Abstract
The opportunistic human fungal pathogen Cryptococcus neoformans has tremendous impact on global health, causing 181,000 deaths annually. Current treatment options are limited, and the frequent development of drug resistance exacerbates the challenge of managing invasive cryptococcal infections. In diverse fungal pathogens, the essential molecular chaperone Hsp90 governs fungal survival, drug resistance, and virulence. Therefore, targeting this chaperone has emerged as a promising approach to combat fungal infections. However, the role of Hsp90 in supporting C. neoformans pathogenesis remains largely elusive due to a lack of genetic characterization. To help dissect the functions of Hsp90 in C. neoformans, we generated a conditional expression strain in which HSP90 is under control of the copper-repressible promoter CTR4-2. Addition of copper to culture medium depleted Hsp90 transcript and protein levels in this strain, resulting in compromised fungal growth at host temperature; increased sensitivity to stressors, including the azole class of antifungals; altered C. neoformans morphology; and impaired melanin production. Finally, leveraging the fact that copper concentrations vary widely in different mouse tissues, we demonstrated attenuated virulence for the CTR4-2p-HSP90 mutant specifically in an inhalation model of Cryptococcus infection. During invasion and establishment of infection in this mouse model, the pathogen is exposed to the relatively high copper concentrations found in the lung as compared to blood. Overall, this work generates a tractable genetic system to study the role of Hsp90 in supporting the pathogenicity of C. neoformans and provides proof-of-principle that targeting Hsp90 holds great promise as a strategy to control cryptococcal infection.
Keywords: Cryptococcus, Hsp90, virulence, azole, melanin, stress tolerance, genetic tool
Introduction
Invasive human fungal pathogens impose tremendous economic and health burdens on society, in part due to rising populations of at-risk individuals with impaired immune function secondary to an expanding array of medical interventions or HIV infection (Brown et al. 2012). Despite the severe health threat posed by fungal pathogens, we only have three major classes of antifungal drugs to combat invasive infections: polyenes, azoles, and echinocandins (Robbins et al. 2017). Polyenes sequester plasma membrane ergosterol, which is the functional equivalent of cholesterol, the membrane sterol present in mammalian cells. Azoles inhibit ergosterol biosynthesis by targeting the lanosterol demethylase enzyme encoded by ERG11, and echinocandins target the biosynthesis of (1,3)-β-glucan, a basic building block of the fungal cell wall (Lee et al. 2021). Resistance, fueled by the widespread prophylactic use of antifungals both in medicine and in agriculture, has been documented to each of these drug classes in human fungal pathogens (Fisher et al. 2018). To make matters worse, limited resources and inefficient diagnostic tools are contributing factors responsible for the co-occurrence of lethal fungal infections with COVID-19 (Nargesi et al. 2021). In particular, the emergence among COVID-19 patients of deadly mucormycosis, caused by zygomycetous Mucorales sp. (Raut and Huy 2021), further highlights the threat these pathogens pose to human health.
Species of Candida, Aspergillus, and Cryptococcus represent the dominant invasive fungal pathogens responsible for the majority of deaths due to fungal infection in humans. Specifically, the basidiomycete Cryptococcus neoformans infects approximately 223,100 individuals per year, with the resulting cryptococcal meningitis responsible for 181,100 deaths annually (Rajasingham et al. 2017). C. neoformans is intrinsically resistant to echinocandins, and therefore treatment options remain limited to polyenes, azoles, and the pyrimidine analog flucytosine that targets RNA and DNA synthesis (Denning 2003; Iyer et al. 2021). Notably, due to the frequent emergence of drug resistance, flucytosine is never used as a monotherapy and is only used in combination (Day et al. 2013). Monotherapy with fluconazole also results in poor outcome relative to amphotericin B and flucytosine combination therapy due to the lack of early fungicidal activity for fluconazole resulting in the development of resistance (Bicanic et al. 2009; Hope et al. 2019). C. neoformans is also able to develop resistance to all other current antifungals by employing a plethora of genetic, genomic, and adaptive biological strategies (Iyer et al. 2021). Thus, a pressing need exists to develop novel antifungal drugs and therapeutic strategies to treat Cryptococcus infections more effectively.
A promising approach to enhance the efficacy of current antifungals is to use them in combination with inhibitors of core cellular stress responses. The molecular chaperone Hsp90 is conserved throughout eukaryotes, where it regulates the function of diverse meta-stable signal transducers involved in sensing and responding to stress (Taipale et al. 2010; O’Meara et al. 2017). Previous work has shown that targeting Hsp90 is a powerful therapeutic strategy to combat fungal disease caused by Candida and Aspergillus species (Cowen and Lindquist 2005; Cowen et al. 2009). Hsp90 inhibition abrogates antifungal drug resistance and potentiates antifungal activity through numerous mechanisms, most notably by impairing signaling through the protein phosphatase calcineurin and the Pkc1 cell wall integrity pathway (Cowen and Lindquist 2005; LaFayette et al. 2010; Singh-Babak et al. 2012; Caplan et al. 2018). Hsp90 also modulates key virulence traits in C. albicans, such as the ability to undergo morphogenesis, and in animal models, genetic compromise of Hsp90 function leads to clearance of otherwise lethal Candida or Aspergillus infections (Cowen et al. 2009; Shapiro et al. 2009). In C. neoformans, Hsp90 has been suggested to play crucial roles in controlling key virulence traits, such as thermotolerance and capsule production (Cordeiro et al. 2016; Chatterjee and Tatu 2017). However, these studies have been limited to the use of pharmacological inhibitors as a genetically tractable system for studying C. neoformans Hsp90 has yet to be generated.
In this study, we constructed a regulatable expression system for C. neoformans HSP90 utilizing the copper-suppressible promoter CTR4-2, which has been used previously to study function of the essential genes FAS1 and FAS2 in vivo (Chayakulkeeree et al. 2007). In the CTR4-2-HSP90 strain, supplementation of medium with copper suppressed both HSP90 transcript and protein levels. Using this genetically modified strain as a tool, we found that depletion of C. neoformans HSP90 caused a severe growth defect at 37°C, increased sensitivity to the azole fluconazole, conferred hypersensitivity to several environmental stresses, triggered a cytokinesis defect resulting in the formation of multinucleated elongated cells, and impaired production of melanin, a key virulence trait. Finally, we evaluated the pathogenicity of the copper-regulated HSP90 mutant in two different mouse models of infection, selected because they were expected to expose the strain to either high or low concentrations of copper during the initial phase of infection. We observed attenuation in virulence only in an inhalation model where C. neoformans is exposed to the relatively high copper concentrations found in lung tissue during invasion and establishment of infection. Overall, this study describes the generation of a greatly needed genetic tool to study Hsp90 function in C. neoformans and provides genetic proof-of-principle that targeting Hsp90 could provide a powerful therapeutic strategy for the treatment of cryptococcosis.
Materials and methods
Strains and culture conditions
Strains and plasmids used in this study are listed in Supplementary Table S1. All strains were frozen at −80°C in 25% glycerol and actively maintained on solid yeast extract peptone (YPD: 1% yeast extract, 2% bactopeptone, 2% glucose, and 2% agar) at 4°C for no longer than one month. The CTR4-2p-HSP90 mutants were grown on YPD agar medium supplemented with 100 µg/ml nourseothricin (NAT). To suppress HSP90 expression, mutants were grown in liquid YPD medium supplemented with 25 µM CuSO4 at 30°C overnight.
Construction CTR4-2p-HSP90 mutants
To generate the CTR4-2p-HSP90 mutant, 111 base pairs upstream of the first intron preceding the HSP90 start codon were replaced with a nourseothricin-resistance (NAT) cassette and the CTR4-2 promoter utilizing the TRACE method (Fan and Lin 2018). The CTR4-2p-HSP90 construct was cloned into plasmid pXL1 to yield pCTR4-2p-HSP90 utilizing the Gibson cloning method (Gibson et al. 2009). 898 bp-upstream and 979 bp-downstream of the 111 bps targeted sequence were PCR amplified using oLC8626/oLC8631 and oLC8625/oLC8623 from KN99α genomic DNA. The CTR4-2 promoter was PCR amplified using oLC8624/oLC8629 from plasmid pCTR4-2p (Ory et al. 2004). The NAT cassette was PCR amplified using oLC8630/oLC8632 from plasmid pAI3 (Idnurm et al. 2004), and the plasmid backbone was PCR amplified using oLC8633/oLC8634 from plasmid pXL1 (Hsueh et al. 2009). Overlapping PCR products shared a 20 bp-homologous sequence, and all PCR products were assembled together to generate plasmid pCTR4-2p-HSP90. The plasmid was sequenced to confirm no mutations were introduced during the cloning steps.
