Abstract
Meiotic recombination is a critical process for sexually reproducing organisms. This exchange of genetic information between homologous chromosomes during meiosis is important not only because it generates genetic diversity, but also because it is often required for proper chromosome segregation. Consequently, the frequency and distribution of crossovers are tightly controlled to ensure fertility and offspring viability. However, in many systems, it has been shown that environmental factors can alter the frequency of crossover events. Two studies in flies and yeast point to nutritional status affecting the frequency of crossing over. However, this question remains unexplored in mammals. Here, we test how crossover frequency varies in response to diet in Mus musculus males. We use immunohistochemistry to estimate crossover frequency in multiple genotypes under two diet treatments. Our results indicate that while crossover frequency was unaffected by diet in some strains, other strains were sensitive even to small composition changes between two common laboratory chows. Therefore, recombination is both resistant and sensitive to certain dietary changes in a strain-dependent manner and, hence, this response is genetically determined. Our study is the first to report a nutrition effect on genome-wide levels of recombination. Moreover, our work highlights the importance of controlling diet in recombination studies and may point to diet as a potential source of variability among studies, which is relevant for reproducibility.
Keywords: recombination, crossover frequency, interference, synaptonemal complex, sperm motility, collaborative cross founder strains, diet
Introduction
Meiotic recombination, or the exchange of genetic material between homologous chromosomes that occurs during meiosis, has been extensively studied since the early 20th century due to its important role in generating genetic variation and as an essential tool for genetic mapping; later, it was also found that recombination is required for proper chromosome segregation during meiosis in many organisms (Morgan 1913; Hassold and Hunt 2001). Alterations in the number or distribution of crossovers can result in chromosome missegregation and aneuploidy, with implications in fertility and offspring health (Hassold and Hunt 2001; Ottolini et al. 2015). This delicate balance between the selective benefits of genetic variation, proper chromosome segregation, and reproductive success has been achieved through a tight regulation of the crossing over process, both at the genetic and the epigenetic levels.
Although the control of crossover number and distribution is a complex and yet not fully understood process, it results in three common observations: (1) each homolog must have at least one crossover (the obligate crossover or crossover assurance) (Mather 1937; Pardo-Manuel de Villena and Sapienza 2001; Dumont 2017); (2) crossovers are not independent, because the occurrence of one interferes with the occurrence of a second one nearby (positive crossover interference) (Sturtevant 1915; Muller 1916); (3) crossover numbers can be maintained in spite of variations in the number of double-strand breaks they originate from (crossover homeostasis) (Cole et al. 2012; Baier et al. 2014; Hunter 2015). Consequently, the recombination rate is constrained within species, although considerable variation is observed between species (Dumont and Payseur 2008; Segura et al. 2013; Dumont 2017).
Nevertheless, recombination rate still can vary between individuals of the same species: for instance, up to 30% variation has been reported between mice of different strains and hence, of different genetic background (Koehler et al. 2002; Dumont and Payseur 2011a; Baier et al. 2014; Dumont 2017). In addition, crossover frequency is higher in females than in males in both human and mouse, and this difference has been associated to the length of the synaptonemal complex (SC), the proteinaceous scaffold that forms between chromosomes during meiotic prophase and mediates crossover formation (Tease and Hulten 2004; Petkov et al. 2007). A positive correlation between number of crossovers and SC length has also been observed between different inbred strains of mice (Lynn et al. 2002; Baier et al. 2014). These intraspecific studies suggest that genetic differences, as well as chromatin packaging changes, underlie differences in crossover frequency (de la Casa-Esperon and Sapienza 2003; Kleckner et al. 2003; Kleckner 2006; de la Casa-Esperon 2012; Baier et al. 2014). Among the few loci identified in mice that control recombination rate (de la Casa-Esperon et al. 2002; Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010; Balcova et al. 2016), Prdm9 codes for a histone methyltransferase that determines recombination hotspots. These and other studies in several species conclude that recombination is genetically and epigenetically controlled.
In spite of this control, age and certain external factors, such as temperature changes (Plough 1917; Bomblies et al. 2015; Lloyd et al. 2018), diet (Neel 1941), stress (Belyaev and Borodin 1982), toxicants (Susiarjo et al. 2007; Vrooman et al. 2015; Gely-Pernot et al. 2017), and infections (Singh 2019), are capable of modifying the recombination rate. In mice, the best-studied case is that of bisphenol A (BPA) exposure, a component of epoxy resins and polycarbonate plastics used in a wide variety of consumer products. BPA is an endocrine disruptor capable of binding estrogen receptors (Susiarjo et al. 2007; Alonso-Magdalena et al. 2012). Accidental intake of BPA from damaged mouse cages led to increased meiotic disturbances and aneuploidy in female oocytes (Hunt et al. 2003). Subsequent studies showed that BPA exposure also altered the levels of recombination in both female and male meiosis (Susiarjo et al. 2007; Brieño-Enriquez et al. 2011; Vrooman et al. 2015).
From the studies of BPA exposures in mice, we have learnt several lessons: first, the effects on recombination depend on sex and genetic background. BPA exposures resulted in increased recombination in C57BL/6 females, but not in males of the same inbred strain (Susiarjo et al. 2007; Vrooman et al. 2015). However, in males of the CD-1 outbred strain, BPA was able to induce the opposite effect (a reduction of crossover frequency) (Vrooman et al. 2015). Second, BPA also induces other alterations in the germline—e.g., in diverse processes during spermatogenesis, resulting in reduced sperm production (Liu et al. 2013; Wisniewski et al. 2015; Xie et al. 2016). Third, BPA is also capable of eliciting heritable changes and has been associated with epigenetic modifications of the germline (Manikkam et al. 2013; Susiarjo et al. 2013; Wolstenholme et al. 2013; Susiarjo et al. 2015; Ziv-Gal et al. 2015; Rahman et al. 2020).
However, BPA is not unique: other estrogenic substances are also capable of inducing meiosis and recombination changes (Vrooman et al. 2015; Gely-Pernot et al. 2017; Horan et al. 2017, 2018). Interestingly, the meiotic disturbances caused by BPA on metaphase II mouse oocytes can be prevented by a diet rich in phytoestrogens which, in turn, can elicit abnormalities in absence of BPA (Muhlhauser et al. 2009); phytoestrogens are also capable of counteracting methylation changes induced by BPA (Dolinoy et al. 2007). Isoflavone phytoestrogens, mainly genistein and daidzein, are natural compounds abundant in soy and other legumes.
These observations open the question as to whether not just toxicants, but also diets, could affect the recombination rate in mouse. As dietary options could be infinite, we have focused our attention on two categories of diet. First, we were interested in diets that could modify the germline epigenome based on the lessons learnt from BPA studies. Second, we were interested in common diets. With respect to the former, diets that have shown to induce heritable epigenetic changes in the male germline are low-protein, high fat, or caloric restriction diets (Donkin and Barres 2018; Siddeek et al. 2018). For instance, in utero 50% caloric restriction can cause metabolic disturbances in the F1 and F2 generations in mice, as well as methylation changes in the transmitting sperm (Martínez et al. 2014; Radford et al. 2014). Male mouse undernourishment can reduce paternal sperm methylation and fertility and have a negative impact on the health of their offspring (Anderson et al. 2006; McPherson et al. 2016). Hence, we decided to test whether paternal undernourishment could affect meiotic recombination rates in a mouse model. This treatment is of particular interest given its relevance to humans, where undernourishment is a burden for many.
With respect to our interest in common diets, it is important to know if these diets have significant effects on recombination and other reproductive phenotypes from a reproducibility perspective. In our animal facility, two diets are regularly used, which differ in their protein, energy, and phytoestrogen content (see Materials and Methods). As previously discussed, these three dietary factors have been shown to cause meiotic or epigenetic changes in the germline, which leads to the possibility that content differences among common mouse diets may affect recombination as well.
Therefore, we analyzed whether differences between common diets as well as undernourishment can affect recombination rates in adult males. We performed our study in diverse genetic backgrounds, given the variability in crossover frequency, as well as variation in the effects of environmental exposures on recombination and spermatogenesis, reported between different mouse strains (Spearow et al. 1999; Thigpen et al. 2007; Vrooman et al. 2015). We observed that common diets can trigger recombination rate changes in adult male mice. These changes are strain specific and, thus, depend on the genetic background. In addition, these diets can elicit sperm motility changes, but no major spermatogenesis disturbances were observed. Therefore, we propose that recombination could be particularly sensitive to certain alterations, potentially epigenetic, caused by diverse effectors such as diet; hence, recombination could be a biomarker of environmentally induced perturbations in the germline. Moreover, our data compellingly show that diet composition must be taken into account when performing recombination and sperm studies.
Materials and methods
Mouse strains and diets
C57BL/6J (B6), PWK/PhJ, and MOLF/EiJ mice were obtained from Jackson Laboratory through Charles River and were bred in our facilities for several generations under the same diet and environmental conditions before the studies began (see Supplementary Reagent Table for a summary of strains and reagents providers). All experimental procedures used in this study were approved by the Committee of Ethics in Animal Care of the University of Castilla‐La Mancha. Mouse chow diets were provided by Harlan Laboratories and Capsumlab. Teklad Global 18% Protein Rodent Diet is designed to support gestation, lactation, and growth of rodents and, therefore, fed to pregnant and nursing female mice; hence, it will be referred as the “breeding” diet from here on. Teklad Global 14% Protein Rodent Maintenance Diet (and its equivalent Capsumlab Maintenance Complete Chow, used only in the initial set of experiments) is designed to promote longevity and normal body weight in rodents and, therefore, the routinely “maintenance” diet used in many facilities like ours. Description of both diets can be found in Supplementary Table S1.
We performed two studies: in the initial one, adult males from the three strains were analyzed for the effect of two diets on recombination (undernourishment and breeding diets) provided during 24 days relative to a control group kept ad libitum with maintenance diet. Animals switched to breeding diet had free access to the chow, but those of the “undernourishment” group were fed with 50% (2.25 g of maintenance diet) of the regular daily intake (4.5 g, according to Bachmanov et al. 2002). Each diet group had three adult mice (average 5.8 months), except the B6 control and B6 breeding groups, each with two mice. Health and weight of the animals were regularly monitored. The second study was aimed to verify the differences observed between breeding and maintenance diets in B6 mice and expand the study to testes and sperm phenotypes. Hence, two groups of five B6 mice (average 5.9 months) were fed ad libitum during 24 days with each of the two diets. In both studies, all animals were housed in the same room and conditions and treated almost simultaneously, so that each day we processed a mouse of a different treatment group, in order to avoid differences in uncontrolled environmental exposures (temperature, chemicals, etc.) between animals, as well as other sources of experimental bias.
Collaborative Cross (CC) founder mice were obtained from the Jackson Laboratory and reared on a different diet (Laboratory Rodent Diet 5001) in the Biological Research Facility at North Carolina State University. All animals were housed in the same room and were thus subject to the same environmental conditions. At 8 weeks of age, MLH1 immunohistochemistry analysis (see below) was performed in three animals per strain (two in NZO/HILtj) and 25 spermatocytes were analyzed per mouse. All experimental protocols were approved by the Institutional Animal Care and Use Committee of North Carolina State University.
Tissue collection and processing for histochemistry and sperm analyses
Dates for mouse euthanasia, sample collection, and processing were randomized to avoid experimental artifacts, and the diet group of the processed samples and resulting images were blinded until all measurements were completed to avoid subjective bias during the analyses.
After the 24-day diet period, adult male mice were euthanized by cervical dislocation and weighed. After removing and weighing the testes, chromosome spreads for immunostaining were prepared from one testicle as described below. The other testicle was submerged in Bouin’s solution and processed for histochemistry. Fixed and paraffin-embedded tissues were sectioned and stained with hematoxylin and eosin. Histological analysis of the composition and distribution of the diverse cell types of the seminiferous tubules was performed as previously described (Ahmed and de Rooij 2009; Borg et al. 2010).
