7077. doi: 10.1074/jbc.273.12.7072. [DOI] [PubMed] [Google Scholar]
- 30.Ostermeyer A G, Anderson S M. Induction of growth factor-independence and GM-CSF secretion by the v-src oncogene in murine myeloid cells does not require membrane association of pp60v-src. Exp Hematol. 1996;24:176–184. [PubMed] [Google Scholar]
- 31.Park W Y, Ahn J H, Feldman R A, Seo J S. c-Fes tyrosine kinase binds to and activates STAT3 after granulocyte-macrophage colony-stimulating factor stimulation. Cancer Lett. 1998;129:29–37. doi: 10.1016/s0304-3835(98)00077-9. [DOI] [PubMed] [Google Scholar]
- 32.Racke F K, Lewandowska K, Goueli S, Goldfarb A N. Sustained activation of the extracellular signal-regulated kinase mitogen-activated protein kinase pathway is required for megakaryocytic differentiation of K562 cells. J Biol Chem. 1997;272:23366–23370. doi: 10.1074/jbc.272.37.23366. [DOI] [PubMed] [Google Scholar]
- 33.Read R D, Lionberger J M, Smithgall T E. Oligomerization of the Fes tyrosine kinase: evidence for a coiled-coil domain in the unique N-terminal region. J Biol Chem. 1997;272:18498–18503. doi: 10.1074/jbc.272.29.18498. [DOI] [PubMed] [Google Scholar]
- 34.Rogers J A, Cheng H Y, Smithgall T E. SH2 domain substitution modulates the kinase and transforming activities of the Fes protein-tyrosine kinase. Cell Growth Differ. 2000;11:581–592. [PubMed] [Google Scholar]
- 35.Rogers J A, Read R D, Li J, Peters K L, Smithgall T E. Autophosphorylation of the Fes tyrosine kinase: evidence for an intermolecular mechanism involving two kinase domain tyrosine residues. J Biol Chem. 1996;271:17519–17525. doi: 10.1074/jbc.271.29.17519. [DOI] [PubMed] [Google Scholar]
- 36.Sensis Y, Zirngibl R, McVeigh J, Haman A, Hoang T, Greer P A. Targeted disruption of the murine fps/fes proto-oncogene reveals that Fps/Fes kinase activity is dispensible for hematopoiesis. Mol Cell Biol. 1999;19:7436–7446. doi: 10.1128/mcb.19.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smithgall T E, Rogers J A, Peters K L, Li J, Briggs S D, Lionberger J M, Cheng H, Shibata A, Scholtz B, Schreiner S, Dunham N A. The c-Fes family of protein-tyrosine kinases. Crit Rev Oncogen. 1998;9:43–62. doi: 10.1615/critrevoncog.v9.i1.40. [DOI] [PubMed] [Google Scholar]
- 38.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 39.Whalen A M, Galasinski S C, Shapiro P S, Nahreini T S, Ahn N G. Megakaryocytic differentiation induced by constitutive activation of mitogen-activated protein kinase kinase. Mol Cell Biol. 1997;17:1947–1958. doi: 10.1128/mcb.17.4.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G, Smithgall T E, Glazer R I. K562 leukemia cells transfected with the human c-fes gene acquire the ability to undergo myeloid differentiation. J Biol Chem. 1989;264:10276–10281. [PubMed] [Google Scholar]