The guide RNA (gRNA) sequence targets 154 bp upstream of the HSP90 start codon. The U6 promoter for the gRNA cassette was PCR amplified using oLC8640/oLC8641 from JEC21α genomic DNA. The gRNA scaffold was PCR amplified using oLC8639/oLC8642 from plasmid pDD162, and the gRNA cassette was then PCR amplified using oLC8641/oLC8642 via overlap PCR. The CAS9 cassette was PCR amplified using oLC8637/oLC8638 from plasmid pXL1-HYG-CAS9 (Fan and Lin 2018).
To transform C. neoformans, pCTR4-2p-HSP90 was digested with SacI and NruI, and the DNA fragment containing the CTR4-2p-HSP90 construct was gel purified. The CTR4-2p-HSP90 construct, CAS9 cassette, and gRNA cassette were then transformed into KN99α via electroporation (Fan and Lin 2018). Transformants were selected on YPD-NAT agar medium and genotyped via PCR for the correct integration of the CTR4-2 promoter. Two PCR-validated CTR4-2p-HSP90 mutants derived from a single transformation were used in the subsequent experiments described in this manuscript. Primers used in this study are listed in Supplementary Table S2.
RNA extraction and real time-PCR
Wild-type KN99α and CTR4-2p-HSP90 strains were grown in liquid YPD and YPD supplemented with either 25 µM CuSO4 or 200 µM bathocuproinedisulphonic acid (BCS) (Sigma) at 30°C or 37°C overnight. Cells were diluted to an OD600 of 0.1 in the same liquid media and cultured under the same conditions for 5 h or until in mid-log phase. Cells were pelleted, washed once with 1X phosphate buffered saline (PBS), and flash frozen in liquid nitrogen. Cells were lysed by bead beating four times for one minute with chilling on ice for one minute in between cycles. RNA was extracted using the QIAGEN RNeasy kit and DNase treated using the QIAGEN RNase free DNase. cDNA synthesis was carried out using the iScript cDNA synthesis kit (BioRad). Real time-PCR was performed in technical triplicates using the BioRad CFX-384 Real Time System using Fast SYBR Green Master Mix (Applied Biosystems) following manufacturer’s protocol. The PCR cycle started with 95°C for 3 min followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The melting curve was performed with the following cycle conditions: 95°C for 10 s and 65°C for 10 s with an increase of 0.5°C per cycle up to 95°C. The experiment was performed in biological duplicate to confirm data reproducibility. T-test with Welch’s correction was used to determine statistical significance. Primers used are listed in Supplementary Table S2.
Protein extraction and western blot
Wild-type and CTR4-2p-HSP90 strains were cultured and prepared under the same conditions as used to extract RNA. Protein extraction and western blotting were performed as previously described with minor modifications (Hossain et al. 2020). In brief, pelleted cells were flash frozen in liquid nitrogen and lysed in 650 µl lysis buffer (0.42 M NaOH and 1.9% β-mercaptoethanol). Protein was precipitated with 150 µl 50% trichloroacetic acid (TCA), pelleted at 14,000 g for 10 min at 4°C, and washed once with 500 µl ice-cold acetone. Pellets were resuspended in 150 µl loading buffer [40 mM Tris-HCl (pH 6.8), 5% (w/v) SDS, 100 mM NaEDTA, and 8.3 M Urea], incubated at 42°C for 10 min, and spun down by centrifugation at 14,000 g for 2 min. Supernatant were collected and protein levels were determined by DC protein assay (BioRad). For each sample, 10 µg total protein and PageRuler pre-stained protein ladder (Invitrogen) were separated by SDS-PAGE using precast 15-well NuPAGETM 4-12% Bis-Tris gel (Invitrogen). Separated proteins were electro-transferred to nitrocellulose membrane for an hour at 300 mA at room temperature. Blots were blocked in 10% skim milk in TBS-T [10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% Tween-20] for 1 h at room temperature. Hsp90 was detected using a polyclonal rabbit anti-Candida Hsp90 antibody [1:5000 in 5% skim milk in TBS-T, gift from B. Larson (Burt et al. 2003)] overnight at 4°C with gentle agitation. Blots were then washed with TBS-T three times, incubated with horseradish peroxidase (HRP) conjugated goat anti-rabbit antibody (1:3000 in 5% skim milk in TBS-T, Rockland Immunochemicals, Inc.) for an hour at room temperature with gentle agitation, and then washed with TBS-T three times. Chemiluminescent signals were detected using Clarity ECL HRP substrate kit (BioRad) following manufacturer’s instructions. Lastly, blots were stained with Coomassie stain (0.1% w/v Coomassie BB R-250, 50% v/v methanol, 10% v/v glacial acetic acid) to examine relative loading of all samples. The Hsp90 antibody recognizes two bands; the top band has been validated as Hsp90 using synthesized C. neoformans Hsp90 protein(Marcyk et al. 2021). Hsp90 protein levels were quantified using Fiji (Schindelin et al. 2012), normalized against the nonspecific bottom band, and relative levels were compared to wild type grown in YPD. All western blot experiments were performed in biological duplicate to confirm data reproducibility.
Spot dilution assay
Wild-type and CTR4-2p-HSP90 strains were grown in liquid YPD overnight at 30°C. Cells were washed with sterile water once, diluted to an OD600 of 0.5, and then 10-fold serially diluted four times. Serial dilutions were spotted on YPD agar and YPD agar supplemented with 25 µM CuSO4 or 200 µM BCS. The following antifungals and stress-inducing reagents were added individually to solid agar media: 0.5 µg/ml amphotericin B, 2 µg/ml fluconazole, 5 µM radicicol, 2 or 5 µM HSP990, 1 M NaCl, 1 M sorbitol, 15 mM dithiothreitol (DTT), 0.1 µg/ml tunicamycin, 0.02% SDS, 0.5 mg/ml calcofluor white (CFW), 3.5 mM H2O2, 25 µM MG132, 4 µg/ml benomyl, 100 mM hydroxyurea, and 0.04% methylmethanesulfonate. For low pH-sensitivity, YPD medium was buffered with 150 mM HEPES and pH adjusted to 4 and 6 with HCl prior to autoclave. Due to poor solubility of CuSO4 in alkaline solution (typically below 1 µM at pH 8), high pH sensitivity was not tested. Plates were incubated at 30°C or 37°C for 2 days and then imaged using ChemiDoc (BioRad). All experiments were performed in biological duplicate to confirm data reproducibility.
E-test strip assay
Wild-type and CTR4-2p-HSP90 strains were grown in liquid YPD at 30°C overnight, washed once with sterile water, and diluted to a cell density of 108 cells/ml. Cell suspensions (100 μL) were spread on YPD agar and YPD agar supplemented with 5 µM CuSO4 using a sterile cotton swab. Amphotericin B and fluconazole E-test strips (bioMérieux) were placed in the middle of agar plates. Plates were incubated at 37°C for 6 days and were then imaged using a ChemiDoc (BioRad).
Fluconazole cidality assay
Wild-type and CTR4-2p-HSP90 strains were grown in liquid YPD at 30°C overnight, washed once with sterile water, and diluted to a cell density of 106 cells/ml in YPD alone or YPD supplemented with 1 µg/ml fluconazole, 2 µM radicicol, 2 µM HSP990, 25 µM CuSO4, or combinations as indicated. Cells were incubated at 37°C with shaking and CFU were determined by plating cells on solid YPD media at 0-, 4-, 8-, and 24-h time points.