Mature spermatozoa were collected from the caudae epididymides in 500 µl modified TYH buffer (in mM: 135 NaCl, 4.7 KCl, 1.7 CaCl2, 1.2 KH2PO4, 1.2 MgSO4, 5.6 glucose, 10 HEPES, pH 7.4 adjusted at 37°C with NaOH). Then, the sperm motility was assessed using a computer-aided sperm analyzer (Sperm Class Analyzer® CASA System, Microptic; Barcelona, Spain). Aliquots of 5 µl sperm/sample were placed on a prewarmed (37°) Leja chamber and examined in a phase-contrast microscope (Nikon Eclipse 80i, Tokyo, Japan) equipped with a warmed stage (37°) and a Basler A302fs digital camera (Basler Vision Technologies, Ahrensburg, Germany), which is connected to a computer by an IEEE 1394 interface. Evaluations were made at 10× magnification and at least 10 fields or 200 spermatozoa were recorded for each sample. Settings were adjusted to mouse spermatozoa. Recorded parameters were total motility (%), progressive motility (%), curvilinear velocity (VCL, μm/s), straight-line velocity (VSL, μm/s), average path velocity (VAP, μm/s), linearity (LIN; %), straightness (STR, %), wobble (WOB; %), lateral head displacement (ALH, μm), and beat cell frequency (BCF, Hz).
Sperm viability was assessed by mixing 5 µl of sperm diluted in THY buffer with 10 µl of eosin-nigrosin for 30 s and spreading the mix on a slide. The percentage of viable sperm was evaluated under the microscope, as eosin stains only the dead sperm, whereas live sperm remains white.
Chromatin stability was assessed using the Sperm Chromatin Structure Assay (SCSA), a flow cytometric test where sperm DNA breaks are evaluated indirectly by analyzing DNA denaturability (Evenson et al. 1980). The assay measures the susceptibility of sperm DNA to acid-induced DNA denaturation, detected by staining with the fluorescent dye acridine orange (AO). Samples were diluted with TNE buffer (0.15 M NaCl, 0.01 M Tris–HCl, 1 mM EDTA; pH 7.4) at a final sperm concentration of 2 × 106 cells and mixed with 400 μl of an acid-detergent solution for 30 s. Then, 1.2 ml of AO was added, and samples were evaluated 2 min later with a Cytomics FC500 flow cytometer (Beckman Coulter, Brea, CA, USA). AO was excited with a 488 nm argon laser. A total of 5000 spermatozoa per sample were evaluated. We expressed the extent of DNA denaturation in terms of DNA fragmentation index (DFI), which is the ratio of red to total (red plus green) fluorescence intensity, i.e., the level of denatured DNA over the total DNA. The DFI value was calculated for each sperm cell in a sample, and the resulting DFI frequency profile was obtained. Total DNA fragmentation index was defined as the percentage of spermatozoa with a DFI value over 25. High DNA stainability (HDS), which offers a measure of the percentage of immature sperm cells, was defined as the percentage of spermatozoa with green fluorescence higher than channel 600 (of 1024 channels).
Immunostaining, microscopy, and scoring
Chromosome spreads were prepared from spermatocytes as previously described (Anderson et al. 1999; de Boer et al. 2009; Milano et al. 2019). Briefly, one of the two testes was decapsulated in hypotonic extraction buffer (HEB: 30 mM Tris, pH 8.2, 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 0.5 mM DTT, and 0.5 mM PMSF). Seminiferous tubule fragments were minced in 100 mM sucrose and then fixed onto slides with 1% paraformaldehide containing 0.15% Triton X-100 in a humidified chamber. Slides were washed in 1× PBS with Photo-Flo 200 (Kodak), dried and processed for immunostaining, or stored at −80°C until use.
MLH1 immunostaining allows for identification of about 90% of mammalian crossover sites (Anderson et al. 1999; Cole et al. 2012). For immunostaining, chromosome spreads were washed in 1× PBS with 0.4% Photo-Flo 200 (Kodak) and 1× PBS with 0.1% Triton X-100. The slides were blocked in 10% antibody dilution buffer (ADB: 3% bovine serum albumin, 0.05% Triton, 10% goat serum in 1× PBS). Then, they were incubated overnight at room temperature with primary antibodies: mouse anti-human MLH1 (BD Biosciences) diluted 1:100 and rabbit anti-SCP3 (Abcam) diluted 1:1000 in ADB. Slides were washed as previously and incubated for 1 h at 37° with secondary antibodies: Alexa Fluor 488 goat anti-mouse IgG and goat anti-rabbit conjugated with Alexa Fluor 555 (Life Technologies) diluted 1:1000 and 1:2000 in ADB, respectively. Slides were washed in 0.4% Photo-Flo and mounted with Prolong Gold Antifade Reagent with DAPI (Life Technologies Limited).
All slides were imaged on a Zeiss LSM 710 confocal microscope and analyzed using Zeiss Zen lite software. Only mid and mid-late pachytene stage spermatocytes with fully synapsed autosomes, little synapsis or just end-to-end association of the X and Y chromosomes and characteristic sex-body formation were scored (Anderson et al. 1999, Ashley et al. 2004); cells with poor staining or other scoring difficulties were excluded. In the first study, 25 spermatocytes were analyzed per animal, while more (22–40, average 37.3) were examined per mice in the second. For each spermatocyte, we counted the number of foci localizing to the SC of the 19 autosomes because the appearance and disappearance of foci on the XY bivalent and on the autosomes are temporally uncoupled (Anderson et al. 1999); total SC length was also measured in autosomes only. Autosomal SC length was initially measured by manually tracing the length of the SYCP3 signal. Given the large dataset of our second experiment, we developed an ImageJ Macro (named “Synaptonemal & CO analyzer”) for SC semiautomatic measuring. This imageJ macro works in four steps: (1) intensity threshold selection of SC signals; (2) automatic detection of each SC, which is reduced to its central skeleton line; (3) manual editing and (4) automatic measuring of the resulting SCs skeletons. For full description, open access, and validation, see J. Soriano, A. Belmonte, and E. de la Casa-Esperon (in preparation). Distance between MLH1 foci was measured in bivalents with two or more foci. The diet group of the samples was blinded until after focus counts and measurements were determined, and reviewed by a second observer; any cells with discrepant or ambiguous MLH1 number were discarded.
Statistical analysis
Comparisons in the average numbers of foci between different strains and/or diets were tested by ANOVA or Student t-test analysis, pooling the results from multiple mice of each group following previous examples (Cole et al. 2012; Vrooman et al. 2015; Zelazowski et al. 2017). Welch ANOVA was applied when homogeneity of variances could not be assumed. These analyses have been successfully employed in comparable studies despite MLH1 foci not following a normal distribution, because of the robustness of ANOVA analysis (Baier et al. 2014; Dumont 2017). Similar conclusions about statistical significance were obtained if nonparametric tests were performed. For statistically significant differences (P < 0.05), a Tukey’s post hoc honestly significant difference (HSD) test was performed to infer which groups differed. A Chi-square test was used to determine significance in the number of bivalents classified according to their foci number (E0–E3) between diet groups. Weight, sperm count and SCSA data were analyzed by Student t-test. Total motility, progressive motile spermatozoa, VCL, VSL, VAP, LIN, ALH, BCF, and sperm viability were evaluated by a factorial ANOVA in mice fed with different diets. When the variables were significant (P < 0.05), post hoc comparisons with Bonferroni correction were carried out. Analyses were performed using SPSS Statistics software.
We also used a generalized linear model to compare the average numbers of foci between different strains and/or diets. The full model includes effects of strain, diet, animal, and interaction effects. This was implemented in JMP Pro version 14.
Results
Recombination levels depend on the genetic background
Variation in crossover frequency, as well as variability in the effects of chemical exposures on recombination, have been observed among mouse strains (Koehler et al. 2002; Dumont and Payseur 2011b; Baier et al. 2014; Vrooman et al. 2015). To investigate if diets have an impact on recombination levels, we selected three mouse inbred strains of diverse genetic background: C57BL/6J (B6), PWK/PhJ (PWK), and MOLF/EiJ (MOLF). B6 is a classical inbred strain widely used in recombination studies (Koehler et al. 2002; Dumont and Payseur 2011b; Baier et al. 2014), which is mostly of Mus musculus domesticus origin (93% of autosomal sequences (Yang et al. 2011)); PWK was derived from wild mice of M. m. musculus subspecies (Gregorova and Forejt 2000) (94% of autosomal sequences of M. m. musculus origin according to Yang et al. (2011)); MOLF is representative of the Japanese Mus musculus molossinus subspecies, which is the result of the hybridization between M. m. musculus and Mus musculus castaneus (Silver 1995).
Our first goal was to determine the baseline crossover frequencies of the three strains with regular maintenance diet. Our analysis reveals a significant effect of strain on MLH1 focus count (P ≪ 0.0001, ANOVA). B6 values are the lowest and similar to those obtained in previous studies (Baier et al. 2014; Vrooman et al. 2014; Balcova et al. 2016). B6 males have 23.54 ± 1.97 MLH1 foci per spermatocyte (mean ± SD), while MOLF males have significantly more foci per spermatocyte (24.85 ± 2.57; P = 0.015, HSD). MLH1 focus counts are substantially higher in PWK males than both B6 and MOLF males (29.15 ± 2.87; P ≪ 0.0001, both comparisons HSD) (Figure 1A and Supplementary Figure S1 and Table 1). Hence, genetic differences between the selected strains have an impact on the levels of recombination.
Figure 1.
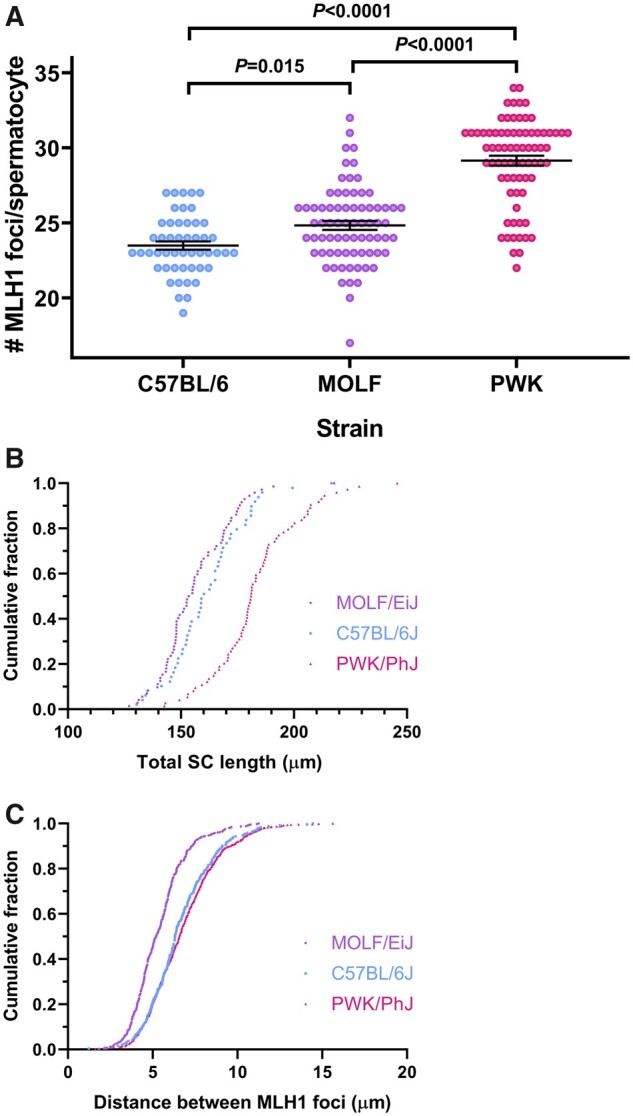
Strain effects on recombination rates, SC length and interference. Recombination levels depend on the genetic background. Autosomal MLH1 foci counts in pachytene spermatocytes are shown in A for mice of the three strains fed ad libitum with maintenance diet. Significant differences were observed between the three strains, as explained in Table 1. Each dot represents the focus count of a single nucleus. Black bars represent means ± SEM. The cumulative fraction of the total autosomal SC length and the intercrossover distances measured in micrometers are represented in B and C, respectively. PWK spermatocytes have significantly longer SC, while MLH1 interfocus distances in MOLF are significantly shorter than those of the other two strains (Table 1).