Propidium iodide staining and microscopy
Morphology of cells from overnight cultures of wild-type and CTR4-2p-HSP90 strains at 37°C was observed and imaged using the microscope Axio Imager.M1 (Carl Zeiss). To observe nuclear morphology, cells were fixed in 70% ethanol at 4°C overnight, washed once with NS buffer [10 mM Tris-HCl (pH 7.5), 250 mM sucrose, 1 mM NaEDTA (pH 8.0), 1 mM MgCl2, 0.1 mM CaCl2, 0.1 mM ZnCl2, 0.4 mM phenylmethylsulfonyl fluoride (PMSF), 7 mM β-mercaptoethanol], and then stained in NS buffer with propidium iodide (10 µg/ml) and RNase (1 mg/ml) at room temperature for 2 h. Propidium iodide signal was imaged using X-cite series 120 light source with the DsRed channel.
Melanin production
Melanin production was tested in biological duplicate on dopamine medium [1 g L-asparagine, 1 g glucose, 3 g KH2PO4, 0.122 g MgSO4, 1 mg thiamine, 5 µg biotin, and 100 mg L-DOPA (3,4-dihydroxy-L-phenyalanine) per liter]. Wild-type and CTR4-2p-HSP90 strains were grown in liquid YPD at 30°C overnight, washed once with sterile water, and diluted to a density of 108 cells per ml. Wild-type and mutant cells were 10-fold serially diluted and spotted on dopamine medium and dopamine medium supplemented with 5 µM CuSO4. Wild-type cells were also 10-fold serially diluted and spotted on dopamine medium with or without 0.5 µM HSP990, 2 µM HSP990, 0.5 µM RAD, or 2 µM RAD. Agar plates were incubated at 30°C or 37°C for 2 days. Melanin production was imaged using ChemiDoc (BioRad).
Capsule production
Capsule production was measured in biological duplicate as previously described (Zaragoza and Casadevall 2004). In brief, wild-type and CTR4-2p-HSP90 strains were grown in liquid Sabouraud medium (Sigma) supplemented with 25 µM CuSO4 overnight at 30°C, washed once with sterile water, and 30 µl of cells were inoculated in 10% Sabouraud medium in 50 mM HEPES (pH 7.3) with or without 25 µM CuSO4. After shaking for 48 h at 30°C, 500 µl of cells were collected and concentrated in 20 µl H2O, mixed with one drop of India ink, and 4 µl of cells were prepared on a glass slide and observed and imaged using the microscope Axio Imager.M1 (Carl Zeiss). For each sample, capsule thickness was measured for 50 cells from at least five different images using Fiji (Schindelin et al. 2012). T-test with Welch’s correction was used to determine statistical significance.
In vivo gene expression
Female 8-week-old A/J (Jax) mice were inoculated with KN99α or CTR4-2p-HSP90 either intravenously or intranasally as described below (n = 3 mice per group). Eight days post-inoculation, lungs were harvested from intranasally inoculated mice and brains were harvested from intravenously inoculated mice. Organs were freeze dried and homogenized with 2.3 mm zirconia/silica beads on a Mini-BeadBeater (BioSpec Products, Inc.). RNA was extracted using TRIzol (Invitrogen) and chloroform, then purified using the RNeasy Plus kit (Qiagen) following manufacturer’s instructions. cDNA was synthesized from 500 ng of RNA with iScript cDNA synthesis kit (BioRad) and 25 ng of resulting cDNA was used in each reaction. Gene expression was measured by qRT-PCR using iQ SYBR Green Supermix (BioRad) with a CFX96 Real Time system (BioRad) then HSP90 expression was normalized to TEF1. To ensure primers amplified C. neoformans genes specifically, we used cDNA from mock-infected mice administered PBS in lieu of fungal cells. All primer pairs tested failed to amplify cDNA from mock-infected tissues. T-test with Welch’s correction was used to determine statistical significance.
Mouse model of cryptococcal infection
For pulmonary virulence studies, 8-week-old female A/J (Jax) mice were inoculated with 5 × 104 CFU of KN99α or CTR4-2p-HSP90 (n = 10 mice per group) in 50 µl PBS via intranasal instillation. For systemic virulence studies, 8-week-old female A/J (Jax) mice were inoculated with 1 × 104 CFU of KN99α or CTR4-2p-HSP90 (n = 10 mice per group) in 200 µl PBS by tail vein injection. Mice were monitored for weight loss and disease symptoms daily and were euthanized when they reached endpoint criteria, including 10% weight loss. Mice were housed 5 per cage with access to food and water. Percent survival was plotted on Kaplan-Meier curve and Log-rank analysis was used to determine statistical significance.
Ethics statement
All animal experiments were performed following the Guide for the Care and Use of Laboratory Animals of the National Research Council. The animal experiment protocols were approved by Institutional Animal Care and Use Committee at the University of Iowa (Protocol: 0092064).
Data and reagent policy
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Results
Generation of a regulatable expression system for Cryptococcus neoformans HSP90
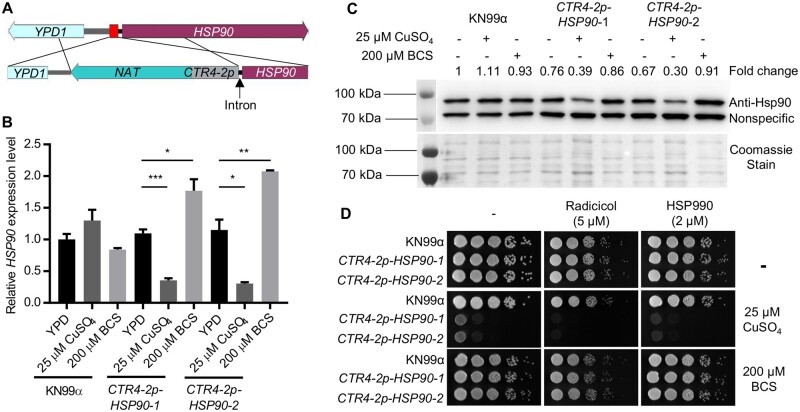
To characterize the function of Hsp90 in C. neoformans, we constructed a conditional expression allele of HSP90 under control of the copper-suppressible promoter CTR4-2 (Figure 1A and Supplementary Figure S1). Prior attempts to generate a regulatable expression system for C. neoformans HSP90 had been unsuccessful, but upon close examination of the locus, we noted the gene immediately upstream of HSP90, YPD1, is also essential, with the intergenic sequence between the two being only 443 base pairs long (Supplementary Figure S1) (Lee et al. 2011). Preceding the HSP90 coding sequence, an intron, which could influence HSP90 expression, was also noted (Janbon 2018). To avoid disruption of YPD1 and the intron, we replaced 111 base pairs in front of the HSP90 intron with the NAT-CTR4-2p construct (Supplementary Figure S1). Transformants from a single transformation were genotyped to confirm correct integration of the promoter, and two colonies were selected for further characterization (Supplementary Figure S1). To verify the functionality of the CTR4-2 promoter, we measured HSP90 transcript levels in cells growing at the host-range temperature of 37°C in the absence and presence of added copper. While supplementation with 25 µM CuSO4 had no significant effect on HSP90 expression in the wild-type control, it significantly reduced HSP90 expression in both transformants. In addition, supplementation with the copper chelator bathocuproinedisulphonic acid (BCS) significantly increased HSP90 expression compared to YPD medium alone (Figure 1B), consistent with the chelator inactivating copper normally present in the YPD medium (typically ∼1 µM) that was impacting gene expression (Attar et al. 2020). To assess whether alterations in transcript level manifested in changes in protein level, we performed western blot analysis. Relative Hsp90 level remained unchanged upon treatment with either copper or BCS in the wild type, while the addition of copper to the CTR4-2p-HSP90 strains resulted in a 60%–70% reduction relative to wild-type levels of Hsp90 after culture at either 37°C or 30°C (Figure 1C and Supplementary Figure S2A). Supplementation with BCS resulted in Hsp90 protein levels in the mutant isolates similar to those in wild-type and YPD-only controls (Figure 1C and Supplementary Figure S2A). Overall, this characterization confirmed the successful generation of a regulatable expression system for controlling HSP90 expression in C. neoformans.
Figure 1.