Table 1.
Strain effects on the number of autosomal MLH1 foci and total autosomal SC length per pachytene spermatocyte, and on the length of the SC between MLH1 foci
| Strain | B6 | MOLF | PWK |
|---|---|---|---|
| MLH1 foci: | 23.54 ± 1.97a | 24.85 ± 2.57b | 29.15 ± 2.87c |
| SC length (μm): | 162.2 ± 17.1d | 156.6 ± 16.0d | 183.3 ± 19.4e |
| Interfocus distance (μm): | 6.63 ± 2.00f | 5.43 ± 1.57g | 6.83 ± 2.06f |
| %Interfocus/ SC length: | 64.0 ± 12.9h | 56.2 ± 12.8i | 63.1 ± 13.3h |
| μmSC/MLH1 foci | 6.9 | 6.3 | 6.3 |
Comparison of the three strains fed with control maintenance diet shows significant differences in the average number of MLH1 foci (F = 81.5, P ≪ 0.0001), SC length (F = 45.4, P ≪ 0.0001) and intercrossover distance (F = 73.4, P ≪ 0.0001). Post hoc analysis reveals significant differences in MLH1 focus frequency between the three strains (ab, P = 0.015; bc, P ≪ 0.0001; ac, P ≪ 0.0001). In addition to displaying the highest crossover frequency, PWK has the longest SC length (de, P ≪ 0.0001 when compared to any of the two other strains). In contrast, MOLF has the shortest interfocus distance per se (fg, P ≪ 0.0001 when compared to any of the two other strains) or calculated as a percentage of SC length of the corresponding bivalent (hi, P ≪ 0.0001 again when MOLF was compared to any of the two other strains). Data were obtained from 50 B6, 75 MOLF and 75 PWK spermatocytes, with 244, 455, and 765 interfocus distance measurements respectively. Analyses were performed by ANOVA and significant differences between groups were assessed using Tukey’s post hoc tests. Values are shown as mean ± SD.
Recombination variability in genetically diverse mice: the CC and Diversity Outbred stock founder strains
C57BL/6 is one of the most widely used mouse strains, but interest for PWK mice, at the other extreme of the crossover frequency, is increasing as a representative strain of the M. m. musculus subspecies in many genetic studies and resources, such as the CC and Diversity Outbred (DO) population. Both resources are the result of crosses between eight founder strains that include B6 and PWK, but also A/J, 129S1/SvImJ, NOD/LtJ, NZO/HlLtJ, CAST/EiJ and WSB/EiJ. These eight strains were selected because they capture most of the genetic diversity present in Mus musculus and, therefore, CC and DO mice have become instrumental for multiple genetic studies (Churchill et al. 2004; Roberts et al. 2007; Chesler et al. 2008; Threadgill et al. 2011; Collaborative Cross Consortium 2012; Svenson et al. 2012). Indeed, analysis of CC mice has enabled the characterization of several loci and mechanisms that control recombination (Liu et al. 2014). However, the crossover rate had not been described for all the founder strains (including PWK). Therefore, we decided to characterize the crossover frequency of the eight strains under the same developmental and environmental conditions for the benefit of two purposes: first, for future genetic studies with the CC and DO mice, including further analyses of loci involved in recombination; and second, for comparing the crossover variation of the strains selected for our study relative to the extent of recombination variability present in Mus musculus.
We observe 22.21 ± 1.86 MLH1 foci per spermatocyte in CAST/EiJ mice, 22.80 ± 2.22 in NZO/HlLtJ, 22.89 ± 2.60 in C57BL/6J, 23.40 ± 2.35 in A/J, 23.75 ± 2.61 in 129S1/SvImJ, 24.19 ± 23.36 in WSB/EiJ, 25.48 ± 2.40 in NOD/LtJ, 28.11 ± 3.83 in PWK/PhJ (Figure 2). There are significant differences in the MLH1 foci per spermatocyte between the strains (P ≪ 0.0001, ANOVA) although, as shown in Figure 2, the majority has frequencies similar to that of B6. CAST/EiJ has the least MLH1 foci count per spermatocyte, but not significantly lower than B6. At the other extreme, PWK values are again significantly higher than any of the other strains (P < 0.0001 in all cases), with 27% more crossovers than the CAST/EiJ spermatocytes, a variability in crossover frequency between mice of different strains similar to that reported in other studies (Koehler et al. 2002; Dumont and Payseur 2011b; Baier et al. 2014). Intermediate values are observed in NOD/LtJ and WSB/EiJ. Foci counts are significantly higher in NOD/LtJ than in B6 (P < 0.0001).
Figure 2.
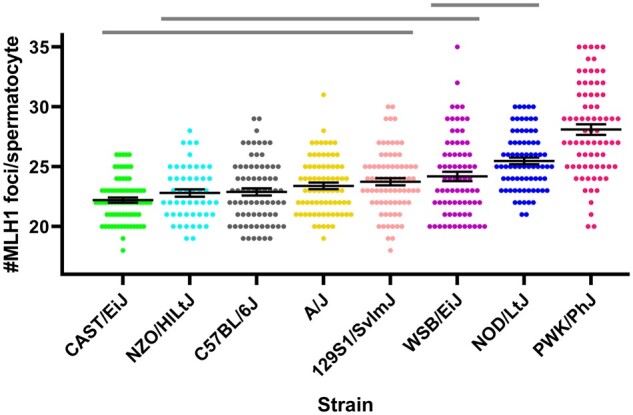
Recombination rate variability among CC founder strains. Autosomal MLH1 foci counts in pachytene spermatocytes are shown for 8 genetically diverse mouse inbred strains, mostly of M. m. domesticus origin, except CAST/EiJ and PWK/PhJ, which are representative of M. m. castaneus and M. m. musculus subspecies, respectively (Yang et al., 2011). Each dot represents the focus count of a single nucleus. Black bars symbolize means ± SEM. A significant strain effect on MLH1 foci counts is observed (P ≪ 0.0001, ANOVA). Horizontal upper lines represent homogeneous subsets from post hoc comparisons using the Tukey’s test (α = 0.01).
Comparing these with our previous data, we observe that B6 and PWK results are not significantly different than those obtained in our study for the same strains fed with control maintenance diet (P = 0.26 and P = 0.06, respectively, t-test) and are located toward the low and at the high ends of the recombination variability distribution, while MOLF values are intermediate. Our results confirm previous observations of the impact of the genetic background on recombination frequency (Liu et al. 2014) and provide new data about the crossover rate of the CC and DO founder strains.
Changes in SC length or interference may underlay recombination differences between strains
Crossover distribution and frequency is limited by crossover interference. As a consequence, mouse spermatocytes chromosomes with a short SC can only undergo one crossover (Sym and Roeder 1994; Lawrie et al. 1995; Tease and Hulten 2004; Petkov et al. 2007). When all chromosomes are considered, cells with longer SCs (measured as μm of immunostained SC) are expected to have more crossovers (Froenicke et al. 2002; Lynn et al. 2002; Kleckner et al. 2003; Dumont and Payseur 2011a). Our data indicate a significant effect of strain on SC length (P ≪ 0.0001, ANOVA). When we compare the total length of the SC of the autosomes per cell (in μm, Table 1 and Figure 1B), we observe significantly longer SC in PWK (183.3 ± 19.4) compared to those observed in B6 (162.2 ± 17.1) and MOLF spermatocytes (156.6 ± 16.0; P ≪ 0.0001 in both cases, HSD). Hence, the larger SC in PWK may explain the higher crossover frequency observed in this strain respect to those of B6 and MOLF. We found no significant difference in SC length between MOLF and B6 spermatocytes (P = 0.20, HSD). This is interesting because these two strains have significant differences in the number of MLH1 foci as reported above. This suggests that factors other than SC length may account for the differences in recombination levels observed between these two strains.
As SC length cannot explain the increase of MLH1 foci in MOLF respect to B6 spermatocytes, we wondered if variation in interference strength could be the cause. As a surrogate for CO interference, we measured the distance between MLH1 foci of bivalents with two or more crossovers. Analysis of variance indicates a significant effect of strain on intercrossover distance (P ≪ 0.0001, ANOVA; similar results were obtained with nonparametric tests). Post hoc tests reveal that the average interfocus distance in MOLF spermatocytes is significantly shorter than in B6 and PWK spermatocytes (P ≪ 0.0001 both comparisons, HSD; Table 1 and Figure 1C). This is also true when the interfocus distances are expressed as percentage of the length of the SC (de Boer et al. 2009) (P ≪ 0.0001 both comparisons, HSD; Table 1 and Supplementary Figure S2). Our data indicate that not SC length, but interference changes may explain the crossover rate increase observed in MOLF mice respect to that of B6. Therefore, different mechanisms appear to lie beneath the recombination variability between diverse mouse strains.
Undernourishment may influence recombination levels in a strain-dependent manner
To test whether diet could affect recombination, we explored if a 50% restriction to food access in adult males could have an impact on meiotic recombination. Previous studies had shown that a 24-day exposure to dietary changes or environmental factors was sufficient to induce spermatogenesis changes in adult rodents (Assinder et al. 2007; Gely-Pernot et al. 2017). Moreover, a 24-day diet would ensure continuous exposure at least since the spermatogonial stage until pachytene, when recombination is analyzed. Therefore, we chose this time period for our studies about the impact of diets on recombination. We fed adult male mice of each of the three strains with 50% of their regular daily intake of maintenance chow, while controls had access to the same maintenance diet ad libitum. At the end of the 24-day treatment, animals were euthanized and testes were processed for crossover analysis. Analysis of the data by a univariate generalized linear model shows that strain (P < 0.0001) and diet (P = 0.035) significantly affect the MLH1 focus frequency. We note that two of the three undernourished PWK animals had to be euthanized before the conclusion of the treatment period due to severe weight loss (20% of body weight); no relationship was observed between the sacrifice timing and MLH1 focus frequency. In contrast, B6 and MOLF were comparatively robust to the effects of undernourishment on body weight. B6 mice often get overweight and can resist well brief periods of food shortage. MOLF is a wild-derived strain of lean mice like PWK, but while the body weight of the latter was reduced 9–20%, MOLF only lost 5–7%.
We observed that PWK spermatocytes, already with high crossover frequency, had a small but significant increase (P = 0.037) in MLH1 foci number when food intake was restricted to 50% (Table 2). We also noticed considerable inter-individual variation in crossover frequency, perhaps associated with incomplete penetrance of the phenotypic effects of diet (Figure 2 and Supplementary Figure S1). Due to the small magnitude of the effect, we could not determine if it was associated with SC length or interference changes. Neither B6 nor MOLF spermatocytes showed significant changes in MLH1 focus count between ad libitum and 50% restricted diets by post hoc tests (Table 2), suggesting an efficient control of the recombination levels when adult males of these two strains face undernutrition.
Table 2.