Generation and characterization of a regulatable expression system for C. neoformans HSP90. (A) Schematic diagram showing the engineering of the CTR4-2p-HSP90 strain. The red box between HSP90 and YPD1 indicates the targeted sequence to be replaced with the NAT-CTR4-2p cassette. (B,C) Wild-type KN99α and two CTR4-2p-HSP90 colonies from a single transformation were grown in liquid YPD medium alone or with CuSO4 or BCS at 37°C overnight. In the morning, strains were sub-cultured in the same liquid media and grown at 37°C for 5 h. Cells were harvested to examine (B) HSP90 expression levels by RT-PCR and (C) Hsp90 protein levels by western blot. (B) Relative level of expression for HSP90 was normalized to GPD1. Expression was normalized to wild type in YPD alone. Error bars represent standard deviation of the mean for technical triplicates. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. (C) Hsp90 protein levels were normalized to nonspecific protein bands recognized by the anti-Hsp90 antibody and set relative to wild-type cells grown in YPD alone. Fold changes of Hsp90 levels are listed above the western blot image and Coomassie stain of the membrane confirming equal loading is shown below, which matches the pattern observed with the nonspecific band. (D) Wild-type and mutant cells from overnight cultures were 10-fold serially diluted and spotted on YPD agar or YPD agar supplemented with CuSO4 or BCS with or without Hsp90 inhibitors. Agar plates were incubated at 37°C and imaged after 2 days.
With the successful generation of a conditional expression mutant, a valuable tool to study essential gene function, we wanted to confirm Hsp90 is essential in C. neoformans. To do so, wild-type and CTR4-2p-HSP90 strains were serially diluted and spotted onto agar plates in the absence or presence of 25 µM CuSO4. Copper-mediated repression of C. neoformans HSP90 caused a significant growth defect at 37°C (Figure 1D) and a marginal defect at 30°C (Supplementary Figure S2B). Addition of the structurally unrelated Hsp90 inhibitors radicicol or HSP990 further exacerbated the growth defect phenotype of the CTR4-2p-HSP90 strains in the presence of copper at both temperatures (Figure 1D and Supplementary Figure S2B), consistent with the principle that reduced dosage of an essential gene renders a strain more susceptible to pharmacological inhibitors that target that gene’s protein product (Xue et al. 2021). Thus, Hsp90 plays a critical role in cellular growth in C. neoformans, and the CTR4-2 promoter system enables the study of HSP90 function in this pathogen.
Hsp90 modulates fluconazole susceptibility of C. neoformans
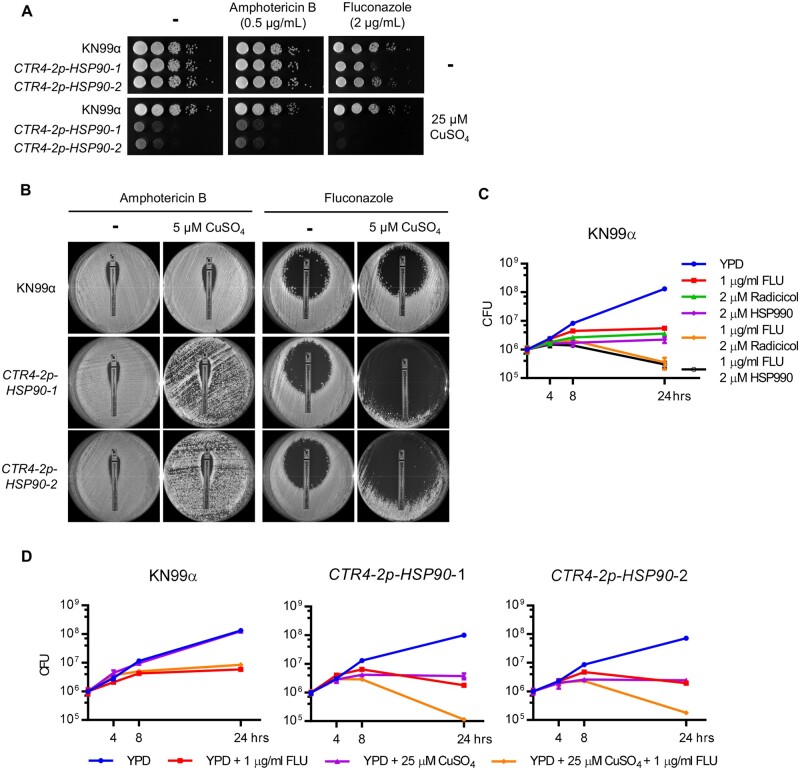
Hsp90 influences antifungal drug susceptibility in Candida and Aspergillus species (Cowen and Lindquist 2005). Therefore, we tested whether suppression of HSP90 potentiates amphotericin B and fluconazole activity in C. neoformans. To do so, the C. neoformans CTR4-2p-HSP90 strains were spotted onto rich medium in the absence and presence of antifungal and copper. While copper alone reduced growth of the HSP90 mutants, depletion of HSP90 increased sensitivity to fluconazole but not to amphotericin B compared with no-copper controls as well as the wild-type strain at both 37°C and 30°C (Figure 2A and Supplementary Figure S3A). Similarly, when strains were plated on rich medium with or without copper in the presence of E-test strips releasing a concentration gradient of either amphotericin B or fluconazole, an enhanced clearing zone was observed in the presence of copper and the fluconazole E-test strip with the CTR4-2p-HSP90 strains (Figure 2B). These results further support the conclusion that depletion of HSP90 enhances fluconazole efficacy in C. neoformans.
Figure 2.
Depletion of HSP90 increases sensitivity to fluconazole. (A) Wild-type and CTR4-2p-HSP90 strains were 10-fold serially diluted and spotted on YPD agar and YPD agar supplemented with 25 µM CuSO4 with or without 0.5 µg/ml amphotericin B or 2 µg/ml fluconazole. Agar plates were incubated at 37°C and imaged after 2 days. (B) Wild-type and CTR4-2p-HSP90 strains were spread on YPD agar with or without 5 µM CuSO4, and an ETEST strip of amphotericin B or fluconazole was placed in the center of the plate to test antifungal drug susceptibility. Plates were imaged after incubation at 37°C for 6 days. (C) Wild-type cells were inoculated at 1 × 106 CFU in liquid YPD medium in the absence or presence of fluconazole, radicicol, HSP990, or a combination as indicated. Cells were cultured at 37°C and plated on YPD agar medium at 0, 4, 8, and 24 h postinoculation for CFU determination. Error bars represent standard deviation of the mean for technical triplicates. (D) Wild-type and CTR4-2p-HSP90 strains were inoculated at 1 × 106 CFU in liquid YPD medium in the absence or presence of fluconazole, CuSO4, or a combination as indicated. Cells were cultured at 37°C and plated on YPD agar medium at 0, 4, 8, and 24 h postinoculation for CFU determination. Error bars represent standard deviation of the mean for technical triplicates.
While the azoles exhibit a fungistatic mode-of-action against yeasts, use of this antifungal in combination with Hsp90 inhibitors in Candida species results in a fungicidal combination (Cowen et al. 2009). To examine whether this effect also occurs in C. neoformans, we cultured wild-type and CTR4-2p-HSP90 strains in the absence or presence of fluconazole and copper, and plated aliquots at various time points to assess viable colony forming units (CFU). Both pharmacological inhibition of Hsp90 (Figure 2C) and genetic suppression of HSP90 (Figure 2D) caused an approximate one Log10 reduction in CFU after 24 h of drug exposure compared to the initial inoculum, indicating that compromise of Hsp90 enables fluconazole to kill Cryptococcus cells. Overall, our results highlight a critical role for Hsp90 in azole tolerance of C. neoformans.
Hsp90 helps maintain protein homeostasis and genome stability
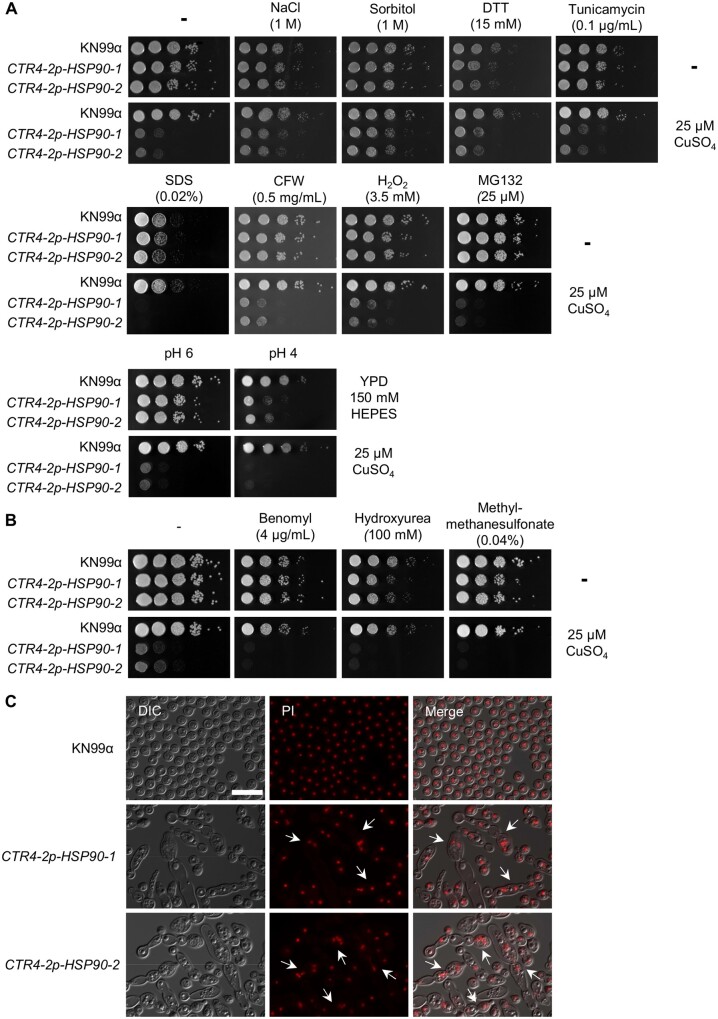
Hsp90 governs cellular responses to a plethora of environmental stressors (Cowen 2009; O’Meara et al. 2019). To assess the impact of depletion of Hsp90 on C. neoformans susceptibility to diverse cellular stress, we tested wild-type strain and copper-regulated HSP90 mutants against a panel of stressors, including osmotic stress (NaCl and sorbitol), ER stress [dithiothreitol (DTT) and tunicamycin], cellular membrane stress (SDS), cell wall stress (CFW), oxidative stress [hydrogen peroxide (H2O2)], proteotoxic stress (proteosome inhibitor MG132), and low pH (pH 4) (Figure 3A and Supplementary Figure S3B). While depletion of HSP90 had little impact on most stresses examined, an increase in sensitivity to SDS, MG132, and low pH was observed at both 37°C (Figure 3A) and 30°C (Supplementary Figure S3). Notably, the sensitivity to the proteosome inhibitor MG132 is consistent with global issues of protein misfolding overwhelming the functional capacity of Hsp90 (O’Meara et al. 2019; Hossain et al. 2020).
Figure 3.
Depletion of HSP90 increases sensitivity to genotoxic stressors. (A) Wild-type and CTR4-2p-HSP90 strains were 10-fold serially diluted and spotted on YPD agar medium or YPD agar supplemented with CuSO4 with or without individual stress-inducing reagents at the concentrations indicated. (B) Wild-type and CTR4-2p-HSP90 strains were 10-fold serially diluted and spotted on YPD agar and YPD agar supplemented with CuSO4 with or without genotoxic stress-inducing reagents at the concentrations indicated. Agar plates were incubated at 37°C and imaged after 2 days. (C) Wild-type and CTR4-2p-HSP90 strains from overnight cultures grown in liquid YPD medium supplemented with 25 µM CuSO4 were stained with propidium iodide (PI) to examine nuclear morphology. Cells with multiple nuclei are highlighted with white arrows. Scale bar represents 20 µm.
In C. albicans, Hsp90 is instrumental for cell cycle progression and stability of the cyclin-dependent kinase Cdc28 such that perturbation of Hsp90 function increases sensitivity to cell cycle inhibitors (Senn et al. 2012). To examine whether this phenotype was conserved in C. neoformans, we also tested the wild-type strain and copper-regulated HSP90 mutants for sensitivity to the genotoxic stress-inducing agents benomyl, hydroxyurea, and methyl methanesulfonate (MMS) (Figure 3B and Supplementary Figure S4A). Depletion of HSP90 increased sensitivity to all three compounds at 37°C (Figure 3A) and 30°C (Supplementary Figure S4A), suggesting that Hsp90 plays a conserved role in cell cycle progression in C. neoformans. Closer examination of the cells via microscopy revealed abnormal polarized growth in the CTR4-2p-HSP90 mutants in the presence of copper (Figure 3C and Supplementary Figure S4B), with cells often displaying multinucleate phenotypes (Figure 3C). Consistent with its role in other organisms, these findings suggest that Hsp90 may play a role in cell-cycle progression in C. neoformans.
Hsp90 acts to support the virulence of C. neoformans
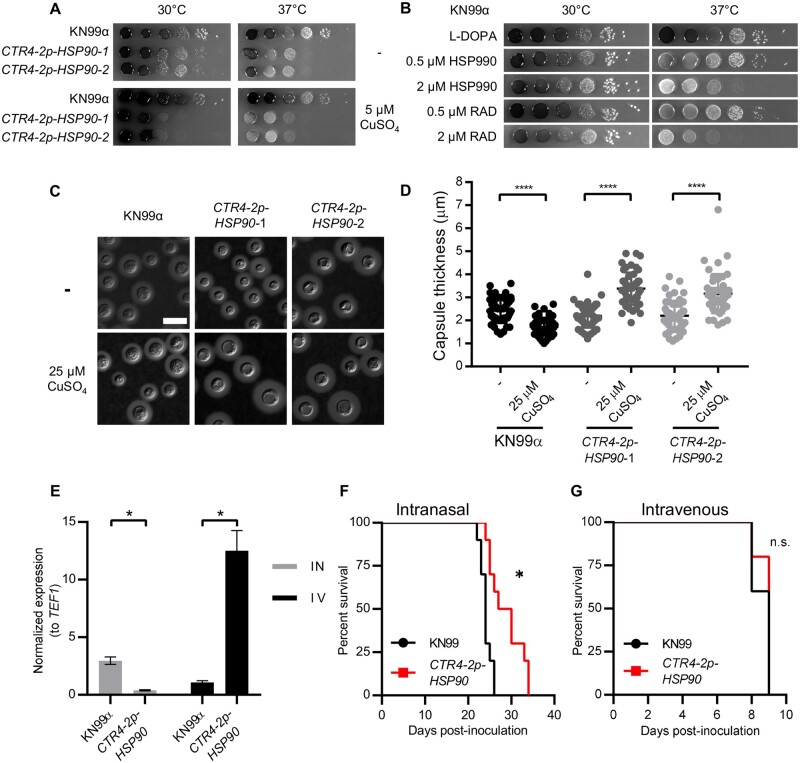
Melanin and capsule production are two major virulence traits in C. neoformans (Kwon-Chung et al. 2014). Thus, we wished to test whether copper-mediated suppression of HSP90 in CTR4-2p-HSP90 mutants would impair their induction. First, we focused on melanin production. We observed a copper-dependent reduction in melanin production in CTR4-2p-HSP90 mutants at 37°C, which was not observed in the wild-type control (Figure 4A), suggesting that Hsp90 modulates melanin production in C. neoformans. To confirm this observation, we tested melanin production in wild-type cells in the presence of Hsp90 inhibitors HSP990 and radicicol. Indeed, pharmacological inhibition of Hsp90 reduced melanin production at 37°C, supporting the role of Hsp90 in regulating this critical virulence phenotype (Figure 4B). Importantly, this occurred at concentrations that had minimal impact on growth, highlighting that the reduction in melanin production was not merely due to growth inhibition.
Figure 4.