Diet effects on MLH1 foci number per spermatocyte.
| Strain | B6 | MOLF | PWK |
|---|---|---|---|
| Maintenance diet | 23.54 ± 1.97 | 24.85 ± 2.57 | 29.15 ± 2.87 |
| 50% diet | 23.79 ± 1.63 | 25.01 ± 2.08 | 30.13 ± 1.95 |
| Breeding diet | 24.70 ± 2.21 | 24.35 ± 1.62 | 29.95 ± 2.41 |
Overall, we observe significant strain and diet effects on MLH1 foci number (P ≪ 0.0001 and P = 0.01, respectively). Analyses within strains reveal significant diet effects in MLH1 foci number in B6 and PWK mice (P = 0.006 and P = 0.033, respectively, with ANOVA tests), but not in MOLF animals. In the B6 strain, significant differences are observed between the breeding diet and both the maintenance and 50% diet (P = 0.008 and P = 0.026, respectively, by Tukey’s tests), but not between these two. In the PWK strain, significant differences are only observed between 50% and maintenance diets (P = 0.037, Tukey’s test, see text for discussion). Data are the result of the analysis of 75 spermatocytes per treatment and strain group, except B6 maintenance and breeding groups, each with 50 cells. Values are shown as mean ± SD. Overall, a significant interaction between strain and diet effects is observed (P = 0.005).
Diet composition can affect recombination levels in a strain-dependent manner
We wondered if common laboratory diets could have an effect on recombination and, consequently, may confound the results of recombination studies. Two chows are routinely used in our and many other animal facilities, depending on the purpose: animal maintenance or breeding (see Materials and Methods and Supplementary Table S1). The breeding diet is aimed to support gestation, lactation and growth, and has 2% more protein and 8% additional energy density than the maintenance chow, devised to promote longevity and normal body weight. In addition, phytoestrogens are present in the breeding diet (150–250 mg isoflavones/kg diet), while avoided in the maintenance one. As previously discussed, these components have been linked to epigenetic or developmental changes in the germline. Hence, we decided to test if crossover frequency could vary in mouse spermatocytes depending on the diet of choice.
Adult mice are routinely fed with maintenance diet. We separated animals of the three strains and provided them with ad libitum access to the breeding diet during 24 days, while others were kept with the maintenance diet. After the 24-day period, spermatocytes were obtained for recombination analysis by immunohistochemistry. To examine which factors are associated with recombination variability, we used a univariate generalized linear model. Our results indicate that diet significantly affects the MLH1 focus frequency (Pdiet = 0.042). We used post hoc tests to determine which diet comparisons were of particular note statistically. We observed no significant effect of the diets on MLH1 focus frequency in MOLF and PWK mice (Table 2). However, a significant increase was observed in B6 mice fed with breeding diet (24.70 ± 2.21) compared to maintenance chow (23.54 ± 1.97, P = 0.008, Table 2). Our results also indicate that strain significantly affects MLH1 frequency (P ≪ 0.0001), and there is a significant strain by diet interaction effect (P = 0.010) as well. These data therefore indicate that, in addition to genetic differences in recombination frequency, diet composition can affect crossover frequency in adult male mice in a strain-dependent manner.
Diet effects on recombination are robust to method of analysis
To test whether our findings were robust to the method of statistical analysis, we analyzed these data in aggregate. That is, we used a generalized linear model on data from control, maintenance and calorie restricted diets to test for effects of strain, diet, animal, and any interaction effects. Our results indicate that our findings are robust to statistical approach, with a strong effect of strain (P ≪ 0.0001) and a modest but significant effect of diet (P = 0.01). We also find an effect of animal (P < 0.001). This model accounts for 62% of phenotypic variance in recombination rate, with the bulk of the variance being accounted for by between strain variations. This is consistent with previous work (e.g., Dumont and Payseur 2011a). Importantly, the proportion of the variance due to within-animal sampling is approximately 6%, which indicates in part the consistency of our approach for scoring MLH1 foci.
Diet effects on recombination levels in C57BL/6 mice are reproducible
We were surprised to find that common chows could affect crossover frequency in B6 mice, while not in the two other strains. In order to find if this was a fortuitous or a consistent observation, we designed a larger experiment (see Materials and Methods). Animals were also subject to maintenance or breeding diet for 24 days and spermatocytes were prepared for analysis immediately after. As shown in Table 3 and Figure 3a, B6 mice kept in maintenance diet had similar MLH1 foci number (23.50 ± 2.17) to those previously observed, and the breeding diet also elicited a significant increase in crossover frequency (24.24 ± 2.31, two-sided Student t-test, P = 0.001). This increase could not be explained by significant changes in SC total length (Table 3).
Table 3.
Diet effects on crossover frequency in C57BL/6 male mice: analyses per spermatocyte and per bivalent
| (A) B6 spermatocytes: | Maintenance diet | Breeding diet |
|---|---|---|
| MLH1 foci | 23.50 ± 2.17a | 24.24 ± 2.31b |
| SC length (μm) | 169.0 ± 21.2 | 164.6 ± 17.9 |
| (B) Number and type of bivalents: | ||
| E0 | 19 (0.5%) | 16 (0.5%) |
| E1 | 2808 (75.4%) | 2417 (71.8%) |
| E2 | 895 (24.0%) | 927 (27.5%) |
| E3 | 2 (0.1%) | 5 (0.1%) |
(A) After analyzing an independent group of B6 mice, a significant increase in MLH1 foci count per spermatocyte was again observed in animals fed with breeding diet respect to those kept in maintenance diet (ab, t-student test, P = 0.001), while no significant differences in SC length were detected (P = 0.20, Mann–Whitney U test). Data are the result of the analysis of 196 spermatocytes in the maintenance group and 177 in the breeding group. (B) When bivalents were classified according to the number of crossovers (E0–E3), a significant change was also observed (P = 0.004, χ2 test). This change was mainly due to an increase of E2 bivalents at the expense of E1 in the breeding group, respect to the maintenance group.
Figure 3.
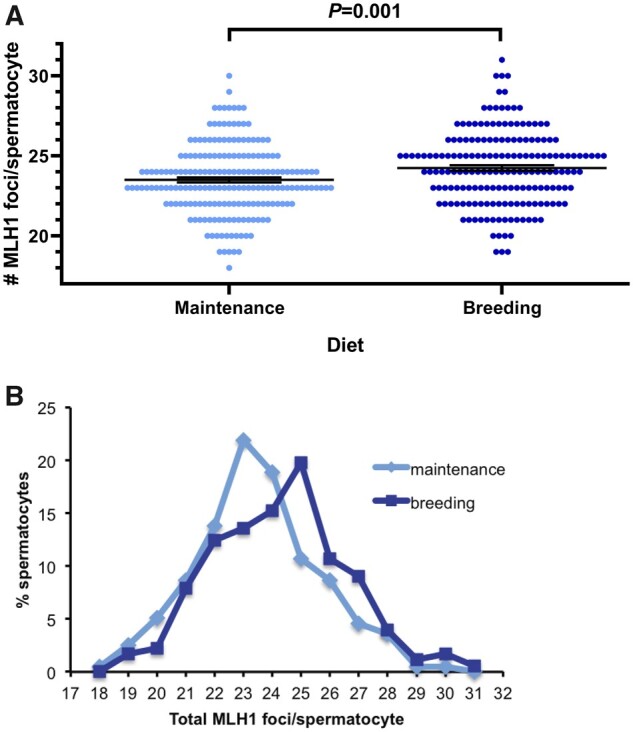
Diet effects on recombination rates. (A) Diet effects on recombination rates are strain-dependent: only C57BL/6 mice show significant differences in recombination rates when fed with maintenance vs breeding diets in two independent experiments (here are depicted the results of the second one, see Table 3). Each dot represents the focus count of a single nucleus. Black bars represent means ± SEM. (B) Spermatocyte distribution according to the total number of MLH1 foci per nucleus in C57BL/6 males subject to maintenance and breeding diets.
The studies of BPA effects on recombination stemmed from the observation of an increment in chromosome missegregation and aneuploidy caused by this endocrine disruptor (Hunt et al. 2003). In mouse as in other species, proper chromosome segregation requires a minimum of one crossover per chromosome; otherwise, crossover failure often results in aneuploidy (Hassold and Hunt 2001). Hence, we decided to explore if the observed diet effect on recombination had any impact on the frequency of nonrecombinant bivalents (E0). We compared the number of bivalents with zero, one, two or three MLH1 foci (E0, E1, E2, and E3, respectively) of each diet group. In both cases, 95% of the spermatocytes had between 20 and 28 MLH1 foci (Figure 3, A and B) mainly located in E1 and E2 bivalents; those with no crossovers were very rare, as were bivalents with three crossovers, representing 0.5% or less each (Table 3). A significant change was observed between the two diet groups, mainly due to an increase of E2 bivalents at the expense of E1 in animals fed with breeding diet respect to those kept in maintenance diet (χ2 = 13.2; P = 0.004, Table 3 and Figure 3B). Therefore, we conclude that, compared to the maintenance diet, the breeding chow induces an increase in MLH1 frequency through a shift of E1 to E2 bivalents, without substantial changes in E0 and the associated risk of aneuploidy.
Diet effects on C57BL/6 spermatogenesis: sperm motility, but not other phenotypes, are affected by diet
We wondered if recombination per se was particularly sensitive to dietary changes, or if these were a byproduct of large spermatogenesis disturbances caused by the switch between common diets. Hence, we explored if diet could also affect other aspects of spermatogenesis that resulted in changes in the quality and number of sperm, as observed in several diet and estrogenic exposure studies (Spearow et al. 1999; Assinder et al. 2007; Horan et al. 2017; Meena et al. 2017; Horan et al. 2018; Nassan et al. 2018; Nätt et al. 2019). The breeding diet is more caloric than the maintenance diet (Supplementary Table S1) and, accordingly, we observed a slight but not significant increase in body and testis weight in animals fed with breeding chow respect to those kept in maintenance diet (Supplementary Table S2). This small difference disappears when testis weight is corrected for body weight (Supplementary Table S2). We examined if testes histology was affected by diet, as observed in animals treated with natural or synthetic estrogens (Spearow et al. 1999; Horan et al. 2017). We did not observe noticeable changes in the number and cell composition of seminiferous tubules between the two diet groups (Supplementary Figure S3). When sperm isolated from the cauda epididymis was analyzed, no differences were detected in sperm count between the two diet groups. Sperm viability was neither significantly affected (Supplementary Table S2).
Sperm acquires progressive motility in the epididymis, characterized by high velocities and symmetrical, low-amplitude flagellar bends. By computer-assisted sperm analysis, we assessed several parameters of epididymal sperm motility: percentage of total motility and of progressively motile spermatozoa, VAP, VCL and VSL, LIN, STR, WOB, ALH, and BCF (see Materials and Methods; Boyers et al. 1989; Mortimer 1997). None of them showed significant differences between the two diet groups, except the proportion of sperm with progressive motility, which was significantly reduced in the breeding diet (18.8 ± 2.0) respect to the maintenance diet (25.8 ± 2.4, mean ± SE; P = 0.038; Supplementary Table S2). Though not significantly, sperm velocity also appeared to decrease in the breeding diet group (measured as VAP, VCL and VSL). The proportion of motile sperm has been associated with fertilization success (Davis et al. 1991).
In addition, we evaluated whether diet had an effect on sperm DNA integrity by SCSA (Evenson et al. 1980). We analyzed both the DFI (average and total, see Materials and Methods), as well as the HDS; the latter offers a measure of the condensation degree of the sperm chromatin and the percentage of immature sperm cells, because this high stainability is considered to be the result of a lack of full protamination and, thus, an increased histone retention (Evenson et al. 2000). None of these parameters showed significant differences between sperm of mice fed with maintenance vs breeding diets (Supplementary Table S2). Therefore, these diets had no significant effect on sperm DNA damage or condensation, as measured by SCSA. Only sperm motility was significantly affected by diet of all the phenotypes assayed in sperm and testes.
Discussion
Sperm quality has declined over the last decades among healthy men (Carlsen et al. 1992; Splingart et al. 2012; Levine et al. 2017; Sengupta et al. 2017). This decline has been associated to chemical exposures and lifestyle changes, including diets and the increase of diet-related diseases such as obesity (Nordkap et al. 2012; Nassan et al. 2018). Changes in sperm count and motility have a direct impact on fertility. But alterations in other underrated sperm features, such as meiotic recombination, are also observed in infertile men (Ferguson et al. 2009; Ren et al. 2016) and can increase the risk of chromosome missegregation and aneuploidy (Hassold and Hunt 2001). Hence, fertility requires a tight control of recombination (Coop and Przeworski 2007; Cole et al. 2012).