Hsp90 modulates melanin and capsule production, and depletion of HSP90 attenuates virulence in a pulmonary model of cryptococcal infection. (A) Wild-type and CTR4-2p-HSP90 cells were 10-fold serially diluted and spotted on L-DOPA agar medium with or without CuSO4. (B) Wild-type cells were 10-fold serially diluted and spotted on L-DOPA agar medium with or without Hsp90 inhibitors at the concentrations indicated. Agar plates were incubated at 30°C or 37°C and imaged after 2 days. (C) Representative images of India ink-stained wild-type and CTR4-2p-HSP90 strains show capsule production. Cells were grown in 10% Sabouraud media in 50 mM HEPES (pH7.3) with or without CuSO4 at 30°C for 48 h. Scale bar represents 10 µm. (D) Capsule thickness was measured (N = 50) for samples shown in A. **** P ≤ 0.0001. (E) Lungs were harvested from intranasally (IN, gray bars) inoculated mice and brains from intravenously (IV, black bars) inoculated mice (n = 3 per group). For each sample, relative gene expression levels were measured by qRT-PCR and expression levels for HSP90 normalized to the expression levels of the constitutively expressed gene TEF1. Results were then normalized to the HSP90 expression level determined for wild-type KN99α in brains of intravenously inoculated mice. * P ≤ 0.05. (F,G) A/J mice were inoculated intranasally (F) or intravenously by tail vein injection (G) with the wild-type or CTR4-2p-HSP90 strain (n = 10 mice per group). Mice were monitored for disease progression. *P = 0.0013, n.s. = not significant.
Next, we examined the effect of depleting Hsp90 on capsule production. Under capsule-inducing conditions, depletion of HSP90 at 37°C resulted in a morphology consistent with a cytokinesis defect, preventing us from assessing Hsp90’s role in capsule production at 37°C (Supplementary Figure S5). Interestingly, at 30°C, addition of copper significantly reduced capsule thickness of the wild-type strain but caused a significant increase in capsule thickness in both CTR4-2p-HSP90 mutants (Figure 4, C and D). This finding is in contrast to previous reports that pharmacological inhibition of Hsp90 by radicicol in nutrient-limited capsule-inducing media inhibits capsule production, which is likely the result of growth defects upon Hsp90 compromise in those conditions (Cordeiro et al. 2016; Chatterjee and Tatu 2017).
Finally, we asked whether Hsp90 contributes to the pathogenic potential of C. neoformans in mice. Although the druggability of Hsp90 has been well established, the poor potency of human-targeted inhibitors against fungi coupled with the toxicity associated with inhibiting host Hsp90 have precluded these compounds from being tested as antifungals in vivo. In C. albicans, genetic depletion of Hsp90 has provided proof-of-principle that targeting this chaperone has great therapeutic potential to treat systemic candidiasis (Cowen et al. 2009), but analogous experiments in C. neoformans have yet to be performed. To assess whether C. neoformans HSP90 is important for virulence in vivo, we took advantage of the fact that copper concentrations vary widely in different tissues within a mammalian host. As evidence, the copper transporter gene CTR4 is expressed by C. neoformans at a constant low level in fungus growing in the lungs of mice, but is expressed at a high level in fungus isolated from the central nervous system due to the different physiological copper concentrations present at these sites (Steen et al. 2003; Ding et al. 2013). Thus, we hypothesized that a CTR4-2p-HSP90 mutant would display growth defects in a mouse lung but not in the brain. Indeed, compared to the parental wild-type strain, HSP90 expression was significantly reduced in the CTR4-2p-HSP90 mutant strain when isolated from the lung (Figure 4E) and we observed significantly attenuated virulence for this mutant relative to the wild-type control upon intranasal inoculation (Figure 4F). By this route of administration, the fungus must first establish infection in the lungs prior to lethal systemic dissemination. In contrast, HSP90 expression was significantly increased in the CTR4-2p-HSP90 mutant when isolated from the brain (Figure 4E) and the mutant displayed no virulence defect compared to wild type upon intravenous inoculation (Figure 4G). By this route of administration, C. neoformans rapidly disseminates directly to the brain. Overall, the results strongly support the importance of Hsp90 as an enabler of virulence in C. neoformans.
Discussion
The human fungal pathogen C. neoformans poses a major burden on human health, with treatment options limited to only two major antifungal classes. Essential genes that are required for survival serve as optimal targets for novel antifungal development (Iyer et al. 2021). In several fungal species, including the model yeast Saccharomyces cerevisiae and the distantly related fungal pathogen C. albicans, it is estimated that ∼20% of the open reading frames in their genomes are essential (Winzeler et al. 1999; Segal et al. 2018). In contrast, only a small number of essential genes have been identified and characterized in C. neoformans due to the technical challenges of genetic manipulation in this organism (Ianiri and Idnurm 2015; Iyer et al. 2021). In this study, we generated the first conditional expression mutant for Hsp90, a critical stress-response regulator that has shown great promise as an antifungal target in Candida and Aspergillus (Cowen and Lindquist 2005; Cowen et al. 2009). The successful generation of conditional isolates provided the opportunity to directly assess the biological functions of Hsp90 in C. neoformans. Although supplementation with even a high concentration of copper did not completely repress HSP90 expression, it was sufficient to cause a severe growth defect upon culturing conditional mutants at the physiological temperature of 37°C. Addition of well-validated Hsp90 inhibitors further exacerbated the growth defect of the strains in the presence of copper, confirming the important role of Hsp90 in supporting the growth and survival of C. neoformans.
While the genetic manipulation of C. neoformans has traditionally required the use of biolistic transformation techniques, the application of CRISPR-Cas9 has largely alleviated the technical challenges associated with mutant construction. Genomic resources including a ∼1500-gene deletion mutant library have been generated in C. neoformans to study phenotypes associated with nonessential genes (Brown et al. 2014), however, methods for studying essential genes in C. neoformans have relied on only a few well-characterized regulatable promoters (Ory et al. 2004; Ruff et al. 2009; Fan and Lin 2018). The copper suppressive promoter CTR4-2 has been employed to study essential gene functions both in vitro and in vivo, and thus was selected to regulate HSP90 expression in this study (Chayakulkeeree et al. 2007). The intergenic environment upstream of HSP90 presented a challenge, as there is a very short intergenic sequence between HSP90 and yet another essential gene, YPD1. Thus, the available genomic space for inserting a drug resistance marker and the CTR4-2 promoter upstream of HSP90 had to be meticulously assessed to avoid unintended disruptions. While construction of this conditional expression mutant has been a valuable breakthrough for the study of Hsp90 in C. neoformans, we were unable to achieve complete transcriptional repression of the chaperone. In the future, it may be worth adapting and optimizing alternative conditional expression systems, including the tetracycline-repressible expression system (Roemer et al. 2003), in order to more tightly regulate HSP90 expression in C. neoformans and avoid copper-induced artifacts.
Utilizing the CTR4-2 promoter, depletion of HSP90 confirmed previous observations based on chemical inhibitors that Hsp90 modulates key virulence phenotypes and antifungal drug tolerance in C. neoformans (Cordeiro et al. 2016; Chatterjee and Tatu 2017). However, the mechanisms by which Hsp90 regulates these traits require further exploration. One of the key traits is the ability of fungal pathogens to survive host temperature (Garcia-Solache and Casadevall 2010), and here we provided clear evidence that Hsp90 is required for growth at host-range temperature in C. neoformans, thereby expanding the repertoire of genes implicated in thermal adaptation in this pathogen (Yang et al. 2017). It remains to be defined exactly how Hsp90 influences the activity of its client proteins to enable thermal adaptation in C. neoformans. Another key trait of fungal pathogens is the ability to develop antifungal drug tolerance and resistance. In C. albicans, Hsp90 modulates cellular responses to antifungals by stabilizing components of the Pkc1-MAPK and calcineurin stress-response pathways (Cowen and Lindquist 2005; Cowen et al. 2009; Shapiro et al. 2009). Calcineurin is a conserved nonessential protein phosphatase critical for morphogenesis, drug resistance, and virulence in multiple Candida species (Cruz et al. 2002; Blankenship et al. 2003; Chen et al. 2011, 2012, 2014; Zhang et al. 2012). In C. neoformans, calcineurin governs both morphogenesis during sexual reproduction and growth at host temperature (Odom et al. 1997; Cruz et al. 2001). Calcineurin inhibitors also synergize with fluconazole against C. neoformans (Del Poeta et al. 2000), and thus, investigating whether Hsp90 stabilizes this regulator to modulate C. neoformans virulence would be an interesting avenue to pursue. Our HSP90 conditional expression system provides an opportunity to elucidate Hsp90 chaperone-mediated regulatory mechanisms in C. neoformans to broaden the antifungal drug development target space.