In spite of this control, several studies have shown that certain environmental factors are able to modify the recombination rate (Plough 1917; Belyaev and Borodin 1982; Susiarjo et al. 2007; Bomblies et al. 2015; Vrooman et al. 2015; Lloyd et al. 2018; Singh 2019). Even if these recombination changes may not be large enough to compromise fertility, they could have important consequences in the transmission and evolution of traits, as well as in genetic mapping studies (Pardo-Manuel de Villena et al. 2000; Dumont and Payseur 2008; Krzywinska et al. 2016; Ritz et al. 2017). To date, only one analysis in flies has reported an effect of nutrition on crossover rate (Mostoufi and Singh 2021; Neel 1941), an effect that has also been suggested in yeast (Abdullah and Borts 2001). Our study was aimed to explore whether diet could not only affect sperm features, but also recombination in mammalian spermatocytes.
Recombination rate variation among mouse inbred strains: crossover frequency is regulated by different mechanisms
Because both crossover rate and the effect of environmental exposures on recombination depend on the genetic background (Koehler et al. 2002; Dumont and Payseur 2011b; Baier et al. 2014; Vrooman et al. 2015), we selected for our study genetically diverse strains representative of three Mus musculus subspecies. We found that crossover frequency was significantly different among them and reported, for the first time, very high values in PWK mice, only comparable to those of the PWD/PhJ strain (29.92 ± 2.51; Dumont and Payseur 2011b); 29.58 (95% CI, 28.66–30.56; Balcova et al. 2016)), also of M. m. musculus origin (Gregorova and Forejt 2000).
For a broader view of recombination variability in mouse, we expanded our analysis to characterize the crossover frequencies of the 8 founder strains of the CC and DO stock, because they capture nearly 90% of the known variation present in laboratory mice (Churchill et al. 2004; Roberts et al. 2007). Again, our results showed that PWK spermatocytes have the highest crossover frequency, while CAST/EiJ mice are at the opposite extreme as previously described (Baier et al. 2014). Our observations confirm the importance of the genetic background on the levels of recombination (Koehler et al. 2002; Dumont and Payseur 2011b; Baier et al. 2014; Liu et al. 2014; Dumont 2017), and provide new data about the relative crossover rate of the CC and DO founder strains, which are relevant for future mapping and recombination studies in mice.
These results also revealed that the three strains selected for our study represent the low (B6), medium (MOLF) and high (PWK) levels of recombination present in Mus musculus. Mus musculus molossinus is considered the result of the hybridization between M. m. castaneus and M. m. musculus, and while strains of these two subspecies (CAST and PWK) have the lowest and highest recombination rate of all those analyzed in this study, we and others observe an intermediate recombination rate in MOLF (Silver 1995; Peterson and Payseur 2021). We wondered if the observed increases in recombination rates could be due to reduced interference or enlarged SC length (Froenicke et al. 2002; Lynn et al. 2002; Kleckner et al. 2003; Tease and Hulten 2004; Petkov et al. 2007; Dumont and Payseur 2011a) and found that both possibilities occurred in the strains under study.
The B6 total autosomal SC length we observe is similar to that previously reported (Vranis et al. 2010). But in PWK, the higher levels of recombination and longer SC respect to the other two strains are consistent with the positive correlation between total SC length and recombination rate reported in mouse and other animals (Lynn et al. 2002; Baier et al. 2014; Ruiz-Herrera et al. 2017). Interestingly, a recent study has identified several loci that affect SC length and some of them also modulate recombination rate (Wang et al. 2019). A previous study proposed a simple linear relationship between crossover rate and total SC length, so that the ratio between SC length and the number of MLH1 foci would be almost constant in mouse spermatocytes (Lynn et al. 2002). The values observed in our B6 animals coincide with those reported in that study (6.9 μm SC/MLH1 foci; Table 1). However, lower values are observed for MOLF and PWK (6.3 μm), which have higher crossover rates. Wang et al. (2019) and others (Vranis et al. 2010) also found the length of SC per MLH1 focus varies among mouse strains and proposed that it could be a consequence of interference variation, but our PWK data suggest this ratio can change independently of interference fluctuations.
In contrast, differences in total SC length cannot explain the intermediate level of recombination found in MOLF spermatocytes. In this case, the shorter intercrossover distance suggests that, compared to B6, a weaker positive interference in MOLF spermatocytes could be the cause of their higher crossover rate. An inverse correlation between interference strength and recombination rate has also been observed in other mammals (Segura et al. 2013) and a locus that affects both interference and recombination levels has been identified in cattle (Wang et al. 2016). Therefore, our results suggest that diverse and, at least to some extent, independent mechanisms determine the breadth of recombination levels present in mice.
Recombination rate is both sensitive and resistant to diets: genetic background determines crossover frequency, even under stressful nutritional conditions
Next, we explored if diets can alter recombination levels in different genetic backgrounds by choosing diets that could affect the male germline epigenome or compromise sperm function, such as undernutrition (Salian et al. 2009; Manikkam et al. 2013; Susiarjo et al. 2013; Martínez et al. 2014; Radford et al. 2014; Rahman et al. 2015; Xin et al. 2015; McPherson et al. 2016; Rahman et al. 2020). Our data suggest that, in PWK spermatocytes, the already high recombination rate increased even more when food intake was limited to 50%. Nutritional deficit causes an increase in crossover frequency in Drosophila melanogaster and Saccharomyces cerevisiae (Neel 1941; Abdullah and Borts 2001). However, we cannot conclude that a reduction in nutrients availability was directly responsible for the observed changes in recombination. Although diverse dietary restrictions have been reported to improve life span (McCay et al. 1935; Fontana et al. 2010), not all mouse strains respond the same; on the contrary, health deterioration and life shortening occur in some (Liao et al. 2010; Radford et al. 2014; Mitchell et al. 2016). Similarly, we found that while B6 and MOLF animals performed well under reduced food intake, some PWK animals suffered severe weight loss. Hence, undernutrition may have generated extreme physiological or metabolic disturbances in PWK males capable of altering the control of the recombination levels. Indeed, diverse types of stress have been reported to affect recombination rate in several organisms (Belyaev and Borodin 1982; Modliszewski and Copenhaver 2017). In studies of mouse stress and Arabidopsis temperature effects, recombination levels increase under extreme and stressful conditions, a trend consistent with our observations (Belyaev and Borodin 1982; Lloyd et al. 2018).
In contrast, recombination rate in the other two strains (MOLF and B6) was not significantly affected by 50% dietary restriction. Hence, we conclude that, in certain genetic backgrounds, recombination levels are tightly controlled even under stressful conditions such as undernutrition or, as reported in other studies, infections (Dumont et al. 2015). As many organisms, including (unfortunately) many humans, face short, seasonal, or long periods of nutrients deprivation, understanding the effects of nutritional changes on recombination, a critically meiotic important process, is important. Given how fundamental diet is to organismal fitness and function, understanding the effect to which diet-induced changes in recombination persist across generations is important as well. Moreover, given that the effects of diet are genotype-specific, more work is needed to comprehend the genetic basis of this interaction.
Common diets can affect male recombination rate in a strain-dependent manner: recombination in mice is more sensitive to environmental exposures than previously expected
We decided to test whether not just toxicants or stressful exposures, but also small differences between common diets can alter crossover frequencies in adult male mice. Hence, we temporarily fed adult males of the three selected strains with the two chows routinely used in our facility. Although they differ in energy, protein, and phytoestrogen content, their compositions are not markedly dissimilar. Hence, we were surprised to find that B6 males showed higher recombination rates when fed with the breeding chow than when kept in the maintenance diet. Again, we observed that the diet effect on recombination was strain-dependent, but now affected to a different strain than undernutrition (PWK), demonstrating the importance of genetic differences in the variable response to diverse diets.
We were also surprised to find that B6, a strain that is insensitive to the effect BPA and other estrogenic substances on male recombination, was precisely the one responsive to our diet differences (Vrooman et al. 2015). However, those results were also unexpected in view of the high estrogenic sensitivity of B6 testes (Spearow et al. 1999) and previous results reporting a B6 female recombination response to BPA (Susiarjo et al. 2007). Moreover, our results (increased recombination in the breeding diet group, which contains phytoestrogens) were in the opposite direction to the reduced crossover frequency observed in CD-1 mouse males after exposure to synthetic estrogenic substances (Vrooman et al. 2015). Hence, we decided to test whether our observation was a spurious result by providing the same dietary regime to an independent and larger group of B6 adult mice. Our results confirmed that crossover frequency is sensitive to small and apparently healthy diet changes.
These results have important implications, especially for recombination studies, although the chows selected for our study are just a small example of the composition variability found among common rodent diets (Ruhlen et al. 2011). We also wondered if the diet-induced recombination changes could also affect meiotic chromosome segregation and aneuploidy studies (Hassold and Hunt 2001; Hunt et al. 2003). Unlike BPA exposures, our results predict that aneuploidy rate and subsequent fertility of B6 mice should not be affected by changes in composition between common chows because the frequency of achiasmate chromosomes remained very low; however, we cannot exclude effects with other diets or genetic backgrounds.
We wondered which mechanisms could explain this diet-induced recombination rate change and if this may be associated to total SC length or interference variation, as we observed for strain-dependent diversity in crossover frequencies. Unfortunately, we were unable to discriminate if interference was affected by diet. While the diet effect on recombination we detect, though reproducible, is small, intercrossover distances are quite variable; consequently, interference analyses by this method are only possible when relatively large effects on recombination are examined, as those observed by strain effects in our study or by mutations in other reports (Roig et al. 2010). But we could analyze if the observed diet effect on recombination was associated to total SC length variation, as described in recombination changes induced by environmental exposures such as temperature in plants (Phillips et al. 2015; Modliszewski and Copenhaver 2017; Lloyd et al. 2018). However, that was not our case. Similarly, Vrooman et al. (2015) did not detect SC length changes associated to crossover frequency variation caused by estrogenic substances in adult mice, neither SC length could explain all the temperature effects on recombination in Arabidopsis (Lloyd et al. 2018).
Recombination can be particularly sensitive to dietary changes
In view of the results, a question emerges: are other aspects of meiosis or spermatogenesis affected? To provide an answer, we examined the testes and sperm of the same mice studied for recombination.
Previous studies had reported changes in spermatogenesis progression, sperm count or motility caused by diets differing in fat, protein, or phytoestrogen contents among others (Assinder et al. 2007; Eustache et al. 2009; Cederroth et al. 2010; Tavares et al. 2016; Matuszewska et al. 2020; Morgan et al. 2020). Our histological analysis of the testes did not reveal any apparent changes in the morphology or cell content of the seminiferous tubules suggestive of spermatogenesis alterations caused by diet. Epididymal sperm count, DNA fragmentation and viability were not significantly affected either, as they were not most of the sperm kinematic parameters analyzed. But interestingly, the percentage of progressively motile spermatozoa decreased after mice transferring to breeding diet, suggesting this diet might be optimal during pregnancy or nursing, but not for male fertility (Davis et al. 1991). Our results are in agreement with those of Nätt et al. (2019) and others (Assinder et al. 2007; Nassan et al. 2018; Salas-Huetos et al. 2018), suggesting sperm is capable of rapidly responding to diet, even to small changes.
Indeed, the composition differences between the chows under our study are relatively small compared with those analyzed in studies of high-fat or low-protein diets effects in testes (Crisostomo et al. 2019; Matuszewska et al. 2020; Morgan et al. 2020). However, they are comparable to other diet studies such as the one performed by Assinder et al. (2007), who observed changes in testes of adult Wistar rats fed during 24 days with low- or high-phytoestrogen diets (112 and 465 µg/g, respectively, compared to the 150–250 µg/g of the breeding diet object of our study). This study suggested that particular components of common diets could have an impact on spermatogenesis and sperm quality.
Our findings raise the question as to whether the two observed diet-induced effects (on recombination levels in pachytene spermatocytes and on epidydimal sperm motility) are related or not and caused by the same of by different dietary components. BPA studies have revealed effects not only on recombination but also on sperm motility and at multiple stages of spermatogenesis (Tiwari and Vanage 2013; Rahman et al. 2015; Vrooman et al. 2015). Though the relation between the different effects is unclear, our results show that the switch between common chows does not cause major disturbances in spermatogenesis that could also perturb recombination. On the contrary, recombination can be particularly sensitive to dietary changes.
Open questions and implications for recombination studies
What is the mechanism that links diet with recombination? It is tempting to speculate that epigenetic changes, such as histone modifications and DNA methylation, could be involved in the diet effect on recombination rate, because crossover frequency and distribution depend on the chromatin architecture and epigenetic marks of the chromosomes (de la Casa-Esperon and Sapienza 2003; Kleckner et al. 2003; Buard et al. 2009; Termolino et al. 2016; Zelkowski et al. 2019). Germline epigenetic modifications have been found to be susceptible to dietary changes (e.g., in energy, protein or phytoestrogen content) and to environmental exposures capable of affecting recombination (e.g., BPA, atrazine) (Manikkam et al. 2013; Xin et al. 2015; Gely-Pernot et al. 2017; Modliszewski and Copenhaver 2017). Indeed, it has been proposed that recombination rate could vary as a consequence of the germline epigenetic response to environmental exposures (Modliszewski and Copenhaver 2017). Epigenetic modifications may also be the underlying cause of the sperm motility differences observed between the two diet groups, as other diets have been reported to elicit both sperm epigenome and motility changes (Siddeek et al. 2018; Nätt et al. 2019).
Although the SCSA results did not suggest large sperm chromatin alterations due to diets, this is not surprising in view of the moderate differences between the diets under study and the reproductive success of the animals fed with them in facilities throughout the world. Moreover, diet-induced epigenetic changes in the male germline have been shown to be heterogeneous among studies (Sharma and Rando 2017; Donkin and Barres 2018; Siddeek et al. 2018); for instance, the nature of the epigenetic marks that result in transgenerational inheritance has been questioned, as they are either of small magnitude, variable sort or even undetectable in some generations (Shea et al. 2015; Xue et al. 2016; Sharma and Rando 2017). Hence, although the sperm epigenome has been proposed as a marker for environmental exposures, the analyses often turn out to be very complicated (Siddeek et al. 2018). In contrast, our results show that crossover rate is sensitive not only to disrupting toxicants (Horan et al. 2018) but also to small changes in diet and could potentially be used as an indicator of environmentally induced perturbations in the germline.
Our results also show that this recombination sensitivity depends on the genetic background, which is also true for many other responses to diverse exposures, including diets (Spearow et al. 1999; Thigpen et al. 2007; Vrooman et al. 2015; Latchney et al. 2018). For this reason, studies about the effects of environmental factors must explore their impact in genetically diverse strains, such as the founders of the CC and DO mice. Disparate results can also result from variability in the doses, timing and duration of exposure, among others. For instance, the effects of BPA on recombination are developmental stage, sex and strain dependent (Susiarjo et al. 2007; Vrooman et al. 2015). We now add a further factor to control in recombination studies: diet.
Future studies will determine which component of the chows (or combination of them) is responsible for the observed changes in recombination, as well as the effective doses. Phytoestrogens are attractive candidates, as they can elicit germline epigenetic as well as sperm motility changes, have estrogenic properties like BPA and can even modulate the effects of this compound in mice (Atanassova et al. 2000; Dolinoy et al. 2007; Muhlhauser et al. 2009; Guerrero-Bosagna and Skinner 2014; Patisaul 2017). But changes in energy content or even minor components of the diets have also shown to affect the germline and constitute interesting candidates (Ruhlen et al. 2011; Ideraabdullah and Zeisel 2018; Nassan et al. 2018; Salas-Huetos et al. 2018; Siddeek et al. 2018; Crisostomo et al. 2019).
Finally, while BPA and other estrogenic compounds affect recombination when provided to female embryos or neonatal males (Susiarjo et al. 2007; Vrooman et al. 2015; Gely-Pernot et al. 2017), our results demonstrate that recombination in adult male mice is sensitive to diet influences. It will be interesting to explore whether earlier developmental stages, particularly those in which the germline epigenetic reprogramming takes place and are particularly vulnerable to exposures such as endocrine disruptors (McCarrey 2014; Ly et al. 2015), as well as female recombination, are also susceptible to diet effects.
In conclusion, our study in mice shows that male recombination rate is sensitive to dietary changes, and this sensitivity depends on the genetic background. This is the first report of a diet effect on genome-wide levels of recombination. Our results send a cautionary note for recombination studies, as diet constitutes a new factor that should be taken into account.
Data availability
Data have been deposited in Dryad (doi:10.5061/dryad.cfxpnvx6d).
Supplementary material is available at GENETICS online.
Supplementary Material
Acknowledgments
We would like to thank Dolores García Olmo and Isabel Blanco Gutierrez for donating animals and materials. We acknowledge Julia Maria Samos Juarez for advice with the diets experimental design, as well as the Albacete UCLM Animal Experimentation Center staff for mouse monitoring and feeding according to the experimental procedure; this was possible thanks to the UCLM Vicerrectorado de Investigacion and J. Julian Garde. We are grateful to Jose Ramon Marin Tebar for capturing the microscope images and to Joaquim Soriano Felipe for support with SC automated analysis. We express our gratitude to the Biobank of Albacete for processing the testicular tissue samples. We also would like to thank Matthieu Falque and Olivier C. Martin for discussions about chromosome interference. We gratefully acknowledge Harry Sedgwick, Beth Dumont, and David Threadgill for their assistance with the analysis of the CC mice.
Funding
E.d.l.C.-E. received financial support through the program “Plan Propio de Investigacion” of the University of Castilla-La Mancha (2018/11744), cofunded by the European Regional Development Fund (FEDER, UE).
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Abdullah MF, Borts RH. 2001. Meiotic recombination frequencies are affected by nutritional states in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 98:14524–14529. doi:10.1073/pnas.201529598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed EA, de Rooij DG. 2009. Staging of mouse seminiferous tubule cross-sections. Methods Mol Biol. 558:263–277. doi:10.1007/978-1-60761-103-5_16. [DOI] [PubMed] [Google Scholar]
- Alonso-Magdalena P, Ropero AB, Soriano S, Garcia-Arevalo M, Ripoll C, et al. 2012. Bisphenol-A acts as a potent estrogen via non-classical estrogen triggered pathways. Mol Cell Endocrinol. 355:201–207. doi:10.1016/j.mce.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Reeves A, Webb LM, Ashley T. 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 151:1569–1579. doi:10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LM, Riffle L, Wilson R, Travlos GS, Lubomirski MS, et al. 2006. Preconceptional fasting of fathers alters serum glucose in offspring of mice. Nutrition. 22:327–331. doi:10.1016/j.nut.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Ashley T, Westphal C, Plug-de Maggio A, de Rooij DG. 2004. The mammalian mid-pachytene checkpoint: meiotic arrest in spermatocytes with a mutation in Atm alone or in combination with a Trp53 (p53) or Cdkn1a (p21/cip1) mutation. Cytogenet Genome Res. 107:256–262. doi:10.1159/000080603. [DOI] [PubMed] [Google Scholar]
- Assinder S, Davis R, Fenwick M, Glover A. 2007. Adult-only exposure of male rats to a diet of high phytoestrogen content increases apoptosis of meiotic and post-meiotic germ cells. Reproduction. 133:11–19. doi:10.1530/rep.1.01211. [DOI] [PubMed] [Google Scholar]
- Atanassova N, McKinnell C, Turner KJ, Walker M, Fisher JS, et al. 2000. Comparative effects of neonatal exposure of male rats to potent and weak (environmental) estrogens on spermatogenesis at puberty and the relationship to adult testis size and fertility: evidence for stimulatory effects of low estrogen levels. Endocrinology. 141:3898–3907. doi:10.1210/endo.141.10.7723. [DOI] [PubMed] [Google Scholar]
- Bachmanov AA, Reed DR, Beauchamp GK, Tordoff MG. 2002. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav Genet. 32:435–443. doi:10.1023/a:1020884312053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier B, Hunt P, Broman KW, Hassold T. 2014. Variation in genome-wide levels of meiotic recombination is established at the onset of prophase in mammalian males. PLoS Genet. 10:e1004125.doi:10.1371/journal.pgen.1004125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcova M, Faltusova B, Gergelits V, Bhattacharyya T, Mihola O, et al. 2016. Hybrid sterility locus on chromosome X controls meiotic recombination rate in mouse. PLoS Genet. 12:e1005906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, et al. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 327:836–840. doi:10.1038/nrg3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaev DK, Borodin PM. 1982. The influence of stress on variation and its role in evolution. Biol Zentral. 100:705–714. [Google Scholar]
- Bomblies K, Higgins JD, Yant L. 2015. Meiosis evolves: adaptation to external and internal environments. New Phytol. 208:306–323. doi:10.1111/nph.13499. [DOI] [PubMed] [Google Scholar]
- Borg CL, Wolski KM, Gibbs GM, O'Bryan MK. 2010. Phenotyping male infertility in the mouse: how to get the most out of a ‘non-performer’. Hum Reprod Update. 16:205–224. doi:10.1093/humupd/dmp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyers SP, Davis RO, Katz DF. 1989. Automated semen analysis. Curr Probl Obstet Gynecol Fertil. 12:165–200. [Google Scholar]
- Brieño-Enriquez MA, Robles P, Camats-Tarruella N, Garcia-Cruz R, Roig I, et al. 2011. Human meiotic progression and recombination are affected by Bisphenol A exposure during in vitro human oocyte development. Hum Reprod. 26:2807–2818. doi:10.1093/humrep/der249. [DOI] [PubMed] [Google Scholar]
- Buard J, Barthes P, Grey C, de Massy B. 2009. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 28:2616–2624. doi:10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen E, Giwercman A, Keiding N, Skakkebaek NE. 1992. Evidence for decreasing quality of semen during past 50 years. BMJ. 305:609–613. doi:10.1136/bmj.305.6854.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederroth CR, Zimmermann C, Beny JL, Schaad O, Combepine C, et al. 2010. Potential detrimental effects of a phytoestrogen-rich diet on male fertility in mice. Mol Cell Endocrinol. 321:152–160. doi:10.1016/j.mce.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, et al. 2008. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 19:382–389. doi:10.1007/s00335-008–9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, et al. ; Complex Trait Consortium. 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 36:1133–1137. doi:10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Cole F, Kauppi L, Lange J, Roig I, Wang R, et al. 2012. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol. 14:424–430. doi:10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Cross Consortium. 2012. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 190:389–401. doi:10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M. 2007. An evolutionary view of human recombination. Nat Rev Genet. 8:23–34. doi:10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- Crisostomo L, Rato L, Jarak I, Silva BM, Raposo JF, et al. 2019. A switch from high-fat to normal diet does not restore sperm quality but prevents metabolic syndrome. Reproduction. 158:377–387. doi:10.1530/REP-19-0259. [DOI] [PubMed] [Google Scholar]