Hsp90 functions as a signaling hub, stabilizing various signal transducers that modulate key fungal virulence phenotypes (Cowen and Lindquist 2005; Shapiro et al. 2009; O’Meara et al. 2019; Hossain et al. 2020). In C. albicans, Hsp90 modulates morphological transitions via the Ras1-PKA signaling pathway, as well as the cyclin-dependent kinase Pho85, cyclin Pcl1, and transcription factor Hms1 (Shapiro et al. 2009, 2012). In C. albicans, Hsp90 client proteins also include regulators that play important roles in cell-cycle progression and morphogenesis (Hossain et al. 2021). In S. cerevisiae, cell cycle is regulated by the oscillatory expression of stage-specific cyclins that bind to and activate the master cyclin-dependent kinase (CDK) Cdc28 (Lew et al. 1997). In C. albicans, inhibition of Hsp90 destabilizes Cdc28 leading to cell cycle arrest and morphogenesis defects (Senn et al. 2012). Filaments induced by compromise of Hsp90 function pharmacologically, genetically, or by increasing proteolytic stress, are multinucleate as a result of mis-regulation of cell cycle progression (Veri et al. 2018; Hossain et al. 2020). Interestingly, we found that depletion of HSP90 in C. neoformans resulted in an apparent cytokinesis defect and the formation of multinucleated elongated cells, suggesting Hsp90 may have a conserved function in cell cycle progression in C. neoformans. Unique to C. neoformans, the cell cycle is differentially regulated during the formation of polyploid titan cells, which are gigantic cells that form inside host lungs and play important roles in pathogenesis, stress responses, and drug tolerance (Okagaki et al. 2010; Zaragoza et al. 2010). The major pathway regulating titan cell formation is the cAMP/PKA/Rim101 signaling cascade (Dambuza et al. 2018; Hommel et al. 2018; Trevijano-Contador et al. 2018), and many signal transducers in these pathways are regulated by Hsp90 in C. albicans (O’Meara et al. 2017). Cell cycle regulators Pho85 and Pcl1 have also been shown to be involved in the endoreplication pathway during auto-diploidization in C. neoformans (Fu et al. 2021). Thus, it is possible that components of Hsp90 regulatory circuitry are involved in titan cell formation during host infection and contribute to C. neoformans virulence.
Here, the CTR4-2 promoter offered, for the first time, the opportunity to directly assess Hsp90’s involvement in C. neoformans virulence. By relying on differential physiological copper concentrations within different tissues, we were able to regulate HSP90 expression in vivo. We observed attenuated virulence in an inhalation infection model where the CTR4-2 promoter repressed HSP90 expression in lung tissue where copper levels are high (Ding et al. 2013). In contrast, in the systemic infection model, HSP90 expression remained high due to the relatively low copper environment found in blood and brain (Champika 2018), and thus, we did not observe attenuated virulence for the mutant strain. Overall, this work provides a valuable genetic tool to assist in the development of Hsp90 inhibitors with activity against Cryptococcus.
Data availability
Supplementary material is available at GENETICS online.
Supplementary Material
Acknowledgments
The authors thank all Cowen lab members for their helpful discussion. They also thank Dr. Joseph Heitman for sharing strains and plasmids for TRACE transformation.
Funding
S.R.B is supported by an F32 Postdoctoral Individual National Research Service Award (AI145160), A.J.J. is supported by a T32 NINDS Institutional Research Training Grant (5T32AI007260), D.J.K. is supported by a National Institute of Health (NIH) R01 grant (R01AI147541-01A1), and L.E.C. is supported by the Canadian Institutes of Health Research (CIHR) Foundation grant (FDN-154288). Work reported here was supported in part by an NIH R01 grant (R01AI120958). L.E.C. is a Canada Research Chair (Tier 1) in Microbial Genomics Infectious Disease and co-Director of the CIFAR Fungal Kingdom: Threats & Opportunities program.
Conflicts of interest
L.E.C. and L.W. are cofounders and shareholders in Bright Angel Therapeutics, a platform company for development of novel antifungal therapeutics. L.E.C. is a consultant for Boragen, a small molecule development company focused on leveraging the unique chemical properties of boron chemistry for crop protection and animal health.
Contributor Information
Ci Fu, Department of Molecular Genetics, University of Toronto, Toronto, ON M5G 1M1, Canada.
Sarah R Beattie, Departments of Pediatrics, Carver College of Medicine, University of Iowa, Iowa City, IA 52242, USA.
Andrew J Jezewski, Departments of Pediatrics, Carver College of Medicine, University of Iowa, Iowa City, IA 52242, USA.
Nicole Robbins, Department of Molecular Genetics, University of Toronto, Toronto, ON M5G 1M1, Canada.
Luke Whitesell, Department of Molecular Genetics, University of Toronto, Toronto, ON M5G 1M1, Canada.
Damian J Krysan, Departments of Pediatrics, Carver College of Medicine, University of Iowa, Iowa City, IA 52242, USA; Microbiology/Immunology, Carver College of Medicine, University of Iowa, Iowa City, IA 52242, USA.
Leah E Cowen, Department of Molecular Genetics, University of Toronto, Toronto, ON M5G 1M1, Canada.
Literature cited
- Attar N, Campos OA, Vogelauer M, Cheng C, Xue Y, et al. 2020. The histone H3-H4 tetramer is a copper reductase enzyme. Science. 369:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicanic T, Muzoora C, Brouwer AE, Meintjes G, Longley N, et al. 2009. Independent association between rate of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: analysis of a combined cohort of 262 patients. Clin Infect Dis. 49:702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG, et al. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot Cell. 2:422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, et al. 2012. Hidden killers: human fungal infections. Sci Transl Med. 4:165rv13. [DOI] [PubMed] [Google Scholar]
- Brown JCS, Nelson J, VanderSluis B, Deshpande R, Butts A, et al. 2014. Unraveling the biology of a fungal meningitis pathogen using chemical genetics. Cell. 159:1168–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt ET, Daly R, Hoganson D, Tsirulnikov Y, Essmann M, et al. 2003. Isolation and partial characterization of Hsp90 from Candida albicans. Ann Clin Lab Sci. 33:86–93. [PubMed] [Google Scholar]
- Caplan T, Polvi EJ, Xie JL, Buckhalter S, Leach MD, et al. 2018. Functional genomic screening reveals core modulators of echinocandin stress responses in Candida albicans. Cell Rep. 23:2292–2298. [DOI] [PubMed] [Google Scholar]
- Champika G. 2018. Clinical features of acute copper sulphate poisoning. J Heavy Met Toxicity Dis. 3:2. doi:10.21767/2473-6457.10021. [Google Scholar]
- Chatterjee S, Tatu U. 2017. Heat shock protein 90 localizes to the surface and augments virulence factors of Cryptococcus neoformans. PLoS Negl Trop Dis. 11:e0005836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayakulkeeree M, Rude TH, Toffaletti DL, Perfect JR. 2007. Fatty acid synthesis is essential for survival of Cryptococcus neoformans and a potential fungicidal target. Antimicrob Agents Chemother. 51:3537–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Brand A, Morrison EL, Silao FG, Bigol UG, et al. 2011. Calcineurin controls drug tolerance, hyphal growth, and virulence in Candida dubliniensis. Eukaryot Cell. 10:803–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Konieczka JH, Springer DJ, Bowen SE, Zhang J, et al. 2012. Convergent evolution of calcineurin pathway roles in thermotolerance and virulence in Candida glabrata. G3 (Bethesda). 2:675–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Yu SJ, Huang HY, Chang YL, Lehman VN, et al. 2014. Calcineurin controls hyphal growth, virulence, and drug tolerance of Candida tropicalis. Eukaryot Cell. 13:844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro RA, Evangelista AJJ, Serpa R, Marques FJF, Melo CVS, et al. 2016. Inhibition of heat-shock protein 90 enhances the susceptibility to antifungals and reduces the virulence of Cryptococcus neoformans/Cryptococcus gattii species complex. Microbiology. 162:309–317. [DOI] [PubMed] [Google Scholar]
- Cowen LE. 2009. Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS Pathog. 5:e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 309:2185–2189. [DOI] [PubMed] [Google Scholar]
- Cowen LE, Singh SD, Kohler JR, Collins C, Zaas AK, et al. 2009. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci USA. 106:2818–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz MC, Fox DS, Heitman J. 2001. Calcineurin is required for hyphal elongation during mating and haploid fruiting in Cryptococcus neoformans. EMBO J. 20:1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, et al. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21:546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambuza IM, Drake T, Chapuis A, Zhou X, Correia J, et al. 2018. The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog. 14:e1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JN, Chau TTH, Wolbers M, Mai PP, Dung NT, et al. 2013. Combination antifungal therapy for cryptococcal meningitis. N Engl J Med. 368:1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Poeta M, Cruz MC, Cardenas ME, Perfect JR, Heitman J. 2000. Synergistic antifungal activities of bafilomycin A(1), fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob Agents Chemother. 44:739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DW. 2003. Echinocandin antifungal drugs. Lancet. 362:1142–1151. [DOI] [PubMed] [Google Scholar]