- Davis RO, Overstreet JW, Asch RH, Ord T, Silber SJ. 1991. Movement characteristics of human epididymal sperm used for fertilization of human oocytes in vitro. Fertil Steril. 56:1128–1135. doi:10.1016/s0015-0282(16)54728-1. [DOI] [PubMed] [Google Scholar]
- de Boer E, Lhuissier FG, Heyting C. 2009. Cytological analysis of interference in mouse meiosis. Methods Mol Biol. 558:355–382. doi:10.1007/978-1-60761-103-5_21. [DOI] [PubMed] [Google Scholar]
- de la Casa-Esperon E. 2012. Nonmammalian parent-of-origin effects. Methods Mol Biol. 925:277–294. doi:10.1007/978-1-62703-011-3_19. [DOI] [PubMed] [Google Scholar]
- de la Casa-Esperon E, Loredo-Osti JC, Pardo-Manuel de Villena F, Briscoe TL, Malette JM, et al. 2002. X chromosome effect on maternal recombination and meiotic drive in the mouse. Genetics. 161:1651–1659. doi:10.1093/genetics/161.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Casa-Esperon E, Sapienza C. 2003. Natural selection and the evolution of genome imprinting. Annu Rev Genet. 37:349–370. doi:10.1146/annurev.genet.37.110801.143741. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 104:13056–13061. doi:10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkin I, Barres R. 2018. Sperm epigenetics and influence of environmental factors. Mol Metab. 14:1–11. doi:10.1016/j.molmet.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL. 2017. Variation and evolution of the meiotic requirement for crossing over in mammals. Genetics. 205:155–168. doi:10.1534/genetics.116.192690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Devlin AA, Truempy DM, Miller JC, Singh ND. 2015. No evidence that infection alters global recombination rate in house mice. PLoS One. 10:e0142266.doi:10.1371/journal.pone.0142266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. 2008. Evolution of the genomic rate of recombination in mammals. Evolution. 62:276–294. doi:10.1111/j.1558–5646.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. 2011a. Evolution of the genomic recombination rate in murid rodents. Genetics. 187:643–657. doi:10.1534/genetics.110.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. 2011b. Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genet. 7:e1002116.doi:10.1371/journal.pgen.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustache F, Mondon F, Canivenc-Lavier MC, Lesaffre C, Fulla Y, et al. 2009. Chronic dietary exposure to a low-dose mixture of genistein and vinclozolin modifies the reproductive axis, testis transcriptome, and fertility. Environ Health Perspect. 117:1272–1279. doi:10.1289/ehp. 0800158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenson DP, Darzynkiewicz Z, Melamed MR. 1980. Relation of mammalian sperm chromatin heterogeneity to fertility. Science. 210:1131–1133. doi:10.1126/science.7444440. [DOI] [PubMed] [Google Scholar]
- Evenson DP, Jost LK, Corzett M, Balhorn R. 2000. Characteristics of human sperm chromatin structure following an episode of influenza and high fever: a case study. J Androl. 21:739–746. doi:10.1002/j.1939–4640.2000.tb02142.x. [PubMed] [Google Scholar]
- Ferguson KA, Leung S, Jiang D, Ma S. 2009. Distribution of MLH1 foci and inter-focal distances in spermatocytes of infertile men. Hum Reprod. 24:1313–1321. doi:10.1093/humrep/dep021. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. 2010. Extending healthy life span–from yeast to humans. Science. 328:321–326. doi:10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froenicke L, Anderson LK, Wienberg J, Ashley T. 2002. Male mouse recombination maps for each autosome identified by chromosome painting. Am J Hum Genet. 71:1353–1368. doi:10.1086/344714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gely-Pernot A, Saci S, Kernanec PY, Hao C, Giton F, et al. 2017. Embryonic exposure to the widely-used herbicide atrazine disrupts meiosis and normal follicle formation in female mice. Sci Rep. 7:3526.doi:10.1038/s41598-017–03738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorova S, Forejt J. 2000. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies—a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol (Praha ) 46:31–41. [PubMed] [Google Scholar]
- Guerrero-Bosagna CM, Skinner MK. 2014. Environmental epigenetics and phytoestrogen/phytochemical exposures. J Steroid Biochem Mol Biol. 139:270–276. doi:10.1016/j.jsbmb.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2:280–291. doi:10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Horan TS, Marre A, Hassold T, Lawson C, Hunt PA. 2017. Germline and reproductive tract effects intensify in male mice with successive generations of estrogenic exposure. PLoS Genet. 13:e1006885.doi:10.1371/journal.pgen.1006885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan TS, Pulcastro H, Lawson C, Gerona R, Martin S, et al. 2018. Replacement bisphenols adversely affect mouse gametogenesis with consequences for subsequent generations. Curr Biol. 28:2948–2954.e3. doi:10.1016/j.cub.2018.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PA, Koehler KE, Susiarjo M, Hodges CA, Ilagan A, et al. 2003. Bisphenol A exposure causes meiotic aneuploidy in the female mouse. Curr Biol. 13:546–553. doi:10.1016/s0960-9822(03)00189-1. [DOI] [PubMed] [Google Scholar]
- Hunter N. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. a016618.doi:10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideraabdullah FY, Zeisel SH. 2018. Dietary modulation of the epigenome. Physiol Rev. 98:667–695. doi:10.1152/physrev.00010.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. 2006. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 115:175–194. doi:10.1007/s00412-006–0055-7. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Storlazzi A, Zickler D. 2003. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet. 19:623–628. doi:10.1016/j.tig.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Koehler KE, Cherry JP, Lynn A, Hunt PA, Hassold TJ. 2002. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics. 162:297–306. doi:10.1093/genetics/162.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinska E, Kokoza V, Morris M, de la Casa-Esperon E, Raikhel AS, et al. 2016. The sex locus is tightly linked to factors conferring sex-specific lethal effects in the mosquito Aedes aegypti. Heredity (Edinb). 117:408–416. doi:10.1038/hdy.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchney SE, Fields AM, Susiarjo M. 2018. Linking inter-individual variability to endocrine disruptors: insights for epigenetic inheritance. Mamm Genome. 29:141–152. doi:10.1007/s00335-017–9729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie NM, Tease C, Hulten MA. 1995. Chiasma frequency, distribution and interference maps of mouse autosomes. Chromosoma. 104:308–314. doi:10.1007/BF00352262. [DOI] [PubMed] [Google Scholar]
- Levine H, Jorgensen N, Martino-Andrade A, Mendiola J, Weksler-Derri D, et al. 2017. Temporal trends in sperm count: a systematic review and meta-regression analysis. Hum Reprod Update. 23:646–659. doi:10.1093/humupd/dmx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. 2010. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. 9:92–95. doi:10.1111/j.1474-9726.2009.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Duan W, Li R, Xu S, Zhang L, et al. 2013. Exposure to bisphenol A disrupts meiotic progression during spermatogenesis in adult rats through estrogen-like activity. Cell Death Dis. 4:e676.doi:10.1038/cddis.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, Morgan AP, Chesler EJ, Wang W, Churchill GA, et al. 2014. High-resolution sex-specific linkage maps of the mouse reveal polarized distribution of crossovers in male germline. Genetics. 197:91–106. doi:10.1534/genetics.114.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Morgan C, H Franklin FC, Bomblies K. 2018. Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics. 208:1409–1420. doi:10.1534/genetics.117.300588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly L, Chan D, Trasler JM. 2015. Developmental windows of susceptibility for epigenetic inheritance through the male germline. Semin Cell Dev Biol. 43:96–105. doi:10.1016/j.semcdb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Lynn A, Koehler KE, Judis LAnn, Chan ER, Cherry JP, et al. 2002. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science. 296:2222–2225. doi:10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. 2013. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One. 8:e55387.doi:10.1371/journal.pone.0055387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez D, Pentinat T, Ribó S, Daviaud C, Bloks VW, et al. 2014. In utero undernutrition in male mice programs liver lipid metabolism in the second-generation offspring involving altered Lxra DNA methylation. Cell Metab. 19:941–951. doi:10.1016/j.cmet.2014.03.026. [DOI] [PubMed] [Google Scholar]
- Mather K. 1937. The determination of position in crossing-over. II. The chromosome length-chiasma frequency relation. Cytologia. 1:514–526. [Google Scholar]
- Matuszewska J, Ziarniak K, Dudek M, Kołodziejski P, Pruszyńska-Oszmałek E, et al. 2020. Effects of short-term exposure to high-fat diet on histology of male and female gonads in rats. Acta Histochem. 122:151558.doi:10.1016/j.acthis.2020.151558. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. 2014. Distinctions between transgenerational and non-transgenerational epimutations. Mol Cell Endocrinol. 398:13–23. doi:10.1016/j.mce.2014.07.016. [DOI] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, Maynard LA. 1935. The effect of retarded growth upon the length of life span and upon the ultimate body size. J Nutr. 10:63–79. doi:10.1093/jn/10.1.63. [PubMed] [Google Scholar]
- McPherson NO, Fullston T, Kang WX, Sandeman LY, Corbett MA, et al. 2016. Paternal under-nutrition programs metabolic syndrome in offspring which can be reversed by antioxidant/vitamin food fortification in fathers. Sci Rep. 6:27010. doi:10.1038/srep27010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena R, Supriya C, Pratap Reddy K, Reddy PS. 2017. Altered spermatogenesis, steroidogenesis and suppressed fertility in adult male rats exposed to genistein, a non-steroidal phytoestrogen during embryonic development. Food Chem Toxicol. 99:70–77. doi:10.1016/j.fct.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Milano CR, Holloway JK, Zhang Y, Jin B, Smith C, et al. 2019. Mutation of the ATPase domain of MutS homolog-5 (MSH5) reveals a requirement for a functional MutSγ complex for all crossovers in mammalian meiosis. G3 (Bethesda). 9:1839–1850. doi:10.1534/g3.119.400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, et al. 2016. Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 23:1093–1112. doi:10.1016/j.cmet.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modliszewski JL, Copenhaver GP. 2017. Meiotic recombination gets stressed out: CO frequency is plastic under pressure. Curr Opin Plant Biol. 36:95–102. doi:10.1016/j.pbi.2016.11.019. [DOI] [PubMed] [Google Scholar]
- Morgan HL, Ampong I, Eid N, Rouillon C, Griffiths HR, et al. 2020. Low protein diet and methyl-donor supplements modify testicular physiology in mice. Reproduction. 159:627–641. doi:10.1530/REP-19–0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH. 1913. Heredity and sex. New York, NY: Columbia University Press. [Google Scholar]
- Mortimer ST. 1997. A critical review of the physiological importance and analysis of sperm movement in mammals. Hum Reprod Update. 3:403–439. doi:10.1093/humupd/3.5.403. [DOI] [PubMed] [Google Scholar]
- Mostoufi SL, Singh ND. 2021. Diet-induced changes in titer support a threshold effect of Wolbachia-associated plastic recombination in Drosophila melanogaster. bioRxiv. doi:10.1101/2021.03.18.436076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlhauser A, Susiarjo M, Rubio C, Griswold J, Gorence G, et al. 2009. Bisphenol A effects on the growing mouse oocyte are influenced by diet. Biol Reprod. 80:1066–1071. doi:10.1095/biolreprod.108.074815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. 1916. The mechanism of crossing-over. Am Nat. 50:193–221, 284–305, 350–366, 421–434. [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, et al. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 327:876–879. doi:10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassan FL, Chavarro JE, Tanrikut C. 2018. Diet and men's fertility: does diet affect sperm quality? Fertil Steril. 110:570–577. doi:10.1016/j.fertnstert.2018.05.025. [DOI] [PubMed] [Google Scholar]
- Nätt D, Kugelberg U, Casas E, Nedstrand E, Zalavary S, et al. 2019. Human sperm displays rapid responses to diet. PLoS Biol. 17:e3000559.doi:10.1371/journal.pbio.3000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel JV. 1941. A relation between larval nutrition and the frequency of crossing over in the third chromosome of Drosophila melanogaster. Genetics. 26:506–516. doi:10.1093/genetics/26.5.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordkap L, Joensen UN, Blomberg Jensen M, Jørgensen N. 2012. Regional differences and temporal trends in male reproductive health disorders: semen quality may be a sensitive marker of environmental exposures. Mol Cell Endocrinol. 355:221–230. doi:10.1016/j.mce.2011.05.048. [DOI] [PubMed] [Google Scholar]
- Ottolini CS, Newnham L, Capalbo A, Natesan SA, Joshi HA, et al. 2015. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet. 47:727–735. doi:10.1038/ng.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, de la Casa-Esperon E, Briscoe TL, Sapienza C. 2000. A genetic test to determine the origin of maternal transmission ratio distortion. Meiotic drive at the mouse Om locus. Genetics. 154:333–342. doi:10.1093/genetics/154.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. 2001. Recombination is proportional to the number of chromosome arms in mammals. Mamm Genome. 12:318–322. doi:10.1007/s003350020005. [DOI] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science. 327:835.doi:10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patisaul HB. 2017. Endocrine disruption by dietary phyto-oestrogens: impact on dimorphic sexual systems and behaviours. Proc Nutr Soc. 76:130–144. doi:10.1017/S0029665116000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AL, Payseur BA. 2021. Sex-specific variation in the genome-wide recombination rate. Genetics. 217:1–11. doi:10.1093/genetics/iyaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Broman KW, Szatkiewicz JP, Paigen K. 2007. Crossover interference underlies sex differences in recombination rates. Trends Genet. 23:539–542. doi:10.1016/j.tig.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Phillips D, Jenkins G, Macaulay M, Nibau C, Wnetrzak J, et al. 2015. The effect of temperature on the male and female recombination landscape of barley. New Phytol. 208:421–429. doi:10.1111/nph.13548. [DOI] [PubMed] [Google Scholar]
- Plough HH. 1917. The effect of temperature on crossingover in Drosophila. J Exp Zool. 24:147–209. doi:10.1002/jez.1400240202 [Google Scholar]
- Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, et al. 2014. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 345:1255903.doi:10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Kwon WS, Lee JS, Yoon SJ, Ryu BY, et al. 2015. Bisphenol-A affects male fertility via fertility-related proteins in spermatozoa. Sci Rep. 5:9169.doi:10.1038/srep09169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Pang WK, Ryu DY, Park YJ, Pang MG. 2020. Multigenerational and transgenerational impact of paternal bisphenol A exposure on male fertility in a mouse model. Hum Reprod. 35:1640–1752. doi:10.1093/humrep/deaa139. [DOI] [PubMed] [Google Scholar]
- Ren H, Ferguson K, Kirkpatrick G, Vinning T, Chow V, et al. 2016. Altered crossover distribution and frequency in spermatocytes of infertile men with azoospermia. PLoS One. 11:e0156817.doi:10.1371/journal.pone.0156817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz KR, Noor MAF, Singh ND. 2017. Variation in recombination rate: adaptive or not? Trends Genet. 33:364–374. doi:10.1016/j.tig.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. 2007. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 18:473–481. doi:10.1007/s00335-007–9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, et al. 2010. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 6:e1001062.doi:10.1371/journal.pgen.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlen RL, Taylor JA, Mao J, Kirkpatrick W, Welshons WV, et al. 2011. Choice of animal feed can alter fetal steroid levels and mask developmental effects of endocrine disrupting chemicals. J Dev Orig Health Dis. 2:36–48. doi:10.1017/S2040174410000711. [Google Scholar]
- Ruiz-Herrera A, Vozdova M, Fernandez J, Sebestova H, Capilla L, et al. 2017. Recombination correlates with synaptonemal complex length and chromatin loop size in bovids-insights into mammalian meiotic chromosomal organization. Chromosoma. 126:615–631. doi:10.1007/s00412-016–0624-3. [DOI] [PubMed] [Google Scholar]
- Salas-Huetos A, Moraleda R, Giardina S, Anton E, Blanco J, et al. 2018. Effect of nut consumption on semen quality and functionality in healthy men consuming a Western-style diet: a randomized controlled trial. Am J Clin Nutr. 108:953–962. doi:10.1093/ajcn/nqy181. [DOI] [PubMed] [Google Scholar]
- Salian S, Doshi T, Vanage G. 2009. Perinatal exposure of rats to Bisphenol A affects the fertility of male offspring. Life Sci. 85:742–752. doi:10.1016/j.lfs.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Segura J, Ferretti L, Ramos-Onsins S, Capilla L, Farre M, et al. 2013. Evolution of recombination in eutherian mammals: insights into mechanisms that affect recombination rates and crossover interference. Proc Biol Sci. 280:20131945.doi:10.1098/rspb.2013.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Dutta S, Krajewska-Kulak E. 2017. The disappearing sperms: analysis of reports published between 1980 and 2015. Am J Mens Health. 11:1279–1304. doi:10.1177/1557988316643383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Rando OJ. 2017. Metabolic Inputs into the Epigenome. Cell Metab. 25:544–558. doi:10.1016/j.cmet.2017.02.003. [DOI] [PubMed] [Google Scholar]
- Shea JM, Serra RW, Carone BR, Shulha HP, Kucukural A, et al. 2015. Genetic and epigenetic variation, but not diet, shape the sperm methylome. Dev Cell. 35:750–758. doi:10.1016/j.devcel.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddeek B, Mauduit C, Simeoni U, Benahmed M. 2018. Sperm epigenome as a marker of environmental exposure and lifestyle, at the origin of diseases inheritance. Mutat Res Rev Mutat Res. 778:38–44. doi:10.1016/j.mrrev.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Silver LM. 1995. Mouse Genetics: Concepts and Applications. New York, USA: Oxford University Press. [Google Scholar]
- Singh ND. 2019. Wolbachia infection associated with increased recombination in Drosophila. G3 (Bethesda). 9:229–237. doi:10.1534/g3.118.200827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearow JL, Doemeny P, Sera R, Leffler R, Barkley M. 1999. Genetic variation in susceptibility to endocrine disruption by estrogen in mice. Science. 285:1259–1261. doi:10.1126/science.285.5431.1259. [DOI] [PubMed] [Google Scholar]
- Splingart C, Frapsauce C, Veau S, Barthelemy C, Royere D, et al. 2012. Semen variation in a population of fertile donors: evaluation in a French centre over a 34-year period. Int J Androl. 35:467–474. doi:10.1111/j.1365–2605.2011.01229.x. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. 1915. The behavior of the chromosomes as studied through linkage. ZVer-erbungslehre. 13:234–287. doi:10.1007/BF01792906. [Google Scholar]
- Susiarjo M, Hassold TJ, Freeman E, Hunt PA. 2007. Bisphenol A exposure in utero disrupts early oogenesis in the mouse. PLoS Genet. 3:e5.doi:10.1371/journal.pgen.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. 2013. Bisphenol A exposure disrupts genomic imprinting in the mouse. PLoS Genet. 9:e1003401.doi:10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susiarjo M, Xin F, Bansal A, Stefaniak M, Li C, et al. 2015. Bisphenol A exposure disrupts metabolic health across multiple generations in the mouse. Endocrinology. 156:2049–2058. doi:10.1210/en.2014–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, et al. 2012. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 190:437–447. doi:10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Roeder GS. 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 79:283–292. doi:10.1016/0092–8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Tavares RS, Escada-Rebelo S, Correia M, Mota PC, Ramalho-Santos J. 2016. The non-genomic effects of endocrine-disrupting chemicals on mammalian sperm. Reproduction. 151: R1–R13. doi:10.1530/REP-15-0355. [DOI] [PubMed] [Google Scholar]
- Tease C, Hulten MA. 2004. Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenet Genome Res. 107:208–215. doi:10.1159/000080599. [DOI] [PubMed] [Google Scholar]
- Termolino P, Cremona G, Consiglio MF, Conicella C. 2016. Insights into epigenetic landscape of recombination-free regions. Chromosoma. 125:301–308. doi:10.1007/s00412-016–0574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thigpen JE, Setchell KD, Padilla-Banks E, Haseman JK, Saunders HE, et al. 2007. Variations in phytoestrogen content between different mill dates of the same diet produces significant differences in the time of vaginal opening in CD-1 mice and F344 rats but not in CD Sprague-Dawley rats. Environ Health Perspect. 115:1717–1726. doi:10.1289/ehp. 10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Miller DR, Churchill GA, de Villena FP. 2011. The Collaborative Cross: a recombinant inbred mouse population for the systems genetic era. ILAR J. 52:24–31. doi:10.1093/ilar.52.1.24. [DOI] [PubMed] [Google Scholar]
- Tiwari D, Vanage G. 2013. Mutagenic effect of Bisphenol A on adult rat male germ cells and their fertility. Reprod Toxicol. 40:60–68. doi:10.1016/j.reprotox.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Vranis NM, Van der Heijden GW, Malki S, Bortvin A. 2010. Synaptonemal complex length variation in wild-type male mice. Genes (Basel)). 1:505–520. doi:10.3390/genes1030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrooman LA, Nagaoka SI, Hassold TJ, Hunt PA. 2014. Evidence for paternal age-related alterations in meiotic chromosome dynamics in the mouse. Genetics. 196:385–396. doi:10.1534/genetics.113.158782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrooman LA, Oatley JM, Griswold JE, Hassold TJ, Hunt PA. 2015. Estrogenic exposure alters the spermatogonial stem cells in the developing testis, permanently reducing crossover levels in the adult. PLoS Genet. 11:e1004949.doi:10.1371/journal.pgen.1004949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, Dumont BL, Jing P, Payseur BA. 2019. A first genetic portrait of synaptonemal complex variation. PLoS Genet. 15:e1008337.doi:10.1371/journal.pgen.1008337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Shen B, Jiang J, Li J, Ma L. 2016. Effect of sex, age and genetics on crossover interference in cattle. Sci Rep. 6:37698.doi:10.1038/srep37698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski P, Romano RM, Kizys MM, Oliveira KC, Kasamatsu T, et al. 2015. Adult exposure to bisphenol A (BPA) in Wistar rats reduces sperm quality with disruption of the hypothalamic-pituitary-testicular axis. Toxicology. 329:1–9. doi:10.1016/j.tox.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Wolstenholme JT, Goldsby JA, Rissman EF. 2013. Transgenerational effects of prenatal bisphenol A on social recognition. Horm Behav. 64:833–839. doi:10.1016/j.yhbeh.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Bu P, Li F, Lan S, Wu H, et al. 2016. Neonatal bisphenol A exposure induces meiotic arrest and apoptosis of spermatogenic cells. Oncotarget. 7:10606–10615. doi:10.18632/oncotarget.7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin F, Susiarjo M, Bartolomei MS. 2015. Multigenerational and transgenerational effects of endocrine disrupting chemicals: a role for altered epigenetic regulation? Semin Cell Dev Biol. 43:66–75. doi:10.1016/j.semcdb.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Schoenrock SA, Valdar W, Tarantino LM, Ideraabdullah FY. 2016. Maternal vitamin D depletion alters DNA methylation at imprinted loci in multiple generations. Clin Epigenet. 8. doi:10.1186/s13148-016–0276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, et al. 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 43:648–655. doi:10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazowski MJ, Sandoval M, Paniker L, Hamilton HM, Han J, et al. 2017. Age-dependent alterations in meiotic recombination cause chromosome segregation errors in spermatocytes. Cell. 171:601–614.e13. doi:10.1016/j.cell.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelkowski M, Olson MA, Wang M, Pawlowski W. 2019. Diversity and determinants of meiotic recombination landscapes. Trends Genet. 35:359–370. doi:10.1016/j.tig.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Ziv-Gal A, Wang W, Zhou C, Flaws JA. 2015. The effects of in utero bisphenol A exposure on reproductive capacity in several generations of mice. Toxicol Appl Pharmacol. 284:354–362. doi:10.1016/j.taap.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in Dryad (doi:10.5061/dryad.cfxpnvx6d).
Supplementary material is available at GENETICS online.