- Ding C, Festa RA, Chen YL, Espart A, Palacios O, et al. 2013. Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe. 13:265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Lin X. 2018. Multiple applications of a transient CRISPR-Cas9 coupled with electroporation (TRACE) system in the Cryptococcus neoformans species complex. Genetics. 208:1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. 2018. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 360:739–742. [DOI] [PubMed] [Google Scholar]
- Fu C, Davy A, Holmes S, Sun S, Yadav V, et al. 2021. 2021 Dynamic genome plasticity during unisexual reproduction in the human fungal pathogen Cryptococcus deneoformans. bioRxiv. doi:10.1101/2021.06.01.446667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Solache MA, Casadevall A. 2010. Global warming will bring new fungal diseases for mammals. mBio. 1:e00061–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd et al. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6:343–345. [DOI] [PubMed] [Google Scholar]
- Hommel B, Mukaremera L, Cordero RJB, Coelho C, Desjardins CA, et al. 2018. Titan cells formation in Cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Pathog. 14:e1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope W, Stone NRH, Johnson A, McEntee L, Farrington N, et al. 2019. Fluconazole monotherapy is a suboptimal option for initial treatment of cryptococcal meningitis because of emergence of resistance. mBio. 10:e02575–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S, Lash E, Veri AO, Cowen LE. 2021. Functional connections between cell cycle and proteostasis in the regulation of Candida albicans morphogenesis. Cell Rep. 34:108781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S, Veri AO, Cowen LE. 2020. The proteasome governs fungal morphogenesis via functional connections with Hsp90 and cAMP-protein kinase A signaling. mBio. 11:e00290–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue C, Heitman J. 2009. A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans. EMBO J. 28:1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianiri G, Idnurm A. 2015. Essential gene discovery in the basidiomycete Cryptococcus neoformans for antifungal drug target prioritization. mBio. 6:e02334–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Reedy JL, Nussbaum JC, Heitman J. 2004. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot Cell. 3:420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer KR, Revie NM, Fu C, Robbins N, Cowen LE. 2021. Treatment strategies for cryptococcal infection: challenges, advances and future outlook. Nat Rev Microbiol. 19:454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G. 2018. Introns in Cryptococcus. Mem Inst Oswaldo Cruz. 113:e170519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Fraser JA, Doering TL, Wang Z, Janbon G, et al. 2014. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 4:a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFayette SL, Collins C, Zaas AK, Schell WA, Betancourt-Quiroz M, et al. 2010. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 6:e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Ko YJ, Kim SY, Bahn YS. 2011. Multiple roles of Ypd1 phosphotransfer protein in viability, stress response, and virulence factor regulation in Cryptococcus neoformans. Eukaryot Cell. 10:998–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Puumala E, Robbins N, Cowen LE. 2021. Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chem Rev. 121:3390–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Weinert T, Pringle JR. 1997. Cell cycle control in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Press. p. 607–695. [Google Scholar]
- Marcyk PT, LeBlanc EV, Kuntz DA, Xue A, Ortiz F, et al. 2021. Fungal-selective resorcylate aminopyrazole Hsp90 inhibitors: optimization of whole-cell anticryptococcal activity and insights into the structural origins of cryptococcal selectivity. J Med Chem. 64:1139–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargesi S, Bongomin F, Hedayati MT. 2021. The impact of COVID-19 pandemic on AIDS-related mycoses and fungal neglected tropical diseases: why should we worry? PLoS Negl Trop Dis. 15:e0009092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, et al. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, et al. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6:e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, O’Meara MJ, Polvi EJ, Pourhaghighi MR, Liston SD, et al. 2019. Global proteomic analyses define an environmentally contingent Hsp90 interactome and reveal chaperone-dependent regulation of stress granule proteins and the R2TP complex in a fungal pathogen. PLoS Biol. 17:e3000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Robbins N, Cowen LE. 2017. The Hsp90 chaperone network modulates Candida virulence traits. Trends Microbiol. 25:809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory JJ, Griffith CL, Doering TL. 2004. An efficiently regulated promoter system for Cryptococcus neoformans utilizing the CTR4 promoter. Yeast. 21:919–926. [DOI] [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, et al. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 17:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raut A, Huy NT. 2021. Rising incidence of mucormycosis in patients with COVID-19: another challenge for India amidst the second wave? Lancet Respir Med. 9:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins N, Caplan T, Cowen LE. 2017. Molecular evolution of antifungal drug resistance. Annu Rev Microbiol. 71:753–775. [DOI] [PubMed] [Google Scholar]
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, et al. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol. 50:167–181. [DOI] [PubMed] [Google Scholar]
- Ruff JA, Lodge JK, Baker LG. 2009. Three galactose inducible promoters for use in C. neoformans var. grubii. Fungal Genet Biol. 46:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal ES, Gritsenko V, Levitan A, Yadav B, Dror N, et al. 2018. Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans. mBio. 9:e02048–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn H, Shapiro RS, Cowen LE. 2012. Cdc28 provides a molecular link between Hsp90, morphogenesis, and cell cycle progression in Candida albicans. Mol Biol Cell. 23:268–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Sellam A, Tebbji F, Whiteway M, Nantel A, et al. 2012. Pho85, Pcl1, and Hms1 signaling governs Candida albicans morphogenesis induced by high temperature or Hsp90 compromise. Curr Biol. 22:461–470. [DOI] [PubMed] [Google Scholar]
- Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, et al. 2009. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. 19:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Babak SD, Babak T, Diezmann S, Hill JA, Xie JL, et al. 2012. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 8:e1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen BR, Zuyderduyn S, Toffaletti DL, Marra M, Jones SJ, et al. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot Cell. 2:1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. 2010. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 11:515–528. [DOI] [PubMed] [Google Scholar]
- Trevijano-Contador N, de Oliveira HC, Garcia-Rodas R, Rossi SA, Llorente I, et al. 2018. Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Pathog. 14:e1007007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veri AO, Miao Z, Shapiro RS, Tebbji F, O’Meara TR, et al. 2018. Tuning Hsf1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PLoS Genet. 14:e1007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906. [DOI] [PubMed] [Google Scholar]
- Xue A, Robbins N, Cowen LE. 2021. Advances in fungal chemical genomics for the discovery of new antifungal agents. Ann N Y Acad Sci. 1496:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Jung KW, Bang S, Lee JW, Song MH, et al. 2017. Rewiring of signaling networks modulating thermotolerance in the human pathogen Cryptococcus neoformans. Genetics. 205:201–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Casadevall A. 2004. Experimental modulation of capsule size in Cryptococcus neoformans. Biol Proced Online. 6:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Garcia-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, et al. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog. 6:e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Silao FG, Bigol UG, Bungay AA, Nicolas MG, et al. 2012. Calcineurin is required for pseudohyphal growth, virulence, and drug resistance in Candida lusitaniae. PLoS One. 7:e44192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary material is available at GENETICS online.